Summary
DNA polymerases can only synthesize nascent DNA from single-stranded DNA (ssDNA) templates. In bacteria, the unwinding of parental duplex DNA is carried out by the replicative DNA helicase (DnaB) that couples NTP hydrolysis to 5′-to-3′ translocation. The crystal structure of the DnaB hexamer in complex with GDP-AlF4 and ssDNA reported here reveals that DnaB adopts a closed spiral staircase quaternary structure around an A-form ssDNA with each C-terminal domain coordinating two nucleotides of ssDNA. The structure not only provides structural insights into the translocation mechanism of superfamily IV helicases, but also suggests that members of this superfamily employ a translocation mechanism that is distinct from other helicase superfamilies. We propose a hand-over-hand mechanism in which sequential hydrolysis of NTP causes a sequential 5′-to-3′ movement of the subunits along the helical axis of the staircase, resulting in the unwinding of two nucleotides per subunit.
Introduction
Helicases couple nucleic acid-dependent NTP hydrolysis to their translocation on and unwinding of nucleic acid duplexes through a RecA-core NTPase domain that belongs to the additional strand catalytic glutamate (ASCE) class of P-loop NTPases (Leipe et al., 2003; Singleton et al., 2007). Among the six superfamilies (SF) of helicases, hexameric helicases are spread among SF3-6, while the remaining SF1-2 contain monomeric helicases (Singleton et al., 2007). The hexameric helicases can be further subdivided into two groups: the AAA+ (ATPases associated with various cellular activities) 3′-to-5′ helicases of SF3 and SF6; and the RecA-type 5′-to-3′ helicases of SF4-5. (Berger, 2008; Singleton et al., 2007).
The DnaB hexamer (DnaB6) is the bacterial replicative DNA helicase that unwinds double-stranded DNA (dsDNA) ahead of the replication fork and provides single-stranded DNA (ssDNA) templates for the DNA polymerase III holoenzyme (Kaplan and Steitz, 1999; LeBowitz and McMacken, 1986; Schaeffer et al., 2005). It is a member of SF4 that also includes DnaB-family G40P helicase of bacteriophage SPP1 and gene 4 helicase from bacteriophage T7 (T7gp4). DnaB monomer consists of an N-terminal helical domain (NTD), a linker domain, and a RecA-core C-terminal helicase domain (CTD) (Figure 1A) (Bailey et al., 2007b; LeBowitz and McMacken, 1986). The unliganded Bacillus stearothermophilus (Bst) DnaB6 and G40P helicases form a flat double-layered ring with the NTD adopting a trimer-of-dimers architecture that stacks on top of a hexameric ring of the CTDs (Bailey et al., 2007a; Wang et al., 2008). Following DnaA and DnaC-dependent loading onto a ssDNA at the replication bubble, DnaB6 not only serves as the replicative helicase, but also provides a nucleation site for replisome assembly through its NTD contact with DnaG primase and its CTD association with the τ subunit of the clamp loader complex, which subsequently recruits DNA polymerase III holoenzyme to the replication fork (Schaeffer et al., 2005).
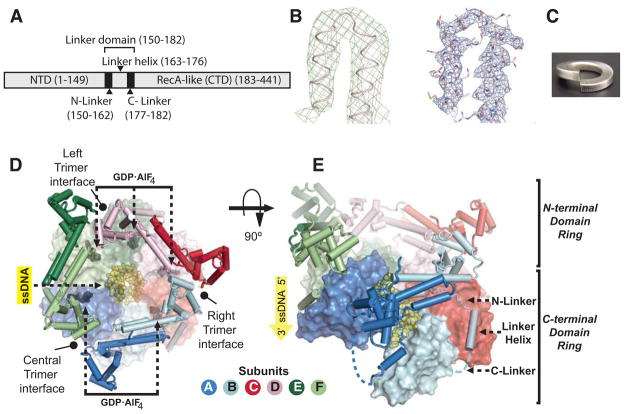
Figure 1. Overall architecture of the cocrystal structure DnaB6 with ssDNA and GDP-AlF4.
A) Domain organization of a DnaB monomer
B) Initial experimental electron density at 5.9 Å resolution contoured at 2σ (green) and final 2Fo-Fc electron density at 3.55 Å resolution contoured at 2σ (blue)
C) A right-handed lockwasher
D) Top view of DnaB6. The N-terminal domain (NTD) trimer-of-dimers and linker domains are shown in cartoon representation, while the C-terminal domain (CTD) staircase is shown in solvent accessible surface representation. The ssDNA and five molecules of GDP-AlF4 are shown as yellow and black spheres respectively.
E) Side view of DnaB6 rotated 90° with respect to (D). The C-linkers that are not visible in the electron density map is modeled as blue dashed lines.
See also Figure S7
The translocation mechanisms of SF4 and SF6 helicases remain unknown, while those of SF3 and SF5 helicases have been elucidated through the crystal structures of papillomavirus E1 helicase complexed with ssDNA and ADP (Enemark and Joshua-Tor, 2006) and Rho helicase complexed with ssRNA and ADP-BeF3 (Thomsen and Berger, 2009), respectively. These structures show that these flat hexameric helicases simultaneously engage their nucleic acid substrate with a stoichiometry of one nucleotide per subunit through their nucleic acid binding loops that adopt a spiral staircase arrangement with the loop height correlating with the nucleotide binding states. A coordinated escort mechanism based on these structures suggests that the nucleic acid substrate is pulled through the helicases as the nucleic acid binding loops move along the translocation axis during the ATP hydrolysis cycle (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009). Comparison of the two structures reveals that the two helicases travel in opposite directions because the order of NTP hydrolysis by their catalytic sites is reversed even though they bind their nucleic acid substrates in the same orientation (Thomsen and Berger, 2009).
To elucidate the translocation mechanism of DnaB6 and begin to understand how dsDNA is unwound at the replication fork, we have determined the cocrystal structure of BstDnaB6 with a ssDNA and five molecules of GDP-AlF4, an NTP analog that mimics the transition state of NTP hydrolysis, that was refined at 3.3 Å resolution (I/σI = 2.27 at 3.55 Å resolution). In striking contrast to the flat ring structures of the E1 and Rho helicase complexes (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009), our structure shows that the subunits of DnaB6 are arranged in a right-handed spiral staircase conformation and bound to two nucleotides of an A-from ssDNA per subunit. We propose that DnaB6 and other SF4 helicases may utilize a hand-over-hand translocation mechanism in which sequential hydrolysis of NTP is coupled to 5′-to-3′ translocation of the subunits with a step size of two nucleotides. In the hand-over-hand mechanism, translocation results from the migration of individual subunits along the helical axis of a ssDNA substrate instead of the movement of nucleic acid binding loops that was suggested for E1 and Rho helicases (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009).
Results
Structure Determination
The structure of DnaB6 in complex with ssDNA and five molecules of GDP-AlF4 was determined using the single isomorphous replacement with anomalous scattering method in space group P2221 with two copies of the complex in the asymmetric unit. The initial experimental phases were determined at 5.9 Å resolution from a Ta6Br142+ derivative prepared from a crystal of DnaB6 with a 40-nucleotide-long ssDNA and GDP-AlF4 (Table 1). These phases were then applied to a 3.55 Å resolution dataset (I/σI = 2.27), obtained from a second crystal containing a 16-nucleotide-long ssDNA and GDP-AlF4. The final model of the second crystal was refined by including data up to 3.3 Å resolution (I/σI = 0.8) to an Rwork/Rfree of 24.25%/28.85% (Figure 1B).
Table 1.
Data collection and refinement statistics
| Data collection statistics
| |||||||
|---|---|---|---|---|---|---|---|
| Data set | Native 1 | Derivative 1 | Derivative 2 | Derivative 3 | Native 2 | ||
| Heavy atom | - | 1.7 mM Ta6Br142+ for 7 days | 1.7 mM Ta6Br142+ for 3 days | 100 μM EMP for 2 hours | - | ||
|
| |||||||
| ssDNA length (nucleotides) | 40 | 16 | |||||
|
| |||||||
| Space group | P2221 | P2221 | P2221 | P2221 | P2221 | P2221 | P2221 |
|
| |||||||
| Unit cell a, b, c (Å) | 153.05 | 153.13 | 153.10 | 153.27 | 153.40 | 152.81 | 149.12 |
| 177.38 | 176.16 | 175.39 | 175.60 | 175.65 | 176.70 | 180.32 | |
| 275.20 | 279.28 | 278.79 | 278.73 | 279.30 | 275.42 | 279.13 | |
|
| |||||||
| Wavelength (Å) | 1.11 | 1.2545 | 1.2545 Peak |
1.2300 Remote |
1.2549 Inflection |
1.11 | 1.11 |
|
| |||||||
| Resolution range ((Å) | 50-5.7 | 50-6.0 | 50-6.0 | 50-6.5 | 50-6.2 | 50-7.25 | 50-3.55 |
|
| |||||||
| Unique reflections | 22,513 | 19,505 | 19,686 | 15,782 | 16,763 | 11,225 | 103,421 |
|
| |||||||
| Redundancy a | 6.5 (6.8) | 3.4 (3.5) | 3.4 (3.3) | 2.4 (2.4) | 3.4 (3.2) | 6.4 (6.2) | 3.0 (3.0) |
|
| |||||||
| Completeness (%)a | 99.8 (100) | 99.9 (100) | 99.8 (99.9) | 97.5 (98.1) | 99.8 (99.8) | 99.9 (100) | 89.4 (87.5) |
|
| |||||||
| I/σa | 22.2 (0.84) | 20.0 (1.2) | 15.0 (1.0) | 13.0 (1.19) | 13.0 (1.2) | 12.4 (2.4) | 32.7 (2.27) |
|
| |||||||
| Rmerge(%)a | 7.6 (>100) | 7.5 (>100) | 10.8 (>100) | 9 (75.7) | 12.3 (96.3) | 16.7 (75.2) | 12.5 (4.85) |
|
| |||||||
| Hexamers in AU | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
|
| |||||||
|
Refinement statistics
| |||||||
| Rwork (%) | 24.25 | ||||||
|
| |||||||
| Rfree (%) | 28.85 | ||||||
|
| |||||||
| RMSD bond (Å)/angle (°) | 0.0106/1.51 | ||||||
|
| |||||||
| Numbers of atoms | 40,304 | ||||||
|
| |||||||
| Figure of merit | 0.775 | ||||||
Number in parenthesis referred to high resolution shell
Overall Architecture
DnaB6 in complex with ssDNA and GDP-AlF4 forms a double-layered, right-handed spiral staircase. The NTD and CTD rings are split at one point and bent into a helical shape, which resembles two spring lockwashers stacked on top of each other (Figure 1C, D and E). In contrast, the unliganded hexamer adopts a double-layered flat ring conformation (Bailey et al., 2007a; Wang et al., 2008). In the ssDNA complex, the NTD trimer-of-dimers is organized around a pseudo 3-fold rotational axis of symmetry, analogous to a 3-step staircase in which each step comprises one dimer (Figure 1E). The CTD ring, on the other hand, is a 6-step staircase with each step consisting of a RecA-like domain. The center of mass of the CTD staircase has a pitch of 28 Å generated from a 5.6 Å rise and a 60° rotation per subunit along a pseudo 6-fold rotational screw symmetry axis (Figure 1E).
The linker domain comprises an N-linker, a linker helix, and a C-linker (Figure 1A and E). The N-linkers join the two layers of DnaB6 because the NTD and CTD of the same subunit make minimal contact with each other through interfaces whose sizes vary from 0 to 500 Å2 as opposed to an average of 500 Å2 in the unliganded hexamer (Bailey et al., 2007a). The only exception is the top CTD subunit that interacts extensively with its NTD subunit through a 1100 Å2 interface. The linker helices and the C-linkers glue the CTD ring together through hydrophobic packing between the linker helix of one subunit and the adjacent CTD subunit. The adjacent CTD subunits make more extensive contacts with each other upon the binding of ssDNA and GDP-AlF4 than they do in the unliganded DnaB6, resulting in a decrease in the diameter of the CTD staircase around the ssDNA without affecting the diameter of the NTD staircase. With the exception of the 18 Å gap at the trimer interface that splits the staircase of the NTD trimer-of-dimers (Figure 1E), the top and bottom steps of the CTD staircase are in close contact with each other through the linker helix and the C-linker (Figure 1E), thereby making the hexamer overall a closed assembly.
Conformation of ssDNA substrate and helicase-DNA interactions
A Fo-Fc difference electron density map showed density for the bases and phosphate backbone of fourteen nucleotides of the ssDNA substrate in the central channel of the CTD staircase prior to the inclusion of the ssDNA in the model (Figure 2C). However, as indicated by their temperature factors, only eleven nucleotides in the center of the ssDNA whose 5′ end projects toward the NTD staircase interact tightly with the helicase (Figure 1E), consistent with the 10-nucleotide footprint of E. coli DnaB6 and Forster Resonance Energy Transfer (FRET) data (Jezewska et al., 1998a, b).
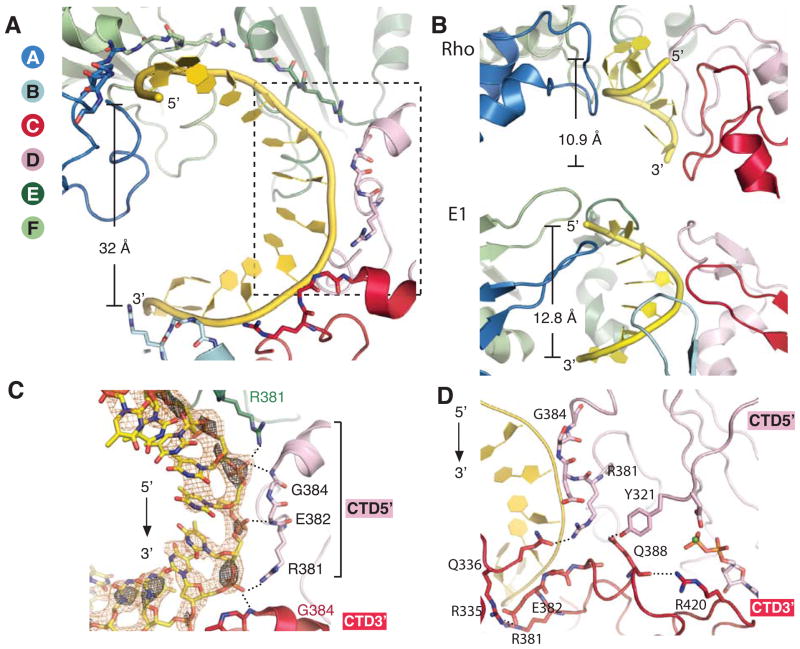
Figure 2. Interactions between an A-form ssDNA and the DNA-binding loops in the CTD staircase.
A) The DNA-binding loops of DnaB6 form a right-handed spiral staircase along the A-form ssDNA with a pitch of 32 Å, which is depicted in yellow cartoon representation. The CTD subunits are shown as cartoon with the residues involved in DNA-binding displayed as sticks.
B) The spiral staircase of nucleic acid-binding loops of Rho (PDB ID: 3ICE) and E1 (PDB ID: 2GXA). One of the RNA binding loops of Rho is removed for clarity. Subunits are colored according to their positions along the spiral staircase as in (A). Each helicase-bound nucleic acid substrate is displayed as yellow cartoon with its pitch shown on the left.
C) The helicase-ssDNA interactions of the boxed region in (A) and representative Fo-Fc electron density for DNA. The Fo-Fc density that was calculated prior to including the ssDNA in the model is shown in orange and black mesh at contour levels 2σ and 3σ, respectively. For a pair of CTD subunits, the CTD5′ is the subunit that interacts with the nucleotides at the 5′ end of the ssDNA and is equivalent to an upper step of the staircase, while CTD3′ is the subunit that contacts the nucleotides at the 3′ end of the ssDNA and acts as a lower step in the staircase. The phosphate backbone of the ssDNA is recognized by the amide groups in the peptide backbone of E382 and E384 as well as the sidechain of R381.
D) The hydrogen bond networks that connect adjacent DNA-binding loops and link the active site to the DNA-binding loop.
See also Figure S2
The phosphate backbone of the ssDNA superimposes on that of A-form DNA with a root mean square deviation (RMSD) of 2.0 Å (Figure 2A, Table 2), and on that of the template strand DNA bound to the β-clamp in the structure of β-clamp:clamp loader complex with its substrate (Kelch et al., 2012) with an RMSD of 0.54 Å. The backbone also exhibits a 20% reduction in the average rise per residue compared to B-form DNA (Table 2), consistent with a single molecule study where it was reported that DNA compaction is involved in the unwinding cycle of DnaB6 (Ribeck et al., 2010).
Table 2. Comparison of the geometry attributes of helicase-bound nucleic acid substrates to standard nucleic acids.
The attributes of E1 and Rho-bound substrates were calculated from PDB IDs 2GXA and 3ICE (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009), respectively.
| Standard B-form DNA | DnaB | E1 | Standard A-form RNA | Rho | |
|---|---|---|---|---|---|
| Step size based on crystal structures (nucleotides) | 2 | 1 | 1 | ||
| Pitch of 12 residues (Å) | 40.6 | 32.1 | 33.7 | ||
| Pitch of 6 residues (Å) | 16.9 | 12.8 | 14.1 | 10.9 | |
| Average rise per residue (Å) | 3.38 | 2.7±1.2 | 2.6 | 2.8 | 2.2±1.6 |
| Deviation from the rise per residue of B-from DNA | −0.7 Å | −0.8 | |||
| −20% | −23% | ||||
| Deviation from the rise per residue of A-from RNA | −0.6 Å | ||||
| −22% | |||||
| RMSD against A-form ssDNA | 2.0 Å | ||||
| RMSD against B-form ssDNA | 2.5 Å | ||||
The six DNA-binding loops of DnaB6 are structurally analogous to the L1 loop of RecA (Chen et al., 2008; Story and Steitz, 1992) and form the right-handed spiral staircase that tracks the phosphate backbone of the bound A-form ssDNA with each loop contacting two phosphate groups via a set of three hydrogen bonds (Figure 2C), suggesting a 2-nucleotide physical step size for DnaB6. In contrast, Rho and E1 coordinate one nucleotide of their nucleic acid substrates per subunit (Figure 2B) (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009).
NTP-bound conformation of DnaB6 active sites
Of the six NTPase active sites that are located between two adjacent RecA-like domains, five contain GDP-AlF4-Ca2+ as revealed by the Fo-Fc electron density map calculated prior to the inclusion of these ligands into the model (Figure 1D, 3A and S1). The metal ions were modeled as Ca2+ ions due to the excess CaCl2 in the crystallization buffer. There is no electron density for GDP-AlF4-Ca2+ at the interface between the top and bottom subunits of the CTD staircase where the ring is split. The base of the GDP does not make any specific interactions with the enzyme, consistent with previous biochemical data that show that the helicase can hydrolyze any NTP (Ayora et al., 2002; Kaplan and Steitz, 1999).
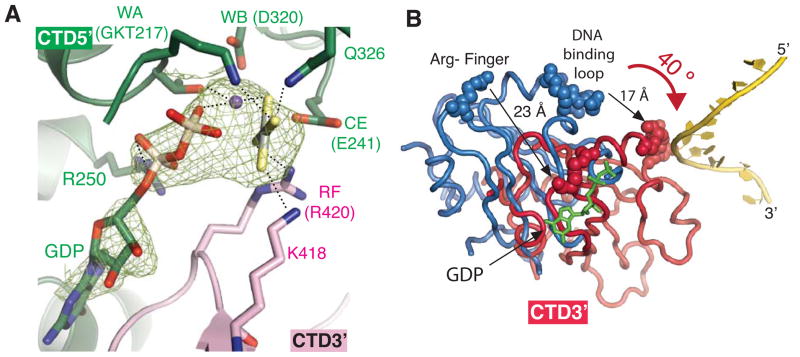
Figure 3. Formation of a NTP-bound active site.
A) Detailed interactions between the active site residues and GDP-AlF4 in a representative active site (between subunits E and D). Representative Fo-Fc electron density of GDP-AlF4 in the NTPase active site calculated prior to including GDP-AlF4-Ca2+ in the model is shown in green mesh at the contour level of 2σ. Nucleotide binding residues from the CTD5′ include Walker A-motif (WA: G215, K216, T217), Walker B motif (WB: D320), a catalytic glutamate (CE: E241), R250 and Q362. R250 coordinates the α-phosphate. Nucleotide binding residues from the CTD3′ include K418 and R420. See Figure S1 for final 2Fo-Fc electron density of GDP-AlF4 and the active site residues.
B) Subunit rotation upon GDP-AlF4 and ssDNA binding. The diagram was generated by superimposing one subunit from the unliganded DnaB6 structure (PDB ID: 2R6A) on the subunit of the current DnaB6 structure that provides the Walker A motif for GDP-AlF4 (CTD5′) by a least square method. Only the adjacent subunits that donate the Arg-finger for NTP coordination (CTD3′) are shown, while the subunits used in the superimposition are omitted. The dark pink subunit is from the DnaB6-ssDNA-GDP-AlF4 complex, while the blue subunit comes from the unliganded DnaB6. DNA-binding loops and Arg-fingers are shown as spheres, while DNA is shown as a yellow cartoon. GDP is represented with green sticks.
The formation of the NTP binding site at the interface between two adjacent RecA-like domains of DnaB6 is similar to those of RecA, F1-ATPase and T7gp4 helicase (Abrahams et al., 1994; Singleton et al., 2000; Story and Steitz, 1992), probably due to the conservation of the key nucleotide binding residues among these systems (Figure 3A). One half of the active site of DnaB6, CTD5′, consists of the Walker A motif (WA), Walker B motif (WB), and a catalytic glutamate (CE); the other half of the active site, CTD3′, is composed of K418 and R420, which stabilize the transition state analog. Alanine mutagenesis of the nucleotide binding residues in DnaB6 and T7gp4 resulted in the loss of the ATPase, helicase, and DNA-binding activities (Crampton et al., 2004; Soultanas and Wigley, 2002).
The conformations of the catalytic residues in the five active sites are identical to one another and are also similar to the ATP-bound states in other RecA-like ATPases (Figure 3A) (Abrahams et al., 1994; Chen et al., 2008; Thomsen and Berger, 2009). Typically, positively charged residues known as phosphate sensors simultaneously engage and neutralize the negative charges of the γ-phosphate of NTPs and the transition state of the ATP hydrolysis reaction (Enemark and Joshua-Tor, 2008). In DnaB6, we observe that four phosphate sensor residues interact with the transition state analog: K216 of the Walker A motif and Q326 from CTD5′, and K418 and R420 from CTD3′.
Our structure also reveals that R420 may serve as an allosteric switch that links ATP binding to an increase in the DNA binding affinity of DnaB6. The interaction between R420 and AlF4 suggests that K418 and R420 sense the presence of γ-phosphate, hence serving as the Lys- and Arg-fingers that have been observed in some GTPase-activating proteins, F1-ATPase, and other helicases (Abrahams et al., 1994; Enemark and Joshua-Tor, 2006; Scheffzek et al., 1997; Thomsen and Berger, 2009). Secondary structure alignment of the Walker A motif of the RecA-like domain of F1-ATPase (Abrahams et al., 1994) onto that of DnaB6 in the ssDNA complex structure further confirms this notion. Movement of the Arg-finger to stabilize the γ-phosphate may also promote DNA binding by aligning the DNA-binding loops along the ssDNA axis (Figure 2D).
Domain rotation upon the binding of ssDNA and GDP-AlF4
The binding of GDP-AlF4 and ssDNA induces the rotation of CTDs that results in the formation of a staircase conformation of the DNA-binding loops (Figure 3B). In the unliganded DnaB6 structure (Bailey et al., 2007a), a conformational change is needed to form NTP binding sites since the Lys- and Arg-fingers are too far (23 Å) away from the Walker A motif with which they form the putative NTPase active site. In the ssDNA complex, the RecA-like domain rotates 40° toward the ssDNA substrate to bring the Lys- and Arg-fingers into the active site to neutralize the negative charge of the AlF4 (Figure 3B). The rotation not only allows the rotating subunit to interact with two nucleotides of the ssDNA by moving the DNA-binding loop 17 Å towards the ssDNA substrate, but also causes the rotating subunit to translate 5.6 Å toward the 3′ end of the ssDNA, creating a spiral CTD staircase with a rise of 5.6 Å per CTD subunit. Taken together, the association of both GDP-AlF4 and ssDNA converts the flat ring conformation of the unliganded DnaB6 that cannot accommodate binding of NTP (Bailey et al., 2007a) into the spiral staircase conformation with assembled NTP binding pockets and an organized DNA-binding loop track that is complementary to the helical conformation of DNA.
Discussion
Binding of nucleic acids to hexameric helicases
DnaB6 has evolved two different mechanisms to interact with a DNA substrate that is twice as long as that of E1 and Rho. Instead of the one nucleotide per subunit stoichiometry seen in the structures of E1 and Rho with their substrates (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009), DnaB6 interacts with two nucleotides of ssDNA per subunit, suggesting that DnaB6 has a physical step size of two nucleotides as previously predicted by a nuclease protection assay (Jezewska et al., 1998b) (Figure 2A and C). Furthermore, the staircase formation strategy of DnaB6 contrasts markedly with those of E1 and Rho. The spiral staircase conformation of the DNA-binding loops of DnaB6 arises from the change in its quaternary structure due to the migration of entire subunits. In contrast, in E1 and Rho, the spiral conformation of nucleic acid binding loops is due to the movement of the loops themselves, while the overall ring remains flat (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009). Hence, the elongated pitch of the DNA-binding loops of DnaB6 allows the helicase to bind a ssDNA substrate with a pitch that is approximately 3-fold longer than those of E1 and Rho (Figure 2A and B, and Table 2). A truncated version of the DnaB-family G40P helicase, which lacks its NTD but retains its helicase activity, also adopts a spiral staircase conformation when co-crystallized with ATPγS in a P61 space group (Mesa et al., 2006; Wang et al., 2008), suggesting that the resulting spiral staircase conformations of DnaB6 and G40P are not due to crystal packing.
The strikingly different quaternary structure of the DnaB6 complex with ssDNA compared to those of E1 and Rho (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009) raises a question of whether the flat ring conformation of DnaB6 could accommodate a 12-nucleotide-long ssDNA. Comparison of the structures of unliganded DnaB6 and G40P (Bailey et al., 2007a; Wang et al., 2008) with the current structure suggests that this would not be possible. Not only are the flat ring structures of DnaB6 and G40P not able to assemble the NTPase active site as previously mentioned, but also the DNA-binding loops of the unliganded DnaB6 are randomly positioned along their putative translocation axes over a range of 20 Å, while those of the unliganded G40P adopt a staircase conformation with a pitch of only 16 Å (Figure S2). If a flat ring DnaB6 were to interact with a 12-nucleotide-long ssDNA, then the pitch of the substrate would be limited to 16–20 Å, resulting in an average rise per residue of only 50% compared to B-form DNA instead of the 80% average rise per residue that is observed in the current structure (Table 2).
Despite the different quaternary structures and step sizes, the structures of DnaB6 and other helicases bound to their nucleic acid substrates (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009) suggest that hexameric helicases share common nucleic acid binding mechanisms that differ from monomeric helicases. Hexameric helicases interact exclusively with the phosphate oxygen atoms of their nucleic acid substrates through nucleic acid-binding loops that are arranged into a right-handed, 6-step spiral staircase (Figure 2A and B). In contrast, monomeric helicases engage both the phosphate backbones and bases of their nucleic acid substrates (Kim et al., 1998; Saikrishnan et al., 2009; Velankar et al., 1999). Moreover, the hexameric helicase-bound nucleic acid substrates exhibit a 20–23% decrease in their average helical rises per residue relative to standard nucleic acids (Table 2).
DnaB6 may utilize a hand-over-hand mechanism for 5′-to-3′ translocation
Two key features of the current DnaB6 structure suggest that DnaB6 most likely uses a different translocation mechanism from E1 and Rho, though one might have expected the mechanism of Rho and DnaB6 to be similar since both are RecA-type hexameric helicases. The pitch of ssDNA bound to DnaB6 is approximately three times longer than that of the substrates of Rho and E1 (Figure 2A and B, and Table 2) (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009). Furthermore, the DNA-binding element of DnaB6 only consists of a 5-amino-acid-long loop, significantly shorter than the 12- and 15-amino-acid-long β-hairpins of E1 and Rho (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009). During the translocation of E1 and Rho, the nucleic acid binding loop must swing from one end of the nucleic acid binding loop staircase to the opposite end in order to engage an incoming nucleotide of the nucleic acid substrate (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009). The combination of the two aforementioned structural elements of DnaB6 suggests that the top DNA-binding loop may not be able to traverse the pitch of the ssDNA substrate to reach the two new nucleotides at the 3′ side of the ssDNA without major quaternary structure rearrangements.
Although we currently lack crystallographic data that establish the position of the first NTP hydrolysis and whether sequential, concerted or stochastic NTP hydrolysis is coupled to translocation (Lyubimov et al., 2011), we have considered three different 5′-to-3′ translocation models for DnaB6, in which ssDNA binding is stabilized by NTP binding while ssDNA release is stimulated by NTP hydrolysis as indicated from biochemical studies of T7gp4 (Hingorani and Patel, 1993).
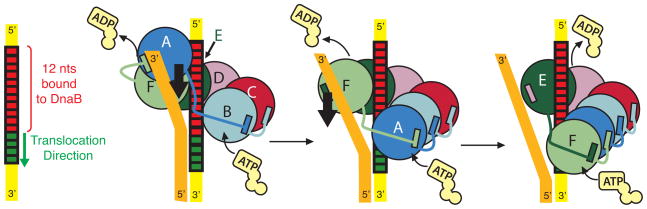
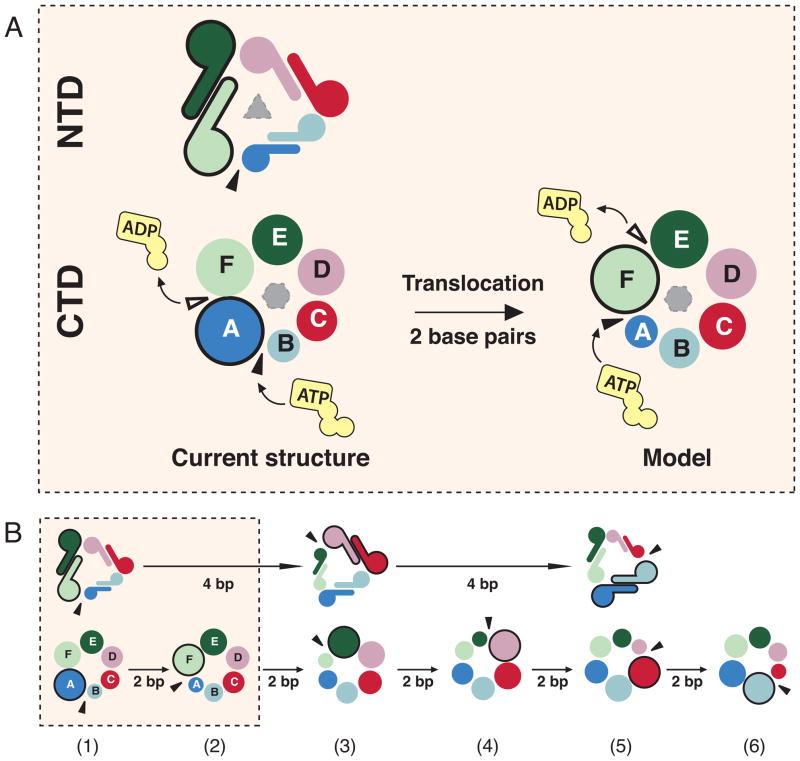
In the first hand-over-hand model, DnaB6 couples the translocation of a subunit to sequential NTP hydrolysis and operates in an analogous manner to a rope climber, whereby the top subunit of the CTD staircase moves towards the 3′ end of the lagging strand (Figure 4 and Movie S1). Upon hydrolysis of the NTP in the top active site, the top subunit dissociates from the two nucleotides at the 5′ end of the ssDNA (Hingorani and Patel, 1993). The translocation or the dissociation of the top CTD subunit from its adjacent subunit is driven by the release of the γ-phosphate that originally bridged the Arg-finger of the top subunit to the Walker A motif of the adjacent subunit. An incoming NTP would then bind to the empty and solvent-exposed Walker A motif of the bottom subunit. As the top subunit dissociates from its neighbor at the top of the staircase, it migrates towards the bottom of the staircase where it provides the Arg-finger to stabilize the newly bound NTP and promotes the unwinding of two base pairs. The cycle is repeated when NTP is hydrolyzed at the top of the staircase. Essentially, the hydrolysis of one ATP and subsequent release of the γ-phosphate (~-12 kcal/mole) is coupled to the unwinding with a physical step size of two base pairs (~+3.6kcal/mol) (von Hippel and Delagoutte, 2001), consistent with the footprint of DnaB6 that is suggestive of the stoichiometry of two nucleotides per subunit (Jezewska et al., 1998b).
Figure 4. The hand-over-hand mechanism of DnaB6.
Subunits are colored as in Figure 1. The lagging strand on which DnaB6 translocates is colored yellow while the leading strand is colored orange. Red nucleotides illustrate residues that interact with the helicase in the crystal structure presented here, while green nucleotides represent those that are entering the hexamer after ATP turnover. The thick black arrow denotes the downward movement of the top CTD subunit towards the 3′ end of the lagging strand. See also Figures S3, S4, S5 and S6 and Movie S1
Although an ensemble kinetic method showed that the kinetic step size of DnaB6 was 1.4±0.2 nucleotides or between 1.2 to 1.6 nucleotides (Galletto et al., 2004), the physical step size based on our structure and the kinetic step size describe two different parameters in the DnaB6 translocation cycle. The physical step size is the average distance moved by DnaB6 along its ssDNA substrate per NTP hydrolysis and is not necessarily equivalent to a kinetic step size, which is defined as an average number of nucleotides translocated between two successive rate-limiting steps in the translocation cycle (Yodh et al., 2004). Additional experiments at the single molecule level are required to confirm the physical step size of DnaB6.
Although the available crystallographic evidence cannot rule out the possibility that NTP hydrolysis is initiated at the bottom active site during the hand-over-hand mechanism, we propose that the top active site is where the first NTP hydrolysis occurs because the hydrolysis of NTP at the bottom active site would result in a helicase with a 3′-to-5′ polarity that is opposite to the observed polarity of DnaB6 (LeBowitz and McMacken, 1986). Moreover, the proposed ATP turnover that propagates from the top to the bottom active site with respect to the 5′ end of the ssDNA is consistent with the direction in which ATP is hydrolyzed by Rho that translocates in the same direction as DnaB6 (Thomsen and Berger, 2009).
The NTP hydrolysis at the top active site could be preferred due to the interactions between the top CTD subunit and its NTD. It has been suggested that the NTD trimer-of-dimers of DnaB-family helicases may also play a role in defining the polarity of DnaB6 since the removal of the NTD from the DnaB-family G40P helicase generated a bidirectional helicase (Mesa et al., 2006). In our structure, the top subunit is the only subunit that interacts extensively with the NTD staircase. It can be postulated that the interactions between the corresponding N- and C-terminal domains may stimulate the NTPase and/or helicase activities of the top RecA-like domain such that NTP is preferentially hydrolyzed at the top active site.
While we have considered two alternative translocation mechanisms (Figure S3 and S4), we favor the hand-over-hand model (Figure 4 and Movie S1). Not only is it consistent with a biochemical study of T7gp4, which indicates that the transfer of ssDNA from one subunit to the next is coupled to sequential NTP hydrolysis in SF4 helicases (Crampton et al., 2006), but also it is the most energetically efficient mechanism with a step size of two nucleotides per NTP hydrolysis, compared to a 2-nucleotide step size per five NTP molecules in the alternative models. Furthermore, since Rho and DnaB6 are both RecA-type 5′-to-3′ hexameric helicases with 33.5% sequence similarity in their RecA-like domains, the NTP hydrolysis cycle of DnaB6 could operate in a sequential manner like that of Rho. Moreover, the hand-over-hand motion allows each individual DnaB hexamer, hence each replication fork, to independently move in opposite directions from oriC, as demonstrated by a single molecule study in live E. coli cells in which sister replisomes travel in opposite directions before returning to the termination site (Reyes-Lamothe et al., 2008).
The hand-over-hand model is not only mechanistically distinct from the existing translocation mechanisms of hexameric helicases, but also thermodynamically feasible. DnaB6 travels with a physical step size of two nucleotides per NTP molecule instead of one nucleotide as proposed for Rho and E1. Furthermore, as required by the larger pitch of the ssDNA bound to DnaB6 (Table 2), the translocation of DnaB6 involves the movements of entire subunits rather than smaller movements of only nucleic acid binding loops as observed in E1 and Rho (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009). Most importantly, the subunit rearrangement is thermodynamically plausible since the energy from ATP hydrolysis (~-12 kcal/mol) can support both the unwinding of two base pairs (~+3.6kcal/mol) (von Hippel and Delagoutte, 2001) and the thermodynamic penalty associated with the migration of the CTD subunit, which is thermodynamically favorable due to the hydrophilic nature of interface between NTD and CTD (Figure S5). While the mechanism is thermodynamically sound, crystallographic evidence alone is not sufficient to determine whether this model represents an active or passive unwinding, which would require a comparison of the rate of unwinding by the helicase and the rate of strand separation by thermal breathing.
The spiral staircase conformer of DnaB6 could translocate on both ssDNA and dsDNA
In addition to ssDNA, DnaB6 can also act as a NTP-dependent dsDNA translocase in vitro (Kaplan, 2000; Kaplan and O’Donnell, 2002). We propose that the spiral staircase quaternary structure, and not the flat ring conformation of unliganded DnaB6 (Bailey et al., 2007a), represents the conformation of DnaB6 that can translocate on both ssDNA and dsDNA. Superposition of the phosphate backbone of the ssDNA substrate of DnaB6 onto the template strand DNA that is bound to the β-clamp in the β-clamp:clamp loader complex gave an RMSD of 0.54 Å (Kelch et al., 2012). While a few minor clashes between loops in the central channel of the CTD of DnaB6 and the primer strand DNA of the β-clamp:clamp loader complex arise as a result of this alignment, only minor conformational changes that enlarge the diameter of the CTD staircase are required to alleviate these clashes. The striking similarity between the structures of the ssDNA bound to DnaB6 and the template DNA of the A-form duplex bound to the β-clamp:clamp loader complex suggests that the spiral conformation of DnaB6 can accommodate and translocate on dsDNA. In contrast, the unliganded DnaB6 is not in a conformation that supports translocation on dsDNA, since the flat ring conformation of DnaB6 lacks the ability to bind NTP.
Maintenance of the hexameric state by the linker domain
The 35-amino acid-long linker domain (Figure 1A) ensures that DnaB6 remains in a hexameric state and continues to encircle the lagging strand during translocation. Our structure demonstrates that the hydrophobic interactions between the linker helix and the adjacent CTD subunit stabilize the packing of the CTD staircase, while also ensuring that the top CTD subunit of the staircase does not disengage from the core hexameric assembly during the hand-over-hand motion. Meanwhile, the N-linkers of the linker domains tether the NTD trimer-of-dimers and the CTD ring together when the helicase adopts the spiral staircase conformation because, except for the CTD at the top of the staircase, the N- and C-terminal domains of the same subunit do not contact each other.
Whether the hand-over-hand mechanism would be applicable to other hexameric helicases, particularly other SF4 helicases, depends on the presence of accessory domains that can maintain the hexameric state of the helicase during the long-range movement of the RecA-like domains (Figure S6). Consistent with the hypothesis that Rho and DnaB6 have evolved different translocation mechanisms, Rho does not possess any accessory domain that is equivalent to the linker domain of DnaB6. In contrast, the presence of accessory domains around the periphery of the ATPase ring of T7gp4 as well as the oligomerization domain that stacks on top of the AAA+ rings of E1 and SV40 large T antigen suggest that these helicases may be able to incorporate the hand-over-hand motion into their translocation mechanisms
Coupling of the NTD staircase formation to translocation of the CTD hexamer
During the hand-over-hand motion of the 6-step CTD staircase, the NTD trimer-of-dimers is proposed to translocate in three steps. Each trimer interface splits once for every two CTD movements, which coincides with the hydrolysis of two molecules of NTP (Figure 5 and Movie S2). Due to the large 2000-Å2 hydrophobic interface within the NTD dimer (Bailey et al., 2007a), the dimer is presumed to remain intact during translocation and thereby act as an individual step in the NTD staircase. We propose that each hand-over-hand motion of the NTD staircase requires the translocating NTD dimer to tilt as it is pulled in the 3′ direction by the N-linker that connects each NTD to the linker helix and CTD (Figure 1A and E). In Movie S2, the tilt of the translocating NTD dimer was modeled as a 2-step process for two reasons. This model ensures that only one trimer interface is broken during translocation and minimizes the distance between the NTD and its linker helix to ~40 Å, which is within the distance of 48 Å that the 13-amino acid long N-linker could stretch.
Figure 5. Synchronization of the NTD staircase formation with the NTP hydrolysis cycle in the CTD staircase.
A) Top view down the 5′ end of the ssDNA substrate. The NTD (top) and CTD (bottom) staircases are shown as dimers and circles that are spiraling into the plane of the paper, respectively. The sizes of the subunits indicate the positions of the subunits along the spiral staircase with the largest symbols representing the top subunits and the smallest symbols representing the bottom subunits. Black triangles denote sites where the NTD and CTD staircases split into the lockwasher. In the CTD panel, the black triangles also illustrate the binding site of incoming NTP while white triangles indicate the site of NTP hydrolysis.
B) The NTD trimer-of-dimers would act as a 3-step staircase that splits after the translocation over 4 base pairs whereas the CTD staircase splits after two basepairs. State 1 is the current structure, while states 3 and 5 are generated by 120° and 240° clockwise rotation of state 1 along the ssDNA axis. See also Movie S2
Stoichiometry of DnaG primase to DnaB6 during translocation
We propose that a single DnaG may interact with a translocating DnaB6 in the replisome because the motion of the NTD and CTD staircases must be coordinated during translocation. The cocrystal structure of unliganded DnaB6 with the helicase-binding domain (HBD) of DnaG shows that each HBD binds across the NTD trimer interface of DnaB6 (Bailey et al., 2007a). The formation of the NTD staircase disrupts the central NTD trimer interface, leaving the left and right intact interfaces as potential DnaG binding platforms (Figure 1D and S7). These two remaining sites are, however, temporally inequivalent during translocation (Movie S2). As the center trimer interface closes, the left trimer interface becomes the next to open (Figure 1D). Hence, the HBD binding site that has remained undisturbed the longest is the right trimer interface, which has been closed for four ATP hydrolysis events and, therefore, during translocation over eight nucleotides (Figure 1D). We propose that this right trimer interface recruits DnaG for primer synthesis.
Our proposal that the preferred stoichiometry of DnaG to DnaB6 is 1:1 in the replisome is supported by a single molecule study which showed that cooperative binding of a 2–3 molar excess of DnaG to DnaB6 led to abortive replication (Tanner et al., 2008). Comparison of the structure of described here with our earlier structure DnaG (Bailey et al., 2007a) provides the basis for such behavior because the binding of multiple molecules of DnaG likely results in a staircase-to-flat ring conversion, reducing the effective concentration of active DnaB6. The 1:1 ratio that we suggest of DnaG to DnaB6 in the replisome implies that excessive DnaG binding could be inhibitory to the processivity of the replisome as previously proposed by Tanner et al, 2008. It is not in conflict with the stimulatory effect of DnaG on DnaB6 enzymatic activity according to an ensemble study in vitro by LeBowitz and McMacken, 1986. In short, our proposed DnaG:DnaB6 stoichiometry focuses on the ratio of DnaG to DnaB6 in a processive replisome, not on the effect of DnaG on the enzymatic activity of DnaB6 or the rate of replication.
The hand-over-hand model implies that DnaG may not necessarily bind the translocating DnaB6 throughout the synthesis of a 9–12 nucleotide-long RNA primer (Kitani et al., 1985) since our model suggests that each NTD trimer interface may only remain closed during translocation over a maximum of eight nucleotides. This hypothesis is consistent with the disassembly model of the primosome (Manosas et al., 2009), which suggests that, although DnaG is required to interact with DnaB6 for localization and enhanced enzymatic activity, DnaG can remain at the primed site through a contact with ssDNA-binding protein (SSB) while DnaB6 continues its translocation (Yuzhakov et al., 1999).
Recognition of A-form DNA by replication proteins in a spiral staircase conformation
Comparison of the current structure of DnaB6 with ssDNA and the structure of β-clamp:clamp loader bound to primer-template DNA (Kelch et al., 2012) suggests that the conversion of ring-shaped replisomal proteins that adopt a flat ring conformation in an unliganded state into a spiral staircase conformation to recognize an A-form DNA may be a common theme in bacterial replication. Transformation into the spiral staircase creates a helical track of DNA binding moieties that is complementary to the helical shape of A-form DNA. The striking similarity of the structure of the DNA bound to DnaB6 and to the β-clamp:clamp loader complex suggest that these proteins shift the equilibrium between A- and B-form DNAs toward the A-form, presumably due to dehydration of the DNA.
Summary
The structure of DnaB6 in complex with a centrally bound ssDNA and GDP-AlF4 provides the first structural insights into the translocation mechanism of an SF4 helicase. Upon its binding to NTP and ssDNA, DnaB6 undergoes a large scale conformational change from a flat ring to a spiral staircase that allows the helicase to coordinate two nucleotides of A-form ssDNA per subunit, consistent with footprinting evidence (Jezewska et al., 1998b). Together with its remarkably distinct quaternary structure, the longer ssDNA substrate and the shorter DNA-binding loops of DnaB6 suggest that DnaB6 employs a hand-over-hand mechanism that is notably different from existing translocation mechanisms proposed for the Rho and E1 hexameric helicases (Enemark and Joshua-Tor, 2006; Thomsen and Berger, 2009).
Comparison of the structure based translocation mechanisms of DnaB6, Rho (Thomsen and Berger, 2009), and E1 (Enemark and Joshua-Tor, 2006) with the biochemically based translocation mechanisms of many pentameric and hexameric helicases (Chemla et al., 2005; Martin et al., 2005; Moffitt et al., 2009; Moreau et al., 2007) suggests that a general mechanism of translocation for all hexameric helicases may not exist. Instead, our structure provides evidence that different helicases may have tailored their translocation mechanisms according to their step sizes and the presence of accessory domains outside of the common RecA-like domains.
Experimental Procedures
Protein Purification, Crystallization and Data Collection
DnaB6 was purified as previously described in Bailey et al, 2007a. To form the complex, 17 μM DnaB6 in buffer A (20 mM Tris pH 7.4, 0.1 M NaCl, 10 mM MgCl2) was incubated with 5 mM NaF, 0.5 mM AlCl3 and a 3-fold molar excess of ssDNA for 20-30 minutes and with 2 mM GDP for an additional 10 minutes at room temperature. Crystals were grown using the sitting-drop vapor-diffusion method at 12-25°C by mixing equal volumes of the complex (diluted to 6.8 μM with buffer A) with the precipitant containing 4-7% PEG3350, 0.2 M CaCl2, 0.1 M MES pH 6.5 and cryoprotected by supplementing to final concentrations of 10% PEG3350, 25% ethylene glycol and 10 mM MgCl2. Diffraction data were collected at beamlines X-25 and X-29 of the National Synchrotron Light Source of the Brookhaven National Laboratory at 100 K and processed using the HKL suite of programs (Table 1) (Otwinowski, 1997).
Structure determination and refinement
Initial phases were determined using the single isomorphous replacement with anomalous scattering method. Ta6Br142+-derivative 1 was prepared from native crystal 1 (DnaB6 with dT40 and GDP-AlF4) with statistics summarized in Table 1. The positions of seventeen clusters of Ta6Br142+ were found using the direct methods procedure as implemented in SHELXD (Sheldrick, 2010). The initial experimental electron density maps were generated using SHELXE (Sheldrick, 2010) to 5.6-Å resolution. These maps have recognizable features for all 12 NTDs and 11 of the 12 CTDs of two copies of the DnaB6 complex in one asymmetric unit, thus allowing for their unambiguous placement using the atomic model of 2R6A (Bailey et al., 2007a). This model provided initial non-crystallographic symmetry (NCS) matrices for multidomain NCS averaging with solvent flattening using DMMULTI (CCP4, 1994). The resulting experimental phases were used to determine additional heavy atom derivative structures from derivatives 2 and 3 (Table 1), as well as re-refine the first heavy atom derivative using MLPHARE (Otwinowski, 1991).
The MLPHARE-generated phases of native crystal 1 were transferred to the 3.55 Å resolution dataset (I/σI = 2.55) of native crystal 2 (DnaB6 with dT16 and GDP-AlF4) for phase extension using CNS density modification procedure (Brunger et al., 1998) and subjected to new runs of multidomain NCS averaging with DMMULTI (CCP4, 1994). The resulting maps show recognizable features for ssDNA and GDP-AlF4. For NCS averaging, matrices were derived from a rigid-body refined crystal 2 model starting with the crystal 1 model using REFMAC5 (CCP4, 1994).
The model for crystal 2 was iteratively rebuilt in Coot and refined by including data up to 3.3 Å resolution (I/σI = 0.8) using REFMAC5 with strong NCS restraints. To remove model bias and facilitate model rebuilding, multidomain NCS averaging in DMMULTI was carried out for model-calculated phases. The final round of refinement included TLS decomposition of all rigid groups corresponding to individual NTD and CTD subunits.
Supplementary Material
Highlights.
Hexameric helicase DnaB forms a spiral staircase in complex with ssDNA and GDP-AlF4.
DnaB binds to an A-form ssDNA with a step size of 2 nucleotides per subunit.
Translocation can be modeled as moving each subunit in a hand-over-hand mechanism.
Subunit movement along DNA is mechanistically distinct from other hexameric helicases.
Acknowledgments
We thank the staff at APS beamline 24-ID and at BNL beamlines X-25 and X-29. This work was supported by NIH RO1 GM-57510 to T.A.S., the Royal Thai Government Scholarship to O.I. and by Steitz Center for Structural Biology, Gwangju Institute of Science and Technology (SCSB-GIST), Republic of Korea.
Footnotes
Accession Numbers: 4ESV
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1- ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Ayora S, Weise F, Mesa P, Stasiak A, Alonso JC. Bacillus subtilis bacteriophage SPP1 hexameric DNA helicase, G40P, interacts with forked DNA. Nucleic Acids Res. 2002;30:2280–2289. doi: 10.1093/nar/30.11.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007a;318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- Bailey S, Eliason WK, Steitz TA. The crystal structure of the Thermus aquaticus DnaB helicase monomer. Nucleic Acids Res. 2007b;35:4728–4736. doi: 10.1093/nar/gkm507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger JM. SnapShot: nucleic acid helicases and translocases. Cell. 2008;134:888–888.e881. doi: 10.1016/j.cell.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- Crampton DJ, Guo S, Johnson DE, Richardson CC. The arginine finger of bacteriophage T7 gene 4 helicase: role in energy coupling. Proc Natl Acad Sci U S A. 2004;101:4373–4378. doi: 10.1073/pnas.0400968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton DJ, Mukherjee S, Richardson CC. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol Cell. 2006;21:165–174. doi: 10.1016/j.molcel.2005.11.027. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J Mol Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Hingorani MM, Patel SS. Interactions of bacteriophage T7 DNA primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry. 1993;32:12478–12487. doi: 10.1021/bi00097a028. [DOI] [PubMed] [Google Scholar]
- Jezewska MJ, Rajendran S, Bujalowski W. Complex of Escherichia coli primary replicative helicase DnaB protein with a replication fork: recognition and structure. Biochemistry. 1998a;37:3116–3136. doi: 10.1021/bi972564u. [DOI] [PubMed] [Google Scholar]
- Jezewska MJ, Rajendran S, Bujalowski W. Functional and structural heterogeneity of the DNA binding site of the Escherichia coli primary replicative helicase DnaB protein. J Biol Chem. 1998b;273:9058–9069. doi: 10.1074/jbc.273.15.9058. [DOI] [PubMed] [Google Scholar]
- Kaplan DL. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J Mol Biol. 2000;301:285–299. doi: 10.1006/jmbi.2000.3965. [DOI] [PubMed] [Google Scholar]
- Kaplan DL, O’Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- Kaplan DL, Steitz TA. DnaB from Thermus aquaticus unwinds forked duplex DNA with an asymmetric tail length dependence. J Biol Chem. 1999;274:6889–6897. doi: 10.1074/jbc.274.11.6889. [DOI] [PubMed] [Google Scholar]
- Kelch BA, Makino DL, O’Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2012;334:1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- Kitani T, Yoda K, Ogawa T, Okazaki T. Evidence that discontinuous DNA replication in Escherichia coli is primed by approximately 10 to 12 residues of RNA starting with a purine. J Mol Biol. 1985;184:45–52. doi: 10.1016/0022-2836(85)90042-7. [DOI] [PubMed] [Google Scholar]
- LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. Evolution and classification of P-loop kinases and related proteins. J Mol Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosas M, Spiering MM, Zhuang Z, Benkovic SJ, Croquette V. Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome. Nat Chem Biol. 2009;5:904–912. doi: 10.1038/nchembio.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Mesa P, Alonso JC, Ayora S. Bacillus subtilis bacteriophage SPP1 G40P helicase lacking the n-terminal domain unwinds DNA bidirectionally. J Mol Biol. 2006;357:1077–1088. doi: 10.1016/j.jmb.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. Isomorphous Replacement and Anomalous Scatterring. In: Wolf W, Evans PR, Leslie AGW, editors. Isomorphous Replacement and Anomalous Scatterring. 1991. [Google Scholar]
- Otwinowski ZaMW. Processing of X-ray diffraction data collected in oscillation mode. New York: Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeck N, Kaplan DL, Bruck I, Saleh OA. DnaB helicase activity is modulated by DNA geometry and force. Biophys J. 2010;99:2170–2179. doi: 10.1016/j.bpj.2010.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- Schaeffer PM, Headlam MJ, Dixon NE. Protein--protein interactions in the eubacterial replisome. IUBMB Life. 2005;57:5–12. doi: 10.1080/15216540500058956. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Soultanas P, Wigley DB. Site-directed mutagenesis reveals roles for conserved amino acid residues in the hexameric DNA helicase DnaB from Bacillus stearothermophilus. Nucleic Acids Res. 2002;30:4051–4060. doi: 10.1093/nar/gkf527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Tanner NA, Hamdan SM, Jergic S, Loscha KV, Schaeffer PM, Dixon NE, van Oijen AM. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998a. [DOI] [PubMed] [Google Scholar]
- Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- Wang G, Klein MG, Tokonzaba E, Zhang Y, Holden LG, Chen XS. The structure of a DnaB-family replicative helicase and its interactions with primase. Nat Struct Mol Biol. 2008;15:94–100. doi: 10.1038/nsmb1356. [DOI] [PubMed] [Google Scholar]
- Yodh JG, Schlierf M, Ha T. Insight into helicase mechanism and function revealed through single-molecule approaches. Q Rev Biophys. 2004;43:185–217. doi: 10.1017/S0033583510000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzhakov A, Kelman Z, O’Donnell M. Trading places on DNA--a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.