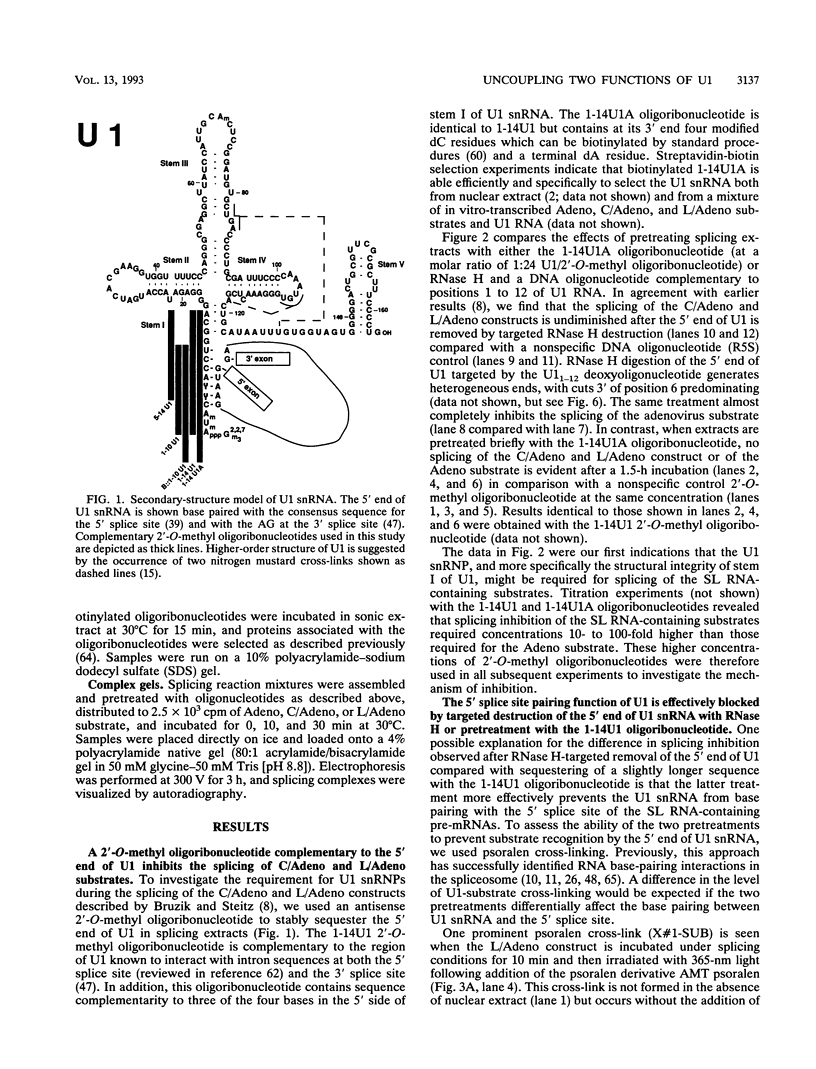
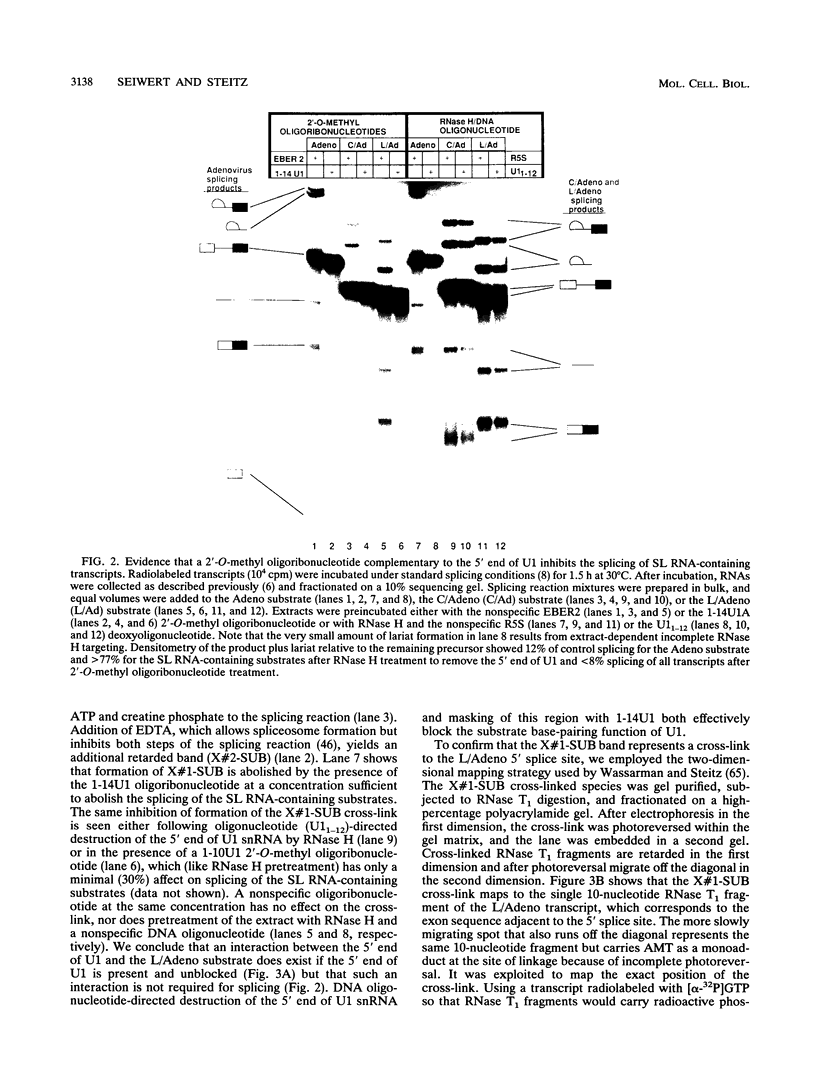
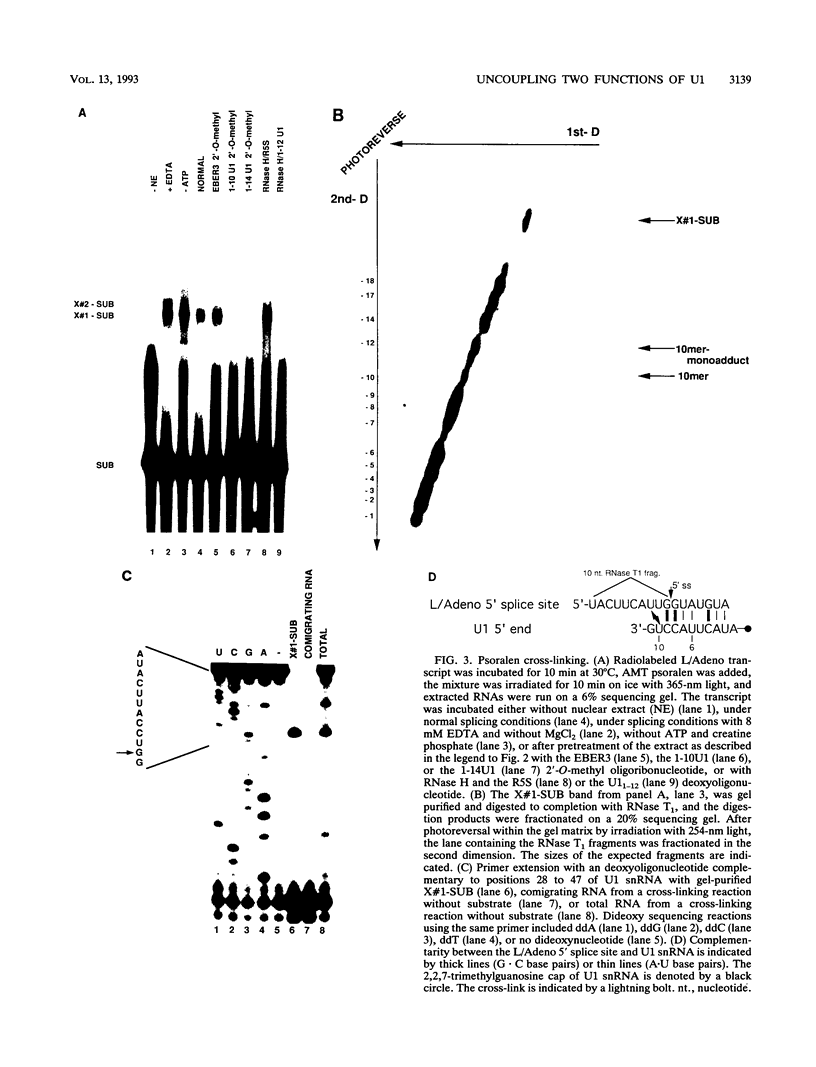
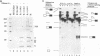Abstract
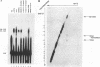
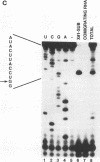
To probe functions of the U1 small nuclear ribonucleoprotein particle (snRNP) during in vitro splicing, we have used unusual splicing substrates which replace the 5' splice site region of an adenovirus substrate with spliced leader (SL) RNA sequences from Leptomonas collosoma or Caenorhabditis elegans. In agreement with previous results (J.P. Bruzik and J.A. Steitz, Cell 62:889-899, 1990), we find that oligonucleotide-targeted RNase H destruction of the 5' end of U1 snRNA inhibits the splicing of a standard adenovirus splicing substrate but not of the SL RNA-containing substrates. However, use of an antisense 2'-O-methyl oligoribonucleotide that disrupts the first stem of U1 snRNA as well as stably sequestering positions of U1 snRNA involved in 5' and 3' splice site recognition inhibits the splicing of both the SL constructs and the standard adenovirus substrate. The 2'-O-methyl oligoribonucleotide is no more effective than RNase H pretreatment in preventing pairing of U1 with the 5' splice site, as assessed by inhibition of psoralen cross-link formation between the SL RNA-containing substrate and U1. The 2'-O-methyl oligoribonucleotide does not alter the protein composition of the U1 monoparticle or deplete the system of essential splicing factors. Native gel analysis indicates that the 2'-O-methyl oligoribonucleotide inhibits splicing by diminishing the formation of splicing complexes. One interpretation of these results is that removal of the 5' end of U1 inhibits base pairing in a different way than sequestering the same sequence with a complementary oligoribonucleotide. Alternatively, our data may indicate that two elements near the 5' end of U1 RNA normally act during spliceosome assembly; the extreme 5' end base pairs with the 5' splice site, while the sequence or structural integrity of stem I is essential for some additional function. It follows that different introns may differ in their use of the repertoire of U1 snRNP functions.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ares M., Jr, Igel A. H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990 Dec;4(12A):2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- Barabino S. M., Blencowe B. J., Ryder U., Sproat B. S., Lamond A. I. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990 Oct 19;63(2):293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Bindereif A., Green M. R. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987 Aug;6(8):2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., Pinto A. L. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol Cell Biol. 1989 Aug;9(8):3350–3359. doi: 10.1128/mcb.9.8.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L., Steitz J. A. Pre-mRNA splicing in vitro requires intact U4/U6 small nuclear ribonucleoprotein. Cell. 1986 Aug 29;46(5):697–704. doi: 10.1016/0092-8674(86)90345-4. [DOI] [PubMed] [Google Scholar]
- Blencowe B. J., Sproat B. S., Ryder U., Barabino S., Lamond A. I. Antisense probing of the human U4/U6 snRNP with biotinylated 2'-OMe RNA oligonucleotides. Cell. 1989 Nov 3;59(3):531–539. doi: 10.1016/0092-8674(89)90036-6. [DOI] [PubMed] [Google Scholar]
- Bruzik J. P., Steitz J. A. Spliced leader RNA sequences can substitute for the essential 5' end of U1 RNA during splicing in a mammalian in vitro system. Cell. 1990 Sep 7;62(5):889–899. doi: 10.1016/0092-8674(90)90264-f. [DOI] [PubMed] [Google Scholar]
- Bruzik J. P., Van Doren K., Hirsh D., Steitz J. A. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature. 1988 Oct 6;335(6190):559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- Calvet J. P., Meyer L. M., Pederson T. Small nuclear RNA U2 is base-paired to heterogeneous nuclear RNA. Science. 1982 Jul 30;217(4558):456–458. doi: 10.1126/science.6178162. [DOI] [PubMed] [Google Scholar]
- Calvet J. P., Pederson T. Base-pairing interactions between small nuclear RNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell. 1981 Nov;26(3 Pt 1):363–370. doi: 10.1016/0092-8674(81)90205-1. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986 Jan 31;44(2):207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987 Nov;1(9):1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Cimino G. D., Gamper H. B., Isaacs S. T., Hearst J. E. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu Rev Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- Datta B., Weiner A. M. Cross-linking of U1 snRNA using nitrogen mustard. Evidence for higher order structure. J Biol Chem. 1992 Mar 5;267(7):4503–4507. [PubMed] [Google Scholar]
- Daugeron M. C., Tazi J., Jeanteur P., Brunel C., Cathala G. U1-U2 snRNPs interaction induced by an RNA complementary to the 5' end sequence of U1 snRNA. Nucleic Acids Res. 1992 Jul 25;20(14):3625–3630. doi: 10.1093/nar/20.14.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubendorff J. W., Whittaker L. J., Eltman J. T., Lipsick J. S. Carboxy-terminal elements of c-Myb negatively regulate transcriptional activation in cis and in trans. Genes Dev. 1992 Dec;6(12B):2524–2535. doi: 10.1101/gad.6.12b.2524. [DOI] [PubMed] [Google Scholar]
- Fradin A., Jove R., Hemenway C., Keiser H. D., Manley J. L., Prives C. Splicing pathways of SV40 mRNAs in X. laevis oocytes differ in their requirements for snRNPs. Cell. 1984 Jul;37(3):927–936. doi: 10.1016/0092-8674(84)90427-6. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Krämer A., Keller W. Different small nuclear ribonucleoprotein particles are involved in different steps of splicing complex formation. Cold Spring Harb Symp Quant Biol. 1987;52:287–298. doi: 10.1101/sqb.1987.052.01.034. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Hall K. B., Konarska M. M. The 5' splice site consensus RNA oligonucleotide induces assembly of U2/U4/U5/U6 small nuclear ribonucleoprotein complexes. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10969–10973. doi: 10.1073/pnas.89.22.10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Dathan N. A., Mattaj I. W. Functional analysis of mutant Xenopus U2 snRNAs. Cell. 1989 Oct 6;59(1):159–169. doi: 10.1016/0092-8674(89)90878-7. [DOI] [PubMed] [Google Scholar]
- Hamm J., Dathan N. A., Scherly D., Mattaj I. W. Multiple domains of U1 snRNA, including U1 specific protein binding sites, are required for splicing. EMBO J. 1990 Apr;9(4):1237–1244. doi: 10.1002/j.1460-2075.1990.tb08231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner T. P., Giglio L. M., Weiner A. M. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev. 1990 Dec;4(12A):2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- Heintz N., Roeder R. G. Transcription of human histone genes in extracts from synchronized HeLa cells. Proc Natl Acad Sci U S A. 1984 May;81(9):2713–2717. doi: 10.1073/pnas.81.9.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison S. F., Crow A., Garcia-Blanco M. A. The spliceosome assembly pathway in mammalian extracts. Mol Cell Biol. 1992 Oct;12(10):4279–4287. doi: 10.1128/mcb.12.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison S. F., Garcia-Blanco M. A. An ATP-independent U2 small nuclear ribonucleoprotein particle/precursor mRNA complex requires both splice sites and the polypyrimidine tract. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5482–5486. doi: 10.1073/pnas.89.12.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B., Kornstädt U., Bach M., Lührmann R. Structure of the small nuclear RNP particle U1: identification of the two structural protuberances with RNP-antigens A and 70K. J Cell Biol. 1992 Feb;116(4):839–849. doi: 10.1083/jcb.116.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987 Jun 19;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Krol A., Westhof E., Bach M., Lührmann R., Ebel J. P., Carbon P. Solution structure of human U1 snRNA. Derivation of a possible three-dimensional model. Nucleic Acids Res. 1990 Jul 11;18(13):3803–3811. doi: 10.1093/nar/18.13.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm G. M., Blencowe B. J., Sproat B. S., Iribarren A. M., Ryder U., Lamond A. I. Antisense probes containing 2-aminoadenosine allow efficient depletion of U5 snRNP from HeLa splicing extracts. Nucleic Acids Res. 1991 Jun 25;19(12):3193–3198. doi: 10.1093/nar/19.12.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Sproat B., Ryder U., Hamm J. Probing the structure and function of U2 snRNP with antisense oligonucleotides made of 2'-OMe RNA. Cell. 1989 Jul 28;58(2):383–390. doi: 10.1016/0092-8674(89)90852-0. [DOI] [PubMed] [Google Scholar]
- Liao X. C., Colot H. V., Wang Y., Rosbash M. Requirements for U2 snRNP addition to yeast pre-mRNA. Nucleic Acids Res. 1992 Aug 25;20(16):4237–4245. doi: 10.1093/nar/20.16.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S., Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991 Dec;5(12B):2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Darzynkiewicz E., Shatkin A. J. Proximity of mRNA5'-region and 18S rRNA in eukaryotic initiation complexes. Nature. 1980 Jul 17;286(5770):226–230. doi: 10.1038/286226a0. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992 Feb 21;68(4):743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Newman A., Norman C. Mutations in yeast U5 snRNA alter the specificity of 5' splice-site cleavage. Cell. 1991 Apr 5;65(1):115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- Pan Z. Q., Prives C. U2 snRNA sequences that bind U2-specific proteins are dispensable for the function of U2 snRNP in splicing. Genes Dev. 1989 Dec;3(12A):1887–1898. doi: 10.1101/gad.3.12a.1887. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rymond B. C., Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. 1986 Nov 27-Dec 3Nature. 324(6095):341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- Reed R., Griffith J., Maniatis T. Purification and visualization of native spliceosomes. Cell. 1988 Jun 17;53(6):949–961. doi: 10.1016/s0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Reich C. I., VanHoy R. W., Porter G. L., Wise J. A. Mutations at the 3' splice site can be suppressed by compensatory base changes in U1 snRNA in fission yeast. Cell. 1992 Jun 26;69(7):1159–1169. doi: 10.1016/0092-8674(92)90637-r. [DOI] [PubMed] [Google Scholar]
- Rinke J., Appel B., Digweed M., Lührmann R. Localization of a base-paired interaction between small nuclear RNAs U4 and U6 in intact U4/U6 ribonucleoprotein particles by psoralen cross-linking. J Mol Biol. 1985 Oct 20;185(4):721–731. doi: 10.1016/0022-2836(85)90057-9. [DOI] [PubMed] [Google Scholar]
- Rosbash M., Séraphin B. Who's on first? The U1 snRNP-5' splice site interaction and splicing. Trends Biochem Sci. 1991 May;16(5):187–190. doi: 10.1016/0968-0004(91)90073-5. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988 Nov 18;242(4881):1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. Pre-mRNA splicing in yeast. Trends Genet. 1991 Mar;7(3):79–85. doi: 10.1016/0168-9525(91)90276-V. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Ryder U., Sproat B. S., Lamond A. I. Sequence-specific affinity selection of mammalian splicing complexes. Nucleic Acids Res. 1990 Dec 25;18(24):7373–7379. doi: 10.1093/nar/18.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H., Abelson J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5' splice site during the splicing reaction in yeast. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11269–11273. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Sharp P. A. "Five easy pieces". Science. 1991 Nov 1;254(5032):663–663. doi: 10.1126/science.1948046. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Steitz J. A. Immunoprecipitation of ribonucleoproteins using autoantibodies. Methods Enzymol. 1989;180:468–481. doi: 10.1016/0076-6879(89)80118-1. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. Exon mutations uncouple 5' splice site selection from U1 snRNA pairing. Cell. 1990 Nov 2;63(3):619–629. doi: 10.1016/0092-8674(90)90457-p. [DOI] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992 Sep 25;257(5078):1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol. 1991 Jul;11(7):3432–3445. doi: 10.1128/mcb.11.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T., Bindereif A. Reconstituted mammalian U4/U6 snRNP complements splicing: a mutational analysis. EMBO J. 1992 Jan;11(1):345–359. doi: 10.1002/j.1460-2075.1992.tb05057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuo C. Y., Weiner A. M. Genetic analysis of the role of human U1 snRNA in mRNA splicing: I. Effect of mutations in the highly conserved stem-loop I of U1. Genes Dev. 1989 May;3(5):697–707. doi: 10.1101/gad.3.5.697. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Kent J. R., Cech T. R. A labile phosphodiester bond at the ligation junction in a circular intervening sequence RNA. Science. 1984 May 11;224(4649):574–578. doi: 10.1126/science.6200938. [DOI] [PubMed] [Google Scholar]
- Zavanelli M. I., Ares M., Jr Efficient association of U2 snRNPs with pre-mRNA requires an essential U2 RNA structural element. Genes Dev. 1991 Dec;5(12B):2521–2533. doi: 10.1101/gad.5.12b.2521. [DOI] [PubMed] [Google Scholar]
- Zillmann M., Rose S. D., Berget S. M. U1 small nuclear ribonucleoproteins are required early during spliceosome assembly. Mol Cell Biol. 1987 Aug;7(8):2877–2883. doi: 10.1128/mcb.7.8.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen V. L., Spritz R. A. Alternative splicing of SV40 early pre-mRNA in vitro. Nucleic Acids Res. 1986 Dec 22;14(24):9911–9926. doi: 10.1093/nar/14.24.9911. [DOI] [PMC free article] [PubMed] [Google Scholar]