Abstract
Milk fat curdle in sewage is one of the refractory materials for active sludge treatment under low temperature conditions. For the purpose of solving this problem by using a bio-remediation agent, we screened Antarctic yeasts and isolated SK-4 strain from algal mat of sediments of Naga-ike, a lake in Skarvsnes, East Antarctica. The yeast strain showed high nucleotide sequence homologies (>99.6%) to Mrakia blollopis CBS8921T in ITS and D1/D2 sequences and had two unique characteristics when applied on an active sludge; i.e., it showed a potential to use various carbon sources and to grow under vitamin-free conditions. Indeed, it showed a biochemical oxygen demand (BOD) removal rate that was 1.25-fold higher than that of the control. We considered that the improved BOD removal rate by applying SK-4 strain was based on its lipase activity and characteristics. Finally, we purified the lipase from SK-4 and found that the enzyme was quite stable under wide ranges of temperatures and pH, even in the presence of various metal ions and organic solvents. SK-4, therefore, is a promising bio-remediation agent for cleaning up unwanted milk fat curdles from dairy milk wastewater under low temperature conditions.
Introduction
Drainage from dairy parlors and milk factories produced in the process of cleaning transport pipes and milking tanks pollute rivers and groundwater with detergents, bactericides, mucus and milk fat are contaminating rivers and underground water [1]. In low temperature conditions, the wastewater is treated by bio-filters [2] and a reed bed system [3], [4]. However, the system is not used widely because of the high running cost and the necessity of a large space. Instead, an activated sludge system is now widely used for industrial treatment of dairy parlor wastewater [5] due to its advantages in maintenance and running cost. However, there is a problem in this system of low temperature conditions in winter having adverse effects on microbial functions.
The use of microorganisms living in polar regions for the purpose of removing nitrogen and phosphorus compounds from wastewater under low temperature conditions has been reported by Chevalier et al. [6] and Hirayama-katayama et al. [7], but it has not yet been applied for milk fat.
In our previous work, we examined 305 isolates of fungi including eight Ascomycetous and six Basidiomycetous species collected from Antarctica and found that they included fungi of the genus Mrakia, in psychrophilic Basidiomycetous yeast, suggesting that Mrakia is a major mycoflora highly adapted to the Antarctic environment (Fujiu, 2010; master's thesis in Graduate School of Science, Hokkaido University). Mrakia spp. and Mrakiella spp. are also common fungal species frequently found in cold climate areas such as Arctic, Siberia, Central Russia, the Alps and Antarctica [8]–[10]. Therefore, we screened our Mrakia isolates for their ability to decompose milk fat under low temperature conditions and evaluated their potential for application to an active sludge system in a region with a cold climate. The results showed that 56 Mrakia spp. exhibited a clear zone according to fat decomposition. Antarctic yeast strain SK-4 had physiological characteristics similar to those of Mrakia blollopis [11].
Here we report that activated sludge containing yeast strain SK-4 has the potential to remove milk fat BOD5. We also describe identification of yeast strain SK-4 and the purification and characterization of the lipase, considered as a major enzyme to degrade milk fat in wastewater.
Materials and Methods
Ethics statement
All necessary permits were obtained for the described field studies. Permission required for field studies was obtained from the Ministry of the Environment of Japan. Sample collection in Antarctica was performed with the permission of the Ministry of the Environment of Japan.
Sample isolation
Algal mat samples were collected from sediments of Naga-ike, a lake in Skarvsnes, located near Syowa station, East Antarctica. The isolate was inoculated on potato dextrose agar (PDA) (DifcoTM, BD Japan, Tokyo, Japan) at 4°C for 1 week. Yeast strain SK-4 was selectively picked for isolation on the basis of its morphology. Yeast strain SK-4 was maintained on PDA plates at 4°C and long-term storage was performed in 40% (w/v) glycerol at −80°C.
Phylogenetic analysis
Phylogenetic analysis was done by sequencing the ITS region including 5.8S rRNA and D1/D2 domain of 26S rRNA. Cells were harvested from 2-weeks-old cultures. DNA was extracted with an ISOPLANT II kit (Wako Pure Chemical Industries, Osaka, Japan) according to the manufacturer's protocol. Extracted DNA was amplified by PCR using KOD-plus DNA polymerase (TOYOBO, Osaka, Japan). The ITS region was amplified by using the following primers: ITS1F (5′-GTA ACA AGG TTT CCG T) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC). The D1/D2 domain was amplified using the following primers: NL1 (5′-GCA TAT CAA TAA GCG GAG GAA AAG) and NL4 (5′-GGT CCG TGT TTC AAG ACG G). Sequences were obtained with an ABI prism 3100 Sequencer (Applied Biosystems, Life Technologies Japan, Tokyo, Japan) using an ABI standard protocol. The ITS region and D1/D2 domain sequences of yeast strain SK-4 are deposited in DNA Data Bank of Japan (BBDJ) (Accession numbers AB630315 and AB691134). Alignment was made using CLUSTAL W (http://clustalw.ddbj.nig.ac.jp/) and corrected manually. Phylogenetic analysis was performed using MEGA software version 4.0 [12] with neighbor-joining analysis of the ITS region containing 5.8S rRNA and maximum parsimony analysis of the D1/D2 domain of 26S rRNA. Bootstrap analysis (1000 replicates) was performed using a full heuristic search.
Physiological characterization
Assimilation of carbon was performed at 15°C on modified Czapek-Dox agar composed by 6.7 g/L of yeast nitrogen base without amino acids (DifcoTM, BD Japan, Tokyo, Japan), 2.0 g/L of sodium nitrate (Wako Pure Chemical Industries, Osaka, Japan), 30 g/L of carbon source and 15.0 g/L of Agar (DifcoTM, BD Japan, Tokyo, Japan). Assimilation of nitrogen and other physiological tests were carried out according to the protocols described by Yarrow [13]. All tests were performed at 15°C after 2 and 4 weeks of inoculation.
Preparation of active sludge and measurement of biochemical oxygen demand
Activated sludge (AS) was cultivated at room temperature with aeration by using cow's milk as the substrate. After one month, the sludge was divided into two parts. One part of the activated sludge was mixed with M. blollopis SK-4 (1.4 g/L, dry weight), and the other part was used as a control. Separated activated sludge was prepared with Mixed Liquor Suspended Solids (MLSS, 3000 mg/L) and cow's milk at 10°C with aeration. One week later, prepared activate sludge was added to cow's milk, and biochemical oxygen demand (BOD5) of waste-treated water was measured after 24 hours. BOD5 assay was carried out using a coulometer (Ohkura Electric, Saitama, Japan).
Inoculum
M. blollopis SK-4 was grown in YPD liquid medium (1% yeast extract, 2% peptone, and 2% glucose) at 15°C for 96 hours at 120 rpm. After 96 hours, M. blollopis SK-4 was collected by centrifugation at 3500×g for 15 min at 4°C. The pellet was transferred to fresh cream liquid medium (0.5% peptone, 0.5% NaCl, 5% fresh cream, pH 7.0) and incubated at 10°C for 14 days at 90 rpm. The resulting culture was used as inoculum.
Lipase production medium
Lipase production medium was composed of 0.2% KH2PO4, 0.29% Na2PO4, 0.02% NH4Cl, 0.04% CaCl2, 0.001% FeCl3, 0.5% yeast extract, and 1% Tween 80. The yeast was cultivated at 10°C for 324 hours at 90 rpm. One mL samples were collected every 24 h and centrifuged at 4°C for 10 min at 20000×g, and then lipase activity was measured.
Assay of lipase activity
Lipase activity was measured by a colorimetric method using p-nitrophenyl-palmitate as a substrate [14]. Forty mL of 50 mM sodium phosphate buffer (pH 7.0) containing 50 mg gum arabic and 0.2 g TritonX-100 was mixed with 3 mL 2-propanol containing 1 mM p-nitrophenyl-palmitate. Eight hundred μL of prepared substrate was added to 200 μL of enzyme solution. The enzyme reaction was carried out at 30°C for 30 min. The released p-nitrophenol was measured at A410. One unit of lipase activity was defined as the activity required to release 1 μmol of free fatty acids per minute at 30°C.
Measurement of protein concentration
Protein concentration was measured by BCA protein assay reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions using bovine serum albumin as a standard.
Purification of lipase
M. blollopis SK-4 lipase was purified by ultrafiltration and Toyopearl-butyl 650 M (Tosho, Tokyo, Japan) hydrophobic interaction chromatography. Four hundred mL of lipase production liquid medium was centrifuged at 4°C for 15 min at 3000×g. The supernatant was filtered through a 0.45-μm of membrane filter (Advantec, Tokyo, Japan). The filtered medium was concentrated by ultrafiltration using an ultracel YM-30 membrane (Millipore, Billerica, MA, USA). The concentrated sample was adsorbed to a Toyopearl butyl 650 M column (2.5×20 cm) containing 1 M sodium chloride and eluted with a linear gradient from 750 mM to 100 mM sodium chloride in 20 mM Tris-HCl buffer (pH 8.5) at a flow rate of 60 mL/h. Fractions of high lipase activity was pooled and concentrated and then stored at 4°C until use. Protein molecular weight was estimated by SDS-PAGE according to Laemmli [15] and stained with CBB R-250. Precision plus protein unstained standards (Bio-Rad Laboratories Japan, Tokyo, Japan) were used as protein molecular weight makers.
Characterization of lipase
Substrate specificity was determined by using substrate as different p-nitrophenyl esters (C4–C18). For determining the effects of metal ions and EDTA on lipase activity, residual lipase activity assays were carried out under standard assay conditions with final concentrations of 1 mM of various bivalent metal ions and EDTA. Lipase activity assay in the absence of metal ions and EDTA was carried out as a control. Optimum pH was measured at 30°C for 30 min and determined at various pH values of 50 mM buffer as follows: sodium citrate (pH 3.0–5.0), sodium phosphate (pH 6.0, 7.0 and 8.0), Tris-HCl (pH 7.5 and 8.5), glycine-NaOH (pH 9.0) and sodium carbonate (pH 9.3, 9.5 and 10.0).
Optimum temperature was measured in 50 mM sodium phosphate buffer (pH 7.0) for 30 min. To determine the pH stability of lipase, the enzyme was preincubated in various buffers for 15 h at 30°C and then adjusted to pH 8.5. The residual enzyme activity was measured by using p-nitrophenyl-palmitate as a substrate at 65°C for 30 min. For determining thermo-stability, lipase was preincubated for 30 min at different temperatures and the residual activity was measured at 65°C for 30 min in 50 mM Tris-HCl (pH 8.5). Effects of organic solvents on lipase activity were determined at 65°C for 30 min in 50 mM Tris-HCl (pH 8.5) containing various organic solvents at final concentration of 5% (v/v). Lipase activity assay in the absence of organic solvents was carried out as a control.
Results
Phylogenetic analysis
As a result of phylogenetic analysis of the ITS region and D1/D2 domain, yeast strain SK-4 was grouped with the clade of Mrakia blollopis CBS 8921T (Fig. 1 A and B). By comparison of the ITS region sequence containing 5.8 S rRNA, yeast strain SK-4 showed high homologies (>99.6%) with M. blollopis CBS8921T. The D1/D2 domain sequence showed no variation with M. blollopis CBS 8921T.
Figure 1. Phylogenetic tree of Mrakia blollopis SK-4 and related species.
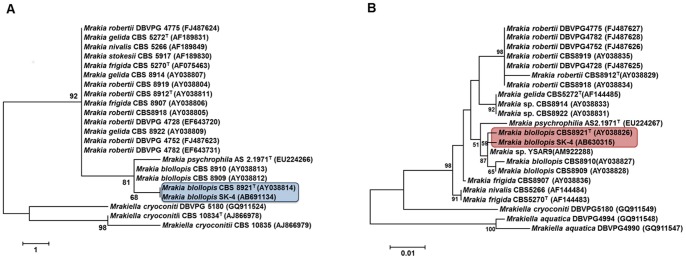
(A) Maximum parsimony analysis of the D1/D2 domain of the 26 rRNA gene sequence. Bootstrap percentages from 1000 replications are shown on the branches. Mrakiella cryoconiti CBS 10834T and Mrakiella cryoconiti CBS 10835 were used as an out group. (B) Neighbor-joining tree of the ITS region containing the 5.8 S rRNA gene sequence. Bootstrap percentages from 1000 replications are shown on the branches. Mrakiella aquatica DBVPG4994 and Mrakiella aquatica DBVPG4990 were used as an out group.
Physiological characterization
Results of assimilation of carbon compounds and other physiological tests of M. blollopis SK-4 are shown in Table 1 with its type strain and related species. Test data for M. blollopis SK-4 are compared with those for M. blollopis CBS8921T [8], M. psychrophila AS2.1971T [16], and M. frigida CBS5270T [17]. Maximum growth temperature of M. blollopis SK-4 was 22°C. Maximum growth temperatures of other related species were lower than 20°C. M. blollopis SK-4 differed from the other strains in substrate utilization as well. The strain could thrive well on lactose, D-arabinose, and inositol medium. Unlike other strains, this strain also grew on vitamin-free medium. A comparison of fermentabilities showed that M. blollopis SK-4 could ferment typical sugars such as glucose, sucrose, galactose, maltose, lactose, raffinose, trehalose and melibiose, while other related species were not able to strongly ferment such as various sugars (Table 1).
Table 1. Comparison of physiological characteristics of Mrakia blollopis SK-4 and other Mrakia species.
| Characteristic | M. blollopis SK-4 | M. blollopis CBS8921T | M. psychrophhila AS2.1971T | M. frigida CBS5270T |
| Maximum growth temperature | 22°C | 20°C | 18°C | 17°C |
| Assimilation of | ||||
| Lactose | + | W | + | V |
| Inositol | + | w/+ | + | V |
| D-arabinose | + | w/− | + | V |
| Ethanol | w/− | + | + | + |
| Growth on 50% glucose | w/− | − | + | − |
| Growth on vitamin-free medium | + | W | + | − |
| Fermentation of | ||||
| Galactose | + | − | − | W |
| Lactose | + | − | − | − |
| Raffinose | + | − | − | W |
| Maltose | + | − | − | W |
Main physiology test results for characteristics of M. blollopis SK-4 and related species are shown. Physiological data were taken from Fell et al. (1969), Xin and Zhou (2007), Thomas-Hall et al. (2010) and this study. +, positive; w, weak; −, negative; v, variable; nd, no data.
Assessment of milk fat decomposition in model wastewater
Model wastewater containing cow's milk as a substrate of milk fat was prepared as an equivalent of BOD sludge loading in standard waste water treatment (0.35 kg-BOD/kg-MLSS·day). Activated sludge containing M. blollopis SK-4 had a BOD removal rate of 83.1%, higher than that in the control (63.8%, Fig. 2A). When BOD volume load was adjusted to 1.5 fold of standard wastewater treatment (0.52 kg-BOD/Kg-MLSS·day), BOD removal rate by activated sludge containing M. blollopis SK-4 was 80.1%, higher that in the control (65.2%, Fig. 2B). Regardless of BOD volume load, activated sludge containing M. blollopis SK-4 had a 1.25-fold higher BOD removal rate than that of the control.
Figure 2. Performance of activated sludge system treating model milking parlor wastewater.
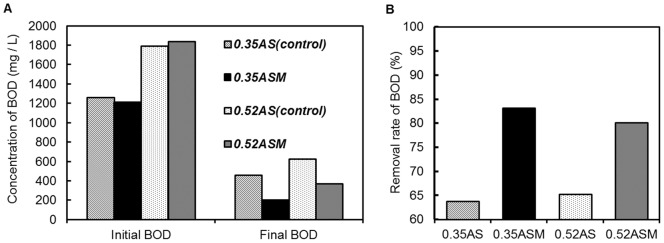
(A) Results of measurement of initial BOD and final BOD. (B) Effect of BOD removal rate by ASM treatment. (0.35AS) for 0.35 BOD loading rate of model parlor wastewater treated by activated sludge (control), (0.35ASM) for 0.35 BOD loading rate of model parlor wastewater treated by activated sludge containing M. blollopis SK-4, (0.52AS) for 0.52 BOD loading rate of model parlor wastewater treated by activated sludge (control) and (0.52ASM) for 0.52 BOD loading rate of model parlor wastewater treated by activated sludge containing M. blollopis SK-4. Abbrebiations for Figure 3 are biochemical oxygen demand (BOD), activated sludge (AS) and activated sludge containing Mrakia blollopis SK-4 (ASM).
Production of lipase from Mrakia blollopis SK-4
Many microorganisms are known to produce lipase using Tween 80 as a substrate [18], [19]. Basidiomycetous yeast M. blollopis SK-4 also produced the enzyme. It is known that the production of lipase from Candida rugosa increased when yeast extract was used as a nitrogen source [20]. The same results as those for M. blollopis SK-4 were obtained. When 0.5% (w/v) yeast extract was used as a nitrogen source in the medium, lipase production by M. blollopis SK-4 lipase increased dramatically after 180 h. Maximum lipase activity of 0.695 U/mL was obtained with 0.5% (w/v) yeast extract, as compared to 0.059U/mL in its absence, after 324 h of inoculation, after which the accumulation of lipase markedly decreased (Spp. Fig. S1).
M. blollopis SK-4 morphology was observed by the fluorescence in situ hybridization (FISH) method during secretion of lipase. Therefore, all of the morphology of M. blollopis SK-4 during secretion of lipase was yeast form (data not shown).
Purification of lipase
Lipase production medium was centrifuged at 3000×for 15 min at 4°C. The supernatant was filtered through a 0.45-μm membrane filter. The filtered solution was concentrated by ultrafiltration. After ultrafiltration, 72.3% of total lipase activities were recovered and 2.2-fold lipase specific activity was obtained. Then, enzyme solution was applied on a Toyopearl butyl-650 M column (2.5×20 cm) and purified by single-step hydrophobic interaction chromatography. Finally, 9.4% of the enzyme was recovered and 20.1-fold of specific activity, compared to crude sample, was obtained with a specific activity of 51.7 U/mg (Table 2). The purified enzyme showed a single band on SDS-PAGE with a molecular mass of 60 kDa (Supp. Fig. S2).
Table 2. Purification of lipase from Mrakia blollopis SK-4.
| Purification step | Total protein (mg) | Total activity (U) | Specific activity (U/mg) | Recovery (%) | Fold |
| Supernatant of culture medium | 108.0 | 278.0 | 2.5 | 100.0 | 1.0 |
| Ultrafilter concentration | 32.2 | 200.9 | 6.2 | 72.3 | 2.2 |
| Toyopearl Butyl-650M | 0.5 | 26.2 | 51.7 | 9.4 | 20.1 |
Characterization of lipase
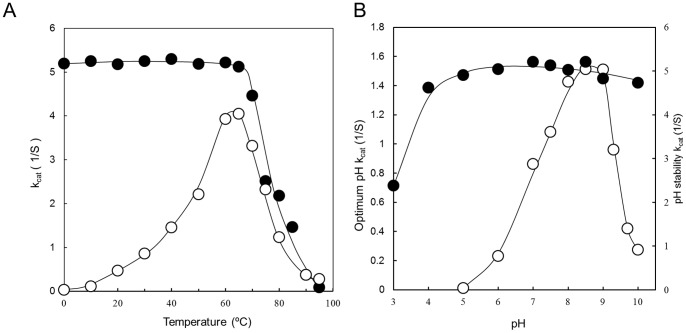
Optimum temperature of lipase activity was 60−65°C (kcat = 3.93 and 4.04 S−1). At temperatures of 80°C and 95°C, 30.5% (kcat = 1.23 S−1) and 6.8% (kcat = 0.27 S−1) of enzyme activity were retained (Fig. 3A). The enzyme showed thermo-stability up to 65°C with 98.3% (kcat = 5.12 S−1) of residual enzyme activity even after 30-min preincubation. At 75°C, 80°C and 85°C, 48.2% (kcat = 2.51 S−1), 41.8% (kcat = 2.18 S−1) and 28.03% (kcat = 1.46 S−1) of enzyme activity remained after 30-min preincubation (Fig. 3A). Optimum pH range of lipase activity was between pH 8.0 and pH 9.0 (kcat = 1.43 and 1.51 S−1), whereas 57.0% (kcat = 0.86 S−1), 63.5% (kcat = 0.96 S−1) and 27.8% (kcat = 0.42 S−1) of the enzyme activity was retained at pH 7.0, pH 9.3 and pH 9.7 compared to 100% at pH 8.5 (Fig. 3B). The enzyme was quite stable over a wide pH range (4.0−10.0) and 45.5% (kcat = 2.38 S−1) of the enzyme activity remained at pH 3.0 even after preincubation for 15 h at 30°C (Fig. 3B). Enzyme activity was affected by metal ions at a concentration of 1 mM, retaining relative activity higher than 80%. There was little inhabitation of lipase activity in the presence of Cu2+ and Pb2+ ions. The metal-chelating agent EDTA did not affect lipase activity (Table 3). One of the most important characteristics of lipase was substrate specificity toward various p-nitrophenyl esters (C4−C18). The substrate specificity was determined in the presence of 1 mM p-nitrophenyl esters as a substrate with 50 mM sodium phosphate (pH 7.0) at 30°C for 30 min. Relative activity toward C4−C14 was more than 100% compared to 100% toward p-nitrophenyl-palmitate. C18 had relative activity of 71.6% (Fig. 4). Various organic solvents (ethanol, methanol, diethyl ether, dimethyl sulfoxide, hexane and N, N- dimethyl formamide) enhanced SK-4 lipase acidity. Solvents such as acetone and chloroform however, had only a slight inhibitory effect in the activity, with relative activities of 77.9% and 70.4%, respectively (Table 3). Organic solvents are known to be severely toxic to most enzymes including lipase [21].
Figure 3. Effects of temperature and pH on SK-4 lipase.
(A) Effects of temperature on lipase activity (○) and thermostability of the lipase (•). For effect of temperature, lipase activity assay was performed at various temperatures in 50 mM sodium phosphate buffer (pH 7.0) for 30 min. For thermostability, lipase was preincubated at various temperatures for 30 min. Remaining lipase activity was examined at 65°C for 30 min in 50 mM Tris-HCl buffer (pH 8.5). The line was fitted by using the Eyring-Arrhenius equation. (B) Effects of pH on lipase activity (○) and pH stability of lipase (•). For effect of pH, lipase activity assay was performed with various buffers at 30°C for 30 min using p-nitrophenyl-palmitate as a substrate. For pH stability, lipase was preincubated in various pH buffers at 30°C for 15 h and then pH of the buffer was adjusted to 8.5. The remaining enzyme activity was examined at 65°C for 30 min using p-nitrophenyl-palmitate as a substrate. The line was fitted by Henderson- Hasselbalch equation.
Table 3. Comparison of the effects of various metal ions, EDTA and various organic solvents on M. blollops SK-4 and Cryptococcus sp. S-2 lipase activity.
| Divalent cation or EDTA or Organic solvents | Relative activity (%) | |
| M. blollopis SK-4 | Cryptococcus sp. S-2 | |
| None | 100.0 | 100.0 |
| EDTA | 106.7 | ND |
| Metal salts | ||
| MgCl2 | 102.2 | >70 |
| MnCl2 | 101.6 | >70 |
| FeCl2 | 90.0 | >70 |
| ZnCl2 | 83.6 | ND |
| CoCl2 | 85.9 | ND |
| CaCl2 | 112.8 | >70 |
| PbCl2 | 61.6 | ND |
| CuCl2 | 67.2 | <70 |
| Ethanol | 107.5 | ND |
| Methanol | 112.3 | 53.3 |
| Diethyl ether | 113.1 | 116.7 |
| 2– Propanol | 95.5 | ND |
| Dimethyl sulfoxide (DMSO) | 134.3 | 121.4 |
| Hexane | 104.9 | 95.2 |
| Acetone | 77.9 | 83.3 |
| Chloroform | 70.4 | 74.8 |
| N, N- dimethylformamide (DMF) | 122.3 | 92.4 |
Figure 4. Substrate specificity of lipase form Mrakia blollopis SK-4.
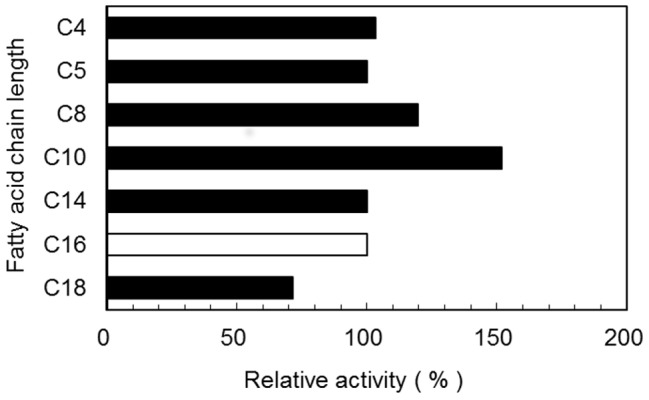
Substrates were p-nitrophenyl esters. Relative activity for each substrate (▪) is expressed as the percentage of activity toward p-nitrophenyl-palmitate (□).
Discussion
The Antarctic yeast strain SK-4, which we identified as M. blollopis, showed several uniques characteristics; for example, it can use various carbon sources as nutrition, it prefers relatively high temperature conditions among allied species and it can be activated even in vitamin-free conditions. These results suggested that strain SK-4 is a potential candidate for a biological agent to decompose various sugars in milk parlor wastewater under low temperature conditions. This expectation was further reconfirmed by the application of strain SK-4 in AS; namely, addition of SK-4 to AS in the model parlor wastewater improved the BOD removal rate.
Improved BOD removal rate of the activated sludge was attributed to lipase activity of SK-4 added. As expected, the lipase purified from M. blollopis SK-4 showed clearly weaker activity at lower temperatures but stronger activity in middle to high temperature conditions compared to activities of those from the other psychrophilic fungi [22]. M. blollopis SK-4 lipase, in addition, was quite stable in wide ranges of temperature and pH conditions and was not affected by the existence of EDTA, various metals ions, or organic solvents. M. blollopis SK-4 is thought to have acquired stable lipases by growing in extreme environments such as the Antarctica.
Comparison of the lipase from M. blollopis SK-4 with that from Cryptococcus sp. S-2, which has actually been used in wastewater treatment [23], [24], revealed that the former was superior to the latter both in thermo-stability and pH stability (Table 4); i.e., the lipase from M. blollopis SK-4 retained 71.1% of the enzyme activity after 30 min at 60°C and was stable for 6 hrs at 30°C in the pH range from 5.0 to 9.0.
Table 4. Comparison of Mrakia blollopis SK-4 lipase characteristics with other yeast lipase characteristics.
| Yeasts | MW in kDa | Optimum pH | pH stability range | Optimum temperature |
| Mrakia blollopis SK-4 | 60 | 8.0∼9.0 | 4.0∼10.0 | 60–65°C |
| Candida antarctaca | 43 (lipaseA) | 7.0 | - | 50°C |
| Candida rugosa ATCC1483 | 60 | 5.0 | - | - |
| Cryptococcus sp. S-2 | 22 | 7.0 | 5.0-9.0 | 37°C |
| Geotrichum candidum | 55 | 5.6∼7.0 | 4.2∼9.8 | 40°C |
| Yarrowia lipolytica | 44 (lip) | 8.0 (lip) | 4.5∼8.0 | 37°C |
| Kurtzmanomyces sp. L-11 | 49 | 1.9∼7.2 | Below 7.1 | 75°C |
| Trichosporon asteroids | 37 | 5.0 | 3.0∼10.0 | 50°C |
In conclusion, M. blollopis SK-4 has the ability to assimilate various carbon compounds and to use various sugars for fermentation. Moreover, the lipase is more tolerant in relatively higher temperature conditions and wider pH ranges, less sensitive to various metal ions and organic solvents, and highly reactive to various chain lengths of substrates. SK-4 lipase, therefore, is a promising biological agent for parlor wastewater treatment even in low temperature regions of the world.
Supporting Information
Effect of yeast extract on lipase production. Mrakia blollopis SK-4 was cultivated by lipase production medium (♦) and lipase production medium without yeast extract (•).
(TIF)
SDS-PAGE of purified lipase from Mrakia blollopis SK-4. Lane 1. Molecular weight marker; 2. Purified lipase.
(TIF)
Acknowledgments
The authors thank Dr. Yoshitomo Kikuchi, researcher AIST and faculty member of the Graduate School of Agriculture, Hokkaido University, for useful technical advice. The authors are also grateful to Dr. SM Singh, Scientist D of the National Centre for Antarctic & Ocean Research, Ministry of Ocean Development (NCAOR) and Dr. Teruo Sano, Professor in the Faculty of Agriculture and Life Science, Hirosaki University for useful suggestions.
Funding Statement
The authors have no support or funding to report.
References
- 1. Healy MG, Rodgers M, Mulqueen J (2007) Treatment of dairy wastewater using constructed wetland and intermittent sand filter. Bioresour Technol. 98: 2268–2281. [DOI] [PubMed] [Google Scholar]
- 2. Shah SB, Bhumbla DK, Basden TJ, Lawrence LD (2002) Cool temperature performance of a wheat straw biofilter for treating dairy wastewater. J Environ Sci Health B. 37: 493–505. [DOI] [PubMed] [Google Scholar]
- 3.Biddlestone AJ, Gray KR, Job GD (1991) Treatment of dairy farm wastewaters in engineered reed bed systems. Process Biochem. 26, 265–268.
- 4.Kato K, Ietsugu H, Koba T, Sasaki H, Miyaji N, et al. (2010) Design and performance of hybrid reed bed systems for treating high content wastewater in the cold climate., 12th International Conference on Wetland Systems for Water Pollution Control. http://reed-net.com/pdf/katoICWS2010.pdf. Accessed 1August 2012.
- 5. Ying C, Umetsu K, Ihara I, Sakai Y, Yamashiro T (2010) Simultaneous removal of organic matter and nitrogen form milking parlor wastewater by a magnetic activated sludge (MAS) process. Bioresour Technol. 101: 4349–4353. [DOI] [PubMed] [Google Scholar]
- 6. Chevalier P, Proulx D, Lessard P, Vincent WF, de la Noue J (2000) Nitrogen and phosphorus removal by high latitude mat-forming cyanobacteria for potential use in tertiary wastewater treatment. J Appl Phycol. 12: 105–112. [Google Scholar]
- 7. Hirayama-Katayama K, Koike Y, Kobayashi K, Hirayama K (2003) Removal of nitrogen by Antarctic yeast cells at low temperature. Polar Bioscience. 16: 43–48. [Google Scholar]
- 8. Thomas-Hall SR, Turchetti B, Buzzini P, Branda E, Boekhout T, et al. (2010) Cold-adapted yeasts from Antarctica and Italian alps-description of three novel species: Mrakia robertii sp. nov., Mrakia blollopis sp. nov. and Mrakiella niccombsii sp. nov. Extremophiles. 14: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bab'eva IP, Lisichkina GA, Reshetova IS, Danilevich VN (2002) Mrakia curviuscula sp. nov.: A New psychrophilic yeast from forest substrates. Microbiology. 71: 449–454. [PubMed] [Google Scholar]
- 10. Margesine R, Fell JW (2008) Mrakiella cryoconiti gen. nov., sp. nov., a psychrophilic, anamorphic, basidiomycetous yeast from alpine and arctic habitats. Int J Syst Evol Microbiol. 58: 2977–2982. [DOI] [PubMed] [Google Scholar]
- 11. Shimohara K, Fujiu S, Tsuji M, Kudoh S, Hoshino T, et al. (2012) Lipolytic activities and their thermal dependence of Mrakia species, basidiomycetous yeast from Antarctica. J water waste. 54: 691–696. [Google Scholar]
- 12.Tamura K, Dudley J, Nei M, Kumar S (2007) Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 24; 1596–1599. [DOI] [PubMed]
- 13.Yarrow D (1998) Methods for the isolation and identification of yeasts. In: Kurtzman CP, Fell JW (eds) The yeast, a taxonomic study 4th edn,. Elsevier, Amsterdam, pp 77–100.
- 14. Berekaa MM, Zaghloul TI, Abdel-Fattah YR, Saeed HM, Sifour M (2009) Production of a novel glycerol-inducible lipase from thermophilic Geobacillus steaorothermophilus strain-5. World J Microbiol Biotechnol. 25: 287–294. [Google Scholar]
- 15. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 16. Xin M, Zhou P (2007) Mrakia psychrophila sp. Nov., a new species isolated from Antarctic soil. J Zhejiang Univ-Sc B 8(4): 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fell JW, Statzell AC, Hunter IL, Phaff HJ (1969) Leucosporidium gen. n., the heterobasidiomycetous stage of several yeasts of the genus Candida . Anton Leeuw Int J G 35: 433–462. [PubMed] [Google Scholar]
- 18. Li CY, Cheng CY, Chen TL (2001) Production of Acinetobacter radioresistens lipase using Tween 80 as the carbon source. Enzyme Microb Technol. 29: 258–26318. [Google Scholar]
- 19. Taoka Y, Nagano N, Okita Y, Izumida H, Sugimoto S, et al. (2011) Effect of Tween 80 on the growth, lipid accumulation and fatty acid composition of Thraustochytrium aureum ATCC 34304. J Biosci Bioeng. 111: 420–424. [DOI] [PubMed] [Google Scholar]
- 20. Fadilog?lu S, Erkmen O (2002) Effect of carbon and nitrogen sources on lipase production by Candida rugosa. Turk J Eng Env Sci. 26: 249–254. [Google Scholar]
- 21. Iizumi T, Nakamura K, Fukase T (1990) Purification and characterization of a thermostable lipase from newly isolated Pseudomonas sp. KWI-56. Agric Biol Chem. 54: 1253–1258. [Google Scholar]
- 22. Hoshino T, Tronsmo AM, Matsumoto N, Sakamoto T, Ohgiya S, et al. (1997) Purification and characterization of a lipolytic enzyme active at low temperature from Norwegian Typhula ishikariensis groupIII strain. Eur J plant Pathol. 103: 357–361. [Google Scholar]
- 23. Iefuji H, Iimura Y, Obata T (1994) Isolation and characterization of a yeast Cryptococcus sp. S-2 that produces raw starch-digesting α-amylase, xylanase and polygalacturonase. Biosci Biotechnol Biochem. 58: 2261–2262. [Google Scholar]
- 24. Kamini NR, Fujii T, Kurosu T, Iefuji H (2000) Production, purification and characterization of an extracellular lipase from the yeast, Cryptococcus sp. S-2. Process Biochem. 36: 317–324. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of yeast extract on lipase production. Mrakia blollopis SK-4 was cultivated by lipase production medium (♦) and lipase production medium without yeast extract (•).
(TIF)
SDS-PAGE of purified lipase from Mrakia blollopis SK-4. Lane 1. Molecular weight marker; 2. Purified lipase.
(TIF)