Abstract
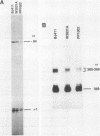
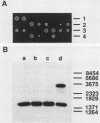
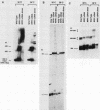
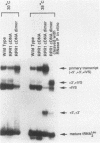
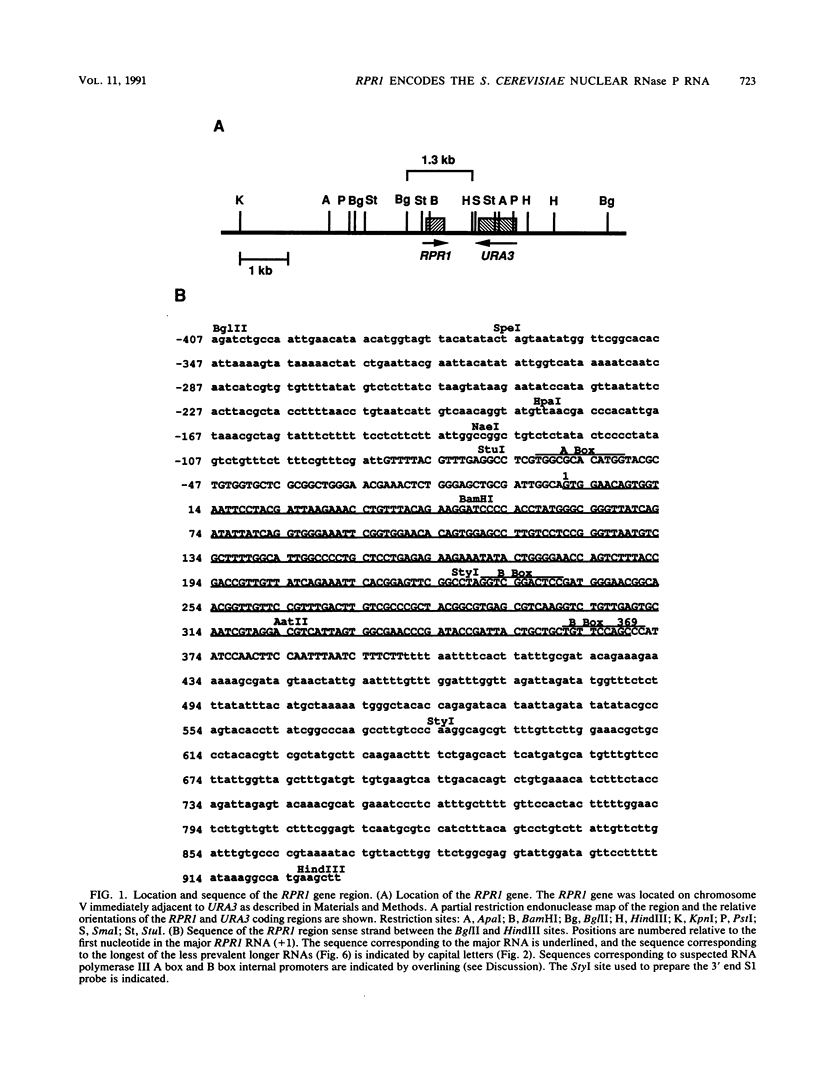
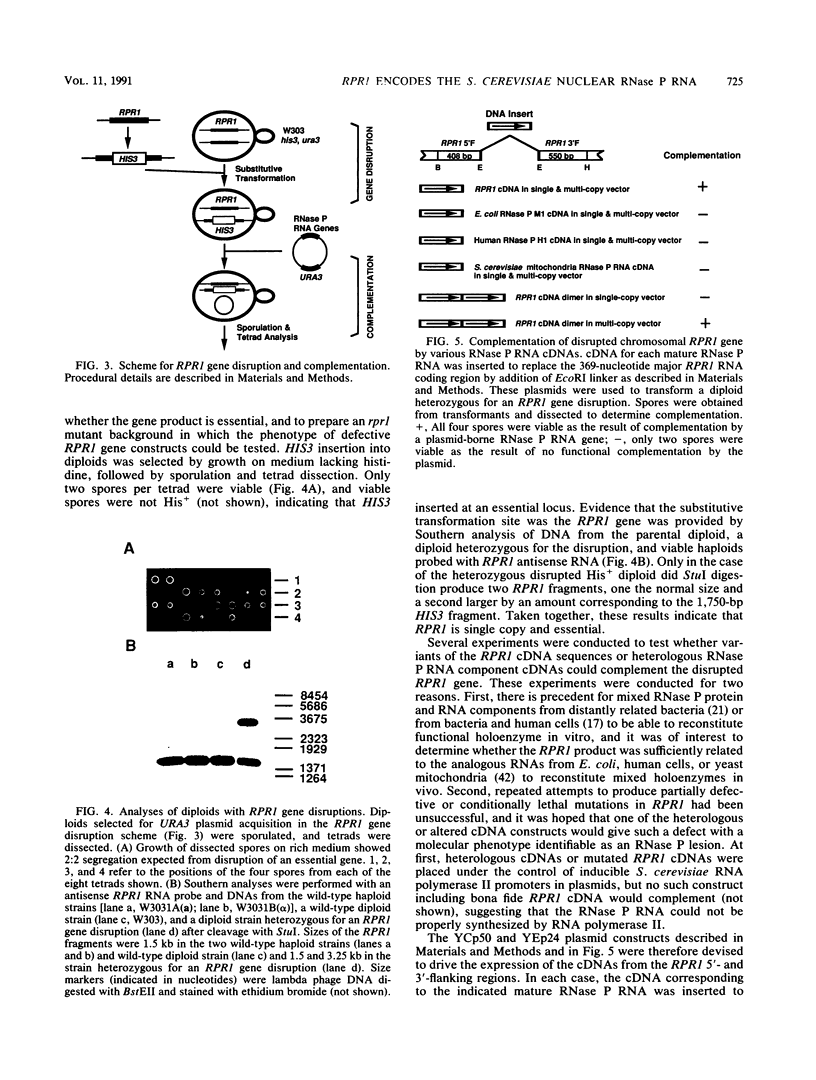
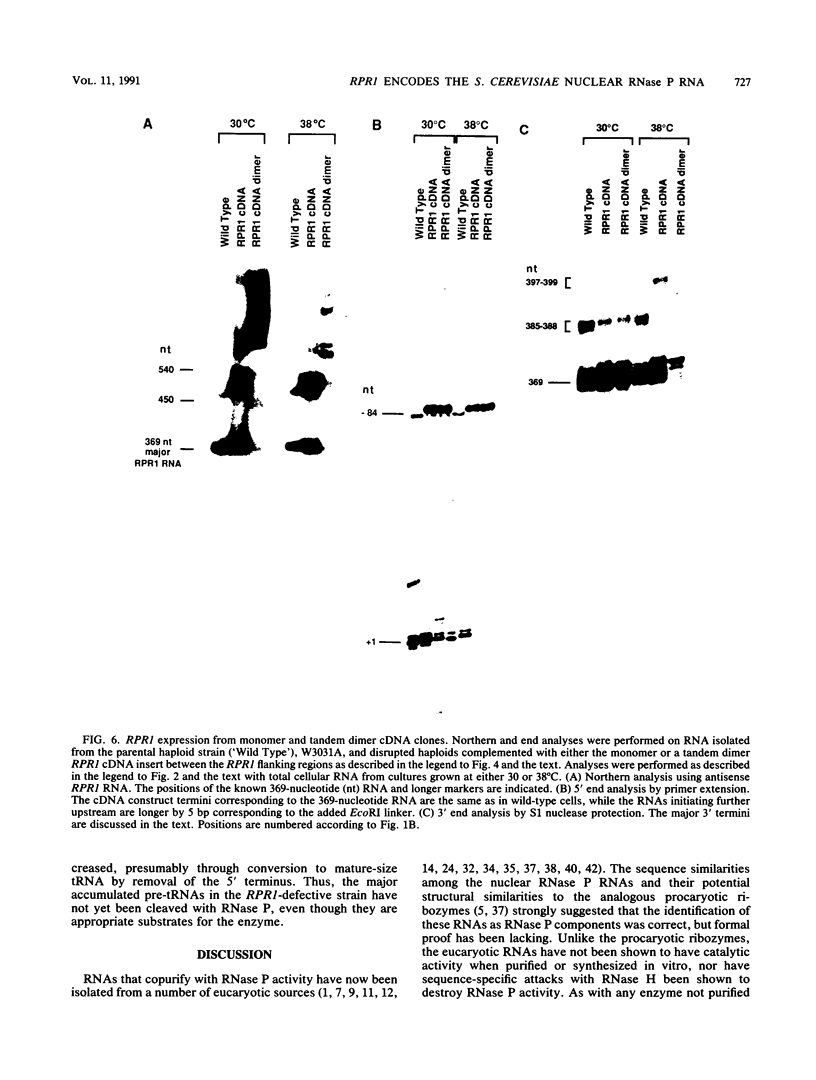
RNA components have been identified in preparations of RNase P from a number of eucaryotic sources, but final proof that these RNAs are true RNase P subunits has been elusive because the eucaryotic RNAs, unlike the procaryotic RNase P ribozymes, have not been shown to have catalytic activity in the absence of protein. We previously identified such an RNA component in Saccharomyces cerevisiae nuclear RNase P preparations and have now characterized the corresponding, chromosomal gene, called RPR1 (RNase P ribonucleoprotein 1). Gene disruption experiments showed RPR1 to be single copy and essential. Characterization of the gene region located RPR1 600 bp downstream of the URA3 coding region on chromosome V. We have sequenced 400 bp upstream and 550 bp downstream of the region encoding the major 369-nucleotide RPR1 RNA. The presence of less abundant, potential precursor RNAs with an extra 84 nucleotides of 5' leader and up to 30 nucleotides of 3' trailing sequences suggests that the primary RPR1 transcript is subjected to multiple processing steps to obtain the 369-nucleotide form. Complementation of RPR1-disrupted haploids with one variant of RPR1 gave a slow-growth and temperature-sensitive phenotype. This strain accumulates tRNA precursors that lack the 5' end maturation performed by RNase P, providing direct evidence that RPR1 RNA is an essential component of this enzyme.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akaboshi E., Guerrier-Takada C., Altman S. Veal heart ribonuclease P has an essential RNA component. Biochem Biophys Res Commun. 1980 Sep 30;96(2):831–837. doi: 10.1016/0006-291x(80)91430-8. [DOI] [PubMed] [Google Scholar]
- Altman S., Guerrier-Takada C. M1 RNA, the RNA subunit of Escherichia coli ribonuclease P, can undergo a pH-sensitive conformational change. Biochemistry. 1986 Mar 25;25(6):1205–1208. doi: 10.1021/bi00354a002. [DOI] [PubMed] [Google Scholar]
- Baer M., Nilsen T. W., Costigan C., Altman S. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res. 1990 Jan 11;18(1):97–103. doi: 10.1093/nar/18.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkiewicz M., Gold H., Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989 Apr;3(4):488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- Boehm S. Similarities between a predicted secondary structure for the M1 RNA ribozyme and the tRNA binding center of 16 S rRNA from E. coli. FEBS Lett. 1987 Aug 17;220(2):283–287. doi: 10.1016/0014-5793(87)80830-x. [DOI] [PubMed] [Google Scholar]
- Bowman E. J., Altman S. Identification of ribonuclease P activity from chick embryos. Biochim Biophys Acta. 1980 Jun 13;613(2):439–447. doi: 10.1016/0005-2744(80)90098-4. [DOI] [PubMed] [Google Scholar]
- Brow D. A., Guthrie C. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev. 1990 Aug;4(8):1345–1356. doi: 10.1101/gad.4.8.1345. [DOI] [PubMed] [Google Scholar]
- Castaño J. G., Ornberg R., Koster J. G., Tobian J. A., Zasloff M. Eukaryotic pre-tRNA 5' processing nuclease: copurification with a complex cylindrical particle. Cell. 1986 Aug 1;46(3):377–385. doi: 10.1016/0092-8674(86)90658-6. [DOI] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doersen C. J., Guerrier-Takada C., Altman S., Attardi G. Characterization of an RNase P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. J Biol Chem. 1985 May 25;260(10):5942–5949. [PubMed] [Google Scholar]
- Engelke D. R., Gegenheimer P., Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1271–1279. [PubMed] [Google Scholar]
- Evans C. F., Engelke D. R. Yeast extracts for transfer RNA gene transcription and processing. Methods Enzymol. 1990;181:439–450. doi: 10.1016/0076-6879(90)81142-h. [DOI] [PubMed] [Google Scholar]
- Garber R. L., Altman S. In vitro processing of B. mori transfer RNA precursor molecules. Cell. 1979 Jun;17(2):389–397. doi: 10.1016/0092-8674(79)90165-x. [DOI] [PubMed] [Google Scholar]
- Gardiner K., Pace N. R. RNase P of Bacillus subtilis has a RNA component. J Biol Chem. 1980 Aug 25;255(16):7507–7509. [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Gold H. A., Altman S. Reconstitution of RNAase P activity using inactive subunits from E. coli and HeLa cells. Cell. 1986 Jan 31;44(2):243–249. doi: 10.1016/0092-8674(86)90758-0. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science. 1984 Jan 20;223(4633):285–286. doi: 10.1126/science.6199841. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. M1 RNA with large terminal deletions retains its catalytic activity. Cell. 1986 Apr 25;45(2):177–183. doi: 10.1016/0092-8674(86)90381-8. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Structure in solution of M1 RNA, the catalytic subunit of ribonuclease P from Escherichia coli. Biochemistry. 1984 Dec 18;23(26):6327–6334. doi: 10.1021/bi00321a006. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Haydock K., Allen L., Altman S. Metal ion requirements and other aspects of the reaction catalyzed by M1 RNA, the RNA subunit of ribonuclease P from Escherichia coli. Biochemistry. 1986 Apr 8;25(7):1509–1515. doi: 10.1021/bi00355a006. [DOI] [PubMed] [Google Scholar]
- Hannig E. M., Thiele D. J., Leibowitz M. J. Saccharomyces cerevisiae killer virus transcripts contain template-coded polyadenylate tracts. Mol Cell Biol. 1984 Jan;4(1):101–109. doi: 10.1128/mcb.4.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth M. J., Martin N. C. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol. 1986 Apr;6(4):1058–1064. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R. Direct identification of small sequence changes in chromosomal DNA. Gene. 1986;44(1):151–158. doi: 10.1016/0378-1119(86)90056-9. [DOI] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R. Direct sequence and footprint analysis of yeast DNA by primer extension. Methods Enzymol. 1991;194:550–562. doi: 10.1016/0076-6879(91)94042-b. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Shimura Y., Sakano H., Ozeki H. Precursor molecules of Escherichia coli transfer RNAs accumulated in a temperature-sensitive mutant. J Mol Biol. 1975 Jul 25;96(1):69–86. doi: 10.1016/0022-2836(75)90182-5. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S. K., Gurevitz M., Apirion D. A small RNA that complements mutants in the RNA processing enzyme ribonuclease P. J Mol Biol. 1982 Dec 15;162(3):515–533. doi: 10.1016/0022-2836(82)90386-2. [DOI] [PubMed] [Google Scholar]
- James B. D., Olsen G. J., Liu J. S., Pace N. R. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell. 1988 Jan 15;52(1):19–26. doi: 10.1016/0092-8674(88)90527-2. [DOI] [PubMed] [Google Scholar]
- Jones E. W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977 Jan;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline L., Nishikawa S., Söll D. Partial purification of RNase P from Schizosaccharomyces pombe. J Biol Chem. 1981 May 25;256(10):5058–5063. [PubMed] [Google Scholar]
- Kole R., Baer M. F., Stark B. C., Altman S. E. coli RNAase P has a required RNA component. Cell. 1980 Apr;19(4):881–887. doi: 10.1016/0092-8674(80)90079-3. [DOI] [PubMed] [Google Scholar]
- Koski R. A., Bothwell A. L., Altman S. Identification of a ribonuclease P-like activity from human KB cells. Cell. 1976 Sep;9(1):101–116. doi: 10.1016/0092-8674(76)90056-8. [DOI] [PubMed] [Google Scholar]
- Krupp G., Cherayil B., Frendewey D., Nishikawa S., Söll D. Two RNA species co-purify with RNase P from the fission yeast Schizosaccharomyces pombe. EMBO J. 1986 Jul;5(7):1697–1703. doi: 10.1002/j.1460-2075.1986.tb04413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence N. P., Altman S. Site-directed mutagenesis of M1 RNA, the RNA subunit of Escherichia coli ribonuclease P. The effects of an addition and small deletions on catalytic function. J Mol Biol. 1986 Sep 20;191(2):163–175. doi: 10.1016/0022-2836(86)90253-6. [DOI] [PubMed] [Google Scholar]
- Leontis N., DaLio A., Strobel M., Engelke D. Effects of tRNA-intron structure on cleavage of precursor tRNAs by RNase P from Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Mar 25;16(6):2537–2552. doi: 10.1093/nar/16.6.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58(2-3):201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Miller D. L., Martin N. C. Characterization of the yeast mitochondrial locus necessary for tRNA biosynthesis: DNA sequence analysis and identification of a new transcript. Cell. 1983 Oct;34(3):911–917. doi: 10.1016/0092-8674(83)90548-2. [DOI] [PubMed] [Google Scholar]
- Motamedi H., Lee K., Nichols L., Schmidt F. J. An RNA species involved in Escherichia coli ribonuclease P activity. Gene cloning and effect on transfer RnA synthesis in vivo. J Mol Biol. 1982 Dec 15;162(3):535–550. doi: 10.1016/0022-2836(82)90387-4. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Ogden R. C., Abelson J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983 Nov;35(1):117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- O'Connor J. P., Peebles C. L. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jan;11(1):425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W., Bellatin J. A., Lockheart A. Altered maturation of sequences at the 3' terminus of 5S gene transcripts in a Saccharomyces cerevisiae mutant that lacks a RNA processing endonuclease. EMBO J. 1983;2(3):353–359. doi: 10.1002/j.1460-2075.1983.tb01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W., Lockheart A., Bellatin J. The effects of protein synthesis inhibition, and of mutations rna1.1 and rna82.1, on the synthesis of small RNAs in yeast. FEBS Lett. 1987 Apr 6;214(1):143–148. doi: 10.1016/0014-5793(87)80030-3. [DOI] [PubMed] [Google Scholar]
- Piper P. W., Stråby K. B. Processing of transcripts of a dimeric tRNA gene in yeast uses the nuclease responsible for maturation of the 3' termini upon 5 S and 37 S precursor rRNAs. FEBS Lett. 1989 Jul 3;250(2):311–316. doi: 10.1016/0014-5793(89)80745-8. [DOI] [PubMed] [Google Scholar]
- Plautz G., Apirion D. Processing of RNA in Escherichia coli is limited in the absence of ribonuclease III, ribonuclease E and ribonuclease P. J Mol Biol. 1981 Jul 15;149(4):813–819. doi: 10.1016/0022-2836(81)90360-0. [DOI] [PubMed] [Google Scholar]
- Reed R. E., Altman S. Repeated sequences and open reading frames in the 3' flanking region of the gene for the RNA subunit of Escherichia coli ribonuclease P. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5359–5363. doi: 10.1073/pnas.80.17.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. E., Baer M. F., Guerrier-Takada C., Donis-Keller H., Altman S. Nucleotide sequence of the gene encoding the RNA subunit (M1 RNA) of ribonuclease P from Escherichia coli. Cell. 1982 Sep;30(2):627–636. doi: 10.1016/0092-8674(82)90259-8. [DOI] [PubMed] [Google Scholar]
- Reich C., Gardiner K. J., Olsen G. J., Pace B., Marsh T. L., Pace N. R. The RNA component of the Bacillus subtilis RNase P. Sequence, activity, and partial secondary structure. J Biol Chem. 1986 Jun 15;261(17):7888–7893. [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sakano H., Yamada S., Ikemura T., Shimura Y., Ozeki H. Temperature sensitive mutants of Escherichia coli for tRNA synthesis. Nucleic Acids Res. 1974 Mar;1(3):355–371. doi: 10.1093/nar/1.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl P., Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M., Anand R., Brown W. R., Fletcher D. S. A model for the separation of large DNA molecules by crossed field gel electrophoresis. Nucleic Acids Res. 1987 Aug 11;15(15):5925–5943. doi: 10.1093/nar/15.15.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M. C., Abelson J. Intron mutations affect splicing of Saccharomyces cerevisiae SUP53 precursor tRNA. Mol Cell Biol. 1986 Jul;6(7):2674–2683. doi: 10.1128/mcb.6.7.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Huber H. E., Richardson C. C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987 Nov 25;262(33):16212–16223. [PubMed] [Google Scholar]
- Wang M. J., Davis N. W., Gegenheimer P. Novel mechanisms for maturation of chloroplast transfer RNA precursors. EMBO J. 1988 Jun;7(6):1567–1574. doi: 10.1002/j.1460-2075.1988.tb02981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]