Abstract
Although copy number variations (CNVs) are expected to affect various diseases, little is known about the association between CNVs and breast cancer susceptibility. Therefore, we investigated this relation. Array comparative genomic hybridization was performed to search for candidate CNVs related to breast cancer susceptibility. Subsequent quantitative real-time polymerase chain reaction was carried out for confirmation. We found seven CNV markers associated with breast cancer risk. The means of the relative copy numbers of patients with a history of breast cancer and women in the control group were 0.8 and 1.8 for Hs06535529_cn on 1p36.12 (P < 0.0001), 2.9 and 2.2 for Hs03103056_cn on 3q26.1 (P < 0.0001), 1.2 and 1.8 for Hs03899300_cn on 15q26.3 (P < 0.0001), 1.0 and 1.5 for Hs03908783_cn on 15q26.3 (P < 0.0001), and 1.1 and 1.7 for Hs03898338_cn on 15q26.3 (P < 0.0001), respectively. Interestingly, nine or more copies of Hs04093415_cn on 22q12.3 were found only in 8/193 (4.1 %) patients with a history of breast cancer and in none of the controls (P = 0.0081). Similarly, 12 or more copies of Hs040908898_cn on 22q12.3 were found only in 7/193 (3.6 %) patients with a history of breast cancer and in none of the controls (P = 0.016). A combination of two CNVs resulted in 80.3 % sensitivity, 80.6 % specificity, 82.4 % positive predictive value, and 78.3 % negative predictive value for the prediction of breast cancer susceptibility. These findings may lead to a new means of risk assessment for breast cancer. Confirmatory studies using independent data sets are needed to support our findings.
Keywords: CNV, Breast cancer susceptibility, CGH, Real-time PCR, Digital PCR
Introduction
Breast cancer is a multifactorial disease caused by genetic and environmental factors [1]. So far, genetic studies have identified four high-penetrance genes (BRCA1, BRCA2, TP53, and PTEN) related to breast cancer [2]. In addition, genetic variations including single-nucleotide polymorphisms (SNPs), small insertion–deletion polymorphisms, and variable numbers of repetitive sequences have been reportedly associated with breast cancer risk, comprising 51 variants in 40 genes graded as a strong relation for 10 variants in 6 genes (ATM, CASP8, CHEK2, CTL4, NBN, and TP53), moderate for 4 variants for 4 genes (ATM, CYP19A1, TERT, and XRCC3), and weak for 37 variants [3].
Another variation in the human genome is that of genomic structural variants including copy number variations (CNVs) [4]. The CNVs involve gains or losses of several to hundreds of kilobases of genomic DNA among phenotypically normal individuals, and at least 11,700 CNV regions larger than 443 bp have been identified [5]. CNVs have been shown to significantly influence messenger RNA expression levels [6, 7], and recent studies have described associations of CNVs with various common disorders [8] as well as with mental illness [9]. As examples, The Wellcome Trust Case Control Consortium identified three CNVs associated with common diseases: IRGM for Crohn’s disease; HLA for Crohn’s disease, rheumatoid arthritis, and type 1 diabetes; and TSPAN8 for type 2 diabetes [10]. In regard to neoplasms, CNVs have recently been reported as factors predisposing individuals to neuroblastoma, prostate cancer, pancreatic cancer, colorectal cancer, and BRCA1-associated ovarian cancer [6, 11–15]. Although CNVs are expected to affect breast cancer risk, little is known about this association except for a previous report in which the proportion of rare CNVs was excessive in patients with hereditary breast cancer without BRCA1/BRCA2 mutations compared with controls [16]. These gaps, in our knowledge, prompted us to study this relation. Here, we report that CNVs significantly affect the susceptibility to breast cancer.
Materials and methods
The study protocol was approved by the institutional review board of Yamaguchi University Graduate School of Medicine, and informed consent was obtained from each patient.
Screening of CNVs by array comparative genomic hybridization
We obtained 30 DNA samples from the peripheral blood of women without a history of breast cancer and 30 DNA samples from the peripheral blood of patients with a history of breast cancer. A pool of blood-derived DNA from the 30 healthy women was used as a reference sample for all hybridizations performed. Assessment of the CNVs in the human genome by oligonucleotide array comparative genomic hybridization (CGH) (human CGH 2.1 M whole-genome tiling array; Roche NimbleGen) was performed according to the manufacturer’s protocol. Array image analysis and normalization were performed with NimbleScan version 2.5 software (Roche NimbleGen). The normalized data were then processed using Nexus Copy Number version 5.0 software (BioDiscovery).
Copy number validation by real-time polymerase chain reaction
Quantitative real-time polymerase chain reaction (PCR) using predesigned TaqMan® Copy Number Assays (Applied Biosystems) containing a primer pair and a FAM dye-labeled minor groove binder (MGB) probe was performed to detect the copy number of the genomic sequence of interest using a larger cohort. For the internal control, a predesigned TaqMan® Copy Number Reference Assay RNase P (Applied Biosystems), which is known to exist in two copies in a diploid genome, was used. We obtained 193 DNA samples from the peripheral blood of patients with a history of breast cancer and 170 DNA samples from age-matched women without a history of breast cancer. The mean age was 57.3 years in the patient group and 55.6 years in the control group. There was no statistical difference in age distribution between the groups. The calibrator sample for quantitative real-time PCR was the DNA pooled from 30 healthy women; the same was used as the reference in the array CGH assay, and the copy number of the calibrator sample was assumed to be 2. The 7900HT system and the StepOnePlus system (Applied Biosystems) were used for the quantitative real-time PCR analysis. The PCRs were carried out according to the manufacturer’s protocol.
TA cloning
To confirm the DNA sequence, a part of the real-time PCR products were gel purified and cloned into the T/A cloning vector pGEM-T Easy (Promega). At least five subclones were isolated and identified by direct sequencing.
Copy number validation by digital PCR
Digital PCR was available for six CNVs including Hs06535529_cn, Hs03899300_cn, Hs03908783_cn, Hs03898338_cn, Hs04090898_cn, and Hs040904315_cn to evaluate absolute copy numbers. Regarding Hs03103056_cn, digital PCR was not available because of difficulties in designing primers and probes for digital PCR. To evaluate the copy number of Hs03899300_cn, we designed forward and reverse primers and a TaqMan® MGB probe of Hs03899300_cn region and hTERT. hTERT was used as the internal control because it is known to exist in two copies in a diploid genome [17]. The primers were 5′-TGCCTGGCACTAAGGTTTAGAGTT-3′ (forward) and 5′-CACTCAGAGGGTTAAGTGAAGTGACA-3′ (reverse) for the Hs03899300_cn region and 5′-GGGTCCTCGCCTGTGTACAG-3′ (forward) and 5′-CCTGGGAGCTCTGGGAATTT-3′ (reverse) for hTERT. The probes were 5′-FAM-TGAGTCGGTGCTTCC-MGB-3′ for the Hs03899300_cn region and 5′-VIC-CACACCTTTGGTCACTC-MGB-3′ for hTERT. We designed these primers and probes to avoid SNPs. Regarding other CNVs, the same Copy Number Assays used in the real-time PCR were available. Reaction mixtures of 20-μL volume comprising 1× ddPCR Master Mix (Bio-Rad), forward and reverse primers and probes for a target and a reference, and DNA were prepared. PCR amplification was performed for a total of 40 cycles with an annealing temperature of 58 °C. Digital PCR was carried out using a QX100 droplet digital PCR system (BioRad) according to the manufacturer’s protocol [18].
Statistical analysis
A Fisher’s exact test, an unpaired t test, a Mann–Whitney test, linear regression analysis, and linear discriminant analysis were used to compare variables. A P value of <0.05 was considered to be significant. Data were analyzed with GraphPad Prism version 4.03, GraphPad InStat version 3.10 (GraphPad Software), and Ekuseru-Toukei 2008 (Social Survey Research Information).
Results
Using array CGH, we found four CNV regions with significant differences in the frequency of copy number changes between the patient group and the control group. The CNV positions were chr1:21,500,972-21,505,481; chr3:162,215,705-162,235,598; chr15:102,029,706-102,034,387; and chr22:37,142,958-37,147,755 (GRCh37/hg19). The CNVs detected by array CGH, however, could be false positives because a poor signal-to-noise ratio of hybridizations leads to considerable variation in the reported CGH ratio [19], and smaller CNVs are much more likely to be false positives than are large CNVs [20]. Therefore, quantitative real-time PCR with a larger cohort was carried out to confirm the CNVs associated with breast cancer susceptibility. We identified seven CNV markers related to breast cancer risk as shown in Table 1. The means of the relative copy numbers of patients with a history of breast cancer and those of women in the control group were 0.8 and 1.8 for Hs06535529_cn on 1p36.12 (P < 0.0001), 2.9 and 2.2 for Hs03103056_cn on 3q26.1 (P < 0.0001), 1.2 and 1.8 for Hs03899300_cn on 15q26.3 (P < 0.0001), 1.0 and 1.5 for Hs03908783_cn on 15q26.3 (P < 0.0001), and 1.1 and 1.7 for Hs03898338_cn on 15q26.3 (P < 0.0001), respectively (Fig. 1). The copy number of the Hs03899300_cn region on 15q26.3 by digital PCR was consistent with that by real-time PCR (Fig. 2), and the decision coefficient (r 2) was 0.9801. Also, copy numbers of other CNVs by digital PCR and by real-time PCR were well correlated: r 2 was 0.9201 for Hs06535529_cn, 0.8450 for Hs03908783_cn, 0.8909 for Hs03898338_cn, 0.9958 for Hs04090898_cn, and 0.9491 for Hs04093415_cn. Interestingly, nine or more copies of Hs04093415_cn on 22q12.3 were found only in eight (4.1 %) patients with a history of breast cancer and in none of the controls (P = 0.0081, Fig. 1 and Table 2). Similarly, 12 or more copies of Hs04090898_cn on 22q12.3 were found only in 7 (3.6 %) patients with a history of breast cancer and in none of the controls (P = 0.0160, Fig. 1 and Table 2). After setting a copy number threshold, we evaluated the relation between the copy number events and breast cancer susceptibility. The sensitivity and specificity were 83.9 and 41.2 % for Hs06535529_cn, 39.4 and 90.0 % for Hs03103056_cn, 76.7 and 70.0 % for Hs03899300_cn, 79.8 and 45.3 % for Hs03908783_cn, 83.4 and 65.9 % for Hs03898338_cn, 4.1 and 100.0 % for Hs04093415_cn, and 3.6 and 100.0 % for Hs04090898_cn (Table 2). Linear discriminant analysis with combination of two CNVs resulted in 80.3 % sensitivity, 80.6 % specificity, 82.4 % positive predictive value, and 78.3 % negative predictive value for the prediction of breast cancer susceptibility. The discriminant score was calculated as follows: Y = −6.9X1 + 3.2X2 + 6.1, where X 1 = the copy number of Hs03899300_cn and X 2 = the copy number of Hs03908783_cn.
Table 1.
CNV markers related to breast cancer risk
| Copy number assay ID | Sequence | Location | Gene | Copy number variation ID |
|---|---|---|---|---|
| (GRCh37/hg19) | (Database of genomic variants) | |||
| Hs06535529_cn | TCGCTGTGCCTGATTTCAGAGCCGGTTTCT | chr1:21,502,843 | EIF4G3 | None |
| GCGGTAAACTCATGGCAAAGCGAAGCCAC | −21,502,924 | |||
| CAACCCCCCCAGAGCGGGACCGG | ||||
| Hs03103056_cn | TGGCAACATCTCAATATCCRCAGAATTTTC | chr3:162,223,478 | None | 2483, 62120, 103483, 115882, |
| ATATTTATCCAGGTAGAATTGATAAACAGA | −162,223,593 | 32527, 37991, 30185, 50989, 2483, | ||
| AAATTCCACAAGAACCATAAATTATTTAAC | 62120, 103483, 115882, 32527, | |||
| ACATACACACACACACTCAAATTTAG | 37991, 30185, 50989 | |||
| Hs03899300_cn | ACTGCCTGGCACTAAGGTTTAGAGTTATGA | chr15:102,028,397 | PCSK6 | 34506, 5327, 3984 |
| GTCGGTGCTTCCCTGTCACTTCACTTAACCC | −102,028,502 | |||
| TCTGAGTGTGCAGTTTGTAGATTTGTTAACT | ||||
| GCACTGAGAGGTCC | ||||
| Hs03908783_cn | GCCTGCCTCCCRGCATGGGCCGCGGCCTCC | chr15:102,030,424 | None | 34506, 66907, 5327, 3984 |
| GCCATGGGCTCCGTGCGGTGGTTTCTCGGG | −102,030,520 | |||
| TACACGCTCGTGAGCCYGGCTGATGCGCCA | ||||
| CATGCCT | ||||
| Hs03898338_cn | ATCGCTGCTGGATCTCTTCTGTCATCCCTCC | chr15:102,031,024 | None | 34506, 5327, 3984 |
| CAGGACCCATTGGTCCTACTGGCCCACTTC | −102,031,100 | |||
| CAGAAAGCAAGCCATC | ||||
| Hs04093415_cn | GTGTCGAGGCTGCTCCTTAAAYGCTTCTTG | chr22:37,143,784 | None | 36022, 36023, 7346, 110470, 36024, |
| CCTGCACGCTGTGCGTGGAAACCCAAAGA | −37,143,858 | 22687, 103172, 23103, 115199, | ||
| AGTGAGAGACGCGAGG | 62002, 6148, 115197, 59075, 79571, | |||
| 22687, 103172, 23103, 115199, | ||||
| 62002, 6148, 115197, 59075, 79571, | ||||
| 110470, 36024, 36022, 36023, 7346 | ||||
| Hs04090898_cn | CTCCTAGTGGGATCCTACAACTCTCAGAAC | chr22:37,145,991 | None | 36022, 36023, 36024, 22687, 103172, |
| AACAGGGTCCCCCTGGACTGTGAGCACAGT | −37,146,097 | 23103, 62002, 6148, 115197, 59075, | ||
| AGAACCAGCTCTTTCTTGGGATTTTAAGAA | 91054, 7347, 79570, 91053, 36022, | |||
| AACAGACAAGCTTCGCG | 36023, 36024, 22687, 103172, | |||
| 23103, 62002, 6148, 115197, 59075, | ||||
| 91054, 7347, 79570, 91053 |
Fig. 1.
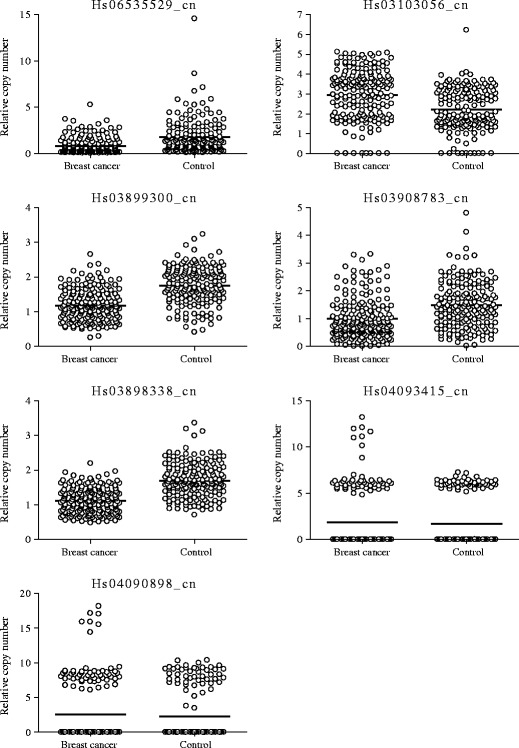
Distribution of copy numbers in patients with a history of breast cancer and in women in the control group. Each sample is indicated by an open circle. The horizontal lines represent the mean copy number in each group
Fig. 2.
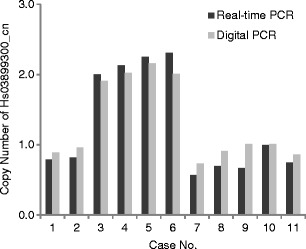
Comparison of Hs03899300_cn copy number between real-time PCR and digital PCR evaluation. Dark and light gray bars represent the copy numbers evaluated by real-time PCR and by digital PCR, respectively
Table 2.
Relation between CNVs and breast cancer susceptibility
| Copy number assay ID | Copy number threshold | Breast cancer | Control | Odds ratio | P value |
|---|---|---|---|---|---|
| (n = 193, %) | (n = 170, %) | ||||
| Hs06535529_cn | <1.5 | 162 (83.9) | 100 (58.8) | 3.7 | <0.0001 |
| Hs03103056_cn | 3.5≤ | 76 (39.4) | 17 (10.0) | 5.8 | <0.0001 |
| Hs03899300_cn | <1.5 | 148 (76.7) | 51 (30.0) | 7.7 | <0.0001 |
| Hs03908783_cn | <1.5 | 154 (79.8) | 93 (54.7) | 3.3 | <0.0001 |
| Hs03898338_cn | <1.5 | 161 (83.4) | 58 (34.1) | 9.7 | <0.0001 |
| Hs04093415_cn | 9.0≤ | 8 (4.1) | 0 (0.0) | 15.6 | 0.0081 |
| Hs04090898_cn | 12.0≤ | 7 (3.6) | 0 (0.0) | 13.7 | 0.0160 |
Discussion
In the current study, we identified CNV loci associated with breast cancer susceptibility. Our results, however, contrast with the study of Craddock et al. [10], who reported that there was no association between CNVs and breast cancer risk. This discrepancy is likely caused by the differences in the array-CGH platforms and analytic tools used. Different calling algorithms in the analytic tools give substantially a different quantity and quality of CNV calls even when identical raw data are used as the input [21]. Differences in preprocessing, labeling, and hybridization protocols, which were performed according to the various manufacturers’ specifications, could contribute to the occurrence of false-negative and false-positive calls [22]. Therefore, comparison of data sets resulting from different platforms and/or different analytic tools will cause problems in association analysis and can create false association signals [21]. To evaluate a copy number exactly, it is necessary to follow a validation study using a different methodology such as that of real-time PCR [22].
In the current study, we found that the copy numbers of Hs03899300_cn, Hs03908783_cn, and Hs03898338_cn, which are located close to each other on 15q26.3, were similar by real-time PCR. These findings were also observed between Hs04093415_cn and Hs040908898_cn on 22q12.3. Furthermore, the copy number of six CNVs including Hs06535529_cn, Hs03899300_cn, Hs03908783_cn, Hs03898338_cn, Hs04090898_cn, and Hs040904315_cn evaluated by digital PCR confirmed the accuracy of the data from the real-time PCR. Thus, false positives and negatives from the real-time PCR could be excluded. To our knowledge, this is the first report to show a distinct relation between CNVs and breast cancer risk.
Interestingly, 9 or more copies of Hs04093415_cn and 12 or more copies of Hs040908898 were observed only in patients with a history of breast cancer, and odds ratios for breast cancer susceptibility were 19.8 and 17.4, respectively. Such high odds ratios suggest strong oncogenic effect in these regions. Because mutations of high-penetrance genes for breast cancer (BRCA1, BRCA2, TP53, and PTEN) have not been tested, and familial history was not available in the present study, further studies are required to elucidate the association of the CNVs and hereditary breast cancer syndromes.
In the current study, some of the CNV regions related to breast cancer susceptibility contained genes such as EIF4G3 and PCSK6. Eukaryotic initiation factor 4 gamma 3 (EIF4G3) is a protein critical for initiation of protein translation [23]. To date, no relation of EIF4G3 with cancer development has been reported. We hypothesize that the decrease in the germline copy number of EIF4G3 may lead to a reduction or failure in translation of some transcripts and possibly give malignant potential to cells. Further examination will be required to elucidate this speculation. Proprotein convertase subtilisin/kexin type 6 (PCSK6) is a member of the protease family of proprotein convertases that activate precursor proteins by cleaving at the specific recognition sequence RXK/RR [24]. The relation between PCSK6 expression and carcinogenesis is controversial. Some investigations reported that overexpression of PCSK6 in immortalized nontumorigenic or papilloma-derived keratinocytes increased their invasiveness [25], whereas other studies linked absent or reduced PCSK6 expression levels to ovarian cancer [26]. Regarding breast cancer, overexpression of prosegment ppPCSK6 resulted in significant enhancement in cell motility, migration, and invasion of collagen in vitro [27]. However, because the effect of the reduced copy number of PCSK6 on normal mammary gland cells has not yet been investigated, further examination will be required to understand the function of PCSK6 in the neoplastic process.
The fact that no genes were mapped to the rest of the CNV regions raises a question as to how such CNVs affect breast cancer development. A possible explanation is that new gene transcripts may exist within the CNVs. Indeed, Diskin et al. found a new gene transcript related to neuroblastoma within the 1q21.1 CNV region where no known genes had been mapped [6]. Another hypothesis is that noncoding RNAs may be involved, such as long intergenic noncoding RNAs that regulate chromatin states and epigenetic inheritance, but knowledge of the molecular mechanisms of their function are still lacking [28]. Because the function of the CNVs is still unknown, further examinations will be required.
In summary, we found several unique CNVs associated with breast cancer. These CNVs may be feasible markers for assessment of the risk of breast cancer. However, as we cannot exclude the possibility that some women without a history of breast cancer may develop breast cancer in the future because the lifetime risk of developing breast cancer in Japan is 6 % [29], confirmatory studies using independent data sets are needed to support our findings.
Acknowledgments
This work was supported by Grants-in-Aid for Science Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan; the Ministry of Economy, Trade and Industry; and the Japan Science and Technology Agency.
Conflicts of interest
None
Abbreviations
- bp
Base pair
- CNV
Copy number variation
- CGH
Comparative genomic hybridization
- PCR
Polymerase chain reaction
- SNPs
Single-nucleotide polymorphisms
References
- 1.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40(1):17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Beeghly-Fadiel A, Long J, Zheng W. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 2011;12(5):477–488. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 5.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 6.Diskin SJ, Hou C, Glessner JT, Attiyeh EF, Laudenslager M, Bosse K, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459(7249):987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanciulli M, Petretto E, Aitman TJ. Gene copy number variation and common human disease. Clin Genet. 2010;77(3):201–213. doi: 10.1111/j.1399-0004.2009.01342.x. [DOI] [PubMed] [Google Scholar]
- 9.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148(6):1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464(7289):713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W, Sun J, Li G, Zhu Y, Zhang S, Kim ST, et al. Association of a germ-line copy number variation at 2p24.3 and risk for aggressive prostate cancer. Cancer Res. 2009;69(6):2176–2179. doi: 10.1158/0008-5472.CAN-08-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucito R, Suresh S, Walter K, Pandey A, Lakshmi B, Krasnitz A, et al. Copy-number variants in patients with a strong family history of pancreatic cancer. Canc Biol Ther. 2007;6(10):1592–1599. doi: 10.4161/cbt.6.10.4725. [DOI] [PubMed] [Google Scholar]
- 13.Thean LF, Loi C, Ho KS, Koh PK, Eu KW, Cheah PY. Genome-wide scan identifies a copy number variable region at 3q26 that regulates PPM1L in APC mutation-negative familial colorectal cancer patients. Gene Chromosome Canc. 2010;49(2):99–106. doi: 10.1002/gcc.20724. [DOI] [PubMed] [Google Scholar]
- 14.Venkatachalam R, Verwiel ET, Kamping EJ, Hoenselaar E, Gorgens H, Schackert HK, et al. Identification of candidate predisposing copy number variants in familial and early-onset colorectal cancer patients. Int J Cancer. 2011;129(7):1635–1642. doi: 10.1002/ijc.25821. [DOI] [PubMed] [Google Scholar]
- 15.Yoshihara K, Tajima A, Adachi S, Quan J, Sekine M, Kase H, et al. Germline copy number variations in BRCA1-associated ovarian cancer patients. Gene Chromosome Canc. 2011;50(3):167–177. doi: 10.1002/gcc.20841. [DOI] [PubMed] [Google Scholar]
- 16.Krepischi AC, Achatz MI, Santos EM, Costa SS, Lisboa BC, Brentani H, et al. Germline DNA copy number variation in familial and early-onset breast cancer. Breast Canc Res. 2012;14(1):R24. doi: 10.1186/bcr3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84(2):1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39(7 Suppl):S16–S21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007;39(7 Suppl):S48–S54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- 21.Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29(6):512–520. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tucker T, Montpetit A, Chai D, Chan S, Chenier S, Coe BP, et al. Comparison of genome-wide array genomic hybridization platforms for the detection of copy number variants in idiopathic mental retardation. BMC Med Genomics. 2011;4:25. doi: 10.1186/1755-8794-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcet-Palacios M, Duggan BL, Shostak I, Barry M, Geskes T, Wilkins JA, et al. Granzyme B inhibits vaccinia virus production through proteolytic cleavage of eukaryotic initiation factor 4 gamma 3. PLoS Pathog. 2011;7(12):e1002447. doi: 10.1371/journal.ppat.1002447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molloy SS, Anderson ED, Jean F, Thomas G. Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 1999;9(1):28–35. doi: 10.1016/S0962-8924(98)01382-8. [DOI] [PubMed] [Google Scholar]
- 25.Mahloogi H, Bassi DE, Klein-Szanto AJ. Malignant conversion of non-tumorigenic murine skin keratinocytes overexpressing PACE4. Carcinogenesis. 2002;23(4):565–572. doi: 10.1093/carcin/23.4.565. [DOI] [PubMed] [Google Scholar]
- 26.Fu Y, Campbell EJ, Shepherd TG, Nachtigal MW. Epigenetic regulation of proprotein convertase PACE4 gene expression in human ovarian cancer cells. Mol Canc Res. 2003;1(8):569–576. [PubMed] [Google Scholar]
- 27.Lapierre M, Siegfried G, Scamuffa N, Bontemps Y, Calvo F, Seidah NG, et al. Opposing function of the proprotein convertases furin and PACE4 on breast cancer cells' malignant phenotypes: role of tissue inhibitors of metalloproteinase-1. Cancer Res. 2007;67(19):9030–9034. doi: 10.1158/0008-5472.CAN-07-0807. [DOI] [PubMed] [Google Scholar]
- 28.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato H, Sobue T, Katanoda K, Saito Y, Tsukuma H, Saruki N, et al. Cancer Statistics in Japan. Tokyo: Foundation for Promotion of Cancer Research; 2012. [Google Scholar]


