Abstract
The human tRNA m5C methyltransferase Misu is a novel downstream target of the proto-oncogene Myc that participates in controlling cell division and proliferation. Misu catalyzes the transfer of a methyl group from S-adenosyl-L-methionine to carbon 5 of cytosines in tRNAs. It was previously shown to catalyze in vitro the intron-dependent formation of m5C at the first position of the anticodon (position 34) within the human pre-tRNALeu(CAA). In addition, it was recently reported that C48 and C49 are methylated in vivo by Misu. We report here the expression of hMisu in Escherichia coli and its purification to homogeneity. We show that this enzyme methylates position 48 in tRNALeu(CAA) with or without intron and positions 48, 49 and 50 in tRNAGly2(GCC) in vitro. Therefore, hMisu is the enzyme responsible for the methylation of at least four cytosines in human tRNAs. By comparison, the orthologous yeast enzyme Trm4 catalyzes the methylation of carbon 5 of cytosine at positions 34, 40, 48 or 49 depending on the tRNAs.
Keywords: tRNA modification enzyme, RNA methyltransferase, 5-methylcytosine, m5C, Misu, NSun2, Trm4
Introduction
Maturation of RNAs is a complex process that involves post-transcriptional chemical modifications, 5′ and 3′ sequence removal and, in some cases, splicing or polyadenylation. These chemical modifications have functions beyond fine-tuning the structure and function of RNA, such as gene regulation at the RNA level, but recent studies have also pointed out that some modifications can be highly dynamic and regulate cellular response to various stress conditions.1-3 tRNAs present the largest variety of chemical modifications and the highest degree of modification among all RNA species. Extensive modification occurring at position 34 of the anticodon or 37, 3′-adjacent to the anticodon, directly affect the codon-anticodon interactions during the decoding of genetic information, and are thus important for maintaining translation fidelity.4,5
Methylations at various base positions and at the 2' hydroxyl group of ribose are the most frequently encountered modifications in tRNAs. Among them, 5-methylcytosine (m5C) is a tRNA stabilizing modification6-8 that is found in eukaryotes and archaea, but not in eubacteria.9 Five distinct families of putative RNA m5C methyltransferases have been identified in eukaryotes.10 In Saccharomyces cerevisiae, m5C formation is catalyzed by a multisite-specific tRNA m5C methyltransferase (MTase), Trm4, which uses S-adenosyl-L-methionine (SAM) as methyl donor.11-13 Depending on the tRNA substrate, the target cytosines of Trm4 are located at positions 34, 40, 48 and/or 49. In eukaryotic cells, some of the pre-tRNAs contain introns that are located one nucleotide downstream from the anticodon and are spliced to form the functional tRNA molecule. Most yeast tRNAs are modified at positions 48 and/or 49 by Trm4, whereas methylations at positions 34 and 40, present only in tRNALeu(CAA) and tRNAPhe(GAA), respectively, occur exclusively in the intron-containing tRNA precursors. In mammals, m5C was found at positions 34, 38, 48, 49, 50 and 72 of tRNAs but not at position 40.14 Recently, the distribution of 5-methylcytosine in human coding and non-coding RNAs was studied in vivo by coupling bisulfite conversion of cellular RNA with next-generation sequencing.15 Using this method, m5C was detected not only at positions 34, 38, 48, 49, 50 and 72 in human tRNAs but also at new sites: 19, 27, 40, 55, 61, 66, 70, 73 and 75.
Yeast Trm4 shows 35% sequence identity with its human ortholog, hMisu (or NSun2). hMisu, expressed in yeast, was shown to methylate C34 only in the intron-containing human pre-tRNALeu(CAA) but it was not able to form m5C at position 48 and 49 of human or yeast precursors.16 It was also more sensitive to the nucleotide sequence surrounding the target position 34 than Trm4. Therefore, it was concluded that hMisu has a much narrower specificity than Trm4. However, using RNA interference technology, Squires et al. recently showed that hMisu has a broader substrate specificity than previously anticipated because it is responsible for methylation of tRNAs at positions 48 and 49, at least in tRNAAsp(GUC).15 Interestingly, methylation of C38 is performed by the enzyme Dnmt2 that bears close similarities to DNA cytosine methyltransferases.17-20
Misu is a nucleolar protein that translocates from the nucleoli in interphase to the spindle in mitosis as an RNA-protein complex that includes 18S rRNA.21 The protein kinase Aurora-B, which is essential for the segregation of eukaryotic chromosomes, has been shown to regulate the methyltransferase activity of hMisu during mitosis.22 Phosphorylation of Ser139 by Aurora-B or the phosphorylation-mimic mutation S139E suppressed the methyltransferase activities of hMisu. The phosphorylation caused hMisu to dissociate from its nucleolar binding protein NMP1, indicating that Aurora-B participates to regulate the assembly of the nucleolar RNA-processing machinery.
The gene encoding murine Misu (mMisu) has been shown to be the target of the proto-oncogene Myc, which plays a direct role in controlling cell division and growth.23 mMisu is expressed at low levels in normal tissues but overexpressed in different types of tumor as a result of Myc activation.23 Gene silencing of mMisu by RNA interference reduced tumor formation in vivo. It was proposed that Misu, together with the nucleolar and spindle-associated protein NuSAP, helps Myc to promote proliferation by stabilizing the mitotic spindle in fast-dividing cells.21 Moreover, it has recently been shown that Misu is required to balance stem cell self-renewal and differenciation in skin24 and that the gene copy number, as well as expression of hMisu, is increased in oral and colorectal cancers.25 Altogether, these results indicate that Misu is a potential target for cancer therapies.
Here, we performed the expression of hMisu in E. coli, its purification to homogeneity, and further explored its substrate specificity using mass spectrometry analysis of digested RNA substrates. We confirm that its substrate specificity is larger than anticipated because, in addition to the intron-dependent C34 methylation, it can methylate cytosines at positions 48, 49 and 50 in tRNAs.
Results
Expression of human Misu in Escherichia coli and purification of the protein
The cDNA of hMisu has previously been cloned into the pYES2 yeast expression vector. This recombinant plasmid was used to transform the yeast strain BY4742, in which the endogeneous TRM4 gene had been deleted.16 The hMisu methyltransferase activity at position 34 was investigated by incubating total protein extracts from the transformed strain with radiolabeled pre-tRNA Leu(CAA) or tRNA Leu(CAA) transcript and analyzing m5C formation after complete hydrolysis of RNA by RNase T2 using thin-layer two-dimensional chromatography and autoradiography.
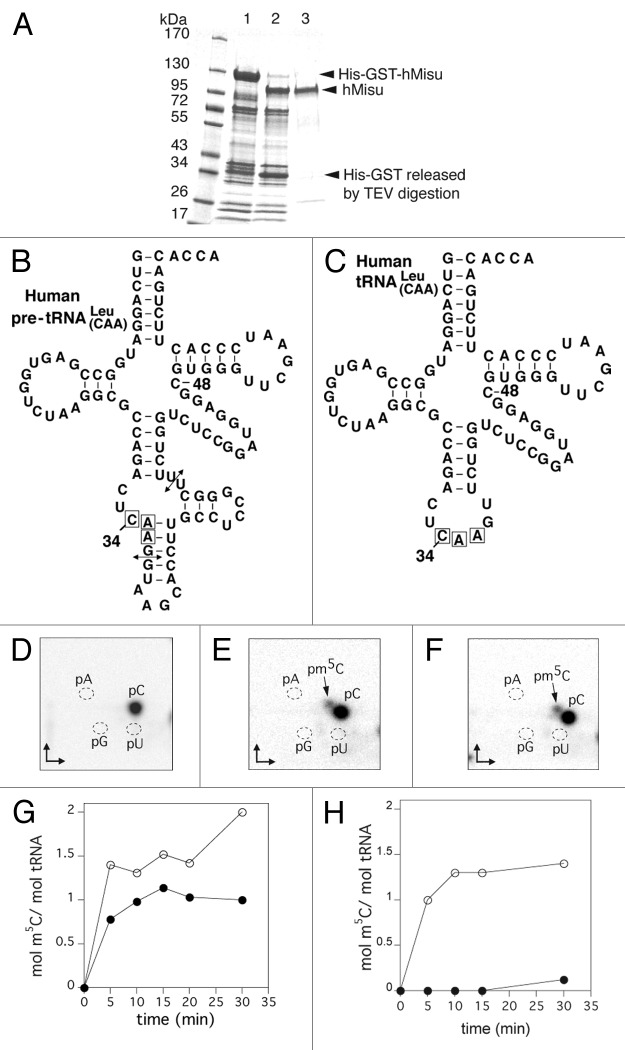
We decided to clone the cDNA of hMisu into an E. coli expression vector that adds a TEV-cleavable His-GST tag at the N-terminus of the protein. After expression and purification of His-GST-hMisu by immobilized-nickel affinity chromatography, the His-GST tag was removed by the TEV protease and nontagged hMisu was further purified by Ni-NTA chromatography. The protein was almost pure, according to the SDS-PAGE analysis (Fig. 1A).
Figure 1. Formation of m5C in human tRNA catalyzed by purified hMisu. (A) 10% SDS PAGE analysis of hMisu purification. Lane 1: Ni-NTA purified His-GST fused hMisu (117 kDa). Lane 2: Proteins mixture after TEV protease digestion of Ni-NTA purified His-GST-hMisu (after digestion, the molecular weight of hMisu is 86 kDa). Lane 3: hMisu after TEV protease digestion and elution of Ni-NTA column with 20 mM imidazole. (B and C) The transcripts used as substrates were the intron-containing human pre- tRNA (B) and intron-less human tRNA . (C) The arrows indicate the position of the intron. (D−F) Thin layer chromatography analyses of pre- tRNA incubated without protein (D) or with hMisu (E), and of tRNA incubated with hMisu (F) in the absence of magnesium. The main spot is C and the minor spot m5C. (G and H) Time course of m5C formation catalyzed by hMisu in pre- tRNA (G) or tRNA (H) in the presence of 50 mM NaCl, and in the absence (open circles) or presence (filled circles) of 10 mM MgCl2.
m5C formation kinetics in human pre-tRNALeu(CAA) or tRNA Leu(CAA)
Human Misu expressed in yeast had been shown to methylate only the cytosine at the first position of the anticodon (position 34) within the human pre-tRNALeu(CAA), in an intron-dependent manner.16 In addition, murine and human Misu21-23 had been shown to incorporate radiolabeled methyl from S-adenosyl-l-methionine (SAM) into total tRNA from E. coli. Total E. coli tRNA can serve as substrate for Misu because m5C is not found in E. coli tRNA sequences.26 Indeed, in this organism, RNA m5C methyltransferases methylate only rRNA.27-30 However, the methyl incorporation in E. coli tRNA by Misu is not intron-dependent because tRNAs do not contain introns in this organism. This led us to investigate whether hMisu can catalyze m5C formation in intron-less human tRNAs. The further study of Misu substrate specificity was performed using the purified enzyme in an in vitro activity test.
We wanted first to verify that hMisu expressed in our system can methylate human pre-tRNALeu(CAA) (Fig. 1B) and not intron-less human tRNALeu(CAA) (Fig. 1C), as shown previously.16 Transcripts obtained by in vitro transcription of tRNA genes in the presence of [α-32P]CTP were incubated with hMisu and SAM (Fig. 1). The time course of m5C formation shows that, in the presence of 10 mM MgCl2, only intron-containing tRNALeu is methylated by hMisu at a ratio of 1 mol m5C/ mol tRNA (Fig. 1G and H). However, in the absence of MgCl2, intron-less tRNALeu becomes substrate of hMisu, as shown by the formation of almost 1 mol m5C/ mol tRNA, whereas 2 mol m5C/ mol tRNA were incorporated in the intron-containing pre-tRNALeu(CAA) (Fig. 1E, F, G and H). Thus, MgCl2 appears to partially inhibit m5C formation in pre-tRNALeu(CAA) and completely in intron-less tRNALeu(CAA). This result led us to study in more detail the inhibition of hMisu activity by magnesium in intron-less tRNALeu(CAA). The hMisu activity was monitored as a function of magnesium concentration with different hMisu to tRNA ratios (46:1 or 1:1, corresponding to the amounts used in the activity tests analyzed by TLC and mass spectrometry, respectively, see below), in the presence of two different salts (NaCl or ammonium acetate) (Fig. S1). Figure S1A shows that in the presence of 50 mM NaCl (such as in Fig. 1), the methylation of intron-less tRNALeu(CAA) by hMisu decreases gradually when the magnesium concentration increases. In the presence of 50 mM ammonium acetate, a less efficient inhibition of hMisu activity by magnesium is observed (Fig. S1D). Interestingly, the enzyme to tRNA ratio has no effect on this inhibition because similar results are obtained at low (Fig S1A and B) and high concentrations of tRNA (Fig. S1C and D).
Human Misu methylates positions 34 and 48 in pre-tRNALeu(CAA) and only position 48 in tRNALeu(CAA)
The position of m5C formed by hMisu in human pre-tRNALeu(CAA) and human tRNALeu(CAA) was identified using MALDI mass spectrometry31 (Figs. 2 and 3). tRNAs incubated with hMisu in the absence of MgCl2 were digested by RNase T1 or RNase A, which specifically cleaves after guanosines and pyrimidines, respectively, and fragments were subjected to MS analysis to reveal potential mass-changing modifications in digestion products. The MS spectra of pre-tRNALeu digests showed an increase of mass of 14 Da corresponding to a methyl only in the RNase T1 fragment AC(32)UC(34)AAGp (Fig. 2) and not in the RNase A fragment AGAC(32)p (Fig. S2), indicating that C34 is methylated. With intron-less tRNALeu(CAA), no methylation was observed in the RNase T1 fragment containing C34 (data not shown). These results confirm that hMisu catalyzes methylation of C34 only in an intron-containing tRNA, pre-tRNALeu(CAA), as shown by Brzezicha et al.16 Another methylation was observed in the RNase A fragment GGAGGC(48)p with tRNALeu(CAA) (Fig. 3) and pre-tRNALeu(CAA) (Fig. S2), showing that C48 is methylated by hMisu independently of the presence of an intron in the anticodon loop. This last methylation was not observed previously,16 probably because MgCl2 was present in the reaction mixture. Accordingly, in the presence of 10 mM MgCl2, as used in the Brzezicha et al. experiment, the methylation of intron-less tRNALeu(CAA) is inhibited almost completely (Fig. 1H; Fig. S1A).
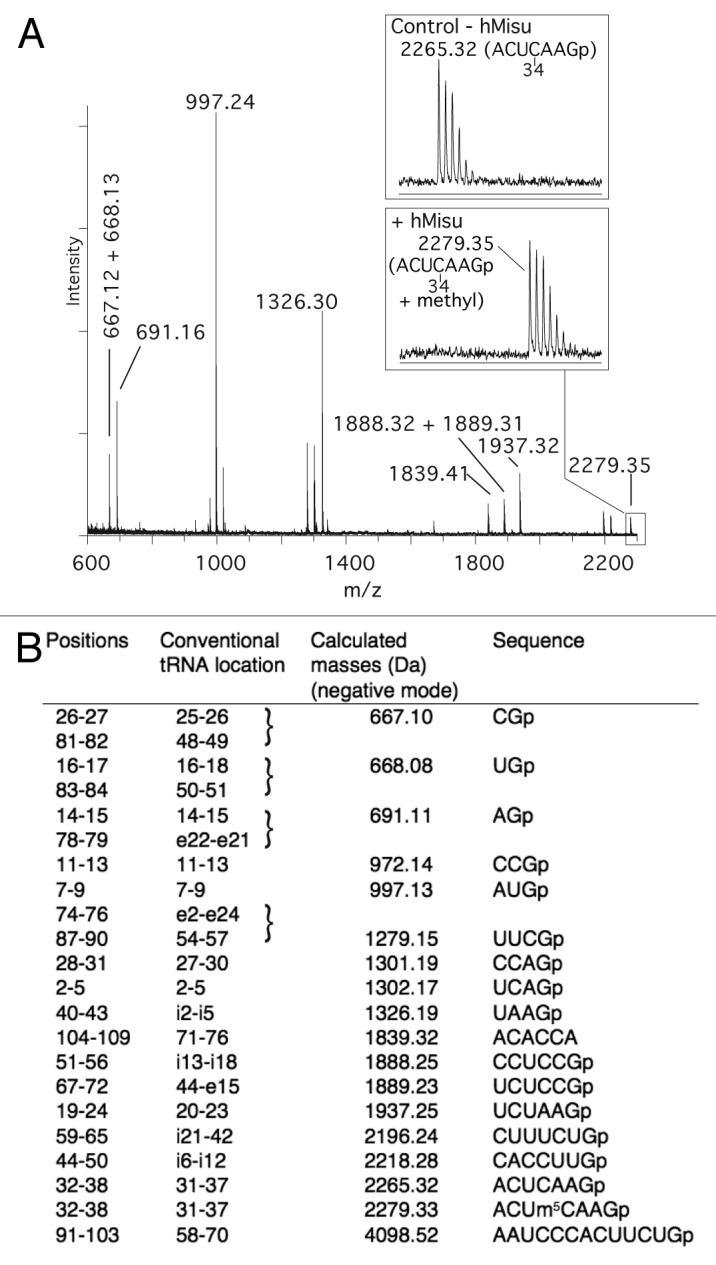
Figure 2. MALDI mass spectrometry analysis of intron-containing human pre- tRNA for methylation at position 34. (A) MALDI mass spectrum of pre- tRNA methylated by hMisu and digested by RNase T1 that cleaves after guanosines. The spectral region around the ACUCAAGp fragment containing C34 is enlarged to show the peak of the nonmethylated ion (m/z 2265.32) in the control without enzyme and the ion methylated by hMisu (m/z 2279.35). (B) List of theoretical masses of RNase T1 fragments of singly protonated pre- tRNA .
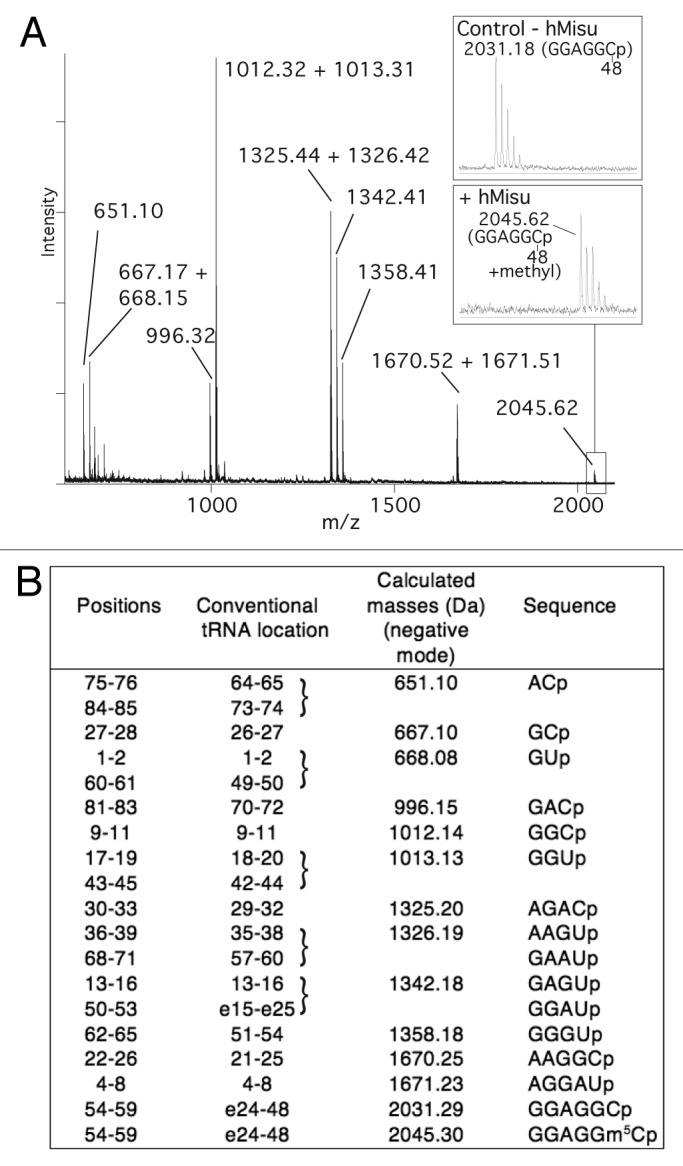
Figure 3. MALDI mass spectrometry analysis of intron-less human tRNA for methylation by hMisu at position 48. (A) MALDI mass spectrum of tRNA methylated by hMisu and digested by RNase A that cleaves after pyrimidines. The spectral region around the fragment GGAGGCp containing C48 is enlarged to show the peak of the nonmethylated ion (m/z 2031.18) from the control without enzyme and the ion methylated by hMisu (m/z 2045.62). (B) List of theoretical masses of RNase A fragments of singly protonated tRNA .
Human Misu methylates also cytosines at positions 49 and 50
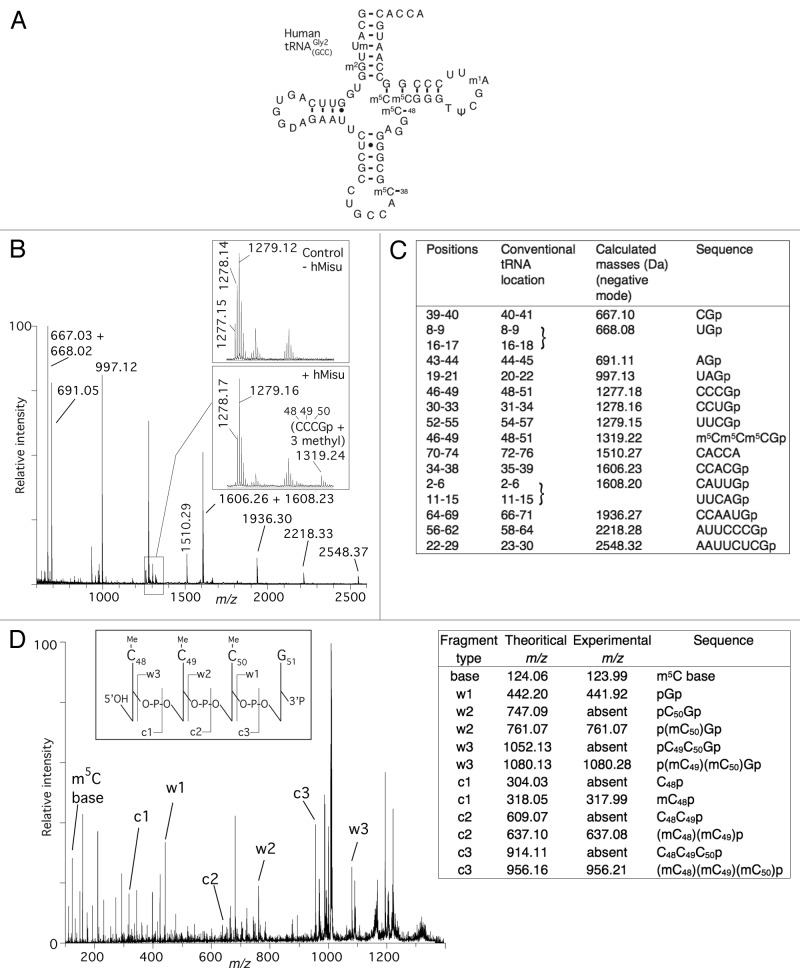
The compilation of the 20 human tRNA sequences present in the database (http://rna-mdb.cas.albany.edu/RNAmods/)14 shows that m5C is also found at positions 38, 49 and 50. Human tRNAGly2(GCC) possesses m5C at positions 38, 48, 49 and 50. The corresponding in vitro transcript was tested as a substrate for hMisu. Figure 4A−C show the MS analyses of the RNase T1 fragments of the T7 transcript of tRNAGly2(GCC) after incubation with hMisu and SAM. Upon incubation with hMisu, the peak corresponding to the fragment CCCGp (m/z 1277.15), adjacent to two peaks corresponding to other fragments (m/z 1278.17 and 1279.16) disappeared almost completely (Fig. 4B; Fig. S3). Concomitantly, a new peak appeared at m/z 1319.24 corresponding to the mass of the CCCGp sequence (nucleotides 48–51) containing three methyl groups. In addition, no peak at m/z 1291.19 or 1305.20 appeared, which would correspond to the mono or dimethylated fragment. Tandem mass spectrometry (MS/MS) was then used to reveal the positions of the modifications at the nucleotide level, as previously reported.32 In this technique, individual oligonucleotides of interest, which have been previously identified by regular MALDI-MS, are isolated and subjected to controlled fragmentation.31 The trimethylated CCCGp ion at m/z 1319.24 was selected and fragmented to determine the positions of the three methylations in this fragment (Fig. 4D). Fragmentation leads to the 5′-phosphorylated w1, w2 and w3 fragments, which correspond to pG51p, pC50G51p and pC49C50G51p, respectively, and to the 5′- non phosphorylated c1, c2 and c3 fragments, which correspond to C48p, C48C49p and C48C49C50p, respectively. As expected, the w1 fragment was not methylated. However, the w2 and w3 fragments possessed one and two methyl groups, respectively, and the c1, c2 and the c3 fragments, one, two and three methyl groups, respectively. Altogether, these results definitely confirm that C48, C49 and C50 of tRNAGly2(GCC) are methylated by hMisu.
Figure 4. MALDI mass spectrometry analysis of human tRNA for methylation by hMisu. (A) Cloverleaf structure of the mature human tRNA indicating its natural posttranscriptional modifications. (B) MALDI mass spectrum of tRNA methylated by hMisu and digested by RNase T1 that cleaves after guanosines. The spectral region around the fragment CCCGp containing C48, C49 and C50 is enlarged to show the peak of the nonmethylated ion (m/z 1277.15) from the control without enzyme and the ion methylated by hMisu (m/z 1319.24), see also Figure S3. (C) List of theoretical masses of RNase T1 fragments of singly protonated tRNA. (D) Identification of the three methylation targets of hMisu at positions 48, 49 and 50 in human tRNA by MS/MS. The fragment at m/z 1319.24 of the methylated sample shown in (B) was selected and fragmented by tandem mass spectrometry. The fragments assignment follows the scheme of McLuckey et al.40 The peaks corresponding to the c-ions and w-ions are generated by loss of the 3′ and 5′ nucleotides, respectively.
Discussion
Misu belongs to a large superfamily of RNA m5C methyltransferases found in organisms from all three domains of life. In eukaryotes, these enzymes were classified in five subfamilies based on their aminoacid sequences10 and nine of them are present in humans. In addition to their enzymatic activity, some of these enzymes have an important cellular role. The function of most of them remains unknown and their substrate specificity has not yet been investigated. As a step toward understanding the importance of these proteins, we have characterized here the substrate specificity of human Misu in vitro, and shown that it is a multi-site specific tRNA methyltransferase responsible for the methylation of C at four different positions (34, 48, 49 and/or 50), depending on the tRNA. Our results confirm the implication of hMisu in modifiying tRNAs at positions 48 and 49, as recently reported,15 and extend its specificity to position 50. Interestingly, Squires et al.15 also detected m5C at numerous unusual sites (19, 27, 40, 55, 61, 66, 70, 73 and 75) in tRNA from Hela cells, where the cancerisation process may have implicated Myc.33 The overexpression of Misu in Myc-induced tumors23 may be responsible of these additional m5C.
Our results are in agreement with the specificity reported for Trm4 from S. cerevisiae11 and that anticipated for Trm4 from M. jannashii,34 two hMisu homologs. These methyltransferases were suggested to recognize, in their tRNA substrates, a stem-bulge-stem structure containing the target cytosine in or close to the bulge. The cytosines methylated by hMisu are also present in a stem-bulge-stem structure. Indeed, in the intron-containing pre-tRNALeu(CAA), C34 belongs to a stem-bulge-stem structure that becomes, after intron splicing, a stem-loop that is no more suitable for methylation by hMisu (Fig. 1B and C). Moreover, the other cytosines methylated by hMisu, C48, C49 and C50, are also located in a stem-bulge-stem structure (Fig. 4A). This hypothesis could explain why C38, present in a stem-loop structure, is not methylated by hMisu. Indeed, it has been shown that C38 of tRNAGly2(GCC) is methylated by Dnmt2.19
Before this work, the inhibition by Mg2+ of m5C tRNA methyltransferase activity had previously been reported for the partially purified rat Misu, concerning the methylation of C48 in E. coli tRNAfMet 35. Melting experiments of tRNAfMet showed that Mg2+ stabilizes its tertiary structure, preventing C48 from methylation by hMisu. On the contrary, the accessibility of C34 in the pre-tRNALeu(CAA) is probably not dependent on tertiary structure interactions, and thus less sensitive to the Mg2+ effect. Here we show that the inhibition by magnesium of the methylation of C48 is less efficient in the presence of ammonium acetate than NaCl. Ammonium acetate could act by destabilizing the tRNA or by disturbing the interaction between magnesium and RNA.
The inhibition by Mg2+ of m5C formation in tRNA has also been observed for other hMisu homologs, such as S. cerevisiae Trm4 (Motorin and Grosjean, personal communication) and PAB1947 enzyme from P. abyssi (our unpublished data). Therefore, it seems that the in vitro inhibition by Mg2+ is a common characteristic of this methyltransferase family.
Conclusion
In addition to cytosine at position 34, the human enzyme Misu expressed in E. coli was shown to be able to modify positions 48, 49 and 50 in human tRNAs. Fifteen out of the 20 human tRNAs that have been sequenced today possess the m5C modification at positions 48 and/or 49 and 6 of them possess m5C50. Therefore, hMisu has not only a much wider specificity than previously thought but it is the main enzyme responsible for methylation of cytosines in tRNAs.
Whereas m5C34 is important for maintaining correct codon-anticodon pairing during translation, as demonstrated in the case of the amber suppressor yeast tRNALeu,36 the function of the modifications at positions 48, 49 and 50 is likely to stabilize the tRNA structure.37 For example, m5C at position 49 acts in synergy with modification m7G46 to stabilize tRNAVal to avoid this tRNA to be degraded through a rapid tRNA degradation pathway.38 The m5C38 modification appears to inhibit the endonucleolytic cleavage of the Dnmt2 substrate tRNAs under stress conditions.19
In addition to positions 34, 38, 48, 49 and 50, m5C was also reported at other positions in human tRNAs (19, 27, 40, 55, 61, 66, 70, 72, 73 and 75).15 These results need to be confirmed by other techniques and in cells different than Hela cells, in which hMisu is overexpressed. It will then be necessary to determine whether cytosines at these positions are methylated by hMisu or by other m5C methyltransferases that remain to be discovered.
Materials and Methods
Cloning, expression and purification of human Misu cDNA in Escherichia coli
Human Misu cDNA was amplified from plasmid pYES216 using AAAACCATGGGGCGGCGGTCGCGGGGTCGG as forward primer and AAAACTCGAGTCACCGGGGTGGATGGACCCCCGCCGG as reverse primer, containing the NcoI and XhoI restriction sites, respectively. The PCR product was digested by NcoI and XhoI and cloned into the E. coli expression vector pETM-30 (a gift from G. Stier, EMBL Heidelberg, Germany). The resulting ORF encodes the human Misu protein with a TEV-cleavable His-GST tag at the N terminus. This fusion protein was expressed in E. coli BL21 star (Invitrogen). The transformed cells were grown at 37°C in 2 l of casamino acid M9CA medium (Interchim) supplemented with 0.2% glucose, 1 mM MgSO4, 0.3 mM CaCl2, 50 μg/ml kanamycin. Expression was induced by 1 mM IPTG (Euromedex) when cells reached an optical density of 0.6 at 600 nm. After 6 h at 20°C, the cells were harvested, resuspended in 5 volumes of lysis buffer (50 mM sodium phosphate pH 8.0, 1 M NaCl, 10 mM imidazole, 10% glycerol, 5 mM β-mercaptoethanol, EDTA-free protease inhibitor cocktail SIGMAFAST (Sigma-Aldrich), and disrupted by sonication.
The cell lysate was cleared by centrifugation at 14,000 g for 30 min at 4°C. The supernatant was loaded on a Ni-NTA superflow resin (2 ml, Qiagen) equilibrated with lysis buffer. The column was washed with wash buffer: 50 mM sodium phosphate pH 8.0, 300 mM NaCl, 20 mM imidazole and 10% glycerol. His-GST-hMisu was eluted with the same buffer containing 150 mM imidazole. The eluted protein was desalted on a PD10 exclusion size column (GE Healthcare) equilibrated with 50 mM sodium phosphate pH 8.0, 100 mM NaCl and 5% glycerol. The His-GST tag was removed by incubating the desalted protein with recombinant His-tagged TEV protease (TEV protease:protein, 1:20, w:w) in 0.1 mM EDTA and 1 mM DTT, overnight at 4°C. After addition of 300 mM NaCl, the TEV digest mixture was loaded on a Ni-NTA column equilibrated with the previous wash buffer without imidazole. Nontagged hMisu, which nonspecifically binds to the column, was eluted with wash buffer containing 20 mM imidazole, whereas His-TEV and His-GST were retained. hMisu was then desalted on a PD10 column equilibrated with 25 mM Tris-HCl pH 7.5, 100 mM NaCl, 10% glycerol, 2 mM DTT and stored as aliquots at -80°C.
Human tRNAGly2GCC gene construction
Two oligonucleotides complementary at their 3′ends were used as primers and templates for PCR synthesis of the human tRNAGly2GCC gene. The oligonucleotides sequences were TATACGACTCACTATAGCATTGGTGGTTCAGTGGTAGAATTCTCGCCTGCCACGCGG (forward primer) and TGTGCATTGGCCGGGAATCGAACCCGGGCCTCCCGCGTGGCAGGCGAGAATTCTACC (reverse primer). The T7 promotor sequence is present at the 5′end of the forward primer to perform in vitro T7 RNA transcription.
T7 transcription of the tRNA genes
Plasmids carrying the genes of human intron-containing pre-tRNA LeuLeu(CAA) and intron-less tRNALeu(CAA)16 were linearized by the MvaI restriction enzyme before transcription. Transcription of human tRNAGly2(GCC) was performed on its PCR gene construct. Radiolabeled tRNAs were transcribed using 20 units of T7 RNA polymerase (Promega) and 10 units of Ribonuclease inhibitor (Sigma-Aldrich) in transcription buffer (Promega), containing 1.25 mM each of ATP, GTP, UTP, 100 μM CTP, 50 μCi of [α-32P] CTP (800 Ci/mmol), 10 mM GMP, 10 mM DTT and 1 μg of linearized plasmid in a total volume of 10 μl at 37°C for 3 h. Nonradioactive transcription was performed using the RiboMAX™ large scale RNA production system-T7 (Promega) and 10 μg of linearized plasmid or PCR synthesized template.
In vitro methylation of radiolabeled tRNA by hMisu
Radiolabeled tRNAs (1 nM) were incubated with 46 nM of hMisu in 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 2 mM DTT, 160 μM SAM in a total volume of 50 μl at 37°C. In some experiments, MgCl2 was added. After phenol extraction with a mixture of phenol pH 4.5:chloroform:isoamylic alcohol (25:24:1), the tRNAs were precipitated with ethanol and subjected to nuclease P1 (Roche) hydrolysis. Nucleotides were resolved by 2D thin layer chromatography on cellulose plates39 and the radiolabeled nucleotides were quantified using a STORM™ system (GE Healthcare).
Mass spectrometry analyses
1.5 μM tRNA was incubated for 30 min at 37°C with 1.5 μM of hMisu in 20 mM Tris-HCl pH 8.0, 50 mM ammonium acetate, 2 mM DTT, 160 μM SAM in a total volume of 50 μl. The methylated tRNA was phenol extracted and concentrated by ethanol precipitation with 0.3 M ammonium acetate pH 5.2. The tRNA pellet was redissolved in 50 μl of water and desalted on a Microspin™ G-25 column (GE Healthcare). The desalted tRNA was divided into two tubes and dried in speed-vac concentrator. The tRNA was digested 5 h at 37°C in 50 mM of 2.5-dihydrobenzoic acid (DHB) containing either 200 units of RNase T1 (Roche) or 2 μg of RNase A (Fermentas) in a total volume of 10 μl. The residual cyclic 2’, 3′-phosphates formed during digestions were removed by adding HCl to a final concentration of 0.2 M and incubating for 30 min at room temperature. MALDI-TOF MS and MALDI-TOF/TOF MS/MS analyses were performed directly on the digestion products using a 4800 MALDI TOF/TOF Analyzer mass spectrometer (Applied Biosystems, Les Ulis, France). This instrument is equipped with an Nd:YAG laser (operating at 355 nm wavelength of < 500 ps pulse and 200 Hz repetition rate). Acquisitions were performed in negative ion mode. For MS/MS experiments, precursor ions were accelerated at 8 kV and the MS/MS spectra were acquired using 2 kV collision energy with CID gas (argon) at a pressure of 3.5 × 10-6 Torr. MS data were processed using DataExplorer 4.4 (Applied Biosystems).
Supplementary Material
Acknowledgments
Funding was provided by the Association pour la Recherche sur le Cancer (to B.G.P.) and the CNRS. We acknowledge the use of the Imagif cloning and proteins expression Platform and thank Laetitia Cormier for performing the cloning experiments and expression tests and David Touboul for critical reading of the manuscript.
Glossary
Abbreviations:
- GST
gluthathione-S-transferase
- IPTG
Isopropyl-beta-D-thiolgalactopyranoside
- TEV
Tobacco Etch Virus
- EDTA
ethylenediaminetetraacetic acid
- DTT
dithiothreitol
- TLC
thin layer chromatography
- PCR
polymerase chain reaction
- MALDI-TOF MS
matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- SAM
S-adenosyl-L-methionine
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental material may be found here: www.landesbioscience.com/journals/rnabiology/article/22180
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/22180
References
- 1.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–5. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 2.Yi C, Pan T. Cellular dynamics of RNA modification. Acc Chem Res. 2011;44:1380–8. doi: 10.1021/ar200057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nawrot B, Sochacka E, Düchler M. tRNA structural and functional changes induced by oxidative stress. Cell Mol Life Sci. 2011;68:4023–32. doi: 10.1007/s00018-011-0773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17:555–60. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 5.Demeshkina N, Jenner L, Yusupova G, Yusupov M. Interactions of the ribosome with mRNA and tRNA. Curr Opin Struct Biol. 2010;20:325–32. doi: 10.1016/j.sbi.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol. 1996;53:79–129. doi: 10.1016/S0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- 7.Basti MM, Stuart JW, Lam AT, Guenther R, Agris PF. Design, biological activity and NMR-solution structure of a DNA analogue of yeast tRNA(Phe) anticodon domain. Nat Struct Biol. 1996;3:38–44. doi: 10.1038/nsb0196-38. [DOI] [PubMed] [Google Scholar]
- 8.Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–44. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 9.Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–30. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavlopoulou A, Kossida S. Phylogenetic analysis of the eukaryotic RNA (cytosine-5)-methyltransferases. Genomics. 2009;93:350–7. doi: 10.1016/j.ygeno.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–18. doi: 10.1017/S1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walbott H, Auxilien S, Grosjean H, Golinelli-Pimpaneau B. The carboxyl-terminal extension of yeast tRNA m5C methyltransferase enhances the catalytic efficiency of the amino-terminal domain. J Biol Chem. 2007;282:23663–71. doi: 10.1074/jbc.M703818200. [DOI] [PubMed] [Google Scholar]
- 13.Walbott H, Husson C, Auxilien S, Golinelli-Pimpaneau B. Cysteine of sequence motif VI is essential for nucleophilic catalysis by yeast tRNA m5C methyltransferase. RNA. 2007;13:967–73. doi: 10.1261/rna.515707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33(Database issue):D139–40. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–33. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA) Nucleic Acids Res. 2006;34:6034–43. doi: 10.1093/nar/gkl765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Göll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–8. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 18.Schaefer M, Lyko F. Solving the Dnmt2 enigma. Chromosoma. 2010;119:35–40. doi: 10.1007/s00412-009-0240-6. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–5. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiagarajan D, Dev RR, Khosla S. The DNA methyltranferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics. 2011;6:103–13. doi: 10.4161/epi.6.1.13418. [DOI] [PubMed] [Google Scholar]
- 21.Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, et al. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol. 2009;186:27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakita-Suto S, Kanda A, Suzuki F, Sato S, Takata T, Tatsuka M. Aurora-B regulates RNA methyltransferase NSUN2. Mol Biol Cell. 2007;18:1107–17. doi: 10.1091/mbc.E06-11-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971–81. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 24.Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011;7:e1002403. doi: 10.1371/journal.pgen.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto M, Hirata S, Sato S, Koga S, Fujii M, Qi G, et al. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012;31:660–71. doi: 10.1089/dna.2011.1446. [DOI] [PubMed] [Google Scholar]
- 26.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39(Database issue):D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tscherne JS, Nurse K, Popienick P, Michel H, Sochacki M, Ofengand J. Purification, cloning, and characterization of the 16S RNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:1884–92. doi: 10.1021/bi981880l. [DOI] [PubMed] [Google Scholar]
- 28.Gu XR, Gustafsson C, Ku J, Yu M, Santi DV. Identification of the 16S rRNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:4053–7. doi: 10.1021/bi982364y. [DOI] [PubMed] [Google Scholar]
- 29.Andersen NM, Douthwaite S. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J Mol Biol. 2006;359:777–86. doi: 10.1016/j.jmb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Purta E, O’Connor M, Bujnicki JM, Douthwaite S. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J Mol Biol. 2008;383:641–51. doi: 10.1016/j.jmb.2008.08.061. [DOI] [PubMed] [Google Scholar]
- 31.Douthwaite S, Kirpekar F. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol. 2007;425:1–20. doi: 10.1016/S0076-6879(07)25001-3. [DOI] [PubMed] [Google Scholar]
- 32.Guelorget A, Roovers M, Guérineau V, Barbey C, Li X, Golinelli-Pimpaneau B. Insights into the hyperthermostability and unusual region-specificity of archaeal Pyrococcus abyssi tRNA m1A57/58 methyltransferase. Nucleic Acids Res. 2010;38:6206–18. doi: 10.1093/nar/gkq381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macville M, Schröck E, Padilla-Nash H, Keck C, Ghadimi BM, Zimonjic D, et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 1999;59:141–50. [PubMed] [Google Scholar]
- 34.Kuratani M, Hirano M, Goto-Ito S, Itoh Y, Hikida Y, Nishimoto M, et al. Crystal structure of Methanocaldococcus jannaschii Trm4 complexed with sinefungin. J Mol Biol. 2010;401:323–33. doi: 10.1016/j.jmb.2010.06.046. [DOI] [PubMed] [Google Scholar]
- 35.Wildenauer D, Gross HJ, Riesner D. Enzymatic methylations: III. Cadaverine-induced conformational changes of E. coli tRNA fMet as evidenced by the availability of a specific adenosine and a specific cytidine residue for methylation. Nucleic Acids Res. 1974;1:1165–82. doi: 10.1093/nar/1.9.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strobel MC, Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol. 1986;6:2663–73. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Squires JE, Preiss T. Function and detection of 5-methylcytosine in eukaryotic RNA. Epigenomics. 2010;2:709–15. doi: 10.2217/epi.10.47. [DOI] [PubMed] [Google Scholar]
- 38.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 39.Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 40.McLuckey SA, Goeringer DE, Glish GL. Collisional activation with random noise in ion trap mass spectrometry. Anal Chem. 1992;64:1455–60. doi: 10.1021/ac00037a026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.