Abstract
The characterization of post-transcriptional gene regulation by small regulatory RNAs of 20–30 nt length, particularly miRNAs and piRNAs, has become a major focus of research in recent years. A prerequisite for the characterization of small RNAs is their identification and quantification across different developmental stages, normal and diseased tissues, as well as model cell lines. Here we present a step-by-step protocol for the bioinformatic analysis of barcoded cDNA libraries for small RNA profiling generated by Illumina sequencing, thereby facilitating miRNA and other small RNA profiling of large sample collections.
Keywords: Bioinformatic analysis, Small RNA, miRNA, Barcoding, Next-generation sequencing, Nucleotide variation
1. Introduction
MicroRNAs (miRNAs) are short 20–23 nucleotide (nt) RNAs that guide sequence-specific post-transcriptional gene regulation in animals and plants. They regulate many critical biological functions including organismal development, normal physiology and tumorigenesis. Thus, miRNA profiling analysis can allow us to gain insights into disease states by characterizing collections of clinical diseased and normal samples.
To facilitate the understanding of miRNA profiling analysis it is helpful to review miRNA biogenesis and structure (reviewed in [1]). Mature miRNAs are excised in a multi-step process from primary transcripts (pri-miRNAs) that contain one or more ~70 nt hairpin miRNA precursors (pre-miRNAs) and have their own promoters or share the promoter with a protein-coding host gene. These hairpin structures are recognized in the nucleus by DGCR8, a double-stranded RNA-binding protein (dsRBP) and RNASEN, also known as RNase III Drosha, and excised to yield pre-miRNAs. These molecules are subsequently transported by XPO5 (exportin 5) to the cytoplasm, where they are further processed by RNase III DI-CER1 (Dicer) in complex with the dsRBPs TARBP2 (TRBP) and/or PRKRA to yield a processing intermediate, composed of a mature miRNA and its complementary miRNA/strand. Some miRNAs bypass the general miRNA processing order and their maturation can be independent of DGCR8 and RNASEN, or are DICER1-independent. DGCR8- and RNASEN-independent miRNAs include mirtrons and tailed mirtrons, which release their pre-miRNA by splicing and exonuclease trimming [2,3].
In the accompanying manuscript we summarize the experimental methodologies for barcoded profiling of small RNAs by next-generation sequencing [4]. We use barcodes to mark individual samples that are processed in sets of up to 20 samples simultaneously. We describe the bioinformatic analysis to reassign the sequence reads from sequenced pools to the individual barcoded samples (referred to as subsamples), analyze their sequence content and compare profiles of subsamples within the same as well as different sequencing runs. The method we describe herein has been used to profile miRNAs in large sample collections in breast cancer [5], liposarcoma [6] and angiosarcoma [7].
miRNA profiling by next generation sequencing not only enables studies of differential expression, but also facilitates determination of nucleotide variation (including RNA editing, 3′ and 5′ modifications) and identification of novel miRNAs. Moreover, sequence read counts, also known as read frequencies, represent a direct measure of global miRNA abundance in any given sample when normalized to reference standards (calibrator oligoribonucleotides) added to each sample in a known amount during small RNA cDNA library preparation. Finally, we address method-specific biases to further clarify miRNA abundance. We recently determined miRNA and calibrator RNA sequence-specific ligation biases by quantifying 770 synthetic miRNAs and 50 calibrator RNAs using the same barcoded adapters and procedure [8]; this provides correction factors for affected miRNAs.
2. Overview of the method
Next generation sequencing outputs are text files which report sequence and a quality score for each sequenced base. These files are processed to (1) trim the 3′ barcoded adapter sequence from each read and assign the read to a specific subsample according to the barcode, (2) generate files with unique (non-redundant) reads for each subsample listing the times each unique read is encountered, (3) remove low complexity sequences and adapter-adapter ligation products, (4) map the unique reads to the genome, and (5) annotate the reads with a specific hierarchy of small RNA annotation databases.
The result is a profile of read frequencies for each miRNA that can be converted to relative read frequencies (normalized against the total miRNA sequence reads for each subsample) or to absolute amounts of input miRNA (by comparing with calibrator oligoribo-nucleotide reads). These miRNA profiles can be grouped into sequence families and genomic clusters and further studied by clustering and comparative expression analysis. miRNA sequence families group miRNAs that display sequence similarity and thus likely target a similar set of mRNAs, while miRNA genomic clusters group miRNAs that are located in close proximity in the genome, and are co-transcribed.
We then describe a curation strategy for miRNAs, an essential step in identifying miRNA candidates for downstream biological assays, by confirming their expression and likelihood of forming prototypical miRNA hairpin structures. We finally discuss our approach for identification of RNA nucleotide variations, and point to approaches that can be used for identification of novel miRNAs.
Sequencing-based miRNA profiles can be deposited at Sequence Read Archive (SRA) (www.ncbi.nlm.nih.gov/sra) both as barcode extracted files for each individual subsample (including sequence reads, read frequencies, and assigned annotation), as well as files including the residual sequence reads that were not uniquely assigned to a barcode for each sequencing run.
3. Materials
A computer workstation or a desktop computer is required to analyze raw small RNA deep sequencing data. Historically, workstations offered higher performance than desktop computers. However today’s desktop computers can be expected to perform as workstations, using widely available operating systems and hardware components. A workstation-type computer may have the following characteristics: memory with error correcting code (ECC) support (a type of data storage that corrects for common kinds of internal data corruption), a larger number of memory sockets using registered modules, multiple processor sockets, powerful CPUs, and a reliable operating system (e.g. Unix-based). In Table 1, we outline the specifications of the computer workstation we currently use to run the small RNA annotation pipeline alongside the recommended minimum for each specification. Processing of a contemporary 20 subsample-containing barcoded library with a total number of sequence reads of ~160 million, required 2 h and 10 min for barcoded adapter trimming and assignment by barcode, and 2 h and 9 min for mapping of reads to the genome and small RNA annotation databases (for a ratio of redundant to unique reads of ~20:1). The required software are modified and compiled from freely available web tools, Perl scripts, and useful Bioconductor (http://bioconductor.org) packages, which are mentioned in the relevant sections of the manuscript.
Table 1.
Computer setup recommendations.
| Our current workstation | Recommendation | |
|---|---|---|
| CPU cores | 32 | ≥8 |
| CPU frequency | 2.4 GHz | Highest possiblea |
| Memory | 132 GB | ≥32 GB |
| Disk space | 1 TB Free space | ≥1 TB Free spaceb |
| Backup | RAID 1 (hardware-based) | RAID 1 or proper external backupc |
| Operating system |
Linux | Linux |
| Support | IT and system administrator |
IT support for installation |
A measure of CPU performance that can be used to compare CPUs within a given family.
Intermediate alignment files typically occupy 3 times the space of the source (uncompressed) raw read fasta or fastq file.
RAID (Redundant Array of Independent Disks) is a storage system that combines multiple disk drives. Data is distributed across the drives in one of several ‘RAID levels’, determined by the required redundancy and performance.
4. Procedure
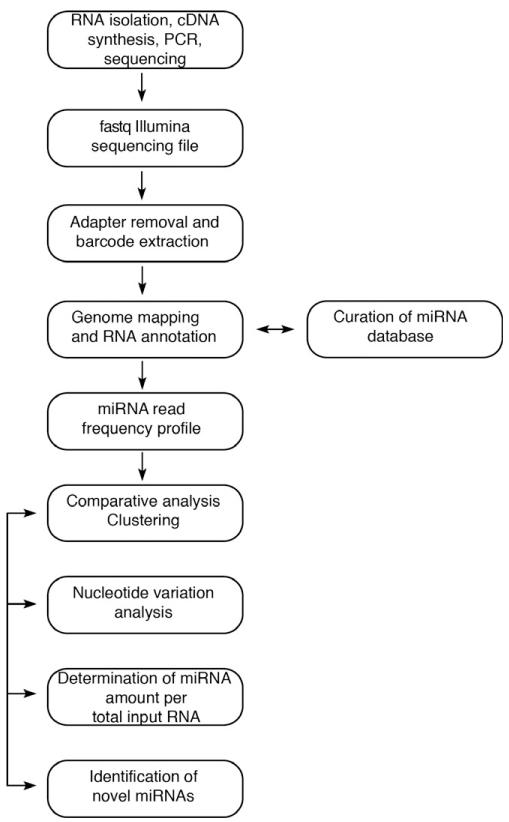
The following step-by-step procedure (Fig. 1) describes the processing of the raw deep sequencing files generated on Illumina HiSeq or Illumina Genome Analyzer II sequencers to obtain miRNA read frequencies. Following image processing, a single deep sequencing run produces a text file in fastq format, including a quality score for each nucleotide call. The size of the fastq files, when compressed, is currently in the multi-GB range; depending on the RNA isolation method and platform used for sequencing, a library can contain from 10 million to over 200 million reads. Based on the quality scores, an initial filtering step may take place (included in platform-specific software packages).
Fig. 1.
General overview of bioinformatic analysis pipeline.
4.1. Database of records containing sample description and experimental processing
A database that is easy to interrogate is important when working with large clinical sample collections. The database should include sample description, experimental processing steps, and sequencing run details.
1. Sample source information
At a minimum the overall database should include information on the sample tissue origin and species. The clinical characteristics of each subsample can be documented in a separate table, but using a common identifier that is also used to link the subsample to its barcode (see example in Table 2). Subsequent unsupervised clustering of subsamples is expected to reflect subsample characteristics and should not occur by barcode, sequencing run, or sample preparation technique. Using our barcoded adapters under unchanging ligation conditions, miRNA expression reflects the sample composition independent of barcode and sequencing run.
Table 2.
Detailed clinical, histologic and pathologic characterization of each specimen listed by unique tissue identifier from [5]. Immunohistochemistry (IHC) for Estrogen Receptor (ESR1/ER), Progesterone Receptor (PGR), Human Epidermal Growth Factor Receptor (HER2/ERBB2); Molecular subtypes, based on mRNA profiles; Clinical follow-up (Overall survival in years, Distant metastasis as first event, Time to follow-up (for patients without metastasis as first event) or Time to metastasis (for patients with metastasis as first event) in years); Tumor cells, percent tumor content in frozen sections used for miRNA determination analysis; NA = not available or not applicable.
| Tissue identifier |
ESR1/ER IHC (1 = positive, 0 = negative) |
PGR/PR IHC (1 = positive, 0 = negative) |
ERBB2/HER2 IHC (1 = positive, 0 = negative) |
Molecular subtypes |
Overall survival (years) |
Distant metastasis as first event (1 = yes, 0 = no) |
Time to follow-up or metastasis (years) |
Tumor cells (%) |
|---|---|---|---|---|---|---|---|---|
| 5 | 1 | 1 | 1 | LumB | NA | NA | NA | 65 |
| 9 | 0 | 0 | 1 | Normal | NA | NA | NA | 70 |
| 16 | 1 | 1 | 1 | LumA | NA | NA | NA | 70 |
| 18 | 0 | 0 | 1 | Her2 | NA | NA | NA | 60 |
| 31 | 1 | 1 | 0 | LumA | NA | NA | NA | 90 |
| 32 | 1 | 1 | 0 | LumA | NA | NA | NA | 90 |
| 36 | 1 | 1 | 0 | LumB | NA | NA | NA | 80 |
| 45 | 1 | 1 | 1 | Her2 | 7 | 0 | 7 | 80 |
| 52 | 0 | 0 | 1 | LumA | 0.8 | 1 | 0.7 | 80 |
| 70 | 1 | 0 | 1 | Her2 | 3.5 | 1 | 2.4 | 70 |
| 75 | 0 | 1 | 1 | Her2 | 12.4 | 0 | 12.4 | 70 |
| 78 | 0 | 0 | 1 | Basal | 8.1 | 0 | 8.1 | 80 |
| 85 | 0 | 0 | 1 | Her2 | 6.4 | 0 | 6.4 | 80 |
| 98 | NA | NA | NA | Normal | NA | NA | NA | NA |
| 103 | NA | NA | NA | Normal | NA | NA | NA | NA |
| 104 | NA | NA | NA | Normal | NA | NA | NA | NA |
| 148 | 0 | 0 | 0 | Basal | 4.4 | 1 | 1.2 | 80 |
| 166 | 0 | 0 | 0 | Basal | NA | NA | NA | 80 |
| 167 | 0 | 0 | 0 | Basal | 6 | 1 | 4.9 | 80 |
| 175 | 0 | 0 | 0 | Basal | 4.4 | 0 | 4.4 | 80 |
2. Experimental procedure information
It is important to document experimental procedures for RNA isolation and generation of each cDNA library. This should include (1) the type of RNA isolation method used, given that different methods vary in small RNA extraction efficiency; (2) the amount of total RNA used for each sample to allow quantitation of the global miRNA amount; (3) the adapters used for the 3′ and 5′ ligation steps; (4) the RNA ligases used for the 3′ and 5′ ligation reactions, which may introduce different biases for each miRNA; (5) the ligation conditions; (6) the presence, type and length of oligoribonucleotide size markers used; (7) the specifics of cDNA library preparation: the expected size range of the library insert, the number of amplification steps (PCR cycles), the extent of adapter–adapter ligation product removal, as judged by agarose gel or bioanalyzer; (8) the calibrator oligoribonucleotide cocktail composition and amount used.
3. Sequencing run information
The database should include information regarding the sequencing run, including the number and description of subsamples included within each sequencing run, the barcode used for each subsample, the sequencing platform used and the unique identifier of the sequencing run (see example in Table 3).
Table 3.
Overview of sequencing run and barcode assignment from [5]. The samples listed as examples in Table 2 and subsequent Tables are included in sequencing run 313.
| Sequencing run |
Reads in sequencing run |
Reads with adapter within size limits |
Reads with barcodes |
Number of samples in sequencing run (samples used for analysis in specific study) |
Sequencing platform used |
|---|---|---|---|---|---|
| 313 | 13809568 | 11262276 | 11126105 | 20 (20) | Illumina GAII |
| 235 | 10834028 | 8679239 | 8584137 | 20 (9) | Illumina GAII |
| 531 | 26532163 | 19873107 | 17640943 | 20 (2) | Illumina GAII |
4.2. Generation of miRNA profiles
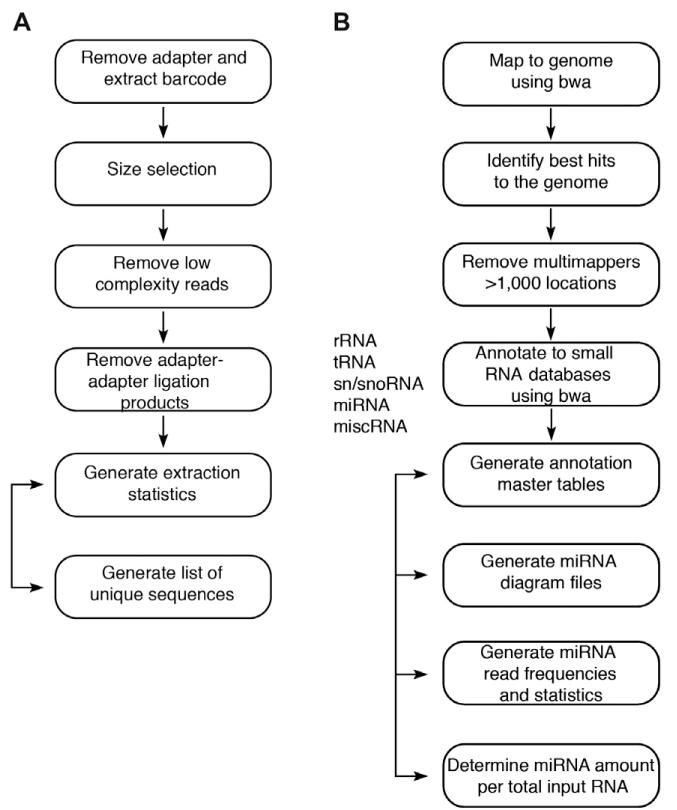
Steps 4–7 describe adapter trimming and assignment by barcode (Fig. 2A), while steps 8-11 describe mapping to the genome and small RNA annotation databases (Fig. 2B). We generated an automated pipeline where a user friendly interface allows information data entry on each sample, uploading of raw fastq or fasta tab-delimited sequencing files, barcoded adapter trimming and subsample extraction, mapping, annotation and selection of subsamples for clustering analysis.
Fig. 2.
Overview of generation of miRNA profiles. RNA isolation, cDNA synthesis and PCR as described in [4]. (A) Barcoded adapter trimming and barcode assignment. (B) Mapping to the genome and annotation to small RNA databases.
4. Barcoded adapter trimming and assignment by barcode
Sequencing is performed unidirectionally. The 5′ adapter sequence serves as primer binding region for sequencing and the first sequenced base corresponds to the first nucleotide of the RNA insert (Fig. 3). The first computational step is therefore to retrieve the sequence corresponding to the original small RNA from the sequence reads by removing the 3′ barcoded adapter sequences and assign the reads to subsamples according to their corresponding barcodes. We use a collection of Perl scripts, derived from Berninger et al. [9], which were further modified to produce files described in 6 and 7 and align the barcoded 3′ adapters to the reads. Alternatives, such as trimLRPatterns, BioBowl script, novoalign are suggested in [10], or AdaptorRemover suggested in [11]. To avoid barcode misassignment we do not allow a mismatch in the first common position of the 3′ adapter next to the barcode, nor do we allow any mismatch, insertion or deletion within the barcode (in other words, reads with imperfect barcodes are discarded). Our decision to not allow mismatches stems from the fact that in order to minimize ligation biases we kept the barcode sequences short (5 nt) and similar in sequence. We require overlap with at least the first four common post-barcode nt of the 3′ adapter, or a minimum of 5 nt if containing one mismatch. No insertions or deletions are allowed. According to these rules, the maximum insert length that can be extracted is N-9 (where N is the length of the sequencing read). For example, for platforms producing 36 nt reads that are barcoded the maximum insert length that can be recovered is 27 nt, whereas for platforms producing 50 nt reads the maximum insert length recovered is 41.
Fig. 3.
Example of initial processing of fastq deep sequencing file. The fastq Illumina file contains 4 lines for each sequence read: (1) a unique read identifier preceded by the ‘@’ symbol, (2) the nucleotide sequence of the read, (3) the same unique read identifier preceded by the ‘+’ symbol (4) quality scores that specify the probability of error in the nucleotide call at each position in the sequence read. Missing nucleotides are usually marked as N or the symbol ‘.’. The fastq file is converted to a fasta file and checked for presence of valid barcode and adapter. The subsample member is subsequently tracked so that the fasta files can be split into individual files by subsample (header contains read identification number and read count).
5. Apply filters on small RNA insert length, low-complexity sequences and adapter–adapter ligation products
We suggest to retain a minimum length of 16 and a maximum length of 25 nt of inserts for annotation, if the primary goal is characterizing miRNA profiles. Initially the Illumina sequence read length was 36, which allowed identification of miRNAs along with the barcode and part of the 3′ adapter sequence (see above). Currently, Illumina provides read lengths of 50 or 100 nt, which may allow full length identification of longer RNA species, such as piRNAs, if desired. To identify longer small RNA species the experimental procedures described in the accompanying manuscript [4] need to be modified by adjusting the size fractionation step during cDNA library preparation. Low complexity reads are defined as mono-, di- and tri-nucleotide repeats and are removed from analysis. Reaction by-products, (such as adapter–adapter ligation products) that are the same length as the desired products containing the size-selected insert RNA, are filtered out using the Needleman–Wunsch alignment algorithm. When these products are identified, these are added to a ‘by-product’ MySQL database table, which is updated in an iterative process to prevent such sequences from entering genomic mapping processes.
6. Generate a list of non-redundant (unique) sequences
The file is in fasta format, with each unique read containing a unique identifier along with the frequency of its occurrence in the header. At this step the quality scores from the fastq file are omitted. Reducing the data to non-redundant reads allows for faster mapping, especially important given the millions of reads generated from each sequencing run. Separate unique sequence files are generated for each subsample (Fig. 3).
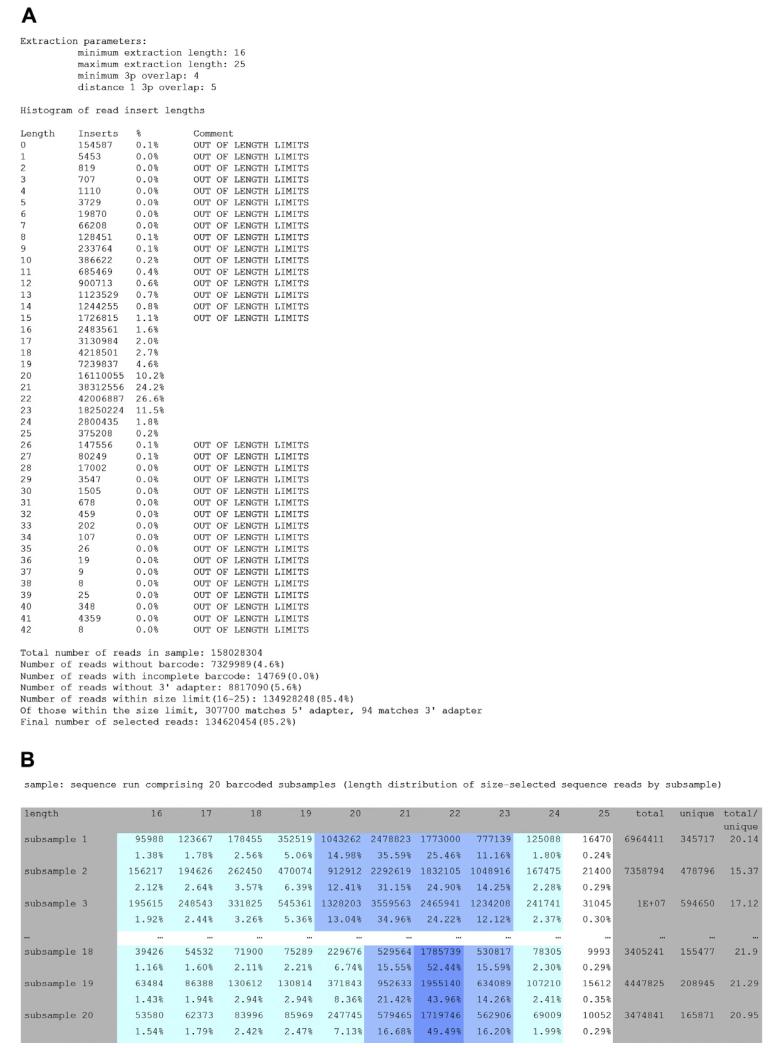
7. Generate barcoded adapter trimming and barcode allocation statistics reports
Steps 4–6 yield the following files, which provide information on the quality of the RNA isolation (e.g. majority of reads should coincide with experimental RNA size selection of 19–24 nt) and the quality of the sequencing (e.g. the majority of reads should include an identifiable barcode and adapter): (1) a barcoded adapter trimming report with a histogram of the length of the insert RNA within the reads in the sequencing run, and categories of reads that were filtered out due to length, absence of barcode, presence of incomplete barcode (defined by the presence of fewer than 4 nt of the adapter), or absence of adapter (Fig. 4A); (2) a histogram of the length of the insert RNA for each subsample (for the chosen length range, e.g. 16–25 nt) with columns summarizing the number of unique reads, as well as the ratio of total reads to unique reads, as a metric for the diversity of RNA species within each subsample and sequencing depth (Fig. 4B); (3) a master table with all unique inserts corresponding to each subsample after barcoded adapter extraction.
Fig. 4.
(A) Summary table representing histogram of small RNA insert length of all reads. (B) Summary table representing histogram of small RNA insert length of all extracted reads by subsample.
8. Map unique filtered sequence reads to the genome (e.g. hg19, mm9)
We use the Burrows–Wheeler aligner (bwa) [12] and suggest allowing up to one error (mismatch, insertion or deletion) while mapping to the genome. We use the bwa (version 0.5.9) default parameters with the exception of parameter n, maximum edit distance, set as 1 or 2 based on the analysis step, mapping to the genome or annotation to small RNA databases, respectively (further explained in later steps). To speed up the mapping process, we set a multi-threading parameter to allow bwa to use multiple cores. Other short read aligners may be used, such as maq, soap, eland, and Bowtie as suggested [10,13,14]. We originally used Oligomap and WU-BLAST, as described in [9]; however, given the increasing sequencing depth, now resulting in many millions of reads for each subsample, the amount of time for annotating each subsample required the use of alternative mapping algorithms.
9. Identify best hits and remove insert sequences that map to >1000 genomic locations
For each small RNA identify the locus/loci with minimum number of errors (mismatch, insertion, deletion) in the insert-to-genome mapping and select the locus with smaller number of errors. Set aside multimappers with hits to >1000 genomic locations. This number can be adjusted based on the small RNA studied, e.g. reduced for profiling miRNAs given that multicopy miRNAs only map to a small number of genomic locations.
10. Map unique filtered insert sequences to small RNA databases using a hierarchy list based on RNA species abundance within the cell
Map small RNAs to RNA annotation databases using bwa, allowing up to 2 errors. We download the small RNA category sequences of the species, from which the small RNAs have been sequenced, from the GenBank repository (www.ncbi.nlm.nih.gov/genbank), and also obtain the annotation of repeat elements in the genome (www.repeatmasker.org). These are the resources used in order of their hierarchy for annotation of unique sequence read inserts (in italics are those included in the summary statistics Table S2 from [5]):
-
-
rRNA – ribosomal RNAs and precursors
-
-
tRNA – transfer RNA
-
-
sn/snoRNA – snRNA (small nuclear RNA) and snoRNA (small nucleolar RNA)
-
-
repeat10_rm – reads that map to >10 locations in a repeat-masked genomic location (interspersed repeats within DNA sequences)
-
-
miRNA – miRBase (www.mirbase.org) or our curated annotation (see following section)
-
-
miscRNA – miscellenous RNA, assigned to any gene that encodes non-coding RNA not included in the other definitions (such as scRNAs – small cytoplasmic RNAs, scRNA-hY1 through 4)
-
-
piRNA – piwi-interacting RNAs
-
-
mis-annotated (doubtful) miRNAs (annotated as miRNAs in miRBase release 16 but did not pass our classification criteria; see following section)
-
-
calibrator oligoribonucleotides
-
-
RNA size marker sequences used during cDNA library preparation
-
-
repeat-masked reads that fall within regions of interest annotated above, including the following types of small RNA: rRNA, tRNA, sn/snoRNA, miscRNA
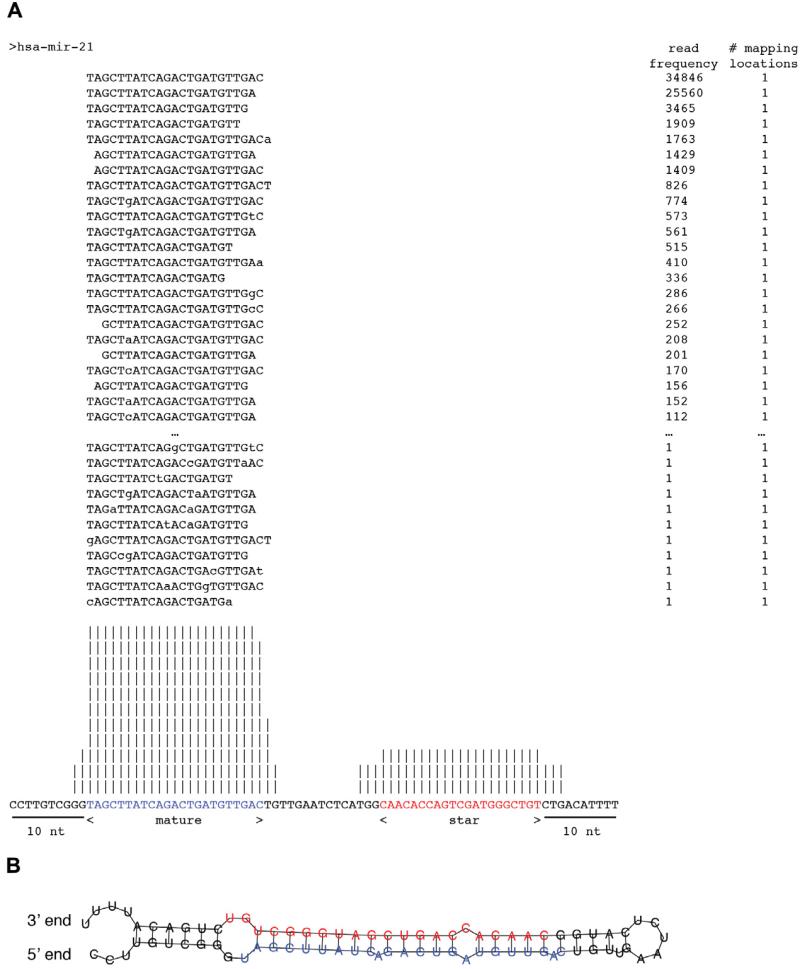
11. Steps 8 through 10 yield the following files, using a collection of Perl scripts, derived from Berninger et al. [9]: (1) annotation master table including the sequence ID, nucleotide sequence, sequence length, number of times a sequence is encountered, its genomic coordinates, the number of mappings to the genome, and the assigned small RNA category (see example in Table 4); (2) mapping statistics, including the number of sequences assigned to each annotation database with 0, 1, or 2 errors, respectively; the oligoribonucleotide calibrator sequences should be removed from these statistics to reflect the biological subsample composition. At the same time, for quality control, the table includes the number of sequences assigned to calibrator oligoribonucleotides with 0, 1, or 2 errors, respectively (see example in Table 5); (3) miRNA precursor read frequency profiles, including the number of unique reads and shared reads (important for assignment of multicopy miRNAs and miRNAs that share extensive sequence similarity) (see example in Table 6); (4) individual miRNA read frequency profiles assigned to their respective genomic location or merged to reflect their indistinguishable mature form (for multicopy miRNAs) with read frequencies (see example in Table 7); (5) diagram files with all the sequences that can be assigned to the miRNA precursor, and their overall distribution along the precursor, specifying the mature and miRNA/sequence (Fig. 5A). This provides insights into patterns related to biogenesis of the hairpin foldback structures of typical miRNAs.
Table 4.
Example of annotation master table. Headings are as follows: (A) Sequence ID; (B) Small RNA annotation category; (C) Length of sequence insert; (D) Sequence of small RNA insert; (E) Number of copies of each sequence; (F) Lowest mapping error to small RNA annotation databases; (G) Lowest mapping error to the genome; (H-M) Number of lowest error mapping to the small RNA annotation database (e.g. rRNA tRNA sn/snoRNA miRNA piRNA, spike) within each category, while in parenthesis the strand and minimum error distance found; (N) Number of lowest error mapping to the genome, while in parenthesis the mapping strand and lowest error distance found, shown for the selected small RNA category according to hierarchy; (O) Genomic coordinates, with only one location listed, even if more were identified.
| SeqID | Annotation | Sequence length |
Sequence | Copies | Annotation error |
Mapping error |
rRNA | tRNA | sn/sno- RNA |
miRNA | piRNA | Spike | Genome | Coordinates |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| seq337 | miRNA | 22 | TAGCT… | 88508 | 0 | 0 | None | None | None | 1(+0) | None | None | 1(+0) | chr17:57918634-57918655[+] |
| seq603 | miRNA | 23 | TAGCT… | 53346 | 0 | 0 | None | None | None | 1(+0) | None | None | 1(+0) | chr17:57918634-57918656[+] |
| seq587 | spike | 22 | CATCG… | 26240 | 0 | N/A | None | None | None | None | None | 1(+0) | None | N/A |
| seq478 | miRNA | 22 | TGAGA… | 15723 | 1 | 1 | None | None | None | 1(+1) | None | None | 1(+1) | chr5:148808541-148808562[+] |
| seq1293 | miRNA | 22 | TGAGG… | 14419 | 0 | 0 | None | None | None | 2(+0) | 1(+0) | None | 1(+0)/1(-0) | chr9:96938635-96938656[+] |
| seq1373 | spike | 22 | TAGCA… | 14324 | 0 | N/A | None | None | None | None | None | 1(+0) | None | N/A |
| seq770 | miRNA | 22 | TGAGG… | 13332 | 0 | 0 | None | None | None | 3(+0) | 1(+0) | None | 2(+0)/1(-0) | chr11:122017276-122017297[-] |
| seq1165 | spike | 22 | TGATA… | 12416 | 0 | N/A | None | None | None | None | None | 1(+0) | None | N/A |
| seq4685 | miRNA | 22 | TTCAA… | 12341 | 0 | 0 | None | None | None | 2(+0) | None | None | 1(+0)/1(-0) | chr12:58218441-58218462[-] |
| seq2037 | spike | 22 | AGGTT… | 12023 | 0 | N/A | None | None | None | None | None | 1(+0) | None | N/A |
Table 5.
Example of mapping statistics for an individual sample from sequencing run 313. Each small RNA annotation category is listed along with the number of reads annotated to each category as a perfect match (distance 0), or including 1 or 2 mismatches (distance 1 or 2). The percentage of reads that belong to each small RNA annotation category is also listed.
| Category | Distance 0 | Distance 1 | Distance 2 | Total | Percentage |
|---|---|---|---|---|---|
| miRNA | 488278 | 137363 | 24576 | 650217 | 78.35 |
| Calibrator | 97546 | 29351 | 5298 | 132195 | 15.93 |
| None | 0 | 0 | 0 | 24000 | 2.89 |
| rRNA | 7431 | 1439 | 248 | 9118 | 1.10 |
| tRNA | 4693 | 1391 | 241 | 6325 | 0.76 |
| repeat_rm | 1501 | 2067 | 0 | 3568 | 0.43 |
| sn/snoRNA | 909 | 364 | 115 | 1388 | 0.17 |
| piRNA | 577 | 105 | 84 | 766 | 0.09 |
| repeat10_rm | 334 | 364 | 0 | 698 | 0.08 |
| marker | 388 | 168 | 25 | 581 | 0.07 |
| tRNA_rm | 147 | 161 | 0 | 308 | 0.04 |
| doubtful_miRNA | 175 | 71 | 39 | 285 | 0.03 |
| miscRNA | 130 | 19 | 2 | 151 | 0.02 |
| sn/snoRNA_rm | 111 | 31 | 0 | 142 | 0.02 |
| miscRNA_rm | 82 | 17 | 0 | 99 | 0.01 |
| rRNA_rm | 53 | 23 | 0 | 76 | 0.01 |
| Total | 602355 | 172934 | 30628 | 829917 | 100.00 |
Table 6.
miRNA precursor read frequency profiles for top 10 expressed miRNA precursors. For each pre-miRNA (~70 nt hairpin miRNA precursor) the number of total mapped reads are listed, reduced to unique reads, and broken down to typical reads (only mapping to the listed miRNA precursor) or shared reads (mapping to additional miRNA precursors; e.g. multicopy miRNAs or miRNAs similar in sequence). Finally, the number of reads is weighted based on the number of typical and shared reads, as well as the number of precursors the shared reads map to.
| Precursor | Total | Unique | Typical | Shared | Weighted |
|---|---|---|---|---|---|
| hsa-mir-21 | 183247 | 1647 | 183247 | 0 | 183247.00 |
| hsa-mir-143 | 45871 | 807 | 45871 | 0 | 45871.00 |
| hsa-mir-141 | 33287 | 912 | 32921 | 366 | 33103.83 |
| hsa-mir-200c | 17582 | 669 | 17075 | 507 | 17328.33 |
| hsa-mir-30a | 12616 | 677 | 12480 | 136 | 12548.00 |
| hsa-mir-126 | 12490 | 664 | 12490 | 0 | 12490.00 |
| hsa-let-7f-2 | 23872 | 504 | 618 | 23254 | 12184.53 |
| hsa-mir-26a-2 | 24156 | 634 | 63 | 24093 | 12090.00 |
| hsa-mir-26a-1 | 24098 | 618 | 5 | 24093 | 12032.00 |
| hsa-let-7f-1 | 23371 | 469 | 71 | 23300 | 11646.13 |
Table 7.
miRNA read frequency and relative read frequency profiles for top 10 expressed miRNAs among all samples reported in [5]. Profiles are reported as read frequencies or relative read frequencies. Table A demonstrates merged mature miRNA profiles, merged to report reads from all genomic locations for multicopy miRNAs (based on mappings to miRNA or miRNA*). Table B demonstrates genomic cluster miRNA profiles, based on mappings to miRNA precursors. Table C demonstrates miRNA sequence family profiles, based on mappings to miRNA or miRNA*.
| miRNA | Read frequency |
Relative read frequency |
Read frequency |
Relative read frequency |
Read frequency |
Relative read frequency |
Read frequency |
Relative read frequency |
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Tissue identifier |
||||||||
| 5 | 9 | 16 | 18 | |||||
| A. Merged mature miRNA profiles | ||||||||
| hsa-miR-21 | 569625.50 | 0.56 | 138351.00 | 0.31 | 226553.00 | 0.28 | 123523.00 | 0.38 |
| hsa-let-7a(3) | 22484.14 | 0.02 | 12651.20 | 0.03 | 31969.65 | 0.04 | 8346.24 | 0.03 |
| hsa-let-7f(2) | 22242.18 | 0.02 | 16545.63 | 0.04 | 27608.55 | 0.03 | 12579.28 | 0.04 |
| hsa-miR-22 | 9871.00 | 0.01 | 9399.00 | 0.02 | 7772.00 | 0.01 | 5259.00 | 0.02 |
| hsa-miR-143 | 31092.00 | 0.03 | 28619.00 | 0.06 | 55555.00 | 0.07 | 10441.00 | 0.03 |
| hsa-miR-26a(2) | 25868.67 | 0.03 | 14067.00 | 0.03 | 27507.67 | 0.03 | 7725.33 | 0.02 |
| hsa-miR-24(2) | 10976.00 | 0.01 | 9224.00 | 0.02 | 12229.00 | 0.02 | 4747.00 | 0.01 |
| hsa-let-7b | 7084.54 | 0.01 | 3120.72 | 0.01 | 6824.97 | 0.01 | 1845.43 | 0.01 |
| hsa-miR-141 | 15311.17 | 0.01 | 16623.50 | 0.04 | 37158.17 | 0.05 | 5338.33 | 0.02 |
| hsa-miR-148a | 7187.00 | 0.01 | 25224.83 | 0.06 | 12809.33 | 0.02 | 6325.50 | 0.02 |
| all other miRNAs | 300341.80 | 0.29 | 171311.12 | 0.38 | 357254.66 | 0.44 | 141659.89 | 0.43 |
| total miRNA reads | 1022084.00 | 1.00 | 445137.00 | 1.00 | 803242.00 | 1.00 | 327790.00 | 1.00 |
| B. Genomic cluster profiles | ||||||||
| cluster-hsa-mir-21(1) | 571602.50 | 0.56 | 138558.00 | 0.31 | 227113.00 | 0.28 | 123752.00 | 0.38 |
| cluster-hsa-mir-98(13) | 63957.13 | 0.06 | 42553.55 | 0.10 | 87725.25 | 0.11 | 29768.55 | 0.09 |
| cluster-hsa-mir-23a(6) | 30687.00 | 0.03 | 27727.00 | 0.06 | 40218.17 | 0.05 | 13378.00 | 0.04 |
| cluster-hsa-mir-143(2) | 36136.00 | 0.04 | 33399.00 | 0.07 | 64212.00 | 0.08 | 12153.00 | 0.04 |
| cluster-hsa-mir-22(1) | 9937.00 | 0.01 | 9487.00 | 0.02 | 7824.00 | 0.01 | 5302.00 | 0.02 |
| cluster-hsa-mir-141(2) | 22870.17 | 0.02 | 23777.00 | 0.05 | 57888.00 | 0.07 | 8503.83 | 0.03 |
| cluster-hsa-mir-17(12) | 10404.00 | 0.01 | 4624.50 | 0.01 | 14919.25 | 0.02 | 7220.50 | 0.02 |
| cluster-hsa-mir-29a(4) | 27680.00 | 0.03 | 8674.00 | 0.02 | 16653.00 | 0.02 | 7467.00 | 0.02 |
| cluster-hsa-mir-199a-1(3) | 7866.00 | 0.01 | 2075.67 | 0.00 | 14351.67 | 0.02 | 4596.33 | 0.01 |
| cluster-hsa-mir-26a-1(2) | 25896.67 | 0.03 | 14080.00 | 0.03 | 27544.67 | 0.03 | 7732.33 | 0.02 |
| all other miRNA clusters | 215685.54 | 0.21 | 140436.28 | 0.32 | 245333.00 | 0.31 | 108134.45 | 0.33 |
| total miRNA reads | 1022722.00 | 1.00 | 445392.00 | 1.00 | 803782.00 | 1.00 | 328008.00 | 1.00 |
| C. Sequence family profiles | ||||||||
| sf-hsa-miR-21(1) | 569625.50 | 0.56 | 138351.00 | 0.31 | 226553.00 | 0.28 | 123523.00 | 0.38 |
| sf-hsa-let-7a-1(12) | 68459.00 | 0.07 | 41386.00 | 0.09 | 87052.00 | 0.11 | 32331.00 | 0.10 |
| sf-hsa-miR-141(5) | 32804.00 | 0.03 | 32049.00 | 0.07 | 70590.00 | 0.09 | 13039.00 | 0.04 |
| sf-hsa-miR-22(1) | 9871.00 | 0.01 | 9399.00 | 0.02 | 7772.00 | 0.01 | 5259.00 | 0.02 |
| sf-hsa-miR-30a(6) | 29655.00 | 0.03 | 11638.33 | 0.03 | 29895.67 | 0.04 | 8081.00 | 0.02 |
| sf-hsa-miR-26a-1(3) | 33000.00 | 0.03 | 17053.00 | 0.04 | 35505.00 | 0.04 | 10743.00 | 0.03 |
| sf-hsa-miR-143(1) | 31092.00 | 0.03 | 28619.00 | 0.06 | 55555.00 | 0.07 | 10441.00 | 0.03 |
| sf-hsa-miR-29a(4) | 27392.33 | 0.03 | 8540.00 | 0.02 | 16478.00 | 0.02 | 7418.33 | 0.02 |
| sf-hsa-miR-148a(3) | 9516.00 | 0.01 | 27602.00 | 0.06 | 15603.00 | 0.02 | 7479.00 | 0.02 |
| sf-hsa-miR-199a-1-3p(3) | 9143.00 | 0.01 | 2153.00 | 0.00 | 16283.00 | 0.02 | 4906.00 | 0.01 |
| all other miRNA seq families | 201526.17 | 0.20 | 128347.00 | 0.29 | 241955.33 | 0.30 | 104569.67 | 0.32 |
| total miRNA reads | 1022084.00 | 1.00 | 445137.33 | 1.00 | 803242.00 | 1.00 | 327790.00 | 1.00 |
Fig. 5.
(A) Diagram file mapping reads to miRNA precursor (with miR-21 reads displayed and both miR-21 and miR-21* reads summarized as histogram in the bottom). Precursor residues in blue represent the mature miR-21 sequence, whereas residues in red represent the miR-21* sequence. The number of reads that map to either the mature or precursor are specified, along with the times that they map to the genome. The precursor is extended 10 nt in both directions from the start of the mature and end of miRNA*. (B) Example of secondary structure predictions for putative miR-21 precursor, exibiting the prototypical fold-back structure. The mature and star sequences line up with the prototypical 1-2 nt overhang, consistent with miRNA biogenesis.
12. Normalize each miRNA profile to relative read frequencies
This normalization method corrects for the variable sequencing depth in each subsample by dividing each miRNA read frequency by the total number of miRNA sequence reads within the subsample in order to facilitate comparison of expression between subsamples (see example in Table 7). Computation of rpm values (reads per million) for each miRNA occurring in the subsample is also frequently used.
13. Consider correcting relative read frequency to derive actual abundance in each subsample for miRNAs that show extensive adapter ligation bias as described in [8]. Ranking miRNAs by abundance is helpful for biological follow-up experiments. For example, for the experimental protocol used in [5] the analysis showed under-representation over 5-fold for miR-193a, miR-193b, miR-26b, miR-29c, and miR-30b.
4.3. Quantitation of absolute miRNA amount
The spiking of subsample RNA with a set of synthetic calibrator RNAs allows for the identification of the total amount n of miRNAs (in mol) relative to the mass of total input RNA (in g). We add a cocktail of 10 calibrator oligoribonucleotides, each at 0.25 fmol quantity, per μg of total RNA [4]. Similar to miRNAs these calibrator RNAs also show sequence specific biases and their read frequencies deviate from their molar ratios [8].
14. Calculate total miRNA amount n in total RNA based on the following formula: , where or is the total amount of miRNA or calibrator (in mol) relative to the mass of total input RNA (in g), FmiR or Fcal is the number of reads (also referred to as read frequency) of miRNA or calibrator RNA, k corresponds to the observed number of miRNAs, and l corresponds to the added number of calibrators in the subsample.
4.4. miRNA sequencing statistics and quality control
15. We suggest summarizing the following characteristics after barcoded adapter trimming, mapping and annotation for each subsample to assess the quality of the sequencing and quality of cDNA library for review by the experimenter: (a) extraction statistics, (b) mapping and annotation statistics, (c) miRNA mapping statistics, (d) calibrator oligoribonucleotide mapping statistics, and (e) global miRNA amount per total input RNA (see example in Table 8).
Table 8.
Summary of sequencing statistics for each sample miRNA profile determination. Information provided includes the sample and barcode used, extraction statistics for each sample, miRNA mapping distance, calibrator mapping distance, mapping and annotation statistics for each biological subsample (excluding the number of calibrator sequences within each susbsample), miRNA amount. Other category includes viral miRNAs, piRNAs, non-prototypical doubtful miRNAs, miscellaneous RNA, repeat masker, sequences that mapped to the genome but have no annotation, sequences that did not map to the genome and have no annotation.
| Sample name and barcode |
Extraction statistics sample |
miRNA mapping distance |
Calibrator mapping distance |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue identifier |
Sample ID |
Barcode | Sequencing run |
Total reads |
Unique reads |
Calibrators | %Calibrator | miRNA dist0 |
miRNA dist1 |
miRNA dist2 |
%Mismatched miRNA |
Calibrator dist0 |
Calibrator dist1 | Calibrator dist2 |
%Mismatched calibrators |
| 5 | 5B | TCGAT | 313 | 1033399 | 79624 | 140346 | 13.58 | 650367.0 | 147354.3 | 23911.7 | 20.84 | 100992 | 33307 | 6047 | 28.04 |
| 9 | 9B | TCCTA | 313 | 546460 | 49465 | 137086 | 25.09 | 284947.5 | 72141.0 | 11327.0 | 22.66 | 98889 | 32404 | 5793 | 27.86 |
| 16 | 16B | TCCGT | 313 | 830273 | 61502 | 135543 | 16.33 | 488281.5 | 139317.2 | 23379.3 | 24.99 | 97546 | 32191 | 5806 | 28.03 |
| 18 | 18B | TCGCG | 313 | 395201 | 48457 | 84104 | 21.28 | 210158.0 | 45818.2 | 6804.2 | 20.03 | 58376 | 21478 | 4250 | 30.59 |
| 31 | 31C | TAATA | 313 | 690661 | 81974 | 123245 | 17.84 | 372568.5 | 76573.2 | 11079.3 | 19.05 | 88401 | 29287 | 5557 | 28.27 |
| 32 | 32C | TAACG | 313 | 582431 | 46028 | 89902 | 15.44 | 372644.5 | 75271.0 | 10683.0 | 18.74 | 63461 | 22394 | 4047 | 29.41 |
| 36 | 36B | TAGAG | 313 | 486599 | 42153 | 88851 | 18.26 | 301547.0 | 55816.7 | 8297.5 | 17.53 | 62937 | 21860 | 4054 | 29.17 |
| 45 | 45C | TGTGT | 313 | 635979 | 50507 | 116145 | 18.26 | 387804.5 | 76286.7 | 11621.8 | 18.48 | 85098 | 26501 | 4546 | 26.73 |
| 52 | 52B | TGATG | 313 | 408261 | 43563 | 103544 | 25.36 | 214713.5 | 47787.0 | 7714.0 | 20.54 | 73868 | 25074 | 4602 | 28.66 |
| 70 | 70B | TTACA | 313 | 433328 | 78220 | 53945 | 12.45 | 63101.0 | 12468.5 | 2055.0 | 18.71 | 39444 | 12244 | 2257 | 26.88 |
| 75 | 75C | TTGGT | 313 | 1323042 | 72000 | 180304 | 13.63 | 892154.5 | 166245.2 | 23871.3 | 17.57 | 131810 | 41153 | 7341 | 26.90 |
| 78 | 78B | TATCA | 313 | 527274 | 53126 | 97431 | 18.48 | 302911.0 | 66232.0 | 9938.5 | 20.09 | 70994 | 22403 | 4034 | 27.13 |
| 85 | 85B | TAGGA | 313 | 564942 | 53282 | 77707 | 13.75 | 356721.5 | 70288.5 | 10235.0 | 18.42 | 55691 | 18623 | 3393 | 28.33 |
| 98 | 98C | TCACT | 313 | 28203 | 8975 | 3246 | 11.51 | 11846.5 | 4710.5 | 830.0 | 31.87 | 2069 | 977 | 200 | 36.26 |
| 103 | 103B | TCATC | 313 | 53717 | 12577 | 7961 | 14.82 | 24923.5 | 8492.5 | 1355.5 | 28.32 | 5823 | 1815 | 323 | 26.86 |
| 104 | 104C | TCCAC | 313 | 384394 | 40186 | 79613 | 20.71 | 197981.0 | 69313.5 | 12053.0 | 29.13 | 56787 | 19253 | 3573 | 28.67 |
| 148 | 148B | TTAAG | 313 | 255665 | 26407 | 144763 | 56.62 | 69489.5 | 17596.0 | 2971.5 | 22.84 | 105897 | 32908 | 5958 | 26.85 |
| 166 | 166B | TCTGA | 313 | 393639 | 38595 | 96248 | 24.45 | 194411.5 | 50403.5 | 8174.0 | 23.15 | 69151 | 22931 | 4166 | 28.15 |
| 167 | 167B | TCTCC | 313 | 672805 | 67575 | 116815 | 17.36 | 385421.0 | 93550.8 | 15994.7 | 22.13 | 83599 | 27987 | 5229 | 28.43 |
| 175 | 175B | TCTAG | 313 | 879832 | 81516 | 124764 | 14.18 | 556194.5 | 103909.7 | 15345.8 | 17.66 | 89081 | 30057 | 5626 | 28.60 |
| Mapping and annotation statistics of biological subsample |
miRNA amount (n) (fmoles/μg of total RNA) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Human miRNAs | %miRNA | rRNA | % rRNA | tRNA | %tRNA | sn/snoRNA | %sn/snoRNA | Other | %other | |
| 821633.0 | 92.00 | 13275 | 1.49 | 11663 | 1.31 | 2828 | 0.32 | 43654.0 | 4.89 | 19.6 |
| 368415.5 | 89.99 | 9588 | 2.34 | 7975 | 1.95 | 1232 | 0.30 | 22163.5 | 5.41 | 11.3 |
| 650978.0 | 93.70 | 9184 | 1.32 | 6630 | 0.95 | 1484 | 0.21 | 26454.0 | 3.81 | 16.1 |
| 262780.3 | 84.47 | 20773 | 6.68 | 4117 | 1.32 | 2446 | 0.79 | 20980.7 | 6.74 | 10.5 |
| 460221.0 | 81.11 | 47285 | 8.33 | 18469 | 3.25 | 6058 | 1.07 | 35383.0 | 6.24 | 12.5 |
| 458598.5 | 93.11 | 6504 | 1.32 | 7716 | 1.57 | 1183 | 0.24 | 18527.5 | 3.76 | 17.1 |
| 365661.2 | 91.93 | 5017 | 1.26 | 6303 | 1.58 | 889 | 0.22 | 19877.8 | 5.00 | 13.8 |
| 475713.0 | 91.51 | 7508 | 1.44 | 10300 | 1.98 | 1304 | 0.25 | 25009.0 | 4.81 | 13.7 |
| 270214.5 | 88.68 | 9316 | 3.06 | 5218 | 1.71 | 1624 | 0.53 | 18344.5 | 6.02 | 8.7 |
| 77624.5 | 20.46 | 153373 | 40.43 | 40923 | 10.79 | 26397 | 6.96 | 81065.5 | 21.37 | 4.8 |
| 1082271.0 | 94.71 | 9957 | 0.87 | 7102 | 0.62 | 1548 | 0.14 | 41860.0 | 3.66 | 20.1 |
| 379081.5 | 88.19 | 10327 | 2.40 | 8364 | 1.95 | 1541 | 0.36 | 30529.5 | 7.10 | 13.0 |
| 437245.0 | 89.74 | 17761 | 3.65 | 5771 | 1.18 | 1825 | 0.37 | 24633.0 | 5.06 | 18.8 |
| 17387.0 | 69.67 | 3200 | 12.82 | 749 | 3.00 | 411 | 1.65 | 3210.0 | 12.86 | 18.9 |
| 34771.5 | 75.99 | 3533 | 7.72 | 1472 | 3.22 | 446 | 0.97 | 5533.5 | 12.09 | 14.6 |
| 279347.5 | 91.66 | 5354 | 1.76 | 3647 | 1.20 | 877 | 0.29 | 15555.5 | 5.10 | 18.1 |
| 90057.0 | 81.20 | 4919 | 4.44 | 2630 | 2.37 | 497 | 0.45 | 12799.0 | 11.54 | 4.2 |
| 252989.0 | 85.07 | 7301 | 2.46 | 6328 | 2.13 | 1044 | 0.35 | 29729.0 | 10.00 | 13.5 |
| 494966.5 | 89.02 | 15123 | 2.72 | 10239 | 1.84 | 1999 | 0.36 | 33662.5 | 6.05 | 14.2 |
| 675450.0 | 89.46 | 27086 | 3.59 | 15469 | 2.05 | 2906 | 0.38 | 34157.0 | 4.52 | 18.1 |
Extraction statistics: Total number of reads assigned to each subsample, number of unique reads, number and percent of reads that are assigned to calibrator oligoribonucleotides. If unexpectedly for certain subsamples the majority of reads are assigned to calibrators, it may indicate faulty concentration determination of input total RNA, ribonuclease contamination, DNA contamination, loss of RNA, etc.
Mapping and annotation statistics of biological subsample: Number and percentage of (1) miRNAs, (2) rRNAs, (3) tRNAs, (4) sn/snoRNAs, (5) other RNA, combining all other categories described above (repeat10_rm, miscRNA, piRNA, mis-annotated miRNA, all other repeat-masked RNAs). For example, in our small RNA profiling protocol, a high percentage of rRNAs is indicative of partial RNA degradation/hydrolysis of the sample.
miRNA mapping statistics: Identify the miRNA sequence reads that map with 0, 1, and 2 errors, and calculate the percent mismatched miRNA reads compared to the perfectly matching miRNA reads.
Calibrator oligoribonucleotide read mapping statistics: Identify the calibrator sequences that map with 0, 1, and 2 errors, and calculate the percent mismatched calibrator reads compared to the perfectly matching calibrator reads. A high percentage of mismatched calibrator sequences signifies a low quality sequencing run.
miRNA amount (n): Reported in fmol amount of total miRNA per lg of subsample input total RNA entering cDNA library construction (step 14).
16. Further quality assessment can be performed by clustering subsamples to ensure that no bias exists due to differences in experimental conditions (e.g. the day the sequencing was performed, barcode used, sequencing platform used). Hafner et al. established that no significant bias can be attributed to the different barcoded adapters [8]. We did observe barcode misassignment due to multiple sequencing errors. Even when a barcoded adapter was absent in an experiment, we found sequence reads assigned to it, albeit at ~1000-fold lower frequency compared to barcoded adapters actually present during cDNA library construction. This small fraction of misassigned reads suggests a minimum read count requirement for a subsample profile to be included in downstream analysis. The profiles of the calibrators can provide an extra level of quality assessment (see step 23).
4.5. Public repositories for small RNA sequences
17. Submit files including barcode-extracted reads, as well as unassigned reads to the SRA (www.ncbi.nlm.nih.gov/sra). The SRA was planned to be phased out by the end of 2011 due to funding constraints and at the time GEO (www.ncbi.nlm.nih.gov/geo) served as a repository for sequencing files; one may find datasets submitted during that time in GEO instead of SRA. NIH support has enabled the SRA continuation and now it will operate as the NIH’s primary archive of high-throughput sequencing data and as part of the international partnership of archives at the NCBI, the European Bioinformatics Institute and the DNA database of Japan. Data submitted to any of the three organizations are shared among them.
Barcode-extracted files provide additional annotation; they are usually text files with the RNA sequence, its read frequency and annotation category.
The repositories also require submission of files listing the remainder of the sequences within the sequencing run that were not assigned to a particular barcode. In order to accurately process this file, one would need to know how many subsamples were included in the sequencing run and their barcode sequences, and not only the ones reported in the manuscript barcode extracted files (see example in Table 3).
4.6. Curated human miRNA entries
The rapid acquisition of deep sequencing small RNA cDNA profiles from many human tissues led to a rapid expansion of entries in miRBase. When we mapped reads obtained in our lab from over 1000 human small RNA cDNA libraries against miRBase entries, we noticed that some read alignments did not correspond to prototypical miRNA biogenesis patterns and were more likely resulting from mapping errors. We therefore use our own curated set of miRNA genes in Table S4 in [5] for annotation of reads. Samples were derived from diverse normal and diseased human tissues and yielded 561,200,705 reads. Verification of miRNAs was performed by analyzing their expression levels and cistronic expression pattern, the mapping pattern of reads to existing miRNA regions, as well as secondary structure prediction for the expected fold-back structure of precursor molecules.
In summary, after mapping all sequence reads to miRBase 16, we filtered out miRNAs with <50 sequence reads, accepted miRNAs with 50–100 reads only if they were part of a precursor miRNA cluster, filtered out multi-mapping miRNAs (>30 genomic locations), and for miRNAs with <30 genomic locations assessed mapping pattern in the miRNA expression histogram (Fig. 5A). We then generated secondary structure files for the remaining miRNAs using RNAfold from the Vienna package (http://www.tbi.univie.ac.at/~ivo/RNA/) [15] and only accepted miRNAs with prototypical fold-back structures (Fig. 5B), with the exception of miR-451-DICER1 and miR-618, which are processed independent of DICER1. Finally, we renamed miRNAs according to read frequency for the 3p or 5p arm. If the reads in either arm constituted <20% of both arms combined, then we considered the minor species as miRNA/; otherwise, we assigned each arm as −5p or −3p.
Of the 1045 miRBase annotated human precursor sequences, 488 failed one or more of our criteria (expression, mapping pattern and RNA fold), with 282 having little or no expression evidence. Further support for the validity of annotated miRNAs is currently obtained by analysis of sequence conservation between species, using sequencing data obtained from macaque small RNA cDNA libraries (unpublished data).
4.7. miRNA read frequency by grouping sequence families and genomic clusters (cistrons)
Grouping of reads based on miRNA sequencing families or coexpressed cistronically organized miRNAs facilitates a biologically relevant interpretation of miRNA profiles and variation between subsamples. Fig. 6 shows examples of miRNA genomic clusters and sequence families. As described earlier, miRNA sequence families group miRNAs that display sequence similarity and thus likely target a similar set of mRNAs, while miRNA expression clusters group miRNAs that are located in close proximity in the genome, and are co-transcribed.
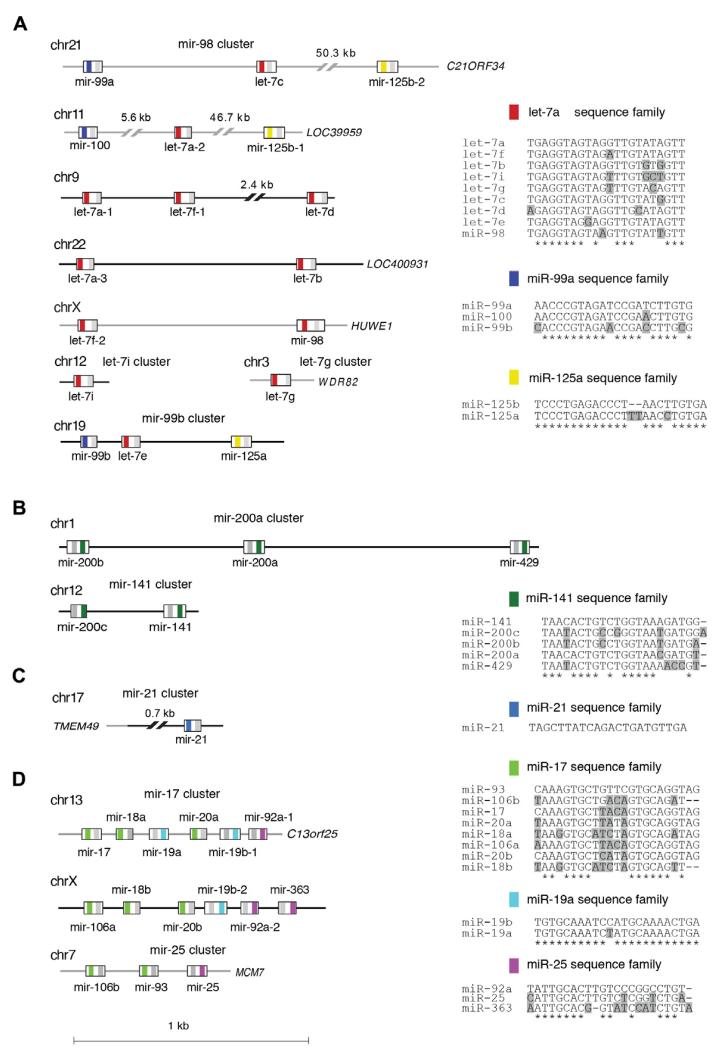
Fig. 6.
miRNA genomic and functional organization. The genomic and functional organization of several miRNA clusters is clarified. The genomic locations for each of the miRNA members are defined. Grey lines denote intronic regions. miRNA mature sequences are color-coded according to the sequence family to which they belong (i.e. in the mir-98 cluster, red significes the let-7a sequence family). The miRNA* sequence is defined with a grey bar. The sequence families are depicted as sequence alignments compared to the most highly expressed miRNA family member shown on top, based on profiles of approximately 1000 human samples (see text). Shaded residues denote differences from the most highly expressed miRNA family member.
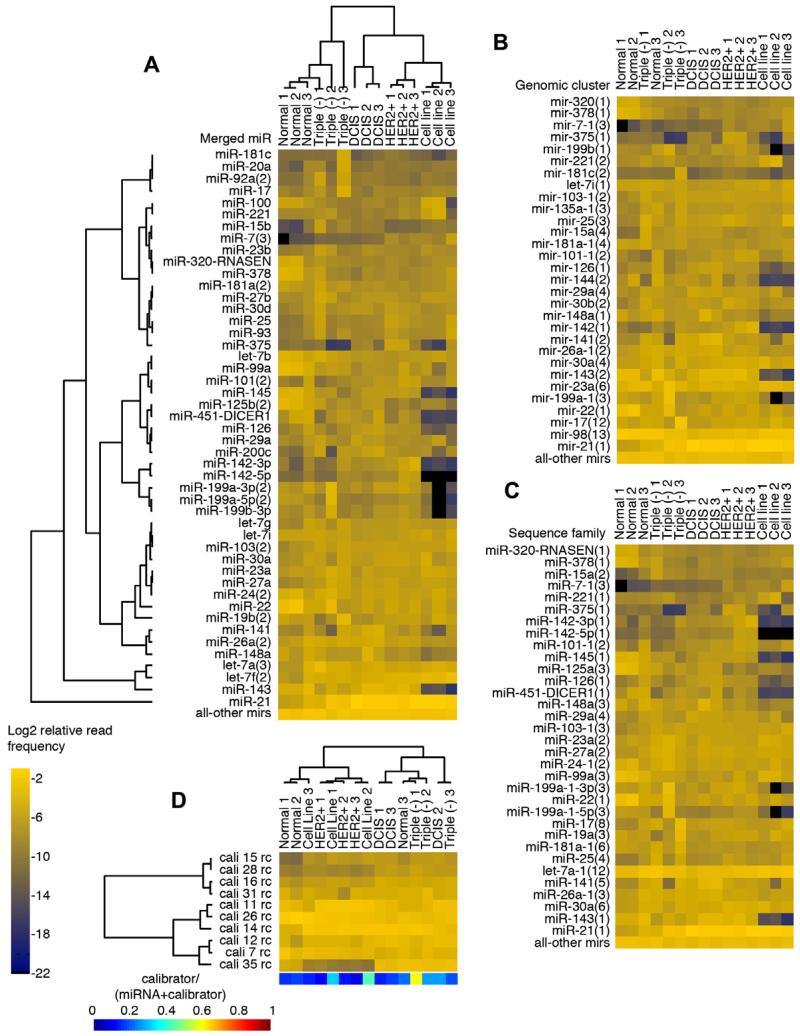
We defined miRNA genomic clusters [5] taking into consideration expressed sequence tag (EST) evidence and levels of miRNA expression from our data (similar to the procedure described in [16]). With the exception of cluster-mir-98(13), typically, the greatest genomic distance between clustered miRNAs was 5 kb. We defined sequence families on the basis of seed sequence similarity (position 2–8) allowing only one transition in these positions, as well as 3′ end similarity (position 9 through 3′ end) allowing up to 50% mismatches, with additional manual curation. Our naming convention depicts the number of miRNAs in a sequence family or expression cluster as the number in parenthesis. Fig. 7 demonstrates the collapsing of mature miRNA profiles to miRNA sequence families and genomic clusters for a subset of breast samples.
Fig. 7.
(A) Merged-miRNA view of an example of miRNA expression for a set of breast cancer samples from [5]. Samples and miRNAs are clustered using the Bayesian approach [18]. Expression is presented as the Log2 relative read frequency of miRNAs, and selected using the top 60% expressed across all samples. (B) miRNA-sequence-family view for the same set of samples. The merged miRNA view was first expanded to include other sequence family members, then collapsed by their sequence family definitions. (C) miRNA-genomic-cluster view for the same set of samples. The merged-miRNA view was first expanded to include other genomic cluster members, then collapsed by their genomic cluster definitions. (D) Clustering based on calibrator oligoribonucleotide reads. Expression is plotted similarly to miRNA expression using the Log2 relative read frequency of calibrators. Although samples cluster mostly based on sequencing run, the calibrator expression is unaffected by the sample content. However, the miRNA profiles cluster according to normal breast or breast tumor subtype, with the exception of one sample (triple negative 1). Breast tumor subtypes include triple negative (Triple (−)), ERBB2 over-expressing (HER2+), and Ductal Carcinoma in situ (DCIS). The color bar below represents the frequency of calibrator reads relative to the sum of the calibrator and miRNA reads. Triple negative 1 has a high calibrator content and may explain why in the sequence family view it clusters with normal breast samples.
18. Condense miRNAs into sequence families and genomic clusters according to definitions in Supplementary Table S4 in [5]. This step allows for data reduction and ease of miRNA profile presentation (see example in Table 7 and Fig. 7).
4.8. Clustering barcoded samples
Various algorithms can be used to perform clustering of samples based on miRNA read frequencies [5,9]. Tools that were originally designed for microarray samples can also be used for Log2-transformed relative read frequency miRNA profiles [17]. Given that miRNAs show a non-normal distribution with a relatively small number of miRNAs highly expressed and most miRNAs expressed at much lower levels, with the lower expressed miRNAs subject to large sampling noise, the algorithm described in [9] uses a Bayesian probability framework and was specifically designed for miRNA read data.
19. Decide on filters to select samples for clustering or differential expression analysis. Filters may be based on read depth, and it is preferable to compare subsamples with similar sequence read depth. We suggest selecting a cutoff for monitoring top expressed miRNAs which when dysregulated trigger tractable changes in target mRNA expression. Suggested filters: (1) miRNAs that comprise a specific percentage of total miRNA reads in at least one sample (e.g. top 85%), (2) fixed miRNA read frequency within each subsample (e.g. miRNAs present with a minimum of 10 reads), (3) requirement for a specific miRNA read frequency across all subsamples or a number of selected subsamples. For example in [5], setting a cutoff of 10 or more reads per miRNA for the pool of all 49,479,978 miRNA sequence reads, we identified a total of 888 mature and miRNA sequences from the curated set of 1033 mature and miRNA/sequences. Resetting the threshold to 5,000 reads, we identified/231 miRNA and miRNA species together constituting 99% of all miRNA reads.
20. Cluster miRNAs and subsamples using algorithms discussed below. To evaluate how well the unsupervised clustering captures clinical, pathological or experimental variables and assess confounding factors, we include annotation labels representing variables of interest next to the dendrograms.
Variations of Bayesian algorithms, such as the one described in [9,18] and used in mirZ/smirnaDB website/database (http://www.mirz.unibas.ch/cloningprofiles/) cluster both miRNAs and subsamples, as in [16]. Note that this website uses a different definition of miRNA sequence groups and expression clusters from the one described in this manuscript. miRNAs from every species were grouped together in sequence groups if they shared the same seed sequence (position 2–7 of the mature miRNA). miRNA precursors were clustered together based on their relative distance in the genome: two precursors were placed in the same cluster if they were <50 kb from each other. Only precursors that could be mapped to the genome assembly were used to construct precursor clusters.
Cluster miRNAs and subsamples using unsupervised hierarchical clustering with complete linkage and Spearman correlation [5]. Note that this clustering is rank-based and thus does not require normalization by the total subsample read count (described in 12).
21. Plot heatmaps of miRNA expression in the subsamples for each miRNA (see Figs. 1 and 2 in [5]). Different types of mappings to a palette of colors for the heatmap can be used to highlight different types of information. A color mapping based on Log2 read frequencies depicts miRNA expression; these subsamples can be sorted from the highest overall expressed miRNA within all subsamples to the lowest overall expressed miRNA to highlight differences in more abundant miRNAs, which may be more biologically relevant. Other color mappings may also depict miRNA read frequencies standardized across each miRNA to accentuate the differences in expression across subsamples assessing one miRNA at a time.
22. It can be useful to compare profiles derived from heterogeneous tissue samples to profiles from a homogeneous cell population, such as cell lines, to identify miRNAs expressed in a specific cell of interest. For example, comparing a cancer tissue biopsy sample, which usually includes not only tumor cells but also immune cells, connective tissue or normal tissue, to a cancer cell line, may identify miRNAs that are specifically expressed in tumor cells within the heterogeneous tissue sample (Fig. 7).
23. Comparing the clustering of profiles of the calibrators and the clustering of subsample profiles excludes techniquebased biases (Fig. 7).
24. Principal component analysis (PCA) of miRNA read counts can be used as another means of data evaluation and reduction, highlighting similarities and differences between samples.
4.9. Differential expression analysis
The assumptions on distribution used by standard Significance Analysis of Microarrays (SAM) [17] tests do not hold for RNA read frequency profiles. Recently the SAM tests have been adapted for sequencing data (SAMseq), utilizing a nonparametric method to measure the significance of features from RNA-Seq data [19,20]. There are two main characteristics of read frequency data that distinguish them from microarray data. Firstly, algorithms for analysis of differential expression of read frequency data need to take into account the lower confidence in the small number of reads obtained for lowly abundant miRNAs. Secondly, due to variable sequencing depth of miRNA read frequency profiles, the variation of the total miRNA read frequency across different samples is much greater than the variation of miRNA read frequencies within each sample. Therefore, we suggest using differential expression statistical tools designed for sequencing read frequency profiles:
25. mirZ: Specifically tailored for miRNA sequencing profiles. It is based on a Bayesian model for computing the posterior probability that the frequency of a miRNA in the total miRNA population differs between two sets of samples [9,18]. This probability is computed assuming a binomial sampling model and integrating over the unknown miRNA frequencies in the samples. This approach takes into account both the variability between sample size and the absolute miRNA counts, but does not account for within-group variability, as group members are summed prior to analysis.
26. edgeR: A Bioconductor (http://bioconductor.org) software package for studying differential expression of replicated count data based on a negative binomial distribution [21]. Replicates may represent subsamples of a specific disease state, such as normal versus tumor. It uses an overdispersed Poisson model to account for both biological and technical variability. Empirical Bayes methods are used to minimize the degree of overdispersion across transcripts, improving the reliability of inference. Results are plotted as Log2 of the fold change of read frequencies between two groups of samples compared as a function of the Log2 of the average miRNA relative read frequencies in the two groups of samples compared. These are called ‘smear’ plots (for example see Fig. 4 in [5]). This allows an interpretation of differential expression analysis in the context of miRNA expression levels. Lower expressed miRNAs likely exert weaker effects on their mRNA targets, unless they show a high degree of complementarity to their target; thus, large changes in expression in lowly expressed miRNAs may not result in changes in mRNA expression.
27. DESeq: Also a Bioconductor package, which similarly to edgeR uses a method based on a negative binomial distribution, with variance and mean linked by local regression to determine differences in expression within read frequency profiles [22].
28. DEGseq: A third Bioconductor package designed to identify differentially expressed genes that uses as input uniquely mapped reads from RNA-Seq data [23]. This method uses a combination of two statistical models: (1) the number of reads derived from a gene follows a binomial distribution that can be approximated by a Poisson distribution, (2) Fisher’s exact test and likelihood ratio test identify differentially expressed genes.
29. SAMseq: A nonparametric method that is less sensitive to outliers (see example in [14]).
4.10. Identification of rare nucleotide variations and mutations
Most frequently encountered nucleotide variations are, (1) A-to-G and C-to-U RNA editing events by dsRNA-specific adenosine deaminases [16,24], and cytidine deaminases [25], respectively. The editing may be tissue-specific; (2) 3′ end terminal variations, such as polyuridylation by terminal uridylyl-transferases (TUTases) and polyadenylation by poly(A) polymerases; (3) DNA-encoded common single-nucleotide polymorphisms (SNPs). Frequent 3′ and 5′ processing variations are also known as isomirs [26]. Because these events can be found to various degrees in most samples, we developed a method for the discovery of rare events such as mutations of miRNAs in specific diseased samples [5].
30. Define specific nucleotide variations for miRNAs if they display (1) ≥10 altered sequence reads for a specific position and (2) are present in ≥25% of the specific position in at least one sample, to focus on somatic mutations and rare SNPs that are present in one allele.
31. To simplify identification of nucleotide variations, we suggest separate analysis for 3′ most terminal variations in the last insert position, because it frequently contains 3′ untemplated nucleotide addition [16]. Despite the removal of the terminal nucleotide, the majority of variations may still comprise variations in the last 2 positions of the predominant mature sequence read, likely representing 3′ terminal events that were insufficiently repressed by this computational approach (the majority of these variations represented changes into A or U). Most such events are single A or U additions. To further focus on 3′ end variations, the data can be re-analyzed to include all positions of the sequence reads, and extended to include additional positions following the end of the predominant mature miRNA sequence. As an alternative approach, we suggest treating separately 3′ tails of unmatched A’s or U’s of any length.
32. Once nucleotide variations are identified with the rules defined above, score as positive each individual sample for the presence of variation in > 10% of the reads. The 10% cutoff avoids cataloging changes due to sequencing error or mis-mapping from abundant miRNAs.
33. Characterize variations according to: (1) distribution of the variation frequency across samples and the number of samples affected, (2) the number of shared reads for miRNA sequence family members and miRNA abundance to determine mis-mapping and (3) location of variation (see example in Table 9).
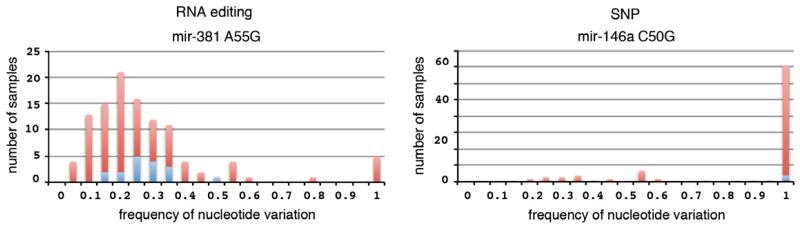
Histogram of nucleotide variations across subsamples (Fig. 8) : Based on the histogram distribution, e.g. unimodal versus multimodal, one can deduce the nature of the variation. For example, a well-represented unimodal distribution of the nucleotide variation frequency is expected for enzymatic deamination events. A trimodal distribution with peaks over 0, 50 and 100% variation frequency likely represents common SNPs. Bimodal distribution likely represents rare somatic mutational events; these somatic mutations could be represented by variations observed in a single patient, usually affecting only one allele.
Mis-mapping from abundant miRNAs: Variable positions in miRNAs with a high degree of sequence similarity should be excluded, due to the high likelihood that variations in these positions for the less abundant miRNA represent mis-mapping from the more abundant miRNA sequence family member.
Location of variation: For ease of presentation we suggest dividing the nucleotide variations into (1) variations present in positions 1 through the position before the two terminal 3′ nucleotides of the mature or miRNA/(2) variations present in the two terminal 3′ nucleotides of the mature or miRNA/.
Table 9.
Nucleotide variation determination. In identifying the variations listed in the table, we required 10 reads with the nucleotide variation. If at least one sample had a nucleotide variation with a frequency of >0.25, the variation was included in the table; samples with the same variations at frequencies >0.1 were subsequently added. Description includes: (A-D) the presence and frequency of the variation among the sample collection: number of samples with each defined nucleotide variation, the highest frequency of the nucleotide variation observed in a sample, the average frequency of the nucleotide variation among all samples that carry it; (E-G) the type and the position of the nucleotide variation in the precursor; (H-I) the position of the variation within the miRNA or miRNA*; (J-M) the expression levels of the miRNA carrying the variation, tabulated either as the maximum relative read frequency for a precursor miRNA in the samples carrying the variation, or as the average relative read frequency for the precursor miRNA among all samples.
| miR/miR* | Samples with nucleotide variation |
Highest frequency of nucleotide variation |
Average frequency of nucleotide variation |
Position of nucleotide variation in precursor |
Type of nucleotide variation |
Known SNP and comments |
Distance to the 5p miR/ miR* start |
Distance to the 3p miR/ miR* start |
Max for precursor with nucleotide variation |
Average for precursor |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Relative read frequency (%) |
Rank | Relative read frequency (%) |
Rank | |||||||||
| let-7c | 5 | 0.37 | 0.18 | 27 | A → g | mis-mapping from let-7b |
17 | -28 | 0.12% | 120 | 0.31% | 63 |
| miR-128-1* | 6 | 1.00 | 0.97 | 27 | T → g | SNP | 17 | -18 | 0.03% | 183 | 0.02% | 166 |
| miR-1304-3p | 1 | 1.00 | 1.00 | 59 | C → a | SNP | 49 | 13 | 0.00% | 281 | 0.00% | 362 |
| miR-146a* | 5 | 1.00 | 0.99 | 50 | C → g | rs2910164 | 40 | 5 | 1.84% | 72 | 0.22% | 79 |
| miR-181a-1 | 13 | 0.56 | 0.34 | 29 | T → g | SNP | 19 | -21 | 0.37% | 144 | 0.32% | 60 |
| miR-181a-2 | 13 | 0.56 | 0.34 | 29 | T → g | SNP | 19 | -19 | 0.30% | 142 | 0.31% | 65 |
| miR-185 | 3 | 0.50 | 0.46 | 26 | T → g | SNP | 16 | -19 | 0.07% | 146 | 0.10% | 106 |
| miR-196a-2* | 42 | 1.00 | 0.83 | 64 | C → t | rs11614913 | 54 | 18 | 0.64% | 174 | 0.07% | 115 |
| miR-200a | 13 | 0.38 | 0.15 | 65 | C → a | mis-mapping from mir-141 |
55 | 17 | 0.62% | 141 | 0.48% | 48 |
| miR-20b | 1 | 0.26 | 0.26 | 11 | C → t | mis-mapping from mir-20a |
1 | -37 | 0.01% | 175 | 0.02% | 164 |
| miR-376a-1-3p | 97 | 1.00 | 0.94 | 53 | A → g | editing | 43 | 6 | 0.30% | 227 | 0.02% | 170 |
| miR-376a-2-3p | 97 | 1.00 | 0.94 | 53 | A → g | editing | 43 | 6 | 0.29% | 226 | 0.02% | 172 |
| miR-376b | 2 | 0.84 | 0.54 | 53 | A → g | editing | 43 | 6 | 0.02% | 174 | 0.01% | 220 |
| miR-376c | 129 | 1.00 | 0.61 | 53 | A → g | editing | 43 | 6 | 1.05% | 191 | 0.06% | 124 |
| miR-381 | 17 | 0.49 | 0.26 | 55 | A → g | editing | 45 | 4 | 0.02% | 223 | 0.01% | 205 |
| miR-449c-3p | 1 | 1.00 | 1.00 | 52 | T → a | SNP | 42 | 5 | 0.00% | 257 | 0.00% | 365 |
| miR-556-3p | 1 | 0.39 | 0.39 | 55 | A → g | SNP | 45 | 6 | 0.01% | 200 | 0.00% | 284 |
| miR-585 | 1 | 1.00 | 1.00 | 50 | G → a | SNP | 40 | 4 | 0.01% | 214 | 0.00% | 358 |
| miR-625-3p | 17 | 0.48 | 0.28 | 54 | A → g | editing | 44 | 7 | 0.10% | 208 | 0.01% | 194 |
| miR-99a | 4 | 0.33 | 0.17 | 26 | T → c | mis-mapping from mir-100 |
16 | -21 | 0.35% | 104 | 0.85% | 29 |
Fig. 8.
Histograms of nucleotide variations representing an RNA editing event for mir-381, manifested by a unimodal distribution and a published SNP in mir-146a, manifested by a trimodal distribution. The blue portion of the bars represents samples that meet the criteria of a minimum of 10 altered sequence reads at the varied position, whereas the red portion of the bars represents samples that do not meet the minimum altered sequence read requirement.
4.11. Identification of novel miRNAs
There are multiple algorithms that can facilitate the discovery of novel miRNAs in sequence read data. One of the most commonly used prediction tools is mirDeep that specifically assesses for the miRNA processing pattern expected from the miRNA foldback structure [27].
Acknowledgments
T.A.F. and I.Z.B. are supported by Grant #UL1 TR000043 from the National Center for Research Resources and the National Center for Advancing Translational Sciences (NCATS), NIH. M.H. is supported by the Deutscher Akademischer Austauschdienst and is currently funded by a fellowship of the Charles Revson, Jr. Foundation. N.R. is supported through a K08 Award (NS072235) from the National Institute of Neurological Disorders and Stroke. T.T. is an HHMI investigator, and this work in his laboratory was supported by NIH Grant 1RC1CA145442, and Starr Foundation Grants. We would like to thank all members of the Tuschl lab for their contribution to miRNA re-annotation and constant feedback, Scott Dewell at the Rockefeller University Genomics Center, as well as our collaborators in the Academic Medical center in Amsterdam (van de Vijver group), Netherlands Cancer Institute (Wessels group), the Biozentrum in Basel (Zavolan group) and Memorial Sloan-Kettering Cancer Center (Sanders group).
References
- [1].Farazi TA, Spitzer JI, Morozov P, Tuschl T. J. Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hafner M, Renwick N, Farazi TA, Mihailovic A, Pena JT, Tuschl T. Methods. 2012 doi: 10.1016/j.ymeth.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Farazi TA, Horlings HM, Ten Hoeve JJ, Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F, van Kouwenhove M, Kreike B, Sie D, Hovestadt V, Wessels LF, van de Vijver MJ, Tuschl T. Cancer Res. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ugras S, Brill E, Jacobsen A, Hafner M, Socci ND, Decarolis PL, Khanin R, O’Connor R, Mihailovic A, Taylor BS, Sheridan R, Gimble JM, Viale A, Crago A, Antonescu CR, Sander C, Tuschl T, Singer S. Cancer Res. 2011;71:5659–5669. doi: 10.1158/0008-5472.CAN-11-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Italiano A, Thomas R, Breen M, Zhang L, Crago AM, Singer S, Khanin R, Maki RG, Mihailovic A, Hafner M, Tuschl T, Antonescu CR. Genes Chromosom. Cancer. 2012;51:569–578. doi: 10.1002/gcc.21943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, Ojo T, Luo S, Schroth G, Tuschl T. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Berninger P, Gaidatzis D, van Nimwegen E, Zavolan M. Methods. 2008;44:13–21. doi: 10.1016/j.ymeth.2007.10.002. [DOI] [PubMed] [Google Scholar]
- [10].Motameny S, Wolters S, Nurnberg P, Schumacher B. Genes. 2010;1:70–84. doi: 10.3390/genes1010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li H, Durbin R. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].McCormick KP, Willmann MR, Meyers BC. Silence. 2011;2:2. doi: 10.1186/1758-907X-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fahlgren N, Sullivan CM, Kasschau KD, Chapman EJ, Cumbie JS, Montgomery TA, Gilbert SD, Dasenko M, Backman TW, Givan SA, Carrington JC. RNA. 2009;15:992–1002. doi: 10.1261/rna.1473809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hofacker IL. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Efron B, Tibshirani R. Genet. Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- [18].Hausser J, Berninger P, Rodak C, Jantscher Y, Wirth S, Zavolan M. Nucleic Acids Res. 2009;37:W266–272. doi: 10.1093/nar/gkp412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li J, Tibshirani R. Stat. Methods Med. Res. 2011 [Google Scholar]
- [20].Li J, Witten DM, Johnstone IM, Tibshirani R. Biostatistics. 2012;13:523–538. doi: 10.1093/biostatistics/kxr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Robinson MD, McCarthy DJ, Smyth GK. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Anders S, Huber W. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang L, Feng Z, Wang X, Wang X, Zhang X. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- [24].Kawahara Y, Nishikura K. FEBS Lett. 2006;580:2301–2305. doi: 10.1016/j.febslet.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. Nat. Struct. Mol. Biol. 2011;18:230–236. doi: 10.1038/nsmb.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ryan BM, Robles AI, Harris CC. Nat. Rev. Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]