Abstract
Neighbor-joining is one of the most widely used methods for constructing evolutionary trees. This approach from phylogenetics is often employed in population genetics, where distance matrices obtained from allele frequencies are used to produce a representation of population relationships in the form of a tree. In phylogenetics, the utility of neighbor-joining derives partly from a result that for a class of distance matrices including those that are additive or tree-like—generated by summing weights over the edges connecting pairs of taxa in a tree to obtain pairwise distances—application of neighbor-joining recovers exactly the underlying tree. For populations within a species, however, migration and admixture can produce distance matrices that reflect more complex processes than those obtained from the bifurcating trees typical in the multispecies context. Admixed populations—populations descended from recent mixture of groups that have long been separated—have been observed to be located centrally in inferred neighbor-joining trees, with short external branches incident to the path connecting their source populations. Here, using a simple model, we explore mathematically the behavior of an admixed population under neighbor-joining. We show that with an additive distance matrix, a population admixed among two source populations necessarily lies on the path between the sources. Relaxing the additivity requirement, we examine the smallest nontrivial case—four populations, one of which is admixed between two of the other three—showing that the two source populations never merge with each other before one of them merges with the admixed population. Furthermore, the distance on the constructed tree between the admixed population and either source population is always smaller than the distance between the source populations, and the external branch for the admixed population is always incident to the path connecting the sources. We define three properties that hold for four taxa and that we hypothesize are satisfied under more general conditions: antecedence of clustering, intermediacy of distances, and intermediacy of path lengths. Our findings can inform interpretations of neighbor-joining trees with admixed groups, and they provide an explanation for patterns observed in trees of human populations.
Keywords: admixture, neighbor-joining, phylogenetics, population genetics
1. Introduction
Distance matrix methods in phylogenetics construct trees of taxa using algorithms applied to matrices that tabulate pairwise evolutionary distances between the taxa.1,2 Among these methods, neighbor-joining3,4 is one of the most popular.5–7 One of its key features is its consistency: if the distance matrix is additive, such that a tree of taxa exists that generates the distances in the matrix, then neighbor-joining recovers this exact tree.5,8,9 Further, neighbor-joining is robust in that theoretical and simulation-based studies have found it to infer sensible trees under a broad range of mathematical and biological conditions.7,9–13
As trees have long been used in population genetics to describe relationships among populations,14,15 the neighbor-joining algorithm has been applied extensively as a population clustering tool, using distance matrices calculated from population-level allele frequencies. In humans, neighbor-joining trees have been and continue to be a regular feature of studies of population relationships.16–20 In population-genetic studies, because migration and admixture sometimes generate evolutionary histories that cannot easily be described by a bifurcating tree of populations, a neighbor-joining tree is treated as a type of population clustering diagram rather than a precise representation of the evolutionary history of the populations.
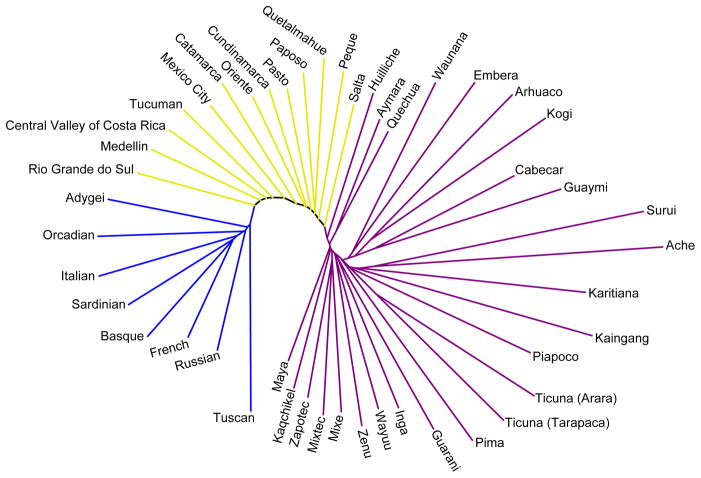
When neighbor-joining has been used with admixed populations—populations recently descended from two or more source groups that have long been separated—particular characteristics of the inferred trees have often been observed (Fig. 1). For example, one simulation study based on human data identified a reduction in the external branch length leading to an admixed population as the strength of gene flow with other populations was increased.21 It has also been suggested on the basis of observed human population trees that a short external branch for a population on a constructed neighbor-joining tree can imply recent admixture of the population, and that admixed populations often appear in the “middle” of a neighbor-joining tree, on branches incident to paths connecting possible source populations.21–25 This pattern is evident in Fig. 2, in which admixed Mestizo populations from Latin America lie on branches incident to the path connecting Native American and European populations. Here, we seek to understand these results on the behavior of admixed populations in the application of the neighbor-joining algorithm. We therefore apply neighbor-joining to populations that satisfy a simple admixture model, first considering the case in which the distance matrix is additive. Next, for the case of n = 4 taxa, we use a mechanistic mathematical investigation to examine three specific properties of neighbor-joining trees involving an admixed population.
Fig. 1.
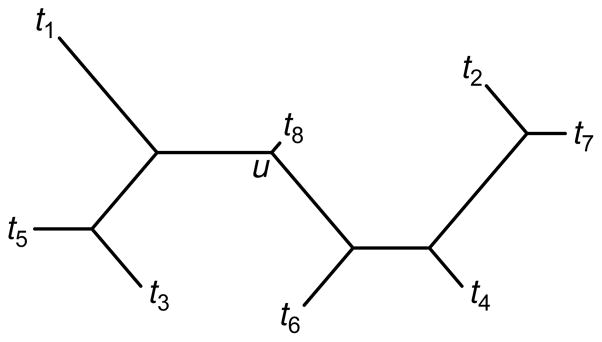
Properties observed for admixed taxa in neighbor-joining trees. Taxon t8 represents an admixture of source populations t1 and t2. The admixed taxon appears on a short external branch incident to the path connecting the source populations. Denoting distances on the tree by d̂ and topological path lengths that count edges separating pairs of taxa by b̂, the tree illustrates the properties of intermediacy of distances (d̂t1,t8 < d̂t1,t2 and d̂t2,t8 < d̂t1,t2, or equivalently, d̂u,t8 < d̂u,t1 and d̂u,t8 < d̂u,t2, where u is the unique node that places t1, t2, and t8 in different subtrees), and intermediacy of path lengths (b̂t1,t8 ≤ b̂t1,t2 and b̂t2,t8 ≤ b̂t1,t2).
Fig. 2.
Neighbor-joining tree of admixed Mestizo populations together with Native American and European populations that represent ancestral source regions for the admixed populations. The tree, obtained using Neighbor and Drawtree in the Phylip package,26 uses data on 678 microsatellite loci in 13 Mestizo, 26 Native American, and 8 European populations.27–29 Allele frequencies were computed from 872 individuals—249 Mestizo, 463 Native American, and 160 European—and distances were computed with Microsat30 using one minus the proportion of shared alleles.31 Mestizo, Native American, and European branches appear in yellow, purple, and blue, respectively. Mestizos lie in the “middle” of the tree, connecting to the path that links the Native Americans and Europeans. External branches for Mestizo populations are shorter on average (0.102) than for Native American (0.146) and European populations (0.109); Mestizo populations have 9 of the 15 shortest external branches.
2. The neighbor-joining algorithm
We briefly review the neighbor-joining algorithm.3,4 Consider a set of n taxa, together with a distance function d computed for each pair of taxa, such that the distance between taxa i and j is denoted dij. The algorithm takes as input the distance matrix D containing entries dij, with i and j ranging from 1 to n, and it outputs a bifurcating unrooted tree. D is symmetric (dij = dji), with zeroes on the diagonals (dii = 0) and nonnegative real entries (dij ≥ 0).
As in other agglomerative algorithms that construct bifurcating trees,2,32 at each of a series of steps, the two nearest taxa according to a selection criterion are connected to a new interior node, becoming “neighbors” on the constructed tree. Branch lengths from the new node to the nodes it agglomerates, as well as the distances to all remaining nodes, are then calculated, and a new distance matrix is obtained. This procedure is repeated iteratively until the last three nodes remain, and these three nodes are then connected to a final interior node. Because the last three nodes are always joined, the number of taxa must exceed three for neighbor-joining to have a nontrivial decision at the first step.
At each step, the key decision is the choice of the pair of taxa that are agglomerated. Neighbor-joining uses an n × n matrix Q, containing entries qij for pairs of taxa (i, j):
| (1) |
The two taxa that are agglomerated are those with the minimal value of qij (choosing randomly in case of ties). If taxa i and j are agglomerated, then their distances to the new node u become
| (2) |
| (3) |
The distances of all remaining nodes k to node u are computed as
| (4) |
The next agglomeration then proceeds from an (n − 1) × (n − 1) distance matrix that replaces distances involving nodes i and j with those involving the single node u.
3. An admixture scenario
We examine a scenario in which one of the taxa is admixed among two of the others. This taxon can be viewed as having been formed from its two source taxa, such that individual members of the taxon have ancestors in both source groups. We label the taxa t1, t2, …, tn. Without loss of generality, let taxon tn be the admixed group, and suppose that it is an admixture of taxa t1 and t2. The relationships among the remaining n − 3 taxa (t3, t4, …, tn−1) and between these taxa and t1, t2, and tn are not specified; we do not consider any additional admixture relationships that might exist among these taxa. We assume n ≥ 4, so that at least one taxon is considered in addition to t1, t2, and tn.
In a standard statistical model of admixture used in population genetics, allele frequencies in an admixed taxon are given by linear combinations of the allele frequencies of the source taxa.33–37 We denote by λ the proportion of the ancestry of taxon tn arising from t1 and by 1 − λ the corresponding proportion arising from t2, where 0 < λ < 1. For any allelic type, if pti denotes the frequency of the specified allele in taxon ti, then
| (5) |
It follows that if for each of the taxa in a pair, a distance function d is linear in each component of the allele frequency vector at a locus, then the distances between the admixed taxon and other taxa are obtained as linear combinations of corresponding distances involving taxa t1 and t2. Therefore, for 1 ≤ i ≤ n − 1,
| (6) |
Eq. 6 continues to hold if for a series of loci, the distance function d is linear for each taxon in each component of the allele frequency vector at each locus, as would occur if the distance between a pair of taxa at a set of loci were computed as the mean of locus-wise distances that were each linear in the components of the allele frequency vector at the specified locus.
We assume that the distance function supplied to neighbor-joining satisfies eq. 6, and that it is symmetric, nonnegative, and zero if and only if it is computed between a taxon and itself; we otherwise do not concern ourselves with the form of the function. While typical population-genetic distance functions often involve nonlinear relationships with allele frequencies and do not necessarily follow eq. 6—consider the nonlinear graphs in Figure 3 of Boca & Rosenberg,38 which illustrate that for the FST measure and an admixed population tn whose frequencies are linear combinations of those of populations t1 and t2, FST (tn, t1) ≠ λFST (t1, t1) + (1 − λ)FST (t2, t1)—eq. 6 is a natural extension of the ubiquitous eq. 5 from allele frequencies to distance functions. For FST, it can be shown from eqs. 1 and 7 of Boca & Rosenberg38 that for small λ, FST (tn, t1) ≈ λFST (t1, t1) + (1 − λ)FST (t2, t1). Thus, we view eq. 6 as a reasonable first approximation for examining properties of neighbor-joining in an admixture scenario.
4. The neighbor-joining algorithm in an admixture scenario
Our goal is to construct a distance matrix according to the admixture rule in eq. 6, mechanistically apply neighbor-joining to the matrix, and characterize the properties of the inference process and the resulting inferred tree. We examine two settings. In the first, arbitrarily many taxa are considered, and their distances produce an additive distance matrix (and therefore satisfy a tree metric39). In the second, a general matrix is investigated, with distances that do not necessarily follow a tree metric, but the matrix includes only four taxa.
4.1. The additive case for n taxa
We first assume that the distance matrix is additive. In this case, by the consistency property of the neighbor-joining algorithm,5,8,9 distances between taxa on the constructed neighbor-joining tree exactly equal those of the input matrix. Denote by d̂ the distance function computed for pairs of nodes in the inferred neighbor-joining tree, such that for taxa ti and tj, d̂titj is the sum of the lengths of the branches on the path connecting ti and tj. Recalling that d represents distance in the input distance matrix, if the matrix is additive, then for all (ti, tj),
| (7) |
Because dt1,tn = (1 − λ)dt1,t2 and dt2,tn = λdt1,t2 by eq. 6,
| (8) |
| (9) |
It follows that d̂t1,tn + d̂t2,tn = d̂t1,t2, from which we can infer that taxa t1, t2, and tn are collinear in the inferred neighbor-joining tree, with tn in the interior of the path from t1 to t2.
We can obtain an even stronger result. Consider a case with at least four taxa: t1, t2, tn, and, without loss of generality, t3 (Fig. 3A). In the inferred neighbor-joining tree, a path of length c connects taxon t3 to some point P on the path from t1 to t2 (including the endpoints). Without loss of generality, we can assume that P lies on the path from t1 to tn (including the endpoints). We denote the distances d̂t1,P and d̂P,tn by nonnegative values y and z, respectively. We denote d̂t1,t2 = dt1,t2 = x, for some nonnegative x.
Fig. 3.
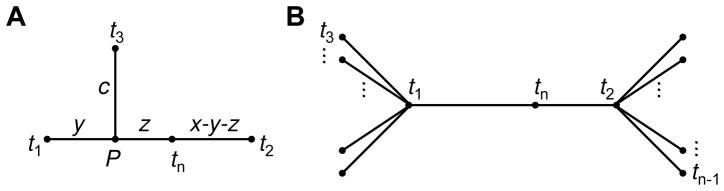
The case of an additive distance matrix for n taxa. (A) Illustration of distances in the model. In the text it is shown that y = 0. (B) The structure required for a tree. Taxa t1 and t2 lie at multifurcating nodes, with tn on the line connecting them. The leaves connected to the multifurcations have labels t3, t4, …, tn−1.
By eq. 7, d̂t1,tn = y + z = dt1,tn = (1 − λ)x. By eqs. 6 and 7,
| (10) |
| (11) |
In other words, c + z = λ(c + y) + (1 − λ)(c + x − y). Together with the relationship y + z = (1 − λ)x and the assumption that λ > 0, eq. 11 implies that y = 0. It then follows that taxon t3 lies on a line with taxa t1, t2, and tn. Further, taxon t3 lies on the side of taxon t1 opposite to taxa t2 and tn; otherwise, by eq. 11, we would have (1 − λ)x − c = λc + (1 − λ)(x − c), which requires c = 0. In turn, c = 0 implies d̂t3,t1 = 0, and hence, dt3,t1 = 0, contradicting the assumption that all pairs of taxa are separated by positive distances in the distance matrix.
We have therefore shown that for an additive tree with taxon tn admixed between t1 and t2, any additional taxon beyond t1, t2, and tn must be collinear with t1, t2, and tn, and must lie exterior to the path connecting t1 and t2. Thus, each additional taxon t3, t4, …, tn−1 is connected to t1 or t2 by an external branch. The admixture model together with the assumption of an additive distance matrix imposes such a strong restriction on the set of allowed distance matrices that it forces all taxa onto a highly constrained tree (Fig. 3B). When we consider the placement of each taxon t3, t4, …, tn−1, we find that this tree has two multifurcating nodes separated by a line that joins taxa t1 and t2, with tn as the only intervening taxon.
The additive case can assist in explaining phenomena observed empirically with admixed populations in the application of neighbor-joining:21–25 in the additive case, tn has external branch length 0, a result compatible with the short external branches detected for admixed taxa. Further, tn lies on the path connecting t1 and t2, compatible with the observation that admixed taxa lie in the “middle” of inferred neighbor-joining trees, with external branches incident to the paths connecting their source taxa. We can thus see that the empirical Fig. 2 resembles Fig. 3B, as the short internal branches among Native Americans and Europeans give rise to a shape with near multifurcations on each side of the admixed Mestizo groups.
4.2. The case of n = 4 taxa, not necessarily additive
The additive case is restrictive and atypical of the population-genetic context, in which migration and admixture generate non-tree-like evolution. We can then consider the more general setting of arbitrary genetic distance matrices with positive entries, examining the smallest nontrivial case, with n = 4 taxa. In this case, the admixed taxon is t4, with source taxa t1 and t2. We set the distances among taxa t1, t2, and t3 to be dt1,t2 = x, dt1,t3 = y and dt2,t3 = z, for some positive x, y, and z. Employing eq. 6, the distance matrix D has the form:
| (12) |
Using eq. 1 to calculate the matrix Q used in deciding which taxa will agglomerate, we obtain
| (13) |
where
| (14) |
| (15) |
| (16) |
Examining the relationships among q1, q2, and q3, we have that
| (17) |
| (18) |
| (19) |
As in the work of Eickmeyer & Yoshida,40 we partition the four-dimensional space of possible values of (λ, q1, q2, q3) according to the tree topologies produced by neighbor-joining.
Three tree topologies are possible with the four taxa (Fig. 4). In Fig. 4A, taxon t4 is separated by three edges from taxa t1 and t2, which themselves are separated by only two edges. In Fig. 4B, t4 is separated by two edges from t2 and by three edges from t1; t1 and t2 are separated by three edges. Taxa t1 and t2 are also separated by three edges in Fig. 4C, but t4 is instead separated by two edges from t1 and by three edges from t2.
Fig. 4.
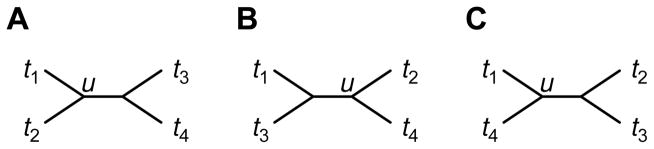
The three possible topologies for n = 4 taxa. Node u is the unique node that places t1, t2, and t4 in different subtrees.
Seven possibilities exist for the smallest entry of Q: (1) q1, (2) q2, (3) q3, (4) q1 and q2 (tied), (5) q1 and q3 (tied), (6) q2 and q3 (tied), and (7) q1, q2, and q3 (all tied). Each choice leads to a particular outcome among the three tree topologies in Fig. 4, with two or more topologies being possible outcomes in cases that involve ties. For each value among q1, q2, and q3, two pairs of taxa produce the same value in the matrix Q. It can be shown that in each case, either choice of which pair is first to agglomerate leads to the same inferred tree. Without loss of generality, we choose the pair that does not include taxon t3.
Four of the seven cases are not possible. In case 1, summing x + z < y and x + y < z in eqs. 17 and 18, we obtain x < 0. In case 4, setting q1 = q2 in eq. 17, x + z = y, from which we obtain x < 0 using x + y < z in eq. 18. Similarly, in case 5, q1 = q3 in eq. 18 produces x < 0 using x + z < y in eq. 17. In case 7, eqs. 17–19 become equalities, leading to x = 0 when x + z = y is substituted into eq. 19. All of these cases contradict the assumption that x > 0.
We consider the three allowable cases (cases 2, 3, and 6). For each of the possible inferred neighbor-joining trees, denote by u the unique interior node that places taxa t1, t2, and t4 in distinct subtrees (Fig. 4). Denote by d̂ the distance between nodes on the inferred tree. In case 2, q2 is smallest, taxa t2 and t4 agglomerate first, and using eqs. 2–4, we obtain
| (20) |
| (21) |
| (22) |
We can show that d̂u,t4 < d̂u,t2 and d̂u,t4 < d̂u,t1. The first of these two inequalities is equivalent to λy < λ(x + z), which holds because λ > 0, and because y < x + z by eq. 17. For the second inequality, note first that y < (1 − 2λ)x + z by eq. 19. Substituting the right-hand side in place of y in eq. 21, d̂u,t4 is less than [2λ(1 − λ)]x/4, which in turn is less than d̂u,t1 because 0 < λ < 1.
In case 3, q3 is smallest, taxa t1 and t4 agglomerate first, and using eqs. 2–4, we obtain
| (23) |
| (24) |
| (25) |
Similarly to case 2, we show d̂u,t4 < d̂u,t1 and d̂u,t4 < d̂u,t2. The first inequality is equivalent to (1 − λ)z < (1 − λ)(x + y), which holds because λ < 1, and because z < x + y by eq. 18. For the second equality, (1 − 2λ)x + z < y by eq. 19. Substituting the left-hand side in place of y in eq. 24, d̂u,t4 is less than [(2λ(1 − λ)]x/4, which in turn is smaller than d̂u,t2 because 0 < λ < 1.
Finally, in case 6, q2 and q3 are tied with the smallest values, and either t2 and t4 agglomerate first as in case 2, or t1 and t4 agglomerate first as in case 3. Neighbor-joining produces the tree in Fig. 4C with probability 1/2, and the tree in Fig. 4B with probability 1/2. With either choice, the same arguments used to demonstrate d̂u,t4 < d̂u,t1 and d̂u,t4 < d̂u,t2 in cases 2 and 3 apply, except that y is equal to (instead of greater than or less than) (1 − 2λ)x + z.
This collection of results demonstrates three phenomena for four-taxon trees built from distance matrices formed according to our admixture model. (1) The admixed taxon agglomerates with one of its two source taxa before the sources agglomerate with each other. Cases 2, 3, and 6 are the only ones allowable, and in these cases, the first neighbor-joining step agglomerates the admixed taxon t4 with one of the sources. (2) Denoting by u the unique node for which the admixed taxon and its source taxa all lie in different subtrees, the distance on the neighbor-joining tree of the admixed taxon to u is smaller than the distances to u of both source taxa. We demonstrated this result in each of the allowed cases, and it therefore holds in general. (3) The number of edges separating the source taxa on the inferred neighbor-joining tree, for each source taxon, is greater than or equal to the number of edges separating the admixed taxon from the source taxon. Only the trees in Figs. 4B and 4C are possible outcomes of neighbor-joining in our model, and the result holds for each of these trees.
5. Properties
Using the four-taxon results, we can formally define three properties of a distance matrix and its resulting neighbor-joining tree. The properties are well-defined for arbitrary n, and it is possible to evaluate whether a given n-taxon distance matrix satisfies them when neighbor-joining is applied. All three properties are possessed by all matrices generated by the four-taxon case of our admixture model.
Property 1: antecedence of clustering
The admixed taxon clusters with one of its source taxa before the source taxa cluster together. Stated precisely, some clade containing t1 but not t2 or tn merges with some clade containing tn but not t1 or t2, or, some clade containing t2 but not t1 or tn merges with some clade containing tn but not t1 or t2, before any clade containing t1 but not t2 or tn merges with any clade containing t2 but not t1 or tn.
Here we allow a clade to have any size, and potentially only a single taxon. In identifying the steps at which t1, t2, and tn merge into the neighbor-joining tree, as in our four-taxon case, to ensure that these taxa do not all merge simultaneously at the final stage, we adopt the convention that if a four-taxon stage is reached in which t1, t2, and tn lie in separate subtrees, we choose to agglomerate two among these three subtrees rather than agglomerating the third one with the unique available subtree that does not contain t1, t2, or tn.
Property 2: intermediacy of distances
The distance on the constructed neighbor-joining tree between the admixed taxon and either of its source taxa is smaller than the corresponding distance between the two source taxa. That is, d̂t1,tn < d̂t1,t2 and d̂t2,tn < d̂t1,t2. Equivalently, if u is the unique node in the constructed neighbor-joining tree for which t1, t2, and tn lie in different subtrees, then d̂u,tn < d̂u,t1 and d̂u,tn < d̂u,t2.
Property 3: intermediacy of path lengths
The number of edges separating the source taxa in the constructed neighbor-joining tree is greater than or equal to the number of edges separating the admixed taxon and either source taxon. If we define b̂ij as the number of edges in the path separating nodes i and j in the inferred tree, then b̂t1,t2 ≥ b̂t1,tn and b̂t1,t2 ≥ b̂t2,tn.
We have already demonstrated that in our admixture model, Properties 2 and 3 hold for all distance matrices in the n-taxon additive case; for Property 2, using eqs. 8 and 9 and 0 < λ < 1, d̂t1,tn < d̂t1,t2 and d̂t2,tn < d̂t1,t2. For Property 3, we have shown that for an n-taxon additive distance matrix, taxon tn lies on the interior of the path connecting t1 and t2, and it is the only taxon so located. Thus, b̂t1,t2 = 2, while b̂t1,tn = b̂t2,tn = 1, and Property 3 holds.
6. Discussion
We have examined neighbor-joining in a model in which an admixed taxon is produced from two source taxa, finding that for a four-taxon scenario, distance matrices and their resulting trees possess three properties: antecedence of clustering, in which the admixed population clusters with one of the sources before the sources cluster with each other; intermediacy of distances, in which the distance on the constructed tree between the admixed taxon and either source taxon is less than the distance between the sources; and intermediacy of path lengths, in which the number of edges separating the admixed taxon and either source taxon is no larger than the number of edges separating the sources. We have further shown that for an arbitrary number of taxa, the latter two properties hold when the distance matrix is additive.
By a mechanistic examination, we have found that our model has features seen in empirical observations of neighbor-joining trees that involve admixed populations. In particular, the placement of admixed populations on short external branches incident to the paths connecting their source populations21–25 matches the demonstration in the additive and four-taxon cases of the intermediacy of distances and intermediacy of path lengths properties. The theoretical approach validates the view that populations that are centrally located on neighbor-joining trees and that possess short external branches might be recently admixed.
Our results suggest a broader investigation of the extent to which the three properties hold with an arbitrary number of taxa. We have not reported a result regarding antecedence of clustering in the n-taxon additive case, nor have we commented on any of the properties for general n-taxon distance matrices that are not necessarily additive. However, we expect that Properties 1–3 will be satisfied by our admixture model considerably more often than in a model in which no special constraints are imposed on distances that involve the nth taxon. As our model also involves an nth taxon with special features, the general analysis of the model might benefit from the “rogue taxon” framework of Cueto & Matsen,41 in which the addition of an nth taxon alters the tree produced for an initial group of n − 1 taxa.
An additional direction is to study alternative admixture models. Distance methods are most sensible when a distance is nearly additive; however, eq. 6 severely restricts the distance matrix, as it forces a structure with two multifurcating nodes. This aspect of the model can be relaxed by assuming that the distance is additive for taxa t1, t2, …, tn−1, and that only distances involving tn satisfy eq. 6. For 1 ≤ i ≤ n − 1, we can then apply the distance
| (26) |
With this distance function, tn is simply placed on the path from t1 to t2 in a preexisting tree relating taxa t1, t2, …, tn−1 (Fig. 5). Properties 2 and 3 continue to hold.
Fig. 5.
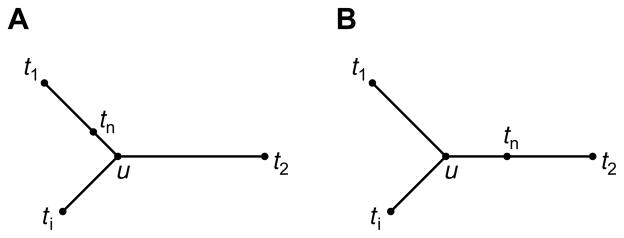
Two possible placements of tn with respect to t1, t2, and ti, illustrating how t1, t2, …, tn can have an additive distance matrix when t1, t2, …, tn−1 have an additive distance matrix. For convenience, we illustrate only one representative taxon ti from among {t3, t4, …, tn−1}.
To obtain eq. 26, we first suppose that t1, t2, …, tn−1 have an additive distance matrix. We wish to place taxon tn on the tree that generates the matrix so that the matrix for t1, t2, …, tn is additive. First, given λ, tn is placed on the path from t1 to t2 such that eqs. 8 and 9 are satisfied. It remains to compute d̂tn,ti for i = 3, 4, …, n − 1. Denote by u the unique node of the tree that places t1, t2, and ti in distinct subtrees (Fig. 5). Then
| (27) |
| (28) |
| (29) |
If d̂t1,tn ≤ d̂t1,u, then tn lies on the path from t1 to u (Fig. 5A), and
| (30) |
If, on the other hand, d̂t1,tn ≥ d̂t1,u, then tn lies on the path from t2 to u (Fig. 5B), and
| (31) |
Applying eqs. 8, 9, and 27–29 together with the fact that d̂ = d for additive distance matrices, we produce the relationship in eq. 26.
Analysis of the three properties using this modified form for the admixture model, or more generally using specific distance functions commonly employed in population genetics, will further illuminate the features of neighbor-joining in admixed populations. Such analyses might also facilitate investigations of the behavior with admixed populations of other tree-building methods, or of phylogenetic network methods42 that are more directly designed to accommodate taxa with non-tree-like evolutionary histories.
Acknowledgments
We thank M. Feldman and four reviewers for comments. This work has been supported by National Institutes of Health grant R01 GM081441, by National Science Foundation grants BCS-1024627 and DBI-1146722, and by the Burroughs Wellcome Fund.
Contributor Information
NAAMA M. KOPELMAN, Porter School of Environmental Studies, Department of Zoology, Tel Aviv University, Ramat Aviv, Israel
LEWI STONE, Porter School of Environmental Studies, Department of Zoology, Tel Aviv University, Ramat Aviv, Israel.
OLIVIER GASCUEL, Méthodes et Algorithmes pour la Bioinformatique, LIRMM-CNRS, Montpellier, France.
NOAH A. ROSENBERG, Department of Biology, Stanford University, Stanford, California, USA.
References
- 1.Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. Phylogenetic inference. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. Sinauer; Sunderland, MA: 1996. pp. 407–514. [Google Scholar]
- 2.Felsenstein J. Inferring Phylogenies. Sinauer; Sunderland, MA: 2004. [Google Scholar]
- 3.Saitou N, Nei M. Mol Biol Evol. 1987;4:406. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 4.Studier JA, Keppler KJ. Mol Biol Evol. 1988;5:729. doi: 10.1093/oxfordjournals.molbev.a040527. [DOI] [PubMed] [Google Scholar]
- 5.Bryant D. J Classif. 2005;22:3. [Google Scholar]
- 6.Gascuel O, Steel M. Mol Biol Evol. 2006;23:1997. doi: 10.1093/molbev/msl072. [DOI] [PubMed] [Google Scholar]
- 7.Mihaescu R, Levy D, Pachter L. Algorithmica. 2009;54:1. [Google Scholar]
- 8.Gascuel O. Concerning the NJ algorithm and its unweighted version, UNJ. In: Mirkin B, McMorris FR, Roberts FS, Rzhetsky A, editors. Mathematical Hierarchies and Biology. American Mathematical Society; Providence: 1997. pp. 149–170. [Google Scholar]
- 9.Atteson K. Algorithmica. 1999;25:251. [Google Scholar]
- 10.Saitou N, Imanishi T. Mol Biol Evol. 1989;6:514. [Google Scholar]
- 11.Kuhner MK, Felsenstein J. Mol Biol Evol. 1994;11:459. doi: 10.1093/oxfordjournals.molbev.a040126. [DOI] [PubMed] [Google Scholar]
- 12.Russo CAM, Takezaki N, Nei M. Mol Biol Evol. 1996;13:525. doi: 10.1093/oxfordjournals.molbev.a025613. [DOI] [PubMed] [Google Scholar]
- 13.Kalinowski ST. Heredity. 2009;102:506. doi: 10.1038/hdy.2008.136. [DOI] [PubMed] [Google Scholar]
- 14.Edwards AWF, Cavalli-Sforza LL. Reconstruction of evolutionary trees. In: Heywood VH, McNeill J, editors. Phenetic and Phylogenetic Classification. Systematics Association; London: 1964. pp. 67–76. [Google Scholar]
- 15.Cavalli-Sforza LL, Edwards AWF. Evolution. 1967;21:550. doi: 10.1111/j.1558-5646.1967.tb03411.x. [DOI] [PubMed] [Google Scholar]
- 16.Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. Nature. 1994;368:455. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 17.Pritchard JK, Stephens M, Donnelly P. Genetics. 2000;155:945. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung H-C, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, Bras JM, Schymick JC, Hernandez DG, Traynor BJ, Simon-Sanchez J, Matarin M, Britton A, van de Leemput J, Rafferty I, Bucan M, Cann HM, Hardy JA, Rosenberg NA, Singleton AB. Nature. 2008;451:998. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 19.Atzmon G, Hao L, Pe’er I, Velez C, Pearlman A, Palamara PF, Morrow B, Friedman E, Oddoux C, Burns E, Ostrer H. Am J Hum Genet. 2010;86:850. doi: 10.1016/j.ajhg.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunley K, Healy M. Am J Phys Anthropol. 2011;146:530. doi: 10.1002/ajpa.21506. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Linares A, Minch E, Meyer D, Cavalli-Sforza LL. Analysis of classical and DNA markers for reconstructing human population history. In: Brenner S, Hanihara K, editors. The Origin and Past of Modern Humans as Viewed from DNA. World Scientific; Singapore: 1995. pp. 123–148. [Google Scholar]
- 22.Bowcock AM, Kidd JR, Mountain JL, Hebert JM, Carotenuto L, Kidd KK, Cavalli-Sforza LL. Proc Natl Acad Sci USA. 1991;88:839. doi: 10.1073/pnas.88.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mountain JL, Lin AA, Bowcock AM, Cavalli-Sforza LL. Phil Trans R Soc Lond B Biol Sci. 1992;337:159. doi: 10.1098/rstb.1992.0093. [DOI] [PubMed] [Google Scholar]
- 24.Lin AA, Hebert JM, Mountain JL, Cavalli-Sforza LL. Gene Geog. 1994;8:191. [PubMed] [Google Scholar]
- 25.Mountain JL, Cavalli-Sforza LL. Proc Natl Acad Sci USA. 1994;91:6515. doi: 10.1073/pnas.91.14.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6 (Department of Genome Sciences. University of Washington; Seattle: 2005. [Google Scholar]
- 27.Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. PLoS Genet. 2005;1:660. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Lewis CM, Jr, Jakobsson M, Ramachandran S, Ray N, Bedoya G, Rojas W, Parra MV, Molina JA, Gallo C, Mazzotti G, Poletti G, Hill K, Hurtado AM, Labuda D, Klitz W, Barrantes R, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Llop E, Rothhammer F, Excoffier L, Feldman MW, Rosenberg NA, Ruiz-Linares A. PLoS Genet. 2007;3:2049. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Ray N, Rojas W, Parra MV, Bedoya G, Gallo C, Poletti G, Mazzotti G, Hill K, Hurtado AM, Camrena B, Nicolini H, Klitz W, Barrantes R, Molina JA, Freimer NB, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Dipierri JE, Alfaro EL, Bailliet G, Bianchi NO, Llop E, Rothhammer F, Excoffier L, Ruiz-Linares A. PLoS Genet. 2008;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minch E, Ruiz Linares A, Goldstein DB, Feldman MW, Cavalli-Sforza LL. MI-CROSAT (version 1.5d): a program for calculating statistics on microsatellite data. Department of Genetics, Stanford University; Stanford, CA: 1998. [Google Scholar]
- 31.Mountain JL, Cavalli-Sforza LL. Am J Hum Genet. 1997;61:705. doi: 10.1086/515510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gascuel O. Mol Biol Evol. 1994;11:961. doi: 10.1093/oxfordjournals.molbev.a040176. [DOI] [PubMed] [Google Scholar]
- 33.Long JC, Smouse PE. Am J Phys Anthropol. 1983;61:411. doi: 10.1002/ajpa.1330610403. [DOI] [PubMed] [Google Scholar]
- 34.Fournier DA, Beacham TD, Riddell BE, Busack CA. Can J Fish Aquat Sci. 1984;41:400. [Google Scholar]
- 35.Rosenberg NA, Li LM, Ward R, Pritchard JK. Am J Hum Genet. 2003;73:1402. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang H, Peng J, Wang P, Risch NJ. Genet Epidemiol. 2005;28:289. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 37.Alexander DH, Novembre J, Lange K. Genome Res. 2009;19:1655. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boca SM, Rosenberg NA. Theor Pop Biol. 2011;80:208. doi: 10.1016/j.tpb.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semple C, Steel M. Phylogenetics. Oxford University Press; Oxford: 2003. [Google Scholar]
- 40.Eickmeyer K, Yoshida R. Lect Notes Comp Sci. 2008;5147:81. [Google Scholar]
- 41.Cueto MA, Matsen FA. Bull Math Biol. 2011;73:1202. doi: 10.1007/s11538-010-9556-x. [DOI] [PubMed] [Google Scholar]
- 42.Huson DH, Rupp R, Scornavacca C. Phylogenetic Networks: Concepts, Algorithms and Applications. Cambridge University Press; Cambridge: 2010. [Google Scholar]