Abstract
Protein kinases represent the most effective class of therapeutic targets in cancer; therefore determination of kinase aberrations is a major focus of cancer genomic studies. Here, we analyzed transcriptome sequencing data from a compendium of 482 cancer and benign samples from 25 different tissue types, and defined distinct ‘outlier kinases’ in individual breast and pancreatic cancer samples, based on highest levels of absolute and differential expression. Frequent outlier kinases in breast cancer included therapeutic targets like ERBB2 and FGFR4, distinct from MET, AKT2, and PLK2 in pancreatic cancer. Outlier kinases imparted sample-specific dependencies in various cell lines as tested by siRNA knockdown and/or pharmacologic inhibition. Outlier expression of polo-like kinases was observed in a subset of KRAS-dependent pancreatic cancer cell lines, and conferred increased sensitivity to the pan-PLK inhibitor BI 6727. Our results suggest that outlier kinases represent effective precision therapeutic targets that are readily identifiable through RNA-sequencing of tumors.
Keywords: Pancreatic Cancer, RNA-Seq, Kinases, Outlier Expression, Personalized Medicine
Introduction
The dependence of cancers on a primary driver, most often a kinase (1, 2), forms the guiding principle of targeted therapy that has had some notable clinical successes, such as imatinib for BCR-ABL-positive chronic myeloid leukemia, trastuzumab and lapatinib for ERBB2-positive breast cancers, gefitinib for lung cancers with kinase domain mutations in EGFR (3, 4), and more recently crizotinib for lung cancers with ALK gene fusions (5). Thus, protein kinases are the mainstay of a majority of the current targeted therapeutic strategies for cancers, and inhibitors of several oncogenic kinases such as AKT, BRAF, CDKs, KIT, RET, SRC, MAPKs, MET, PIK3CA, PLKs, AURKs, S6Ks, and VEGFR are under various stages of clinical use, trials, or development (4, 6). While activating somatic mutations are associated with a few of these genes, overexpression of kinases (resulting from genomic amplification or other underlying somatic aberrations) is often a strong indicator of aberrant activity that may impart dependence of cancer cells.
Pancreatic cancer is the 4th leading cause of cancer related deaths in the U.S., with the worst prognosis (5-year survival < 3%) of all major malignancies (7), attributed to diagnosis of the disease at an advanced, unresectable stage and poor responsiveness to chemo-/radiation-therapy (8, 9). The overarching oncogenic driver of pancreatic cancer is mutant-KRAS that has eluded therapeutic interventions (10, 11), spurring the search for alternative targets (11). The identification of distinct kinases in independent screens for synthetic lethal interactors of KRAS (12-14) led us to systematically explore the expression profiles of all the 468 human kinases (kinome) to identify and test ‘personalized kinase targets’ in a panel of pancreatic cancer cell lines.
Next-generation sequencing of transcriptomes offers significant advantages over microarrays in terms of throughput, elimination of probe bias, and simultaneous monitoring of diverse components of transcriptome biology (15), including gene expression (15-18), alternative splicing (19, 20), chimeric/-read through transcripts (21, 22), and non-coding transcripts (23, 24). Furthermore, transcriptome sequencing affords a direct and quantitative readout of transcript abundance facilitating sample-wise gene expression analyses using a digital metric of normalized fragment reads, which are not possible using microarrays. Here, we set out to use transcriptome data from a compendium of 482 cancer and benign samples from 25 different tissue types to carry out gene expression profiling of the complete complement of kinases in the human genome, the kinome, to identify ‘individual sample-specific outlier kinases’ inspired by the concept of cancer outlier profile analysis (COPA)(25, 26). Importantly, while COPA analysis was used to identify subsets of ‘samples displaying outlier expression of candidate genes’, here we interrogated subsets of ‘outlier genes in individual samples’, focusing on kinases that display the highest levels of absolute expression among all the kinases in a sample and the highest levels of differential expression compared to the median level of expression of the respective gene(s) across the compendium. As proof of concept, we observed outlier expression of the therapeutic target ERBB2 specifically in all the breast cancer cell lines analyzed that are known to be ERBB2-positive. Thus we hypothesized that specific outlier kinases in other samples may also impart ‘dependence’ due to clonal selection for extremely high expression and may thereby represent personalized therapeutic targets.
Here, we analyzed kinome expression profiles of breast and pancreatic cancer samples to identify sample-specific outlier kinases. Next, focusing on cell lines displaying outlier expression of kinases with available therapeutics or pharmacological inhibitors, we tested their dependence on specific outlier kinases compared with non-specific targets using shRNA/siRNA and/or small molecule inhibitors to test their effects on cell proliferation. Using this approach we identified several cell line-specific dependencies as well as kinase targets showing enhanced effects with ERBB2 inhibition in breast and KRAS knockdown in pancreatic cancer cells.
Results
Delineation of cancer-specific kinome outlier profiles using transcriptome sequencing data
Taking advantage of the direct and unbiased readout of gene expression in terms of defined RNA-Seq reads, we carried out a systematic analysis of the human kinome expression in cancer. RNA-Seq based, normalized read-counts of all 468 kinases available in our transcriptome compendium, comprised of 482 samples from 25 different tissue types, revealed distinct kinases expressed at very high levels- both in absolute terms and in the context of their typical range of expression levels- in virtually all samples examined (Supplementary Table S1).
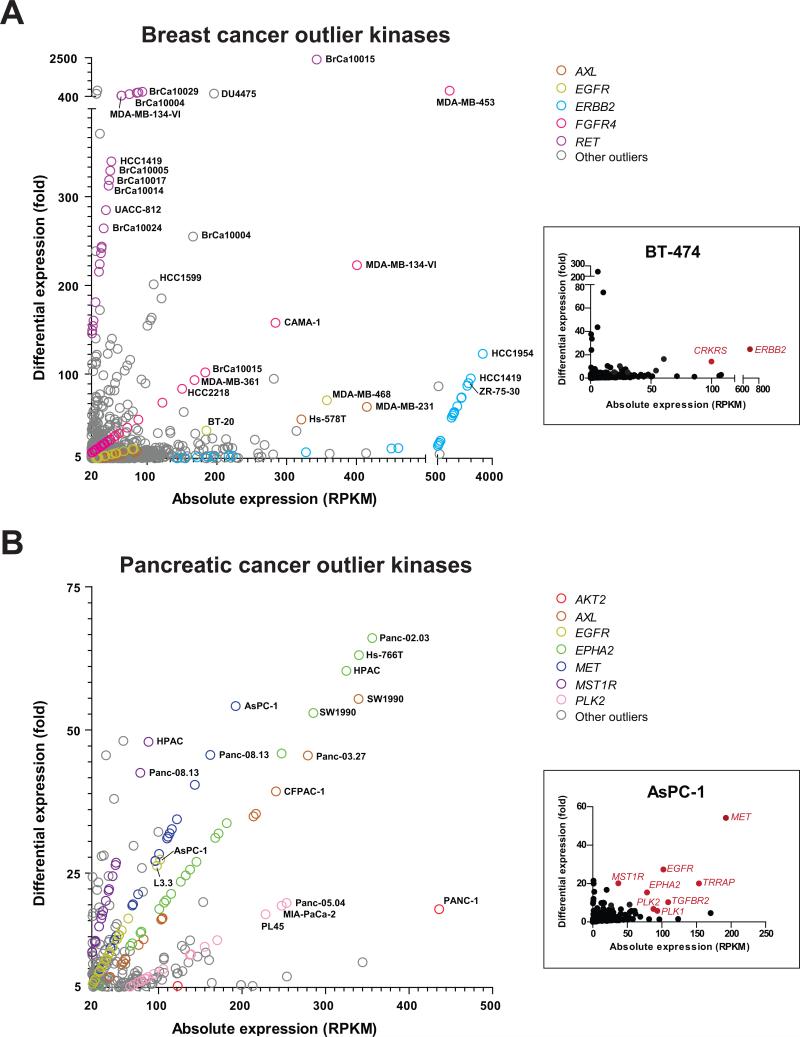
Querying individual breast cancer samples (43 cell lines and 67 tissues) for kinases that display the highest levels of absolute expression (> 20 RPKM) among all the kinases in an individual sample and the highest levels of differential expression compared to the median level of expression of the respective gene across the compendium (> 5 fold), we identified outlier kinases across the cohort of breast cancer samples (Fig. 1A; Supplementary Table S2). Additionally, each of the outliers was assessed for significant Mahalanobis distance from the center of the scatterplot distribution (χ2 test p-value < 0.05) to prioritize sample-specific kinase outliers. For example, in the breast cancer cell line BT-474, ERBB2 is the predominant outlier kinase (Fig. 1A, inset). Remarkably, using this approach, all breast cancer cell lines known to be ERBB2-positive were scored as displaying an outlier expression of ERBB2. Interestingly, many ERBB2-positive cell lines also displayed outlier expression of additional kinases like CRKRS (Fig. 1A, inset), FGFR4, and/or RET, among others (Supplementary Table S2). Similar to the well-known case of ERBB2, we hypothesized that in general, outlier kinases specific to individual cancer samples could represent additional therapeutic avenues and were thus explored further.
Figure 1. Scatter plot representation of outlier kinases in (A) breast and (B) pancreatic cancer samples.
Kinases displaying an absolute expression > 20 RPKM (reads per kilobase transcript per million total reads) and differential expression > 5 fold (versus median value across the compendium) were designated as outliers. The colored circles represent salient kinases displaying outlier expression in multiple samples. Examples of sample-specific kinome profiles are shown in the insets (BT-474 breast cancer and AsPC-1 pancreatic cancer cell lines); kinases with statistically significant outlier expression (absolute expression > 20 RPKM, differential expression > 5 fold, and p-value < 0.05) are highlighted in red.
Likewise, kinome expression data from 22 pancreatic cancer cell lines and 13 pancreatic tissue samples also revealed a set of outlier kinases specifically overexpressed in pancreatic cancers (Fig. 1B; Supplementary Table S3), with the outlier kinase profile of a representative pancreatic cancer cell line AsPC-1 depicted in the inset (Fig. 1B). Assessment of outlier kinases in pancreatic and breast cancer cohorts revealed distinct outlier kinase profiles between the two diseases. For example, common outlier kinases in breast cancer included ERBB2, FGFR4, and RET, while kinases displaying outlier expression across multiple pancreatic cancer samples included EPHA2, MET, PLK2, MST1R, and AKT2. Interestingly, AXL and EGFR demonstrated outlier expression in both pancreatic and breast cancer samples.
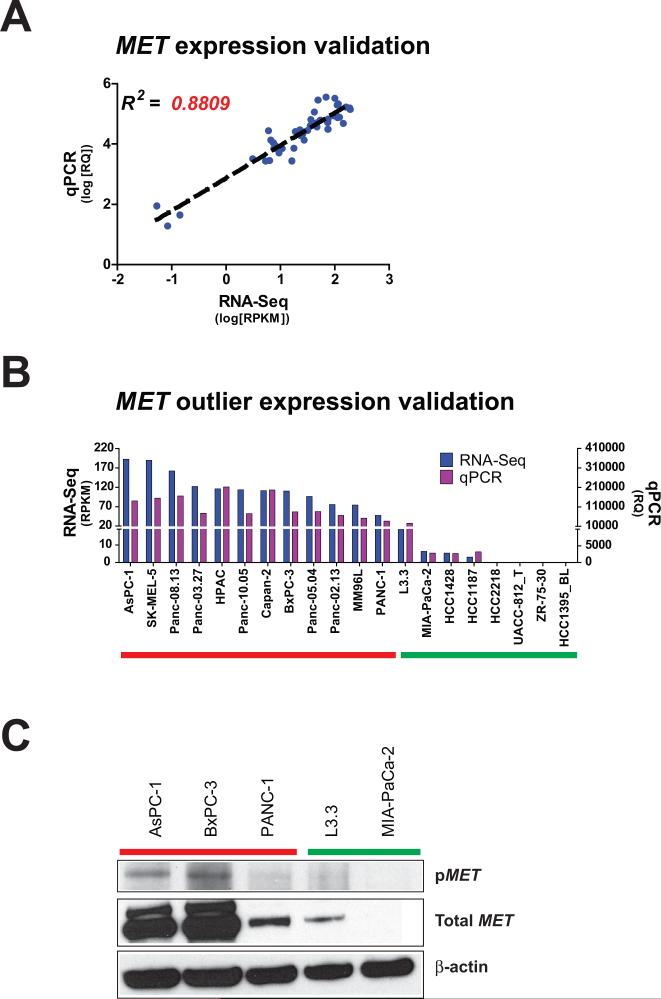
Before proceeding to test outlier kinase specific dependencies in individual cell lines, we validated the gene expression readout provided by the RNA-Seq data. First, comparing the gene expression profiles of a prostate cancer cell line DU145 across four independent RNA-Seq runs, we observed a robust correlation (R2 > 0.96) between the technical replicates (Supplementary Fig. S1A). Next, we analyzed the variance across RNA-Seq data from a breast cancer cell line MCF-7 treated with estrogen (0h, 3h, and 6h) as biological quasi-replicates. Interestingly, here too we observed an overall high correlation (R2 > 0.91), albeit less than the technical replicates (Supplementary Fig. S1B). Next, we validated the expression profiles of kinase genes derived from RNA-Seq by qPCR and western blot analyses. As an example, a strong correlation (R2 > 0.88) was observed between the levels of MET expression by RNA-Seq and qPCR, over a range of expression values across a panel of samples (Fig. 2A). Also, individual samples showing outlier expression of MET by RNA-Seq, showed distinctly higher expression by qPCR, compared to non-outlier samples (Fig. 2B). Similarly, we performed qPCR validation of RNA-Seq data from multiple samples for eight additional kinases, again demonstrating strong, statistically significant correlations with overall gene expression levels (Supplementary Fig. S2) as well as outlier calls (Supplementary Fig. S3). Furthermore, extending the correlation of outlier expression to protein levels, cell lines with outlier expression of MET were found to display higher levels of total as well as phosphorylated MET, compared to cells without outlier expression of MET (Fig. 2C). Finally, to assess the feasibility of identifying outlier kinases in cancer tissue samples in the backdrop of underlying benign stromal, vascular and immune cells, we observed a strong correlation between the RNA-Seq data and outlier calls between a primary tumor-derived xenograft tissue, DS-08-947 and its derivative cell line (Supplementary Fig. S4A, Supplementary Table S4). Similar correlation was observed between BxPC-3 and PANC-1 cell lines and xenograft tissues derived from them (Supplementary Fig. S4B).
Figure 2. Validation of RNA-Seq reads and outlier calls for MET.
(A) Log-transformed RNA-Seq expression for MET, measured as RPKM, is plotted against log-transformed gene expression, measured as relative quantity (RQ) by qPCR. Each point represents a unique sample. Dashed black line represents linear regression. R2, correlation coefficient. (B) RNA-Seq reads (blue) and qPCR gene expression (purple) for MET is plotted for 20 different samples. (C) Western blot for phospho-MET and total MET is shown for 5 samples. Samples with predicted MET outlier expression by RNA-Seq are highlighted by the red bars. Samples with predicted non-outlier expression are highlighted by the green bars.
A subset of ERBB2-positive breast cancer cell lines display outlier expression of FGFR4
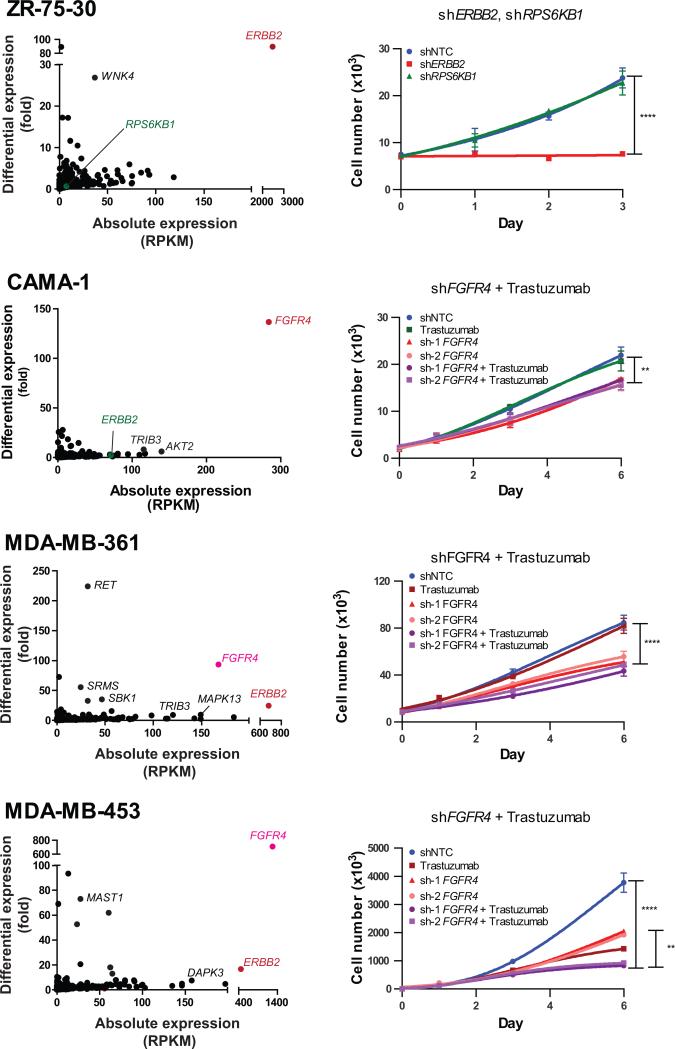
Among the ERBB2-positive breast cancer cell lines analyzed by RNA-Seq, ZR-75-30 exhibited singular outlier kinase expression of ERBB2, whose knockdown resulted in a strong growth inhibition (Fig. 3). However, knockdown of RPS6KB1, another oncogenic kinase on chromosome 17 located near the ERBB2 amplicon and overexpressed in 40-50% of breast cancers, did not affect the proliferation rate of ZR-75-30 cells, which do not show outlier expression of RPS6KB1 (Fig. 3). Many other ERBB2-positive cell lines however displayed outlier expression of additional kinases, frequently including FGFR4, such as MDA-MB-361 and MDA-MB-453 (Fig. 3), as well as MDA-MB-330, HCC202, and HCC1419 (Supplementary Table S2). To assess the dependence on the outlier expression of FGFR4 in the backdrop of ERBB2 overexpression, multiple short hairpin RNA-encoding lentiviral constructs were used to knockdown FGFR4 in MDA-MB-361 and MDA-MB-453 cells exhibiting outlier expression of both ERBB2 and FGFR4, as well as in CAMA-1, with outlier expression of FGFR4 but not ERBB2. Target knockdown for all siRNA and shRNA experiments were assessed by qPCR and/or western blot (Supplementary Fig. S5A-H). Remarkably, knockdown of FGFR4 resulted in decreased cell proliferation in all the three cell lines with FGFR4 outlier expression (Fig. 3), while treatment of these cells with ERBB2 targeting trastuzumab, had no effect the proliferation of CAMA-1 and MDA-MB-361 cells. On the other hand, MDA-MB-453 cells showed diminished cell proliferation rates independently upon FGFR4 knockdown as well as trastuzumab treatment, and showed an additive effect upon combined treatment.
Figure 3. Sample-wise outlier kinases in ERBB2-positive breast cancer cell lines.
(Left) The scatter plots display kinome expression profiles of individual breast cancer cell lines. Kinases with (red/pink) and without (green) outlier expression that were targeted for knockdown are shown in color. Labels in black denote additional kinases with outlier expression. (Right) Growth curves show the effect of targeting outlier (ERBB2) versus non-outlier (RPS6KB1) kinases in ZR-75-30 cells and the effects of trastuzumab and/or knockdown of the outlier FGFR4 in CAMA-1, MDA-MB-361, and MDA-MB-453 cells. Values represent mean ± SD. **, P < 0.01; ****, P < 0.0001.
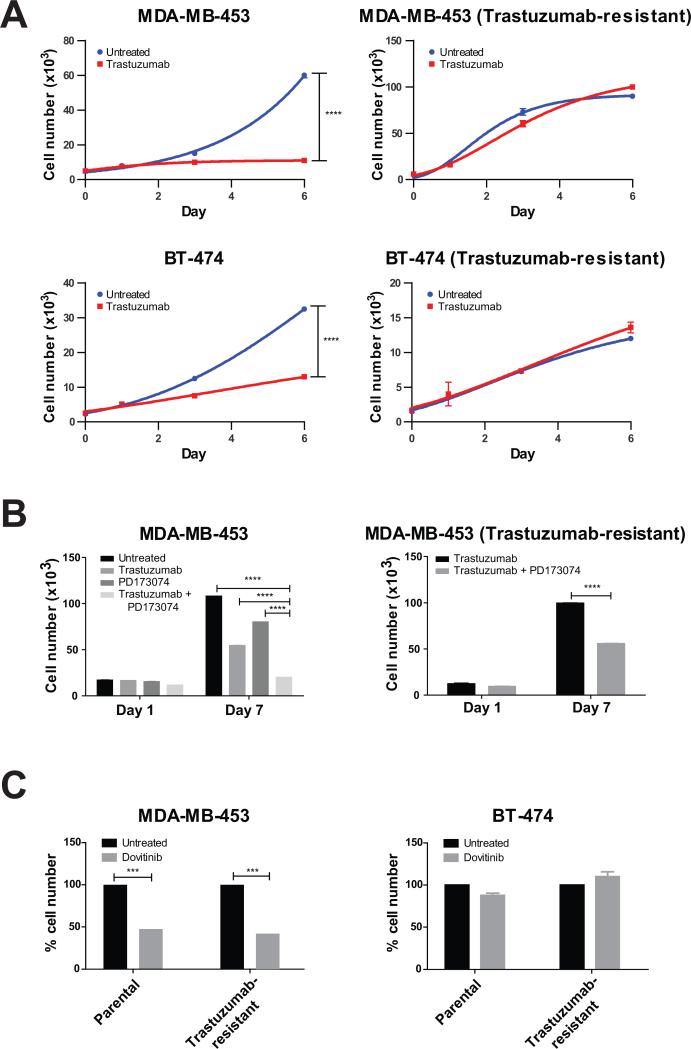
To further examine the dependence of a subset of ERBB2-positive cells on FGFR4, we generated trastuzumab-resistant sub-lines of MDA-MB-453 and BT-474, an ERBB2-positive breast cancer cell line that does not exhibit FGFR4 outlier expression (Fig. 4A). Consistent with the experiments involving trastuzumab and shRNA-mediated knockdown of FGFR4 (Fig. 3), MDA-MB-453 cells were found to be independently responsive to both trastuzumab and PD173074, a small molecule inhibitor of FGFR, while a combined treatment with both of these reagents provided the strongest effect on cell proliferation (Fig. 4B, left). Interestingly, MDA-MB-453 cells, grown to be resistant to trastuzumab, continued to display responsiveness to PD173074 (Fig. 4B, right), suggesting that FGFR4 represents an independent therapeutic target in a subset of ERBB2-positive breast cancer cells. Similar results were obtained with another FGFR inhibitor Dovinitib, which significantly decreased cell proliferation in both the MDA-MB-453 parental and trastuzumab-resistant sub-line (Fig. 4C, left) but did not affect the BT-474 parental or trastuzumab-resistant sub-line, neither of which display FGFR4 outlier expression (Fig. 4C, right). Next, we carried out dose-response experiments using specific pharmacologic inhibitors against outlier kinases (Supplementary Fig. S6A-C). Cell lines exhibiting outlier expression of FGFRs displayed a dose-dependent response to PD173074 and Dovitinib, with significantly lower IC50 values as compared to cell lines without outlier expression (Supplementary Fig. S6A, B). Taken together, these results suggest that a subset of ERBB2-positive breast cancers that display outlier expression of FGFR4 may specifically respond to combined treatment with ERBB2 and FGFR inhibitors more effectively compared to ERBB2-directed therapy alone.
Figure 4. Trastuzumab-resistant cell lines respond to targeting of the outlier kinase FGFR4.
(A) The growth curves show the effect of trastuzumab treatment on MDA-MB-453 and BT-474 (left) and their trastuzumab-resistant sublines (right). (B) The bar graphs demonstrate the individual and combined effects of trastuzumab and the FGFR inhibitor PD173074 on cell proliferation in MDA-MB-453 (left) and its trastuzumab-resistant subline (right). (C) The bar graphs display the effect of the FGFR inhibitor Dovitinib on parental and trastuzumab-resistant sublines of MDA-MB-453 (with FGFR4 outlier expression) and BT-474 (without FGFR4 outlier expression) on day 5. Values represent mean ± SD. ***, P < 0.001; ****, P < 0.0001.
Pancreatic cancer cell lines are sensitive to knockdown of cell-specific outlier kinases
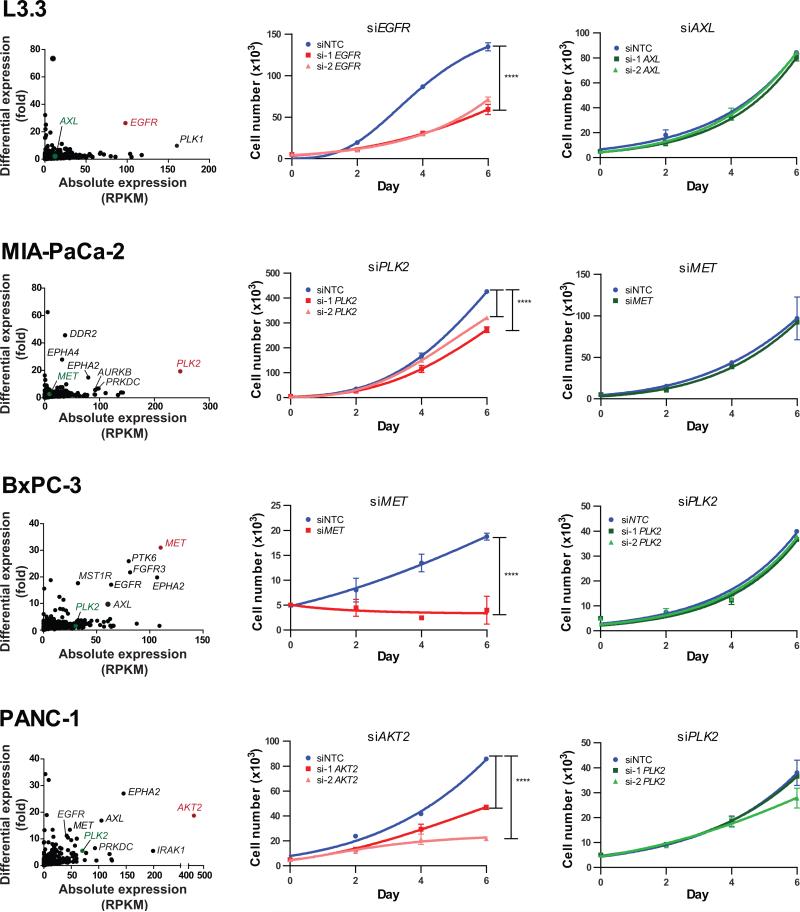
We next extended our kinome outlier analysis to pancreatic cancer, a tumor type critically lacking in rational therapeutic options, particularly in the realm of actionable kinases. Kinome expression profiles of individual pancreatic cancer cell lines were used to identify sample-specific outlier kinases (Fig. 5, left). The pancreatic cancer cell lines were then tested for effects on cell proliferation following siRNA-based knockdown of sample-specific outlier and non-outlier kinases. Knockdown of the sample-specific outlier kinases, for example, EGFR in L3.3, PLK2 in MIA-PaCa-2, MET in BxPC-3 and AKT2 in PANC-1 cells, inhibited the proliferation of respective cells (Fig. 5, middle). A similar growth inhibition was observed following knockdown of MET in HPAC and AXL in Panc-08.13 and PL45 cells (Supplementary Fig. S7). Conversely, knockdown of the non-outlier kinases AXL in L3.3, MET in MIA-Paca-2, PLK2 in BxPC-3 and PANC-1 cells did not significantly affect cell growth (Fig. 5, right). Also, L3.3 cells remained unaffected by knockdown of the non-outlier PLK2 (Supplementary Fig. S7). These observations strongly support the notion that outlier kinases represent specific therapeutic targets in individual cancer samples.
Figure 5. Pancreatic cancer cell lines are sensitive to knockdown of outlier kinases.
(Left) Scatter plots display kinome profiles of select pancreatic cancer cell lines; kinases targeted for knockdown are shown in color (red, outliers; green, non-outliers). Labels in black denote additional kinases with outlier expression. The growth curves display the effects of siRNA-mediated knockdown of sample-specific outliers (middle) and non-outliers (right) for each cell line. Values represent mean ± SD. ****, P < 0.0001.
Notably, knockdown of the outlier kinase PLK2 in MIA-PaCa-2 cells did not have as profound an effect on cell proliferation as outlier kinase-targeting in many other samples (Fig. 5, middle). We hypothesized that this could be due to a pervasive influence of oncogenic KRAS activity in these cells. To test this, next we analyzed the effect of KRAS knockdown in pancreatic cancer cell lines with PLK outlier expression.
Outlier expression of Polo-like Kinases marks a subset of KRAS-dependent pancreatic cancer cells
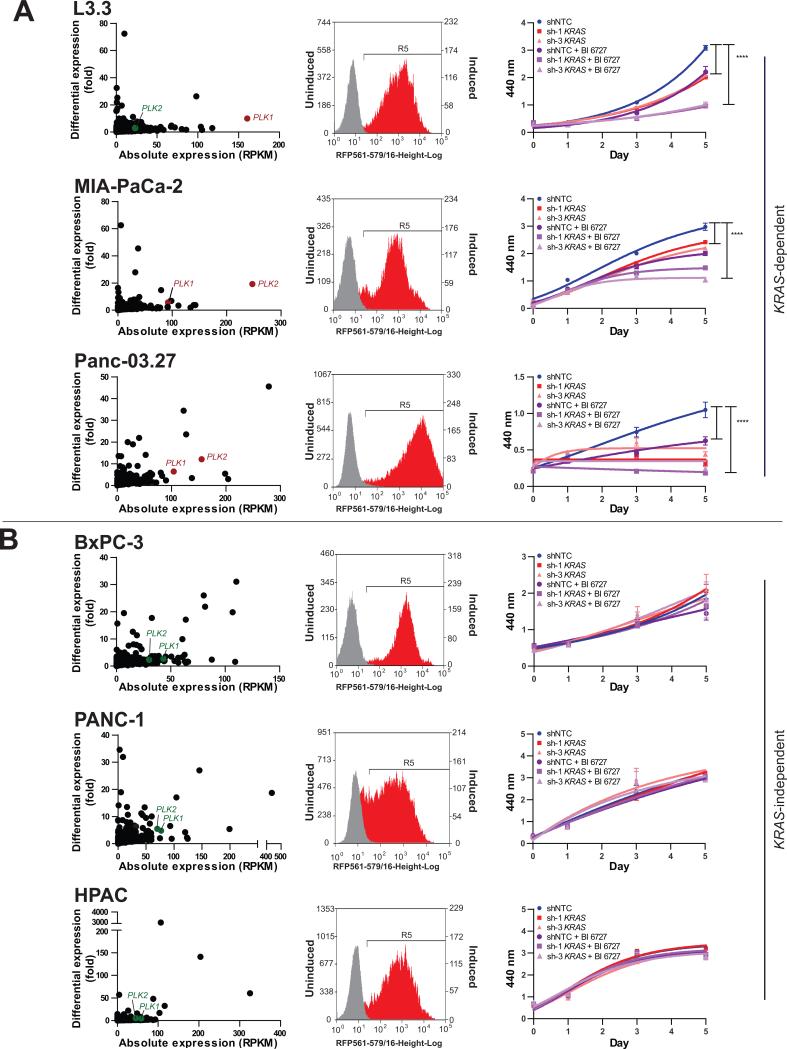
A panel of pancreatic cancer cell lines with and without PLK outlier expression was stably transduced with two independent inducible shRNAs against KRAS and assessed for sensitivity to KRAS knockdown and/or the PLK inhibitor BI 6727 (Fig. 6). Following induction by doxycycline, the cells expressing KRAS shRNAs were distinguished by red fluorescence, resulting from the RFP tag co-expressed with the shRNA (Fig. 6, middle). KRAS knockdown efficiency of approximately 50% or more was obtained in all the cells tested (Supplementary Fig. S5H). Of the cell lines tested, knockdown of KRAS significantly inhibited the proliferation of L3.3, MIA-PaCa-2, and Panc-03.27, which all harbor oncogenic mutations in KRAS and were therefore designated as KRAS-dependent (Fig. 6A). BxPC-3 cells, which have wild-type KRAS, as well as HPAC and PANC-1 cells, which have mutant KRAS, were not affected by KRAS knockdown and were therefore categorized as KRAS-independent (Fig. 6B). Incidentally, all three PLK outlier cell lines tested here, L3.3, MIA-PaCa-2, and Panc-03.27, were found to be in the KRAS-dependent category based on their reduced proliferation upon KRAS knockdown (Fig. 6A). Furthermore, treatment with the PLK inhibitor BI 6727 significantly inhibited proliferation in cell lines with PLK outlier expression (Fig. 6A, right) but had no effect in cell lines without PLK outlier expression (Fig. 6B, right). The decrease in cell proliferation following BI 6727 treatment was associated with increased apoptosis, as measured by flow cytometry of Annexin V/Propidium Iodide-stained cells (Supplementary Fig. S8A). Finally, treatment with BI 6727 in combination with knockdown of KRAS enhanced the inhibition of cell proliferation in the KRAS-dependent, PLK outlier cells (Fig. 6A, right) but had no effect in the KRAS-independent cells without PLK outlier expression (Fig. 6B, right). Investigating the likely reason for the lack of sensitivity to KRAS knockdown in a subset of pancreatic cancer cells harboring oncogenic KRAS, we observed that following KRAS knockdown, the levels of phospho-ERK, one of the major downstream effector proteins in the RAS signaling pathway, were reduced in the KRAS-dependent cell lines, L3.3 and MIA-PaCa-2, but not in the KRAS-independent cell line PANC-1 (Supplementary Fig. S8B), suggesting that ERK activity in PANC-1 cells may be sustained by other convergent pathways. Notably, the KRAS-independent cell lines BxPC-3 and PANC-1 did respond to inhibition of their respective outlier kinases, both in vitro (Fig. 5, middle) as well as in vivo, as described below.
Figure 6. Knockdown of KRAS combined with PLK inhibition reduces cell proliferation in indicated KRAS-dependent cell lines (A) but not in KRAS-independent cell lines (B).
The scatter plots demonstrate the absolute and differential expressions of PLK1 and PLK2 for each cell line (left). The flow cytometric profiles of doxycycline-induced cells expressing KRAS shRNA with RFP expression (red) versus un-induced cells (gray) are displayed (middle).The growth curves show the individual and combined effects of KRAS shRNA and the PLK inhibitor BI 6727, using WST-1 assay measured at absorbance 440 nm (right). Values represent mean ± SD. ****, P < 0.0001.
Inhibition of outlier kinases inhibits the growth of pancreatic cancer cell line xenografts
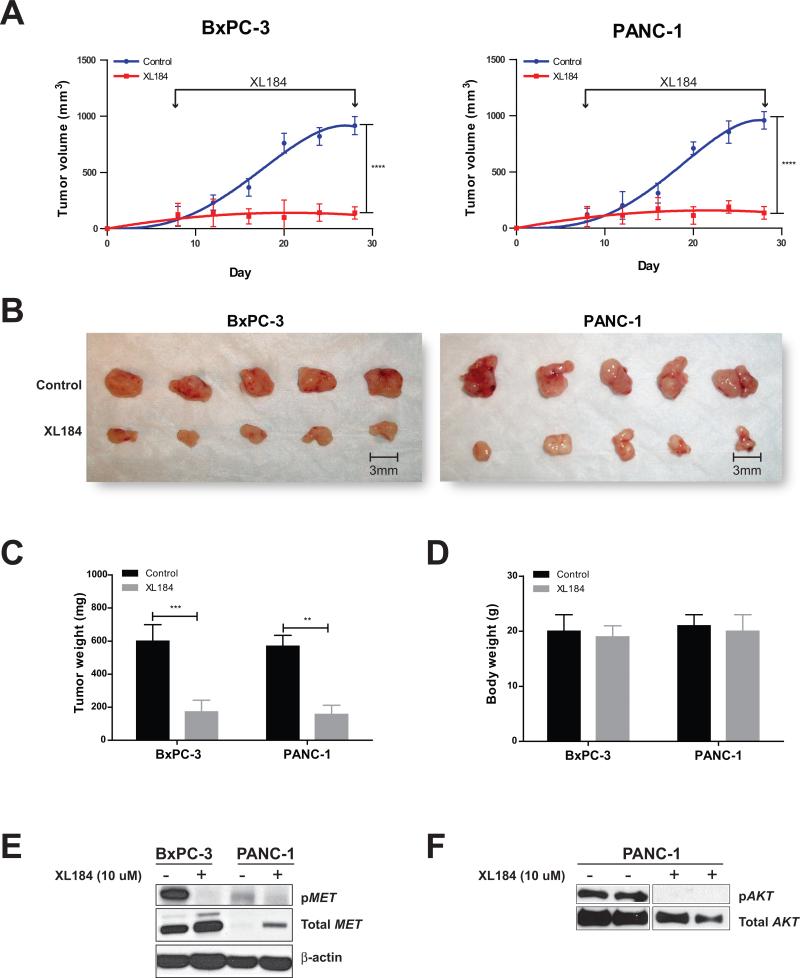
To test the effect of inhibiting sample-specific outlier kinases in vivo, we treated orthotopic tumor xenografts of two KRAS-independent pancreatic cancer cell lines BxPC-3 and PANC-1 established in NOD/SCID mice with the MET inhibitor XL184. BxPC-3 cells and to a lesser but significant degree, PANC-1 cells, were found to have MET outlier expression by RNA-Seq, which was validated by qPCR and western blot analyses (Fig. 2). Notably, both of these cell lines also displayed a dose dependent response to XL184, in vitro, with significantly lower IC50 values compared to L3.3 cell line that does not have outlier expression of MET (Supplementary Fig. S6C). Consistent with our hypothesis of dependence on outlier kinases, growth of both BxPC-3 and PANC-1 xenografts were also significantly inhibited by treatment with XL184, as measured by tumor volume and weight (Fig. 7A-C). Of note, there was no significant difference in body weight of XL184 treated and untreated mice, suggesting that the effective dose of the inhibitor caused no measurable toxicity in vivo (Fig. 7D).
Figure 7. XL184 treatment suppresses tumor growth in BxPC-3 and PANC-1 pancreatic cancer xenografts.
(A) The growth curves demonstrate the effect of the MET inhibitor XL184 on tumor growth in BxPC-3 and PANC-1 xenografts. (B) BxPC-3 and PANC-1 xenograft tumors after 3 weeks of XL184 treatment are shown as compared to the controls. The bar graphs display tumor weight (C) and total body weight (D) after 3 weeks of XL184 treatment. Values represent mean ± SE. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (E) Immunoblot results showing the effect of XL184 treatment on phospho-MET (pMET) in BxPC-3 and PANC-1 cells. (F) Immunoblot results showing the effect of XL184 treatment on phospho-AKT (pAKT) level in the PANC-1 orthotopic xenograft.
The specificity of response to the MET inhibitor XL184 was analyzed by western blot which demonstrated a sharp decrease in phospho-MET levels in BxPC-3 and to a relatively lesser extent in PANC-1 cells following treatment with XL184 (Fig. 7E). Considering that AKT2 represents the predominant outlier kinase in PANC-1 cells (Mahalanobis distance 217.6, p-value ~ 0; Supplementary Table S3), lending significant dependence on AKT2 (Fig. 5), we queried whether the profound inhibitory effect of XL184 on PANC-1 xenografts was also mediated through non-specific targeting of AKT. Western blot analysis of PANC-1 xenograft tumor lysates revealed a markedly decreased level phospho-AKT following XL184 treatment (Fig. 7F). This supports the notion that XL184 suppresses PANC-1 proliferation through inhibition of both AKT and MET signaling. Thus, PANC-1 represents an example of a cancer sample showing dependency on multiple actionable outliers that may respond to a combinatorial therapeutic option or appropriate pan kinase inhibitors.
Discussion
The advent of high-throughput sequencing enables a comprehensive characterization of the genomic and transcriptomic landscape of individual cancer samples, inexorably leading to the challenge of defining and prioritizing clinically relevant findings to translate into improved diagnostic and therapeutic options (27, 28). Clinical sequencing of cancers aims to identify actionable genomic aberrations and match patients with available therapies. Protein kinases, being central to biological and disease processes, including cancer, and being therapeutically targetable, comprise a large proportion of available and potential targets; thus any novel disease-specific kinase aberrations are of great clinical interest. This study proposes and tests the hypothesis that specific kinases showing outlier expression in individual cancer samples impart ‘dependence’ on the cells, which may be targeted in combination with existing treatment modalities. Importantly, a case is made for considering the entire profile of kinome aberrations to prioritize potentially effective targets.
The ‘sample-centric’ analysis of kinome expression revealed unique profiles of outlier kinases that were tested for dependency. The receptor tyrosine kinase ERBB2 overexpressed in 20-30% of breast cancers confers a more aggressive phenotype, increased metastasis, and worse patient prognosis (29, 30). In our outlier kinase analysis, several well-known ‘ERBB2-positive’ breast cancer cell lines including MDA-MB-361 and MDA-MB-453 were found to display outlier expression of ERBB2 as expected, but frequently also an outlier expression of the therapeutic target FGFR4. Notably, a survey of microarray-based gene expression data in Oncomine (31, 32) also displayed a subset of ERBB2-positive primary breast cancer samples with outlier expression of FGFR4 (data not shown), emphasizing the clinical relevance of our observations. Targeting outlier FGFR4 in ERBB2-positive breast cancer samples was found to confer independent as well as additive inhibitory effects upon their combined knockdown (Fig. 3), highlighting the potential of combining two or more outlier kinase targets in treating cancer, even in cases with a predominant driver such as ERBB2. Interestingly, we also observed that the ERBB2-positive MDA-MB-453 cells grown resistant to trastuzumab treatment continued to remain dependent on FGFR4 and responded to FGFR inhibitors (Fig. 4). In clinical trials with ERBB2-positive metastatic breast cancer, 50 to 74% patients have been reported to be not responsive to trastuzumab monotherapy or in combination with chemotherapy (33, 34). Our results suggest that the ERBB2-positive breast cancers may be partly dependent on additional drivers, such as FGFR4, RET, EGFR, and MET, which may sustain these cancers following therapeutic abrogation of ERBB2 activity. Another important corollary to our observations is that combinatorial targeting of ERBB2 and additional outlier kinases at the outset may be much more effective than approaching a single target at a time, a concept that warrants further study. Further, each cancer sample needs to be investigated individually to rationally determine patient-specific unique target combinations.
Next, we extended the approach of nominating sample-specific outlier kinases to pancreatic cancer, which is characterized by a bleak prognosis due to presentation at an advanced stage and resistance to traditional chemotherapy and radiation in the setting of its pancreatic cancer sanctuary, encompassing tumor stroma, extracellular matrix, tumor infiltrating immune cells, and cancer stem cells. Given the paucity of effective targets in pancreatic cancer, the strong response of pancreatic cancer cell lines to knockdown/inhibition of a priori designated outlier kinases is a promising lead. Our results also underscore the importance of matching sample-specific actionable targets with the appropriate therapeutics. For example, targeting MET was found to be more effective in pancreatic cancer cell lines with MET outlier expression compared to non-outlier samples. Notably, many of our experimental results are consistent with several anecdotal studies using kinase inhibitors against EGFR, MET and AKT2 (35-39).
We also examined the effect of targeting sample-specific outlier kinases in conjunction with the oncogenic KRAS mutation that is present in virtually all cases of pancreatic cancer. Consistent with previous reports (40-42), we observed that only a subset of KRAS-mutant cells display KRAS dependency. Using tet-inducible shKRAS stable cell lines, we determined L3.3, MIA-PaCa-2, and Panc-03.27 cells to be KRAS-dependent, while BxPC-3 cells (the only pancreatic cancer cell line in our panel with wild-type KRAS) as well as PANC-1 and HPAC were KRAS-independent. Interestingly, comparing our results with the published literature, we noted a general lack of consensus in the “KRAS-dependence” status of pancreatic cancer cell lines (10, 14, 40-45). For example, while two prior studies using siRNA-mediated knockdown of KRAS in the KRAS-mutant cell line MIA-PaCa-2 designated it as KRAS-dependent, based on reduced cellular proliferation, invasion, and colony formation assays (10, 44), more recently, Collisson et al. observed no significant effect on proliferation in MIA-PaCa-2 cells transduced with shKRAS lentivirus (40). Similarly, PANC-1 was identified as KRAS-dependent in four different studies by both siRNA/shRNA-mediated knockdowns, as assessed by cellular proliferation, colony formation, invasion, and xenograft tumor growth (10, 14, 43, 44), while three studies found PANC-1 to be KRAS-independent by shRNA-mediated knockdown and farnesyl transferase inhibitor treatment using similar in vitro assays (40-42). Conversely, the KRAS-wild type cell line BxPC-3 has been consistently reported to be KRAS-independent (14, 44), similar to our findings. Interestingly, HPAC was described as KRAS-dependent by Collisen at al (40) but was found to be KRAS-independent in our assays. No published references were found for L3.3 and Panc-03.27, which we report as KRAS-dependent.
Several KRAS synthetic lethal screens and DNA microarray analyses have been used to describe genes/gene signatures associated with KRAS-dependence (12-14, 40, 41, 46) and include kinases such as PLK1, MST1R, and SYK (12, 40, 41). Interestingly, we observed outlier expression of PLK to be restricted to KRAS-dependent cells, and these cells showed higher sensitivity to the pan-PLK inhibitor BI 6727 both alone and in combination with KRAS knockdown, as compared to KRAS-independent cells. Previously, Luo et al. identified PLK1 as a RAS synthetic lethal in a lung and a colorectal cancer cell line, although they did not test any pancreatic cancer cell lines. Our results additionally demonstrate that cells only respond to the pan-PLK inhibitor BI 6727 if they have outlier expression of either PLK1 or PLK2 (Fig. 6A, B). This highlights the importance of using therapeutic targets in a sample-specific manner.
Overall, our study provides a generalizable metric to define and prioritize personalized target spectra specific to individual tumors. The recent report of a remarkably successful treatment of a patient with acute lymphoblastic leukemia, with sunitinib targeting “a wildly active” expression of FLT3 kinase identified by RNA-Seq, when whole genome sequencing failed to identify any actionable aberrations (47), provides an anecdotal yet powerful illustration of the potential application of systematic identification of outlier kinases proposed in our study.
Methods
Kinome analysis
Transcriptome sequencing data from 482 cancer and benign samples from 25 different tissue types previously generated on Illumina GA and GAII platforms, were mapped using Bowtie (48) against University of California Santa Cruz (UCSC) Genome Browser genes in the hg18 human genome assembly (49). Unique best match hit sequences normalized for the number of reads per kb transcript per million total reads in the given sequencing run (RPKM) (16) were used to generate a gene expression data matrix for the entire compendium (24). The expression data for the complete list of kinase genes (50) were used to identify “outlier kinases” in individual samples based on their absolute expression within the sample and differential expression (defined as absolute expression divided by median expression level of that gene across the compendium). GraphPad Prism software was used to generate kinome expression profiles for each sample, plotting absolute expression versus differential expression for all kinases.
Statistical significance of outlier expression was quantified using a Mahalanobis distance metric [D2 = (x - μ)’Σ-1(x - μ); Σ = covariance matrix, D = Mahalanobis distance of the point x to the mean μ] (51, 52), to measure the “distance” of each kinase's absolute and differential expression from the center of the scatter plot distribution. P-values were calculated assuming a chi-squared distribution, with 2 degrees of freedom. Kinases with absolute expression > 20 RPKM, differential expression > 5 fold, and p-value < 0.05 were nominated as having “outlier expression”. R language (53) was used to perform statistical analysis.
Cell culture
All human breast and pancreatic cancer and benign epithelial cell lines were purchased from the American Type Culture Collection (ATCC), except the benign immortalized pancreatic epithelial cell line HPDE and the xenograft cell lines derived from primary pancreatic adenocarcinoma tissues, which were provided by D.M.S. The pancreatic adenocarcinoma cell line L3.3 was obtained from the University of Texas MD Anderson Characterized Cell Line Core. All cell lines were grown in recommended culture media and maintained at 37°C in 5% CO2. To ensure cellular identities, a panel of cell lines was genotyped at the University of Michigan Sequencing Core using Profiler Plus (Applied Biosystem) and compared with the short-tandem repeat (STR) profiles of respective cell lines available in the STR Profile Database (ATCC).
Transcript knockdowns and cell proliferation assays
ON-TARGETplus siRNA against AKT2, AXL, EGFR, MET, PLK2, and non-targeting control (siNTC) from Dharmacon (Supplementary Table S5A) were used at 100 nM. Cells were transfected in 6-well plates at a density of 50,000 cells per well using Oligofectamine (Invitrogen), as per the manufacturer's protocol. Transfection was repeated 24 hours later, the cells grown for an additional 48 hours, and re-plated at a density of 5,000 cells per well in 24-well plates. Cells were counted over a period of 1 to 6 days using Beckman Coulter cell counter (Beckman Coulter). Transient transductions with shRNA against ERBB2, RPS6KB1, FGFR4, or non-targeting control (shNTC) were carried out in 6-well plates in the presence of 8 ug/mL hexadimethrine bromide (Polybrene; Sigma). For trastuzumab (Herceptin; Roche) experiments, cells were grown for 3 days in 24-well plates with and without trastuzumab (100 ug/mL), in combination with the FGFR inhibitors PD173074 (TOCRIS Bioscience) at 1 uM or TKI-258 (Dovitinib; Selleck Chemicals) at 0.1 uM. Trastuzumab-resistant cell lines were generated from MDA-MB-453 and BT-474 by maintaining the cells in the continuous presence of 100 ug/mL trastuzumab over 1 month. Cell proliferation assays were carried out over a period of 1 to 7 days using Beckman Coulter cell counter, and growth curves were plotted using GraphPad Prism software. Statistical comparisons were conducted using one-way ANOVA.
Generation of stable cell lines with doxycycline-inducible KRAS-shRNA lentiviral constructs
Doxycycline-inducible shRNAmir-TRIPZ lentiviral constructs targeting KRAS or non-targeting control (shNTC) (Open Biosystems) tagged with red fluorescence protein (RFP) were used to transduce a panel of pancreatic cell lines in the presence of 8 ug/mL Polybrene (Supplementary Table S5A). Forty-eight hours post-transduction, cells were selected in medium containing 1 ug/mL puromycin (Invitrogen) for 4 days. shRNA expression was induced by growing cells in medium containing 1 μg/mL doxycycline (Sigma) for 72 hours. The enrichment of stable cells and efficiency of shRNA induction were assessed by measuring the percentage of cells displaying red fluorescence by flow cytometry (FACSAria Cell Sorter BD Biosciences). Experiments with stable cell lines were performed in the presence of 1 ug/mL doxycycline, refreshed daily. Experiments with the PLK inhibitor BI 6727 (Volasertib; Selleck Chemicals) were carried out with cells plated in 96-well culture plates at a density of 3000 to 4000 cells/well and treated with 10 nM BI 6727 or DMSO. This concentration was selected based on IC50 values calculated from prior proliferation assays using 1 to 500 nM BI 6727 (data not shown). At 0, 1, 3, and 5 days following drug treatment, viable cells were quantified using WST-1 reagent (Roche) and absorbance measured at 440 nm, as per the manufacturer's protocol. Growth curves were plotted using GraphPad Prism software. Statistical comparisons were conducted using oneway ANOVA.
Western Blot analysis
Cell or tissue lysates were separated on 4-12% SDS polyacrylamide gels (Novex) and blotted on PVDF membranes (Amersham) by semi-dry transfer. Antibodies to FGFR4 (Santa Cruz); phospho-AKT, total AKT, phosho-ERK, total ERK, phospho-MET, and total MET (Cell Signaling) were used at 1:1000 dilutions for standard immunoblotting and detection by enhanced chemiluminescence (ECL Prime), as per the manufacturer's protocol. For phospho-MET blots, cells treated with 10 ug XL184 for 12 hours were stimulated with 100 ng/ml human recombinant HGF (Invitrogen) for 1 hour before harvesting in RIPA buffer.
Quantitative real-time PCR assay
RNA was isolated from cell lysates by the RNeasy Micro Kit (Qiagen), and cDNA was synthesized from 1 ug RNA, using SuperScript III (Invitrogen) and Random Primers (Invitrogen), as per the manufacturer's protocol. Quantitative real-time PCR (qPCR) was carried out on StepOne Real Time PCR system (Applied Biosystems) using gene-specific primers designed with Primer-BLAST (Supplementary Table S5B-C) and synthesized by IDT Technologies. Validation of RNA-seq results was carried out using TaqMan Universal PCR Master Mix II with uracil-N-glycosylase (Applied Biosystems) and Universal ProbeLibrary System probes (Roche) following manufacturer's protocol. Validation of siRNA-/shRNA-mediated knockdown was carried out using Fast SYBR Green Master Mix (Invitrogen), as per the manufacturer's protocol. qPCR data were analyzed using relative quantification method and plotted as average fold-change compared to the control. GAPDH was used as an internal reference. For qPCR validation studies, GraphPad Prism software was used to perform linear regression and calculate R2 correlation coefficients.
Dose response
Experiments with FGFR inhibitors PD173074 and Dovitinib and MET inhibitor XL184 were carried out with cells seeded at a density of 3000 to 4000 cells/well plated in 96-well culture plates and treated with concentrations from 100 uM to 0.1 uM. WST-1 assay (Roche) was performed after 72 hours and readings were recorded at 440 nm. GraphPad Prism software was used to generate non-linear regression curves and calculate IC50 values.
Apoptosis assay
Apoptosis assay was carried out using ApoScreen Annexin V Apoptosis Kit (Southern Biotech), as per the manufacturer's protocol. Briefly, cells treated for 48 hours with DMSO or increasing concentrations of BI 6727 were washed with cold PBS, suspended in cold 1X binding buffer, stained with Annexin V and Propidium Iodide (PI), and subjected to flow cytometry by FACSAria Cell Sorter (BD Biosciences). Results were analyzed and plotted using Summit 6.0 Software (Beckman Coulter).
In Vivo Tumorigenicity Assay
Six-week-old male NOD/SCID mice (Taconic) were housed under pathogen-free conditions approved by the American Association for Accreditation of Laboratory Animal Care in accordance with current regulations and standards of the US Department of Agriculture and Department of Health and Human Services. Animal experiments were approved by the University of Michigan Animal Care and Use Committee and performed in accordance with established guidelines. Mice anesthetized with an intra-peritoneal injection of xylazine (9 mg/kg) and ketamine (100 mg/kg body weight) were implanted with 1×106 BxPC-3 or PANC-1 cells suspended in 50 uL 1:1 mixture of Media 199 and Matrigel (BD Biosciences) injected subcutaneously into their flanks using a 30-gauge needle. When tumor size reached 0.4 mm, mice were randomized into control and treatment groups (n = 8 per group). The MET inhibitor XL184 (Exelixis Chemicals) was orally administered at 30 mg/kg body weight twice per week for three weeks. Tumor growth was monitored weekly. Tumor caliper measurements were converted into tumor volumes using the formula: ½[length × (width)2] mm3 and plotted using GraphPad Prism software. At three weeks of treatment, mice were weighed, and then euthanized and the tumors harvested. Statistical comparisons were conducted using one-way ANOVA.
Supplementary Material
Statement of Significance.
Various breast and pancreatic cancer cell lines display sensitivity to knockdown or pharmacological inhibition of sample-specific outlier kinases identified by high-throughput transcriptome sequencing. Outlier kinases represent personalized therapeutic targets that could improve combinatorial therapy options.
Acknowledgments
The authors thank Terrence Barrette, Michael Quist, Robert Lonigro, and Sheeba Powar for bioinformatics help, Mark Hynes for help with animal work, Irfan A. Asangani and Filip Bednar for useful discussions. Trastuzumab (Herceptin; Roche) was kindly provided by Dr. Max Wicha (University of Michigan Cancer Center).
Grant Support
This work was supported in part by the National Institute of Health (NIH) 5-R21-CA-155992-02 (C.K.-S.), NIH 2T32CA009672-21 (I.W.), NIH R01CA131045-01(DMS), NIH P50CA130810-1A (D.M.S.) and the Department of Defense Era of Hope grant BC075023 (A.M.C.). D.M.S. is also supported by the Rich Rogel Fund for Pancreatic Cancer Research. A.M.C. is supported by the Doris Duke Charitable Foundation Clinical Scientist Award. A.M.C. is an American Cancer Society Research Professor and A. Alfred Taubman Scholar. C.K.-S. is supported by University of Michigan GI SPORE Career Development Award.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interests were disclosed.
Authors’ Contributions
Conception and design: Kumar-Sinha C., Chinnaiyan A.M.
Development of methodology: Kumar-Sinha C., Shankar S.
Acquisition of data: Kothari V., Wei I., Shankar S., Kalyana-Sundaram S., Wang L., Ma L.W., Vats P., Grasso C., Robinson D., Wu Y.M., Cao X., Kumar-Sinha C.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Wei I., Shankar S., Kothari V., Kalyana-Sundaram S., Vats P., Grasso C., Kumar-Sinha C.
Writing, review, and/or revision of the manuscript: Kumar-Sinha C., Kothari V., Wei I., Shankar S., Kalyana-Sundaram S., Simeone D.M., Chinnaiyan A.M.
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Kothari V., Wei I., Shankar S., Kalyana-Sundaram S., Grasso C., Chinnaiyan A.M., Kumar-Sinha C.
Study supervision: Kumar-Sinha C., Chinnaiyan A.M.
Carried out all experiments: Kothari V., Wei I., Shankar S., Wang L., Ma L.W., Robinson D., Kumar-Sinha C.
References
- 1.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–80. doi: 10.1158/0008-5472.CAN-07-3293. discussion 80. [DOI] [PubMed] [Google Scholar]
- 2.Baselga J, Arribas J. Treating cancer's kinase ‘addiction’. Nat Med. 2004;10:786–7. doi: 10.1038/nm0804-786. [DOI] [PubMed] [Google Scholar]
- 3.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130–7. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camidge DR, Doebele RC. Treating ALK-positive lung cancer-early successes and future challenges. Nat Rev Clin Oncol. 2012 doi: 10.1038/nrclinonc.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning BD. Challenges and opportunities in defining the essential cancer kinome. Sci Signal. 2009;2:pe15. doi: 10.1126/scisignal.263pe15. [DOI] [PubMed] [Google Scholar]
- 7.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–88. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zanini N, Masetti M, Jovine E. The definition of locally advanced pancreatic cancer. British journal of cancer. 2010;102:1306–7. doi: 10.1038/sj.bjc.6605630. author reply 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardenes HR, Chiorean EG, Dewitt J, Schmidt M, Loehrer P. Locally advanced pancreatic cancer: current therapeutic approach. Oncologist. 2006;11:612–23. doi: 10.1634/theoncologist.11-6-612. [DOI] [PubMed] [Google Scholar]
- 10.Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA. Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-directed therapy. Mol Cancer Res. 2005;3:413–23. doi: 10.1158/1541-7786.MCR-04-0206. [DOI] [PubMed] [Google Scholar]
- 11.Strimpakos A, Saif MW, Syrigos KN. Pancreatic cancer: from molecular pathogenesis to targeted therapy. Cancer Metastasis Rev. 2008;27:495–522. doi: 10.1007/s10555-008-9134-y. [DOI] [PubMed] [Google Scholar]
- 12.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–48. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–12. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–34. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 17.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Lovci MT, Kwon YS, Rosenfeld MG, Fu XD, Yeo GW. Determination of tag density required for digital transcriptome analysis: application to an androgen-sensitive prostate cancer model. Proc Natl Acad Sci U S A. 2008;105:20179–84. doi: 10.1073/pnas.0807121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 20.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106:12353–8. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nature biotechnology. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu Y-M, Cao X, et al. Expressed Pseudogenes in the Transcriptional Landscape of Human Cancers. Cell. 2012;149:13. doi: 10.1016/j.cell.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacDonald JW, Ghosh D. COPA--cancer outlier profile analysis. Bioinformatics. 2006;22:2950–1. doi: 10.1093/bioinformatics/btl433. [DOI] [PubMed] [Google Scholar]
- 26.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 27.Hutchinson L. Personalized cancer medicine: era of promise and progress. Nat Rev Clin Oncol. 2011;8:121. doi: 10.1038/nrclinonc.2011.14. [DOI] [PubMed] [Google Scholar]
- 28.Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011;8:184–7. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- 29.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 30.Yu D, Hung MC. Overexpression of ErbB2 in cancer and ErbB2-targeting strategies. Oncogene. 2000;19:6115–21. doi: 10.1038/sj.onc.1203972. [DOI] [PubMed] [Google Scholar]
- 31.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia (New York, NY. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia (New York, NY. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. The New England journal of medicine. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 34.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–26. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 35.Buck E, Eyzaguirre A, Haley JD, Gibson NW, Cagnoni P, Iwata KK. Inactivation of Akt by the epidermal growth factor receptor inhibitor erlotinib is mediated by HER-3 in pancreatic and colorectal tumor cell lines and contributes to erlotinib sensitivity. Molecular cancer therapeutics. 2006;5:2051–9. doi: 10.1158/1535-7163.MCT-06-0007. [DOI] [PubMed] [Google Scholar]
- 36.Ali S, El-Rayes BF, Sarkar FH, Philip PA. Simultaneous targeting of the epidermal growth factor receptor and cyclooxygenase-2 pathways for pancreatic cancer therapy. Mol Cancer Ther. 2005;4:1943–51. doi: 10.1158/1535-7163.MCT-05-0065. [DOI] [PubMed] [Google Scholar]
- 37.Morgan MA, Parsels LA, Kollar LE, Normolle DP, Maybaum J, Lawrence TS. The combination of epidermal growth factor receptor inhibitors with gemcitabine and radiation in pancreatic cancer. Clin Cancer Res. 2008;14:5142–9. doi: 10.1158/1078-0432.CCR-07-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, Watson DK, et al. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci U S A. 1996;93:3636–41. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miwa W, Yasuda J, Murakami Y, Yashima K, Sugano K, Sekine T, et al. Isolation of DNA sequences amplified at chromosome 19q13.1-q13.2 including the AKT2 locus in human pancreatic cancer. Biochemical and biophysical research communications. 1996;225:968–74. doi: 10.1006/bbrc.1996.1280. [DOI] [PubMed] [Google Scholar]
- 40.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sepp-Lorenzino L, Ma Z, Rands E, Kohl NE, Gibbs JB, Oliff A, et al. A peptidomimetic inhibitor of farnesyl:protein transferase blocks the anchorage-dependent and -independent growth of human tumor cell lines. Cancer Res. 1995;55:5302–9. [PubMed] [Google Scholar]
- 43.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–55. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 44.Nakada Y, Saito S, Ohzawa K, Morioka CY, Kita K, Minemura M, et al. Antisense oligonucleotides specific to mutated K-ras genes inhibit invasiveness of human pancreatic cancer cell lines. Pancreatology. 2001;1:314–9. doi: 10.1159/000055830. [DOI] [PubMed] [Google Scholar]
- 45.Shen YM, Yang XC, Yang C, Shen JK. Enhanced therapeutic effects for human pancreatic cancer by application K-ras and IGF-IR antisense oligodeoxynucleotides. World J Gastroenterol. 2008;14:5176–85. doi: 10.3748/wjg.14.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 47.Kolata G. Treatment for Leukemia, Glimpses of the Future. The New York Times; Jul 7, 2012. Genetic Gamble, New Approaches to Fighting Cancer. [Google Scholar]
- 48.Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics. 2010 doi: 10.1002/0471250953.bi1107s32. Chapter 11:Unit 11 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. http://genome.ucsc.edu.
- 50.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 51.Mahalanobis PC. On the generalised distance in statistics. Proceedings of the National Institute of Sciences of India. 1936;2:49–55. [Google Scholar]
- 52.De Maesschalck R, Jouan-Rimbaud D, Massart DL. The Mahalanobis distance. Chemometrics and Intelligent Laboratory Systems. 2000;50:1–18. [Google Scholar]
- 53. http://www.r-project.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.