Abstract
ERG rearrangements, (most commonly TMPRSS2: ERG [T2:ERG] gene fusions), have been identified in approximately 50% of prostate cancers (PCa). Quantification of T2:ERG in post-DRE urine, in combination with PCA3, improves the performance of serum PSA for PCa prediction on biopsy Here we compared urine T2:ERG and PCA3 scores to ERG+ (determined by immunohistochemistry) and total prostate cancer burden in 41 mapped prostatectomies. Prostatectomies had a median of 3 tumor foci (range: 1–15) and 2.6 cm of summed linear tumor dimension (range: 0.6–7.1 cm). Urine T2:ERG score most correlated with summed linear ERG+ tumor dimension and number of ERG+ foci (rs=0.68 and 0.67, respectively, both p<0.001). Urine PCA3 score showed weaker correlation with both number of tumor foci (rs=0.34, p=0.03) and summed linear tumor dimension (rs=0.26, p=0.10). In summary, we demonstrate a strong correlation between urine T2:ERG score and total ERG+ PCa burden at prostatectomy, consistent with high tumor specificity.
Keywords: TMPRSS2:ERG, prostate cancer, PCA3, urine
Introduction
Recently discovered chromosomal rearrangement in prostate carcinoma (PCa), resulting in fusion of the 5′ translated region of the androgen-regulated gene TMPRSS2 (transmembrane protease, serine 2) with members of the ETS family of transcription factors, including ERG or ETV11, are promising new biomarkers to aid in the detection of PCa2–6. ETS fusions have been reported in approximately 50% of PSA screened prostate cancers and fusions between TMPRSS2 and ERG represent 90% of all ETS fusions; by fluorescence in situ hybridization (FISH) or RT-PCR, ERG rearrangements (as a surrogate for TMPRSS2:ERG fusion) are very specific for prostate cancer or HGPIN immediately adjacent to cancer, and hence, the ability to detect this fusion can potentially be utilized for the detection of prostate carcinoma2–10. More recently, monoclonal antibodies against ERG have been developed which detect the truncated ERG protein product of TMPRSS2:ERG fusions11,12. By immunohistochemistry (IHC), these antibodies are strongly correlated with ERG rearrangement as detected by FISH, and stain ~50% of prostate cancers and ~15% of HGPIN (immediately adjacent to ERG+ cancer), with exceptionally rare staining in non-neoplastic prostate tissue3,11–16.
Recently, our group has evaluated a clinical grade, transcription-mediated amplification (TMA) assay that quantifies TMPRSS2:ERG (T2:ERG) mRNA in post-DRE (digital rectal exam) urine17. This assay is based on the same technology as the PROGENSA PCA3 assay, a urine based assay for the quantification of the non- coding transcript PCA318,19, which is FDA approved for predicting prostate cancer on rebiopsy. In a prospective study of over 1,300 men, we showed that urine T2:ERG score, used in combination with urine PCA3 score, enhances the utility of serum PSA to predict prostate cancer risk on biopsy; urine T2:ERG score was also significantly correlated with the number of involved cores and maximum % core involvement at biopsy, and maximum index tumor dimension at prostatectomy17.
PCA3 encodes a non-translated transcript over-expressed in >95% of all prostate cancers with high prostate specificity20–22. The PROGENSA™ PCA3 Assay has demonstrated utility for predicting prostate biopsy outcome, and urine PCA3 score has shown an association with tumor volume23–28 and multifocality29 in prostatectomy cohorts. However, as PCA3 encodes a non-coding transcript, precluding IHC based detection, only a single in situ based evaluation of PCA3 expression has been reported PCA3 expression in the majority of prostate cancers and HGPIN lesions30.
As FISH and IHC studies have shown that ERG rearrangements and protein expression are exceptionally rare in benign prostate tissue or cancer mimickers, we hypothesized that urine T2:ERG should be strongly correlated with the total ERG+ prostate cancer burden in a given patient. Likewise, published studies indicate that urine PCA3 score is correlated with overall tumor burden. Thus, here we compared urine T2:ERG and PCA3 scores to ERG+ and overall cancer burden at prostatectomy to assess the cancer specificity of these urine biomarkers.
Materials and Methods
Study Cohort
The prostatectomy cohort studied was identified from a cohort of 301 men referred for prostate needle biopsy at the University of Michigan Health System (UMHS), who were all assessed by transcription mediated amplification (TMA) for urine T2:ERG and urine PCA3 scores as described below. Forty one men who subsequently underwent prostatectomy at our institution between 2008 and 2011 were included in the study. None of the patients received preoperative radiation or androgen deprivation therapy. Clinicopathologic characteristics including age of patient, ultrasound volume at biopsy, pre-biopsy PSA levels, PSAD and biopsy details (total number of biopsy cores, number of positive cores and percentage of cores positive) were obtained from our clinical database. All biopsy and prostatectomy cases and urine specimens were obtained with Institutional Review Board approval.
Urine T2:ERG and PCA3 assays
Assessment of urine T2:ERG and PCA3 were performed essentially as described17. Urine specimens were obtained immediately after attentive DRE, refrigerated, and processed within 4 hours by mixing with an equal volume of urine transport medium and stored above −70 C until analysis. Amounts of T2:ERG and PSA mRNA were determined by transcription mediated amplification (TMA). To generate a T2:ERG score, the amount of T2:ERG mRNA is normalized to the amount of PSA mRNA, which is calculated utilizing the formula: 100,000 × average urine TMPRSS2:ERG copies/ml)/ (average urine PSA copies/ml). Samples with average PSA copies/ml > 20,000 were considered informative. Patients in the current study were assessed with a third generation, final clinical TMA assay, which is highly correlated to first and second generation assays (Spearman correlation (rs) = 0.86, p<0.001) described in 17. The PROGENSA PCA3 assay (Gen-Probe Inc, San Diego, CA, USA) similarly quantitates PCA3 and PSA mRNA in post- DRE urine. The PCA3 score was calculated utilizing the formula: 1,000 × (average urine PCA3 copies/ml)/ (average urine PSA copies/ml). Samples with average PSA copies/ml > 7,000 were considered informative. Identical primers for quantifying PSA are used in the PROGENSA PCA3 assay and T2:ERG assay.
Prostatectomy Evaluation
Fresh prostates removed after surgery were weighed, measured, inked, and fixed in 10% neutral formalin. Seminal vesicles, apex, and base were amputated, and the remaining prostate was serially sectioned at 4 mm to 5 mm intervals perpendicular to the long axis of the gland from the base to apex and quartered. All prostatectomy specimens were reviewed by the study pathologists. Tumor maps were generated by tracking each section and reconstructing them as a whole-mount section. A cancer focus was considered spatially separate or multifocal if it was 3 mm or more from the closest cancer in any single section or 4mm or more from the closest cancer on the adjacent section above or below, as described previously31. The largest tumor focus was designated as the index tumor and additional smaller tumors were labeled as multifocal tumors. For each prostatectomy, the total number of tumor foci, linear dimension and Gleason score of the index focus, and linear tumor dimension and Gleason score of all tumor foci was documented. As the greatest linear dimension of the index focus is reported clinically at UMHS rather than index focus volume, we used the greatest linear dimension of all foci as a cancer volume measurement, which has been validated previously32. Immunohistochemistry for ERG (see below) was performed on sections representing all index and multifocal foci from each case. As ERG staining was uniformly nuclear, strong and diffuse except as noted, we assigned all tumor foci as ERG+ or ERG−, and tumor foci with heterogeneous ERG staining were considered ERG+. The index tumor focus showed the highest Gleason score in the majority of cases. In the rare cases where a smaller multifocal focus had a higher Gleason score compared to the index tumor, the smaller multifocal tumor focus with the highest Gleason score was considered as the index tumor. The summed linear tumor dimension was calculated by summing the largest dimension of the index focus and the largest dimension of all multifocal tumor foci. Likewise, the summed ERG+ linear tumor dimension was calculated by summing the largest dimension of all ERG+ tumor foci, including the index tumor when ERG+.
ERG Immunohistochemistry (IHC)
IHC on unstained formalin fixed, paraffin-embedded levels of all tumor foci from the prostatectomy specimen blocks was performed using a monoclonal antibody against ERG, clone EPR 3864 (Epitomics, Burlingame, CA), using the automated Discovery XT staining platform (Ventana Medical Systems, Tucson, AZ) as described12,33. ERG staining was evaluated by the study pathologists. Staining of vessels was used as a positive control and slides without staining of vessels were excluded from further analysis. All immunostains were reviewed by study pathologists.
Statistical Analysis
Associations between urine T2:ERG score, urine PCA3 score and clinicopathological data were assessed using GraphPad Prism 5 (Graph Pad Software). Comparisons of the number of ERG+ and ERG− foci, and summed ERG+ and total tumor dimension per case, were assessed by paired t-tests. Correlations between urine T2:ERG and PCA3 scores and continuous and categorical clinicopathologic variables were assessed with Spearman’s rho (rs) and the Wilcoxon rank-sum test, respectively. Linear regression analysis was also performed to assess the association between urine biomarkers, and between urine biomarkers and ERG+ and total tumor volume. Urine T2:ERG scores were log transformed (log(T2:ERG+1) to minimize the impact of outliers, which resulted in increased R2 values for all associations compared to non-transformed T2:ERG scores. Two-tailed tests were used for all comparisons and p values <0.05 were considered statistically significant.
Results
Prostatectomy cohort
The 41 prostatectomies included in the study had a median of 3 tumor foci (range 1–15) and 2.6 cm of summed linear tumor dimension (range 0.6–7.1 cm). The index focus showed the highest Gleason score in 39/41 (95%) cases. In two cases (cases 12 and 38), a smaller multifocal focus showed higher Gleason grade than the larger index focus ( 4+3 and 3+4 in the smaller multifocal focus vs. 3+3 respectively) and was considered as the index focus for analysis. The vast majority of cases in this study were confined to prostate (pT2, 37/41, 90%), with index tumor Gleason scores of 7 (31/41, 76%). Pathological data for all cases are shown in Table 1.
Table 1.
Pathological data and urine TMPRSS2:ERG (T2:ERG) and PCA3 scores for prostatectomy patients (n=41)
| Case | Summed tumor dimension (cm) | Summed ERG+ tumor dimension (cm) | Index nodule dimension (cm) | ERG status of index nodule | Gleason score of index nodule | Total number of tumor nodules | Total number of ERG+ nodules | Highest Gleason score of multifocal nodules | Urine T2:ERG score | Urine PCA3 score |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3.4 | 0.0 | 1.2 | Negative | 3+3 | 3 | 0 | 3+3 | 0 | 35 |
| 2 | 2.6 | 0.0 | 1.4 | Negative | 3+4 | 3 | 0 | 3+4 | 0.1 | 27 |
| 3 | 3.2 | 0.0 | 1.8 | Negative | 4+3 | 2 | 0 | 3+4 | 0.5 | 3 |
| 4 | 0.8 | 0.0 | 0.8 | Negative | 3+3 | 1 | 0 | NA | 1.3 | 66 |
| 5 | 5.1 | 0.0 | 1.0 | Negative | 3+4 w/ 5 | 5 | 0 | 3+4 | 2.4 | 60 |
| 6 | 2.9 | 0.0 | 1.5 | Negative | 4+3 | 5 | 0 | 3+3 | 3.7 | 19 |
| 7 | 1.4 | 0.0 | 0.7 | Negative | 3+4 | 2 | 0 | 3+3 | 6.1 | 74 |
| 8 | 0.9 | 0.0 | 0.6 | Negative | 3+4 | 2 | 0 | 3+4 | 96.4 | 50 |
| 9 | 0.9 | 0.1 | 0.8 | Negative | 3+3 | 2 | 1 | 3+3 | 96.3 | 13 |
| 10 | 3.4 | 0.2 | 2.8 | Negative | 4+3 w/5 | 3 | 1 | 3+3 | 39.6 | 74 |
| 11 | 2.4 | 0.4 | 2.0 | Negative | 3+4 | 3 | 2 | 3+3 | 860.8 | 42 |
| 12 | 2.4 | 0.0 | 1.9 | Negative | 4+3 | 2 | 0 | 3+3 | 2.6 | 86 |
| 13 | 2.4 | 0.5 | 0.7 | Negative | 3+3 | 6 | 2 | 3+3 | 14.1 | 22 |
| 14 | 0.6 | 0.6 | 0.6 | Positive | 4+4 | 1 | 1 | NA | 0.1 | 3 |
| 15 | 0.9 | 0.6 | 0.6 | Positive | 3+3 | 3 | 1 | 3+3 | 21.5 | 83 |
| 16 | 1.8 | 0.9 | 0.9 | Negative | 3+4 | 2 | 1 | 3+3 | 25.2 | 15 |
| 17 | 2.6 | 0.9 | 0.7 | Negative | 3+4 | 6 | 3 | 3+4 | 1951.5 | 72 |
| 18 | 4.3 | 1.0 | 1.6 | Negative | 3+4 | 5 | 2 | 3+3 | 6.1 | 55 |
| 19 | 2.2 | 1.1 | 1.1 | Positive | 3+4 | 3 | 1 | 3+3 | 40.7 | 6 |
| 20 | 1.1 | 1.1 | 1.1 | Positive | 3+4 | 1 | 1 | NA | 53.0 | 8 |
| 21 | 2.0 | 1.2 | 1.1 | Positive | 3+4 | 5 | 2 | 3+3 | 19.2 | 38 |
| 22 | 1.9 | 1.3* | 1.2 | Positive | 3+4 | 5 | 2 | 3+3 | 97.7 | 12 |
| 23 | 1.3 | 1.3 | 1.3 | Positive | 3+4 | 1 | 1 | NA | 288.7 | 31 |
| 24 | 3.8 | 1.5 | 1.6 | Negative | 3+4 | 3 | 1 | 3+4 | 21.6 | 17 |
| 25 | 2.6 | 1.5 | 0.8 | Positive** | 3+4 | 5 | 2 | 3+4 | 446.5 | 104 |
| 26 | 2.3 | 1.9 | 1.0 | Positive | 3+3 | 6 | 3 | 3+3 | 31.1 | 95 |
| 27 | 3.2 | 2.1 | 2.1 | Positive | 4+3 | 2 | 1 | 3+4 | 343.6 | 36 |
| 28 | 2.3 | 2.3 | 1.0 | Positive | 3+3 | 4 | 2 | 3+3 | 288.3 | 37 |
| 29 | 2.7 | 2.4 | 1.6 | Positive | 3+4 | 4 | 2 | 3+3 | 48.9 | 126 |
| 30 | 2.6 | 2.6 | 1.9 | Positive | 3+4 | 2 | 2 | 3+4 | 37.4 | 32 |
| 31 | 4.9 | 2.6 | 2.0 | Positive** | 3+4 | 6 | 4 | 3+4 | 167.4 | 75 |
| 32 | 3.2 | 2.7 | 2.1 | Positive | 3+4 | 4 | 3 | 3+3 | 37.9 | 11 |
| 33 | 5.5 | 2.7 | 1.5 | Positive | 3+4 | 4 | 2 | 3+4 | 34.1 | 47 |
| 34 | 2.9 | 2.8 | 1.0 | Positive | 3+4 | 8 | 7 | 3+3 | 112.9 | 33 |
| 35 | 4.9 | 3.3 | 1.6 | Positive** | 3+4 | 5 | 3 | 3+4 | 147.6 | 69 |
| 36 | 3.5 | 3.4 | 2.4 | Positive | 3+4 | 3 | 2 | 3+3 | 707.7 | 42 |
| 37 | 3.7 | 3.7 | 2.0 | Positive | 3+4 | 3 | 3 | 3+3 | 6031.6 | 11 |
| 38 | 3.8 | 3.8* | 0.7 | Positive | 4+3 | 7 | 6 | 3+3 | 236.3 | 43 |
| 39 | 3.8 | 3.8 | 1.9 | Positive | 3+4 | 3 | 3 | 3+4 | 762.5 | 105 |
| 40 | 7.1 | 4.6 | 0.8 | Positive | 3+4 | 15 | 8 | 3+3 | 1261.4 | 186 |
| 41 | 5.0 | 5.0 | 2.1 | Positive | 4+3 | 3 | 3 | 3+4 | 301.8 | 40 |
A tumor focus was lost on deeper sections, precluding IHC staining for ERG status.
Heterogenous ERG staning, designated ERG+ for analysis.
A total of 159 tumor foci were evaluated for ERG staining (including index foci), of which 78 tumor foci (49%) were ERG+. Tumor foci, when positive, showed strong nuclear staining with ERG in all cancerous glands within the tumor focus, except for 3 foci where the index tumor showed heterogeneous ERG expression (moderate to strong staining, considered ERG+ for analysis). ERG was expressed in cancerous glands in 32 of 41 cases (78%) within at least one tumor focus, while the remaining 9 of 41 cases (22%) lacked ERG expression in all tumor foci. ERG expression in the index tumor was noted in 24 (73%) of cases. Representative ERG+ and ERG− foci are shown in Figure 1. The pathological data, ERG IHC, and urine T2:ERG and PCA3 scores are summarized in Table 1.
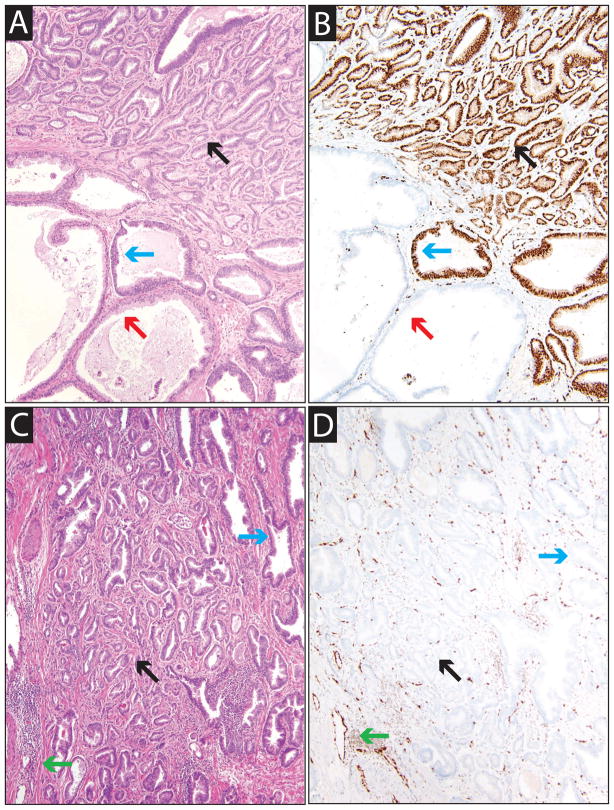
Figure 1. Mapping ERG+ and ERG− tumor foci in prostatectomy specimens.
Prostatectomy specimens (n=41) were mapped, and the index focus and all multifocal foci were identified (see Methods). Immunohistochemistry for ERG was performed on sections representing all index and multifocal foci from each case, and each focus was classified as ERG+ or ERG−. Staining of vessels was used as a positive control and sections without staining of vessels were excluded and staining repeated. A&B. Hematoxylin and eosin (H&E, A) and ERG (B) stained sections of an ERG+ index focus (case 31). Areas of benign glands, HGPIN and carcinoma are indicated by red, blue and black arrows, respectively. C&D. H&E (C) and ERG (D) stained sections of an ERG− multifocal focus (case 6). Endothelial cells and lymphocytes serve as an internal positive control (green arrows). All images are 10× original magnification.
The median summed linear dimension of ERG+ cancer was 1.2 cm (range 0–5.0 cm). There was no significant difference between the number (mean 1.9 vs. 2.0, paired t-test, p=0.89) or summed linear tumor dimension (mean 1.6 cm vs. 1.3 cm, paired t-test, p=0.52) of ERG+ and ERG− foci per case. Given the low frequency of >T2 disease and Gleason scores < or >7, association of index focus ERG status and Gleason grade and stage were not assessed.
Overall, across 169 total sections, ERG staining was extremely specific for prostatic adenocarcinoma. Although vessels and lymphocytes stain with ERG, this represents expression of wild-type ERG, and does not represent ERG rearrangements leading to TMPRSS2:ERG over-expression12. When positive for ERG staining, HGPIN was always present adjacent to ERG+ cancer, with the exception of one ERG+ HGPIN focus not immediately adjacent to ERG+ cancer (this HGPIN gland was 0.4 cm from ERG positive cancer). ERG positivity in benign glands was extremely rare, with ~ 35 ERG+ glands noted in 8 cases across 169 total sections. Only one focus composed of two benign acini was greater than 0.4 cm from ERG positive cancer. Using the estimated number of benign glands per prostatectomy section by Furusato et al.34, the overall specificity of ERG staining for prostate cancer is >99.99%. Representative ERG+ HGPIN and benign glands not immediately adjacent to ERG+ carcinoma are shown in Figure 2.
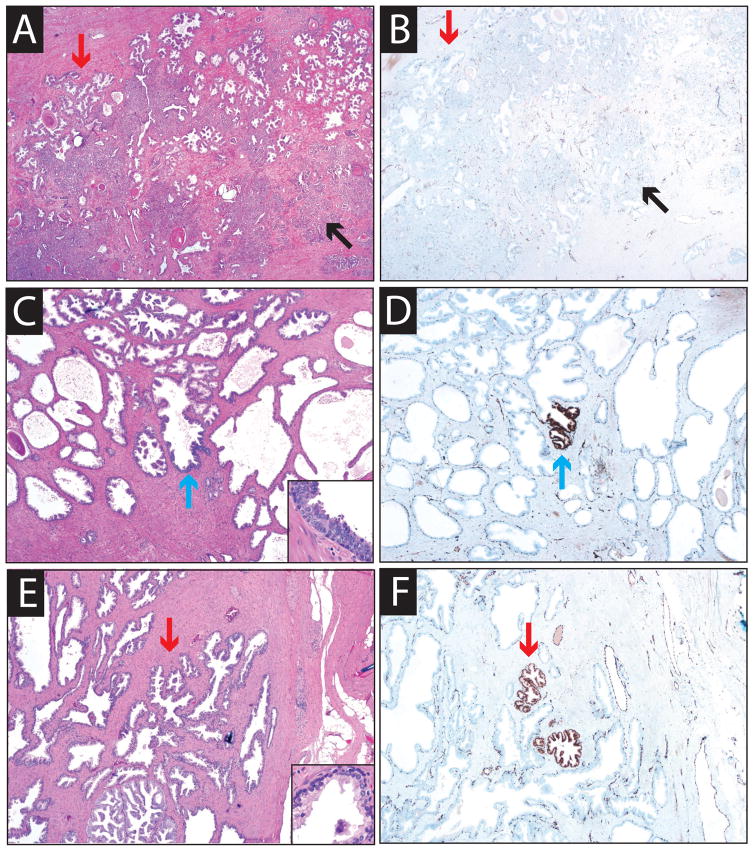
Figure 2. ERG staining is specific for PCa and immediately adjacent HGPIN.
ERG staining was evaluated in 169 prostatectomy sections as described in Fig. 1. Across all 169 sections, only one focus of ERG+ high grade prostatic intraepithelial neoplasia (HGPIN) and approximately 2 ERG+ benign glands not immediately adjacent to ERG+ carcinoma were identified. A&B. Hematoxylin and eosin (H&E, A) and ERG (B) stained sections from an ERG− cancer focus showing no expression in cancerous or benign glands. C&D. H&E (C) and ERG (D) stained sections from case 38 demonstrating ERG+ HGPIN not immediately adjacent to cancer. E&F. H&E (E) and ERG (F) stained sections from case 35 demonstrating ERG+ morphologically benign glands not immediately adjacent to cancer. Areas of benign glands, HGPIN and carcinoma are indicated by red, blue and black arrows, respectively. Staining of vessels was used as a positive control.
Urine T2:ERG and PCA 3
All 41 patients had sufficient urine PSA mRNA expression to evaluate the T2:ERG score. The median T2:ERG score was 41 (range 0–6,032). As shown in Table 2, of patients with 0 cm, >0.1 to 1.0 cm, 1.1 to 2.0 cm and >2.0 cm of summed ERG+ linear tumor dimension, 1/9 (11%), 4/9 (44%), 6/8 (75%) and 15/15 (100%), respectively, had urine T2:ERG scores >30, which is associated with an ~70–75% risk of prostate cancer on biopsy17. Of patients with 0 cm, >0.1 to 1.0 cm, 1.1 to 2.0 cm and >2.0 cm of summed ERG+ linear tumor dimension, 1/9 (11%), 7/9 (78%), 8/8 (100%) and 15/15 (100%), respectively, had urine T2:ERG scores >10, below which is associated with an ~30–35% risk of prostate cancer on biopsy17.
Table 2.
Association of summed ERG+ and total tumor dimension with urine TMPRSS2:ERG (T2:ERG ) and PCA3 scores
| Summed ERG+ tumor dimension (cm)
|
||||
|---|---|---|---|---|
| 0 cm | 0.1–1.0 cm | 1.1–2.0 cm | >2.0 cm | |
| T2:ERG score >10 | 1/9 (11%) | 7/9 (78%) | 8/8(100%) | 15/15 (100%) |
| T2:ERG score > 30 | 1/9 (11%) | 4/9 (44%) | 6/8 (75%) | 15/15 (100%) |
| Summed tumor dimension (cm)
|
||||
|---|---|---|---|---|
| 0.1–1.0 cm | 1.1–2.5 cm | 2.6–3.5 | >3.5 cm | |
| PCA3 score >25 | 3/5 (60%) | 7/12 (58%) | 10/13 (77%) | 9/11 (82%) |
| PCA3 score >35 | 3/5 (60%) | 6/12 (50%) | 6/13 (46%) | 9/11 (82%) |
All 41 patients had sufficient urine PSA mRNA expression to evaluate the PCA3 score. The median PCA3 score was 40 (range 3–187). As shown in Table 2, of patients with 0.1 to 1.0 cm, 1.1 to 2.5 cm, 2.6 to 3.5 cm and >3.5 cm of summed total linear tumor dimension, 3/5 (60%), 6/12 (50%), 6/13 (46%) and 9/11 (82%), respectively, had urine PCA3 scores>35, which has been proposed as an optimal cutoff for the detection of cancer on biopsy35. Of patients with 0.1 to 1.0 cm, 1.1 to 2.5 cm, 2.6 to 3.5 cm and >3.5 cm of summed total linear tumor dimension, 3/5 (60%), 7/12 (58%), 10/13 (77%) and 9/11 (82%), respectively had urine PCA3 score>25, the FDA approved cutoff for predicting the presence of prostate cancer after initial negative biopsy.
We next addressed our hypothesis that urine T2:ERG should be strongly correlated with total ERG+ prostate cancer burden, and urine PCA3 should be strongly correlated with total prostate cancer burden. As shown in Table 3, comparing correlations of urine T2:ERG with various clinicopathological parameters, urine T2:ERG most significantly correlated with the summed linear dimension of ERG+ cancer and the number of ERG+ tumor foci (rs=0.68 and rs= 0.67, respectively, both p<0.0001). There was no significant association with other parameters, including summed total linear tumor dimension (rs=0.24, p=0.13) and urine PCA3 score (rs=0.22, p=0.18). Similarly, summed linear dimension of ERG+ cancer was most significantly associated with the total number of ERG+ tumor foci and urine T2:ERG score (rs=0.86 and rs= 0.68, respectively, both p<0.0001), as shown in Table 4. Urine T2:ERG was significantly associated with ERG+ vs. ERG− index focus status (median 130 vs. 6.1, Wilcoxon rank-sum test, p=0.002), and a trend of association with index Gleason score >6 vs. 6 was observed (median 51 vs. 22, p=0.11), however the small number of Gleason 6 cases limited this analysis.
Table 3.
Clinicopathological data and associations with urine TMPRSS2:ERG (T2:ERG ) and PCA3 scores for prostatectomy patients (n=41)
| Parameter | (n ) | Prostatectomy Patients | T2:ERG score | PCA3 score | ||
|---|---|---|---|---|---|---|
|
| ||||||
| rs (95% CI) | p | rs (95% CI) | p | |||
| Sum ERG+ tumor dimension (cm) | 41 | 1.2 (0.2–2.6) | 0.68 (0.46 to 0.82) | < 0.0001 | 0.14 (−0.19 to 0.44) | 0.39 |
| ERG+ tumor foci (n) | 41 | 2 (1–3) | 0.67 (0.45 to 0.81) | < 0.0001 | 0.24 (−0.08 to 0.52) | 0.13 |
| Prostate Weight (g) | 40 | 50 (42–58) | −0.25 (−0.53 to 0.08) | 0.12 | 0.02 (−0.30 to 0.34) | 0.91 |
| Sum tumor dimension (cm) | 41 | 2.6 (2–3.7) | 0.24 (−0.08 to 0.52) | 0.13 | 0.26 (−0.06 to 0.53) | 0.10 |
| Tumor foci (n) | 41 | 3 (2–5) | 0.23 (−0.09 to 0.51) | 0.15 | 0.34 (0.03 to 0.59) | 0.03 |
| T2:ERG score | 41 | 41 (14–288) | NA | NA | 0.22 (−0.11 to 0.50) | 0.18 |
| PCA3 score | 41 | 40 (19–72) | 0.22 (−0.11 to 0.50) | 0.18 | NA | NA |
| Index tumor dimension (cm) | 41 | 1.2 (0.8–1.9) | 0.21 (−0.12 to 0.49) | 0.19 | −0.06 (−0.37 to 0.26) | 0.73 |
| PSAD (ng/ml / g)1 | 40 | 0.096 (0.074–0.136) | 0.19 (−0.14 to 0.48) | 0.24 | 0.01 (−0.31 to 0.33) | 0.94 |
| Bx maximum core + (%) | 41 | 50% (20%–70%) | 0.18 (−0.14 to 0.47) | 0.26 | −0.16 (−0.46 to 0.16) | 0.31 |
| Age (yr) | 41 | 59 (55–63) | 0.17 (−0.15 to 0.46) | 0.28 | 0.06 (−0.26 to 0.37) | 0.70 |
| PSA (ng/ml) | 41 | 5.4 (4.5–6.4) | 0.10 (−0.22 to 0.41) | 0.52 | −0.02 (−0.33 to 0.30) | 0.92 |
| Bx cores + (%) | 41 | 33% (14%–54%) | 0.08 (−0.25 to 0.38) | 0.63 | −0.02 (−0.33 to 0.30) | 0.90 |
| Parameter | n | Prostatectomy Patients | T2:ERG score | PCA3 score | ||
|---|---|---|---|---|---|---|
|
| ||||||
| median (IQR) | p | median (IQR) | p | |||
| Index tumor status: | ||||||
| ERG − | 41 | 17 (41%) | 6.1 (1.9–68) | 0.002 | 42 (18–69) | 0.78 |
| ERG + | 24 (59%) | 130 (38–330) | 39 (17–81) | |||
| Index tumor Gleason score: | ||||||
| 6 | 41 | 7 (17%) | 22 (1.3–96) | 0.11 | 37 (22–83) | 0.74 |
| >6 | 34 (83%) | 51 (16–310) | 41 (17–72) | |||
| Race: | ||||||
| White | 41 | 33 (80%) | 41 (20–290) | 0.88 | 40 (14–74) | 0.99 |
| Not White | 8 (20%) | 64 (6.1–290) | 39 (25–65) | |||
| Bx Epstein status2: | ||||||
| Insignificant | 41 | 5 (12%) | 31 (11–270) | 0.58 | 83 (20–100) | 0.31 |
| Significant | 36 (88%) | 45 (8.1–292) | 39 (17–68) | |||
Number (n) of patients with data and median (IQR) or # (%) at prostatectomy, or biopsy (bx), are given. Spearman’s rho (rs) (95% CI) and p value for correlation with urine T2:ERG or PCA3 score, or median T2:ERG or PCA3 score (95% CI) and p values from Wilcoxon Rank-Sum tests are shown.
Serum PSA/prostatectomy weight.
Epstein criteria: any >T1c, PSAD>=0.15ng/ml, Gl >6, >=3 cores + or >50% greatest core as significant.
Table 4.
Associations of summed ERG+ and total tumor dimension for prostatectomy patients (n=41)
| Parameter | (n ) | Summed ERG+ tumor dimension | Summed tumor dimension | ||
|---|---|---|---|---|---|
| Rs (95% CI) | P | Rs (95% CI) | P | ||
| ERG+ tumor foci (n) | 41 | 0.86 (0.75 to 0.92) | < 0.0001 | 0.44 (0.15 to 0.66) | 0.004 |
| T2:ERG score | 41 | 0.68 (0.46 to 0.82) | < 0.0001 | 0.24 (−0.09 to 0.52) | 0.13 |
| Sum tumor dimension (cm) | 41 | 0.51 (0.23 to 0.71) | 0.0007 | NA | NA |
| Sum ERG+ tumor dimension (cm) | 41 | NA | NA | 0.51 (0.23 to 0.71) | 0.0007 |
| Tumor foci (n) | 41 | 0.38 (0.07 to 0.62) | 0.02 | 0.50 (0.22 to 0.71) | 0.0008 |
| Bx maximum core + (%) | 41 | 0.33 (0.01 to 0.58) | 0.04 | 0.43 (0.13 to 0.65) | 0.005 |
| Index tumor dimension (cm) | 41 | 0.32 (0.00 to 0.58) | 0.04 | 0.53 (0.26 to 0.72) | 0.0004 |
| Bx cores + (%) | 41 | 0.29 (−0.03 to 0.55) | 0.07 | 0.45 (0.15 to 0.67) | 0.004 |
| PSAD (ng/ml / g) | 40 | 0.28 (−0.05 to 0.55) | 0.09 | 0.41 (0.10 to 0.64) | 0.009 |
| Prostate Weight (g) | 40 | −0.17 (−0.47 to 0.15) | 0.28 | 0.01 (−0.31 to 0.33) | 0.95 |
| PCA3 score | 41 | 0.14 (−0.19 to 0.44) | 0.39 | 0.26 (−0.06 to 0.53) | 0.10 |
| PSA (ng/ml) | 41 | 0.13 (−0.19 to 0.43) | 0.41 | 0.30 (−0.02 to 0.56) | 0.06 |
| Age (yr) | 41 | 0.07 (−0.25 to 0.38) | 0.65 | 0.13 (−0.19 to 0.43) | 0.40 |
Number (n) of patients with data and Spearman’s rho (rs) (95% CI) and p value for correlation with sum of ERG+ tumor dimension and total tumor dimension are shown.
Serum PSA/prostatectomy weight.
Urine PCA3 score was most correlated with the total number of tumor foci and summed total linear tumor dimension (rs=0.34, p=0.03 and rs=0.26, p=0.10), however these correlations were substantially weaker than the correlation between urine T2:ERG score and the number of ERG+ tumor foci or summed ERG+ linear tumor dimension (Table 3). There was no significant correlation between urine PCA3 score and urine T2:ERG score or other clinical parameters (Table 3). Likewise, as shown in Table 4, summed total linear tumor dimension was significantly correlated with a number of indicators of total tumor burden (as well as ERG+ tumor burden) at biopsy and prostatectomy, however, there was no significant correlation with urine PCA3 or T2:ERG scores, which showed nearly equal correlations (0.26 and 0.24, respectively). Lastly, urine PCA3 score was not significantly associated with ERG+ vs. ERG− index focus status (median 42 vs. 039, Wilcoxon rank-sum test, p=0.78) or index Gleason score >6 vs. 6 (median 37 vs. 41, p=0.74).
Linear regression analysis also supported the above associations of urine T2:ERG and PCA3 with ERG+ and total tumor burden. T2:ERG was not significantly associated with summed total linear tumor dimension (R2=0.07, p=0.10), while PCA3 showed a statistically significant correlation with summed total linear tumor dimension, however it explained little of the variation in total tumor dimension (R2=0.15, p=0.01). T2:ERG showed a strong correlation with summed ERG+ linear tumor dimension (R2=0.43, p<0.0001), while PCA3 was not significantly associated with summed ERG+ linear tumor dimension (R2=0.08, p=0.08). Lastly, there was no significant association between T2:ERG and PCA3 scores (R2=0.06, p=0.12).
Discussion
In this study, through assessing both urine and prostatectomy tissues, we have demonstrated a strong correlation between urine T2:ERG score and total ERG+ tumor burden, supporting the very high cancer specificity of this biomarker in urine and tissue. Recurrent T2:ERG fusions, which occur in approximately 50% of PSA-screened PCa, result in over-expression of a truncated ERG protein 4,11,12,16,33. This rearrangement can be confidently detected at the chromosomal level by FISH studies, as has been demonstrated in numerous studies (reviewed in 2,4,6). We and others have more recently evaluated ERG protein expression in prostatectomy specimens using a monoclonal antibody against ERG (EPR3864) and documented excellent concordance of ERG staining by IHC compared with FISH for ERG rearrangements in >1,000 tumors3,12–16,33,36,37. Similar concordance between another monoclonal antibody against ERG and FISH for ERG rearrangement has been reported recently3,11,16. Likewise, we have previously confirmed the high concordance (92%) between the TMA T2:ERG assay and the presence of ERG rearrangements by FISH in prostate needle biopsy cores17. Importantly, by IHC, ERG expression is extremely rare in benign prostatic acini11,12,33 and is nearly 100% specific for prostate cancer or HGPIN immediately adjacent to prostate cancer in tissue studies3,11–16,33,36,37, which we confirmed here (>99.99% specificity for cancer). Although, the protein product of T2:ERG fusion cannot be detected in serum, our group has recently evaluated a clinical grade, urine based TMA assay for quantifying T2:ERG fusion mRNA17. This assay is based on the same technology as the FDA approved urine-based PROGENSA PCA3 assay which in conjunction with serum PSA has proven to be useful for prostate cancer detection and has been shown to be correlated with features of clinically significant disease18,19,25–29,35,38–41.
We have previously shown that urine T2:ERG and PCA3 scores show moderate, but significant correlation with the greatest linear dimension of the index focus at prostatectomy (n=187, rs=0.26 and rs=0.30, both p<0.001)17. However, PCa is frequently multifocal and shows multiple separate tumor foci in addition to the index tumor31. Heterogeneity amongst the multifocal tumor foci with respect to both histology and Gleason grade has been well described42,43. Similar to previous results42, we found that the majority of our cases (37/41, 90%) had multifocal tumor foci, with 18/37 (49%) cases with multifocal tumors demonstrating heterogeneity in Gleason scores between index and multifocal tumor foci. Recently, multiple groups have confirmed the heterogeneity of ETS gene fusions status (as indicated by FISH for TMPRSS2 or ERG rearrangements) between tumor foci in multifocal PCa44–46. For example, our group analyzed 93 multifocal PCa foci from 43 radical prostatectomy specimens and found that 70% of cases harbor TMPRSS2 rearrangement, of which 70% were discordant in at least one tumor focus, consistent with multifocal PCa arising from multiple, independent clonal expansions46.
Although we have previously shown significant correlation between urine T2:ERG and the greatest linear dimension of the index tumor at prostatectomy (rs=0.26)17, we hypothesized that measuring all multifocal tumor foci and stratifying ERG+ and ERG− tumor foci would demonstrate more significant correlation between urine T2:ERG score and ERG+ tumor burden. Importantly, our study confirms a strong correlation between urine T2:ERG and the summed total dimension and number of ERG+ tumor foci (rs=0.68 and 0.67). In our present study, there was no significant correlation between urine T2:ERG score and greatest dimension of the index tumor focus (rs=0.21, p=0.19) or summed total linear tumor dimension (rs=0.24, p=0.13), however the correlation coefficients are similar to those seen in our previous study regarding index tumor size17, suggesting that the smaller size of our current cohort may explain the lack of statistical significance. Linear regression analysis also demonstrated similar findings. Importantly, we recently evaluated ERG protein expression in a full spectrum of lesions encountered in routine diagnostic prostate needle biopsies, including diagnostically challenging biopsies, and showed ERG positivity in 44% of PCa and 18% of HGPIN; ERG expression was not observed in benign mimics of cancer such as adenosis and partial atrophy and was also exceedingly rare in benign glands33. These results are consistent with those observed in our current prostatectomy cohort and other studies showing that nearly the entire burden of ERG+ prostate tissue (as a surrogate for ERG rearrangements and TMPRSS2:ERG transcript) is carcinoma or HGPIN adjacent to carcinoma3,11–16,33,36,37. Thus, the total amount of ERG+ protein in a given prostate is nearly entirely ERG+ cancer, which our study demonstrates is strongly correlated with the urine T2:ERG score. Thus, while a limitation of T2:ERG as a biomarker is that it is not expressed in all tumor foci, it is extremely specific for prostate cancer, and there is no known mechanism for markedly elevated T2:ERG in the urine other than prostate cancer.
Interestingly, the correlation between urine PCA3 score and the summed total linear tumor dimension (rs=0.26, p=0.10) is weaker than the correlation between urine T2:ERG score and summed linear ERG+ tumor dimension (rs=0.68, p<0.0001). Although the small size of our current cohort may explain the lack of statistical significance between urine PCA3 score and summed total linear tumor dimension, we observed a similar correlation between urine PCA3 score and the largest dimension of the index tumor at prostatectomy in our previous study (rs=0.30, p<0.001)17, and our current results are consistent with previously published correlations of urine PCA3 scores and total tumor volume at prostatectomy (r and rs= 0.27–0.41)24–26,28. Linear regression analysis also identified a significant correlation between PCA3 and summed total linear tumor dimension, however PCA3 scores accounted for a limited amount of the variation in total tumor dimension (~15%).
PCA3 has been shown to be markedly over-expressed in >95% of prostate cancers and gene expression studies support prostate specificity, suggesting that the lower correlation between urine PCA3 and total cancer burden compared to urine T2:ERG and ERG+ cancer burden is not due to lack of (or variation in) PCA3 expression in some prostate cancer foci. Importantly, given that PCA3 is a non-coding transcript that does not produce a protein product that can be detected by an antibody, there is a lack of studies that have systematically evaluated the specificity of PCA3 across precursor lesions and benign mimics of prostate cancer at the tissue level, unlike ERG. Only one reported study has evaluated PCA3 expression in situ in prostatic tissues. Evaluating 24 and 26 cases by chromogenic and radioactive in situ hybridization detection methods, respectively, Popa et al. showed that while the majority of PCa (92–96%) showed at least focal cytoplasmic PCA3 expression, the majority of HGPIN (71–96%) as well as a subset of benign glands (29–33%) also showed PCA3 expression30. Hence, in a single study, PCA3 tissue expression appears similar to that of AMACR, a sensitive and specific prostate cancer marker with utility in tissue based diagnosis, but also with expression in the majority of HGPIN foci47–53. Additional studies will be needed to determine if PCA3, like AMACR, is expressed in a subset of benign mimickers of prostate cancer as well48,54,55.
The expression of PCA3 in the majority of HGPIN lesions (based on a single tissue based study) may contribute to the lower correlation of urine PCA3 scores and total tumor burden compared to urine T2:ERG scores and ERG+ cancer burden. A number of studies have correlated urine PCA3 score with the presence of HGPIN at biopsy, with conflicting results. For example, Deras et al. found no difference in PCA3 score for HGPIN vs. no evidence of abnormal pathology18, while Haese et al. found increased PCA3 scores in men with HGPIN56; these two studies yielded equivalent diagnostic accuracy for biopsy-detectable cancer. However, these studies were based on HGPIN identified at biopsy, and did not assess the entire prostatic HGPIN burden. Nevertheless, urine PCA3 has clear utility in predicting the presence of prostate cancer at biopsy and is significantly associated with indicators of aggressive disease at prostatectomy, and multiplexing urine PCA3 and T2:ERG will likely allow for more complete assessment of prostate cancer risk and evaluation of prostate cancer burden17,24–26,28,29,35,39–41,57,58.
The current study has some limitations. As our series is rather small, does not include the full spectrum of pathology (i.e. Gleason scores and stage) seen at prostatectomy and lacks long term follow up, associations with outcome measures are limited and will require additional studies. Additionally, although urine was prospectively collected prior to biopsy, our study does not represent a prospectively defined prostatectomy cohort. Lastly, although ERG expression by IHC has been highly correlated to ERG rearrangement by FISH, and TMPRSS2 is the 5′ partner in the vast majority of ERG rearranged prostate cancers, other 5′ partners can pair with ERG, such as NDRG159, which would not be detected by the urine T2:ERG assay but would result in ERG protein expression. However, a strength of our study, was the ability to directly compare urine T2:ERG and PCA3 scores to tissue based ERG+ and total cancer burden, and correlations observed between urine T2:ERG scores and index tumor dimension and PCA3 and index dimension and total tumor volume are consistent with previous reports17,25,26,28. Although the clearly stronger correlation of urine T2:ERG with total ERG+ cancer burden supports the very high specificity of ERG (and urine T2:ERG) for T2:ERG positive prostate cancer, our findings will need to be validated in larger series.
In summary, by comparing urine T2:ERG and PCA3 scores to ERG+ and total cancer burden at prostatectomy, our results confirm the extraordinary specificity of prostatic tissue ERG expression for prostate cancer (>99.99%) and demonstrate strong concordance of total ERG+ prostate cancer burden with urine T2:ERG score. This strong correlation supports the potential utility of T2:ERG in a variety of clinical situations which can now be prospectively addressed, including risk stratifying men with elevated serum PSA, those with prior negative biopsy and those considering active surveillance (as high urine T2:ERG scores is strongly associated with a large volume of ERG+ prostate cancer). Urine T2:ERG score may also have utility in predicting upgrading on prostatectomy, as high urine T2:ERG score but low tumor volume on biopsy may indicate undetected ERG+ cancer.
Acknowledgments
The authors thank Gary Pestano (Ventana Medical Systems) for providing the ERG antibody and IHC reagents, Jack Groskopf (Gen-Probe, Inc.) for providing T2:ERG and PCA3 assays, and Angela Fullen and Amy Gursky for technical assistance. Supported in part by the Early Detection Research Network (U01 CA111275 and U01 CA113913) and NIH S.P.O.R.E. (P50 CA69568). A.M.C. is supported by the Prostate Cancer Foundation and the Doris Duke Foundation, and is an American Cancer Society Clinical Research Professor and an A. Alfred Taubman Scholar.
Funding:
Supported in part by the Early Detection Research Network (U01 CA111275 and U01 CA113913) and NIH S.P.O.R.E. (P50 CA69568).
Footnotes
Conflict of interest:
The University of Michigan has been issued a patent on the detection of ETS gene fusions in prostate cancer, on which A.M.C. and S.A.T. are listed as co-inventors. The University of Michigan licensed the diagnostic field of use to Gen-Probe, Inc, which sublicensed rights to Ventana Medical Systems, Inc. Neither company played a role in data collection, interpretation or analysis, and did not participate in the study design or the decision to submit for publication. N.P. has served as a consultant for Ventana Medical Systems. A.M.C. has served as consultant to Gen-Probe, Inc. and Ventana Medical Systems. S.A.T. has received honoraria from Ventana Medical Systems.
References
- 1.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosen P, Sesterhenn IA, Brassell SA, et al. Clinical potential of the ERG oncoprotein in prostate cancer. Nat Rev Urol. 2012 doi: 10.1038/nrurol.2012.10. [DOI] [PubMed] [Google Scholar]
- 4.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomlins SA, Mehra R, Rhodes DR, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 6.Tomlins SA, Bjartell A, Chinnaiyan AM, et al. ETS Gene Fusions in Prostate Cancer: From Discovery to Daily Clinical Practice. Eur Urol. 2009;56:275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 7.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han B, Mehra R, Lonigro RJ, et al. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–1093. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosquera JM, Perner S, Genega EM, et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008;14:3380–3385. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perner S, Mosquera JM, Demichelis F, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 11.Furusato B, Tan SH, Young D, et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13:228–237. doi: 10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park K, Tomlins SA, Mudaliar KM, et al. Antibody-Based Detection of ERG Rearrangement-Positive Prostate Cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He H, Magi-Galluzzi C, Li J, et al. The diagnostic utility of novel immunohistochemical marker ERG in the workup of prostate biopsies with “atypical glands suspicious for cancer”. Am J Surg Pathol. 2011;35:608–614. doi: 10.1097/PAS.0b013e31820bcd2d. [DOI] [PubMed] [Google Scholar]
- 14.van Leenders GJ, Boormans JL, Vissers CJ, et al. Antibody EPR3864 is specific for ERG genomic fusions in prostate cancer: implications for pathological practice. Mod Pathol. 2011;24:1128–1138. doi: 10.1038/modpathol.2011.65. [DOI] [PubMed] [Google Scholar]
- 15.Yaskiv O, Zhang X, Simmerman K, et al. The utility of ERG/P63 double immunohistochemical staining in the diagnosis of limited cancer in prostate needle biopsies. Am J Surg Pathol. 2011;35:1062–1068. doi: 10.1097/PAS.0b013e318215cc03. [DOI] [PubMed] [Google Scholar]
- 16.Braun M, Goltz D, Shaikhibrahim Z, et al. ERG protein expression and genomic rearrangement status in primary and metastatic prostate cancer-a comparative study of two monoclonal antibodies. Prostate Cancer Prostatic Dis. 2012 doi: 10.1038/pcan.2011.67. [DOI] [PubMed] [Google Scholar]
- 17.Tomlins SA, Aubin SM, Siddiqui J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deras IL, Aubin SM, Blase A, et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. J Urol. 2008;179:1587–1592. doi: 10.1016/j.juro.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 19.Groskopf J, Aubin SM, Deras IL, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52:1089–1095. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 20.Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 21.de Kok JB, Verhaegh GW, Roelofs RW, et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–2698. [PubMed] [Google Scholar]
- 22.Hessels D, Klein Gunnewiek JM, van Oort I, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8–15. doi: 10.1016/s0302-2838(03)00201-x. discussion 15–16. [DOI] [PubMed] [Google Scholar]
- 23.Auprich M, Chun FK, Ward JF, et al. Critical assessment of preoperative urinary prostate cancer antigen 3 on the accuracy of prostate cancer staging. Eur Urol. 2011;59:96–105. doi: 10.1016/j.eururo.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 24.Ploussard G, Durand X, Xylinas E, et al. Prostate cancer antigen 3 score accurately predicts tumour volume and might help in selecting prostate cancer patients for active surveillance. Eur Urol. 2011;59:422–429. doi: 10.1016/j.eururo.2010.11.044. [DOI] [PubMed] [Google Scholar]
- 25.Durand X, Xylinas E, Radulescu C, et al. The value of urinary prostate cancer gene 3 (PCA3) scores in predicting pathological features at radical prostatectomy. BJU Int. 2012 doi: 10.1111/j.1464-410X.2011.10682.x. [DOI] [PubMed] [Google Scholar]
- 26.Nakanishi H, Groskopf J, Fritsche HA, et al. PCA3 molecular urine assay correlates with prostate cancer tumor volume: implication in selecting candidates for active surveillance. J Urol. 2008;179:1804–1809. doi: 10.1016/j.juro.2008.01.013. discussion 1809–1810. [DOI] [PubMed] [Google Scholar]
- 27.van Poppel H, Haese A, Graefen M, et al. The relationship between Prostate CAncer gene 3 (PCA3) and prostate cancer significance. BJU Int. 2012;109:360–366. doi: 10.1111/j.1464-410X.2011.10377.x. [DOI] [PubMed] [Google Scholar]
- 28.Whitman EJ, Groskopf J, Ali A, et al. PCA3 score before radical prostatectomy predicts extracapsular extension and tumor volume. J Urol. 2008;180:1975–1978. doi: 10.1016/j.juro.2008.07.060. discussion 1978–1979. [DOI] [PubMed] [Google Scholar]
- 29.Vlaeminck-Guillem V, Devonec M, Colombel M, et al. Urinary PCA3 score predicts prostate cancer multifocality. J Urol. 2011;185:1234–1239. doi: 10.1016/j.juro.2010.11.072. [DOI] [PubMed] [Google Scholar]
- 30.Popa I, Fradet Y, Beaudry G, et al. Identification of PCA3 (DD3) in prostatic carcinoma by in situ hybridization. Mod Pathol. 2007;20:1121–1127. doi: 10.1038/modpathol.3800963. [DOI] [PubMed] [Google Scholar]
- 31.Wise AM, Stamey TA, McNeal JE, et al. Morphologic and clinical significance of multifocal prostate cancers in radical prostatectomy specimens. Urology. 2002;60:264–269. doi: 10.1016/s0090-4295(02)01728-4. [DOI] [PubMed] [Google Scholar]
- 32.Siu W, Dunn RL, Shah RB, et al. Use of extended pattern technique for initial prostate biopsy. J Urol. 2005;174:505–509. doi: 10.1097/01.ju.0000165385.53652.7a. [DOI] [PubMed] [Google Scholar]
- 33.Tomlins SA, Palanisamy N, Siddiqui J, et al. Antibody Based Detection of ERG Rearrangements in Prostate Core Biopsies, Including Diagnostically Challenging Cases. Arch Pathol Lab Med. doi: 10.5858/arpa.2011-0424-OA. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furusato B, Tan SH, Young D, et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010 doi: 10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de la Taille A, Irani J, Graefen M, et al. Clinical evaluation of the PCA3 assay in guiding initial biopsy decisions. J Urol. 2011;185:2119–2125. doi: 10.1016/j.juro.2011.01.075. [DOI] [PubMed] [Google Scholar]
- 36.Hoogland AM, Jenster G, van Weerden WM, et al. ERG immunohistochemistry is not predictive for PSA recurrence, local recurrence or overall survival after radical prostatectomy for prostate cancer. Mod Pathol. 2011 doi: 10.1038/modpathol.2011.176. [DOI] [PubMed] [Google Scholar]
- 37.Falzarano SM, Zhou M, Carver P, et al. ERG gene rearrangement status in prostate cancer detected by immunohistochemistry. Virchows Arch. 2011;459:441–447. doi: 10.1007/s00428-011-1128-4. [DOI] [PubMed] [Google Scholar]
- 38.Wu AK, Reese AC, Cooperberg MR, et al. Utility of PCA3 in patients undergoing repeat biopsy for prostate cancer. Prostate Cancer Prostatic Dis. 2012;15:100–105. doi: 10.1038/pcan.2011.52. [DOI] [PubMed] [Google Scholar]
- 39.Day JR, Jost M, Reynolds MA, et al. PCA3: from basic molecular science to the clinical lab. Cancer Lett. 2011;301:1–6. doi: 10.1016/j.canlet.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 40.Roobol MJ, Schroder FH, van Leeuwen P, et al. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. Eur Urol. 2010;58:475–481. doi: 10.1016/j.eururo.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 41.Aubin SM, Reid J, Sarno MJ, et al. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: validation in the placebo arm of the dutasteride REDUCE trial. J Urol. 2010;184:1947–1952. doi: 10.1016/j.juro.2010.06.098. [DOI] [PubMed] [Google Scholar]
- 42.Arora R, Koch MO, Eble JN, et al. Heterogeneity of Gleason grade in multifocal adenocarcinoma of the prostate. Cancer. 2004;100:2362–2366. doi: 10.1002/cncr.20243. [DOI] [PubMed] [Google Scholar]
- 43.Ruijter ET, Miller GJ, van de Kaa CA, et al. Molecular analysis of multifocal prostate cancer lesions. J Pathol. 1999;188:271–277. doi: 10.1002/(SICI)1096-9896(199907)188:3<271::AID-PATH359>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 44.Barry M, Perner S, Demichelis F, et al. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology. 2007;70:630–633. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furusato B, Gao CL, Ravindranath L, et al. Mapping of TMPRSS2-ERG fusions in the context of multi-focal prostate cancer. Mod Pathol. 2008;21:67–75. doi: 10.1038/modpathol.3800981. [DOI] [PubMed] [Google Scholar]
- 46.Mehra R, Han B, Tomlins SA, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 47.Beach R, Gown AM, De Peralta-Venturina MN, et al. P504S immunohistochemical detection in 405 prostatic specimens including 376 18-gauge needle biopsies. Am J Surg Pathol. 2002;26:1588–1596. doi: 10.1097/00000478-200212000-00006. [DOI] [PubMed] [Google Scholar]
- 48.Epstein JIHerawi M. Prostate needle biopsies containing prostatic intraepithelial neoplasia or atypical foci suspicious for carcinoma: implications for patient care. J Urol. 2006;175:820–834. doi: 10.1016/S0022-5347(05)00337-X. [DOI] [PubMed] [Google Scholar]
- 49.Jiang Z, Woda BA, Rock KL, et al. P504S: a new molecular marker for the detection of prostate carcinoma. Am J Surg Pathol. 2001;25:1397–1404. doi: 10.1097/00000478-200111000-00007. [DOI] [PubMed] [Google Scholar]
- 50.Jiang Z, Wu CL, Woda BA, et al. P504S/alpha-methylacyl-CoA racemase: a useful marker for diagnosis of small foci of prostatic carcinoma on needle biopsy. Am J Surg Pathol. 2002;26:1169–1174. doi: 10.1097/00000478-200209000-00007. [DOI] [PubMed] [Google Scholar]
- 51.Luo J, Zha S, Gage WR, et al. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- 52.Rubin MA, Zhou M, Dhanasekaran SM, et al. alpha-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. Jama. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 53.Wu CL, Yang XJ, Tretiakova M, et al. Analysis of alpha-methylacyl-CoA racemase (P504S) expression in high-grade prostatic intraepithelial neoplasia. Hum Pathol. 2004;35:1008–1013. doi: 10.1016/j.humpath.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 54.Przybycin CG, Kunju LP, Wu AJ, et al. Partial atrophy in prostate needle biopsies: a detailed analysis of its morphology, immunophenotype, and cellular kinetics. Am J Surg Pathol. 2008;32:58–64. doi: 10.1097/PAS.0b013e318093e3f6. [DOI] [PubMed] [Google Scholar]
- 55.Yang XJ, Wu CL, Woda BA, et al. Expression of alpha-Methylacyl-CoA racemase (P504S) in atypical adenomatous hyperplasia of the prostate. Am J Surg Pathol. 2002;26:921–925. doi: 10.1097/00000478-200207000-00011. [DOI] [PubMed] [Google Scholar]
- 56.Haese A, de la Taille A, van Poppel H, et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol. 2008;54:1081–1088. doi: 10.1016/j.eururo.2008.06.071. [DOI] [PubMed] [Google Scholar]
- 57.Ploussard G, de la Taille A. Urine biomarkers in prostate cancer. Nat Rev Urol. 2010;7:101–109. doi: 10.1038/nrurol.2009.261. [DOI] [PubMed] [Google Scholar]
- 58.Aubin SM, Reid J, Sarno MJ, et al. Prostate cancer gene 3 score predicts prostate biopsy outcome in men receiving dutasteride for prevention of prostate cancer: results from the REDUCE trial. Urology. 2011;78:380–385. doi: 10.1016/j.urology.2011.03.033. [DOI] [PubMed] [Google Scholar]
- 59.Pflueger D, Rickman DS, Sboner A, et al. N-myc downstream regulated gene 1 (NDRG1) is fused to ERG in prostate cancer. Neoplasia. 2009;11:804–811. doi: 10.1593/neo.09572. [DOI] [PMC free article] [PubMed] [Google Scholar]