Abstract
The function of neural circuits is an emergent property that arises from the coordinated activity of large numbers of neurons. To capture this, we propose launching a large-scale, international public effort, the Brain Activity Map Project, aimed at reconstructing the full record of neural activity across complete neural circuits. This technological challenge could prove to be an invaluable step toward understanding fundamental and pathological brain processes.
“The behavior of large and complex aggregates of elementary particles, it turns out, is not to be understood in terms of a simple extrapolation of the properties of a few particles. Instead, at each level of complexity entirely new properties appear.” –More Is Different, P.W. Anderson
“New directions in science are launched by new tools much more often than by new concepts. The effect of a concept-driven revolution is to explain old things in new ways. The effect of a tool-driven revolution is to discover new things that have to be explained.” –Imagined Worlds, Freeman Dyson
Emergent Properties of Brain Circuits
Understanding how the brain works is arguably one of the greatest scientific challenges of our time. Although there have been piecemeal efforts to explain how different brain regions operate, no general theory of brain function is universally accepted. A fundamental underlying limitation is our ignorance of the brain’s microcircuitry, the synaptic connections contained within any given brain area, which Cajal referred to as “impenetrable jungles where many investigators have lost themselves” (Ramón y Cajal, 1923). To explore these jungles, neuroscientists have traditionally relied on electrodes that sample brain activity only very sparsely—from one to a few neurons within a given region. However, neural circuits can involve millions of neurons, so it is probable that neuronal ensembles operate at a multineuronal level of organization, one that will be invisible from single neuron recordings, just as it would be pointless to view an HDTV program by looking just at one or a few pixels on a screen.
Neural circuit function is therefore likely to be emergent—that is, it could arise from complex interactions among constituents. This hypothesis is supported by the well-documented recurrent and distributed architecture of connections in the CNS. Indeed, individual neurons generally form synaptic contacts with thousands of other neurons. In distributed circuits, the larger the connectivity matrix, the greater the redundancy within the network and the less important each neuron is. Despite these anatomical facts, neurophysiological studies have gravitated toward detailed descriptions of the stable feature selectivity of individual neurons, a natural consequence of single-electrode recordings. However, given their distributed connections and their plasticity, neurons are likely to be subject to continuous, dynamic rearrangements, participating at different times in different active ensembles. Because of this, measuring emergent functional states, such as dynamical attractors, could be more useful for characterizing the functional properties of a circuit than recording receptive field responses from individual cells. Indeed, in some instances where large-scale population monitoring of neuronal ensembles has been possible, emergent circuit states have not been predictable from responses from individual cells.
Emergent-level problems are not unique to neuroscience. Breakthroughs in understanding complex systems in other fields have come from shifting the focus to the emergent level. Examples include statistical mechanics, nonequilibrium thermodynamics, and many-body and quantum physics. Emergent-level analysis has led to rich branches of science describing novel states of matter involving correlated particles, such as magnetism, superconductivity, superfluidity, quantum Hall effects, and macroscopic quantum coherence. In biological sciences, the sequencing of genomes and the ability to simultaneously measure genome-wide expression patterns have enabled emergent models of gene regulation, developmental control, and disease states with enhanced predictive accuracy.
We believe similar emergent-level richness is in store for circuit neuroscience. An emergent level of analysis appears to us crucial for understanding brain circuits. Likewise, the pathophysiology of mental illnesses like schizophrenia and autism, which have been resistant to traditional, single-cell level analyses, could potentially be transformed by their consideration as emergent-level pathologies.
The Brain Activity Map as the Functional Connectome
To elucidate emergent levels of neural circuit function, we propose to record every action potential from every neuron within a circuit—a task we believe is feasible. These comprehensive measurements must be carried out over time-scales on which behavioral output, or mental states, occur. Such recordings could represent a complete functional description of a neural circuit: a Brain Activity Map (BAM). This mapping will transcend the “structural connectome,” the static anatomical map of a circuit. Instead, we propose the dynamical mapping of the “functional connectome,” the patterns and sequences of neuronal firing by all neurons. Correlating this firing activity with both the connectivity of the circuit and its functional or behavioral output could enable the understanding of neuronal codes and their regulation of behavior and mental states. This emergent level of understanding could also enable accurate diagnosis and restoration of normal patterns of activity to injured or diseased brains, foster the development of broader biomedical and environmental applications, and even potentially generate a host of associated economic benefits.
Imaging Every Spike from Every Neuron
To achieve this vision, one clearly needs to develop novel technologies. To date, it has not been possible to reconstruct the full activity patterns of even a single region of the brain. While imaging technologies like fMRI or MEG can capture whole-brain activity patterns, these techniques lack single-cell specificity and the requisite temporal resolution to permit detection of neuronal firing patterns. To preserve single-cell information while recording the activity of complete circuits, vigorous efforts must be launched to massively upscale the capabilities of both imaging and nanoprobe sensing.
Over the last two decades, neuroscientists have made transformational advances in techniques to monitor the activity of neuronal ensembles. Optical techniques are minimally invasive and can provide great spatial and temporal flexibility, have single-cell resolution, and can be applied to living preparations, even awake behaving ones (Helmchen et al., 2011). Calcium imaging can measure the multineuronal activity of a circuit (Yuste and Katz, 1991) (Figure 1), and despite a limited time resolution, this technique can partially reconstruct firing patterns of large (>1,000) populations of neurons in vitro or in vivo (Grienberger and Konnerth, 2012).
Figure 1. Large-Scale Calcium Imaging of Neuronal Activity.

(A) Living brain slice from primary visual cortex of a mouse stained with the calcium indicator fura-2 AM. More than a thousand neurons are labeled and can be imaged with a two-photon microscope. From Yuste et al. (2011).
(B) The calcium concentration in the soma of a neuron (bottom) faithfully tracks the electrical firing pattern of the cell (top). From Smetters et al. (1999).
(C) Reconstructed “raster plot” of the spontaneous spiking activity of 754 cells from a similar experiment. From Cossart et al., 2003.
Calcium imaging, while useful, can only approximate the real functional signals of neurons, and it is preferable to capture the complete activity of a circuit by voltage imaging (Peterka et al., 2011). Current methods for voltage imaging in vertebrate circuits, however, cannot capture action potentials at a large scale with single-cell resolution. Novel voltage sensors with better signal-to-noise, less photodamage, and faster temporal resolution are needed. Continued improvements are being made in voltage indicators, and particularly promising are nanoparticles, small inorganic compounds that have large absorption and highly efficient emission. These are robust during extended illumination and can be very sensitive to the external electric field. Zero-dimensional nanoparticles, i.e., quantum dots, could be directly used to measure voltage in neurons. Other nanoparticles, such as nanodiamonds (Mochalin et al., 2012), may provide an even higher sensitivity to magnetic and electric fields. In addition, by acting as “antennas” for light, nanoparticles can greatly enhance optical signals emitted by more traditional voltage reporters.
But regardless of the method chosen for imaging neuronal activity, to capture all spikes from all neurons, one needs to increase the number of imaged neurons and extend the depth of the imaged tissue. A variety of recent advancements in optical hardware and computational approaches could overcome these challenges (Yuste, 2011). Novel methods include powerful light sources for two-photon excitation of deep tissue, faster scanning strategies, scanless approaches using spatio-light-modulators to “bathe” the sample with light, high-numerical aperture objectives with large fields of view, engineered point spread functions and adaptive optics corrections of scattering distortions, light-field cameras to reconstruct signals emanating in 3D, and, finally, advances in computational optics and smart algorithms that use prior information of the sample. A combination of many of these novel methods may allow simultaneous 3D imaging of neurons located in many different focal planes in an awake animal. In addition, GRIN fibers and endoscopes allow imaging deeper structures, such as the hippocampus, albeit with some invasiveness.
Large-Scale Electrical Recordings with Nanoprobes
Electrical recording of neuronal activity is now becoming possible on a massively parallel scale by harnessing novel developments in silicon-based nanoprobes (Figure 2). Silicon-based neural probes with several dozen electrodes are already available commercially; it is now feasible to record from dozens of sites per silicon neural probe, densely, at a pitch of tens of μm (Du et al., 2009a). Stacking of two-dimensional multishank arrays into three-dimensional probe arrays would provide the potential for hundreds of thousands of recording sites. There are technical hurdles to be surmounted, but when the technology is perfected, recording from many thousands of neurons is conceivable with advanced spike-sorting algorithms. The “Holy Grail” will be to record from millions of electrodes, keeping the same bandwidth, reducing the electrode pitch down to distances of ~15 μm, and increasing the probe length to cortical dimensions of several centimeters. This will require significant innovation in systems engineering.
Figure 2. Nanoprobes, Wireless, and Synthetic Biology Technologies for the BAM Project.
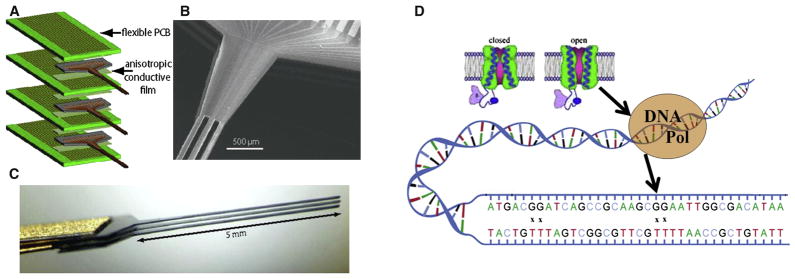
(Left) Silicon nanoprobe arrays (after Du et al., 2009b). (A) Flip-chip assembly scheme for connecting the silicon devices with printed circuit boards. (B) SEM micrograph of the rear section of a 50-μm-thick shaft array showing the multilayer stacked structure. Adjacent layers have a spacing of 100 μm, which is set by the thickness of the flexible cable. (C) Side view of the 50-μm-thick shaft array showing that the shafts are stress balanced and are able to retain approximately constant relative spacing.
(Right) Synthetic biology approaches. (D) A voltage sensitive calcium channel influences the error rate of an engineered DNA polymerase. X marks sites of mismatch between “T” in the template strand (lower) and “G” new copy strand. Note scale of the various devices and cells.
Wireless and Synthetic Biology Approaches
We also envision techniques for wireless, noninvasive readout of the activity of neuronal populations (Figure 2). These might include wireless electronic circuits based on silicon very large-scale integration (VLSI), synthetic biological components, or their hybrids. It is easy to underestimate the potential of today’s microelectronic technology, and we think that it will ultimately become feasible to deploy small wireless microcircuits, untethered in living brains, for direct monitoring of neuronal activity, although there are significant technological challenges.
As an alternative to silicon VLSI, synthetic biology might provide an interesting set of novel techniques to enable noninvasive recording of activity (Figure 2). This could be considered a wireless option, albeit a radically different one. For example, DNA polymerases could be used as spike sensors since their error rates are dependent on cation concentration. Prechosen DNA molecules could be synthesized to record patterns of errors corresponding to the patterns of spikes in each cell, encoded as calcium-induced errors, serving as a “ticker-tape” record of the activity of the neuron. The capability of DNA for dense information storage is quite remarkable. In principle, a 5-μm-diameter synthetic cell could hold at least 6 billion base pairs of DNA, which could encode 7 days of spiking data at 100 Hz with 100-fold redundancy.
A BAM Project Roadmap and Choice of Species
For any given circuit, the reconstruction of activity might proceed in three steps. First, initial mapping could be done using calcium imaging with spiking reconstruction carried out at 100 Hz. This could be performed with improvements to existing methods. The second step would involve voltage imaging of action potentials (and subthreshold electrical activity), ideally with a temporal resolution of 1 kHz. These first two steps could be carried out in 3D yet they would be limited to superficial structures (<2 mm deep). In a third step, similar reconstructions of neuronal activity, but penetrating deep into brain circuits, could be performed. These would first be achieved with massively multiplexed nanoprobes, later complemented by novel wireless approaches.
But which circuits should be worked on, and in which order? We envision parallel efforts on several different preparations—progressing from reconstructing the activity of small, simple circuits to more complicated, larger ones. For example, in the short term (5 years), one could reconstruct the activity of a series of small circuits, all less than 70,000 neurons, from model organisms. C. elegans is the only complete connectome (302 neurons and 7,000 connections) (White et al., 1986), and all of its neurons could be imaged simultaneously with two-photon imaging and genetic calcium indicators. In addition, one could reconstruct the entire activity pattern of a discrete region of the Drosophila brain, such as the medulla, with ~15,000 neurons. The Drosophila connectome is currently 20% complete at the mesoscale (Chiang et al., 2011), and could be finalized within three years. Finally, for vertebrate circuits, short-term goals could include imaging the activity of all the ganglion cells in a mouse retina (~50,000 neurons), the mitral cells in the mouse olfactory bulb (~70,000), or a mouse neocortical brain slice (~40,000, optically accessible neurons).
For midterm goals (10 years), one could image the entire Drosophila brain (135,000 neurons), the CNS of the zebra-fish (~1 million neurons), or an entire mouse retina or hippocampus, all under a million neurons. One could also reconstruct the activity of a cortical area in a wild-type mouse or in mouse disease models. Finally, it would also be interesting to consider mapping the cortex of the Etruscan shrew, the smallest known mammal, with only a million neurons.
For a long-term goal (15 years), we would expect that technological developments will enable the reconstruction of the neuronal activity of the entire neocortex of an awake mouse, and proceed toward primates. We do not exclude the extension of the BAM Project to humans, and if this project is to be applicable to clinical research or practice, its special challenges are worth addressing early. Potential options for a human BAM Project include wireless electronics, safely and transiently introducing engineered cells to make tight (transient) junctions with neurons for recording and possibly programmable stimulation, or a combination of these approaches.
Computational Analysis and Modeling
Our stated goal of recording every spike from every neuron raises the specter of a data deluge, so development of proactive strategies for data reduction, management, and analysis are important. To estimate data storage capacities required for the BAM we consider the anatomical connectome. Bock et al. (2011) have reconstructed 1,500 cell bodies with 1 × 1013 pixels (Bock et al., 2011). By analogy we can estimate that 7 × 106 mouse cortical cells would require ~5 × 1016 bytes. This is less data than the current global genome image data. Some might argue that analogies to genomics are limited in that brain activity is of much higher dimensionality than linear genomics sequences. But high-dimensional, dynamic transcriptome, immunome, and whole-body analyses are increasingly enabled by plummeting costs.
Brains are complex dynamical systems with operations on a very wide range of timescales, from milliseconds to years. Brain activity maps, like the broader “omics” and systems biology paradigms, will need (1) combinatorics, (2) the state dependence of interactions between neurons, and (3) neuronal biophysics, which are extremely varied, adapted, and complex. We envision the creation of large data banks where the complete record of activity of entire neural circuits could be freely downloadable. This could spur a revolution in computational neuroscience, since the analysis and modeling of a neural circuit will be possible, for the first time, with a comprehensive set of data. As the Human Genome Project generated a new field of inquiry (“Genomics”), the generation of these comprehensive data sets could enable the creation of novel fields of neuroscience.
Data Access and Ethical Considerations
We feel strongly that an effort such as the BAM Project should be put squarely in the public domain. Because it will require large-scale coordination between many participants, and because the information will benefit mankind in many ways, it makes sense for this project to be run as a public enterprise with unrestricted access to its resulting data.
There are also potential ethical ramifications of the BAM Project that will arise if this technology moves as swiftly as genomics has in the last years. These include issues of mind-control, discrimination, health disparities, unintended short- and long-term toxicities, and other consequences. Well in advance, the scientific community must be proactive, engaging diverse sets of stakeholders and the lay public early and thoughtfully.
Outcomes and Anticipated Benefits
The BAM Project will generate a host of scientific, medical, technological, educational, and economic benefits to society. Indeed, the widespread effect of this research underscores the need for it to be controlled by the public.
In terms of anticipated scientific benefits, the generation of a complete functional description of neural circuits will be invaluable to address many outstanding questions in neuroscience for which emergent functional properties could be key (Table 1). Together, answers to these questions can open the doors to deciphering the neural code, as well as unlocking the possibility of reverse-engineering neural circuits.
Table 1.
Outstanding Questions in Neuroscience for Which Emergent Functional Properties Could Be Key
| Are there circuit attractors? |
| What is the functional connectivity diagram of a circuit? |
| What detailed computations take place locally? |
| What are the real-time, multiple, long-range interactions that underlie cognitive functions and behavior? |
| How do local computations and long-range interactions influence each other? |
| What are the paths of information flow? |
| Do alternative pathways produce similar outputs? |
| When the brain “organizes” itself during development, or “reorganizes” itself after an injury, what is actually happening to activity locally and globally? |
| When pharmacoactive drugs alter behavior, what are the local and global effects on activity? |
| When memories are transferred from one brain region to another over time, how do activity patterns change? |
| What design principles can be discerned in how the brain functions? |
| Is there an underlying functional architecture to the brain’s networks? |
| What are the true functional underpinnings of perception, recognition, emotion, understanding, consciousness, and subconscious processes? |
In addition to promoting basic research, we anticipate that the BAM Project will have medical benefits, including novel and sensitive assays for brain diseases, diagnostic tools, validation of novel biomarkers for mental disease, testable hypotheses for pathophysiology of brain diseases in animal models, and development of novel devices and strategies for fine control brain stimulation to rebalance diseased circuits. Not least, we might expect novel understanding and therapies for diseases such as schizophrenia and autism.
Many technological breakthroughs are bound to arise from the BAM Project, as it is positioned at the convergence of biotechnology and nanotechnology. These new technologies could include optical techniques to image in 3D; sensitive, miniature, and intelligent nanosystems for fundamental investigations in the life sciences, medicine, engineering, and environmental applications; capabilities for storage and manipulation of massive data sets; and development of biologically inspired, computational devices.
As in the Human Genome Project, where every dollar invested in the U.S. generated $141 in the economy (Battelle, 2011), technological and computing innovations developed in the course of the BAM project will provide economic benefits, potentially leading to the emergence of entirely new industries and commercial ventures. If the Genome Project was “arguably the single most influential investment to have been made in modern science” (Battelle, 2011), the BAM Project, we believe, will have comparable ramifications.
Finally, we should not underestimate the repercussions that such a project could have for education. The proposed activities are broadly interdisciplinary and will lead to the training of a new generation of scientists and the opening up of new strategies for evaluating pedagogical effectiveness.
A Call for a Community Effort
To succeed, the BAM Project needs two critical components: strong leadership from funding agencies and scientific administrators, and the recruitment of a large coalition of interdisciplinary scientists. We believe that neuroscience is ready for a large-scale functional mapping of the entire brain circuitry, and that such mapping will directly address the emergent level of function, shining much-needed light into the “impenetrable jungles” of the brain.
Acknowledgments
This collaboration arose from a workshop held at Chicheley Hall, the Kavli Royal Society International Centre, supported by The Kavli Foundation, the Gatsby Charitable Foundation, and the Allen Institute for Brain Science. We also thank A.S. Chiang, K. Deisseroth, S. Fraser, C. Koch, E. Marder, O. Painter, H. Park, D. Peterka, S. Seung, A. Siapas, A. Tolias, and X. Zhuang—participants at a smaller, subsequent Kavli Futures Symposium, where initial ideas were jointly refined. We acknowledge support from the DOE (A.P.A.), NHGRI (G.M.C.), NIH and the Mathers Foundation (R.J.G.), NIH and Fondation pour la Recherche et l’Enseignement Superieur, Paris (M.L.R.), and the Keck Foundation and NEI (R.Y.).
Footnotes
A more extensive version of this paper and additional documents about the BAM can be found at http://hdl.handle.net/10022/AC:P:13501.
References
- Battelle. Economic impact of the human genome project. 2011 http://www.battelle.org/publications/humangenomeproject.pdf.
- Bock DD, Lee WC, Kerlin AM, Andermann ML, Hood G, Wetzel AW, Yurgenson S, Soucy ER, Kim HS, Reid RC. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, Shih CT, Wu JJ, Wang GT, Chen YC, et al. Curr Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Cossart R, Aronov D, Yuste R. Nature. 2003;423:283–288. doi: 10.1038/nature01614. [DOI] [PubMed] [Google Scholar]
- Du J, Riedel-Kruse IH, Nawroth JC, Roukes ML, Laurent G, Masmanidis SC. J Neurophysiol. 2009a;101:1671–1678. doi: 10.1152/jn.90992.2008. [DOI] [PubMed] [Google Scholar]
- Du J, Roukes ML, Masmanidis SC. Dual-side and three-dimensional micro-electrode arrays fabricated from ultra-thin silicon substrates. J Micromech Microeng. 2009b;19 doi: 10. 1088/0960-1317/19/7/075008. [DOI] [Google Scholar]
- Grienberger C, Konnerth A. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Konnerth A, Yuste R. Imaging in Neuroscience: a Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Press; 2011. [Google Scholar]
- Mochalin VN, Shenderova O, Ho D, Gogotsi Y. Nat Nanotechnol. 2012;7:11–23. doi: 10.1038/nnano.2011.209. [DOI] [PubMed] [Google Scholar]
- Peterka DS, Takahashi H, Yuste R. Neuron. 2011;69:9–21. doi: 10.1016/j.neuron.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón y Cajal S. Recuerdos de mi vida: Historia de mi labor científica. Madrid: Alianza Editorial; 1923. [Google Scholar]
- Smetters DK, Majewska A, Yuste R. Detecting action potentials in neuronal populations with calcium imaging. Methods. 1999;18:215–221. doi: 10.1006/meth.1999.0774. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Yuste R. Imaging. Cold Spring Harbor, New York: Cold Spring Harbor Press; 2011. [Google Scholar]
- Yuste R, Katz LC. Neuron. 1991;6:333–344. doi: 10.1016/0896-6273(91)90243-s. [DOI] [PubMed] [Google Scholar]
- Yuste R, McLean JM, Vogelstein J, Paninski L. Cold Spring Harb Protoc. 2011;2011:985–989. doi: 10.1101/pdb.prot5650. [DOI] [PubMed] [Google Scholar]


