Abstract
mRNA vaccines combine desirable immunological properties with an outstanding safety profile and the unmet flexibility of genetic vaccines. Based on in situ protein expression, mRNA vaccines are capable of inducing a balanced immune response comprising both cellular and humoral immunity while not subject to MHC haplotype restriction. In addition, mRNA is an intrinsically safe vector as it is a minimal and only transient carrier of information that does not interact with the genome. Because any protein can be expressed from mRNA without the need to adjust the production process, mRNA vaccines also offer maximum flexibility with respect to development. Taken together, mRNA presents a promising vector that may well become the basis of a game-changing vaccine technology platform. Here, we outline the current knowledge regarding different aspects that should be considered when developing an mRNA-based vaccine technology.
Keywords: mRNA, adjuvant, vaccine, mRNA production, mRNA design, mRNA uptake, formulation, protein expression
Introduction
RNA is considered as notoriously unstable making its therapeutic use a provocative idea. Despite the sensitivity of the molecule to the virtually omnipresent ribonucleases (RNases),1 mRNA as a therapeutic was first promoted in 1989 after the development of a broadly applicable in vitro transfection technique.2 Only a couple of years later, mRNA was advocated as a vaccine platform, perhaps being ideal in the sense that it brings together the immunological features of live attenuated vaccines such as endogenous antigen expression and T cell induction with those of killed or subunit vaccines like defined composition and safety.3,4
Particularly compared with DNA as a therapeutic or more specifically as a vaccine, mRNA offers strong safety advantages.5 As the minimal genetic construct, it harbors only the elements directly required for expression of the encoded protein. Moreover, while recombination between single-stranded RNA molecules may occur in rare cases,6,7 mRNA does not interact with the genome. Thus, potentially detrimental genomic integration is excluded. Finally, this lack of genomic integration in combination with mRNA being non-replicative as well as metabolically decaying within a few days8 makes mRNA a merely transient carrier of information.
mRNA as the technological basis of therapeutics and vaccines is characterized by a great flexibility with respect to production and application. Any protein can be encoded and expressed by mRNA, in principle enabling the development of prophylactic and therapeutic vaccines fighting diseases as diverse as infections and cancer as well as protein replacement therapies. Since changes of the encoded protein just alter the sequence of the RNA molecule, leaving its physico-chemical characteristics largely unaffected, diverse products can be manufactured using the same established production process without any adjustment, saving time and reducing cost compared with other vaccine platforms. In terms of efficacy, mRNA-based therapeutics profit from the fact that they do not need to cross the nuclear envelope as opposed to DNA. In contrast to peptides, mRNA vaccines lack MHC haplotype restriction. In addition, mRNA binds to pattern recognition receptors and mRNA vaccines may be designed to be self-adjuvanting,9 a property which peptide- and protein-based vaccines lack.
All in all, mRNA presents a promising, even if challenging, class of therapeutic molecules that has the potential to become the basis of a “disruptive technology.”10 In the following we are casting light on what has to be considered when developing an mRNA-vaccine technology touching important topics such as mRNA manufacturing and quality, mRNA format and formulation as well as antigen/protein expression and immunological properties of mRNA-vaccines.
mRNA Production
mRNA synthesis
Functional synthetic mRNA may be obtained by in vitro transcription of a cDNA template, typically plasmid DNA (pDNA), using a bacteriophage RNA polymerase.11 Hence, the preparation of pDNA is the first step in the production of mRNA. Manufacture of mRNA might thus appear to require more effort than manufacture of pDNA. However, unpolished pDNA contains traces of bacterial genomic DNA and three forms of pDNA (supercoiled, relaxed circle or linear) in variable proportions. Hence, the reproducible preparation of pure and invariant pDNA, as required for a vaccine, is demanding. Remains of bacterial DNA and the heterogeneity of pDNA are not a concern, on the other hand, if linearized pDNA is transcribed using bacteriophage RNA polymerase,5 because all DNA is removed during further processing steps (see below).
Synthetic mRNA contains a protein-encoding open reading frame (ORF) flanked at the minimum by two elements essential for the function of mature eukaryotic mRNA: a “cap,” i.e., a 7-methyl-guanosine residue joined to the 5′-end via a 5′-5′ triphosphate,12 and a poly(A) tail at the 3′-end.13 Accordingly, a pDNA template for in vitro transcription contains at least a bacteriophage promoter, an ORF, optionally a poly(d(A/T)) sequence transcribed into poly(A) and a unique restriction site for linearization of the plasmid to ensure defined termination of transcription (the cap is not encoded by the template).
The linearized pDNA template is transcribed into mRNA in a mixture containing recombinant RNA polymerase (T7, T3 or SP6) and nucleoside triphosphates. It is possible to obtain capped mRNA by transcription. To this end, a cap analog like the dinucleotide m7G(5′)-ppp-(5′)G (called “regular cap analog” in the following) may be included in the reaction.14 If the cap analog is in excess of GTP, transcription initiates with the cap analog rather than GTP, yielding capped mRNA.15 Alternatively, the cap may be added enzymatically post transcription. A poly(A) tail may also be added post transcription if it is not provided by the pDNA template. Following transcription, the pDNA template as well as contaminating bacterial DNA is digested by DNase.
mRNA purification
At this point, the sample contains the desired mRNA transcript within a complex mixture including various nucleotides, oligodeoxynucleotides, short abortive transcripts from abortive cycling during initiation,16 as well as protein. These contaminants may be removed from the sample by a combination of precipitation and extraction steps.
However, the sample includes additional contaminating RNA species that cannot be separated from the correct transcript by simple means: Shorter than designated transcripts arise from premature termination during elongation. Longer than designated transcripts arise from template DNA linearized with an enzyme that leaves a 3′-overhang17 or from traces of nonlinearized template DNA. Undesirable transcripts are also produced due to the RNA-dependent RNA polymerase activity of bacteriophage polymerases.18 Accordingly, to be used as a drug substance, mRNA will have to be purified further to remove such contaminating transcripts.
A single chromatographic step that separates mRNA according to size removed both shorter and longer transcripts, yielding a pure single mRNA product.19 Implementation of such a chromatographic purification within a GMP production process for mRNA increased the activity of mRNA molecules several-fold in terms of protein expression in vivo.8
Increased protein expression as a result of stringent purification of mRNA was also observed when transcripts coding for luciferase or erythropoietin were purified by HPLC.20 The increase in protein expression was much higher than would be expected simply based on the removal of incorrect transcript. The authors demonstrated that increased protein expression after HPLC purification was also due to the removal of contaminating, e.g., double-stranded, RNA that activates innate immune sensors, thereby reducing protein expression.
mRNA Design
Synthetic mRNA for therapy is in general designed following the blueprint of eukaryotic mRNA. Cap and poly(A) tail are essential elements because they are required for efficient translation.2,15,21 Positioned at the very 5′- and 3′-end of mRNA, Cap and poly(A) tail are also required to stabilize mRNA in the cytosol, where decay is catalyzed predominantly by exonucleases.22,23
However, to further increase both translation and stability, mRNA requires 5′ and 3′ untranslated regions (UTRs) to flank the ORF.24-27 UTRs have to be carefully chosen because they may also impair translation or mRNA stability.28 In particular, specific cis-acting destabilizing sequences like AU-rich elements29 and miRNA binding sites30,31 mostly reside in UTRs, although they may also be found in ORFs.32 Care must be taken to avoid such destabilizing signals.
Following these considerations, efforts have been made to identify beneficial mRNA elements in order to improve translation and stability of synthetic mRNA molecules inside the cell. Improved mRNA formats thus identified will likely also yield better mRNA vaccines, as it is widely assumed that the efficacy of an mRNA vaccine will rise as protein expression is increased and prolonged.
Cap
mRNA may be capped during transcription by including a cap analog in the reaction. However, it has been found that the regular cap analog is often incorporated in the reverse orientation so that the m7G nucleotide does not constitute the cap but is instead the first transcribed nucleotide. As a result, about one third of mRNA molecules are not methylated at their cap.33 Such mRNA lacking methylation of the cap base is not translated.34
In order to avoid unmethylated cap by reverse orientation, mRNA may be transcribed without cap analog and subsequently capped using the vaccinia virus capping complex.35 This complex with triphosphatase, guanylyltransferase and (guanine-7-)methyltransferase activity adds a natural cap to the 5′-triphosphate of an RNA molecule. However, an additional enzymatic step may complicate production, particularly at an industrial scale.
Alternatively, a cap exclusively in the correct orientation is obtained with the use of “anti-reverse” cap analogs (ARCAs). In the most common ARCA, 3′-O-methylation of the base-methylated guanosine only allows addition of a nucleotide at the non-methylated guanosine. ARCA-capped mRNA was translated at more than doubled efficiency in rabbit reticulocyte lysate compared with mRNA capped by regular cap analog.34 In addition, it has been shown that mRNA transcribed in vitro with ARCA also has a longer half-life in cultured cells.36 In an independent study, ARCA-capped mRNA has been reported to both increase and prolong protein expression in cultured cells.37
Protein expression from in vitro transcribed, enzymatically capped mRNA can be further increased by enzymatic 2’-O-methylation of the first transcribed nucleotide, resulting in protein expression comparable to that from mRNA capped with ARCA co-transcriptionally.38
ARCAs have been further modified within the triphosphate linkage in order to inhibit decapping of the corresponding mRNA and increase binding of eukaryotic initiation factor 4E involved in the recruitment of ribosomes. Modifications either substituted for a bridging oxygen (e.g., (methylenebis)phosphonate and imidodiphosphate) or a non-bridging oxygen (e.g., phosphorothioate, phosphoroselenoate and boranophosphate).39 Phosphorothioate-modified ARCAs yielded mRNA with both further increased translation efficiency and elongated half-life in cultured cells compared with ARCA.40 However, phosphorothioate-modified ARCAs are obtained as a mixture of two diastereomers that must be separated after synthesis because of their different biological activity.
Poly(A) tail
When the poly(A) tail was unveiled to enhance translation initiation, it was noted that the efficiency of polysome formation increased with increasing length of the poly(A) tail up to 68 residues.15 Translation of in vitro transcribed mRNA transfected into cultured cells still increased slightly by lengthening the poly(A) tail from 54 to 98 residues.41 This study was further extended by investigating the effect of even longer poly(A) tails on protein expression.38 The peak protein level, reached one day after electroporation of mRNA into cells, was doubled when the poly(A) tail was extended from 64 to 150 residues. Further extension of the poly(A) tail by enzymatic polyadenylation led to an additional moderate increase in peak expression. By contrast, upon transfection of UMR-106 cells, protein levels 16 h post transfection increased with increasing length of the poly(A) tail only up to 60 residues, but declined with further increasing poly(A) tail length.42 In practical terms, it is noteworthy that maintenance of long poly(d(A/T)) sequences is demanding and strongly dependent on the bacterial strain.42
UTRs
Already early on, in vitro transcribed mRNA contained 5‘- and 3‘-UTRs, specifically those of the β globin gene of Xenopus.11 Both the Xenopus β globin 5′- and 3′-UTRs were demonstrated to impart much greater translational efficiency on heterologous mRNA in the mouse NIH 3T3 fibroblast cell line.2 A combination of the β globin 5′-UTR, improving translation and the α globin 3′-UTR, known to stabilize mRNA,27 has been used in the construction of a library from amplified tumor-derived cRNA for use as vaccines against metastatic melanomas.43 Globin UTRs are still in widespread use in in vitro transcribed mRNA including RNA for immune therapy.38,44,45
UTRs from non-globin genes have also been included in in vitro transcribed mRNAs used for investigations of the therapeutic value of mRNA. The 5′-UTR of tobacco etch virus enhances translation of in vitro transcribed mRNA in mammalian cells46 and has been included in mRNA expressing erythropoietin in different cell types20 and mice.47 Furthermore, a structure of the 5′-UTR of human heat shock protein 70 enhanced translation of mRNA in mammalian cells and was predicted to be valuable in the context of genetic vaccination.48
Inclusion of an internal ribosomal entry site (IRES) in in vitro transcribed mRNA can be an alternative and/or complementary means to achieve expression of therapeutic proteins. For instance, the EMCV IRES was included in mRNAs coding for four transcription factors used to reprogram fibroblasts to pluripotent stem cells.49 The EMCV IRES has even been used successfully to direct protein expression from mRNA lacking a cap.50 Vaccination with dendritic cells transfected with such IRES-containing, cap-less mRNA protected mice from metastasis upon intravenous injection of melanoma cells.
Completely novel UTRs may be provided by screening whole transcriptomes for sequence elements that either increase translation or mRNA stability.51
ORF
Codon usage is also considered as a factor affecting the efficiency of translation in many species. However, in humans codon usage bias does not correlate with tRNA levels and gene expression.52,53 In conclusion, codon optimization cannot be expected to (generally) improve mRNA translation in humans, particularly if the ORF is already of human (or even mammalian) origin.
Obviously, the start codon should be part of a Kozak sequence54 and the sequence surrounding the stop codon may be optimized.55 In addition, no upstream start codons, preceding the correct start codon, should be present in the mRNA.
Combinatorial design
In order to obtain effective vaccine platforms, different beneficial mRNA elements have been joined.
Capping with ARCA has been combined with a long transcribed poly(A) tail of 100 residues. Such luciferase-encoding mRNA was tested in immortalized cell lines (JawsII, HepG2, HeLa) as well as immature and mature human dendritic cells.56 Compared with mRNA capped with regular cap analog and ending with a shorter A64 poly(A) tail, a very substantial improvement in protein expression was seen in all tested cell types. The magnitude of the rise in protein level afforded by either element alone or their combination was strongly dependent on the cell type.
Sahin and coworkers combined two consecutive β globin 3‘-UTRs, a rather long transcribed poly(A) tail of 120 residues57 and a phosphorothioate modified anti-reverse cap.58 This resulted in increased and prolonged protein expression in transfected dendritic cells. Upon injection of mRNA into the lymph node, protein expression peaked at 8 h and was demonstrated up to 72 h after mRNA injection.
Using our proprietary mRNA technology modifying coding and noncoding parts of the molecule, we were able to improve both the level and duration of expression, increasing total protein expression by several orders of magnitude.59 Upon intradermal mRNA injection, strongly prolonged translation gives maximum protein levels 24 to 48 h after mRNA injection and lasts for many more days (Fig. 1).
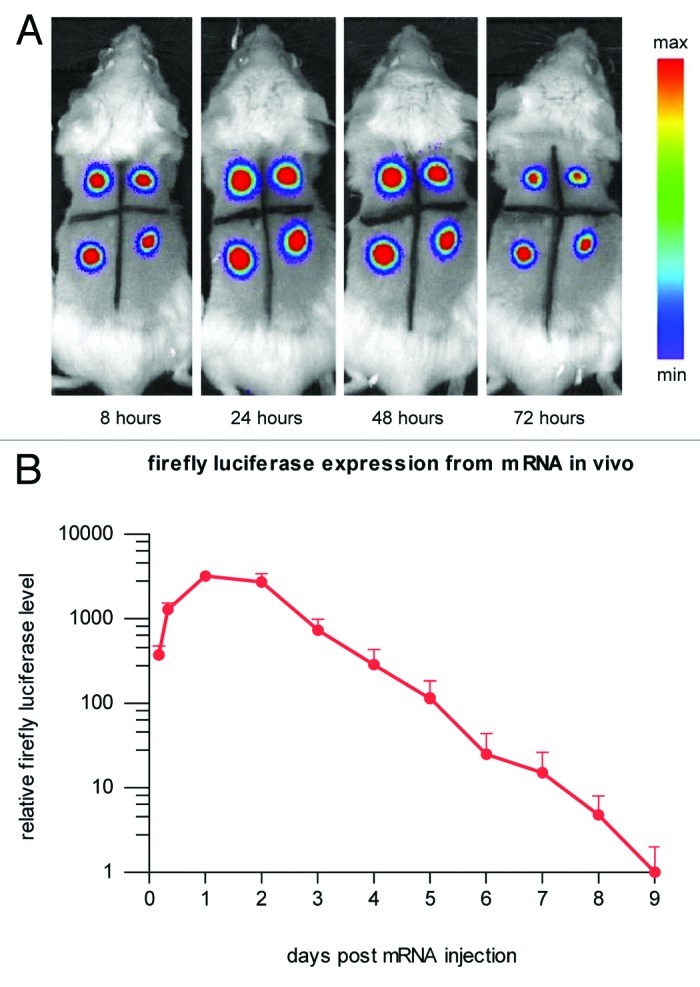
Figure 1. Protein expression in vivo is strongly prolonged using CureVac’s proprietary mRNA technology and lasts for many days. Firefly luciferase-encoding mRNA, optimized for translation and stability, was injected intradermally in a BALB/c mouse (4 injection sites). At various time points after mRNA injection, luciferase expression was visualized in the living animal by optical imaging. (A) Visualization of luciferase expression at selected time points, showing maximal protein levels 24 to 48 h after mRNA injection. (B) Time course of luciferase expression until 9 d after mRNA injection. Background signal was set to 1.
mRNA Uptake
To be translated and elicit an antigen-specific immune response, an mRNA-vaccine has to reach the cytosol of target cells. However, as opposed to DNA vaccines, RNA vaccines only have to cross the plasma membrane, but not the nuclear envelope which may improve the probability of successful in vivo transfection.60 As early as 1990, the uptake of mRNA by mouse muscle cells upon simple injection, i.e., without any additional help from special delivery systems, was demonstrated.61 Later on, numerous studies confirmed that locally administered naked mRNA is taken up by cells in target tissues.8,62-65 The mechanism by which naked mRNA enters cells remained unclear initially. However, elucidating and understanding the uptake route is important to facilitate the development of more efficient mRNA-vaccines.
A plethora of studies investigated the cellular entry of nucleic acids. Most of them looked into the uptake routes of pDNA, DNA oligonucleotides, siRNA or long dsRNA and a complex picture emerged. The molecules entered cells by diffusion controlled mechanisms or diverse endocytic pathways, often strongly dependent on the respective cell type or species and frequently showed a vesicular localization, i.e., an entrapment in endocytic or lysosomal compartments.66-73 However, mRNA differs from these types of molecules due to its unique combination of physico-chemical and structural parameters. In contrast to DNA, mRNA contains uridine instead of deoxythymidine, preferentially adopts a C3′-endo conformation and is hydroxylated at the 2′-position of the ribose. The single-stranded nature lets mRNA fold into complex secondary and tertiary structures, completely unknown from double-stranded DNA and RNA molecules, respectively. Finally, its length of a few hundred to several thousand nucleotides distinguishes mRNA from other single-stranded RNAs like antisense RNA or aptamers.
First insight into the uptake mechanism of naked mRNA was gained by a mouse study investigating intradermal administration by injection.8 Local entry into cells of the dermis which were not exclusively professional antigen presenting cells (pAPCs) turned out to be saturable, improvable by calcium and associated with the movement of vesicles. More elaborate work in vitro revealed that uptake of naked mRNA is a widespread phenomenon among primary cells and cell lines of diverse types.74 These efforts confirmed saturability of uptake and demonstrated that it is also temperature and dose dependent. Most of the mRNA appeared to enter cells via caveolae/lipid rafts,74 most likely mediated by (a) scavenger-receptor(s) which are known to concentrate in caveolae and to preferentially recognize and facilitate internalization of negatively charged macromolecules.75-78
To a minor degree, macropinocytosis also appeared to be involved in mRNA uptake of different primary cells and cell lines.74 By contrast, macropinocytosis apparently predominates mRNA uptake by dendritic cells upon intranodal injection.79 The picture becomes even more complicated when looking at formulated mRNA vaccines. For instance, a recently developed two-component vaccine consisting of naked and protamine-complexed mRNA reveals different routes and kinetics of uptake for the two components, albeit both are taken up via an endosomal pathway.9,80
mRNA uptake and expression in vivo is quite efficient (much more efficient than spontaneous uptake by cells in vitro) and comparable even with cells transfected in vitro under optimal conditions.8,61 In part, hydrodynamic pressure may contribute to target cell transfection in case of local injections81 as it does upon intravenous administration.82 However, the correlation between pressure and transfection efficiency/protein expression may not be linear but show an optimum.83 Anyway, a large amount of the mRNA appears to stay trapped in endosomal vesicles. Hence, mRNA vaccines may profit strongly from approaches increasing the fraction of mRNA that reaches the cytosol.
Formulation of mRNA
mRNA is threatened by rapid degradation by ubiquitous extracellular ribonucleases before being taken up by cells.84 Thus, the efficacy of mRNA vaccines may benefit significantly from complexing agents which protect RNA from degradation. Complexation may also enhance uptake by cells and/or improve delivery to the translation machinery in the cytoplasm. To this end, mRNA is often complexed with either lipids or polymers.
Importantly, not all complexing agents that promote transfection of DNA are suitable for complexation of mRNA. Different large polycations, all proven DNA transfection reagents, were shown to strongly inhibit translation of mRNA in cell-free translation systems as well as inside cells. Only much smaller polycations allowed for efficient translation. Likely, mRNA is not released in the cytosol if bound to large polycations.85 Interestingly, DNA may be released in the cytosol from large polycations by endogenous RNA.86
In line with the general conception that mRNA should be protected and uptake enhanced, the first report demonstrating the induction of an immune response upon direct injection of mRNA in vivo used mRNA encapsulated in liposomes.87 Common is the use of cationic lipids, for instance used for the intradermal and intravenous injection of antigen-encoding mRNA.88 However, complexation of mRNA with protamine, a small arginine-rich nuclear protein which stabilizes DNA during spermatogenesis, was shown to also efficiently stabilize mRNA against degradation by serum components.63
In addition, complexing agents rationally designed to further improve delivery of nucleic acids to the cytosol have been used for formulation of mRNA vaccines. Hemagglutinating virus of Japan (HVJ)–liposomes have been reported to deliver their cargo directly into the cytoplasm of host cells in vivo by means of a virus–cell fusion mechanism. Such liposomes were used to inject mRNA (replicating in this case) encoding melanoma antigen gp100 into the spleen of mice.89 Alternatively, vectors improving cytosolic nucleic acid delivery by means of permeation of endosomal membranes due to their high histidine content have been used to formulate antigen-encoding mRNA.90
Complexing agents may have to be tailored to the specific route of delivery. Due to the abundance of professional antigen presenting cells in the skin,91 this organ may be particularly suitable for vaccination. However, delivery of a DNA vaccine into mouse skin by tattooing failed when formulated into cationic nanoparticles but was successful upon PEGylation of the nanoparticles to shield their surface charge.92 Likely, adsorption of the cationic nanoparticles to the negatively charged extracellular matrix in the skin prevented their uptake by cells. In addition, the use of complexing agents in vivo is often hampered by toxicity, particularly for high molecular weight compounds.93 Still, progress in the drug delivery field is steady,94,95 including innovative approaches to targeting of drugs to particular cell types.96 Looking ahead, improved delivery is certain to contribute to increased efficacy of mRNA vaccines.
mRNA-Mediated Protein Expression
As an intermediate carrier of genetic information, endogenous mRNA is used as template for protein expression. Hence, like DNA, mRNA is at least in principle an attractive means to force cells to produce proteins of interest by introducing exogenous nucleic acid molecules. For mRNA, this concept was first applied in the early 1970s. Microinjection of preparations of rabbit hemoglobin mRNA and encephalomyocarditis virus RNA, respectively, into oocytes from Xenopus laevis provided clear evidence that these molecules gave rise to the expression of RNA-encoded proteins.97,98 More than ten years later, in vitro transcribed RNA from brome mosaic virus (BMV) and poliovirus cDNA were shown to be infectious, an unequivocal indication of protein expression from those RNAs.99,100 However, at that time viable techniques allowing use of mRNA as a general tool for protein expression were still missing. This changed with the adaptation of efficient transfection methods such as electroporation and cationic lipofection for the delivery of RNA.2,101 Further developments and insights into mRNA biology enabled significant overexpression of proteins after delivery.102 Finally, the in vitro use of mRNA culminated in the establishment of cell reprogramming protocols that may be of some medical relevance in the future.49,103
Whereas all these examples cover mRNA-mediated protein expression exclusively taking place in vitro, meanwhile, cell based approaches of mRNA-mediated protein expression have expanded into in vivo settings. On the one hand, mRNA injection into fertilized oocytes or early embryos became a well-established tool in developmental biology.104 On the other hand, loading of dendritic cells with antigen-encoding mRNA originally described by Boczkowski et al.105 became a widely used approach in immunology and was investigated in several clinical trials in humans (see section mRNA-based vaccines).
Since these semi-in vivo applications introducing the mRNA ex vivo are laborious and technically very demanding, scientists were interested in direct in vivo application early on. First efforts demonstrated that local injection of naked mRNA can lead to expression of different proteins in mouse muscle tissue.61,62 In an attempt to improve mRNA delivery, a particle-mediated administration via gene gun was developed and demonstrated to give rise to protein expression in liver and epidermis.106 Later, successful protein expression upon intradermal injection in mice was proven.63 Using this administration route, it was shown that (perhaps various) MHC class II-negative non-pAPCs take up and express mRNA.8 Together, these findings suggest that mRNA can be taken up and expressed by different cell types in vivo, which is consistent with in vitro data.74
These results conclusively show that mRNA-mediated protein expression in vivo is generally possible. In addition, they demonstrate that expression is sufficient to raise detectable immune responses. However, raising an effective immune response and, even more, achieving a therapeutic effect by mRNA-mediated protein supply may be more demanding in terms of the required level of protein expression. Using our proprietary mRNA-technology, we could demonstrate that a single intramuscular injection of erythropoietin (Epo)-encoding mRNA led to a biologically relevant increase of reticulocytes in mice (Fig. 2). Therapeutic effects using Epo-mRNA were confirmed by two independent studies.47,65 The potency of mRNA-mediated protein expression was further underlined by an analysis of protein complementation in a surfactant protein B-deficient mouse model.65 However, in contrast to our work, these studies deployed mRNA harboring modified nucleotides to increase protein expression. While such modifications can enhance translation of the mRNA107,108 and may be beneficial for protein replacement therapies, they interfere with the design of mRNA-vaccines with self-adjuvanticity, an important feature required for a potent vaccine (see next section).
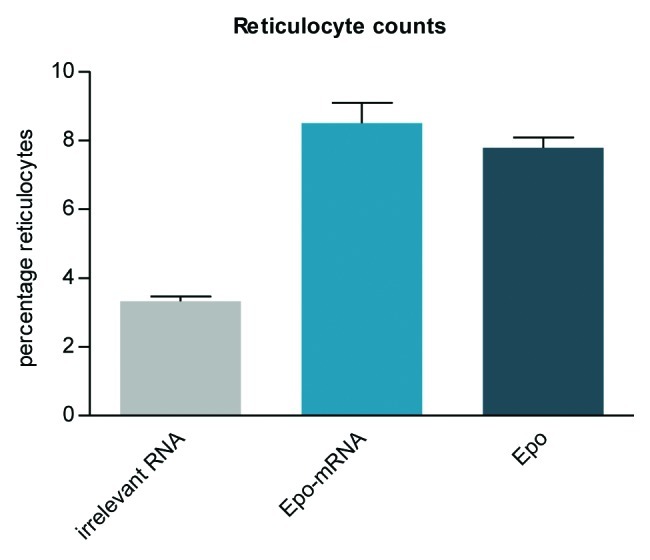
Figure 2. A biologically relevant increase of reticulocytes is induced in mice using CureVac’s proprietary mRNA technology. A single intramuscular injection in BALB/c mice of erythropoietin (Epo)-encoding mRNA, optimized for translation and stability, causes the expression of functional Epo. Reticulocyte levels are raised comparably by mRNA and recombinant protein injected intramuscularly.
Adjuvanticity of mRNA (Vaccines)
To be efficient, vaccines should contain a strong adjuvant supplying a danger signal for the initiation and support of the adaptive immune response in addition to an appropriate antigen.109 The immunostimulatory properties of RNA were first discovered by the observation of interferon induction upon exposure of cells to exogenous RNA extracted from viruses.110 Further support came from synthetic double-stranded RNA inducing interferon upon intravenous injection into rabbits.111 However, severe side effects of these early RNA adjuvants soon limited their further use.112 The idea of synthetic RNA, mainly produced by in vitro transcription, as immunostimulant was then re-stimulated particularly during the last decade bringing forth a plethora of studies.
In 2004, in vitro transcribed mRNA was shown to serve as an adjuvant, if it was stabilized by either complexation or chemical modification.113 One year later, a strong danger signal was ascribed to protamine-condensed mRNA leading to TNFα and IFNα secretion by various cells.114 A thorough analysis of complexes of single-stranded RNA and protamine indicated that cell activation in terms of cell selectivity and induced cytokine pattern may depend on particle size.115 Recently, research on protamine-complexed RNA culminated in a simplified vaccine approach, combining naked and protamine-formulated mRNA.9 The resulting mRNA vaccine consists of two components complementing each other; while antigen supply is mainly driven by the naked mRNA, the protamine complexes contribute a strong immunostimulatory signal. Of note, protamine-formulated RNA can also confer adjuvanticity to, e.g., protein vaccines.116
Among potent adjuvant targets, RNA-sensing receptors are a particularly diverse class of molecules evolved to detect and counteract viral infections by orchestrating the innate and adaptive arms of the immune system.117,118 Single- and double-stranded RNAs are recognized by toll-like receptors (TLR) 7/8 and 3, respectively, in the endosome.119-121 However, TLR3 is not only activated by double-stranded RNA, an intermediary for many viruses, but also by mRNA either released from cells or produced by in vitro transcription.122 Protamine-complexation of RNA appears to preserve its interaction with TLRs as indicated by the stimulation of several blood cell types.114 For mRNA vaccines it was demonstrated that activation of TLR7 and potentially TLR3 is critical for priming immune responses.9,45 Notably, optimal sequence motifs for receptor binding could be identified for single-stranded RNA.123-126
Other pattern recognition receptors may be important for the functionality of an RNA-based adjuvant (and vaccine) as well. The cytosolic helicase RIG-I recognizes uncapped RNA molecules harboring a 5′-triphosphate moiety.127-129 Together with the homologous proteins MDA5 and LGP2, RIG-I forms a receptor family whose members can all bind double-stranded RNA,130-132 but do have additional recognition patterns. For example, MDA5 is involved in the discrimination of RNAs based on the ribose 2′-O-methylation status of the cap structure.133 For the sake of completeness, the cytoplasmic RNA sensors PKR and 2′-5′-oligoadenylate synthetase, inhibiting translation by phosphorylation of eIF-2α and activating RNase L, respectively, should be mentioned here as well.134,135 However, the contribution of these non-TLR RNA-sensors to the immunostimulation by RNA-based adjuvants (and vaccines) is still a matter of debate. Notably, as far as investigated, the interaction between endosomal as well as cytoplasmic receptors and RNA is impaired if the RNA harbors distinct nucleotide modifications.108,136,137 As a consequence, such modified RNA impairs the design of self-adjuvanting mRNA-vaccines.
mRNA-Based Vaccines
After in vivo administration of mRNA was proven to be feasible,61 the concept of using mRNA as a basis for vaccines was pursued almost immediately. First success was reported in 1993 when subcutaneous injection of liposome-encapsulated mRNA encoding the nucleoprotein (NP) of influenza virus was demonstrated to elicit NP-specific cytotoxic T cells (CTLs).87 By contrast, naked mRNA failed to raise specific CTLs in this setting. Shortly afterwards, the use of naked mRNA triggered the induction of antigen-specific antibodies in response to a heterologous prime-boost schedule (repeated intramuscular RNA vaccination, challenge with tumor cells).62 However, none of the animals was protected against tumor challenge. An antigen-specific antibody response induced solely with mRNA was demonstrated first using particle-mediated mRNA delivery into mouse epidermis.106
In 2000, the field of mRNA vaccines was advanced by introducing a new protocol for vaccination allowing the administration of naked mRNA via intradermal injection.63 This basic vaccination design did not require any transfection reagents, special equipment or heterologous boost, yet could elicit a complete adaptive immune response consisting of antigen-specific antibodies and T cells with lytic activity against the model antigen β-galactosidase. Directly thereafter, intradermal injection of total RNA isolated from the S1509 tumor cell line was shown to induce immunity to a subsequent challenge with the tumor.138 Tumor growth inhibition was also achieved by intradermal as well as intravenous injection of in vitro transcribed and lipid-complexed mRNA encoding the model antigen ovalbumin (OVA).88 However, analogous vaccination with mRNA coding for a model tumor/self-antigen was not sufficient to break tolerance to this self-antigen in TRAMP mice. A similar approach using histidylated lipopolyplexes for systemic injection revealed that MART1 mRNA could not only prevent B16 melanoma from progression but also from metastasis.90
In a comparison of different administration routes for the delivery of naked mRNA vaccines good immunogenicity against ovalbumin and influenza A virus hemagglutinin could be demonstrated after repeated and frequent injections into the lymph node.45 In order to optimize the vaccine’s potency, the authors engineered the antigen by adding an MHC class I molecule trafficking signal for increased antigen presentation.44 Unlike with intranodal injection, the authors could not elicit such immune responses upon perinodal, subcutaneous or intradermal injections.45 Recently, an alternative, simplified approach was described leading to successful immunization by intradermal injection.9 Combining naked mRNA with protamine-formulated mRNA results in a two-component vaccine capable of inducing strong immune responses and tumor protection in prophylactic as well as therapeutic settings in mice. In this vaccine, the two components fulfill complementary functions: while the naked mRNA confers optimal antigen expression, the protamine-complexed mRNA contributes strong immunostimulatory effects. Notably, for tumor treatment this new type of mRNA vaccine can be combined with other, standard, therapies such as chemotherapy, thereby achieving improved effects as compared with each treatment alone.59
As an alternative to direct injection of mRNA, an immune response may also be induced by vaccination with pAPCs transfected with mRNA ex vivo. mRNA-transfected murine dendritic cells (DCs) were shown to elicit anti-tumor immunity in EG.7-OVA and B16 melanoma models.105 Tumor growth was also significantly reduced upon injection of epidermal cells enriched for Langerhans cells, which belong to the group of pAPCs, that had been transfected with total RNA derived from tumor cells.138 Using human DCs, transfection with mRNA encoding CEA or the E6 antigen of human papillomavirus type 16 induced a primary CTL response in vitro.139 Today, ex vivo mRNA transfection of pAPCs is the most frequently used approach for mRNA vaccination in the clinic. For instance, a clinical trial utilizing telomerase mRNA-transfected DCs demonstrated the capability of such applications to stimulate antigen-specific cellular immune responses.140 However, the underlying procedure is very time consuming, laborious and needs patient-specific (autologous) cell preparations.
Very few clinical studies of the direct administration of mRNA-based vaccines have been published. The first trial deployed autologous mRNA libraries derived from melanoma lesions, whereas in a later study a cocktail of protamine-complexed mRNAs encoding six different antigens was given intradermally using an intensified treatment regimen.43,141,142 A further clinical trial with patients with renal cell carcinoma stage IV included the administration of GM-CSF as adjuvant 24 h after vaccination with six antigens,143 an approach that will be discussed in more detail in the next section.
Adjuvanted mRNA-Based Vaccines
As discussed, mRNA-vaccines can be designed to possess self-adjuvanticity contributing to their excellent performance. Although pDNA vaccines also show native immunogenicity, great efforts were made to improve the immune response by co-delivery or, more elegantly, co-expression of co-stimulatory molecules and cytokines. Indeed, inclusion of adjuvant molecules encoded as DNA could enhance DNA vaccines. Co-injection of DNA encoding tumor or viral antigens and the cytokine granulocyte macrophage colony-stimulating factor (GM-CSF) improved T and B cell responses.144-147 As a further example, DNA expressing a recombinant soluble multi-trimeric TNF superfamily ligand enhanced the immune response to an HIV-1 Gag DNA vaccine.148 Moreover, DNA encoding the cytokine IL-2 was demonstrated to affect the polarization of the immune response, an important vaccine parameter the optimum of which differs among indications. While a DNA vaccine for Helicobacter pylori elicited a strong Th2 response, it was shifted toward a Th1-biased response by IL-2.149 In addition to directly administering a DNA encoding the adjuvant molecule, more indirect approaches are also feasible. For instance, the co-delivery of a CD40-expressing plasmid induced anti-CD40 antibodies, part of which were capable of activating CD40 which in turn led to an improved immune response to an HBV DNA vaccine.150
In an analogous fashion, RNA vaccines supplemented with additional adjuvant molecules have been the object of various investigations. In mice, recombinant GM-CSF enhanced the immune response to the model antigen β-galactosidase and affected polarization of immunity by shifting a Th2 to a Th1 response.64 In addition, GM-CSF as a supplement of an mRNA vaccine was already tested in a clinical trial.143 An improved anti-tumor effect of a naked mRNA vaccine in mice was demonstrated for human FLT3 ligand protein fused to human IgG4-Fc fragment.151 However, in light of getting good protein expression upon mRNA administration in vitro and in vivo (see section mRNA-mediated protein expression), providing auxiliary adjuvant molecules via mRNA rather than as protein appears to be a feasible and much more elegant approach. First support for this idea came from a study investigating the effect of GM-CSF, IL-2 and CD80, all encoded by mRNA, on the potency of model mRNA vaccines.88 Among these adjuvants, GM-CSF mRNA improved the induction of CTL activity in a dose-dependent manner. Moreover, a more durable CTL response indicated an enhanced generation of memory cells. We have recently investigated the anti-tumor effect of an accessory adjuvant molecule encoded by mRNA. To this end, we included CD40 ligand as a co-stimulatory molecule activating pAPCs152 which may establish an autoregulatory circuit improving the immune response by enhancing antigen presentation. CD40 ligand supplied as mRNA significantly improved tumor growth reduction as compared with the non-adjuvanted two-component mRNA vaccine (Fig. 3). In summary, these data suggest that the combination of mRNA vaccines and additional mRNAs encoding auxiliary adjuvant molecules is a very promising approach which, however, remains challenging in terms of arriving at suitable, i.e., effective and practical, treatment regimens.
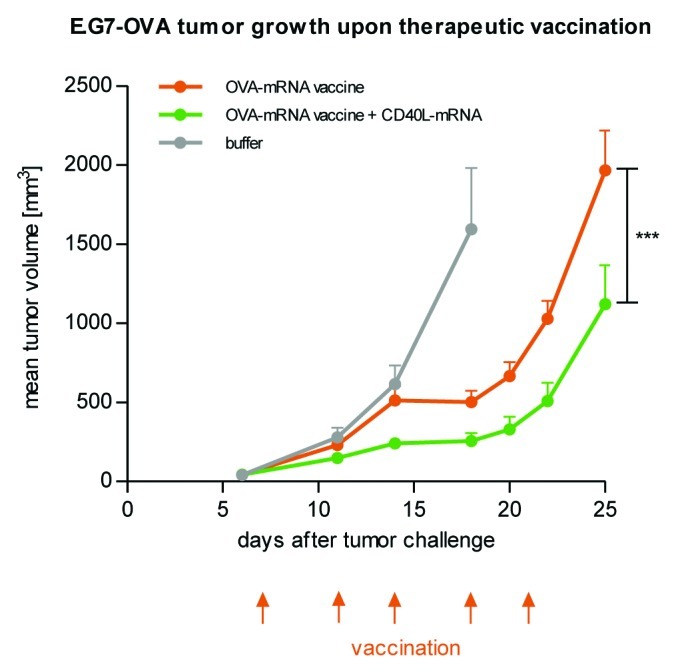
Figure 3. CD40 ligand as an accessory adjuvant molecule encoded by mRNA increases the anti-tumor effect of a two-component mRNA vaccine. Mice (n = 8 per group) were challenged subcutaneously with syngenic E.G7-OVA tumor cells on day 0. Commencing on day 7, mice were vaccinated intradermally with either OVA-mRNA vaccine alone or in combination with mRNA coding for CD40 ligand according to the indicated schedule. Mice treated with buffer served as the control. The combination of CD40 ligand-encoding mRNA together with OVA mRNA vaccination increases the efficacy of the therapeutic anti-tumor mRNA vaccination.
Concluding Remarks
About two decades after the first successful administration of mRNA in vivo, mRNA-based vaccines promise to become a game-changing vaccine technology platform for therapeutic as well as prophylactic applications. Today, the scientific community is eagerly waiting for first clinical efficacy data. But there is still a wide field for further development/improvements of mRNA-based vaccines. As discussed, the format and uptake of the mRNA are critical parameters for efficient antigen expression which can be influenced by novel RNA designs as well as mRNA formulation and administration. However, any changes to these parameters may have major implications on mRNA production and/or its interactions with RNA-sensors and should be carefully considered early on. For instance, in addition to the previously mentioned nucleotide modifications, novel delivery modes may severely affect vaccine adjuvanticity. While direct delivery into the cytosol would certainly enhance antigen expression, the lack of interaction with endosomal RNA receptors may severely weaken immunostimulation by the vaccine and this issue would likely have to be addressed. The inclusion of accessory mRNA molecules into an mRNA vaccine may be an interesting option for achieving optimal effects in case of particularly challenging treatments. Moreover, the combination with other anti-tumor therapies will most likely yield the greatest potency. However, this would increase the complexity of the vaccine and/or the treatment regimen making the development more challenging. Taken together, mRNA offers a promising vaccine vector in the light of being flexible, effective and safe. Hence, it could become a “disruptive technology” not just for cancer immunotherapy, but also for vaccination, either prophylactic or therapeutic, against infectious diseases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank our colleague Birgit Scheel for helpful comments and critical reading of the manuscript.
Glossary
Abbreviations:
- ARCA
anti-reverse cap analog
- cDNA
complementary DNA
- CTL
cytotoxic T cell
- dsRNA
double-stranded RNA
- EMCV
encephalomyocarditis virus
- GMP
good manufacturing practice
- HPLC
high performance liquid chromatography
- IRES
internal ribosome entry site
- mRNA
messenger RNA
- MHC
major histocompatibility complex
- miRNA
microRNA
- ORF
open reading frame
- pAPC
professional antigen presenting cell
- pDNA
plasmid DNA
- PEG
polyethylenglycol
- siRNA
small interfering RNA
- tRNA
transfer RNA
- TLR
toll-like receptor
- UTR
untranslated region
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/22269
References
- 1.Sorrentino S. Human extracellular ribonucleases: multiplicity, molecular diversity and catalytic properties of the major RNase types. Cell Mol Life Sci. 1998;54:785–94. doi: 10.1007/s000180050207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci U S A. 1989;86:6077–81. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilleman MR. Recombinant vector vaccines in vaccinology. Dev Biol Stand. 1994;82:3–20. [PubMed] [Google Scholar]
- 4.Liu MA. Immunologic basis of vaccine vectors. Immunity. 2010;33:504–15. doi: 10.1016/j.immuni.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Pascolo S. Vaccination with messenger RNA. Methods Mol Med. 2006;127:23–40. doi: 10.1385/1-59745-168-1:23. [DOI] [PubMed] [Google Scholar]
- 6.Jäschke A, Helm M. RNA sex. Chem Biol. 2003;10:1148–50. doi: 10.1016/j.chembiol.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Chetverin AB. Replicable and recombinogenic RNAs. FEBS Lett. 2004;567:35–41. doi: 10.1016/j.febslet.2004.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Probst J, Weide B, Scheel B, Pichler BJ, Hoerr I, Rammensee HG, et al. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007;14:1175–80. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 9.Fotin-Mleczek M, Duchardt KM, Lorenz C, Pfeiffer R, Ojkić-Zrna S, Probst J, et al. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J Immunother. 2011;34:1–15. doi: 10.1097/CJI.0b013e3181f7dbe8. [DOI] [PubMed] [Google Scholar]
- 10.Kaslow DC. A potential disruptive technology in vaccine development: gene-based vaccines and their application to infectious diseases. Trans R Soc Trop Med Hyg. 2004;98:593–601. doi: 10.1016/j.trstmh.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Krieg PA, Melton DA. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984;12:7057–70. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee AK. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990;15:277–81. doi: 10.1016/0968-0004(90)90054-F. [DOI] [PubMed] [Google Scholar]
- 14.Konarska MM, Padgett RA, Sharp PA. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–6. doi: 10.1016/0092-8674(84)90268-X. [DOI] [PubMed] [Google Scholar]
- 15.Munroe D, Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol Cell Biol. 1990;10:3441–55. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong P, Martin CT. Mechanism of instability in abortive cycling by T7 RNA polymerase. J Biol Chem. 2006;281:23533–44. doi: 10.1074/jbc.M604023200. [DOI] [PubMed] [Google Scholar]
- 17.Schenborn ET, Mierendorf RC., Jr. A novel transcription property of SP6 and T7 RNA polymerases: dependence on template structure. Nucleic Acids Res. 1985;13:6223–36. doi: 10.1093/nar/13.17.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnaud-Barbe N, Cheynet-Sauvion V. Oriol G, Mandrand B, Mallet F. Transcription of RNA templates by T7 RNA polymerase. Nucleic Acids Res. 1998;26:3550–4. doi: 10.1093/nar/26.15.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pascolo S. Messenger RNA-based vaccines. Expert Opin Biol Ther. 2004;4:1285–94. doi: 10.1517/14712598.4.8.1285. [DOI] [PubMed] [Google Scholar]
- 20.Karikó K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–16. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 22.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–7. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita A, Chang T-C, Yamashita Y, Zhu W, Zhong Z, Chen C-YA, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–63. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989;9:5134–42. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991;1:111–5. [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–8. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol. 1999;19:4552–60. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–50. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–50. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang Z, Rajewsky N. The impact of miRNA target sites in coding sequences and in 3’UTRs. PLoS One. 2011;6:e18067. doi: 10.1371/journal.pone.0018067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasquinelli AE, Dahlberg JE, Lund E. Reverse 5′ caps in RNAs made in vitro by phage RNA polymerases. RNA. 1995;1:957–67. [PMC free article] [PubMed] [Google Scholar]
- 34.Stepinski J, Waddell C, Stolarski R, Darzynkiewicz E, Rhoads RE. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA. 2001;7:1486–95. [PMC free article] [PubMed] [Google Scholar]
- 35.Venkatesan S, Gershowitz A, Moss B. Modification of the 5′ end of mRNA. Association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7-)methyltransferase complex from vaccinia virus. J Biol Chem. 1980;255:903–8. [PubMed] [Google Scholar]
- 36.Grudzien E, Kalek M, Jemielity J, Darzynkiewicz E, Rhoads RE. Differential inhibition of mRNA degradation pathways by novel cap analogs. J Biol Chem. 2006;281:1857–67. doi: 10.1074/jbc.M509121200. [DOI] [PubMed] [Google Scholar]
- 37.Zohra FT, Chowdhury EH, Tada S, Hoshiba T, Akaike T. Effective delivery with enhanced translational activity synergistically accelerates mRNA-based transfection. Biochem Biophys Res Commun. 2007;358:373–8. doi: 10.1016/j.bbrc.2007.04.059. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Moon E, Carpenito C, Paulos CM, Liu X, Brennan AL, et al. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer Res. 2010;70:9053–61. doi: 10.1158/0008-5472.CAN-10-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rydzik AM, Kulis M, Lukaszewicz M, Kowalska J, Zuberek J, Darzynkiewicz ZM, et al. Synthesis and properties of mRNA cap analogs containing imidodiphosphate moiety--fairly mimicking natural cap structure, yet resistant to enzymatic hydrolysis. Bioorg Med Chem. 2012;20:1699–710. doi: 10.1016/j.bmc.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 40.Grudzien-Nogalska E, Jemielity J, Kowalska J, Darzynkiewicz E, Rhoads RE. Phosphorothioate cap analogs stabilize mRNA and increase translational efficiency in mammalian cells. RNA. 2007;13:1745–55. doi: 10.1261/rna.701307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng J, Schoenberg DR. mRNA with a <20-nt poly(A) tail imparted by the poly(A)-limiting element is translated as efficiently in vivo as long poly(A) mRNA. 2005; 11:1131-40. [DOI] [PMC free article] [PubMed]
- 42.Elango N, Elango S, Shivshankar P, Katz MS. Optimized transfection of mRNA transcribed from a d(A/T)100 tail-containing vector. Biochem Biophys Res Commun. 2005;330:958–66. doi: 10.1016/j.bbrc.2005.03.067. [DOI] [PubMed] [Google Scholar]
- 43.Carralot J-P, Weide B, Schoor O, Probst J, Scheel B, Teufel R, et al. Production and characterization of amplified tumor-derived cRNA libraries to be used as vaccines against metastatic melanomas. Genet Vaccines Ther. 2005;3:6. doi: 10.1186/1479-0556-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreiter S, Selmi A, Diken M, Sebastian M, Osterloh P, Schild H, et al. Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals. J Immunol. 2008;180:309–18. doi: 10.4049/jimmunol.180.1.309. [DOI] [PubMed] [Google Scholar]
- 45.Kreiter S, Selmi A, Diken M, Koslowski M, Britten CM, Huber C, et al. Intranodal vaccination with naked antigen-encoding RNA elicits potent prophylactic and therapeutic antitumoral immunity. Cancer Res. 2010;70:9031–40. doi: 10.1158/0008-5472.CAN-10-0699. [DOI] [PubMed] [Google Scholar]
- 46.Gallie DR, Tanguay RL, Leathers V. The tobacco etch viral 5′ leader and poly(A) tail are functionally synergistic regulators of translation. Gene. 1995;165:233–8. doi: 10.1016/0378-1119(95)00521-7. [DOI] [PubMed] [Google Scholar]
- 47.Karikó K, Muramatsu H, Keller JM, Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther. 2012;20:948–53. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vivinus S, Baulande S, van Zanten M, Campbell F, Topley P, Ellis JH, et al. An element within the 5′ untranslated region of human Hsp70 mRNA which acts as a general enhancer of mRNA translation. Eur J Biochem. 2001;268:1908–17. doi: 10.1046/j.1432-1327.2001.02064.x. [DOI] [PubMed] [Google Scholar]
- 49.Yakubov E, Rechavi G, Rozenblatt S, Givol D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem Biophys Res Commun. 2010;394:189–93. doi: 10.1016/j.bbrc.2010.02.150. [DOI] [PubMed] [Google Scholar]
- 50.Tan X, Wan Y. Enhanced protein expression by internal ribosomal entry site-driven mRNA translation as a novel approach for in vitro loading of dendritic cells with antigens. Hum Immunol. 2008;69:32–40. doi: 10.1016/j.humimm.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Goodarzi H, Najafabadi HS, Oikonomou P, Greco TM, Fish L, Salavati R, et al. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature. 2012;485:264–8. doi: 10.1038/nature11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol. 2001;53:290–8. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- 53.Duret L. Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev. 2002;12:640–9. doi: 10.1016/S0959-437X(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 54.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–92. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 55.Liu Q. Comparative analysis of base biases around the stop codons in six eukaryotes. Biosystems. 2005;81:281–9. doi: 10.1016/j.biosystems.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Mockey M, Gonçalves C, Dupuy FP, Lemoine FM, Pichon C, Midoux P. mRNA transfection of dendritic cells: synergistic effect of ARCA mRNA capping with Poly(A) chains in cis and in trans for a high protein expression level. Biochem Biophys Res Commun. 2006;340:1062–8. doi: 10.1016/j.bbrc.2005.12.105. [DOI] [PubMed] [Google Scholar]
- 57.Holtkamp S, Kreiter S, Selmi A, Simon P, Koslowski M, Huber C, et al. Modification of antigen-encoding RNA increases stability, translational efficacy and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–17. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 58.Kuhn AN, Diken M, Kreiter S, Selmi A, Kowalska J, Jemielity J, et al. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010;17:961–71. doi: 10.1038/gt.2010.52. [DOI] [PubMed] [Google Scholar]
- 59.Fotin-Mleczek M, Zanzinger K, Heidenreich R, Lorenz C, Thess A, Duchardt KM, et al. Highly potent mRNA based cancer vaccines represent an attractive platform for combination therapies supporting an improved therapeutic effect. J Gene Med. 2012;14:428–39. doi: 10.1002/jgm.2605. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto A, Kormann M, Rosenecker J, Rudolph C. Current prospects for mRNA gene delivery. Eur J Pharm Biopharm. 2009;71:484–9. doi: 10.1016/j.ejpb.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 61.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–8. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 62.Conry RM, LoBuglio AF, Wright M, Sumerel L, Pike MJ, Johanning F, et al. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55:1397–400. [PubMed] [Google Scholar]
- 63.Hoerr I, Obst R, Rammensee HG, Jung G. In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol. 2000;30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 64.Carralot JP, Probst J, Hoerr I, Scheel B, Teufel R, Jung G, et al. Polarization of immunity induced by direct injection of naked sequence-stabilized mRNA vaccines. Cell Mol Life Sci. 2004;61:2418–24. doi: 10.1007/s00018-004-4255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kormann MSD, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol. 2011;29:154–7. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 66.Budker V, Budker T, Zhang G, Subbotin V, Loomis A, Wolff JA. Hypothesis: naked plasmid DNA is taken up by cells in vivo by a receptor-mediated process. J Gene Med. 2000;2:76–88. doi: 10.1002/(SICI)1521-2254(200003/04)2:2<76::AID-JGM97>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 67.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–7. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 68.Lingor P, Michel U, Schöll U, Bähr M, Kügler S. Transfection of “naked” siRNA results in endosomal uptake and metabolic impairment in cultured neurons. Biochem Biophys Res Commun. 2004;315:1126–33. doi: 10.1016/j.bbrc.2004.01.170. [DOI] [PubMed] [Google Scholar]
- 69.Wolff JA, Budker V. The mechanism of naked DNA uptake and expression. Adv Genet. 2005;54:3–20. doi: 10.1016/S0065-2660(05)54001-X. [DOI] [PubMed] [Google Scholar]
- 70.Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, et al. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–5. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- 71.Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 72.Saleh M-C, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncb1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–57. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 74.Lorenz C, Fotin-Mleczek M, Roth G, Becker C, Dam TC, Verdurmen WPR, et al. Protein expression from exogenous mRNA: uptake by receptor-mediated endocytosis and trafficking via the lysosomal pathway. RNA Biol. 2011;8:627–36. doi: 10.4161/rna.8.4.15394. [DOI] [PubMed] [Google Scholar]
- 75.Peiser L, Mukhopadhyay S, Gordon S. Scavenger receptors in innate immunity. Curr Opin Immunol. 2002;14:123–8. doi: 10.1016/S0952-7915(01)00307-7. [DOI] [PubMed] [Google Scholar]
- 76.Greaves DR, Gordon S. Thematic review series: the immune system and atherogenesis. Recent insights into the biology of macrophage scavenger receptors. J Lipid Res. 2005;46:11–20. doi: 10.1194/jlr.R400011-JLR200. [DOI] [PubMed] [Google Scholar]
- 77.Janeway CA, Jr., Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 78.Graf GA, Connell PM, van der Westhuyzen DR, Smart EJ. The class B, type I scavenger receptor promotes the selective uptake of high density lipoprotein cholesterol ethers into caveolae. J Biol Chem. 1999;274:12043–8. doi: 10.1074/jbc.274.17.12043. [DOI] [PubMed] [Google Scholar]
- 79.Diken M, Kreiter S, Selmi A, Britten CM, Huber C, Türeci Ö, et al. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011;18:702–8. doi: 10.1038/gt.2011.17. [DOI] [PubMed] [Google Scholar]
- 80.Probst J, Fotin-Mleczek M, Schlake T, Thess A, Kramps T, Kallen K-J. Messenger RNA Vaccines. Gene Vaccines. Vienna: Springer Vienna, 2012:223-45. [Google Scholar]
- 81.González-González E, Ra H, Spitler R, Hickerson RP, Contag CH, Kaspar RL. Increased interstitial pressure improves nucleic acid delivery to skin enabling a comparative analysis of constitutive promoters. Gene Ther. 2010;17:1270–8. doi: 10.1038/gt.2010.74. [DOI] [PubMed] [Google Scholar]
- 82.Herweijer H, Wolff JA. Gene therapy progress and prospects: hydrodynamic gene delivery. Gene Ther. 2007;14:99–107. doi: 10.1038/sj.gt.3302891. [DOI] [PubMed] [Google Scholar]
- 83.Danialou G, Comtois AS, Matecki S, Nalbantoglu J, Karpati G, Gilbert R, et al. Optimization of regional intraarterial naked DNA-mediated transgene delivery to skeletal muscles in a large animal model. Mol Ther. 2005;11:257–66. doi: 10.1016/j.ymthe.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 84.Probst J, Brechtel S, Scheel B, Hoerr I, Jung G, Rammensee H-G, et al. Characterization of the ribonuclease activity on the skin surface. Genet Vaccines Ther. 2006;4:4. doi: 10.1186/1479-0556-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bettinger T, Carlisle RC, Read ML, Ogris M, Seymour LW. Peptide-mediated RNA delivery: a novel approach for enhanced transfection of primary and post-mitotic cells. Nucleic Acids Res. 2001;29:3882–91. doi: 10.1093/nar/29.18.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huth S, Hoffmann F, von Gersdorff K, Laner A, Reinhardt D, Rosenecker J, et al. Interaction of polyamine gene vectors with RNA leads to the dissociation of plasmid DNA-carrier complexes. J Gene Med. 2006;8:1416–24. doi: 10.1002/jgm.975. [DOI] [PubMed] [Google Scholar]
- 87.Martinon F, Krishnan S, Lenzen G, Magné R, Gomard E, Guillet JG, et al. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur J Immunol. 1993;23:1719–22. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 88.Hess PR, Boczkowski D, Nair SK, Snyder D, Gilboa E. Vaccination with mRNAs encoding tumor-associated antigens and granulocyte-macrophage colony-stimulating factor efficiently primes CTL responses, but is insufficient to overcome tolerance to a model tumor/self antigen. Cancer Immunol Immunother. 2006;55:672–83. doi: 10.1007/s00262-005-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou WZ, Hoon DS, Huang SK, Fujii S, Hashimoto K, Morishita R, et al. RNA melanoma vaccine: induction of antitumor immunity by human glycoprotein 100 mRNA immunization. Hum Gene Ther. 1999;10:2719–24. doi: 10.1089/10430349950016762. [DOI] [PubMed] [Google Scholar]
- 90.Mockey M, Bourseau E, Chandrashekhar V, Chaudhuri A, Lafosse S, Le Cam E, et al. mRNA-based cancer vaccine: prevention of B16 melanoma progression and metastasis by systemic injection of MART1 mRNA histidylated lipopolyplexes. Cancer Gene Ther. 2007;14:802–14. doi: 10.1038/sj.cgt.7701072. [DOI] [PubMed] [Google Scholar]
- 91.Mathers AR, Larregina AT. Professional antigen-presenting cells of the skin. Immunol Res. 2006;36:127–36. doi: 10.1385/IR:36:1:127. [DOI] [PubMed] [Google Scholar]
- 92.van den Berg JH, Oosterhuis K, Hennink WE, Storm G, van der Aa LJ, Engbersen JFJ, et al. Shielding the cationic charge of nanoparticle-formulated dermal DNA vaccines is essential for antigen expression and immunogenicity. J Control Release. 2010;141:234–40. doi: 10.1016/j.jconrel.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 93.Thomas M, Ge Q, Lu JJ, Chen J, Klibanov AM. Cross-linked small polyethylenimines: while still nontoxic, deliver DNA efficiently to mammalian cells in vitro and in vivo. Pharm Res. 2005;22:373–80. doi: 10.1007/s11095-004-1874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Soliman M, Nasanit R, Abulateefeh SR, Allen S, Davies MC, Briggs SS, et al. Multicomponent synthetic polymers with viral-mimetic chemistry for nucleic acid delivery. Mol Pharm. 2012;9:1–13. doi: 10.1021/mp200108q. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Y, Satterlee A, Huang L. In vivo gene delivery by nonviral vectors: overcoming hurdles? Mol Ther. 2012;20:1298–304. doi: 10.1038/mt.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiao Z, Levy-Nissenbaum E, Alexis F, Lupták A, Teply BA, Chan JM, et al. Engineering of targeted nanoparticles for cancer therapy using internalizing aptamers isolated by cell-uptake selection. ACS Nano. 2012;6:696–704. doi: 10.1021/nn204165v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gurdon JB, Lane CD, Woodland HR, Marbaix G. Use of frog eggs and oocytes for the study of messenger RNA and its translation in living cells. Nature. 1971;233:177–82. doi: 10.1038/233177a0. [DOI] [PubMed] [Google Scholar]
- 98.Laskey RA, Gurdon JB, Crawford LV. Translation of encephalomyocarditis viral RNA in oocytes of Xenopus laevis. Proc Natl Acad Sci U S A. 1972;69:3665–9. doi: 10.1073/pnas.69.12.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ahlquist P, French R, Janda M, Loesch-Fries LS. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc Natl Acad Sci U S A. 1984;81:7066–70. doi: 10.1073/pnas.81.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van der Werf S, Bradley J, Wimmer E, Studier FW, Dunn JJ. Synthesis of infectious poliovirus RNA by purified T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986;83:2330–4. doi: 10.1073/pnas.83.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Callis J, Fromm M, Walbot V. Expression of mRNA electroporated into plant and animal cells. Nucleic Acids Res. 1987;15:5823–31. doi: 10.1093/nar/15.14.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Karikó K, Kuo A, Barnathan E. Overexpression of urokinase receptor in mammalian cells following administration of the in vitro transcribed encoding mRNA. Gene Ther. 1999;6:1092–100. doi: 10.1038/sj.gt.3300930. [DOI] [PubMed] [Google Scholar]
- 103.Warren L, Manos PD, Ahfeldt T, Loh Y-H, Li H, Lau F, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cho KW, Morita EA, Wright CV, De Robertis EM. Overexpression of a homeodomain protein confers axis-forming activity to uncommitted Xenopus embryonic cells. Cell. 1991;65:55–64. doi: 10.1016/0092-8674(91)90407-P. [DOI] [PubMed] [Google Scholar]
- 105.Boczkowski D, Nair SK, Snyder D, Gilboa E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J Exp Med. 1996;184:465–72. doi: 10.1084/jem.184.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qiu P, Ziegelhoffer P, Sun J, Yang NS. Gene gun delivery of mRNA in situ results in efficient transgene expression and genetic immunization. Gene Ther. 1996;3:262–8. [PubMed] [Google Scholar]
- 107.Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther. 2008;16:1833–40. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–92. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pulendran B, Ahmed R. Translating innate immunity into immunological memory: implications for vaccine development. Cell. 2006;124:849–63. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 110.Isaacs A, Cox RA, Rotem Z. Foreign nucleic acids as the stimulus to make interferon. Lancet. 1963;2:113–6. doi: 10.1016/S0140-6736(63)92585-6. [DOI] [PubMed] [Google Scholar]
- 111.Field AK, Tytell AA, Lampson GP, Hilleman MR. Inducers of interferon and host resistance. II. Multistranded synthetic polynucleotide complexes. Proc Natl Acad Sci U S A. 1967;58:1004–10. doi: 10.1073/pnas.58.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Absher M, Stinebring WR. Toxic properties of a synthetic double-stranded RNA. Endotoxin-like properties of poly I. poly C, an interferon stimulator. Nature. 1969;223:715–7. doi: 10.1038/223715a0. [DOI] [PubMed] [Google Scholar]
- 113.Scheel B, Braedel S, Probst J, Carralot J-P, Wagner H, Schild H, et al. Immunostimulating capacities of stabilized RNA molecules. Eur J Immunol. 2004;34:537–47. doi: 10.1002/eji.200324198. [DOI] [PubMed] [Google Scholar]
- 114.Scheel B, Teufel R, Probst J, Carralot J-P, Geginat J, Radsak M, et al. Toll-like receptor-dependent activation of several human blood cell types by protamine-condensed mRNA. Eur J Immunol. 2005;35:1557–66. doi: 10.1002/eji.200425656. [DOI] [PubMed] [Google Scholar]
- 115.Rettig L, Haen SP, Bittermann AG, von Boehmer L, Curioni A, Krämer SD, et al. Particle size and activation threshold: a new dimension of danger signaling. Blood. 2010;115:4533–41. doi: 10.1182/blood-2009-11-247817. [DOI] [PubMed] [Google Scholar]
- 116.Parvanova I, Rettig L, Knuth A, Pascolo S. The form of NY-ESO-1 antigen has an impact on the clinical efficacy of anti-tumor vaccination. Vaccine. 2011;29:3832–6. doi: 10.1016/j.vaccine.2011.03.073. [DOI] [PubMed] [Google Scholar]
- 117.Sadler AJ, Williams BRG. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–68. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McCartney SA, Colonna M. Viral sensors: diversity in pathogen recognition. Immunol Rev. 2009;227:87–94. doi: 10.1111/j.1600-065X.2008.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–8. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 120.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–31. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 121.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–9. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 122.Karikó K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem. 2004;279:12542–50. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 123.Diebold SS, Massacrier C, Akira S, Paturel C, Morel Y, Reis e Sousa C. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur J Immunol. 2006;36:3256–67. doi: 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- 124.Hornung V, Barchet W, Schlee M, Hartmann G. RNA recognition via TLR7 and TLR8. Handb Exp Pharmacol. 2008;183:71–86. doi: 10.1007/978-3-540-72167-3_4. [DOI] [PubMed] [Google Scholar]
- 125.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–7. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83:4174–84. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–7. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 128.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 129.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 130.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 131.Wilkins C, Gale M., Jr. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–7. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bruns AM, Horvath CM. Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol. 2012;47:194–206. doi: 10.3109/10409238.2011.630974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–43. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hovanessian AG. On the discovery of interferon-inducible, double-stranded RNA activated enzymes: the 2′-5’oligoadenylate synthetases and the protein kinase PKR. Cytokine Growth Factor Rev. 2007;18:351–61. doi: 10.1016/j.cytogfr.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 135.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–8. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 136.Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–75. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 137.Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–38. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Granstein RD, Ding W, Ozawa H. Induction of anti-tumor immunity with epidermal cells pulsed with tumor-derived RNA or intradermal administration of RNA. J Invest Dermatol. 2000;114:632–6. doi: 10.1046/j.1523-1747.2000.00929.x. [DOI] [PubMed] [Google Scholar]
- 139.Nair SK, Boczkowski D, Morse M, Cumming RI, Lyerly HK, Gilboa E. Induction of primary carcinoembryonic antigen (CEA)-specific cytotoxic T lymphocytes in vitro using human dendritic cells transfected with RNA. Nat Biotechnol. 1998;16:364–9. doi: 10.1038/nbt0498-364. [DOI] [PubMed] [Google Scholar]
- 140.Su Z, Dannull J, Yang BK, Dahm P, Coleman D, Yancey D, et al. Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8+ and CD4+ T cell responses in patients with metastatic prostate cancer. J Immunol. 2005;174:3798–807. doi: 10.4049/jimmunol.174.6.3798. [DOI] [PubMed] [Google Scholar]
- 141.Weide B, Carralot J-P, Reese A, Scheel B, Eigentler TK, Hoerr I, et al. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J Immunother. 2008;31:180–8. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 142.Weide B, Pascolo S, Scheel B, Derhovanessian E, Pflugfelder A, Eigentler TK, et al. Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J Immunother. 2009;32:498–507. doi: 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- 143.Rittig SM, Haentschel M, Weimer KJ, Heine A, Muller MR, Brugger W, et al. Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Mol Ther. 2011;19:990–9. doi: 10.1038/mt.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xiang Z, Ertl HC. Manipulation of the immune response to a plasmid-encoded viral antigen by coinoculation with plasmids expressing cytokines. Immunity. 1995;2:129–35. doi: 10.1016/S1074-7613(95)80001-8. [DOI] [PubMed] [Google Scholar]
- 145.Iwasaki A, Stiernholm BJ, Chan AK, Berinstein NL, Barber BH. Enhanced CTL responses mediated by plasmid DNA immunogens encoding costimulatory molecules and cytokines. J Immunol. 1997;158:4591–601. [PubMed] [Google Scholar]
- 146.Geissler M, Gesien A, Tokushige K, Wands JR. Enhancement of cellular and humoral immune responses to hepatitis C virus core protein using DNA-based vaccines augmented with cytokine-expressing plasmids. J Immunol. 1997;158:1231–7. [PubMed] [Google Scholar]
- 147.Loudon PT, Yager EJ, Lynch DT, Narendran A, Stagnar C, Franchini AM, et al. GM-CSF increases mucosal and systemic immunogenicity of an H1N1 influenza DNA vaccine administered into the epidermis of non-human primates. PLoS One. 2010;5:e11021. doi: 10.1371/journal.pone.0011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kanagavelu SK, Snarsky V, Termini JM, Gupta S, Barzee S, Wright JA, et al. Soluble multi-trimeric TNF superfamily ligand adjuvants enhance immune responses to a HIV-1 Gag DNA vaccine. Vaccine. 2012;30:691–702. doi: 10.1016/j.vaccine.2011.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen J, Lin L, Li N, She F. Enhancement of Helicobacter pylori outer inflammatory protein DNA vaccine efficacy by co-delivery of interleukin-2 and B subunit heat-labile toxin gene encoded plasmids. Microbiol Immunol. 2012;56:85–92. doi: 10.1111/j.1348-0421.2011.00409.x. [DOI] [PubMed] [Google Scholar]
- 150.Xu H, Zhao G, Huang X, Ding Z, Wang J, Wang X, et al. CD40-expressing plasmid induces anti-CD40 antibody and enhances immune responses to DNA vaccination. J Gene Med. 2010;12:97–106. doi: 10.1002/jgm.1412. [DOI] [PubMed] [Google Scholar]
- 151.Kreiter S, Diken M, Selmi A, Diekmann J, Attig S, Hüsemann Y, et al. FLT3 ligand enhances the cancer therapeutic potency of naked RNA vaccines. Cancer Res. 2011;71:6132–42. doi: 10.1158/0008-5472.CAN-11-0291. [DOI] [PubMed] [Google Scholar]
- 152.Caux C, Massacrier C, Vanbervliet B, Dubois B, Van Kooten C, Durand I, et al. Activation of human dendritic cells through CD40 cross-linking. J Exp Med. 1994;180:1263–72. doi: 10.1084/jem.180.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]


