Abstract
Development of well-defined nanomedicines is critical for their successful clinical translation. A simple synthesis and purification procedure is established for chemically cross-linked polyion complexes of Cu/Zn superoxide dismutase (SOD1) or catalase with a cationic block copolymer, methoxy-poly(ethylene glycol)-block-poly(L-lysine hydrochloride) (PEG-pLL50). Such complexes, termed cross-linked nanozymes (cl-nanozymes) retain catalytic activity and have narrow size distribution. Moreover, their cytotoxicity is decreased compared to non-cross-linked complexes due to suppression of release of the free block copolymer. SOD1 cl-nanozymes exhibit prolonged ability to scavenge experimentally induced reactive oxygen species (ROS) in cultured brain microvessel endothelial cells and central neurons. In vivo they decrease ischemia/reperfusion-induced tissue injury and improve sensorimotor functions in a rat middle cerebral artery occlusion (MCAO)model after a single intravenous (i.v.) injection. Altogether, well-defined cl-nanozymes are promising modalities for attenuation of oxidative stress after brain injury.
Keywords: Antioxidant enzymes, Block ionomer complexes, Blood–brain barrier, Cross-linked nanozymes, Stroke therapy
1. Introduction
Attenuation of oxidative stress and inflammation is a promising strategy to prevent brain tissue damage and treat central nervous system (CNS)-related disorders, including Parkinson’s disease (PD), Alzheimer’s disease, amyotrophic lateral sclerosis [1,2], as well as traumatic brain injury, stroke and transient ischemic attacks [3-5]. Antioxidant enzymes like copper–zinc superoxide dismutase (Cu/Zn SOD, also known as SOD1) and catalase are potent scavengers of ROS. However, their delivery to the brain represents a major challenge because of their proteolytic degradation, immunogenicity, short circulation half-life and poor permeability across the blood–brain barrier (BBB) [6]. Even though the BBB can be partially compromised in stroke [7], it still remains the key impediment for CNS transport of enzymes [8]. Several strategies were explored to improve delivery of antioxidant enzymes including PEGylation [9,10], use of fusion constructs with protein transduction domains [11-14], encapsulation in liposomes [15-17], or poly(d,l-lactide-co-glycolide) (PLGA) nanoparticles [18], lecithinization [19-21], or conjugation with antibodies (immunotargeting) (see supplementary information for the complete list of references [22–46]). PEP-1-SOD1 and PEP-1-catalase fusion constructs were developed using a genetic engineering approach and attenuated ischemic neuronal damage in vivo upon intraperitoneal (i.p.) injection [11,13]. However, such constructs have relatively low stability in circulation which may be a likely reason i.p. was the preferred route of administration and can also induce immune responses in patients [18]. The circulation time and stability of proteins can be increased to several hours by PEGylation — modification of the protein with poly(ethylene glycol) (PEG). However, such modification also drastically decreases permeability of proteins across the brain microvessels and entry into brain cells, hindering therapeutic effects of PEGylated SOD1 after cerebral ischemia/reperfusion injury [10,47].
Muzykantov and coworkers exploited the constitutive vascular expression of platelet-endothelial adhesion molecule (PE-CAM)-1 and intercellular adhesion molecule (ICAM)-1 for catalase delivery to the endothelium using multivalent conjugates of catalase with anti-PECAM or anti-ICAM antibodies [22,25,37,41] or coating catalase-loaded nanoparticles with these antibodies [30]. These conjugates or nanoparticles were internalized by endothelial cells, remained functionally active and protected pulmonary vasculature against acute oxidative stress [25,37]. More recently, the same group reported that SOD1 and catalase immobilized in magnetic nanoparticles were stable against proteolytic degradation, transported into endothelial cells in vitro and rescued these cells from H2O2-induced oxidative stress [24].
In another approach SOD1 incorporated into PLGA nanoparticles was shown to reduce ischemic brain injury after intracarotid (i.c.) injection [18]. However, PLGA-matrix hinders access of the substrate to the enzyme active sites. Furthermore, instability of proteins in such formulations, especially upon PLGA hydrolysis may limit their utility [48]. Finally, cationic liposome-entrapped SOD1 was shown to reduce infarction upon cerebral ischemia in rats, but lowstability of this formulation and possible toxicity impeded its further use [18,49].
Our laboratory has developed a distinct class of catalytic nanoparticles based on polyion complexes (also known as “block ionomer complexes”, BICs) of enzymes and cationic block copolymers (nanozymes) based on spontaneous self-assembly of oppositely-charged polyions – a simple approach that results in stoichiometric complexes due to complete complexation (100% loading efficiency) compared to limitations in case of SOD1 (~32%) and catalase (~21%) encapsulated in cationic liposomes [50]. We also demonstrated their potential to treat PD (using catalase nanozymes loaded in cell carriers), Angiotensin-II hypertension (using SOD1 nanozymes), and delivery of active butyrylcholinesterase enzyme to the brain in healthy mice [51-55]. More recently, we reported that covalently stabilized cl-nanozymes of SOD1 improved stability (in the blood and brain tissue) and delivered SOD1 to the brain parenchyma in healthy mice [56]. We hypothesized that purification of cl-nanozymes (removal of non-cross-linked species) will result in a homogenous sample with well-defined chemical composition and physicochemical characteristics, which may further increase its stability against dissociation, improve its in vivo disposition and thus enhance overall efficacy of the delivery process. We believe that the translational potential of such cl-nanozymes is contingent on their production as “pharmaceutical-grade” entities. Here we synthesized and characterized well-defined antioxidant cl-nanozymes containing SOD1 or catalase, studied behavior of selected formulations in an in vitro model of cultured brain microvessel endothelial cells and central neurons, and demonstrated a proof-of-concept of the therapeutic efficacy of SOD1 cl-nanozymes in vivo in a rat MCAO model of ischemia/reperfusion injury.
2. Materials and methods
2.1. Materials
SOD1 (from bovine erythrocytes), hydrogen peroxide (H2O2), 2,3,5-triphenyltetrazolium chloride (TTC) and copper standards for Inductively Coupled Plasma Mass Spectroscopy (ICP-MS) – TraceCERT®, 1000 mg/L Cu in nitric acid – were from Sigma-Aldrich (St. Louis, MO). Catalase (from bovine liver) was from Calbiochem (Gibbstown, NJ). PEG-pLL50 was from Alamanda Polymers™ (Huntsville, AL). Its molecular mass determined by gel permeation chromatography was 13,000 Da and polydispersity index was 1.09; the PEG molecular mass was 4600 Da and the degree of polymerization of pLL block was 51. Cross-linkers 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP) and bis(sulfosuccinimidyl)suberate (BS3) were from Thermo Fisher Scientific (Rockford, IL). NAP™ desalting columns and HiPrep 16/60 Sephacryl S-400 HR column were from GE Healthcare (Piscataway, NJ). Criterion Tris–HCl gels and Precision Plus Protein™ All Blue Standards were from Bio-Rad (Hercules, CA). SYPRO® Ruby protein gel stain and cell culture reagents were purchased from Invitrogen (Carlsbad, CA). CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) was from Promega (Madison, WI). LumiMax Superoxide Anion Detection Kit was from Agilent Technologies, Inc. (Santa Clara, CA). All other reagents and supplies were from Fisher Scientific (Pittsburgh, PA) unless noted otherwise.
2.2. Synthesis and purification of cl-nanozymes
Enzyme and polymer stock solutions were prepared in 10 mM HEPES (pH 7.4) and 10 mM HEPES-buffered saline (HBS; pH 7.4) for SOD1 and catalase, respectively. Non-cross-linked BICs (Z+/−=2) were prepared as described earlier [56]. Targeted degree of cross-linking was defined as the molar ratio between cross-linker (DTSSP/BS3) and pLL amines. Pre-calculated amount of the respective cross-linker was dissolved in the reaction buffer (HEPES/HBS), quickly added to BICs, and the reaction mixture was briefly vortexed and incubated for 2 h on ice. Unreacted cross-linker was desalted using NAP™ columns following manufacturer’s instructions. Cl-nanozymes were purified using size exclusion chromatography (SEC) (small/intermediate scale) or centrifugal filtration (large scale). SEC was carried out using an ÄKTA™ Fast Protein Liquid Chromatography (FPLC) (Amersham Biosciences, Piscataway, NJ) system. DTSSP cl-nanozymes were lyophilized overnight, reconstituted in de-ionized water (DW), loaded onto a HiPrep 16/60 Sephacryl™ S-400 HR column and eluted using 10 mM HBS (pH 7.4) at a flow rate of 0.5 mL/min. Prior to each experiment, the column was pre-conditioned with free block copolymer (PEG-pLL50) to minimize non-specific adsorption of the cl-nanozymes [57]. Fractions spanning each distinct peak were pooled, concentrated using Amicon® Ultra-4 Centrifugal Filter Units with a molecular weight cutoff (MWCO) of 3000 Da and desalted using NAP™ columns as needed. Protein concentration was determined using Micro BCA™ Protein Assay Kit. In large-scale preparations, cl-nanozymes were purified by centrifugal filtration using Macrosep™ Centrifugal Device (Pall Life Sciences, Ann Arbor, MI) with a MWCO of either 100 kDa (for SOD1) or 1000 kDa (for catalase). Briefly, unreacted cross-linker in cross-linked BICs was desalted using NAP™ columns and eluate was collected in 10 mM HEPES containing 0.3 M NaCl (pH 7.4). Samples were loaded onto the centrifugal device and concentrated to 10% original volume by centrifuging at 4500 rpm. Two rounds of purification were done in 10 mM HEPES buffer containing 0.3 M NaCl (pH 7.4) and the third round was done in 10 mM HEPES buffer (pH 7.4). The concentrate was collected and desalted using NAP™ columns to remove excess NaCl.
2.3. Dynamic light scattering (DLS)
Intensity-mean z-averaged particle diameter (Deff), polydispersity index (PDI), and ζ-potential were measured using a Zetasizer Nano ZS (Malvern Instruments Ltd., MA) [56]. Both size and ζ-potential measurements were conducted in low ionic strength buffer (10 mM HEPES, pH 7.4) unless indicated otherwise. Wherever indicated, catalase BICs were desalted to remove excess NaCl before measuring ζ-potential. Data is represented as mean values (n=3).
2.4. ICP-MS
Copper (Cu2+) content in SOD1 samples was determined using ICP-MS. Standards/samples were diluted in double distilled nitric acid and measurements were performed in 10 replicates using a PerkinElmer Nexion 300Q ICP Mass Spectrometer. The data were analyzed using the total quantity method. Concentration of the predominant isotope, 63C was calculated from the standard curve generated using copper standards.
2.5. Enzyme activity
SOD1 enzyme activity was determined using two independent methods — inhibition of PG autoxidation by added SOD1 (indirect method) [58] and scavenging of experimentally generated superoxide radicals (O2· −) by added SOD1 using electron paramagnetic resonance (EPR) spectroscopy (direct method) [52] (see supplementary information). Wherever indicated, SOD1 activity measured using PG assay was normalized to Cu2+ content determined using ICP-MS. Enzyme activity of catalase was measured by following decomposition of H2O2 as previously reported [59]. Slope (reaction rate of H2O2 decomposition) was calculated as ΔA240/min. Catalytic activity of catalase among the different samples was compared in terms of the slope of linear regression. Activity was expressed as percent (%) relative to native enzyme. Enzyme activity of SOD1 (reported by Sigma-Aldrich) was ~4000 U/mg protein and catalase (reported by Calbiochem) was ~46,500 U/mg protein.
2.6. Gel retardation assay
Formation of cl-nanozymes was confirmed by their compromised ability to migrate in a polyacrylamide gel under denaturing conditions. Five or 3 μg protein (SOD1 or catalase) was denatured in the sample buffer (no reducing agent added), loaded on an 18/10% Criterion Tris–HCl gel and electrophoresed at 200 V (100 mA) for 1 h, and stained using SYPRO® Ruby protein gel stain and imaged on a Typhoon gel scanner (Amersham Biosciences Corporation, Piscataway, NJ) at 100 μ pixel size.
2.7. Sedimentation equilibrium analysis
Molecular weight of purified SOD1 cl-nanozymes was determined by sedimentation equilibrium analysis using a Beckman Optima XL-I analytical ultracentrifuge and an AN-60Ti rotor as previously reported [60]. Data analysis was performed using the Beckman XL-A/XL-I software package with Microcal, ORIGIN v4 software.
2.8. Cell culture
Immortalized bovine brain microvessel endothelial cells containing a Middle T-antigen gene (TBMECs) and CATH.a neuronal cell line (CRL-11179™) were from American Type Culture Collection (Manassas, VA) and were cultured as described earlier [52,61].
2.9. Cytotoxicity assay
TBMECs were seeded at 5000 cells/well in a collagen, fibronectin-coated 96 well plate and cultured until 100% confluence. CATH.a cells were seeded at 10,000 cells/well and differentiated into neurons as described earlier [52]. Cells were incubated with indicated concentrations of samples (in case of the free polymer control, cells were treated with equivalent concentrations of PEG-pLL50 that would be present in non-cross-linked BICs) diluted in complete growth medium for 24 h and cell viability was determined using a commercially available MTS assay kit. Percent (%) cell viability was calculated using the formula (Asample/Auntreated cells)×100. Data represents average±SD (n=3).
2.10. O2· − scavenging in vitro in cell cultures
TBMECs/CATH.a cells were grown in 48 well plates and were treated with SOD1/formulated SOD1 (50 μg/mL) diluted in complete growth medium for 2 h. Treatment mixture was removed and cells were further incubated with fresh medium for different times: 0, 1, 2, 4 or 12 h. Post-incubation, cells were washed using PBS and lysed using 1× cell lysis buffer (Cell Signaling Technology, Boston, MA). Hypoxanthine and xanthine oxidase were used to generate O2· − in cell lysates and LumiMax Superoxide Anion Detection Kit was used to determine percent (%) O2· − remaining (relative to untreated cells). Data represent average±SD (n=4).
2.11. Rat MCAO model and experimental details
Young adult male Sprague–Dawley rats (250–300 g) were anesthetized with ketamine/xylazine cocktail and isoflurane. Right common carotid artery was incised, and a filament with bulbous tip was inserted through this incision into internal carotid artery and further until bifurcation of middle cerebral artery (MCA). Bulbous tip occluded the entrance to MCA and blocked blood supply to part of the right brain hemisphere of the rat. Filament was carefully withdrawn after 2 h and immediately incision on MCA was permanently closed and 0.5 mL saline, native SOD1 or purified SOD1 cl-nanozyme was administered i.v. via the tail vein at a dose of 10 kU/kg body weight. Post-surgery rats were returned to their cages for 22 h. Sensorimotor functions of rats (response to touch of a side of a trunk, touch of vibrissae on one side, forelimbs outreach, floor walking and climbing of a cage wall) were evaluated 24 h after the beginning of ischemic episode using a reported method [62]. After the evaluation, rats were euthanized and brains were dissected. Dissected brains were sectioned (6 sections 2 mm thick each) and sections were stained with TTC to visualize the infarct region. Stained sections were photographed and digital images were quantified using ImageJ software (National Institute of Health, Bethesda, MD). Infarct areas were outlined and determined (in conditional units) as follows: [(infarct area #1)/(entire hemisphere area #1)+…. (infarct area #6)/(entire hemisphere area #6)]=infarct index of brain #1. Data is represented as infarct index average (5–6 animals/group)±standard error of mean (SEM).
2.12. Statistical analysis
Statistical comparisons between two groups were made using Student’s t-test while comparisons between multiple groups were done using non-parametric one-way ANOVA with multiple comparisons (Kruskal–Wallis) using Origin 8.5 software (Northampton, MA). P-value <0.05 was considered statistically significant.
3. Results
3.1. Synthesis, characterization and purification of cl-nanozymes
cl-Nanozymes containing either SOD1 or catalase were synthesized as described earlier. A graphical representation of the cl-nanozymes is shown in Scheme 1. Samples are denoted as follows throughout: nS or nC — native SOD1 or catalase; S1 or C1 — BICs of SOD1 or catalase (Z=2), S2 (S2p) or C2 (C2p) — DTSSP-cross-linked cl-nanozymes; and S3 (S3p) or C3 (C3p) — BS3-cross-linked cl-nanozymes. Subscript index “p” refers to purified samples. The targeted degree of cross-linking defined earlier was optimized for each enzyme and cross-linking chemistry to ensure that the enzyme retained at least 75% of the initial activity of the native enzyme. The optimal targeted degrees of cross-linking for SOD1 and catalase cl-nanozymes were determined to be 0.5 and 1.0, respectively. Cross-linking was confirmed by retarded enzyme migration in denaturing gel electrophoresis (Supplementary data Fig. s1). DTSSP produced cross-links that contained cleavable disulfide bonds, while BS3 produced non-cleavable cross-links. Dithiothreitol (DTT) treatment cleaved disulfide cross-links, noted as a decrease in the high molecular mass band density corresponding to DTSSP-cl-nanozymes (Supplementary data Fig. s1). In contrast, there were no changes in band density in the case of BS3-cl nanozymes with non-cleavable cross-links. The physicochemical characteristics (hydrodynamic diameters, PDI, ζ-potential) and enzyme activity of samples are listed in Table 1. The values of Deff for native SOD1 and catalase were in good agreementwith the theoretical hydrodynamic diameters estimated using the Protein Utilitiesmodule in the Malvern Zetasizer Nano software (5.2 and 12.5 nm for SOD1 and catalase, respectively). In the SOD1 formulations, there was nearly 2-fold increase in the particle size upon BIC formation, accompanied by a change in the ζ-potential from weakly-negative (native enzyme) to a positive value. The positive ζ-potential of this BIC could be due to some excess of amino groups of either the protein or pLL incorporated into the complex. Notably the size measurements in this case were carried out in low ionic strength buffer, since addition of 0.15 M NaCl favored dissociation of BICs, as noted by the decrease in the particle size (not shown). The size increased further three-fold after the BICs were cross-linked suggesting that such cl-nanozymes contained multiple SOD1 protein molecules. Interestingly, after the cross-linking the ζ-potential decreased and again became weakly negative, which may be indicative of consumption of the protein and/or pLL amino groups that reacted with the cross-linking reagents. The size measurements with catalase BIC were quite interesting in comparison with those of SOD1. Here the sizes of BIC practically did not change compared to the free catalase suggesting that the BIC contained only one catalase protein molecule. However, after cross-linking the sizes increased by about 3.7-fold indicating that multiple catalase molecules were assembled in the cl-nanozymes. We were unable to directly measure the ζ-potential of the catalase or its BIC since they were not stable at low ionic strength and were prepared in 10 mM HBS buffer, pH 7.4, containing 0.15 M NaCl. However, we rapidly desalted the pre-formed BIC and determined its ζ-potential, which was positive. This was in contrast to the native catalase that was negative, suggesting that BICs were indeed formed notwithstanding the lack of the size changes. Finally, similar to the previous case the ζ-potential of cl-nanozymes was lower (and negative) than that of the non-cross-linked BIC.
Scheme 1.
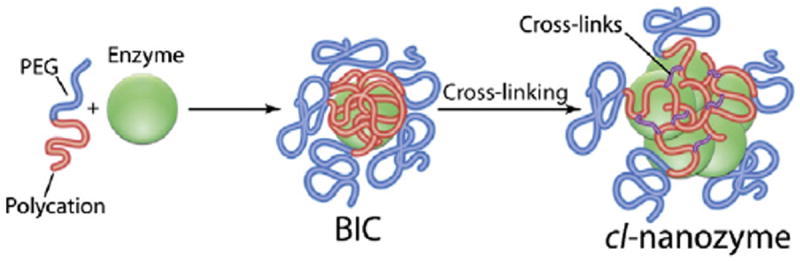
Schematic representation of spontaneous formation of BICs through electrostatic binding of a negatively charged enzyme with a cationic block copolymer followed by covalent cross-linking to obtain cl-nanozymes. The scheme implies 1) each BIC contains one protein globule, and 2) the particle size may further increase upon cross-linking.
Table 1.
Characteristics of BICs and cl-nanozymes.
| Samplea,b | Deff, nm | PDIc | ζ-Potential, mV | Enzyme activity,d % of initial |
|---|---|---|---|---|
| nS | 5.2 | 0.10 | −1.3 | 100 |
| S1e | 9.8 | 0.14 | +7.8 | 97 |
| S2 | 31.0 | 0.13 | −0.1 | 91 |
| S3 | 29.8 | 0.20 | −1.9 | 77 |
| nC | 14.9 | 0.40 | −16.3f | 100 |
| C1e | 13.1 | 0.26 | +16.3f | 121 |
| C2 | 55.6 | 0.28 | −11.0f | 100 |
| C3g | n.a. | n.a. | n.a. | n.a. |
nS or nC — native SOD1 or catalase, S1 or C1 — non-cross-linked BICs of SOD1 or catalase, S2 or C2 — cleavable (DTSSP cross-linked) cl-nanozymes, and S3 or C3 — non-cleavable (BS3 cross-linked) cl-nanozymes.
All SOD1-containing samples (1 mg/mL) were in low ionic strength buffer, 10 mM HEPES, pH 7.4, while the catalase samples, 0.5 mg/mL were in 10 mM HBS, pH 7.4 (unless noted otherwise).
Polydispersity index.
Enzyme activity data is representative of >5 independent experiments. Measurement error was typically <5%.
Z=2.
The measurements were carried out in 10 mM HEPES, pH 7.4 after desalting the samples.
Not available since BS3 did not cross-link the catalase BICs.
Denaturing gel electrophoresis (Fig. 1, Supplementary data Fig. s1) showed that the cross-linked samples contained considerable amounts of free enzymes (two main bands corresponding to the monomer (16 kDa) and dimer (32 kDa) forms of SOD1 and several bands corresponding to the monomer (62 kDa), dimer (124 kDa) and tetramer (247 kDa) forms of catalase). The free PEG-pLL50 may have also been present in these samples as it did not enter the gel and remained in the wells similar to the cl-nanozymes (SYPRO Ruby also stains basic amino acids like lysines). As it follows the sample homogeneity was improved by SEC purification. After purification, almost the entire sample (S2p and C2p) remained in or near the wells, while only a minor portion of proteins (mainly their respective monomers) migrated through the polyacrylamide gel (Fig. 1).
Fig. 1.
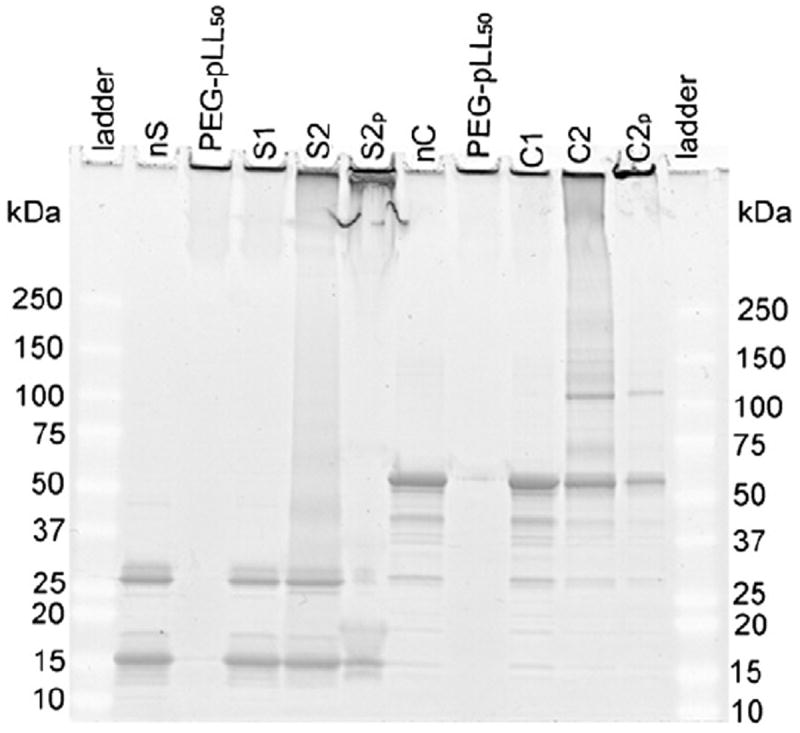
Gel retardation analysis. SOD1 (S) (5 μg/lane) and catalase (C) (3 μg/lane) were loaded on a denaturing polyacrylamide gel and protein bands were stained using SYPRO Ruby. nS or nC – native enzyme – SOD1 or catalase, PEG-pLL50 – free block copolymer, S1 or C1 – non-cross-linked BIC, S2 or C2 – DTSSP-cross-linked cl-nanozymes; subscript ‘p’ refers to the respective purified forms.
The catalytic activity of SOD1 was determined by following inhibition of PG autoxidation by SOD1 [63] and a typical dose response curve is shown in Fig. s2a (Supplementary data). Inhibitory effect of SOD1 on PG autoxidation among the different samples was compared in terms of an IC50 value, defined as the concentration of SOD1 that inhibited PG autoxidation by 50% [58]. Catalytic activity of catalase was determined using the standard H2O2 decomposition assay [59] and a typical dose response plot is shown in Fig. s2b (Supplementary data). Both non-cross linked BICs (S1 or C1) and cl-nanozymes (S2, S3, and C2) retained relatively high activities (77–100%) of the unmodified enzyme.
3.2. Purification and further characterization of cl-nanozymes
Since sample homogeneity is crucial for the pharmaceutical protein formulations, we purified the cl-nanozymes (S2, C2) by separating them from the non-cross linked BIC components using SEC (Fig. 2). Based on the area-under-the-curve (AUC) analysis the cross-linked particles comprised ca. 24% and 39% of the non-purified samples of SOD1 and catalase cl-nanozymes, respectively. The rest was mostly the free enzyme (nS, nC) and a minor portion of non-cross-linked BIC (S1, C1) that did not dissociate during chromatography. The elution volumes of the three fractions – cl-nanozymes (C2p 41 mL, S2p 53 mL), non-cross-linked BICs (C1 87 mL, S1 91 mL) and free enzymes (nC 107 mL, nS 106 mL) – were in logical agreement with the respective particle sizes (Table 1). Particle sizes measured using DLS demonstrated that after purification the Deff of SOD1 cl-nanozyme practically did not change while the Deff of catalase cl-nanozyme increased ca. 60% (Fig. 3). Incidentally the PDI of both samples considerably decreased. The purified cl-nanozymes (S2p and C2p) retained their spherical morphology observed under AFM (Supplementary data Fig. s3), albeit they were more uniform compared to non-purified cl-nanozymes (S2 and C2).
Fig. 2.
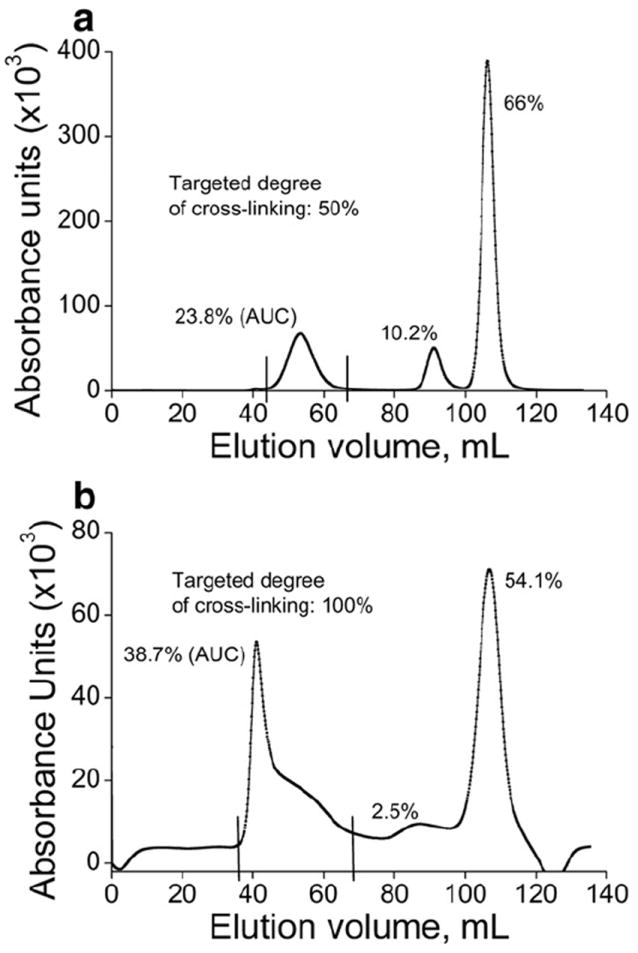
FPLC chromatogram profiles depicting purification of DTSSP-cross-linked (a) SOD1 cl-nanozyme (S2) and (b) catalase cl-nanozyme (C2). S2 or C2 was loaded onto a HiPrep 16/60 Sephacryl™ S-400 HR column and eluted using 10 mM HBS (pH 7.4) at a flow rate of 0.5 mL/min. AUC analysis determined the proportion of each fraction in the sample.
Fig. 3.
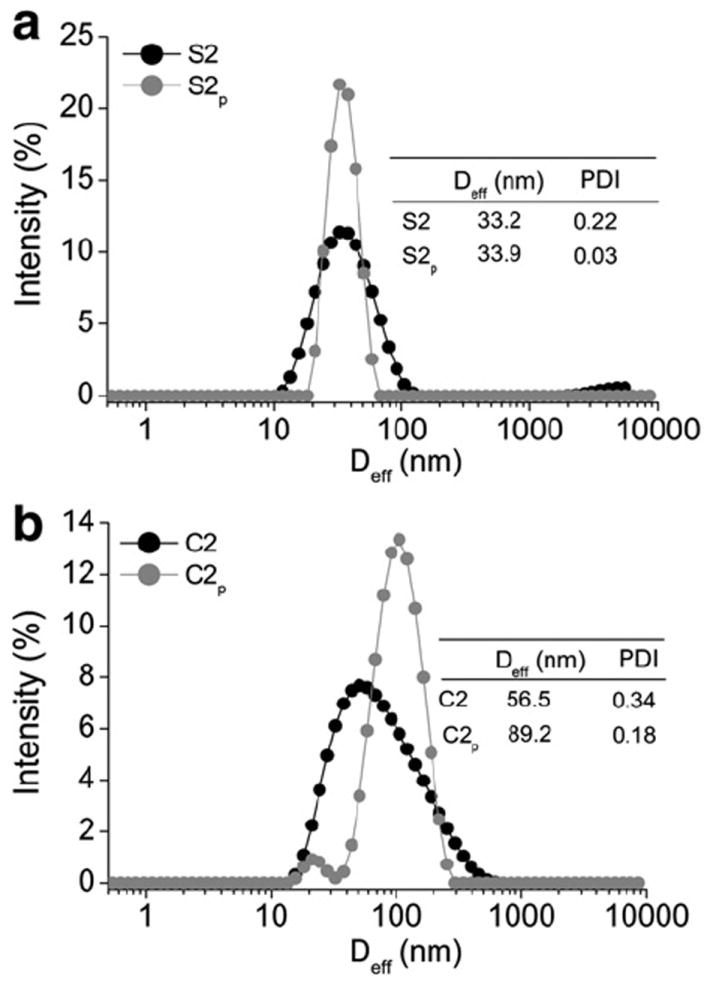
Representative DLS plots showing effects of purification on size distribution of DTSSP-cross-linked (a) SOD1 cl-nanozymes (S2) and (b) catalase cl-nanozymes (C2). Subscript ‘p’ refers to the respective purified forms. Tables in the inset show Deff and PDI measured at 0.1 mg/mL enzyme concentration in 10 mM HBS (pH 7.4).
Table 2 lists the enzyme activity retained by purified clnanozymes. SOD1 concentration in cl-nanozyme samples was normalized to Cu2+ content determined by ICP-MS, and the activity was assayed by PG autoxidation as described before. After purification this cl-nanozyme (S2p) retained only ca. 47% of the activity of the non-purified cl-nanozyme (S2) and native SOD1 (nS) samples. This result was generally consistent with the SOD1 activity measurements using EPR spectroscopy (Supplementary data Table s1) although EPR results showed slightly higher activity for S2p. However, the purified catalase cl-nanozyme (C2p) was nearly as active as the non-purified cl-nanozyme (C2) and native catalase (nC) in H2O2 decomposition assay. A more detailed analysis, however, suggested that the purified cl-nanozyme (C2p) had somewhat lower kcat compared with the non-purified sample (C2), albeit the value was nearly similar to native catalase (nC) (Supplementary data Table s2). However, the increase in the kcat of non-purified cl-nanozyme (C2) compared to native catalase was offset by ca. 1.8-fold increase in its Km value. As a result the catalytic efficiency (kcat/Km) of the non-purified cl-nanozyme (C2) and native enzyme (nC) were nearly the same whereas the kcat/Km of purified cl-nanozyme (C2p) was ca. 48 and 53% lower than nC and C2, respectively.
Table 2.
Enzyme activity of purified cl-nanozymes.
| Sample | Enzyme activity % of initial |
|---|---|
| S2 | 96 |
| S2p | 45 |
| C2 | 100 |
| C2p | 100 |
S2 or C2 — cleavable (DTSSP cross-linked) cl-nanozymes containing SOD1 or catalase and subscript ‘p’ refers to their respective purified forms. Data presented is representative of >5 independent experiments. Measurement error was typically <5%.
The molecular mass and aggregation state of the SOD1 cl-nanozyme was determined using analytical ultracentrifuge sedimentation equilibrium analysis (Fig. 4). The analysis was carried out in 10 mM HBS, which favors dissociation of the non-cross-linked BIC. As expected, this method revealed the presence of mixture of cl-nanozymes and native enzyme in the non-purified sample. The molecular masses determined by this method were in a reasonable agreement with the theoretical estimate of 4.4 MDa for cl-nanozymes (calculated assuming formation of a stoichiometric complex with a Deff of 30 nm) and in excellent agreement with the theoretical value of 32 kDa for native SOD1. Purified cl-nanozymes (S2p) showed an experimental molecular weight of ca. 1.2 MDa, which suggests that they contained ca. 30 SOD1 globules. This observation also points out that the purified sample contained no aggregate(s), which is consistent with the DLS data. Sedimentation equilibrium analysis has a molecular mass range from 2500 Da to 1.5×106 Da; therefore the catalase cl-nanozymes could not be analyzed using this technique.
Fig. 4.
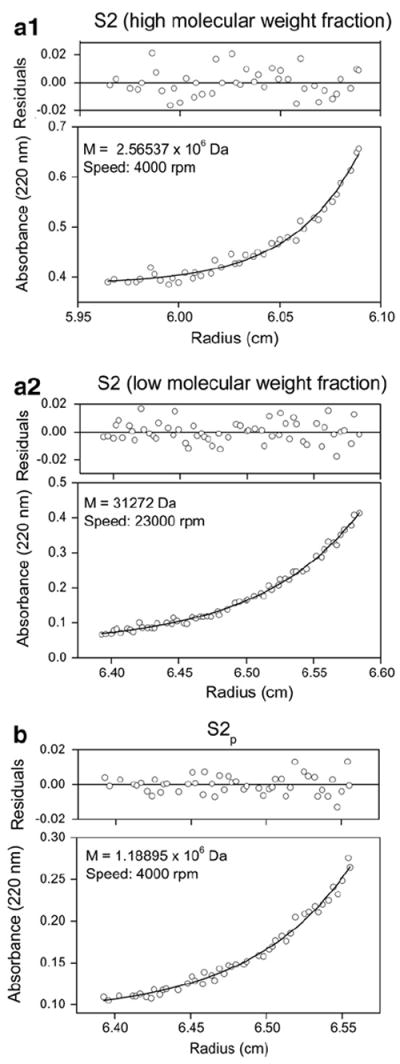
Sedimentation equilibrium analysis of DTSSP-cross-linked SOD1 cl-nanozymes (a1 and a2) before and (b) after purification. The sample concentration in HBS at equilibrium is shown as a function of radius. The solid lines are theoretical curves and figure insets show molecular weight and speed at which equilibrium was attained.
3.3. In vitro studies
In vitro experiments were conducted using two cell line models. TBMEC monolayers were used as an in vitro model of brain microvessel endothelial cells (BMECs). This cell line retains morphological and biochemical features of primary BMECs and has been described as a suitable in vitro model for BBB studies [61]. CATH.a cells were differentiated into neurons [52] and were used as a model of central neurons. It should be noted that only SOD1 formulations were used in all studies henceforth.
3.3.1. Cytotoxicity of formulations
Cytotoxicity of SOD1 formulations was evaluated in both TBMEC monolayers and CATH.a neurons (Fig. 5). The IC50 values of the free block copolymer (PEG-pLL50) and non-cross-linked BIC (S1) were ~35 and 28 μg/mL (TBMEC) and 59 and 113 μg/mL (CATH.a neurons), respectively. This data suggests that the toxicity of non-cross-linked BIC (Z=2) may be due to the admixture or PEG-pLL50 release, which interacts with negatively charged cellular membranes and other macro-molecules through its polycation chain, a well-documented phenomenon for polycations [64,65]. In contrast, purified cl-nanozymes (S2p and S3p) displayed significantly lower toxicity with cell viabilities of 60–70% (TBMEC) and 83–100% (CATH.a neurons) at the highest concentration tested (500 μg/mL). Hence, no IC50 value could be determined for purified cl-nanozymes in the tested range of concentrations. In general, cells treated with DTSSP-cl-nanozymes (S2p) at concentrations ≥25 μg/mL showed slightly lower cell viabilities compared with those treated with BS3-cl-nanozymes (S3p). The difference was more pronounced in CATH.a neurons where the cell viability was ca. 26% less for S2p compared with S3p. It is worth mentioning that non-purified cl-nanozymes appeared to be slightly more toxic than the purified samples (Supplementary data, Fig. s4). Therefore, purification and in particular removal of the free PEG-pLL50 is an important factor decreasing cellular toxicity.
Fig. 5.
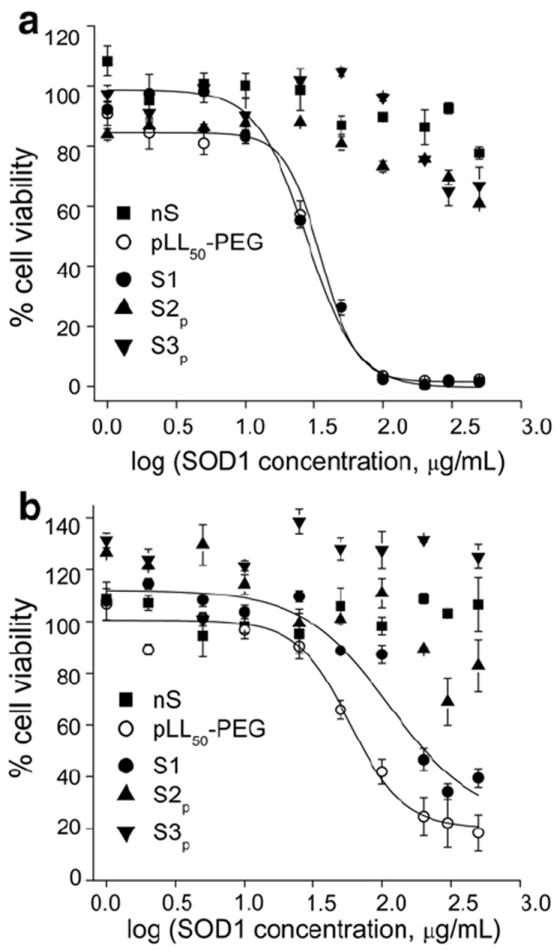
Cytotoxicity of SOD1 formulations determined in (a) TBMEC monolayers and (b) CATH.a neurons. nS – native enzyme, PEG-pLL50 – free block copolymer, S1 – non-cross-linked BIC, S2p and S3p – DTSSP- and BS3-cross-linked purified cl-nanozymes. Cells were treated for 24 h as indicated following which cell viability was determined using a MTS assay kit.
3.4. Superoxide scavenging capability of SOD1 formulations in cultured cells
We determined the ability of SOD1 formulations to scavenge experimentally induced O2· − in cells pre-treated with different formulations (Fig. 6). Both TBMEC and CATH.a neurons treated with purified cl-nanozymes (S2p and S3p) displayed greater ability to scavenge O2· − radicals, compared to cells treated with native SOD1 and non-cross-linked BIC (nS and S1). This effect lasted for at least 12 h post-treatments with S2p and S3p.
Fig. 6.
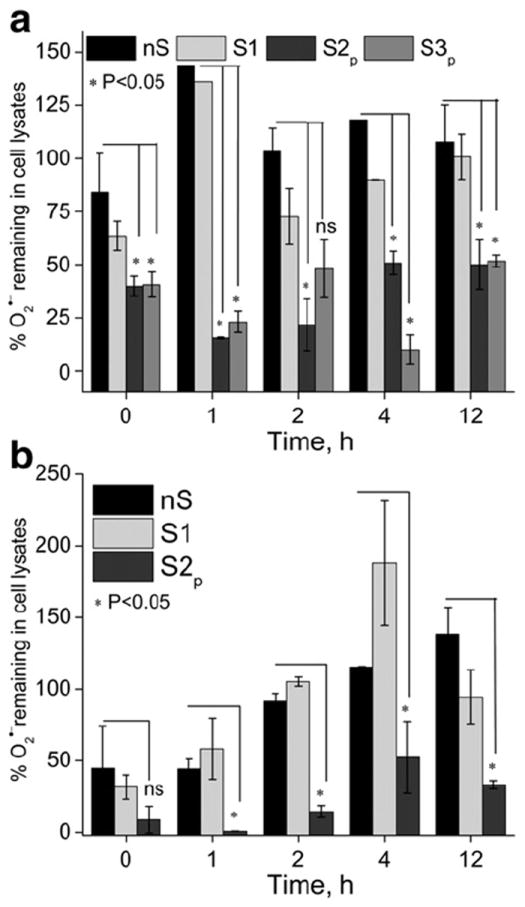
Superoxide scavenging by SOD1 formulations in (a) TBMEC monolayers and (b) CATH.a neurons. Cells were treated for 2 h with native SOD1 or its formulations diluted in complete culture medium, washed and then incubated in fresh medium for different times. nS (native enzyme), S1 (non-cross-linked BIC), S2p and S3p (DTSSP- and BS3-cross-linked-purified cl-nanozymes). O2· − levels are expressed as % relative to untreated cells. *P<0.05: decreases in O2· − levels are statistically significant in cells treated with S2p and S3p (a) and S2p (b) compared with those treated with nS and S1 (ns – not significant).
3.5. Therapeutic efficacy in vivo in a rat MCAO model of ischemia/reperfusion injury
The proof of therapeutic efficacy was shown in a rat MCAO model of stroke. In this model ischemia/reperfusion injury is associated with overproduction of ROS that predominantly cause tissue damage [18]. Hence, we posit that ROS scavenging by purified SOD1 cl-nanozymes can result in attenuation of oxidative damage and produce a therapeutic response. To assess the extent of brain injury after different treatments we used TTC staining as a simple and quick method for determining the infarct size [66]. In the viable brain tissue TTC is enzymatically reduced by dehydrogenases to a red formazan product while pale staining corresponds to infarct areas (Fig. 7a). Rats treated with purified cl-nanozymes (S2p) showed decreased apparent infarct size in the ipsilateral hemisphere, compared with those treated with saline/native SOD1 (nS). Image analysis and quantification of the brain slices indicated a 59% reduction in infarct volume (Fig. 7b). Furthermore, the analysis of the sensorimotor functions of rats revealed a significant 70% improvement in the functional outcomes (Fig. 7c) after a single i.v. injection of purified cl-nanozymes at a dose of 10 kU/kg compared with native SOD1. The administered dose corresponds to a SOD1 concentration of 0.138 mg/mL in case of S2p which is well below the highest concentration (0.5 mg/mL) used in the in vitro cytotoxicity studies.
Fig. 7.
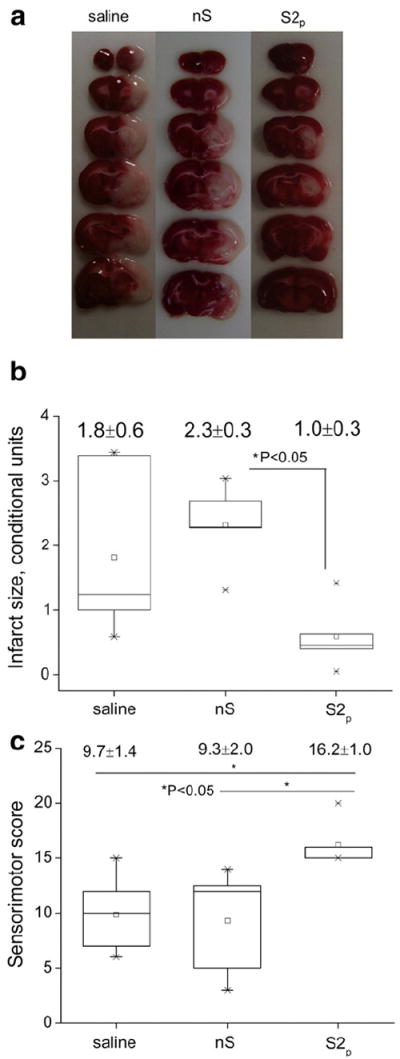
Therapeutic efficacy in a rat MCAO model. (a) TTC staining of brain slices (b) quantitative representation of infarct size and (c) functional outcomes assessed using a sensorimotor score. nS (native SOD1); S2p (DTSSP-cross-linked purified cl-nanozymes). Treatment was administered at the onset of reperfusion (following a 2 h ischemia) i.v. via the tail vein at a dose of 10 kU/kg and sensorimotor functions were evaluated 22 h post-reperfusion before dissecting the brains for TTC staining.
4. Discussion
Previously we reported cl-nanozymes with the cross-linked polyelectrolyte complex core stabilized by amide bonds between the carboxylic groups of SOD1 and the amino groups of PEG-pLL50. Such cl-nanozymes display improved delivery to the brain and are more stable in blood and brain tissues compared to the non-cross-linked SOD1 BICs [56]. Our current design involved cross-linking the amino groups in the polycation template, although we cannot exclude that some protein amino groups may also be cross-linked. To minimize the possibility of these side reactions we used an excess of the polycation (Z=2), which based on ζ-potential measurements results in formation of BICs containing some excess of pLL amines. These amines can react with the cross-linking agents, e.g. DTSSP or BS3. This approach should result in lower extent of modification of enzyme reactive groups compared to the core cross-linking strategy. Indeed SOD1 cl-nanozymes retained ≥90% activity suggesting that protein lysines were mostly spared during the cross-linking reaction as their modification is known to inactivate the enzyme [67]. Moreover, using a different cross-linker, dimethyl 3,3′-dithiobispropionimidate (DTBP) led to a loss of 30 to 80% of SOD1 activity (Supplementary data Table s3), possibly due to extensive modification of protein lysines.
Gel retardation analysis indicated that DTSSP was more efficient in cross-linking than BS3. DTSSP is more hydrophilic than BS3 — their octanol-water partition coefficients (log P) are −2.1 and −1, respectively. This alone may result in better reactivity of DTSSP towards the hydrophilic amino groups. There was an additional indication that different cross-linkers result in different cl-nanozyme formats: while the particle size increased after cross-linking with DTSSP or BS3 (Table 1) it did not change after cross-linking with DTBP (Supplementary data Table s3). Interestingly, a published report [68] indicated that DTSSP-cross-linked pLL/pDNA polyplexes also demonstrated increased particle size, while no such increase was observed in case of DTBP. The sizes may increase due to cross-linking of multiple BIC particles although this does not seem to be reflected in AFM images that display separated spheres for both non-cross-linked and cross-linked BICs. Needless to say the BICs are dynamic formations, which constantly exchange their polyionic components [69]. In the presence of the cross-linker such polyion components may become covalently immobilized in the “host” BICs resulting in particle growth. This should depend on the reactivity of the cross-linker — the growth is more likely for less reactive agents, which form longer living “transitory states”, than for highly reactive agents that tend to rapidly fix the existing structures. The higher reactivity of DTBP compared to DTSSP and BS3 could therefore be responsible for lack of particle enlargement as well as loss of enzyme activity.
Physicochemical characteristics such as particle size, surface charge and morphology influence in vivo disposition of nanoparticles [70,71]. Purification of cl-nanozymes resulted in improved homogeneity of the samples as demonstrated by gel retardation, DLS and AFM. Purified SOD1 cl-nanozyme showed a PDI of <0.05 indicating near monodisperse particles. Purified catalase cl-nanozyme also had particles with unimodal distribution and narrower PDI compared to non-purified cl-nanozyme, albeit in this case we did not achieve a similar degree of homogeneity like SOD1. We expect that the small size (<100 nm), narrower size distribution and chemical homogeneity of the purified cl-nanozymes will decrease their uptake by the reticuloendothelial system (RES), increase their in vivo stability and decrease clearance (Brynskikh et al., manuscript in preparation). Sedimentation equilibrium analysis also demonstrated that purified cl-nanozymes contained no aggregates, which favors avoidance of RES uptake. Native SOD1 has a short circulation half-life of 6 min [72] and is rapidly cleared from circulation in addition to inactivation by ubiquitous proteases. SOD1 cl-nanozyme may circulate longer and be protected against proteolytic degradation. Overall, these well-defined nanozymes showed a good batch-to-batch reproducibility in physicochemical properties (hydrodynamic size and enzyme activity). In addition, their biological properties (cytotoxicity and superoxide scavenging ability) were comparable in studies conducted using nanozymes from different preparations.
The cell line models in our studies represent key cell types of the neurovascular unit. They are likely targets in treating cerebrovascular diseases [73] including stroke given that both neurons and microvessels respond equally rapidly to the ischemic insult [74]. Decreased cytotoxicity of purified SOD1 cl-nanozymes in these cells may potentially allow administration of their higher doses. Appropriate choice of the cross-linking agent also seems to be important since the BS3-cross-linked cl-nanozymes are less toxic than DTSSP-cross-linked cl-nanozymes. We speculate that subcellular reduction of disulfide bonds in DTSSP links may lead to release of the polycationic species that display toxicity. However, it should be noted that upon complete degradation of the block copolymer, lysines will be metabolized to acetyl-coenzyme A (acetyl-CoA) or acetoacetyl-CoA in vivo and PEG is biocompatible.
The prolonged antioxidant effect of purified SOD1 cl-nanozyme in cells (up to 12 h post-withdrawal of the treatment) is most likely due to its improved stability. We believe that the PEG-pLL50 chains in the BIC can sterically protect SOD1molecules against degradation by intracellular proteases. This is further supported by the fact that in spite of the lower uptake of cl-nanozymes compared to non-cross-linked BIC (not shown), the internalized fraction remained more catalytically active over time. This also relateswell to our earlier published observation [56] that cl-nanozyme not only delivered higher amounts of SOD1 to the brain parenchyma, but was also retained to a higher extent in brain capillaries in healthy mice. Higher retention in the brain capillaries may allow us to use such nanoparticles to treat cerebrovascular disorders like stroke where rescue of the BBB from oxidative damage could result in therapeutic outcomes [73]. This, in fact, led us to test the potential of purified SOD1 cl-nanozymes to treat stroke in a rat MCAO model.
Using this model we noted a clear decrease in the infarct volume concomitant with significant improvement in the sensorimotor function after i.v. injection of a single dose of purified SOD1 cl-nanozyme. Its therapeutic efficacy may be due to ROS scavenging both at the level of the brain microvessels and brain parenchyma. The former could be explained by the increased retention and stability of the SOD1 cl-nanozyme in the BBB. In addition, the compromised integrity of the BBB, a well-known phenomenon in CNS pathologies (including stroke [75]), may also improve delivery of the cl-nanozymes to neurons and supportive cells (astrocytes, glial cells and resident inflammatory cells) resulting in their protection from oxidative stress. Thus, both improved accumulation of the SOD1 cl-nanozymes due to BBB permeability and increased retention of active enzyme in the brain microvessels could contribute to decreased brain injury upon stroke.
Notwithstanding the therapeutic effect of cationic liposomes [17,76], their translational significance might be limited due to low stability as pharmaceutical formulations [49]. In contrast to SOD1 liposomes, covalently stabilized cl-nanozymes are stable, and in this regard represent innovative formulations. Toxicity is another concern for cationic carriers (including cationic liposomes) and this is addressed in this work by developing nearly electroneutral forms of cl-nanozymes with considerably decreased toxicity to brain endothelial cells and neurons. While cellular entry of PEG-SOD1 is a limitation [10], cl-nanozyme enters cells of the neurovascular unit, which is likely to be beneficial for treatment.
In contrast to PLGA nanoparticles that gradually release encapsulated SOD1 over days and weeks, SOD1 in cl-nanozyme formulations is fully and immediately available for O2· − scavenging. While PLGA hydrolysis may affect stability of encapsulated proteins [48], SOD1 remains stable in cl-nanozyme formulation as indicated by the sustained decomposition of O2· − in the in vitro studies. Noteworthy, in contrast to PLGA particles that were injected i.c. the cl-nanozymes demonstrated therapeutic efficacy after i.v. injection.
In this regard it is also important that we observed an improvement in functional outcomes, which is a main goal in treating stroke patients [66]. It is worth mentioning that often the preclinical efficacy of stroke drugs in animal experiments do not translate well into clinical benefits since only gross histological techniques are being used in efficacy determination [66]. Administration of purified cl-nanozyme suppressed brain tissue damage and also improved sensorimotor functions. This encourages further investigation of cl-nanozymes and their development for clinical applications.
5. Conclusion
We developed a simple method to prepare well-defined cross-linked antioxidant nanozymes containing SOD1 or catalase and characterized their physicochemical properties. We validated the ability of such constructs to scavenge O2· − radicals in vitro in two cell culture models, cultured brain microvessel endothelial cells and central neurons. As a proof-of-concept, we demonstrated that SOD1 cl-nanozymes can attenuate oxidative damage, induce tissue protection and improve functional outcomes in a rat MCAO model of ischemia/reperfusion injury.
Supplementary Material
Acknowledgments
This work was supported by the United States National Institutes of Health (NIH) grant no. RR021937, the Center for Biomedical Research Excellence (COBRE) Nebraska Center for Nanomedicine, grant no. 1RO1 NS057748 (to E V Batrakova), and the Ministry of Education and Science of Russian Federation grant no. 11.G34.31.0004. We would like to thank the COBRE Nanomaterials Core Characterization Facility (Mr. Nazar Filonov), UNMC AFM core facility, EPR Spectroscopy Facility, and members of Kabanov’s laboratory Dr. Xiang Yi and Mr. Marc R. Ueda for helpful discussions and Dr. Daria Y. Filonova (Alakhova) for help with schematic representation.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jconrel.2012.07.044
References
- 1.Barnham KJ, Masters CL, Bush AI. Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 2.Gilgun-Sherki Y, Melamed E, Offen D. Oxidative stress induced-neurodegenerative diseases: the need for antioxidants that penetrate the blood brain barrier. Neuropharmacology. 2001;40:959–975. doi: 10.1016/s0028-3908(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 3.Chrissobolis S, Miller AA, Drummond GR, Kemp-Harper BK, Sobey CG. Oxidative stress and endothelial dysfunction in cerebrovascular disease. Front Biosci. 2011;16:1733–1745. doi: 10.2741/3816. [DOI] [PubMed] [Google Scholar]
- 4.Kaur J, Arora S, Singh B, Thakur LC, Gambhir J, Prabhu KM. Role of oxidative stress in pathophysiology of transient ischemic attack and stroke. Int J Biol Med Res. 2011;2:611–615. [Google Scholar]
- 5.Pan J, Konstas AA, Bateman B, Ortolano GA, Pile-Spellman J. Reperfusion injury following cerebral ischemia: pathophysiology, MR imaging, and potential therapies. Neuroradiology. 2007;49:93–102. doi: 10.1007/s00234-006-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banks WA. Delivery of peptides to the brain: emphasis on therapeutic development. Biopolymers. 2008;90:589–594. doi: 10.1002/bip.20980. [DOI] [PubMed] [Google Scholar]
- 7.Sood R, Yang Y, Taheri S, Candelario-Jalil E, Estrada EY, Walker EJ, Thompson J, Rosenberg GA. Increased apparent diffusion coefficients on MRI linked with matrix metalloproteinases and edema in white matter after bilateral carotid artery occlusion in rats. J Cereb Blood Flow Metab. 2009;29:308–316. doi: 10.1038/jcbfm.2008.121. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Pardridge WM. Conjugation of brain-derived neurotrophic factor to a blood–brain barrier drug targeting system enables neuroprotection in regional brain ischemia following intravenous injection of the neurotrophin. Brain Res. 2001;889:49–56. doi: 10.1016/s0006-8993(00)03108-5. [DOI] [PubMed] [Google Scholar]
- 9.Beckman JS, Minor RL, Jr, White CW, Repine JE, Rosen GM, Freeman BA. Superoxide dismutase and catalase conjugated to polyethylene glycol increases endothelial enzyme activity and oxidant resistance. J Biol Chem. 1988;263:6884–6892. [PubMed] [Google Scholar]
- 10.Veronese FM, Caliceti P, Schiavon O, Sergi M. Polyethylene glycol-superoxide dismutase, a conjugate in search of exploitation. Adv Drug Deliv Rev. 2002;54:587–606. doi: 10.1016/s0169-409x(02)00029-7. [DOI] [PubMed] [Google Scholar]
- 11.Eum WS, Kim DW, Hwang IK, Yoo KY, Kang TC, Jang SH, Choi HS, Choi SH, Kim YH, Kim SY, Kwon HY, Kang JH, Kwon OS, Cho SW, Lee KS, Park J, Won MH, Choi SY. In vivo protein transduction: biologically active intact pep-1-superoxide dismutase fusion protein efficiently protects against ischemic insult. Free Radic Biol Med. 2004;37:1656–1669. doi: 10.1016/j.freeradbiomed.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 12.Grey M, Yainoy S, Prachayasittikul V, Bulow L. A superoxide dismutase-human hemoglobin fusion protein showing enhanced antioxidative properties. FEBS J. 2009;276:6195–6203. doi: 10.1111/j.1742-4658.2009.07323.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim DW, Jeong HJ, Kang HW, Shin MJ, Sohn EJ, Kim MJ, Ahn EH, An JJ, Jang SH, Yoo KY, Won MH, Kang TC, Hwang IK, Kwon OS, Cho SW, Park J, Eum WS, Choi SY. Transduced human PEP-1-catalase fusion protein attenuates ischemic neuronal damage. Free Radic Biol Med. 2009;47:941–952. doi: 10.1016/j.freeradbiomed.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 14.Lu M, Gong X, Lu Y, Guo J, Wang C, Pan Y. Molecular cloning and functional characterization of a cell-permeable superoxide dismutase targeted to lung adenocarcinoma cells. Inhibition cell proliferation through the Akt/p27kip1 pathway. J Biol Chem. 2006;281:13620–13627. doi: 10.1074/jbc.M600523200. [DOI] [PubMed] [Google Scholar]
- 15.Corvo ML, Boerman OC, Oyen WJ, Van Bloois L, Cruz ME, Crommelin DJ, Storm G. Intravenous administration of superoxide dismutase entrapped in long circulating liposomes. II. In vivo fate in a ratmodel of adjuvant arthritis. Biochim Biophys Acta. 1999;1419:325–334. doi: 10.1016/s0005-2736(99)00081-4. [DOI] [PubMed] [Google Scholar]
- 16.Freeman BA, Young SL, Crapo JD. Liposome-mediated augmentation of superoxide dismutase in endothelial cells prevents oxygen injury. J Biol Chem. 1983;258:12534–12542. [PubMed] [Google Scholar]
- 17.Imaizumi S, Woolworth V, Fishman RA, Chan PH. Liposome-entrapped superoxide dismutase reduces cerebral infarction in cerebral ischemia in rats. Stroke. 1990;21:1312–1317. doi: 10.1161/01.str.21.9.1312. [DOI] [PubMed] [Google Scholar]
- 18.Reddy MK, Labhasetwar V. Nanoparticle-mediated delivery of superoxide dismutase to the brain: an effective strategy to reduce ischemia-reperfusion injury. FASEB J. 2009;23:1384–1395. doi: 10.1096/fj.08-116947. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi R, Hoshino J, Ochiai A, Morizawa Y, Mizushima Y. Lecithinized superoxide dismutase enhances its pharmacologic potency by increasing its cell membrane affinity. J Pharmacol Exp Ther. 1994;271:1672–1677. [PubMed] [Google Scholar]
- 20.Ishihara T, Tanaka K, Tasaka Y, Namba T, Suzuki J, Okamoto S, Hibi T, Takenaga M, Igarashi R, Sato K, Mizushima Y, Mizushima T. Therapeutic effect of lecithinized superoxide dismutase against colitis. J Pharmacol Exp Ther. 2009;328:152–164. doi: 10.1124/jpet.108.144451. [DOI] [PubMed] [Google Scholar]
- 21.Koo DD, Welsh KI, West NE, Channon KM, Penington AJ, Roake JA, Morris PJ, Fuggle SV. Endothelial cell protection against ischemia/reperfusion injury by lecithinized superoxide dismutase. Kidney Int. 2001;60:786–796. doi: 10.1046/j.1523-1755.2001.060002786.x. [DOI] [PubMed] [Google Scholar]
- 22-46.Supplementary information.
- 47.Francis JW, Ren J, Warren J, Brown RH, Jr, Finklestein SP. Postischemic infusion of Cu/Zn superoxide dismutase or SOD:Tet451 reduces cerebral infarction following focal ischemia/reperfusion in rats. Exp Neurol. 1997;146:435–443. doi: 10.1006/exnr.1997.6547. [DOI] [PubMed] [Google Scholar]
- 48.Jiang W, Schwendeman SP. Stabilization of tetanus toxoid encapsulated in PLGA microspheres. Mol Pharm. 2008;5:808–817. doi: 10.1021/mp800027f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinha J, Das N, Basu MK. Liposomal antioxidants in combating ischemia-reperfusion injury in rat brain. Biomed Pharmacother. 2001;55:264–271. doi: 10.1016/s0753-3322(01)00060-9. [DOI] [PubMed] [Google Scholar]
- 50.Yusa T, Crapo JD, Freeman BA. Liposome-mediated augmentation of brain SOD and catalase inhibits CNS O2 toxicity. J Appl Physiol. 1984;57:1674–1681. doi: 10.1152/jappl.1984.57.6.1674. [DOI] [PubMed] [Google Scholar]
- 51.Batrakova EV, Li S, Reynolds AD, Mosley RL, Bronich TK, Kabanov AV, Gendelman HE. A macrophage-nanozyme delivery system for Parkinson’s disease. Bioconjug Chem. 2007;18:1498–1506. doi: 10.1021/bc700184b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenbaugh EG, Roat JW, Gao L, Yang RF, Manickam DS, Yin JX, Schultz HD, Bronich TK, Batrakova EV, Kabanov AV, Zucker IH, Zimmerman MC. The attenuation of central angiotensin II-dependent pressor response and intra-neuronal signaling by intracarotid injection of nanoformulated copper/zinc superoxide dismutase. Biomaterials. 2010;31:5218–5226. doi: 10.1016/j.biomaterials.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaydess A, Duysen E, Li Y, Gilman V, Kabanov A, Lockridge O, Bronich T. Visualization of exogenous delivery of nanoformulated butyrylcholinesterase to the central nervous system. Chem Biol Interact. 2010;187:295–298. doi: 10.1016/j.cbi.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haney MJ, Zhao Y, Li S, Higginbotham SM, Booth SL, Han HY, Vetro JA, Mosley RL, Kabanov AV, Gendelman HE, Batrakova EV. Cell-mediated transfer of catalase nanoparticles from macrophages to brain endothelial, glial and neuronal cells. Nanomedicine (Lond) 2011;6:1215–1230. doi: 10.2217/nnm.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Y, Haney MJ, Klyachko NL, Li S, Booth SL, Higginbotham SM, Jones J, Zimmerman MC, Mosley RL, Kabanov AV, Gendelman HE, Batrakova EV. Polyelectrolyte complex optimization for macrophage delivery of redox enzyme nanoparticles. Nanomedicine (Lond) 2011;6:25–42. doi: 10.2217/nnm.10.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klyachko NL, Manickam DS, Brynskikh AM, Uglanova SV, Li S, Higginbotham SM, Bronich TK, Batrakova EV, Kabanov AV. Cross-linked antioxidant nanozymes for improved delivery to CNS. Nanomed Nanotechnol Biol Med. 2012;8:119–129. doi: 10.1016/j.nano.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 58.Yi X, Zimmerman MC, Yang R, Tong J, Vinogradov S, Kabanov AV. Pluronic-Modified Superoxide Dismutase 1 (SOD1) attenuates Angiotensin II-induced increase in intracellular superoxide in neurons. Free Radic Biol Med. 2010;49:548–558. doi: 10.1016/j.freeradbiomed.2010.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Schellhorn HE. Rapid kinetic microassay for catalase activity. J Biomol Tech. 2007;18:185–187. [PMC free article] [PubMed] [Google Scholar]
- 60.Sorgen PL, Duffy HS, Spray DC, Delmar M. pH-dependent dimerization of the carboxyl terminal domain of Cx43. Biophys J. 2004;87:574–581. doi: 10.1529/biophysj.103.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yazdanian M, Bormann BJ. Immortalized Brain Endothelial Cells. Boehringer Ingelheim Pharmaceuticals, Inc.; Ridgefield, Conn., United States: 2000. [Google Scholar]
- 62.Sun H, Zhao H, Sharpe GM, Arrick DM, Mayhan WG. Effect of chronic alcohol consumption on brain damage following transient focal ischemia. Brain Res. 2008;1194:73–80. doi: 10.1016/j.brainres.2007.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marklund S, Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur J Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 64.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials. 2001;22:471–480. doi: 10.1016/s0142-9612(00)00203-9. [DOI] [PubMed] [Google Scholar]
- 65.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 66.Benedek A, Moricz K, Juranyi Z, Gigler G, Levay G, Harsing LG, Jr, Matyus P, Szenasi G, Albert G. Use of TTC staining for the evaluation of tissue injury in the early phases of reperfusion after focal cerebral ischemia in rats. Brain Res. 2006;1116:159–165. doi: 10.1016/j.brainres.2006.07.123. [DOI] [PubMed] [Google Scholar]
- 67.Cocco D, Rossi L, Barra D, Bossa F, Rotilio G. Carbamoylation of Cu, Zn-superoxide dismutase by cyanate Role of lysines in the enzyme action. FEBS Lett. 1982;150:303–306. doi: 10.1016/0014-5793(82)80756-4. [DOI] [PubMed] [Google Scholar]
- 68.Oupicky D, Carlisle RC, Seymour LW. Triggered intracellular activation of disulfide crosslinked polyelectrolyte gene delivery complexes with extended systemic circulation in vivo. Gene Ther. 2001;8:713–724. doi: 10.1038/sj.gt.3301446. [DOI] [PubMed] [Google Scholar]
- 69.Li Y, Bronich TK, Chelushkin PS, Kabanov AV. Dynamic properties of block ionomer complexes with polyion complex cores. Macromolecules. 2008;41:5863–5868. [Google Scholar]
- 70.Choi HS, Ipe BI, Misra P, Lee JH, Bawendi MG, Frangioni JV. Tissue- and organ-selective biodistribution of NIR fluorescent quantum dots. Nano Lett. 2009;9:2354–2359. doi: 10.1021/nl900872r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, Bawendi MG, Frangioni JV. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis AS, Zhao H, Sun GH, Sapolsky RM, Steinberg GK. Gene therapy using SOD1 protects striatal neurons from experimental stroke. Neurosci Lett. 2007;411:32–36. doi: 10.1016/j.neulet.2006.08.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.del Zoppo GJ. Stroke and neurovascular protection. N Engl J Med. 2006;354:553–555. doi: 10.1056/NEJMp058312. [DOI] [PubMed] [Google Scholar]
- 74.Mabuchi T, Lucero J, Feng A, Koziol JA, del Zoppo GJ. Focal cerebral ischemia preferentially affects neurons distant from their neighboring microvessels. J Cereb Blood Flow Metab. 2005;25:257–266. doi: 10.1038/sj.jcbfm.9600027. [DOI] [PubMed] [Google Scholar]
- 75.Nagaraja TN, Keenan KA, Fenstermacher JD, Knight RA. Acute leakage patterns of fluorescent plasma flow markers after transient focal cerebral ischemia suggest large openings in blood–brain barrier. Microcirculation. 2008;15:1–14. doi: 10.1080/10739680701409811. [DOI] [PubMed] [Google Scholar]
- 76.Chan PH, Longar S, Fishman RA. Protective effects of liposome-entrapped superoxide dismutase on posttraumatic brain edema. Ann Neurol. 1987;21:540–547. doi: 10.1002/ana.410210604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


