Abstract
Far from being simple “bags” of enzymes, bacteria are richly endowed with ultrastructures that challenge and expand standard definitions of the cytoskeleton. Here we review rods, rings, twisted pairs, tubes, sheets, spirals, moving patches, meshes and composites, and suggest defining the term “bacterial cytoskeleton” as all cytoplasmic protein filaments and their superstructures that move or scaffold (stabilize/position/recruit) other cellular materials. The evolution of each superstructure has been driven by specific functional requirements. As a result, while homologous proteins with different functions have evolved to form surprisingly divergent superstructures, those of unrelated proteins with similar functions have converged.
Defining the bacterial cytoskeleton
The word “skeleton” is defined as the basic frame or supporting structure of an object. The term “cytoskeleton” was coined after a network of long, skinny, cell-shape-determining structures was discovered in the cytoplasm of eukaryotic cells. These structures were later found to consist of actin, tubulin and intermediate filament (IF) proteins that move objects through their own growth and disassembly, act as stationary tracks for auxiliary motors, and/or serve as connectors and scaffolds to position and stabilize other materials. The discovery of bacterial homologs of actins, tubulins and IF proteins then led to a new understanding that bacteria, too, were organized and shaped by a cytoskeleton [reviews: 1,2,3]. Bacterial filaments have also been found to push and pull objects and/or bind them together as connectors and scaffolds, but have not yet been found to act as tracks for other motor proteins. In bacteria (and archaea), the eukaryotic notion of the cytoskeleton is challenged however by both the small size of the cell, which causes filament bundles to sometimes be nearly as wide as they are long (and therefore seem much less obviously “filamentous”), and by diverse new non-actin, non-tubulin, and non-IF superstructures (higher-order assemblies) that exhibit many cytoskeletal characteristics and have already been called “cytoskeletal” in the literature. Thus as one of the purposes of this review, here we explore an expansive definition of the bacterial cytoskeleton that includes all cytoplasmic protein filaments and their superstructures that either move or scaffold (stabilize/position/recruit) materials within the cell. Recent progress is highlighted with emphasis on how specific functions drove the evolution of different superstructures.
Superstructures of the bacterial cytoskeleton
Individual rods
Protein polymerization may possibly have first evolved to regulate enzymatic activity, but the resulting filaments might have then proven to be useful as scaffolds as well [review: 4]. The actin homolog MamK may exemplify such simple rods, as it polymerizes into what appear by electron cryotomography (ECT) of intact cells [review: 5] to be simple short rods flanking magnetosome chains (Figure 1A) [6-10]. Despite the apparently simple structure, MamK shows dynamic behavior, promoted by the regulators MamJ and LimJ [9], and plays a role in a pole-to-midcell translocation of the magnetosome chain prior to cell division [10], so there is much complexity to be understood.
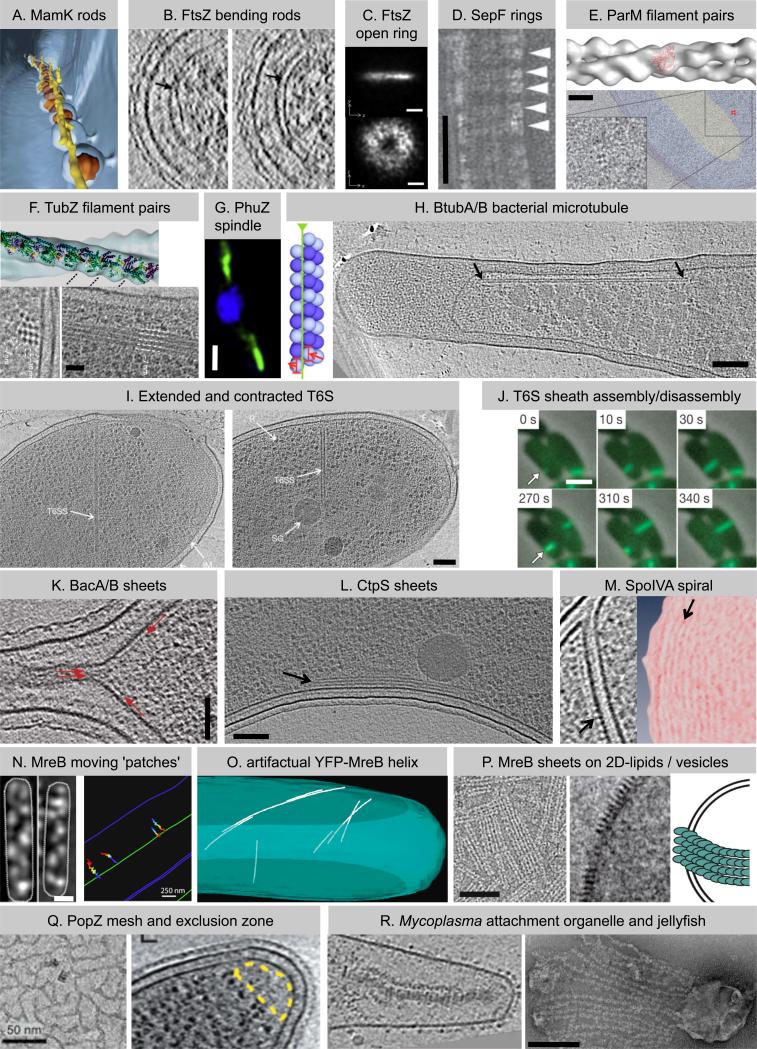
Figure 1. Superstructures of the bacterial cytoskeleton.
(A) Individual MamK rods (yellow in shown segmentation) organize the magnetosome chain [7]. (B) ECT of wildtype cells identified the cell division protein FtsZ (arrows) as straight (left) or bent (right) rods [13]. (C) Superresolution fLM observed FtsZ as a ring-shaped signal with uneven density (y, cell length axis) [19]. (D) SepF bundles FtsZ filaments (running vertical) in vitro by forming rings (arrowheads) around them [25]. (E) Plasmid segregating ParM forms twisted filament pairs in vitro (upper [30]) and small bundles of filament pairs in vivo (lower: perpendicular view [33]). (F) TubZ also segregates plasmids, assembles twisted filament pairs in vitro (upper) and bundles in vivo (lower: perpendicular (left) and longitudinal (right) view of overexpressed TubZ) [42]. (G) PhuZ forms a spindle-like structure (green) that positions phage DNA at midcell (blue, phage/bacterial DNA) [44]. (H) BtubA/B assemble bacterial microtubules (left: model) which localize close to the cytoplasmic membrane in vivo (right, arrows) [50]. (I) Type VI secretion (T6S) requires a contractile phage tail-like sheath which is found in extended (left) and contracted (right) confirmation [59]. (J) The tubular T6S sheath is dynamic and cycles between assembly (upper row), quick contraction, and disassembly (lower row) [59]. (K) Bactofilins [65] and (L) CTP synthase [67] both form sheets (arrows) at the cytoplasmic membrane, where they recruit other proteins. (M) Densities thought to represent SpoIVA (left, arrow) form concentric rings (right panel shows density projections, arrow) on the mother side of the outer spore membrane, where SpoIVA recruits spore coat proteins [79]. (N) MreB forms small patches moving circumferentially around the cell driven by the cell wall synthesis machinery (left: fLM images, right: traced patches) [87,88]. (O) Extended MreB helices in E. coli (segmentation shown) are an artifact of an N-terminal YFP tag [90]. (P) MreB forms filaments and sheets on 2D-lipids (left) and vesicles (middle and model on right) [91]. (Q) PopZ establishes a ribosome exclusion zone (right, yellow) in vivo presumably by the formation of a 3-D mesh (left, in vitro) [93,94]. (R) Gliding motility requires multi-element composites such as the attachment organelle in Mycoplasma pneumoniae (left, [96]) or the jellyfish-like multi-element complex in Mycoplasma mobile (right, [98]). Images are adapted with permissions from references listed in legend. Bars: 200nm in C, R; 50nm in D, K, Q; 100nm in E, H, I, L, P; 25nm in F; 1μm in G, J, N (left); 250nm in N (right).
Another element of the bacterial cytoskeleton that appears to act as an isolated rod in vivo is the tubulin homolog FtsZ, the major cell division protein in almost all bacteria. FtsZ acts as both a scaffold to recruit other proteins to the septum and as a “cytomotive” filament (one that pushes or pulls objects) [reviews: 11,12]. Through ECT of dividing cells, FtsZ filaments were seen to be either straight or curved, isolated filaments typically shorter than the diameter of the cell slightly displaced from the membrane (Figure 1B) [13]. More recent studies showed that fluorescently labeled FtsZ with a short membrane targeting sequence can self-assemble into contractile rings inside liposomes [14]. These in vitro studies have shown that FtsZ alone can generate constrictive forces, that FtsZ protofilaments have a preferred direction of bending [15], and that GTP-hydrolysis and filament remodeling are necessary for continuous constriction [16]. Molecular dynamics simulations have supported a hypothesis that FtsZ protofilament bending alone could generate sufficient force to constrict membranes [17]. Super-resolution fluorescence light microscopy (fLM) confirmed the ECT result by showing that FtsZ was unevenly distributed around the ingression “ring” (Figure 1C) [18-20]. Present data therefore favors (though not clearly [11,21]) an “iterative pinching” model in which FtsZ polymerizes into straight filaments in its GTP-bound state, is tethered to the membrane, and then bends upon GTP hydrolysis, pulling the membrane inwards [13,22]. Cell-wall-remodeling enzymes recruited by FtsZ apparently then add new peptidoglcan behind the advancing membrane, preventing it from relaxing when the FtsZ filament depolymerizes.
FtsZ is tethered to the membrane by the actin homolog FtsA [23]. FtsA was recently shown to polymerize into actin-like filaments and 2D sheets in vitro [24], but its structure and position with respect to other divisome components in vivo remain unknown. Mutants disrupted in polymerization have an elongated cell division phenotype, however, suggesting that polymer formation is important for FtsA function, and truncated FtsA lacking its membrane-binding helix formed straight protofilaments in vivo. Overexpression of full-length FtsA resulted in membrane distortions and the formation of polymer-coated lipid tubes.
Rings
Without evolving any additional binding interfaces, but given a special curvature, individual filaments can loop around and bind themselves to form complete rings. SepF may be an example. Thick-walled bacteria such as Gram-positives and cyanobacteria purportedly have special requirements regarding the arrangement of FtsZ filaments [25]. Proper division of these cells requires SepF, which forms ~50-nm diameter rings in vitro (Figure 1D) and is thought to bundle FtsZ in vivo. If SepF does indeed bundle FtsZ filaments by forming rings around them, it might provide the benefits of filament bundling without the need for intrinsic lateral interactions between FtsZ filaments that might otherwise interfere with FtsZ's conformational changes. While simple linear SepF “crosslinks” might also be able to bundle FtsZ, the authors point out that a ring superstructure can limit the bundle width, promoting the growth of FtsZ filaments lengthwise.
Twisted filament pairs
A superstructure one step more complex than individual or loosely-bundled filaments is a tightly coupled pair. In fact this appears to be a highly effective superstructure, as a diverse set of cytomotive filaments have evolved to form twisted filament pairs that propel other structures throughout the cell. Filament pairs require an additional lateral binding interface, but offer increased stiffness and the possibility of staying attached to an object even while one of the filaments is growing or shrinking. In contrast to FtsZ's bending mechanism, these cytomotive filaments evolved dynamic instability or treadmilling in order to generate motility.
For plasmid segregation, the cytomotive filament usually acts in concert with a centromere-like region of DNA and a DNA-binding adaptor, as exemplified by the ParMRC system [review: 26]. Dynamically instable filaments of the actin homolog ParM cycle between growth and shrinkage until they are stabilized by binding ParR-parC complexes at both ends. Plasmids are then pushed to opposite cell poles by bidirectional elongation of the ParM filaments. Like F-actin, ParM assembles into a polar double-helical filament pair in vitro (Figure 1E), though with a different handedness [27-31]. Erickson has made an interesting argument that ParM may form an apolar, antiparallel double-helical structure in vivo, however, since this could better explain bidirectional filament growth [32]. ECT of cells harboring the ParMRC system found that in vivo, 3-5 presumably double-stranded filaments aligned in a bundle (Figure 1E)[33]. Bundling provides the stiffness that is likely needed to segregate the large plasmid-adaptor complexes, but it is unclear how many filaments in a bundle are attached to plasmids. Like ParM, AlfA (for actin-like filament) [34-36] and various Alps (for actin-like proteins) [37,38] are also involved in plasmid segregation. AlfA is thought to treadmill in vitro and the double-filament has a much higher twist, however, suggesting a different mechanism.
In addition to the above actin homologs, a diverse set of homologs of tubulin (TubZ, phage TubZ and PhuZ) also form DNA-positioning twisted filament pairs. In combination with an adaptor (TubR) and a centromere (tubC), TubZ segregates plasmids through treadmilling [39-41]. Electron microscopy (EM) of in vitro and in vivo TubZ filaments suggests a double-helical, ParM-like structure (Figure 1F) [42]. Well-ordered bundles of presumably double-stranded filaments were observed in cryotomograms of E. coli cells overproducing TubZ (Figure 1F). Strikingly, two recent studies showed that tubulin homologs are also involved in bacteriophage DNA positioning. Upon cell entry, the Clostridium botulinum c-st phage genome is recircularized into a plasmid-like entity and segregated by a phage TubZ, which again acts together with an adaptor and a centromere, but also an additional modulator [43]. In vitro, phage TubZ polymerizes into double-helical filaments that coalesce into bundles. A second phage tubulin homolog, PhuZ from bacteriophage 201! 2-1, is required during the lytic phage cycle [44]. A single-cell assay monitored by fLM revealed that dynamic PhuZ filaments form a spindle that positions phage DNA at the cell center (Figure 1G), enhancing phage production. Like the other DNA positioning filaments, PhuZ forms twisted filament pairs in vitro, and the authors speculate that these generate pushing forces in vivo to hold the phage DNA at midcell. Surprisingly simple changes in cell length or filament number or dynamics can shift whether filament pairs localize objects at the poles or the center of cells [45].
Another class of proteins involved in DNA and protein positioning are Walker A “cytoskeletal” ATPases (WACA) [reviews: 46,47,48]. Two prominent members are ParA and MinD, which segregate DNA and position the septum, respectively. While WACA polymerize into filaments in vitro; whether they form filaments in vivo (and really are therefore “cytoskeletal”) remains unclear.
Tubes
Given lateral interactions between filaments, more complex structures like tubes and sheets can form. Compared to simple rods and twisted pairs, tubes offer enhanced stiffness and mechanical strength, allowing them to act as firm tracks for motors, for instance.
Do bacteria contain microtubules? There are indeed scattered reports of bacterial microtubule-like structures [review: 49]; but later sequencing has revealed that tubulin genes are absent from the organisms described. Hence, microtubules were considered exclusively eukaryotic until a recent study on bacterial tubulins BtubA and BtubB in the phylum Verrucomicrobia [50]. Early on, it was thought that BtubA/B might have taken over the function of its distant homolog FtsZ, since Verrucomicrobia are the closest relatives of FtsZ-lacking Chlamydiae and Planctomycetes, but FtsZ was later found in the same genomes as BtubA/B [51,52]. The sequences and crystal structures of BtubA/B are more similar to eukaryotic tubulins than to FtsZ [50,51,53-55], but negative stain EM suggested that BtubA/B polymerize into protofilament bundles rather than tubes in vitro [54,56]. ECT showed, however, that in vivo BtubA/B assembles into slender tubes (Figure 1H) which run the length of the cell close to the cytoplasmic membrane in bundles of up to four [50]. Interestingly and in contrast to eukaryotic tubulins, BtubA/B can fold without chaperones and they can be heterologously expressed in E. coli; the recombinant proteins also form tubes in vitro and in vivo. Because BtubA/B tubes had the same basic architecture as eukaryotic microtubules (but with only five protofilaments), they were termed ‘bacterial microtubules (bMTs)’ (Figure 1H) [50]. Their 5-protofilament architecture might be ancient, since BtubA/B most probably arose from early tubulin intermediates [50,55]. Interestingly, a gene with low similarities to kinesin light chains is part of the operon encoding bMTs [57], but it remains unclear whether bMTs act as tracks for a motor, cytomotive tubes or scaffolds.
An amazing capability of tubes is illustrated by the bacterial type VI secretion (T6S) system, which injects effector proteins into neighboring cells [review: 58]. A combination of ECT and fLM recently revealed that T6S functions like a spring-loaded molecular dagger (Video 1 and Figure 1I, J) [59]: an inner rod (the dagger) is propelled out of the cell by rapid contraction of a tubular outer sheath (the spring). Because the sheath is a dynamic, proteinaceous, cytoplasmic tube involved in moving material, we include it here as a novel cytomotive element of the bacterial cytoskeleton. Sheath contraction provides the energy needed to move the inner rod [59], and sheath disassembly relies on a AAA+ ATPase [60-62]. T6S genes are highly abundant and widespread among diverse phyla and are involved in competition, defense, pathogenesis and symbiosis [review: 63]. Interestingly, the T6S mechanism and some of its components are homologous to the contractile bacteriophage tail [64]. Phage and bacterium both take advantage of the tube geometry, which is effective to collect the forces needed to transport materials across membranes.
Sheets
In addition to tubes, lateral interactions between filaments can also lead to sheets, which can provide mechanical support, separate compartments, or act as high-surface-area scaffolds. The bactofilins BacA/B in Caulobacter, for instance, localize as sheets at the poles, where they assist stalk morphogenesis by recruiting a peptidoglycan synthase (Figure 1K) [65]. Although bactofilins are almost universally conserved among bacteria, they have a variety of functions. A Myxococcus strain mutated in one bactofilin paralogue had impaired social motility for instance [65], while mutation of another (BacM) led to altered cell morphology [66]. Based on fLM and in vitro experiments the authors suggested that instead of sheets, BacM forms fibers which bundle into helical cables throughout the cell [66], but such a structure has not yet been directly visualized.
The metabolic enzyme CTP synthase (CtpS) also forms sheets in some species. CtpS is conserved in all domains of life, and some bacterial and eukaryotic homologs have been shown to form filaments [review: 3]. While the function of polymerization might typically be the regulation of enzymatic activity, in Caulobacter crescentus CtpS polymers are also involved in cell shape determination [67]. Caulobacter CtpS forms stacks of sheets that lie along the inner curvature of the crescent-shaped cells (Figure 1L) [67,68], where it appears to regulate the activity of another shape-determining polymer, crescentin (CreS) [67].
CreS was the first characterized bacterial intermediate-filament-like protein [69]. CreS is thought to induce curvature in Caulobacter cells by applying asymmetric tension to the cell wall, inhibiting new cell wall growth on one side [70]. fLM, EM, and biochemical studies suggest that CreS forms a substantial ribbon-like structure with properties similar to eukaryotic intermediate filaments [70-73]. No such ribbon-like structure was seen by ECT [68], however, so the in vivo superstructure remains unclear. Like CtpS, CreS localizes to the inner curvature of Caulobacter cells where the two have apparently antagonistic effects: CtpS decreases CreS-induced curvature [67]. One of the distinguishing characteristics of intermediate filament proteins are extended coil-coil domains. In addition to CreS, multiple other coiled-coil rich (Ccrp) proteins have been identified in bacteria and found to be involved in cell shape, rigidity and motility [74-77]. Ccrps assemble into various polymerization condition-dependent superstructures in vitro, but again their superstructures in vivo remain unknown.
Spirals
While the lateral interactions of linear filaments can produce sheets, so can circular coiling. During Bacillus endospore formation, the protein SpoIVA localizes to the mother-side of the outer spore membrane and recruits spore coat proteins [78]. SpoIVA was observed to form polymers in vitro and contains a Walker A box, so it should be considered a WACA (it has not yet been described as such). In fact, in contrast to other better-known WACAs there is strong evidence that SpoIVA actually forms a cytoskeletal structure in vivo: cryotomograms of sporulating Acetonema longum cells showed concentric, ring-shaped densities on the outer spore membrane (Figure 1M) which were most likely SpoIVA [79]. As a filamentous cytoplasmic scaffold, these rings/spirals should also be considered part of the bacterial cytoskeleton.
Moving ‘Patches’(?)
Perhaps the most well-known supposed element of the bacterial cytoskeleton is the actin homolog MreB, which is present in most non-spherical bacteria and is essential for cell shape determination [80,81]. Early fLM suggested that MreB polymerizes into extended helical filaments that encircle the cell just inside the cytoplasmic membrane [82-85], but extended helices were not found in cryotomograms of diverse rod-shaped cells, even after careful computational searches [86]. In 2011 three groups then reported that instead of forming helices, MreB formed small ‘patches’ that moved circumferentially around cells (Video 2 and Figure 1N), driven by cell wall synthesis rather than MreB's intrinsic ATPase activity [87-89]. ECT has now shown that at least in E. coli, the previously reported helices were an artifact caused by an N-terminal YFP tag (Figure 1O) [90]. This is not surprising given the recent findings that E. coli MreB is anchored to the membrane by an N-terminal amphipathic helix (which is most likely blocked by N-terminal tags), and that membrane binding is essential for MreB's role in cell shape determination [91]. Indeed Thermotoga MreB forms double filaments and sheets on a 2D lipid monolayer and it distorts lipid vesicles by the formation of small sheets (Figure 1P). At high protein levels, MreB also distorts membranes in vivo. Thus while the structure and function of MreB in vivo has yet to be determined, the data available today suggest that MreB forms small sheet-like polymers or ’patches’ on the membrane together with cell wall synthetic machinery. Small patches could conceivably be effective in maximizing surface area without overly inhibiting movement along the interface of a complex membrane and a crowded cytoplasm.
Meshes
While lateral binding interactions lead to tubes and sheets, 3-way junctions can lead to 3-D meshes, which allow for cell compartmentalization. Biochemical analyses, in vitro polymerization (Figure 1Q), and cellular ECT suggest that the polar organizing protein Z (PopZ) forms such a mesh in vivo [92-94]. PopZ provides an anchor for the parS/ParB chromosomal centromere at the Caulobacter cell pole and later recruits various proteins for stalked pole development. PopZ also establishes a special ribosome-excluding zone (Figure 1Q).
Multi-element composite
Finally, multiple cytoskeletal elements can come together in multi-element composites. Gliding motility, cell division, and attachment of different Mycoplasma species, for instance, depend on composite superstructures that have also been described as cytoskeletal. Their composition and structure is species-dependent and can include elements termed jellyfish, cap, oval, bowl, bulge, ankle, thick and thin rods, terminal button, etc. (Figure 1R) [95-97, and review 98].
Conclusion
Function dictates superstructure
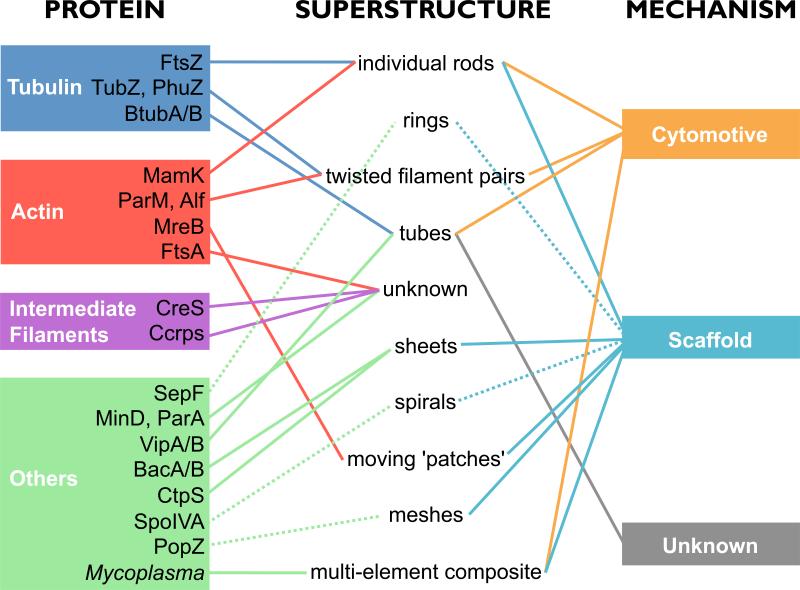
Each superstructure of the bacterial cytoskeleton has special advantages that optimize its role in moving objects, arranging materials, or both (Figure 2). Similar functions lead even unrelated proteins to evolve similar superstructures – actins and tubulins segregate plasmids as twisted filaments, CTP synthase and bactofilins (and possibly MreB) recruit proteins to the membrane as sheets. Homologs with different functions, however, form surprisingly different superstructures – tubulins are found as twisted filament pairs (PhuZ/TubZ), bending rods (FtsZ), and tubes (BtubA/B) (and perhaps more - the superstructures of FtsZ-like proteins [99] and archaeal “artubulins” [100] remain unknown).
Figure 2. The bacterial cytoskeleton: proteins, superstructures and mechanism of function.
Bacterial cytoskeletal elements evolved a variety of superstructures, each adapted to the protein's function in moving (cytomotive) or stabilizing/positioning/recruiting (scaffold) objects within the cell. Connections with weaker evidence are represented as dashed lines.
Boundaries of our definition
Here we have included as bacterial cytoskeletal elements all cytoplasmic filamentous superstructures with either a cytomotive or scaffolding function. As with all definitions, there are ambiguities at the boundaries. Although PhuZ and phage TubZ, for instance, are not truly bacterial proteins, they were included in this review because they act in the bacterial cytoplasm. Phage tails and capsids, however, act mostly outside the cell and were excluded just like flagella and their motors, whose main components are also located outside the cytoplasm. Chemoreceptor arrays and carboxysome shells were not discussed either, since their superstructures are hexagonal or polyhedral, rather than higher-order assemblies of filaments, but these fine distinctions illustrate the growing ambiguities the bacterial “cytoskeleton” is presenting: bactofilin and CtpS sheets may also be hexagonal, some carboxysomes are so long that they could be considered filamentous at the superstructural level [101], and the in vivo superstructures of many widely discussed members of the bacterial “cytoskeleton” like MreB, CreS, FtsA, SepF, most WACA, Ccrps, and SepF remain unknown! Such ambiguities emphasize the importance of molecular-resolution, in vivo imaging of wildtype cells [review: 5]: almost all proteins will polymerize under some condition in vitro (as proven by the success of X-ray crystallography), and fluorescence signals can be deceiving (as shown by the case of MreB [90] and likely other supposedly “helical” filaments). Thus while the growing diversity of superstructures produces some semantic challenges, it is nevertheless amazing and beautiful (who would have imagined membrane-bending pyramids, as recently seen in archaea [102]!), and there is surely much left to discover.
Supplementary Material
Video 1. Type VI secretion functions like a spring-loaded dagger. This narrated video summarizes ECT and fLM results and shows an animation of the type VI secretion mechanism in Vibrio cholerae (from [59]).
Video 2. MreB patches move circumferentially around Bacillus cells. Shown are timelapse fLM stacks for three MreB paralogs (from [88]).
Acknowledgments
Thanks to Matt Swulius, Ariane Briegel, Morgan Beeby and Elitza Tocheva for helpful discussions and comments on the manuscript. Bacterial cytoskeleton research was supported by the Howard Hughes Medical Institute, NIH grant R01 GM094800B, the Center for Environmental Biology Interactions at Caltech and the Bayerische Forschungsstiftung.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Graumann PL. Dynamics of bacterial cytoskeletal elements. Cell Motil Cytoskeleton. 2009;66:909–914. doi: 10.1002/cm.20381. [DOI] [PubMed] [Google Scholar]
- 2.Cabeen MT, Jacobs-Wagner C. The bacterial cytoskeleton. Annu Rev Genet. 2010;44:365–392. doi: 10.1146/annurev-genet-102108-134845. [DOI] [PubMed] [Google Scholar]
- 3.Ingerson-Mahar M, Gitai Z. A growing family: the expanding universe of the bacterial cytoskeleton. Fems Microbiology Reviews. 2012;36:256–266. doi: 10.1111/j.1574-6976.2011.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry RM, Gitai Z. Self-assembling enzymes and the origins of the cytoskeleton. Curr Opin Microbiol. 2011 doi: 10.1016/j.mib.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pilhofer M, Ladinsky MS, McDowall AW, Jensen GJ. Bacterial TEM: new insights from cryo-microscopy. Methods Cell Biol. 2010;96:21–45. doi: 10.1016/S0091-679X(10)96002-0. [DOI] [PubMed] [Google Scholar]
- 6.Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schüler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- 7.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 8.Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schuler D. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol Microbiol. 2010;77:208–224. doi: 10.1111/j.1365-2958.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 9*.Draper O, Byrne ME, Li Z, Keyhani S, Barrozo JC, Jensen G, Komeili A. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol Microbiol. 2011;82:342–354. doi: 10.1111/j.1365-2958.2011.07815.x. [Refuting previous assumptions, this study showed that MamK filaments are dynamic in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Katzmann E, Muller FD, Lang C, Messerer M, Winklhofer M, Plitzko JM, Schuler D. Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol Microbiol. 2011;82:1316–1329. doi: 10.1111/j.1365-2958.2011.07874.x. [This study showed that besides organizing the magnetosome chain, MamK also plays a role in its pole-to-midcell translocation.] [DOI] [PubMed] [Google Scholar]
- 11.Erickson HP, Anderson DE, Osawa M. FtsZ in Bacterial Cytokinesis: Cytoskeleton and Force Generator All in One. Microbiol Mol Biol Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkpatrick CL, Viollier PH. New(s) to the (Z-)ring. Current Opinion in Microbiology. 2011;14:691–697. doi: 10.1016/j.mib.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osawa M, Anderson DE, Erickson H. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osawa M, Anderson DE, Erickson HP. Curved FtsZ protofilaments generate bending forces on liposome membranes. Embo Journal. 2009;28:3476–3484. doi: 10.1038/emboj.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Osawa M, Erickson HP. Inside-out Z rings--constriction with and without GTP hydrolysis. Mol Microbiol. 2011;81:571–579. doi: 10.1111/j.1365-2958.2011.07716.x. [This study suggests that GTP hydrolysis is necessary for continuous membrane constriction by FtsZ, which argues against a closed FtsZ ring structure.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Hsin J, Gopinathan A, Huang KC. Nucleotide-dependent conformations of FtsZ dimers and force generation observed through molecular dynamics simulations. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9432–9437. doi: 10.1073/pnas.1120761109. [Molecular dynamics simulations are used to argue that FtsZ protofilament bending could generate enough force to constrict membranes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo F, Tao HA, Buss J, Coltharp C, Hensel Z, Jie XA. In Vivo Structure of the E. coli FtsZ-ring Revealed by Photoactivated Localization Microscopy (PALM). Plos One. 2010;5 doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biteen JS, Goley ED, Shapiro L, Moerner WE. Three-Dimensional Super-Resolution Imaging of the Midplane Protein FtsZ in Live Caulobacter crescentus Cells Using Astigmatism. Chemphyschem. 2012;13:1007–1012. doi: 10.1002/cphc.201100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 3D-SIM Super Resolution Microscopy Reveals a Bead-Like Arrangement for FtsZ and the Division Machinery: Implications for Triggering Cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milam SL, Osawa M, Erickson HP. Negative-Stain Electron Microscopy of Inside-Out FtsZ Rings Reconstituted on Artificial Membrane Tubules Show Ribbons of Protofilaments. Biophys J. 2012;103:59–68. doi: 10.1016/j.bpj.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu C, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. Journal of Bacteriology. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol.Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 24**.Szwedziak P, Wang Q, Freund SMV, Lowe J. FtsA forms actin-like protofilaments. Embo Journal. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [This study showed for the first time that polymerization of FtsA is important for proper function in vivo, suggesting it might act as a novel cytoskeletal element.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Gundogdu ME, Kawai Y, Pavlendova N, Ogasawara N, Errington J, Scheffers DJ, Hamoen LW. Large ring polymers align FtsZ polymers for normal septum formation. Embo Journal. 2011;30:617–626. doi: 10.1038/emboj.2010.345. [This study discovered the protein SepF, which might bundle FtsZ protofilaments in vivo by forming a ring-shaped superstructure.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salje J, Gayathri P, Lowe J. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol. 2010;8:683–692. doi: 10.1038/nrmicro2425. [DOI] [PubMed] [Google Scholar]
- 27.van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orlova A, Garner EC, Galkin VE, Heuser J, Mullins RD, Egelman EH. The structure of bacterial ParM filaments. Nat Struct Mol Biol. 2007;14:921–926. doi: 10.1038/nsmb1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popp D, Narita A, Oda T, Fujisawa T, Matsuo H, Nitanai Y, Iwasa M, Maeda K, Onishi H, MaAcda Y. Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 2008;27:570–579. doi: 10.1038/sj.emboj.7601978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galkin VE, Orlova A, Rivera C, Mullins RD, Egelman EH. Structural polymorphism of the ParM filament and dynamic instability. Structure. 2009;17:1253–1264. doi: 10.1016/j.str.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galkin VE, Orlova A, Egelman EH. Are ParM Filaments Polar or Bipolar? J Mol Biol. 2012;423:482–485. doi: 10.1016/j.jmb.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erickson HP. Bacterial Actin Homolog ParM: Arguments for an Apolar, Antiparallel Double Helix. J Mol Biol. 2012 doi: 10.1016/j.jmb.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salje J, Zuber B, Lowe J. Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science. 2009;323:509–512. doi: 10.1126/science.1164346. [DOI] [PubMed] [Google Scholar]
- 34.Becker E, Herrera NC, Gunderson FQ, Derman AI, Dance AL, Sims J, Larsen RA, Pogliano J. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 2006;25:5919–5931. doi: 10.1038/sj.emboj.7601443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polka JK, Kollman JM, Agard DA, Mullins RD. The Structure and Assembly Dynamics of Plasmid Actin AlfA Imply a Novel Mechanism of DNA Segregation. Journal of Bacteriology. 2009;191:6219–6230. doi: 10.1128/JB.00676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popp D, Narita A, Ghoshdastider U, Maeda K, Maeda Y, Oda T, Fujisawa T, Onishi H, Ito K, Robinson RC. Polymeric Structures and Dynamic Properties of the Bacterial Actin AlfA. Journal of Molecular Biology. 2010;397:1031–1041. doi: 10.1016/j.jmb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, Patterson PC, Pogliano J. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol. 2009;73:534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivera CR, Kollman JM, Polka JK, Agard DA, Mullins RD. Architecture and assembly of a divergent member of the ParM family of bacterial actin-like proteins. J Biol Chem. 2011;286:14282–14290. doi: 10.1074/jbc.M110.203828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anand SP, Akhtar P, Tinsley E, Watkins SC, Khan SA. GTP-dependent polymerization of the tubulin-like RepX replication protein encoded by the pXO1 plasmid of Bacillus anthracis. Mol Microbiol. 2008;67:881–890. doi: 10.1111/j.1365-2958.2007.06100.x. [DOI] [PubMed] [Google Scholar]
- 41*.Ni L, Xu W, Kumaraswami M, Schumacher MA. Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc Natl Acad Sci U S A. 2010;107:11763–11768. doi: 10.1073/pnas.1003817107. [This study gave mechanistic insights into how TubZ acts in concert with an adaptor and a centromorer-like region in order to segregate plasmids.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aylett CH, Wang Q, Michie KA, Amos LA, Lowe J. Filament structure of bacterial tubulin homologue TubZ. Proc Natl Acad Sci U S A. 2010;107:19766–19771. doi: 10.1073/pnas.1010176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Oliva MA, Martin-Galiano AJ, Sakaguchi Y, Andreu JM. Tubulin homolog TubZ in a phage-encoded partition system. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7711–7716. doi: 10.1073/pnas.1121546109. [This study discovered that a tubulin homolog is used by a bacteriophage to segregate its DNA in the bacterial cytoplasm.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Kraemer JA, Erb ML, Waddling CA, Montabana EA, Zehr EA, Wang HN, Nguyen K, Pham DSL, Agard DA, Pogliano J. A Phage Tubulin Assembles Dynamic Filaments by an Atypical Mechanism to Center Viral DNA within the Host Cell. Cell. 2012;149:1488–1499. doi: 10.1016/j.cell.2012.04.034. [This interdisciplinary study discovered that a phage tubulin homolog is utilized to position phage DNA at midcell.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45*.Drew KR, Pogliano J. Dynamic instability-driven centering/segregating mechanism in bacteria. Proc Natl Acad Sci U S A. 2011;108:11075–11080. doi: 10.1073/pnas.1018724108. [Observations of a synthetic system and computer simulations show that a cytomotive filament can be shifted from pole- to midcell-positioning by surprisingly simple changes in filament length, number, or dynamics.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lowe J, Amos LA. Evolution of cytomotive filaments: The cytoskeleton from prokaryotes to eukaryotes. Int J Biochem Cell Biol. 2008;41:323–329. doi: 10.1016/j.biocel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 47.Szardenings F, Guymer D, Gerdes K. ParA ATPases can move and position DNA and subcellular structures. Curr Opin Microbiol. 2011;14:712–718. doi: 10.1016/j.mib.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Lutkenhaus J. The ParA/MinD family puts things in their place. Trends Microbiol. 2012 doi: 10.1016/j.tim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bermudes D, Hinkle G, Margulis L. Do Prokaryotes Contain Microtubules. Microbiol Rev. 1994;58:387–400. doi: 10.1128/mr.58.3.387-400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. Microtubules in bacteria: ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. Plos Biol. 2011;9:e1001213. doi: 10.1371/journal.pbio.1001213. [Reports the discovery of a microtubules in bacteria built from BtubA/B, and argues that their novel 5-protofilament architecture is likely ancient.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pilhofer M, Rosati G, Ludwig W, Schleifer KH, Petroni G. Coexistence of tubulins and ftsZ in different Prosthecobacter species. Mol Biol Evol. 2007;24:1439–1442. doi: 10.1093/molbev/msm069. [DOI] [PubMed] [Google Scholar]
- 52.Pilhofer M, Rappl K, Eckl C, Bauer AP, Ludwig W, Schleifer KH, Petroni G. Characterization and evolution of cell division and cell wall synthesis genes in the bacterial phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and phylogenetic comparison with rRNA genes. J Bacteriol. 2008;190:3192–3202. doi: 10.1128/JB.01797-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jenkins C, Samudrala R, Anderson I, Hedlund BP, Petroni G, Michailova N, Pinel N, Overbeek R, Rosati G, Staley JT. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc Natl Acad Sci U S A. 2002;99:17049–17054. doi: 10.1073/pnas.012516899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schlieper D, Oliva MA, Andreu JM, Lowe J. Structure of bacterial tubulin BtubA/B: Evidence for horizontal gene transfer. Proc Natl Acad Sci U S A. 2005;102:9170–9175. doi: 10.1073/pnas.0502859102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin-Galiano AJ, Oliva MA, Sanz L, Bhattacharyya A, Serna M, Yebenes H, Valpuesta JM, Andreu JM. Bacterial tubulin distinct loop sequences and primitive assembly properties support its origin from a eukaryotic tubulin ancestor. J Biol Chem. 2011 doi: 10.1074/jbc.M111.230094. doi:10.1074/jbc.M1111.230094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sontag CA, Staley JT, Erickson HP. In vitro assembly and GTP hydrolysis by bacterial tubulins BtubA and BtubB. J Cell Biol. 2005;169:233–238. doi: 10.1083/jcb.200410027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pilhofer M, Bauer AP, Schrallhammer M, Richter L, Ludwig W, Schleifer KH, Petroni G. Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel Two-Step Gene Walking method. Nucleic Acids Res. 2007;35:e135. doi: 10.1093/nar/gkm836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jani AJ, Cotter PA. Type VI Secretion: Not Just for Pathogenesis Anymore. Cell Host & Microbe. 2010;8:2–6. doi: 10.1016/j.chom.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59**.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [A combination of fluorescence microscopy and electron cryotomography reveal that bacterial type VI secretion systems act like spring-loaded molecular daggers.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. Embo Journal. 2009;28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pietrosiuk A, Lenherr ED, Falk S, Bonemann G, Kopp J, Zentgraf H, Sinning I, Mogk A. Molecular Basis for the Unique Role of the AAA(+) Chaperone ClpV in Type VI Protein Secretion. Journal of Biological Chemistry. 2011;286:30010–30021. doi: 10.1074/jbc.M111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Basler M, Mekalanos JJ. Type 6 Secretion Dynamics Within and Between Bacterial Cells. Science. 2012 doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 64.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65**.Kuhn J, Briegel A, Morschel E, Kahnt J, Leser K, Wick S, Jensen GJ, Thanbichler M. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 2010;29:327–339. doi: 10.1038/emboj.2009.358. [Bactofilins are described as novel bacterial cytoskeletal elements and shown to form sheets which recruit a particular cell wall synthase.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koch MK, McHugh CA, Hoiczyk E. BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Molecular Microbiology. 2011;80:1031–1051. doi: 10.1111/j.1365-2958.2011.07629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67**.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol. 2010;12:739–746. doi: 10.1038/ncb2087. [Shows that CTP synthase forms sheets in Caulobacter that counteract the curvature-inducing effects of crescentin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol. 2006;62:5–14. doi: 10.1111/j.1365-2958.2006.05355.x. [DOI] [PubMed] [Google Scholar]
- 69.Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 70.Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 2009 doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Charbon G, Cabeen MT, Jacobs-Wagner C. Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin. Genes Dev. 2009;23:1131–1144. doi: 10.1101/gad.1795509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Esue O, Rupprecht L, Sun SX, Wirtz D. Dynamics of the bacterial intermediate filament crescentin in vitro and in vivo. PLoS One. 2010;5:e8855. doi: 10.1371/journal.pone.0008855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73*.Cabeen MT, Herrmann H, Jacobs-Wagner C. The domain organization of the bacterial intermediate filament-like protein crescentin is important for assembly and function. Cytoskeleton (Hoboken) 2011;68:205–219. doi: 10.1002/cm.20505. [This study investigated the domain organization of crescentin and suggests that it is the basis for crescentin's IF-like characteristics in vitro.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bagchi S, Tomenius H, Belova LM, Ausmees N. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol Microbiol. 2008;70:1037–1050. doi: 10.1111/j.1365-2958.2008.06473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waidner B, Specht M, Dempwolff F, Haeberer K, Schaetzle S, Speth V, Kist M, Graumann PL. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS Pathog. 2009;5:e1000669. doi: 10.1371/journal.ppat.1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fiuza M, Letek M, Leiba J, Villadangos AF, Vaquera J, Zanella-Cleon I, Mateos LM, Molle V, Gil JA. Phosphorylation of a novel cytoskeletal protein (RsmP) regulates rod-shaped morphology in Corynebacterium glutamicum. J Biol Chem. 2010;285:29387–29397. doi: 10.1074/jbc.M110.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Specht M, Schatzle S, Graumann PL, Waidner B. Helicobacter pylori possesses four coiled-coil-rich proteins that form extended filamentous structures and control cell shape and motility. J Bacteriol. 2011;193:4523–4530. doi: 10.1128/JB.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramamurthi KS, Losick R. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell. 2008;31:406–414. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79*.Tocheva EI, Matson EG, Morris DM, Moussavi F, Leadbetter JR, Jensen GJ. Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell. 2011;146:799–812. doi: 10.1016/j.cell.2011.07.029. [ECT images of sporulation in a Gram-negative species provide the strongest evidence that a WACA (SpoIVA) forms a cytoskeletal structure in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Divakaruni A, Baida C, White C, Gober J. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- 81.Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, et al. A widespread family of bacterial cell wall assembly proteins. Embo Journal. 2011;30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jones LJF, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 83.Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. Embo Journal. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shih YL, Le T, Rothfield L. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci U S A. 2003;100:7865–7870. doi: 10.1073/pnas.1232225100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Figge RM, Divakaruni A, Gober J. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 86**.Swulius MT, Chen S, Jane Ding H, Li Z, Briegel A, Pilhofer M, Tocheva EI, Lybarger SR, Johnson TL, Sandkvist M, et al. Long helical filaments are not seen encircling cells in electron cryotomograms of rod-shaped bacteria. Biochem Biophys Res Commun. 2011;407:650–655. doi: 10.1016/j.bbrc.2011.03.062. [Careful computational searches of electron cryotomograms fail to find extended helical structures in diverse rod-shaped bacteria, challenging the numerous claims of their existence from fluorescence studies.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87*.Dominguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Soldner R, Carballido-Lopez R. Processive Movement of MreB-Associated Cell Wall Biosynthetic Complexes in Bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. See 88.
- 88*.Garner EC, Bernard R, Wang WQ, Zhuang XW, Rudner DZ, Mitchison T. Coupled, Circumferential Motions of the Cell Wall Synthesis Machinery and MreB Filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [Refs. #87-89 are all fluorescence microscopy studies showing that MreB forms ‘patches’ driven around the circumference of rod-shaped cells by cell-wall-synthetic machinery.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89*.van Teeffelen S, Wang SY, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. See 88.
- 90*.Swulius MT, Jensen GJ. The helical MreB cytoskeleton in E. coli MC1000/pLE7 is an artifact of the N-terminal YFP tag. J Bacteriol. 2012 doi: 10.1128/JB.00505-12. [Electron cryotomography shows that the MreB helices previously reported in E. coli were an artifact of an N-terminal YFP tag.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91*.Salje J, van den Ent F, de Boer P, Lowe J. Direct Membrane Binding by Bacterial Actin MreB. Molecular Cell. 2011;43:478–487. doi: 10.1016/j.molcel.2011.07.008. [Shows that E. coli MreB binds membranes directly through an N-terminal amphipathic helix, and that MreB can induce membrane curvature by forming sheet-like structures of short filaments.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bowman GR, Comolli LR, Gaietta GM, Fero M, Hong SH, Jones Y, Lee JH, Downing KH, Ellisman MH, McAdams HH, et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol. 2010;76:173–189. doi: 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seybert A, Herrmann R, Frangakis AS. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J Struct Biol. 2006;156:342–354. doi: 10.1016/j.jsb.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 96.Henderson GP, Jensen GJ. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol Microbiol. 2006;60:376–385. doi: 10.1111/j.1365-2958.2006.05113.x. [DOI] [PubMed] [Google Scholar]
- 97.Nakane D, Miyata M. Cytoskeletal asymmetrical dumbbell structure of a gliding mycoplasma, Mycoplasma gallisepticum, revealed by negative-staining electron microscopy. J Bacteriol. 2009;191:3256–3264. doi: 10.1128/JB.01823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miyata M. Unique centipede mechanism of Mycoplasma gliding. Annu Rev Microbiol. 2010;64:519–537. doi: 10.1146/annurev.micro.112408.134116. [DOI] [PubMed] [Google Scholar]
- 99.Makarova KS, Koonin EV. Two new families of the FtsZ-tubulin protein superfamily implicated in membrane remodeling in diverse bacteria and archaea. Biology Direct. 2010;5:33. doi: 10.1186/1745-6150-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yutin N, Koonin EV. Archaeal origin of tubulin. Biol Direct. 2012;7:10. doi: 10.1186/1745-6150-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ. Organization, Structure, and Assembly of alpha-Carboxysomes Determined by Electron Cryotomography of Intact Cells. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.11.019. doi:10.1016/j.jmb.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fu CY, Wang K, Gan L, Lanman J, Khayat R, Young MJ, Jensen GJ, Doerschuk PC, Johnson JE. In vivo assembly of an archaeal virus studied with whole-cell electron cryotomography. Structure. 2010;18:1579–1586. doi: 10.1016/j.str.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1. Type VI secretion functions like a spring-loaded dagger. This narrated video summarizes ECT and fLM results and shows an animation of the type VI secretion mechanism in Vibrio cholerae (from [59]).
Video 2. MreB patches move circumferentially around Bacillus cells. Shown are timelapse fLM stacks for three MreB paralogs (from [88]).