SUMMARY
Cytosine methylation is involved in various biological processes such as silencing of transposable elements (TEs) and imprinting. Multiple pathways regulate DNA methylation in different sequence contexts, but the factors that regulate DNA methylation at a given site in the genome largely remain unknown. Here we have surveyed the methylomes of a comprehensive list of 86 Arabidopsis gene silencing mutants by generating single-nucleotide resolution maps of DNA methylation. We find that DNA methylation is site specifically regulated by different factors. Furthermore, we have identified additional regulators of DNA methylation. These data and analyses will serve as a comprehensive community resource for further understanding the control of DNA methylation patterning.
INTRODUCTION
The Arabidopsis genome is methylated in CG, CHG, and CHH (where H = A, T, or C) sequence contexts (Law and Jacobsen, 2010). It is understood that distinct pathways regulate methylation in each of the three sequence contexts. CG methylation is maintained by METHYLTRANSFERASE 1 (MET1), the plant homolog of mammalian DNA (cytosine-5)-methyltransferase 1 (DNMT1), and CHG methylation is maintained by CHROMOMETHYLASE 3 (CMT3). KRYPTONITE (KYP/SUVH4), SUVH5, and SUVH6 are the primary H3K9 methyltransferases and are required for CMT3 activity (Ebbs and Bender, 2006; Jackson et al., 2002; Lindroth et al., 2001). DOMAINS REARRANGED METHYLTRANSFERASES 1 (DRM1) and 2 (DRM2), plant homologs of mammalian DNMT3, are responsible for CHH methylation through the RNA-directed DNA methylation (RdDM) pathway, which involves two plant specific RNA polymerases, RNA Pol IV and Pol V, as well as 24 nucleotide (24 nt) small interfering RNAs (siRNAs). Although these three main DNA methylation pathways exist, it is notable that the chromatin remodeler DECREASE IN DNA METHYLATION 1 (DDM1) is required for the maintenance of CG and non-CG methylation (Jeddeloh et al., 1999; Vongs et al., 1993). Some interplay between DNA methylation pathways has been reported; however, the extent is largely unknown due to the lack of genome-wide analyses. Studies thus far on DNA methylation have usually been restricted to a few selected loci or insensitive methods such as immunostaining.
Whole-genome bisulfite sequencing (BS-seq) enables determination of methylation levels at single-nucleotide resolution (Cokus et al., 2008; Lister et al., 2008). Here, we have generated high-coverage genome-wide maps of the Arabidopsis methylome in 86 mutants in the same genetic background and tissue type. Along with the current view that distinct pathways control CG, CHG, and CHH methylation, we also found that DNA methylation is regulated in a site-specific manner involving interplays between different pathways. Our results provide a comprehensive view of the regulation of DNA methylation patterning in the Arabidopsis genome. In addition, our results revealed several unexpected features. Close examination of RNAi mutants suggested that specific sites in the genome might be regulated by RNAi factors not involved in the DRM1/2 pathway. Mutation in the chromatin assembly factor 1 (CAF-1) complex, which was thought not to regulate DNA methylation, induced CHG hypermethylation. We also found that RNA Pol II is required for DNA methylation largely independent of Pol IV and Pol V, suggesting an additional pathway for heterochromatin formation in plants. Finally, we found that one of the Su(var)3-9 related genes, SUVR2, is involved in RdDM, revealing a new component in the pathway. Our results open new areas of future research, and our data set allows one to determine the factor(s) involved in controlling DNA methylation at a given cytosine in the genome, and thus will serve as a platform for further studies.
RESULTS AND DISCUSSION
Single-Nucleotide Resolution Maps of DNA Methylation
We performed whole-genome BS-seq on 86 mutants utilizing tissue of the same developmental stage (3-week-old leaves) and in a single ecotype (Columbia) so that we could carefully detect methylation differences due to genotype. By deeply sequencing each mutant, we obtained an average coverage of 43-fold (Table S1). The methylation data are displayed in a modified UCSC genome browser (http://genomes.mcdb.ucla.edu/AthBSseq/). Differentially methylated regions (DMRs) were determined by comparing methylation levels in each mutant to three independent wild-type replicates in 100 base pair tiles throughout the genome (see Experimental Procedures).
Regulation of CG Methylation
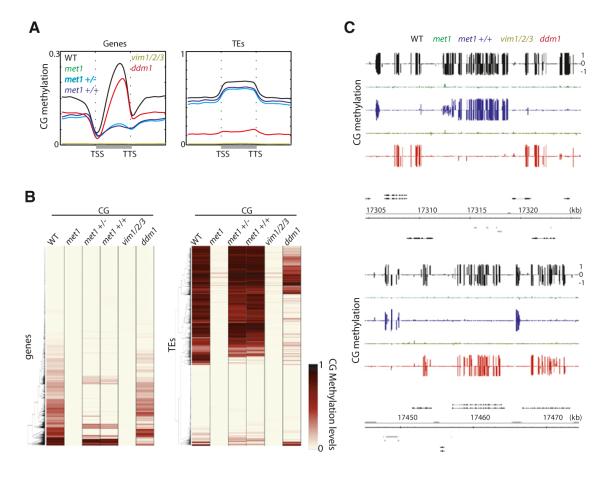
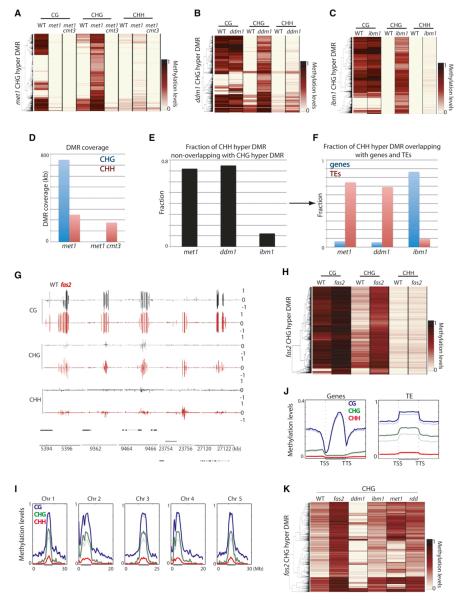
CG methylation is the most abundant type of DNA methylation. CG methylation is present over heterochromatic regions enriched with transposable elements (TEs) and repeats, as well as genic regions (Cokus et al., 2008; Lister et al., 2008). This is in contrast to CHG and CHH methylation, which are almost exclusively present in heterochromatin (Cokus et al., 2008; Lister et al., 2008). Mutation of the CG methyltransferase MET1 results in elimination of CG methylation throughout the genome (Figures 1A and 1B; Figures S1A and S1B available online) (Cokus et al., 2008; Lister et al., 2008). VARIANT IN METHYLATION 1 (VIM1), VIM2, and VIM3 are orthologous to mammalian UBIQUITIN-LIKE, CONTAINING PHD AND RING FINGER DOMAINS 1 (UHRF1) and have been shown to regulate CG methylation (Feng et al., 2010; Woo et al., 2008). In vim1 vim2 vim3 (vim1/2/3), CG methylation was strongly reduced resembling met1 (Figures 1A and 1B; Figures S1A and S1B). Notably, vim1, vim2, and vim3 individually did not affect CG methylation, indicating complete functional redundancy in regulating CG methylation (Figure S1C). Either met1 +/+ or +/− progeny of met1 +/− heterozygous plants have morphological defects, which led us to investigate their methylomes. We found that although TEs largely had wild-type methylation levels, genic methylation was severely impaired (Figures 1A–1C). Hence, our results suggest that genic methylation cannot be restored once lost and is consistent with previous studies suggesting that siRNAs (which are exclusively associated with heterochromatin) are required for restoration of DNA methylation in mutants of chromatin remodeler DDM1 (Teixeira et al., 2009). In ddm1, some heterochromatic DNA methylation has been shown to be reduced (Lippman et al., 2004), and DNA methylation is lost progressively upon inbreeding (Kakutani et al., 1996). We tested 7th generation homozygous ddm1 and found that heterochromatic DNA methylation is severely lost in ddm1; however, genic methylation remained largely intact (Figures 1A–1C; Figure S1A). Hence, DDM1 controls DNA methylation specifically at heterochromatin (Lippman et al., 2004).
Figure 1. CG Methylation.
(A) Average distribution of CG methylation over protein-coding genes (left) and TEs (right). Flanking regions are the same length as the gene or TE body (middle region). TSS = transcription start site. TTS = transcription termination site. (B) Heatmap of CG methylation levels (black, 1; white, 0) within all genes and TEs in chromosome 1. Columns represent data for each indicated genotype, and rows represent the genes/TEs. The rows were sorted by complete linkage hierarchical clustering with Euclidean distance as a distance measure. (C) Genome browser views of CG methylation in chromosome 1. Genes (black bars) and TEs (gray bars) are shown below. See also Figure S1 and Table S1.
Regulation of CHG Methylation
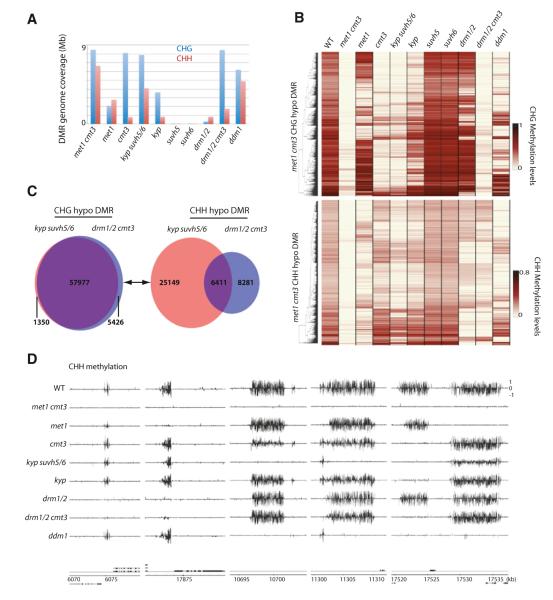
CMT3 is the main CHG methyltransferase in Arabidopsis (Law and Jacobsen, 2010). Indeed, we observed a strong depletion of CHG methylation in cmt3 (Figure 2A). H3K9 methyltransferases KYP, SUVH5, and SUVH6 have been shown to be required for CMT3-dependent CHG methylation (Ebbs and Bender, 2006). Loss of CHG methylation in kyp suvh5 suvh6 (kyp suvh5/6) closely mimicked the loss of CHG methylation in cmt3 (Figure 2B; Figure S2A). Hence, KYP SUVH5/6 regulate CHG methylation through CMT3 genome-wide. We further tested redundancies between KYP, SUVH5, and SUVH6. We found that KYP was solely responsible for certain CHG methylation sites in the genome, whereas mutations in SUVH5 or SUVH6 alone did not show any detectable alterations in CHG methylation (Figure 2B).
Figure 2. Non-CG Methylation.
(A) Genome coverage of defined CHG and CHH hypomethylation DMRs. (B) Heatmap of methylation levels within 17,437 met1 cmt3 CHG (top) and 13,776 CHH (bottom) hypomethylation DMRs. (C) Overlap between kyp suvh5/6 and drm1/2 cmt3 hypomethylation DMRs. (D) Genome browser views of CHH methylation in chromosome 1. Genes (black bars) are shown below. See also Figure S2 and Table S1.
Although a strong depletion of CHG methylation was observed in cmt3, there were patches of DNA methylation in the genome that were not affected (Figure 2B). We found that CHG methylation at these sites often depended on DRM1/2 (Figure 2B). Although DRM1/2 are suggested to be CHH methyltransferases, our results confirm that they also regulate CHG methylation (Cao and Jacobsen, 2002a). DRM1/2 regulate CHG methylation at specific sites in the genome, 60.4% of which sites are nonoverlapping with sites regulated by CMT3 (Figure S2A). To test the degree of redundancy between CMT3 and DRM1/2, we profiled drm1/2 cmt3 triple mutants. We did not observe many additional losses of CHG methylation in drm1/2 cmt3 (Figure S2B), suggesting that CMT3 and DRM1/2 regulate CHG methylation in a mostly nonredundant fashion.
Regulation of CHH Methylation
CHG methylation and CHH methylation highly colocalize in the wild-type genome (Cokus et al., 2008; Lister et al., 2008). Hence, we tested whether loss of CHG methylation is associated with loss of CHH methylation. In cmt3, loss of CHG methylation is only partially associated with loss of CHH methylation (Figure 2B; Figure S2M). Hence, although CMT3 is required for the majority of CHG methylation in the genome, it is required for a relatively small proportion of CHH methylation. In contrast, in drm1/2, loss of CHG methylation was always associated with loss of CHH methylation (Figure S2N). Loss of CHH methylation, however, was not always coupled with loss of CHG methylation (Figure S2N). Thus, CHH methylation maintenance appears more reliant on CHG methylation than CHG methylation is on CHH methylation.
Interestingly, whereas kyp and cmt3 showed very similar losses in CHH methylation, kyp suvh5/6 showed much stronger losses of CHH methylation compared to cmt3 (Figures 2A–2C; Figure S2C). KYP SUVH5/6-dependent clusters of CHH methylation were generally nonoverlapping with DRM1/2-dependent CHH methylation (Figures 2B–2D; Figure S2C). Hence, although KYP SUVH5/6 control CHG methylation through CMT3, our results suggest that KYP SUVH5/6 strongly regulate CHH methylation through a different pathway. Notably, mutations in factors responsible for siRNA biogenesis such as RNA-dependent RNA polymerase 2 (RDR2) and DICER-LIKE 2, 3 and 4 (DCL2/3/4) (discussed more in detail below) did not disrupt CHH methylation at most KYP SUVH5/6-regulated sites (Figure S2O). Thus, KYP SUVH5/6 regulate CHH methylation in a siRNA-independent manner.
Different methylation pathways appeared to target different classes of TEs (Figure S2R). One insight was that both cmt3 and kyp CHH DMRs were overrepresented by LTR/Copia type TEs. The overlap between cmt3 and kyp was high; 71.0% of cmt3 CHH TE DMRs overlapped with kyp CHH TE DMRs (Figure S2S).
Interdependence of CG and Non-CG Methylation
Previous studies based on immunostaining and single-loci ChIP analyses have suggested that mutation in MET1 causes loss of H3K9m2 at certain sites (Soppe et al., 2002; Tariq et al., 2003). Consistent with these findings, we found loss of CHG methylation at certain sites in met1 (Figure 2B). Comparing met1 and vim1/2/3, we found 85.1% overlap between sites that lose CHG methylation, suggesting that CG methylation is required for proper CHG methylation at those sites (Figure S1B). Loss of CHG methylation was observed at a subset of sites in met1 +/+ and met1 +/− progenies of met1 +/− (Figure S2P). These sites corresponded to the subset of heterochromatic sites that did not restore CG methylation (Figure S2P), further supporting the notion that CG methylation is required for maintaining CHG methylation. Loss of CHG methylation in met1 largely occurred at certain KYP SUVH5/6 and CMT3-dependent CHG sites (Figure 2B; Figures S2E and S2G). However, loss of CHH methylation in met1 largely did not overlap with KYP SUVH5/6 and CMT3-dependent CHH sites (Figures S2F and S2H). Hence, although MET1 regulates CHG methylation through KYP SUVH5/6 and CMT3, it regulates CHH methylation mostly through a different pathway.
Although met1 CHH DMRs were much more abundant compared to drm1/2 CHH DMRs, 63.0% of drm1/2 CHH DMRs overlapped with met1 CHH DMRs (Figure 2B; Figures S2I and S2J). This overlap was significantly higher than observed for CMT3 and KYP SUVH5/6-dependent sites (11.4% and 26.7%, respectively). This and the fact that drm1/2 has minimal disruption of CG methylation (Figure S2N) suggest a strong tendency for DRM1/2 targeted methylation to depend on CG methylation. Wild-type CG methylation levels at CG-methylation-dependent and -independent DRM1/2 target sites were similar (Figure S2Q). Therefore, the features that determine whether a DRM1/2 site is dependent on CG methylation or not is unclear.
An additional insight was that met1 cmt3 caused strong reduction in both CHG and CHH methylation. In fact, met1 cmt3 most severely affected CHH methylation of all the mutants we tested (Figure 2A). met1 cmt3 reduced CHH methylation at many additional sites compared to met1 or cmt3 alone (Figures S2K and S2L), suggesting that MET1 and CMT3 cooperatively regulate the bulk of CHH methylation in the genome. Our results indicate a strong genome-wide dependence of asymmetric CHH methylation on symmetrical CG and CHG methylation.
Mutation in DDM1 also disrupted CHG and CHH methylation, where loss of DNA methylation generally occurred at sites regulated by KYP SUVH5/6 rather than sites regulated by DRM1/2 (Figure 2B). Only 27.3% of drm1/2 CHG DMRs and 23.1% of drm1/2 CHH DMRs overlapped with corresponding ddm1 DMRs. Hence, unlike MET1, DDM1 is largely not required for DRM1/2-dependent methylation.
We also found that CG methylation is dependent on non-CG methylation at certain sites. Loss of CG methylation was associated with loss of non-CG methylation at a subset of sites in kyp suvh5/6, cmt3, and drm1/2 (Figures S2M and S2N). For example, although loss of methylation in drm1/2 mostly occurred in CHH and CHG contexts, 18.5% of drm1/2 CHH DMRs were associated with loss in CG methylation. Among sites where CHH methylation was lost in both drm1/2 and kyp suvh5/6, sites that lost CG methylation in drm1/2 and kyp suvh5/6 were largely overlapping (77.8% of those in drm1/2 overlapped with kyp suvh5/6). This confirms that it is likely that the loss of non-CG methylation causes the loss of CG methylation; 71.8% of DRM1/2 CHH DMRs that also lost CG methylation were sites where CG methylation was required for CHH methylation. These sites are interesting as CG methylation and non-CG methylation become interdependent.
Comparison of Regions Methylated by KYP SUVH5/6, CMT3, and DRM1/2
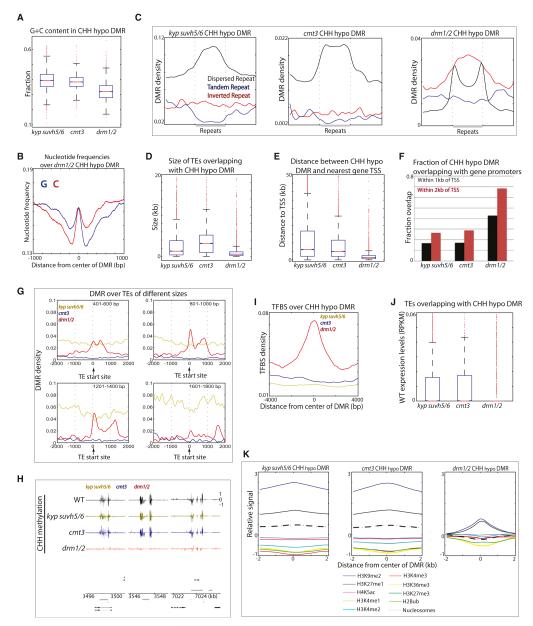
We further examined the characteristics of sites affected in kyp suvh5/6, cmt3, and drm1/2. DRM1/2 target sites were associated with relatively lower G+C sequence composition (Figure 3A; Figure S3A) and showed a tendency of GC skewing (Figure 3B; Figure S3B). Association with inverted repeats was also a unique feature of drm1/2 DMRs (Figure 3C; Figure S3C). Although siRNAs are produced throughout most heterochromatic sites in the genome, it is unclear how DRM1/2 are specifically targeted to a subset of these sites. Our results suggest that sequence composition may be one of the factors that determines which methylation pathway is targeted at a given site in the genome.
Figure 3. Characteristics of CHH Sites Regulated by KYP SUVH5/6, CMT3, and DRM1/2.
(A) G+C content ((G+C)/(G+C+A+T)) in CHH hypomethylation DMRs. Red lines, median; edges of boxes, 25th (bottom) and 75th (top) percentiles; error bars, minimum and maximum points within 1.5×IQR (interquartile range); red dots, outliers. (B) Base composition over drm1/2 CHH hypomethylation DMRs. (C) Average distribution of CHH hypomethylation DMRs (DMR per bp) over different repeats. Flanking regions are the same length as the repeat (middle region). (D) Boxplots of sizes of TEs that overlap with CHH hypomethylation DMRs. (E) Boxplots of distances between CHH hypomethylation DMRs and the closest gene TSS. (F) Fraction of CHH hypomethylation DMRs that are within 1 kb or 2 kb from TSS. (G) Average distribution of DMRs over TEs of indicated sizes. Negative x axis scale is outside of TEs, and positive x axis scale is toward the body of TEs. (H) Genome browser views showing loss of methylation spikes at boundaries of TEs in drm1/2 in chromosome 1. Genes (black bars) and TEs (gray bars) are shown below. (I) TFBS (TFBS per bp) over CHH hypomethylation DMRs. (J) Wild-type expression levels of TEs overlapping with CHH hypomethylation DMRs. (K) Average histone modification and nucleosome distributions over CHH hypomethylation DMRs. See also Figure S3 and Table S1.
We found that DRM1/2 targets small TEs, whereas KYP SUVH5/6 and CMT3 target large TEs (Figure 3D; Figure S3D) (Tran et al., 2005). TEs regulated by DRM1/2 were proximal to promoters of genes, where around ~70% of drm1/2 DMRs fell within 2 kb of transcription start sites of protein-coding genes (Figures 3E and 3F, Figures S3E, F) (Zhong et al., 2012). These proximal genes were not significantly associated with particular biological processes (data not shown). Another distinction was that DRM1/2 specifically methylated the boundaries of TEs (Figures 3G and 3H; Figures S3G and S3H). We next examined the levels of enrichment of transcription factor binding sites (TFBS) at DMRs. We found enrichment of TFBS over drm1/2 DMRs but not over kyp suvh5/6 and cmt3 DMRs (Figure 3I; Figures S3I and S3L). Hence, DRM1/2 target sites tend to be regulatory sites. We then sought to measure the expression levels of TEs in wild-type by performing RNA sequencing. We found that DRM1/2 targeted TEs are more silent compared to CMT3-targeted TEs (Figure 3J; Figure S3J), presumably because they are closer to genes and thus potentially harmful if expressed.
Finally, we examined the distribution of nucleosomes and known histone modifications over DMRs. Consistent with the notion that CMT3 is dependent on H3K9 methylation, kyp suvh5/6 and cmt3 DMRs were associated with higher levels of H3K9me2 compared to drm1/2 DMRs (Figure 3K; Figure S3K). kyp suvh5/6 and cmt3 DMRs were also associated with higher levels of nucleosome occupancy compared to levels in drm1/2 DMRs (Figure 3K; Figure S3K). Hence, different methylation pathways regulate sites with distinct genomic and epigenomic characteristics.
RNA-Directed DNA Methylation
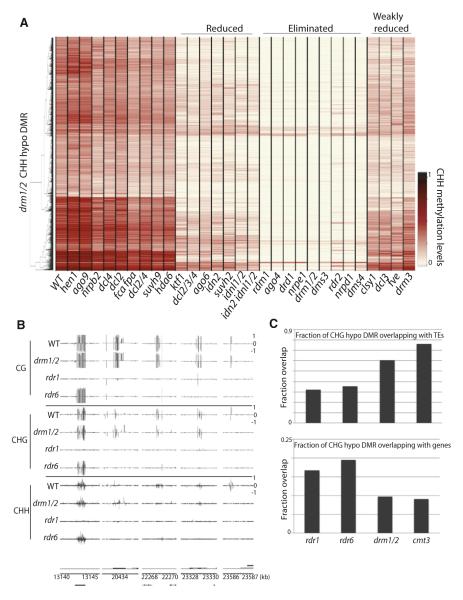
The RdDM pathway involves many accessory factors that guide DNA methylation by DRM2 (Law and Jacobsen, 2010). We sought to examine whether disruption of components of the pathway result in similar DNA methylation defects. We tested 29 mutants previously suggested to affect RdDM (Gu et al., 2011; He et al., 2009; Henderson et al., 2006; Law and Jacobsen, 2010; Zheng et al., 2009). By examining methylation levels at DRM1/2-dependent CHH sites, we found that there are differential effects when disrupting components of the RdDM pathway (Figure 4A). Broadly, there are four classes of RdDM components: those where mutation in the gene eliminates DRM1/2-dependent methylation, those where mutation reduces methylation, those where mutation weakly reduces methylation, and those that only affect a very small proportion of sites (some of which we describe below) (Figure 4A). Importantly, AGO4 and AGO6 were suggested to be partially redundant (Zheng et al., 2007). However, our results suggest that mutation in AGO4 alone is sufficient to eliminate DRM1/2-dependent methylation.
Figure 4. RNA-Directed DNA Methylation.
(A) Heatmap of methylation levels within 4,949 drm1/2 CHH hypomethylation DMRs. Genotypes (columns) have also been clustered. (B) Genome browser views of DNA methylation in chromosome 1. Genes (black bars) are shown below. (C) Overlap of rdr1 and rdr6 CHG hypomethylation DMRs with TEs and genes. See also Figure S4 and Table S1.
The flowering-time regulators FCA and FPA were suggested to be responsible for DNA methylation at certain RdDM sites (Baürle et al., 2007). We did not observe global reduction of DNA methylation at RdDM sites in fca fpa but did find minor alterations in methylation (Figure S4A), in which 69 out of 86 (80.3%) defined fca fpa CHH DMRs overlapped with drm1/2 DMRs. Consistent with the overlap, fca fpa DMRs were associated with promoters of genes (Figure S4B). Analyses of fca and fpa single mutants revealed partial redundancies (Figure S4C).
Although DICER-LIKE 3 (DCL3) cleaves double-stranded RNA (dsRNA) 24 nt siRNA and thus functions RdDM, DCL2 and DCL4 cleaves dsRNA into 22 nt and 21 nt siRNA, respectively, and function in other biological processes (Voinnet, 2008). However, functional redundancies between DCL3 and DCL2/4 have been suggested some loci (Henderson et al., 2006 Indeed, although dcl3 was categorized as a “weakly reduced” mutant, dcl2/3/4 was categorized as a “reduced” mutant (Figure 4A). Hence, in the absence DCL3, DCL2 and DCL4 can mediate DNA methylation at most RdDM sites.
RNAi Factors Are Involved in DNA Methylation
We further examined whether mutants of known RNAi components not implicated in the canonical RdDm pathway, including AGOs, DCLs, HEN1, RDRs, SDEs, and SGS3, affected DNA methylation. Of all mutants tested, RDR1 and RDR6, which are involved in pathways that yield 21 and 22 nt siRNAs, showed the strongest loss of DNA methylation (Figure 4B); 38.4% of rdr6 DMRs overlapped with rdr1 DMRs (Figure S4D). Only 60 out of 215 sites (27.9%) were also DMRs in drm1/2. Furthermore, unlike mutations in RdDM components, where DNA methylation was largely affected in non-CG contexts (Figure S2N), DNA methylation was lost in all three cytosine contexts in rdr1 and rdr6 (Figures S4E and S4F). One characteristic of these sites was that they were more likely to be associated with genes compared to DRM1/2 sites (Figure 4C). Another characteristic of these sites was that 21 and 22 nt siRNA levels in wild-type, measured by small RNA sequencing (Lee et al., 2012), were somewhat enriched compared to levels of 24 nt siRNAs (Figure S4G). Hence, RDR1 and RDR6 may be involved in DNA methylation independent of the DRM1/2 pathway.
Ectopic Gain of CHG Methylation
Gain of DNA methylation has been reported in several mutants, including DNA and histone demethylases as well as DNA methyltransferases. In the DNA demethylase mutant, ros1 dml2 dml3 (rdd), we found that hypermethylation occurred in all three cytosine contexts; however, ros3, which has been suggested to act in the same genetic pathway, showed only very limited hypermethylation (Figure S5A). In contrast, other tested mutants exhibited hypermethylation mostly in CHG contexts and to a lesser extent in the CHH contexts (Figures 5A–5C; Figure S5B). Hence, ectopic hypermethylation in these mutants occurs through a different mechanism (Figure S5C). Importantly, CHG hypermethylated sites in met1 were not necessarily hypermethylated in vim1/2/3 (Figure S5D) and vice versa, suggesting that the hypermethylation phenotype may not be explained directly by loss of CG methylation. This is in contrast to CHG hypomethylation, where met1 and vim1/2/3 caused loss of CHG methylation at very similar sites (Figure S1B). One interpretation for this result may be that CHG hypermethylation occurs stochastically (Mathieu et al., 2007), whereas CHG hypomethylation is a direct consequence of loss of CG methylation. It is worth noting that met1 +/+ and met1 +/− progeny from met1 +/− showed limited CHG hypermethylation and this did not occur at sites that lost CG methylation (Figure S5E). Collectively, our results suggest that CHG hypermethylation in met1 is not likely a phenomenon that compensates for the loss of CG methylation as previously proposed (Cokus et al., 2008; Lister et al., 2008). In met1 cmt3, the CHG hypermethylation is eliminated, indicating that the CHG hypermethylation phenotype in met1 is dependent on CMT3 (Figure 5A).
Figure 5. Ectopic Hypermethylation.
(A) Heatmap of methylation levels within 4,773 met1 CHG hypermethylation DMRs. (B) Heatmap of methylation levels within 2,695 ddm1 CHG hypermethylation DMRs. (C) Heatmap of methylation levels within 13,588 ibm1 CHG hypermethylation DMRs. (D) Genome coverage of defined CHG and CHH hypermethylation DMRs. (E) Fraction of CHH hypermethylation DMRs nonoverlapping with CHG hypermethylation DMRs. (F) Fraction of CHH hypermethylation DMRs nonoverlapping with CHG hypermethylation DMRs that overlap with genes and TEs. (G) Genome browser views of DNA methylation in wild-type and fas2 in chromosome 1. Genes (black bars) and TEs (gray bars) are shown below. (H) Heatmap of methylation levels within 1,572 fas2 CHG hypermethylation DMRs. (I) Chromosomal views of methylation in wild-type (faded lines) and fas2 (solid lines). Regions of pericentromeric heterochromatin are indicated by back bars below the graphs. (J) Average distribution of methylation levels over genes and TEs in wild-type (faded lines) and fas2 (solid lines). Upstream and downstream regions are the same length as the gene/TE (middle region). (K) Heatmap of methylation levels within fas2 CHG hypermethylation DMRs. See also Figure S5 and Table S1.
CHG hypermethylation in ddm1 occurred at distinct sites compared to met1 (Figure S5F). CHG hypermethylation in met1 and vim1/2/3 predominantly occurred in normally unmethylated regions (Figure 5A; Figure S5B); however, CHG hypermethylation in ddm1 occurred predominantly at regions only CG methylated (Figure 5B). This suggests that CHG hypermethylation in ddm1 is not likely due to loss of CG methylation and likely occurs through a different mechanism. Because ddm1 loses a significant amount of both CG and non-CG methylation, a speculation is that loss of non-CG methylation induces CHG hypermethylation at distinct loci in the genome. Histone H3K9 demethylase IBM1 has been suggested to protect CG methylated genes from becoming CHG methylated (Miura et al., 2009). We have confirmed these findings with BS-seq (Figure 5C). Notably, 49.1% of ddm1 CHG hypermethylation DMRs overlapped with those of ibm1 (Figure S5G). Because IBM1 transcripts are largely unaffected in ddm1 (Figure S5H), CHG hypermethylation in ddm1 is not likely due to impaired IBM1. These results suggest a relationship between the hypermethylation occurring in ddm1 and ibm1.
We next examined whether RdDM mutants show any CHG hypermethylation. We defined 79 CHG hypermethylated sites in drm1/2, and found that they tend to occur at sites immediately flanking TEs (Figure S5I). Because of the relatively small number of DMRs, we cannot rule out stochastic variations; however, we observed CHG hypermethylation across RdDM mutants at these same sites (with some variation) (Figure S5J). In drm1/2 cmt3, the CHG hypermethylation was suppressed (Figure S5J), suggesting that CMT3 is again required for CHG hypermethylation. Consistent with the fact that DNA methylation by DRM1/2 is largely regulated by MET1 (Figure S2I and S2J), 51.9% of drm1/2 CHG hypermethylation DMRs overlapped with those of met1, compared to 20.2% of those in ibm1. In summary our results suggest that loss of DNA methylation induces CHG hypermethylation through CMT3.
Ectopic Gain of CHH Methylation
We found that CHG hypermethylated sites were generally associated with CHH hypermethylation in the mutants tested (Figures 5A–5C; Figure S5K). In met1 cmt3, the CHH hypermethylation associated with CHG hypermethylation was suppressed, suggesting that CMT3 is responsible for CHH methylation at these sites (Figure S5K). However, met1 cmt3 exhibited comparable genome-wide CHH hypermethylation levels as met1 (Figure 5D). Interestingly, whereas 65.1% of CHH hypermethylation DMRs were associated with CHG hypermethylation in ibm1, this was the case for only 20.6% and 19.4% in met1 and ddm1, respectively (Figure 5E). Hence, in met1 and ddm1, the bulk of CHH hypermethylation occurs at distinct sites compared to CHG hypermethylation. CHH hypermethylation decoupled from CHG hypermethylation might be explained by transcriptional reactivation of TEs in these mutants, where TE transcripts become processed into siRNA, which then directs CHH methylation. Consistent with this idea, CHH hypermethylation DMRs not overlapping with CHG hypermethylation DMRs corresponded predominantly to TEs in met1 and ddm1 (Figure 5F). TEs that get CHH hypermethylated were overrepresented with LTR/Gypsy type TEs (Figure S5L). In summary, loss of global DNA methylation induces CHH hypermethylation that is largely distinct from the CHG hypermethylation phenomenon.
Mutation in the CAF-1 Complex Induces CHG Hypermethylation
The CAF-1 complex is required for proper heterochromatin formation. FASCIATA 1 (FAS1) and FAS2 are subunits of the CAF-1 complex, and their disruption results in reduced heterochromatin without disturbing DNA methylation at certain repeats (Schönrock et al., 2006). We studied the CAF-1 complex by testing fas2. We found that fas2 tended to exhibit hypermethylation in CHG contexts (1,572 defined sites) (Figures 5G and 5H). Whole-chromosomal views and average plots over TEs suggested modest genome-wide elevation of DNA methylation (Figures 5I and 5J). There was relatively little overlap between fas2 CHG hypermethylated DMRs and those of rdd, met1, ddm1, and ibm1 (4.5%, 18.4%, 3.9%, and 0.9%, respectively) (Figure 5K). CHG hypermethylation DMRs tended to overlap with TEs (60.2%) but not genes (14.0%). TEs that get CHG hypermethylated were somewhat overrepresented with LTR/Gypsy type TEs (Figure S5M). Hence, FAS2 is likely involved in an independent pathway to prevent hypermethylation of TEs.
RNA Pol II Is Involved in DNA Methylation Independent of Pol IV and Pol V
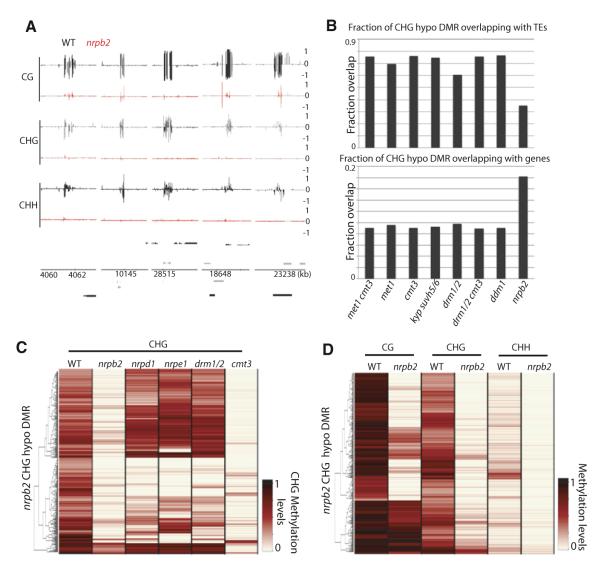
Pol IV and Pol V have likely evolved from Pol II and specifically function in RdDM (Haag and Pikaard, 2011). Pol II was suggested to be involved in regulating DNA methylation at certain intergenic sites by recruiting Pol IV and Pol V (Zheng et al., 2009). Using a weak Pol II mutant allele, nrpb2-3, we confirmed that nrpb2 has reduced DNA methylation at certain sites (Figure 6A). We found a tendency of nrpb2 DMRs to overlap with genic regions compared to drm1/2 DMRs (Figure 6B). Intriguingly, we found that 64.4% and 66.6% of nrpb2 DMRs did not overlap with nrpd1 and nrpe1 DMRs, respectively (Figure 6C), suggesting that for the most part Pol II regulates DNA methylation independently of Pol IV and Pol V. Furthermore, unlike nrpd1 and nrpe1, loss of DNA methylation in nrpb2 occurred in all three cytosine contexts (Figure 6D). Because we are limited to analyzing a weak Pol II allele, since null mutations in Pol II are lethal (Onodera et al., 2008), it is possible that Pol II regulates a larger proportion of DNA methylation in the genome. Hence, we provide evidence that Pol II itself is involved in a pathway that regulates DNA methylation.
Figure 6. RNA Pol II-Directed DNA Methylation.
(A) Genome browser views of DNA methylation in wild-type and nrpb2 in chromosome 1. Genes (black bars) and TEs (gray bars) are shown below. (B) Fraction of CHG hypomethylation DMRs overlapping with TEs and genes. (C and D) Heatmap of methylation levels within 413 nrpb2 CHG hypomethylation DMRs. See also Table S1.
Relationship between Histone Modifications and DNA Methylation
There is growing evidence regarding interplays between histone modifications and DNA methylation (Cedar and Bergman, 2009). In Arabidopsis, H3K9 methylation is required for CHG methylation (Law and Jacobsen, 2010). Loss of DNA methylation is accompanied by ectopic gain in H3K27me3 and H3K4me3 in plants and animals (Hon et al., 2012; Weinhofer et al., 2010;Zhang et al., 2009). We utilized several mutants known to regulate particular histone modifications, and examined the impact on DNA methylation. We tested mutations that alter histone modifications normally localized at DNA hypomethylated sites (H3K4me3 and H3K27me3) as well as those that are normally localized at DNA methylated sites (H3K36me3 and H3K27me1). We tested sdg8, which has reduced H3K36me3 (Xu et al., 2008); atxr5/6, which has reduced H3K27me1 (Jacob et al., 2009); sdg2, which has reduced H3K4me3 (Berr et al., 2010; Guo et al., 2010); and ref6, which has gain of H3K27me3 (Lu et al., 2011). We did not observe notable changes in DNA methylation in these mutants (Figures S6A–S6D). Hence, although H3K36me3 and H3K27me1 colocalize with DNA methylation, loss of these marks does not affect DNA methylation. And although loss of DNA methylation causes gain of H3K4me3 and H3K27me3, alteration of these marks does not affect DNA methylation. These results suggest that H3K9 methylation is the main histone modification that regulates DNA methylation in Arabidopsis.
Histone Deacetylase 6 Regulates DNA Methylation at Promoters
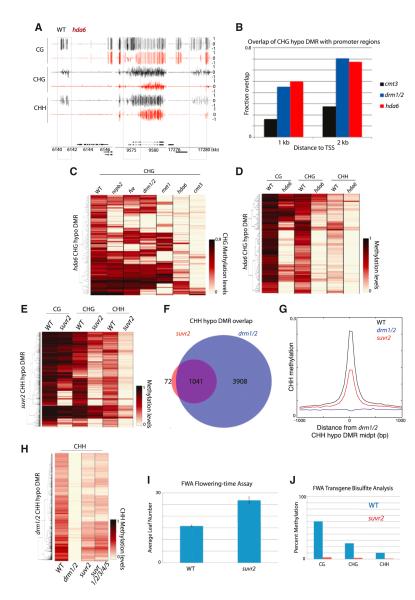
Previous studies have suggested that the histone deacetylase 6 (HDA6) plays a role in RdDM (Aufsatz et al., 2002; He et al., 2009). Studies of rDNA suggested that loss of HDA6 causes reduction in CG/CHG methylation as well as CHH hypermethylation (Earley et al., 2010). In addition, other studies have reported that HDA6 physically interacts with MET1 (Liu et al., 2012) and FVE (Gu et al., 2011), and transcriptionally regulates a similar subset of genes as MET1 (To et al., 2011). We analyzed hda6 and found that, unlike met1, the methylome was largely unaltered, except at particular sites of the genome (Figure 7A). These hypomethylated sites tended to be at promoters of genes, similar to the extent seen in drm1/2 (Figure 7B). However, only 27.5% of hda6 hypomethylation DMRs overlapped with those of drm1/2. Rather, 91.7% of hda6 DMRs corresponded with those methylated by CMT3 (Figure 7C). However, unlike drm1/2 and cmt3, where loss of methylation was largely restricted to non-CG contexts (Figures S2M and S2N), DNA methylation was lost in all three cytosine contexts (Figure 7D). This was similar to what was seen in nrpb2 (Figure 6D). Furthermore, 27.5% of hda6 DMRs overlapped with nrpb2 DMRs despite the fact that nrpb2 affects many fewer sites compared to drm1/2. Hence, although HDA6 has been suggested to interact with MET1 and FVE, our results suggest that HDA6 regulates DNA methylation at subsets of promoters through an independent mechanism. Further, at least in part, HDA6 appears to be associated with Pol II-directed DNA methylation.
Figure 7. Chromatin Modifiers Involved in DNA Methylation.
(A) Genome browser views of DNA methylation in wild-type and hda6 in chromosome 1. Genes (black bars) are shown below. (B) Fraction of CHG hypomethylation DMRs that are within 1 kb or 2 kb from TSS. (C and D) Heatmap of methylation levels within 120 hda6 CHG hypomethylation DMRs. (E) Heatmap of methylation levels within 1,113 suvr2 CHH hypomethylation DMRs. (F) Overlap between suvr2 and drm1/2 CHH hypomethylation DMRs. (G) Average distribution of CHH methylation over drm1/2 CHH hypomethylation DMRs. (H) Heatmap of methylation levels within 4,949 drm1/2 CHH hypomethylation DMRs. (I) FWA flowering-time assay. Approximately 20 plants were measured in each population. Data are represented as mean ± SEM. (J) FWA transgene bisulfite analysis. DNA methylation of the transgenic copy of FWA in FWA transformed plants was analyzed. See also Figure S6 and Table S1.
SUVR2 Is Involved in the DRM1/2 Pathway
Su(var)3-9 is a conserved factor required for gene silencing through H3K9 methylation (Schotta et al., 2003). Arabidopsis has 15 Su(var)3-9 homologs: ten SUVH genes and five SUVR genes (Baumbusch et al., 2001; Pontvianne et al., 2010). We performed BS-seq on each mutant except for suvr4 (due to lack of a knockout allele in a Col background). Interestingly, we found large losses of methylation in the suvr2 mutant, especially at CHH sites. We found 1,113 sites that CHH hypomethylated in suvr2, which, as in drm1/2 (Figure S2N), were often associated with loss of CHG and to a small extent loss in CG (Figure 7E). A total of 1,041 (93.5%) of suvr2 CHH DMRs overlapped with those in drm1/2 (Figure 7F). Comparison of methylation levels between suvr2 and drm1/2 suggested that suvr2 is a weak RdDM mutant (Figure 7G), with suvr2 falling into the “weakly reduced” (Figure 4A) class of RdDM mutants (data not shown). We also tested methylation levels in a suvr1/2/3/4/5 quintuple mutant (into which a Nossen ecotype allele of suvr4 had been introgressed) and did not observe additional methylation loss compared with suvr2 alone (Figure 7H), ruling out functional redundancies with other SUVR genes. Methylation analysis by Southern blot at the known RdDM target, MEDEA-INTERGENIC SUBTELOMERIC REPEATS (MEA-ISR), supported observations seen at the genome-wide level (Figure S6E).
In addition to their role in DNA methylation maintenance, RdDM pathway components also carry out DNA methylation establishment—or de novo methylation (Cao and Jacobsen, 2002b; Chan et al., 2004; Greenberg et al., 2011). Given its potential role as an effector of RdDM, we wanted to test whether SUVR2 also is required for de novo methylation. In order to do so, we utilized the FLOWERING WAGINENGEN (FWA) transgenic system. In the vegetative tissue of wild-type plants, FWA expression is repressed in a DNA methylation-dependent manner at tandem repeats in its 5′ UTR (Soppe et al., 2002). When FWA transgenes are introduced into wild-type plants, the repeats are targeted for de novo DNA methylation and silenced. However, in RdDM mutants, the transgene fails to be methylated, causing ectopic expression that leads to a late-flowering phenotype (Ausin et al., 2009; Chan et al., 2004; Greenberg et al., 2011). When we transformed suvr2 with the FWA transgene, the mutant plants flowered significantly later than wild-type controls (Figure 7I). Consistently, bisulfite analysis of the FWA transgene showed that DNA methylation was virtually absent in all three cytosine contexts (Figure 7J). Taken together, these results strongly indicate that SUVR2 is a canonical RdDM factor that is required for both DRM2 establishment and maintenance methylation.
In order to further place SUVR2 in the RdDM pathway, we performed small RNA northern blots (Figure S6F). RdDM proteins that act downstream of 24 nt siRNA biogenesis—such as NRPE1—only affect siRNA accumulation at a subset of targets, known as type I loci, but not type II loci (Zheng et al., 2009). We found that suvr2 behaved similarly to nrpe1, indicating that SUVR2 is not required for generation of siRNAs. Consistent with the methylation analysis, higher-order suvr mutants did not impact siRNA levels any more than suvr2 alone. In summary, our results indicate that SUVR2 is a new regulator of the DRM2 pathway that acts downstream of siRNA biogenesis.
CONCLUSION
In summary, by generating single-nucleotide resolution maps of the Arabidopsis methylome for a comprehensive list of mutants, we found interplays between different pathways and found additional regulators of DNA methylation. All DNA methylation data generated in this study can be viewed at our genome browser along with various epigenomic data. These genome-wide data sets and tools should serve as a community resource for further understanding DNA methylation patterning in Arabidopsis.
EXPERIMENTAL PROCEDURES
Detailed experimental and analysis methods can be found in the Extended Experimental Procedures.
Plant Material
All mutant lines used in this study were in the Columbia background. Exceptions are the ros1 dml2 dml3 line where each allele was introgressed into Col (Penterman et al., 2007), and suvr1/2/3/4/5, where suvr4 allele was in Nossen. First generation homozygous plants of met1, 2nd generation plants of ibm1 and vim1/2/3, and 7th generation ddm1 plants were used. Plants were grown under continuous light, and three-week-old leaves were used for all experiments.
Whole-Genome Bisulfite Sequencing
A total of 0.5~1 μg of genomic DNA was used to generate BS-seq libraries. Libraries were generated with premethylated adapters as previously described (Feng et al., 2011). Libraries were single-end sequenced on a HiSeq 2000 generating 50mer reads. Sequenced reads were base-called with the standard Illumina software. BS-seq reads were mapped to the TAIR10 genome with BS-seeker (Chen et al., 2010) allowing two mismatches. Identical reads were collapsed into one read. Methylation levels were calculated by the ratio of #C/(#C+#T). DMRs for each mutant were defined by comparing their methylation levels in each cytosine contexts with those of three independent wild-type data.
RNA Sequencing
Libraries were generated and sequenced following manufacturer instructions (Illumina).
Histone Modification Data
Previously published histone ChIP data (Bernatavichute et al., 2008; Roudier et al., 2011; Zhang et al., 2009) were used for analyses. Nucleosome positioning data were mapped by Micrococcal nuclease sequencing.
Southern Blot, FWA Transgene Assay, and Small RNA Northern Blot
Southern blot, FWA transgene assay, and small RNA northern blot were performed as previously described (Greenberg et al., 2011).
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Akhavan for Illumina sequencing; S. Cokus, W. Yan, and M. Pellegrini for help with the UCSC browser; X. Zhang for MNase protocols; and H. Vaucheret, X. Chen, C. Dean, M. Matzke, R. Amasino, X. Zhong, and L. Johnson for seeds. Sequencing was performed at the UCLA BSCRC Bio-Sequencing Core Facility. This work was supported by NIH grant GM60398 and NSF grants 0701745 and 1121245. H.S. was supported by a Fred Eiserling and Judith Lengyel Graduate Doctorate Fellowship. M.V.C.G. was supported by US Public Health Service National Research Service Award GM07104 and a UCLA Dissertation Year Fellowship. S.F. is a Special Fellow of the Leukemia and Lymphoma Society. S.E.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBER The GEO accession numbers for the bisulfite sequencing data reported in this paper are GSE39901 and GSE38286.
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures, six figures, and one table and can be found with this article online at http://dx. doi.org/10.1016/j.cell.2012.10.054.
REFERENCES
- Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat. Struct. Mol. Biol. 2009;16:1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baürle I, Smith L, Baulcombe DC, Dean C. Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science. 2007;318:109–112. doi: 10.1126/science.1146565. [DOI] [PubMed] [Google Scholar]
- Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, McCallum EJ, Ménard R, Meyer D, Fuchs J, Dong A, Shen WH. Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell. 2010;22:3232–3248. doi: 10.1105/tpc.110.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA. 2002a;99(Suppl 4):16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002b;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 2010;24:1119–1132. doi: 10.1101/gad.1914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18:1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Rubbi L, Jacobsen SE, Pellegrini M. Determining DNA methylation profiles using sequencing. Methods Mol. Biol. 2011;733:223–238. doi: 10.1007/978-1-61779-089-8_16. [DOI] [PubMed] [Google Scholar]
- Greenberg MV, Ausin I, Chan SW, Cokus SJ, Cuperus JT, Feng S, Law JA, Chu C, Pellegrini M, Carrington JC, Jacobsen SE. Identification of genes required for de novo DNA methylation in Arabidopsis. Epigenetics. 2011;6:344–354. doi: 10.4161/epi.6.3.14242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Jiang D, Yang W, Jacob Y, Michaels SD, He Y. Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet. 2011;7:e1002366. doi: 10.1371/journal.pgen.1002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Yu Y, Law JA, Zhang X. SET DOMAIN GROUP2 is the major histone H3 lysine [corrected] 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2010;107:18557–18562. doi: 10.1073/pnas.1010478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X., Lu, C., Johnson L, Meyers BC, Green PJ, Jacobsen SE. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 2009;16:763–768. doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. USA. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TF, Gurazada SG, Zhai J, Li S, Simon SA, Matzke MA, Chen X, Meyers BC. RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics. 2012;7:781–795. doi: 10.4161/epi.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu CW, Duan J, Luo M, Wang K, Tian G, Cui Y, Wu K. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 2012;158:119–129. doi: 10.1104/pp.111.184275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Jenuwein T, Cao X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 2011;43:715–719. doi: 10.1038/ng.854. [DOI] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 2009;28:1078–1086. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Nakagawa K, Haag JR, Pikaard D, Mikami T, Ream T, Ito Y, Pikaard CS. Sex-biased lethality or transmission of defective transcription machinery in Arabidopsis. Genetics. 2008;180:207–218. doi: 10.1534/genetics.108.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Pikaard CS. Arabidopsis Histone Lysine Methyltransferases. Adv. Bot. Res. 2010;53:1–22. doi: 10.1016/S0065-2296(10)53001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F, Ahmed I, Bérard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönrock N, Exner V, Probst A, Gruissem W, Hennig L. Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J. Biol. Chem. 2006;281:9560–9568. doi: 10.1074/jbc.M513426200. [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Reuter G. SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing. Genetica. 2003;117:149–158. doi: 10.1023/a:1022923508198. [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA. 2003;100:8823–8827. doi: 10.1073/pnas.1432939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- To TK, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Tanaka M, Endo T, Kakutani T, Toyoda T, et al. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 2011;7:e1002055. doi: 10.1371/journal.pgen.1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran RK, Zilberman D, de Bustos C, Ditt RF, Henikoff JG, Lindroth AM, Delrow J, Boyle T, Kwong S, Bryson TD, et al. Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol. 2005;6:R90. doi: 10.1186/gb-2005-6-11-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 2008;13:317–328. doi: 10.1016/j.tplants.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Weinhofer I, Hehenberger E, Roszak P, Hennig L, Köhler C. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 2010;6:e1001152. doi: 10.1371/journal.pgen.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Dittmer TA, Richards EJ. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 2008;4:e1000156. doi: 10.1371/journal.pgen.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 2008;28:1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat. Struct. Mol. Biol. 2012;19:870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.