Summary
Most patients with acute lymphoblastic leukemia (ALL) fail current treatments highlighting the need for better therapies. Since oncogenic signaling activates a p53-dependent DNA-damage response and apoptosis, leukemic cells must devise appropriate countermeasures. We show here that growth factor independence 1 (Gfi1) can serve such a function, since Gfi1 ablation exacerbates p53 responses, and lowers the threshold for p53-induced cell death. Specifically, Gfi1 restricts p53 activity and expression of pro-apoptotic p53 targets such as Bax, Noxa (Pmaip1) and Puma (Bbc3). Subsequently, Gfi1 ablation cures mice from leukemia and limits the expansion of primary human T-ALL xenografts in mice. This suggests that targeting Gfi1 could improve the prognosis of patients with T-ALL or other lymphoid leukemias.
Introduction
Many patients with acute lymphoid leukemia (ALL) and lymphoma die from tumor relapse (Gokbuget and Hoelzer, 2009). Experiments with mouse models have shown that T-ALL like diseases can be accelerated by the overexpression of the transcriptional repressor Gfi1, which is a well-established nuclear zinc finger protein and regulator of lymphoid development (Gilks et al., 1993; Zornig et al., 1996) (Li et al., 2010; Pargmann et al., 2007; Spooner et al., 2009; Yucel et al., 2003). Germline Gfi1 deletion in mice modestly reduces thymic cellularity, with an accumulation of cells between double negative 1 (DN1) and DN2 stages as well as a skew from CD4+ to CD8+ (Yucel et al., 2003). In contrast, the thymus is relatively normal when Gfi1 is deleted after the DN stage (Zhu et al., 2006), which suggests that Gfi1 mainly acts during early steps of T lymphopoiesis. Gfi1’s ability to accelerate leukemogenesis in mice and its function in lymphoid development prompted us to explore the role of Gfi1 ablation in the initiation or maintenance of lymphoid malignancies.
Results
GFI1 is associated with a subgroup of human T-ALL and accelerates NOTCH1 induced T-ALL in mice
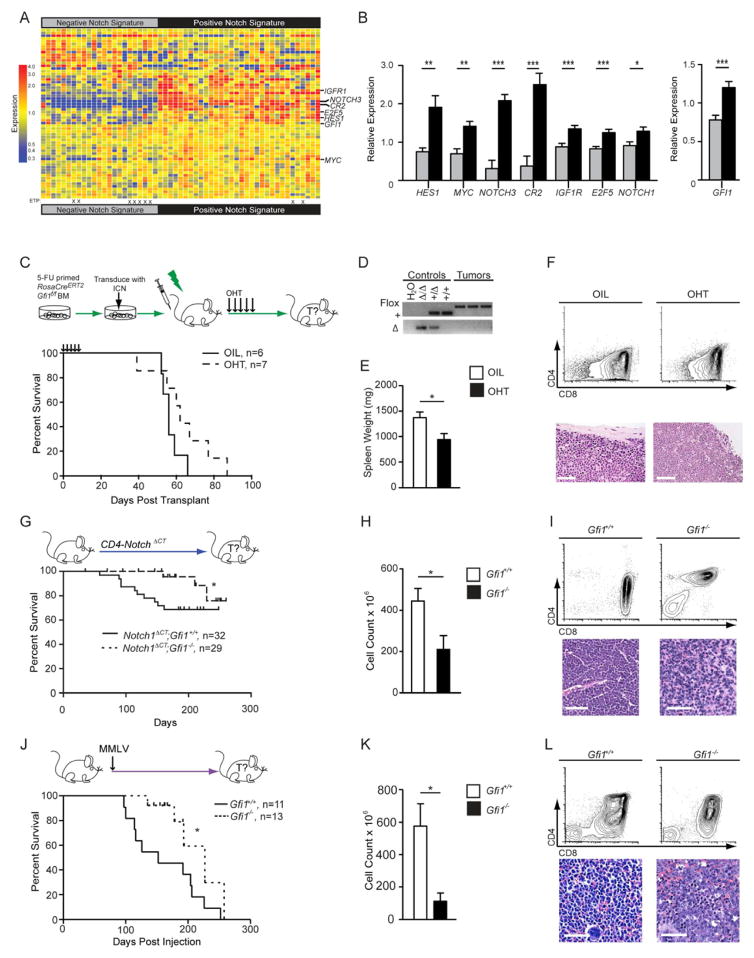
Although the oncogenic impact of high-level Gfi1 expression in murine T-cell leukemogenesis is well established, an association of GFI1 with human T-ALL has not been clearly shown. Since over 50% of human T-ALL display mutated NOTCH1 (Weng et al., 2004) or Notch1 regulatory proteins (O’Neil et al., 2007; Thompson et al., 2007) resulting in overexpression of Notch1 target genes (Palomero et al., 2006; Sharma et al., 2006; Weng et al., 2006), we performed hierarchical clustering of microarray data from independent cohorts of T-ALL patients using NOTCH1 mutation status (Ferrando et al., 2002), Notch1 target gene activation (Palomero et al., 2006; Van Vlierberghe et al., 2008), or early T-cell precursor (ETP)-ALL diagnosis (Coustan-Smith et al., 2009) and examined GFI1 expression (Figures 1A, B, Figures S1A–F).
Figure 1. Gfi1 associates with NOTCH1 in human T-ALL and deletion delays the development of disease.
(A) Heatmap of expression of published Notch1 target genes used to classify gene expression array data from 55 T-ALL patients (GSE8879) into two groups: “Negative Notch Signature” (left) and “Positive Notch Signature” (right). ETP-ALL diagnosis is designated by an “X”.
(B) Quantification of relative expression of NOTCH1, GFI1 and Notch1 target genes HES1, MYC, NOTCH3, CR2, IGF1R, and E2F5 in 55 T-ALL patients with either a “Negative Notch Signature” (gray) or a “Positive Notch Signature” (black).
(C) Top: RosaCreERT2;Gfi1f/f bone marrow cells were transduced with vectors expressing ICN and then transplanted. Mice were given vehicle or tamoxifen to induce Cre activity. Bottom: Kaplan-Meier curve.
(D) PCR genotype analysis of the Gfi1 locus in control tissues (Gfi1Δ/Δ, Gfi1+/Δ, Gfi1+/+) and in representative tumors from mice either treated with vehicle or OHT. FLOX, Gfi1f allele; +, the wild type allele, Δ, deleted allele.
(E, F) Spleen weights (E, n=6 each group) and flow cytometric analysis of thymic tumors (F, top panels) and spleen sections stained with H&E (F, bottom panels) collected during post-mortems from indicated transplant groups. Scale bars represent 50 μm.
(G) Top: Notch1ΔCT;Gfi1+/+ and Notch1ΔCT;Gfi1−/− mice were monitored for tumor development and survival. Bottom: Kaplan-Meier curve.
(H, I) Spleen weights (H) and flow cytometric analysis (I, top panels) and histological sections (I, bottom panels) of Notch1ΔCT;Gfi1+/+ (n=7) and Notch1ΔCT; Gfi1−/− (n=3) tumors.
(J) Top: Gfi1+/+ and Gfi1−/− newborn mice were injected with MMLV. Bottom: Kaplan-Meier curve.
(K) Thymic tumor cell numbers of Notch1ΔCT induced tumors.
(L) Flow cytometric analysis (top panels) and histological section (bottom panels) of MMLV-induced Gfi1+/+ versus Gfi1−/− tumors.
All scale bars represent 50 μm. | in all Kaplan-Meier curve plots indicate censored mice.
Mean and ±SEM are shown unless stated otherwise. * p<0.05, **p<0.01, ***p<0.001.
We observed that patients with ETP-ALL had low levels of GFI1 expression compared to those with a positive NOTCH1 signature (Figure 1B, Figure S1D, E), suggesting a functional role for Gfi1 in NOTCH1-dependent human T-ALL. However, GFI1 is unlikely a Notch1 target, since intracellular Notch1 (ICN) does not occupy the GFI1 locus nor was GFI1 expression altered by gamma-secretase inhibitors (GSIs) based on our own results as well as published data (Figure S1F)(Margolin et al., 2009; Medyouf et al., 2011). Also, we can show that mice transplanted with bone marrow cells over-expressing ICN and Gfi1 developed leukemia faster than mice transplanted with cells only over-expressing ICN (Figures S1G–I), corroborating previous reports on the function of Gfi1 in T-cell leukemogenesis (Schmidt et al., 1998; Zornig et al., 1996) and extending it to human ICN-mediated T-ALL.
Gfi1 deletion delays the development of T-ALL
To test whether ablation of Gfi1 could inhibit the onset of T-ALL, we used five different mouse models, in which we could temporally delete Gfi1. First, we transplanted ICN-expressing bone marrow cells from mice carrying a tamoxifen (OHT)-inducible Rosa26 Cre-recombinase transgene (CreERT2) (Hameyer et al., 2007) enabling inducible deletion of floxed Gfi1 alleles (Gfi1f/f) (Horman et al., 2009; Velu et al., 2009) (Figure 1C). While vehicle- treated animals died within 66 days, OHT-treated recipients developed leukemia within 87 days with similar T-ALL characteristics (Figures 1C–F). However, all tumors emerging after OHT-treatment had intact Gfi1 alleles (Figure 1D), suggesting that ICN-induced T-ALL selects for Gfi1.
To confirm this, we used a T-cell specific Cre transgene (LckCre+) and Gfi1f/Δ transgenic mice, in which Rosa26-locus-mediated expression of ICN and eGFP is blocked by a floxed STOP cassette (Rosa ICNLSL) (Murtaugh et al., 2003). We injected these mice with N-Ethyl-N-Nitrosourea (ENU), which induces T-cell leukemia and shortens the latency of leukemogenesis (Kundu et al., 2005; Yuan et al., 2001). Approximately 50% of all tumors arising in LckCre+;Rosa ICNLSL;Gfi1+/+ mice were eGFP+ (i.e. expressing ICN and Gfi1, Figures S1J, K). However, ENU-induced tumors that arose in LckCre+;Rosa ICNLSL;Gfi1f/Δ mice were always eGFP negative (i.e. ICN negative and Gfi1 wild-type, Figure S1K,) also suggesting that ICN-mediated tumorigenesis selects for Gfi1. In yet another Notch-driven leukemogenesis model, in which constitutive absence of Gfi1 was coupled with a CD4-promoter-driven mutant Notch1 transgene (Notch1ΔCT, (Priceputu et al., 2006), T-ALL development was substantially decreased and delayed (Figures 1G–I).
To explore the impact of Gfi1 loss in mouse models of T-ALL that are not initiated by Notch, we either infected Gfi1+/+ and Gfi1−/− newborn mice with Murine Moloney Leukmia (MMLV) (Scheijen et al., 1997) or injected adolescent mice with ENU. All MMLV-infected Gfi1+/+ mice developed lymphoid malignancies whereas only 40% of MMLV-infected Gfi1−/− mice did. The remaining mice were censored due to neurological problems consistent with reports on older Gfi1−/− mice (unpublished data). Notably, Gfi1−/− lymphoid malignancies were significantly less robust than Gfi1+/+ tumors (Figures 1J–L). Similarly, >85% of the ENU-injected Gfi1+/+ mice, but only 20% of Gfi1−/− mice, developed T-cell leukemia (Figure S1L, the remaining mice succumbed to ENU-induced toxicity). As in other models, ENU-initiated Gfi1−/− tumors developed slower and were significantly less robust than Gfi1+/+ tumors (Figure S1L–N). Neither Gfi1+/+ nor Gfi1−/− ENU-induced tumors were found to harbor Notch1 mutations in the HD or PEST domain (Table S1). Thus, results from these five independent T-ALL models, initiated by various oncogenic pathways, led us to conclude that ablation of Gfi1 delays, impedes, or is counter-selected during T-ALL formation.
T-ALL disease maintenance is Gfi1-dependent
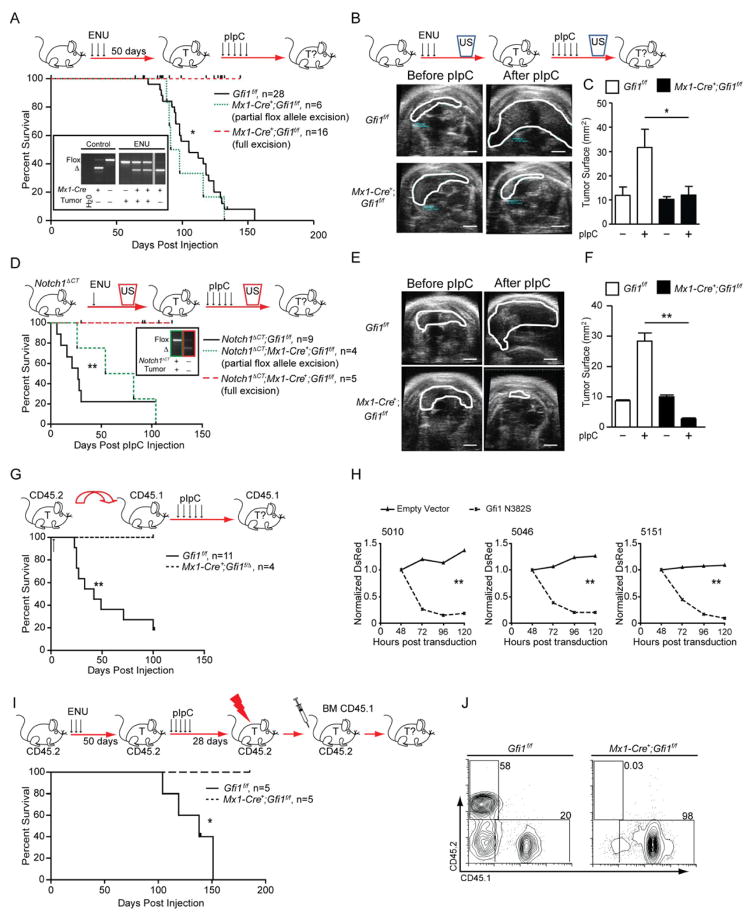
Mx1-Cre+;Gfi1f/f or Gfi1f/f mice were treated with ENU to elicit T-cell leukemia. After 50 days, both groups were injected with pIpC (Horman et al., 2009). All Gfi1f/f mice developed T-ALL, but Mx1-Cre+;Gfi1f/f mice separated into two different sub-groups following pIpC injection. One sub-group remained healthy until the study was terminated (Figure 2A, Mx1-Cre+;Gfi1f/f, full excision), or died of ENU toxicity. The second sub-group displayed partial Gfi1 deletion, and succumbed to T-cell leukemia similar to ENU/pIpC treated Gfi1f/f mice (Figure 2A, Mx1-Cre+;Gfi1f/f partial excision).
Figure 2. Gfi1 is required for T-cell leukemia maintenance.
(A) Top: Gfi1f/f or Mx1-Cre+;Gfi1f/f mice were treated with ENU and subsequently with pIpC. Bottom: Kaplan-Meier curve. Insert: PCR analysis of the Gfi1 locus in control tissues and in representative tumors (T) for Gfi1 flox and excised (Δ) alleles.
(B) Top: Gfi1f/f or Mx1-Cre+;Gfi1f/f mice were treated with ENU and followed for tumors by ultrasound (US). Next, mice were treated with pIpC and tumor development was determined by ultrasound. Bottom: Representative ultrasound images; scale bar = 20 mm
(C) Change of thymic surface area before and after treatment with pIpC for mice (see B).
(D) Top: Notch1ΔCT;Gfi1f/f or Notch1ΔCT;Mx1-Cre+;Gfi1f/f mice were treated with ENU and subsequently monitored by ultrasound for tumor (T) development. Upon appearance of a mass, mice were injected with pIpC and followed for tumor progression or regression by ultrasound. Bottom: Kaplan-Meier curve. Insert: PCR analysis of allele excision (Δ).
(E) Representative ultrasound images of tumors before and after pIpC injection; scale bar =20 mm.
(F) Change of thymic surface area before and after treatment with pIpC (see E).
(G) Top: Gfi1f/f tumors or tumors that had one Gfi1 allele deleted (Mx1-Cre+;Gfi1f/Δ) were transplanted into CD45.1 recipient mice, which were then treated with pIpC. Bottom: Kaplan-Meier curve.
(H) T-ALL cell lines 5151, 5046 and 5010 were transduced with retrovirus vectors expressing Gfi1N382S and DsRed or DsRed alone. DsRed was measured over time by FACS and normalized to the level at 48 hours. 1 of 3 representative experiments is shown.
(I) Top: Gfi1f/f or Mx1-Cre+;Gfi1f/f mice were injected with ENU. 50 days later they were treated with pIpC. 28 days later they were irradiated, and transplanted with wild-type CD45.1 bone marrow cells, then followed for survival. Bottom: Kaplan-Meier curve. One Gfi1f/f mice was sacrificed for morbidity unrelated to leukemia.
(J) Bone marrow of mice in (I) at the end of observation were examined for contribution of the CD45.2 (host bone marrow) and CD45.1 (donor bone marrow).
| in all Kaplan-Meier curve plots indicate censored mice.
Mean and ±SEM are shown unless stated otherwise. * p<0.05, ** p<0.01.
See also Figure S2.
To investigate whether loss of Gfi1 was causing tumor regression or preventing tumor formation, we used ultrasound imaging. Upon detection of a tumor (Figure 2B), Gfi1 deletion was induced with pIpC. All ENU-induced tumors in Gfi1f/f mice clearly showed increases in tumor size whereas tumors that developed in Mx1-Cre+;Gfi1f/f animals showed variable changes in size (Figure 2C, Figure S2A). Following pIpC injection, disease-free survival, tumor growth and blast cell detection all directly correlated with the degree of Gfi1 deletion in the tumor (Figures 2B, 2C, Figure S2B), since we found that Gfi1 deletion was incomplete in tumors that progressed, but was complete in tumors that regressed (Figure S2A).
We verified this observation in a second T-ALL model, in which disease was induced by Notch1 activation and accelerated by ENU injection. Mice were monitored by ultrasound and upon tumor detection, treated with pIpC (Figure 2D). While all pIpC injected Notch1ΔCT;Gfi1f/f mice died, all pIpC injected Notch1ΔCT;Mx1-Cre+;Gfi1f/f tumors with complete deletion of Gfi1 regressed and the mice survived (Figures 2D–F). This regression also correlated with lower numbers of blast cells in the blood of pIpC treated Notch1ΔCT;Mx1-Cre+;Gfi1f/f mice compared to Notch1ΔCT;Gfi1f/f controls (Figure S2C).
Next, Gfi1f/f or Mx1-Cre+;Gfi1f/Δ tumor cells were transplanted into syngeneic recipients. In recipients that did not receive pIpC, only tumors with an intact floxed Gfi1 allele emerged (data not shown). However, when recipient mice were treated with pIpC, all mice that received Gfi1f/f tumors died whereas mice receiving Mx1-Cre+;Gfi1f/Δ tumors survived tumor free (Figure 2G). To demonstrate that loss of Gfi1 specifically leads to tumor regression in a cell autonomous manner, we inhibited Gfi1 function in three Tal1-transformed murine TALL cell lines (Cullion et al., 2009) by overexpressing a DsRed-marked Gfi1 dominant negative mutant (Gfi1N382S) (Horman et al., 2009; Person et al., 2003; Zarebski et al., 2008). Two days after the initial measurement of transduction, and in contrast to empty-vector transduced cells, only 15–20% of cells transduced with DsRed+ Gfi1N382S expressing vectors were still DsRed+ (Figure 2H).
To determine the clinical potential of targeting Gfi1, we injected Gfi1f/f and Mx1-Cre+;Gfi1f/f mice (CD45.2+) with ENU, waited 50 days to allow tumor initiation, and then treated with pIpC to delete Gfi1. Four weeks after the first pIpC injection, both groups of mice were sublethally irradiated and transplanted with syngenic CD45.1+ bone marrow cells (BMT) to prevent bone marrow failure associated with ENU (Figures 2I–J). The combination therapy was not sufficient to cure the mice of T-ALL as 80% of ENU treated Gfi1f/f mice still succumbed to disease (one died of non-tumor related reasons). However, when therapy was combined with Gfi1 deletion, complete tumor remission was observed in every transplant recipient (Figures 2I–J). Taken together, our data strongly implicate Gfi1 in the maintenance of established T cell malignancies, their ability to kill secondary hosts, and potentially in improving therapy.
Maintenance of B-cell lymphoma is dependent on Gfi1
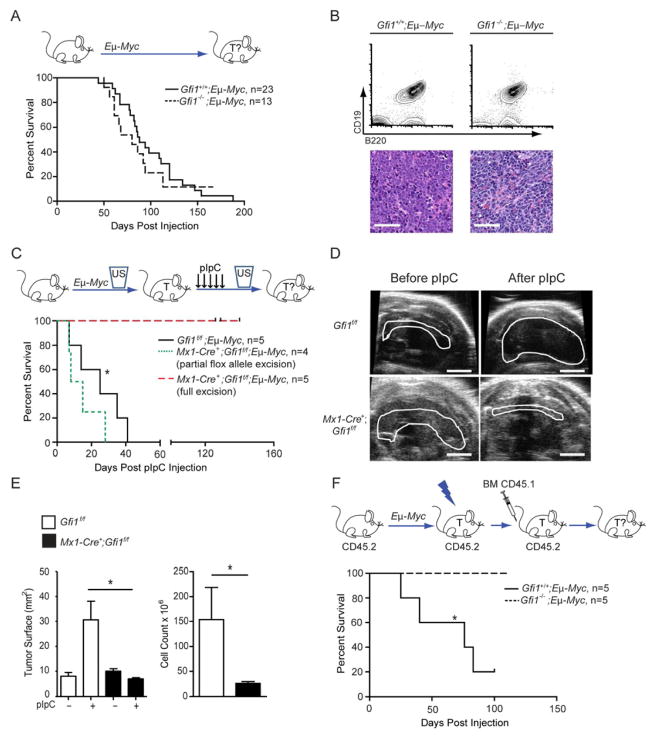
To test whether other lymphoid malignancies were also dependent on Gfi1, we used Eμ-Myc transgenic mice, which develop clonal B cell lymphomas (Adams et al., 1985). Loss of Gfi1 did not affect the latency, incidence, or pathology of tumor initiation (Figures 3A–B), but completely blocked the ability of lymphoma to kill secondary recipients (Figure S3A). Thus, similar to the T cell models, Gfi1 is required for robust tumorigenesis. To determine whether Gfi1 is required for B cell lymphoma maintenance, we used an inducible model (Zhu et al., 2006) to delete Gfi1f/f after a lymphoma had formed. Although pIpC injection had no effect on progression of disease in Gfi1f/f;Eμ-Myc mice, it led to tumor regression and a significant reduction of leukemic blasts in the peripheral blood of Mx1-Cre;Gfi1f/f;Eμ-Myc mice (Figures 3C–E, S3B) suggesting that Gfi1 is indeed necessary to maintain a B cell lymphoma. Similar to the results with our T-ALL models, loss of Gfi1 significantly improved the outcome of Gfi1−/−;Eμ-Myc mice treated with sublethal irradiation and BMT after detection of a tumor, whereas Gfi1+/+;Eμ-Myc animals died of tumor relapse (Figure 3F). These data suggest that targeting Gfi1 could also be beneficial for treating B cell lymphoma.
Figure 3. Gfi1 is required for maintenance of B-cell lymphoma.
(A) Top: Gfi1+/+;Eμ-Myc andGfi1−/−;Eμ-Myc mice were monitored for tumor development and survival. Bottom: Kaplan-Meier curve.
(B) Flow cytometric analysis (top) and histological sections (bottom) of Eμ-Myc-induced Gfi1+/+ and Gfi1−/− tumors. Scale bars represent 50 μm.
(C) Top: Mx1-Cre+;Gfi1f/f;Eμ-Myc and Gfi1f/f;Eμ-Myc were observed by ultrasound for appearance of B-cell lymphoma. Upon appearance of a mass, mice were injected with pIpC and monitored for tumor progression and survival. Bottom: Kaplan-Meier curve.
(D) Representative ultrasound images of tumors before and after pIpC injection. Scale bar = 20 mm.
(E) Change of tumor surface area (left) before and after treatment with pIpC for mice with the indicated genotypes as well as cellularity of mediastinal tumor after treatment (right).
(F) Top: Gfi1+/+;Eμ-Myc and Gfi1−/−;Eμ-Myc animals were observed until enlarged lymph nodes evidenced tumor development, then irradiated and transplanted with CD45.1 bone marrow cells and monitored for survival. Bottom: Kaplan-Meier curve.
| in all Kaplan-Meier curve plots indicate censored mice.
Mean and ±SEM are shown unless stated otherwise. *p<0.05.
See also Figure S3.
Gfi1 integrates the cellular transcriptional response to DNA damage/p53 induction
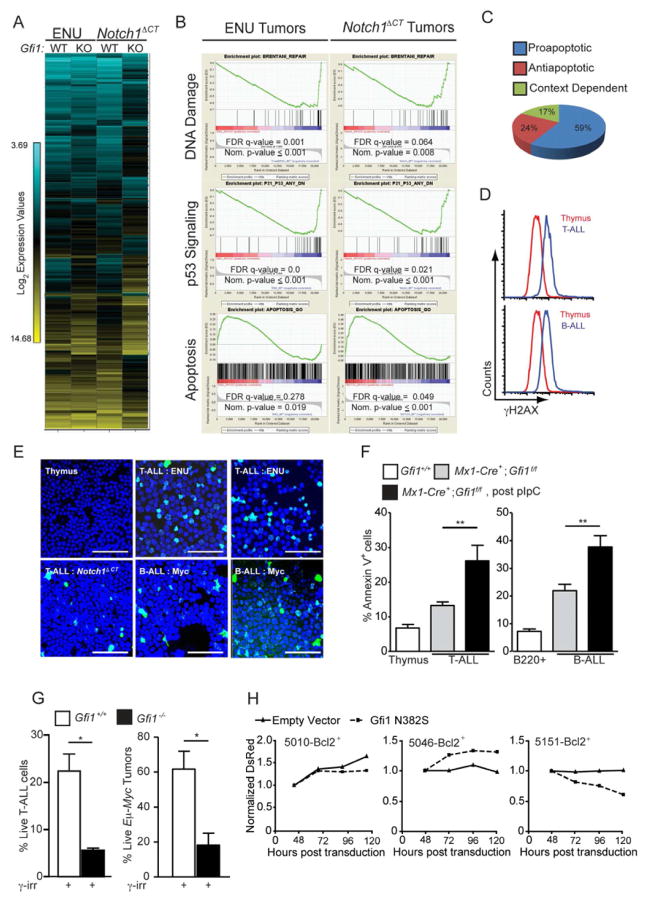
To investigate how loss of Gfi1 induces tumor regression, we compared gene expression profiles of T-cell leukemia from two different models (Figures 2A & 2D) upon inducible deletion of Gfi1 (Figure 4A). Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) demonstrated significant deregulation of multiple key leukemic pathways including cell cycle progression, NFκB signaling, and basal transcription among others (Table S2 and data not shown). Normal thymocytes do not disappear upon loss of Gfi1 as the tumors do. Therefore, to identify mechanisms that might explain tumor regression, we focused on those pathways that were similarly deregulated in both ENU and Notch1ΔCT induced tumors from Gfi1−/− and Gfi1+/+ mice, but were not enriched in normal non-malignant Gfi1−/− versus Gfi1+/+ thymocytes. We noticed a striking number of shared GSEA signatures that included deregulated p53 signaling, DNA damage/repair pathways and a pro-apoptotic response (Figure 4B–C, Table S2), suggesting that an accelerated cell death program might be initiated in tumor cells that lack Gfi1.
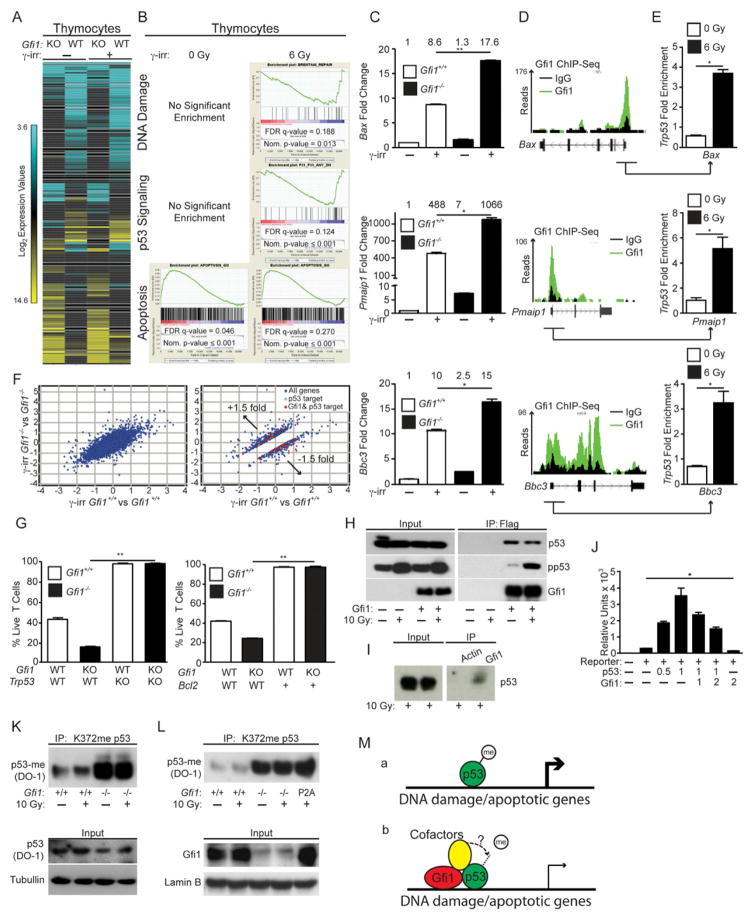
Figure 4. Gfi1 mediates DNA damage and p53 signaling to control apoptosis.
(A) Unsupervised hierarchical clustering of the averaged normalized Log2 gene expression values from ENU (n=3) or ENU/Notch1ΔCT (n=2) induced T-ALL arising in Gfi1f/f (WT) or Mx1-Cre+;Gfi1f/f (KO) pIpC treated mice (ENU WT n=3, ENU KO n=3, ENU/Notch1ΔCT WT n=2, ENU/Notch1ΔCT KO n=2).
(B) GSEA butterfly plots for pathways related to DNA damage, p53 signaling or apoptosis found in both ENU and Notch1ΔCT initiated tumor signatures from (A).
(C) Classification of genes in the leading edge of the GSEA Apoptosis signature in (B) as proapoptotic, antiapoptotic or context dependent.
(D–E) Determination of γH2AX levels in normal tissue as well as in B and T-cell leukemia by FACS (D) and immunofluorescence (E). One experiment was performed. Scale bars = 50 μm.
(F) Level of spontaneous apoptosis in the indicated tissues and tumors before and after Gfi1 deletion. T-ALL: Gfi1+/+, n=4; Gfi1f/f, n=17; Gfi1Δ/Δ, n=5. B-ALL: Gfi1+/+, n=4; Gfi1f/f n=13; Gfi1Δ/Δ, n=4.
(G) Gfi1+/+ (n=7), Gfi1−/− and Gfi1f/Δ(one constitutive Gfi1 KO tumor and two tumors, in which Gfi1 has been deleted with more than 50 % excision, n=3,) thymic tumor cells and Gfi1+/+;Eμ-Myc+ (n=7) and Gfi1−/−;Eμ-Myc+ (n= 3) lymphomas were explanted and irradiated (6 Gy), and examined for AnnexinV staining by FACS.
(H) T-ALL cell lines 5151, 5046, and 5010 were transduced with retrovirus vectors MSCV-Bcl-2, expanded and then transduced with vectors encoding Gfi1N382S and DsRed or DsRed alone. DsRed was measured over time by FACS and normalized to the level at 48 hours. One of 3 representative experiments is shown.
Mean and ±SEM are shown unless stated otherwise. *p<0.05, **p<0.01.
See also Table S2.
An emerging concept proposes that oncogenic signaling induces uncoordinated cell division, generating collapsed replication forks and DNA double strand breaks, which in turn initiate a DNA damage response, activating p53 and inducing apoptosis. Therefore, tumor cells must counteract cell death in order to survive (Bartek et al., 2007; Bartkova et al., 2007; Di Micco et al., 2006; Halazonetis et al.). In agreement with this theory, leukemic cells from our tumor models displayed increased levels of phosphorylated H2AX (γH2AX), indicating DNA double strand breaks, and higher levels of spontaneous apoptosis than untransformed thymocytes (Figures 4D–F). We also noted that the number of apoptotic cells was further increased in those tumors where Gfi1 was inducibly deleted (Figure 4F). Additionally, when we irradiated Gfi1−/− leukemic cells, we observed decreased survival compared to Gfi1+/+ tumors (Figures 4G). Finally, when we over-expressed Bcl2 in Tal1-transformed T-cell lines, counter-selection of the dominant negative mutant Gfi1N382S was either absent or delayed (compare Figure 4H to Figure 2H). These data demonstrate that Gfi1 is required in lymphoid tumors to counter DNA-damage induced death and suggest that DNA damage/p53 induced signals are dominant effectors of Gfi1 loss-of-function apoptotic phenotypes in T-ALL.
In contrast to Gfi1-deleted tumors, Gfi1−/− thymocytes display only mildly increased levels of apoptosis of c-Kit+ subsets (compared to Gfi1+/+) (Yucel et al., 2003). In agreement with this observation, we noted that while GSEA analysis of gene expression data of Gfi1−/− versus Gfi1+/+ thymocytes is enriched for apoptotic signatures, the DNA damage and p53 signatures, which drive the execution of apoptosis, were not enriched (Figures 5A–B). Thus, we hypothesized that the introduction of a DNA damage signal (inherent to tumors) to Gfi1−/− thymocytes may elicit the same increased apoptotic phenotype in thymocytes that was found in tumors. Indeed, gene expression analysis revealed that a comparison between γ-irradiated (to induce DNA damage) Gfi1−/− versus Gfi1+/+ thymocytes recapitulated the exaggerated Gfi1−/− GSEA DNA damage and p53 signatures found in leukemia cells (compare Figures 4B and 5B). Moreover, DNA damage induced by Daunorubicin, Etoposide, or by various doses of γ-irradiation resulted in significantly decreased Gfi1−/− thymocyte survival and mitochondrial potential (Figures S4A–E). While, Gfi1−/− thymocytes showed similar levels of γH2AX, p53 induction and p53 phosphorylation compared to Gfi1+/+ controls (Figures S4F–G), Gfi1−/− thymocytes displayed increased cleaved Caspase 3 and PARP (Figures S4H–I). These data indicate that Gfi1 antagonizes DNA-damage-induced apoptotic pathways downstream of DNA-damage detection, but upstream of caspase and PARP1 cleavage.
Figure 5. Gfi1 restricts p53 dependent induction of apoptosis.
(A) Unsupervised hierarchical clustering of the averaged normalized Log2 gene expression values from Gfi1+/+ (WT) and Gfi1−/− (KO) thymocytes with or without irradiation (Gfi1+/+ control n=2, Gfi1−/− control n=2, Gfi1+/+ irradiated n=3, Gfi1−/− irradiated n=3).
(B) GSEA butterfly plots for pathways related to DNA damage, p53 signaling or apoptosis enriched in Gfi1 deficient tumors (Figure 4B) that emerge in Gfi1−/− T cells only after irradiation.
(C) Expression of Bax, Pmaip1 (Noxa) and Bbc3 (Puma) in Gfi1+/+ and Gfi1−/− thymocytes before and after irradiation. One representative experiment out of at least two experiments is shown. The numbers above the bars represent the mean values of the measurements.
(D) Peaks across the Bax, Pmaip1 and Bbc3 loci from Gfi1 ChIP-Seq of murine hematopoietic progenitor cells immortalized by retroviral transduction of an MLL-ENL expression vector.
(E) ChIP of p53 using primers from Gfi1-bound regions (underscored with arrow in (D)) of Bax, Pmaip1, and Bbc3 before and after irradiation. Represented are the mean and standard deviation of the fold difference compared to IgG control from one experiment with three technical repeats.
(F) Log2 values of the fold change of the irradiated vs. unirradiated gene expression values of all genes (left) or 1.5-fold differentially regulated (right) between Gfi1+/+ and Gfi1−/− thymocytes. Gfi1-bound (identified in (D)) p53 target genes are shown in red.
(G) Percentage of live Gfi1+/+;Trp53+/+, Gfi1−/−;Trp53+/+, Gfi1+/+;Trp53−/−, and Gfi1−/−;Trp53−/− thymocytes after ex vivo γ-irradiation (left, n=3). Percentage of live Gfi1+/+, Gfi1−/−, Gfi1+/+;Vav-Bcl2, and Gfi1−/−;Vav-Bcl2 thymocytes after ex vivo γ-irradiation (right, n=3).
(H) Immunoblot of total cell lysate (left) and immunoprecipitation (right) were performed using p53, phospho- p53 or Gfi1 antibodies on lysates from untreated or irradiated 293T cells transfected as indicated with FLAG-tagged Gfi1 constructs. One representative experiment from at least two experiments is shown.
(I) Immunoblot of total cell lysate (left) and immunoprecipitation using either Gfi1 or an isotype control (actin) antibody (right) were performed using phospho-p53 (Ser15) antibody on lysates from irradiated thymocytes cells. One representative experiment from at least two experiments is shown.
(J) Reporter expression assay using the Bax promoter and various amounts (μg) of transfected vectors encoding p53 or Gfi1.
(K) Thymocytes from the indicated mice were irradiated or left untreated. After 30 minutes, total cell lysates were immunoprecipitated with an anti-mono-methyl K372 p53 antibody, then immunoblotted with an anti-p53 antibody. p53 and tubulin in total cell lysates are also shown. One experiment out of at least two experiments is shown.
(L) Thymocytes nuclear extracts from the indicated mouse strains were immunoprecipitated with an anti-mono-methyl K372 p53 antibody, then immunoblotted with an anti-p53 antibody. Input control shows the level of Gfi1 in thymocytes from Gfi1+/+, Gfi1−/− and Gfi1P2A/P2A mice and the loading control LaminB. One experiment out of at least two experiments is shown.
(M) Schematic representation showing methylated p53 binds to DNA and robustly activates the expression of target genes (a) and Gfi1 co-occupancy of a subset of p53 targets tethers a Gfi1 SNAG-dependent cofactor, which demethylates p53 to dampen the expression of p53 target gene (b).
Mean and ±SEM are shown unless stated otherwise. *p<0.05, **p<0.01.
See also Figure S4.
To analyze this in more detail, the expression of cell-death-associated p53 targets such as Bax, Pmaip1 (Noxa), and Bbc3 (Puma) was tested and found to be further induced in irradiated Gfi1−/− thymocytes compared to Gfi1+/+ controls (Figure 5C). These genes appear to be direct Gfi1 targets as interrogation of Gfi1 ChIP-Seq data showed enriched Gfi1 binding in the regulatory regions of Bax, Pmaip1, and Bbc3 compared to IgG controls (Figure 5D). These data suggest that Gfi1 co-occupies p53-responsive genes and regulate their expression. Interestingly, significant p53 binding to these same Gfi1-bound regions within the promoters (underscored in Figure 5D) of Bax, Pmaip1 and Bbc3 was observed in thymocytes after induction of p53 by irradiation (Figure 5E). To assess whether Gfi1 and p53 globally regulate the expression of pro-apoptotic p53 effector genes, we examined the Leading Edge of the GSEA Gfi1−/− irradiated thymocyte signature and found >70% of the apoptotic genes were in fact proapoptotic effectors (Figure S4J). Moreover, combining the gene expression and ChIP-Seq analyses revealed that Gfi1 occupies fifty-five of seventy-seven p53-effector genes (>70%) deregulated in irradiated Gfi1−/− thymocytes (Figure 5F). We next validated the ChIP-Seq data with ChIP-qPCR using primer sets for 14 of the 55 genes. These genes were (i) occupied by Gfi1 according to Chip-Seq data with reads over 100 compared to Ig controls; (ii) at least 1.5-fold differentially expressed between Gfi1−/− and Gfi1+/+ thymocytes after irradiation; and (iii) known p53 effector genes according to empirically tested data in the Molecular Signature Database (MSigDB). ChIP-qPCR confirmed binding of Gfi1 in irradiated thymocytes with an enrichment of >1.5 fold in 10 of the 14 genes tested, suggested Gfi1 binding in 3 genes with an enrichment of 1.3–1.5, and demonstrated little to no binding in only 1 of the 14 primer sets tested (Figure S4K). Co-occupation of the same loci by Gfi1 and p53 was found in the majority of genes tested (9 of 14, Figure S4L). A time dependent analysis on 4 of the 14 loci (Bax, Pmaip1, Bbc3 and Cdkn1a) revealed that a co-occupation by Gfi1 and p53 is maintained over time, but that p53 occupation clearly dominates at 120 min after the initial DNA damage signal over Gfi1 (Figure S4M). This suggests that during the immediate response after DNA damage Gfi1 and p53 co-regulate target genes, but if the DNA damage signal persists, a p53 dominated regulation prevails.
We investigated the involvement of the p53-activated apoptosis pathway in Gfi1−/− thymocyte survival after DNA damage. To do so, we deleted Trp53 or over-expressed Bcl2 and found that either condition completely rescued the exaggerated Gfi1−/− thymocyte apoptosis upon DNA damage signaling (Figure 5G). Further investigation into the underlying mechanism demonstrated that Gfi1 and p53 can physically interact in transfected cells and in irradiated thymocytes (Figure 5H–I), and that Gfi1 was able to repress p53-mediated transcriptional activation of a model reporter gene (Figure 5J). Notably, methylation of p53 at K372 leads to increased stability of chromatin-bound p53 and to the activation of p53 target genes, whereas demethylation of K372 has an inhibitory effect on p53 (West and Gozani, 2011). Immunoprecipitation and immunoblot experiments with Gfi1+/+ and Gfi1−/− thymocytes showed that absence of Gfi1 leads to a substantial increase of p53-K372me, irrespective of irradiation (Figure 5K). Moreover, thymocytes from knockin mice expressing only a Gfi1P2A mutant (Fiolka et al., 2006) that lacks the ability to bind LSD1 (Saleque et al.) also displayed a substantial increase of p53-K372me (Figure 5L). These data suggest that Gfi1 restricts p53 activity through Gfi1 SNAG-dependent cofactor recruitment and p53 demethylation (Figure 5M).
Targeting GFI1 in human ALL leads to tumor death
To test whether Gfi1 could be a suitable target for therapy of human leukemia, we used human T-ALL cell lines and reduced Gfi1 expression either by transduction of previously described shRNA expressing lentiviral vectors (Velu et al., 2009) or Vivo-Morpholinos (Morcos et al., 2008) specifically designed against GFI1. In both cases, reduction of Gfi1 impeded the growth of T-ALL cell lines, which correlated with a higher level of apoptosis (Figures 6A–C, Figures S5A–C), suggesting that T-ALL is sensitive to the induction of apoptosis. When we used the pan-Bcl2 inhibitors Obatoclax and ABT-263 on three independent T-ALL lines, we observed IC50s approximately ten-fold lower than those observed in AML, where the use of these drugs is currently in clinical trials (Figure 6D). Inhibition of Gfi1 further increased the efficiency of both Obatoclax treatment (Figure 6E) and radiation therapy (Figures S5D). To demonstrate the contribution of p53 to Gfi1 loss-of-function apoptosis, we used Vivo-Morpholinos to first antagonize p53 expression then Gfi1 expression. We observed a significant decrease in the ability of the Gfi1 Vivo-Morpholino to induce apoptosis after p53 Vivo-Morpholino pretreatment (Figure S5E). Similar results were obtained using p53 targeting shRNA lentiviruses followed by Gfi1 Vivo-Morpholino treatment (data not shown).
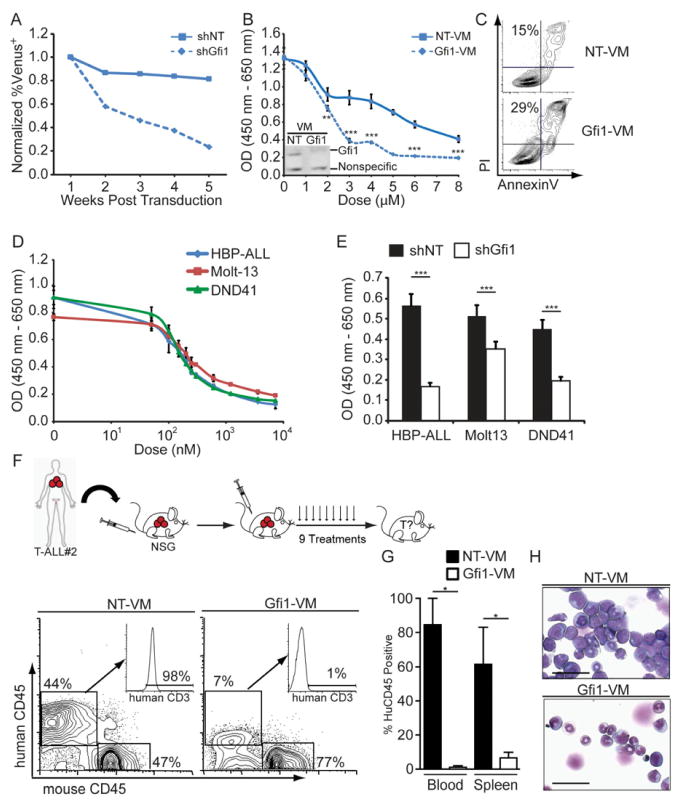
Figure 6. Gfi1 as a target to treat human leukemia.
(A) HBP-ALL cells were transduced with Venus-marked shRNA expressing lentiviral vectors targeting Gfi1 (shGfi1, dotted line) or non-targeting control (shNT, solid line). Expression of Venus was measured by FACS 72 hours post transduction, which was set as 1, and subsequent measurements were taken by FACS over a 5 week period and normalized to the first reading, p=0.058.
(B) Growth of HBP-ALL cells treated with Gfi1 or NT Vivo-Morpholinos (VM) as measured by WST assay for 48 hours. Inset: Immunoblot for Gfi1 in HBP-ALL cells treated with NT or Gfi1-VM (4 μM) for 16 hours.
(C) AnnexinV and PI staining of HBP-ALL cells after 16 hours of Gfi1 or NT VM treatment (4μM).
(D) Growth of T-ALL cell lines treated with indicated doses of the Obatoclax as measured by WST assay for 48 hours.
(E) Gfi1 knockdown was combined with Obatoclax treatment (200 nM) and growth was measured by WST assay for 48 hours. 1 representative experiment is shown; experiments were repeated 2–3 times (A–E).
(F) Top: Primary patient T-ALL samples were transplanted in NSG mice and then mice were injected with Gfi1 or NT VM 3 times per week for 3 weeks. Bottom: FACS analysis of human CD45 and human CD3 of NT (n=2) or Gfi1-treated (n=3) mice.
(G) Quantification of total human CD45+ cells in the blood and spleen from mice in (F).
(H) Cytospins of the BM from mice in (F). Scale bars = 50 μm.
Mean and ±SEM are shown unless stated otherwise. *p<0.05, **p<0.01, ***p<0.001.
See also Figure S5.
Next, we examined Gfi1 inhibition in primary patient samples. Due to the significant limitations of in vitro systems to support primary T-ALL cells survival, we transplanted primary patient specimens into immune deficient Nod/Scid/IL2Rγ−/− (NSG) mice then tested whether targeting Gfi1 using morpholinos is a viable approach to treat leukemia. The cells were allowed to engraft and expand for four days before the mice were injected with Vivo-Morpholinos over a three-week period and monitored for survival. Gfi1 Vivo-Morpholino treated animals showed a trend towards increased survival after only three injections (Figures S5F–I). We repeated the assay with samples from a patient who failed to respond to current therapies, but increased the treatment frequency. When control morpholino (NT) treated mice became moribund, we analyzed the tissues of all of the transplanted mice for the presence of human T-ALL cells. Targeting Gfi1 significantly impeded the expansion of the human leukemia in the bone marrow, peripheral blood and the spleen of the transplanted NSG mice (Figures 6F–H), whereas treatment of healthy mice with the same dosing regimen did not lead to adverse effects (Figure S5J).
Discussion
Important roles for Gfi1 in normal lymphoid development and acceleration of murine T-cell leukemia have previously been established (Blyth et al., 2001; Chakraborty et al., 2008; Dabrowska et al., 2009; Gilks et al., 1993; Scheijen et al., 1997; Schmidt et al., 1996; Uren et al., 2008; Yucel et al., 2003). Yet, important questions remained whether Gfi1 was required for the development or maintenance of human lymphoid leukemia. In the current study, we found that ablation of Gfi1 leads to regression of already established murine lymphoid neoplasms occurring through the induction of p53-dependent apoptotic pathways. Our results indicate that leukemic cells in general require Gfi1, because the ablation of Gfi1 led to lymphoid tumor regression and host survival independently of the transforming pathway or tumor etiology. It is thus conceivable that Gfi1 is an “oncorequisite” factor, a normal cellular protein upon which malignant cells uniquely depend for their survival. This offers a different paradigm for cancer therapeutics and suggests that normal cellular proteins, independent of their mutation status in human tumors, can be excellent targets for clinical intervention.
Our findings are surprising given the recently identified function of Gfi1 in myeloproliferative disease (MPD) and AML, where Gfi1 loss-of-function de-represses HoxA9, Meis1 and Pbx1, and can cooperate with other oncogenic lesions to transform myeloid progenitors (Horman et al., 2009). Furthermore, a single nucleotide polymorphism in the human GFI1 deregulates HOXA9 expression and increases the risk for human AML by 60% (Khandanpour et al., 2012); however, further experimentation is still necessary to incisively define a role for Gfi1 in human AML. HoxA9-signaling is present in mixed lineage leukemia, but is active in less than 10% of T-ALL patients (Ferrando et al., 2002). Thus, rare HoxA9-active-T-ALL patients may not benefit from receiving Gfi1-targeting therapies. Therefore, careful molecular pathology will likely be important to stratify patients for Gfi1-targeted therapeutics.
Recent work suggested that oncogenic signaling in general causes uncoordinated cell division resulting in collapsed replication forks and the initiation of p53 dependent DNA damage responses causing cell death. (Halazonetis et al.) (Bartek et al., 2007; Bartkova et al., 2007; Di Micco et al., 2006). Tumor cells have to counteract this “oncogenic stress” signal to avoid cell death, for instance by mutating TP53. However, TP53 mutations are rare in TALL, hence leukemic cells have to devise other measures to circumvent apoptosis. Our data offer an explanation as to how lymphoid malignancies can overcome p53 activation and why they are dependent on Gfi1. We propose that DNA damage, initiated by oncogenic stress during malignant transformation, induces p53 activity. High Gfi1-expressing subclones can thus be selected during transformation to enable global restriction of p53-mediated apoptosis. Gfi1 exerts this function by (i) co-occupying p53 target genes such as Bax, Pmaip1 and Bbc3, (ii) binding to p53-bound transcriptional complexes and (iii) limiting the methylation of p53 at K372 thereby restricting the activity of p53 and the activation of p53 target genes.
The function of Gfi1 to limit p53-K372 methylation (p53-K369 in murine cells)(Kurash et al., 2008) appears to be dependent on its ability to bind SNAG-dependent cofactors such as LSD1. It is known that demethylation of p53 at K370 is mediated by LSD1 and prevents p53 association with co-activators such as p53BP1 (Huang et al., 2007). We propose that leukemic cells use a Gfi1-LSD1 or a Gfi1-SNAG-dependent cofactor complex to demethylate p53 at K372, which prevents a full activation of p53 and its prop-apoptotic target genes. However, we cannot exclude the possibility that loss of Gfi1-SNAG-dependent transcriptional repression leads to the activation of factors, which may directly affect p53 activation/methylation status. In either case, ablation of Gfi1 leads to an accumulation of more active methylated p53, to a more efficient transactivation of pro-apoptotic p53 target genes, and as a consequence to accelerated cell death. Several independent lines of evidence support this notion including reporter gene assays, Chip-Seq data, biochemical analyses and expression data and offer a mechanistic explanation why Gfi1 ablation leads to regression of murine lymphomas and cause an inhibition of primary human T-ALL cell expansion in immune deficient mice.
Our findings have direct implications for current ALL treatments, which consist of chemotherapy and irradiation. Both are non-specific and highly toxic, damaging host and tumor tissues. These therapies function mainly through the induction of DNA damage and the initiation of p53-dependent DNA damage response pathways that cause cell death. Even when effective, patients can suffer dramatic side effects from standard ALL treatments. Therefore, reducing chemotherapeutic or irradiation dose and thus their side effects while maintaining their efficacy would directly benefit patients. The main result from our study suggest that this goal can be achieved by inhibiting the function of Gfi1 in T-ALL patients, because ablation of Gfi1 accelerates p53 induced cell death in leukemic cells. According to our data, leukemic cells lacking Gfi1 will be more sensitive to DNA damage inducing chemo- or irradiation therapy and undergo accelerated apoptosis. It is thus conceivable that targeting Gfi1 will not only significantly improve response rates but may in particular allow lower effective doses of chemotherapeutic agents or irradiation. In summary, our findings suggest that Gfi1 represents an Achilles Heel of lymphoid leukemias and our approach to target Gfi1 may soon move to clinical trials.
Experimental Procedures
Mice
LckCre+, Mx1-Cre+, C57BL/6, CD45.1, Trp53−/− and NSG mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Regarding other mouse strains, please refer to the supplementary information. Mice were housed in either single ventilated cages with top filters or microisolator cages. The Institutional Animal Care, Use and Ethical Committees responsible for Cincinnati Children’s Hospital Medical Center, (CCHMC), for the Institut de recherches cliniques de Montreal, (IRCM) and for the University Clinic Essen (UKE) reviewed and approved all animal experimentation.
Xenograft transplants and Morpholino Treatment
Diagnostic patient samples were obtained after informed consent according to Helsinki declaration and with approval from the Institutional Review Boards at the IRCM, at CCHMC and at UKE for the described experiments. One million T-ALL cells were transplanted (i.v) into NSG mice which were injected four days later (i.v.) with Vivo Morpholinos (Gene Tools, LLC) as described in the figures with 25nM of Control (“NT-VM”, 5′-CCTCTTACCTCAGTTACAATT TATA-3′) or Gfi1-specific (“Gfi1-VM”, 5′-ATGGTGGTCCGGCACTTTCCCCACT-3′) Vivo Morpholino per injection.
In vivo deletion of Gfi1 and ultrasound observation
Gfi1f/f or RosaCreERT2 Gfi1f/f mice were injected (i.p) with 1mg OHT (Sigma-Aldrich) dissolved in 100 μL of corn oil the first five days following transplantation. Gfi1f/f or Mx1-Cre+;Gfi1f/f mice were either injected (i.p) four weeks after the last ENU injection or three days after the transplantation of the tumor cells with 500mg pIpC (Sigma-Aldrich) seven times every other day. PCR validation of in vivo deletion was performed as previously described (Horman et al., 2009). Ultra sound observation was performed on anesthetized mice and thymic tumors were measured using the Visualsonic ultrasound machine and the Vev0770 imaging software (Toronto, Canada). A tumor was called present if the thymic surface area measured in the horizontal and sagittal plane was larger than 8 mm2, as average thymic surface of age matched, untreated Gfi1f/f control mice is 4 mm2, and if the tumor exhibited growth of more than 50% during the last two weeks of observation.
Statistics
Graph-Pad Prism software (GraphPad software, La Jolla, CA, USA) was used for most statistical analysis. Kaplan-Meier curves were analyzed using Log-rank tests. p≤0.05 were considered significant for all analyses. Differences in incidences of leukemia or lymphoma among the different groups were determined using Fisher’s exact test. Two-tailed unpaired Student t-tests were used to calculate the differences in the gene expression of patient data, WBC and spleen weights of transplanted mice, as well as the differences in cell number or tumors in ENU and MMLV treated mice. The Mann Whitney test was used to determine significance in counter-selection assays. Two-way ANOVAs were used to calculate significance of Vivo Morpholino dose responsive curves. Differences in AnnexinV staining of Bcl2-transgenic Gfi1−/− mice and Trp53p53−/−Gfi1−/− were calculated using one-way ANOVAs. GSEA FDR Q-values of <0.25 were used as a cut-off for enriched signatures.
Supplementary Material
Significance.
Chemotherapy is non-specific and highly toxic, damaging both host and tumor tissues. Even when effective, patients suffer dramatic side effects from standard treatments. Molecular-based targeted therapies have shown great promise, but lack broad applicability due to the heterogeneity of oncogenic pathways mutated during transformation. Here, we demonstrate that ablation of Gfi1 broadly leads to lymphoid tumor regression and host survival independent of the transforming pathway. We demonstrate that Gfi1 limits the pro-apoptotic functions of the endogenous gatekeeper p53. Gfi1 inhibition amplifies p53-dependent pro-apoptotic responses driven by oncogenic stress; consequently, transformed lymphoid tissues are uniquely susceptible to Gfi1 inhibition. Thus, in combination with current therapies, Gfi1 inhibition may allow the use of lower cytotoxic doses, which would benefit patients directly.
HIGHLIGHTS.
High Gfi1 expression is associated with a subgroup of T-ALL.
Gfi1 is required for the development and maintenance of both T and B cell tumors.
Lymphoid tumors need Gfi1 to counteract DNA-damage induced, p53-mediated apoptosis.
Inhibiting GFI1 impedes the expansion of primary human T-ALL in xenograft model.
Acknowledgments
We thank David Hildeman, Anil Jegga, Patrick Zweidler-McKay, Michelle Kelliher, Tom Look and Paul Jolicouer for expertise, for kindly providing plasmids, cell lines, reagents and mice. CK was supported by a fellowship of the Cole foundation, the IFZ fellowship of the University Clinic of Essen and a Max-Eder fellowship from the German Cancer fund. JDP is a Pelotonia Fellow and was supported by the University of Cincinnati Cancer Therapeutics T32 training grant (T32-CA117846). JS was supported by a Gordon Piller PhD studentship from Leukaemia and Lymphoma Research UK, SRH by a fellowship from CancerFree Kids, JZ and WEP by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases. HLG was supported by the Leukemia and Lymphoma Society of America, NIH CA105152, CA159845, Alex’s Lemonade Stand and thanks the Center of Excellence in Molecular Hematology P30 award (DK090971). TM was supported by a Canada Research Chair (Tier 1) and grants from the Canadian Institutes of Health Research (CIHR, MOP – 84238, MOP – 111011).
Footnotes
Accession Numbers
ChIP-Seq data have been submitted to the NCBI archive under accession number GSE31657. Arrays data are accessible under GEO-Omnibus accession GSE32910. All other experimental procedures can be found in the online Supplemental Information.
Author contribution:
CK and JDP designed and performed experiments, analyzed data and wrote the manuscript. M-CG, LV, JS, RC, SRH, JK, BG performed experiments and assisted analyses. JZ and WEP provided the Gfi1f/f mouse strain. UD provided funding and oversaw research. HLG and TM were responsible for concept and design of experiments, oversaw research, wrote the manuscript and provided funding.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Sehested M, Nesland JM, Rajpert-De Meyts E, Skakkebaek NE, Stucki M, Jackson S, Lukas J, Bartek J. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene. 2007;26:7414–7422. doi: 10.1038/sj.onc.1210553. [DOI] [PubMed] [Google Scholar]
- Blyth K, Terry A, Mackay N, Vaillant F, Bell M, Cameron ER, Neil JC, Stewart M. Runx2: a novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene. 2001;20:295–302. doi: 10.1038/sj.onc.1204090. [DOI] [PubMed] [Google Scholar]
- Chakraborty J, Okonta H, Bagalb H, Lee SJ, Fink B, Changanamkandat R, Duggan J. Retroviral gene insertion in breast milk mediated lymphomagenesis. Virology. 2008;377:100–109. doi: 10.1016/j.virol.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, Nikov G, Krishnamoorthy V, Majumder PK, Kelliher MA. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009;113:6172–6181. doi: 10.1182/blood-2008-02-136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowska MJ, Dybkaer K, Johnsen HE, Wang B, Wabl M, Pedersen FS. Loss of MicroRNA targets in the 3′ untranslated region as a mechanism of retroviral insertional activation of growth factor independence 1. J Virol. 2009;83:8051–8061. doi: 10.1128/JVI.00427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland DG, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Fiolka K, Hertzano R, Vassen L, Zeng H, Hermesh O, Avraham KB, Duhrsen U, Moroy T. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep. 2006;7:326–333. doi: 10.1038/sj.embor.7400618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks CB, Bear SE, Grimes HL, Tsichlis PN. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol Cell Biol. 1993;13:1759–1768. doi: 10.1128/mcb.13.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokbuget N, Hoelzer D. Treatment of adult acute lymphoblastic leukemia. Semin Hematol. 2009;46:64–75. doi: 10.1053/j.seminhematol.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hameyer D, Loonstra A, Eshkind L, Schmitt S, Antunes C, Groen A, Bindels E, Jonkers J, Krimpenfort P, Meuwissen R, et al. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol Genomics. 2007;31:32–41. doi: 10.1152/physiolgenomics.00019.2007. [DOI] [PubMed] [Google Scholar]
- Horman SR, Velu CS, Chaubey A, Bourdeau T, Zhu J, Paul WE, Gebelein B, Grimes HL. Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood. 2009;113:5466–5475. doi: 10.1182/blood-2008-09-179747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- Khandanpour C, Krongold J, Schutte J, Bouwman F, Vassen L, Gaudreau MC, Chen R, Calero-Nieto FJ, Diamanti E, Hannah R, et al. The human GFI136N variant induces epigenetic changes at the Hoxa9 locus and accelerates K-RAS driven myeloproliferative disorder in mice. Blood. 2012;120:4006–4017. doi: 10.1182/blood-2011-02-334722. [DOI] [PubMed] [Google Scholar]
- Kundu M, Compton S, Garrett-Beal L, Stacy T, Starost MF, Eckhaus M, Speck NA, Liu PP. Runx1 deficiency predisposes mice to T-lymphoblastic lymphoma. Blood. 2005;106:3621–3624. doi: 10.1182/blood-2005-04-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- Li H, Ji M, Klarmann KD, Keller JR. Repression of Id2 expression by Gfi-1 is required for B-cell and myeloid development. Blood. 2010;116:1060–1069. doi: 10.1182/blood-2009-11-255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Palomero T, Sumazin P, Califano A, Ferrando AA, Stolovitzky G. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc Natl Acad Sci U S A. 2009;106:244–249. doi: 10.1073/pnas.0806445106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medyouf H, Gusscott S, Wang H, Tseng JC, Wai C, Nemirovsky O, Trumpp A, Pflumio F, Carboni J, Gottardis M, et al. High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. The Journal of experimental medicine. 2011;208:1809–1822. doi: 10.1084/jem.20110121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcos PA, Li Y, Jiang S. Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. Biotechniques. 2008;45:613–614. 616, 618. doi: 10.2144/000113005. passim. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O’Neil J, Neuberg D, Weng AP, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pargmann D, Yucel R, Kosan C, Saba I, Klein-Hitpass L, Schimmer S, Heyd F, Dittmer U, Moroy T. Differential impact of the transcriptional repressor Gfi1 on mature CD4+ and CD8+ T lymphocyte function. Eur J Immunol. 2007;37:3551–3563. doi: 10.1002/eji.200737130. [DOI] [PubMed] [Google Scholar]
- Person RE, Li FQ, Duan Z, Benson KF, Wechsler J, Papadaki HA, Eliopoulos G, Kaufman C, Bertolone SJ, Nakamoto B, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priceputu E, Bouallaga I, Zhang Y, Li X, Chrobak P, Hanna ZS, Poudrier J, Kay DG, Jolicoeur P. Structurally distinct ligand-binding or ligand-independent Notch1 mutants are leukemogenic but affect thymocyte development, apoptosis, and metastasis differently. J Immunol. 2006;177:2153–2166. doi: 10.4049/jimmunol.177.4.2153. [DOI] [PubMed] [Google Scholar]
- Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Scheijen B, Jonkers J, Acton D, Berns A. Characterization of pal-1, a common proviral insertion site in murine leukemia virus-induced lymphomas of c-myc and Pim-1 transgenic mice. J Virol. 1997;71:9–16. doi: 10.1128/jvi.71.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Karsunky H, Gau E, Zevnik B, Elsasser HP, Moroy T. Zinc finger protein GFI-1 has low oncogenic potential but cooperates strongly with pim and myc genes in T-cell lymphomagenesis. Oncogene. 1998;17:2661–2667. doi: 10.1038/sj.onc.1202191. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Zornig M, Beneke R, Moroy T. MoMuLV proviral integrations identified by Sup-F selection in tumors from infected myc/pim bitransgenic mice correlate with activation of the gfi-1 gene. Nucleic Acids Res. 1996;24:2528–2534. doi: 10.1093/nar/24.13.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, Krishnamoorthy V, Bhasin M, Capobianco AJ, Kelliher MA. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, Lagcher W, Sie D, Tanger E, Cox T, et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ, Stubbs A, Cools J, Nagata K, Fornerod M, et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood. 2008;111:4668–4680. doi: 10.1182/blood-2007-09-111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720–4728. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West LE, Gozani O. Regulation of p53 function by lysine methylation. Epigenomics. 2011;3:361–369. doi: 10.2217/EPI.11.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, Burel SA, Lagasse E, Weissman IL, Akashi K, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci U S A. 2001;98:10398–10403. doi: 10.1073/pnas.171321298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel R, Karsunky H, Klein-Hitpass L, Moroy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med. 2003;197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarebski A, Velu CS, Baktula AM, Bourdeau T, Horman SR, Basu S, Bertolone SJ, Horwitz M, Hildeman DA, Trent JO, et al. Mutations in growth factor independent-1 associated with human neutropenia block murine granulopoiesis through colony stimulating factor-1. Immunity. 2008;28:370–380. doi: 10.1016/j.immuni.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jankovic D, Grinberg A, Guo L, Paul WE. Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc Natl Acad Sci U S A. 2006;103:18214–18219. doi: 10.1073/pnas.0608981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zornig M, Schmidt T, Karsunky H, Grzeschiczek A, Moroy T. Zinc finger protein GFI-1 cooperates with myc and pim-1 in T-cell lymphomagenesis by reducing the requirements for IL-2. Oncogene. 1996;12:1789–1801. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.