Abstract
Impaired brain glucose uptake and metabolism precede the appearance of clinical symptoms in Alzheimer disease (AD). Neuronal glucose transporter 3 (GLUT3) is decreased in AD brain and correlates with tau pathology. However, what leads to the decreased GLUT3 is yet unknown. In this study, we found that the promoter of human GLUT3 contains three potential cAMP response element (CRE)-like elements, CRE1, CRE2 and CRE3. Overexpression of CRE-binding protein (CREB) or activation of cAMP-dependent protein kinase significantly increased GLUT3 expression. CREB bound to the CREs and promoted luciferase expression driven by human GLUT3-promoter. Among the CREs, CRE2 and CRE3 were required for the promotion of GLUT3 expression. Full-length CREB was decreased and truncation of CREB was increased in AD brain. This truncation was correlated with calpain I activation in human brain. Further study demonstrated that calpain I proteolysed CREB at Gln28–Ala29 and generated a 41-kDa truncated CREB, which had less activity to promote GLUT3 expression. Importantly, human brain GLUT3 was correlated with full-length CREB positively and with activation of calpain I negatively. These findings suggest that overactivation of calpain I caused by calcium overload proteolyses CREB, resulting in a reduction of GLUT3 expression and consequently impairing glucose uptake and metabolism in AD brain.
INTRODUCTION
One of the main features of Alzheimer’s disease (AD) is the severe reduction of glucose uptake and metabolism in the specific brain areas (1–4). In vivo imaging demonstrates consistent and progressive reduction of cerebral metabolic rate for glucose in AD patients (5–8). This reduction correlates with the severity of clinical symptom of AD (9–11). Importantly, glucose metabolic reduction has been observed before the onset of the disease in several groups of at risk individuals, including patients with mild cognitive impairment (MCI) (12), pre-symptomatic individuals carrying mutations for early-onset familial AD (13–15), normal and middle-aged carriers of apolipoprotein E4 (16–20) and in those with a maternal family history of AD (21,22). These studies strongly suggest that the impairment of glucose uptake/metabolism is a cause of neurodegeneration or is mechanistically involved in the pathogenesis of AD.
It is well known that neurons mainly depend on glucose as a fuel for providing energy and glucose cannot be synthesized by or stored in the neuron. Brain neurons take glucose from blood stream. However, glucose cannot pass through the blood–brain barrier or cell membrane freely and requires glucose transporters (GLUTs) to assistant the transport. To date, 14 members of GLUTs have been reported in the human tissue (23). Among them, GLUT3 is the major neuronal GLUT and determines the efficiency of glucose transport into the neuron (24). The level of GLUT3 correlates positively with regional cerebral glucose utilization (25–27). GLUT3, different from other GLUTs, has a higher affinity for glucose and at least 5-fold greater transport capacity (28), which allows it to transport glucose effectively even when its level is very low. In AD brain, GLUT3 level is decreased (29,30) and correlated with the extent of hyperphosphorylation of tau and with the density of neurofibrillary tangles (NFTs) in AD (31). However, what leads to the decrease in GLUT3 in AD brain remains elusive.
It has been reported that the expression of the mouse GLUT3 gene may be regulated by several transcription factors, including HIF-1 (32), Sp1, Sp3 (33,34) and cAMP response element (CRE)-binding protein (CREB) (34). However, the regulation of human GLUT3 expression is not well understood. To understand the mechanism of GLUT3 expression, we analyzed the promoter of human GLUT3 and found that it contains three CRE-like elements. CREB is originally described as a transcription factor that binds to an 8-bp element, TGACGTCA, known as a CRE, in the somatostatin gene promoter (35). Upon binding to CRE, CREB regulates the expression of target genes. CREB is composed of a C-terminal promoter-binding domain and an N-terminal transcription regulation domain, in which PKA (cAMP-dependent protein kinase) phosphorylates Ser133 (36). Importantly, Ser133 phosphorylation is required for its activity to regulate gene expression (37).
In this study, we investigated the regulation of GLUT3 expression by CREB and found that CREB bound to the promoter region of human GLUT3 and regulated its expression. In AD brain, CREB was truncated due to proteolysis by calpain I and the truncated form of CREB had less activity to promote GLUT3 expression, which resulted in a reduction of GLUT3 expression, leading to impaired glucose uptake and metabolism. These results provide a novel insight into the pathogenesis of AD.
MATERIALS AND METHODS
Human brain tissues
Frontal cortices from seven AD and seven age-matched normal human brains used for this study (Table 1) were obtained from the Sun Health Research Institute Donation Program (Sun City, AZ, USA). All brain samples were confirmed histopathologically and stored at −70°C until used. The use of frozen human brain tissue was in accordance with the National Institutes of Health guidelines and was approved by our institute’s institutional review committee.
Table 1.
Human brain tissue of AD and control (Con) cases used in this study
| Case | Age at death (years) | Gender | PMIa (h) | Braak stageb | Tangle scoresc |
|---|---|---|---|---|---|
| AD 1 | 89 | F | 3 | V | 14.5 |
| AD 2 | 80 | F | 2.25 | VI | 14.5 |
| AD 3 | 85 | F | 1.66 | V | 12.0 |
| AD 4 | 78 | F | 1.83 | VI | 15.0 |
| AD 5 | 95 | F | 3.16 | VI | 10.0 |
| AD 6 | 86 | M | 2.25 | VI | 13.5 |
| AD 7 | 91 | F | 3 | V | 8.50 |
| Mean ± SD | 86.29 ± 5.99 | 2.45 ± 0.61 | 12.57 ± 2.51 | ||
| Con 1 | 85 | M | 25 | II | 4.25 |
| Con 2 | 86 | F | 2.5 | III | 5.00 |
| Con 3 | 81 | M | 2.75 | III | 6.41 |
| Con 4 | 88 | F | 3 | II | 2.00 |
| Con 5 | 90 | F | 3 | III | 4.50 |
| Con 6 | 88 | F | 3.5 | III | 2.50 |
| Con 7 | 88 | F | 3 | IV | 4.50 |
| Mean ± SD | 86.6 ± 2.9 | 2.89 ± 0.39 | 4.17 ± 1.50 |
aPMI, post-mortem interval.
bNeurofibrillary pathology was staged according to Braak and Braak (38).
cTangle score was a density estimate and was designated as none, sparse, moderate or frequent (0, 1, 2 or 3 for statistics), as defined according to CERAD AD criteria (39). Five areas (frontal, temporal, parietal, hippocampal and entorhinal) were examined and the scores were combined for a maximum of 15.
Plasmids, proteins and antibodies
Mammalian expression vectors pCI/CREB tagged with HA (hemagglutinin) at N-terminus were constructed and their sequences were confirmed. pGL3-basic, pRL-Tk (thymidine kinase promoter driven Renilla luciferase) and dual luciferase assay kit were bought from Promega (Madison, WI, USA). Luciferase driven by different length or site-mutated promoter of human GLUT3 in pGL3-basic was constructed and confirmed by sequencing. Calpain I, polyclonal anti-calpain I, monoclonal anti-HA, anti-CREB1 and anti-α-tubulin were bought from Sigma (St. Louis, MO, USA). Polyclonal anti-CREB (middle region) and anti-pS133-CREB were from Cell Signaling Technology (Danvers, MA, USA). Maltose-binding protein (MBP)-CREB, Magna CHIP™A/G kit, polyclonal anti-CREB (against amino acid 5–24) and monoclonal anti-PP2A catalytic subunit were purchased from Millipore/Merck KgaA (Darmstadt, Germany). Polyclonal anti-GLUT3 and anti-GAPDH were purchased from Santa Cruz (Santa Cruz, CA, USA). RL2 was bought from Affinity BioReagents (Golden, CO, USA). Peroxidase-conjugated anti-mouse and anti-rabbit IgG were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). ECL Kit was from Thermo Scientific (Rockford, IL, USA), and [γ-32P]ATP was from Amersham Biosciences (Piscataway, NJ, USA).
Cell culture and transfection
HEK-293T cells (ATCC, Manassa, VA, USA) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) at 37°C. SH-SY5Y cells were maintained in DMEM/F12 supplemented with 10% fetal bovine serum at 37°C. Differentiation of SH-SY5Y cells (ATCC) was induced by 10 µM all-trans retinoid acid (ATRA) for 3 days (for transfection) or 7 days (for forskolin treatment). All transfections were performed in triplicate with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) or FuGENE HD (Roch Diagnostics, Indianapolis, IN, USA) according to manufacturer’s manuals.
Reverse-transcription PCR and real-time quantitative PCR
Total RNA was isolated from the frontal cortical or from cultured cells using the RNeasy Mini kit (Qiagen, Valencia, CA, USA) and used for first-strand cDNA synthesis with Oligo-(dT)15–18 by using the RT2 First-Strand Kit (Invitrogen) according to the manufacturer’s instructions. The cDNA of GLUT3 or GAPDH was amplified by PCR using PrimeSTARTM HS DNA Polymerase (Takara Bio Inc., Otsu, Shiga, Japan) at 98°C for 3 min, 98°C for 10 s and at 68°C for 40 s for 30 cycles and then at 68°C for 10 min for extension. The PCR products were resolved on 1.5% agarose gels and visiblized by using the Molecular Imager system (Bio-Rad, Hercules, CA, USA).
For quantitative PCR (qPCR), cDNA of GLUT3, brain-derived neurotrophic factor (BDNF) and GAPDH were amplified by using Brilliant II SYBR® Green QPCR Master Mix (Agilent Technologies, Inc. Santa Clara, CA, USA) in a Agilent Mx3000p PCR detection system under the condition: 95°C for 10 min, 95°C for 30 s and 60°C for 1 min, for 40 cycles. Relative levels of target mRNAs were calculated by the comparative CT (threshold cycle) method (2−ΔΔCT). The PCR specificity was examined by 3% agarose gel using 5 µl from each reaction. The primers used for this study are as follows: GLUT3 forward 5′TCCCCTCCGCTGCTCACTATTT3′ and reverse 5′ATCTCCATGA CGCCGTCCTTTC3′; BDNF forward 5′GCCTTTGGAGCCTCCTCTTCTC3′ and reverse 5′TTTTT GTCTGCCGCCGTTACCC3′; GAPDH forward 5′GGTGGTCTCCTCTGACTTCAACA3′ and reverse 5′GTTGCTGTAGCCAAATTCGTTGT3′.
Plasmid construction of luciferase reporter driven by human GLUT3 promoter and luciferase assay
A ∼2.2-kb fragment of the human GLUT3 genomic DNA from −2000 to +241 bp was amplified by PCR and cloned into pGL3-basic (Promega) by Kpn I and Xho I (New England Biolabs, Ipswich, MA, USA) to generate pGL3/GLUT3−2000. Subsequently, serial deletion mutations and site mutations of CRE2 and CRE3 of pGL3/GLUT3 constructs were generated by using a PCR-based strategy employing primers listed in Table 2. The sequence and orientation of the individual clones were confirmed by DNA sequence analysis.
Table 2.
List of primers used for generating mutated promoter of GLUT3 constructs
| Construct | Forward primer | Reverse primer |
|---|---|---|
| pGL3/GLUT3−1580 | 5′cggggtaccatattaataatcttttccagttatg3′ | 5′ccgctcgagcgctgtaatctaattcaagtcttcaagaaag3′ |
| pGL3/GLUT3−1000 | 5′cggggtaccagaattcagaatgggtgatatggc3′ | 5′ccgctcgagcgctgtaatctaattcaagtcttcaagaaag3′ |
| pGL3/GLUT3−500 | 5′cggggtaccctcaggctgtctttccctcccct3′ | 5′ccgctcgagcgctgtaatctaattcaagtcttcaagaaag3′ |
| pGL3/GLUT3−400 | 5′cggggtaccctaacaaccttaaatctctgat3′ | 5′ccgctcgagcgctgtaatctaattcaagtcttcaagaaag3′ |
| pGL3/GLUT3−300 | 5′cggggtacctgcgagagcataaaatagtgt3′ | 5′ccgctcgagcgctgtaatctaattcaagtcttcaagaaag3′ |
| pGL3/GLUT3−200 | 5′cggggtaccggagtaaggatgagcttttg3′ | 5′ccgctcgagcgctgtaatctaattcaagtcttcaagaaag3′ |
| pGL3/GLUT3−126 | 5′cggggtacctaagagaggggggagggagggcgt3′ | 5′ccgctcgagcgctgtaatctaattcaagtcttcaagaaag3′ |
| pGL3/GLUT3−85 | 5′cggggtaccgggcgggggtagttctgataac3′ | 5′ccgctcgagcgctgtaatctaattcaagtcttcaagaaag3′ |
| pGL3/GLUT3+1 | 5′cggggtaccgtggggtggggtggggctgggggct3′ | 5′ccgctcgagcgctgtaatctaattcaagtcttcaagaaag3′ |
| pGL3/GLUT3+122 | 5′cggggtaccgctgctgagaaggacattttgaag3′ | 5′ccgctcgagcgctgtaatctaattcaagtcttcaagaaag3′ |
| pGL3/GLUT3M1 | 5′gggagggagggcaatattgtctgtgg3′ | 5′ccacagacaatattgccctccctccc3′ |
| pGL3/GLUT3M2 | 5′agggcgttattgaatgtggggcgggg3′ | 5′ccccgccccacattcaataacgccct3′ |
| pGL3/GLUT3M3 | 5′gagaggggggagttagggcgttattg3′ | 5′caataacgccctaactcccccctctc3′ |
| pGL3/GLUT3M4 | 5′aggggggagggagttcgttattgtc3′ | 5′gacaataacgaactccctcccccct3′ |
| pGL3/GLUT3M5 | 5′ctttggatccttcctttggacgtggagaaaacttgctgctgagaag3′ | 5′gttttctccacgtccaaaggaaggatccaaagtctcaccattacag3′ |
| pGL3/GLUT3M6 | 5′gatccttcctgaggaaatggagaaaacttgctgctgagaaggacat3′ | 5′agcaagttttctccatttcctcaggaaggatccaaagtctcacca3′ |
HEK-293T cells were co-transfected with pCI/CREB, pGL3/GLUT3 or their control vectors and pRL-Tk for 48 h. SH-SY5Y cells were transfected with these plasmids under differentiation condition (culture medium containing 10 µM ATRA). The cells were lysed using 0.1 ml of passive lysis buffer (Promega). The luciferase activity was measured by the dual luciferase assay kit (Promega) according to manufacturer’s manuals. The firefly luciferase activity and Renilla luciferase activity were measured subsequently and the firefly luciferase activity was normalized with Renilla luciferase activity.
Immunoprecipitation
HEK-293T cells were transfected with pCI/HA-CREB for 48 h. The cells were washed twice with PBS and lysed with lysate buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 0.1% Nonidet P-40, 0.1% Triton X-100, 0.2% sodium deoxycholate, 2 mM EDTA, 5 mM AEBSF, 10 µg/ml aprotinin, 10 µg/ml leupeptin and 10 µg/ml pepstatin). After the insoluble materials were removed by centrifugation, the supernatant was incubated with anti-HA pre-conjugated protein G beads overnight at 4°C. The beads were washed with lysate buffer twice and with TBS twice, and bound proteins were subjected to further studies.
In vitro proteolysis of CREB
For the proteolysis of CREB in the brain extract, we homogenized human brain tissue in nine volumes of buffer consisting of 50 mM Tris–HCl, pH 7.4, 8.5% sucrose, 10 mM β-mercaptoethanol, 2.0 mM EDTA, followed by centrifugation at 15 000g at 4°C for 10 min. The obtained supernatants were incubated in the presence or absence of various concentrations of Ca2+ and/or protease inhibitors for 10 min at 30°C. The reactions were terminated by addition of Laemmli buffer, followed by boiling in water for 5 min. The proteolysed products were analyzed by western blots developed with antibodies against calpain I and CREB.
For the proteolysis of purified CREB by calpain I in vitro, we incubated recombinant MBP–CREB or immunopurified HA-CREB from CREB-transfected-HEK-293T cells as described above with various concentrations of calpain I (Sigma, USA) in proteolysis buffer (50 mM Tris–HCl, pH 7.4, 1 mM CaCl2) for 10 min at 30°C. The proteolysed products were subjected to further studies.
Chromatin immunoprecipitation
SH-SY5Y cells were transfected with pCI/HA-CREB for 72 h under differentiation condition and subjected for chromatin immunoprecipitation (ChIP) by using Magna ChIPTM A/G kit (Millipore) according to manufacturer’s manuals. Briefly, the cells were fixed with 1% formaldehyde in PBS for 10 min at room temperature and terminated by adding glycine solution to incubate for 5 min. After washing with ice-cold PBS containing 1× protease inhibitor cocktail, the cells were lysed with Lysis Buffer containing protease inhibitor cocktail on ice 15 min. The crude nuclei fraction was collected by centrifugation at 800g for 5 min at 4°C. The nuclei were suspended with Nuclear Lysis Buffer and sonicated to break DNA to ∼200–450 bp. The debris was removed by centrifugation at 10 000g for 10 min at 4°C. After adding Dilution Buffer into the supernatant, anti-HA or normal mouse IgG and proteinA/G magnetic beads were added into the supernatant and incubated for 1 h at 4°C with rotation. The ProteinA/G magnetic bead-antibody/chromatin complex was extensively washed sequentially with low-salt wash buffer, high-salt wash buffer, LiCl wash buffer and TE, and eluted with Elution Buffer with proteinase K. The cross-linking was reversed by incubation at 62°C for 2 h and at 95°C for 10 min and the supernatant was collected carefully. The total DNA was extracted from the supernatant with Wizard SV Gel and PCR Clean-Up System (Promega). CRE1, CRE2 and CRE3 containing sequences were amplified with PCR by using three sets of primers, set 1 (forward 5′TATTTTCTTCTCCTGCTTAGCT3′, reverse 5′AGTCATT TATAGTGTTTCCCTTC3′), set 2 (forward, 5′CCCAGGGTGGAGAGAGTGGAAG3′, reverse 5′TTATAATCTCCGCAAAGGGTGGAG3′) and set 3 (forward 5′GTCATATCCCAGCGAGACCC AG3′, reverse 5′C GCTGTAATCTAATTCAAGTCTTCAAG3′), respectively.
Electrophoretic mobility shift assay
CREB expressed HEK-293T cells were harvested and lysed with lysis buffer (10 mM HEPES, pH 7.9, 0.6% Nonidet P-40, 10 mM KCl, 0.1 mM EDTA, 1 mM DTT and cocktail of protease inhibitors) on ice for 20 min, and then pellet from 15 000g for 3 min centrifugation was suspended in the buffer containing 20 mM HEPES, pH 7.9, 0.4 M NaCl, 0.2 mM EDTA, 20% glycerol and 1 mM DTT. Nucleic proteins were extracted by centrifugation at 15 000g for 10 min. Complementary double-stranded DNA (dsDNA) oligomers containing CRE1 (5′CAGATACT TATGTAACACTTTT3′ and 5′AAAGTGTTACATAAGTATTG3′), CRE2 (5′GGAGGGAGGGCGTT ATTGTCT3′ and 5′AGACAAAACGCCCTCCCTCC3′), CRE3 (5′TCCTGAGGACGTGGAGAAAA C3′ and 5′GTTTTCTCCACGTCCTCAGGA3′) and CRE consensus sequence (5′TCAGCCTGAC GTCATACATCG3′ and 5′CGATGTATGACGTCAGGCTGA3′) as a positive control were annealed and labeled with [γ-32P]ATP (3000 Ci/mM) (Amersham Biosciences, Piscataway, NJ, USA) using T4 polynucleotide kinase (New England Biolabs, USA) and subsequently purified with MicroSpin G-25 column (Amersham Biosciences). Then above nucleic proteins were mixed with 32P-labeled double-stranded CRE1, CRE2, CRE3 or CRE consensus sequence. The reaction mixtures were incubated at 30°C for 40 min and subjected to 6% non-denaturing polyacrylamide gel pre-run at 100 V for 10 min. After electrophoresis with TBE buffer (89 mM Tris–borate, 2 mM EDTA) at 100 V for 60 min, the gel was dried and autoradiographed with a PhosphorImager (BAS-1500, Fujifilm, Japan).
N-terminal sequencing
Recombinant MBP–CREB was proteolysed by calpain I as described earlier. The reaction products were separated by 10% SDS–PAGE and electric blotted onto a PVDF membrane. After staining with Coomassie blue, the 41-kDa truncated CREB was dissected and subjected for N-terminal Edman Sequencing by Protein facility-Protein Sequencing Service, Iowa State University of Science and Technology.
Statistical analysis
Where appropriate, the data are presented as mean ± SD. Data points were compared with the unpaired two-tailed Student’s t-test, and the calculated P-values are indicated in the figures. For the analysis of the correlation between CREB truncation and calpain I activation or GLUT3 level in human brain homogenates, the Pearson correlation coefficient r was calculated.
RESULTS
CREB promotes the expression of GLUT3
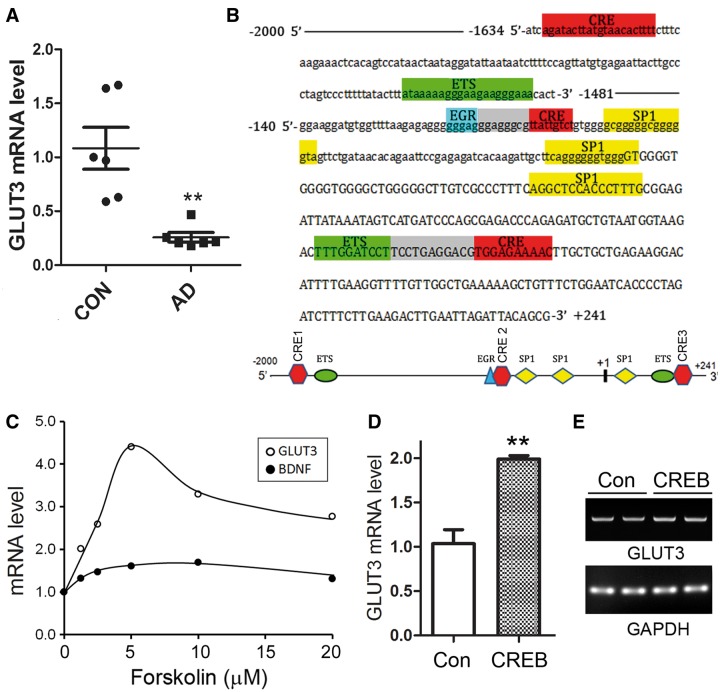
GLUT3 protein level is decreased in AD brain (29–31). Here, we measured GLUT3 mRNA levels by qPCR in frontal cortices from six AD and six control cases. We observed that GLUT3 mRNA was significantly decreased in AD brain (Figure 1A), suggesting that the transcription of GLUT3 is down-regulated in AD brain.
Figure 1.
Expression of GLUT3 is regulated by PKA/CREB. (A) The level of GLUT3 mRNA was reduced in AD brain. Total RNA was extracted from frontal cortices of AD and control cases. The levels of GLUT3 and GAPDH mRNA were measured by qPCR and presented as means ± SD (n = 12); **P < 0.01. (B) Human GLUT3 promoter region has three potential CRE-like elements. The promoter of human GLUT3 was analyzed by Matlnspector software. Cis-elements are labeled with different color and gray boxes show the overlap sequences of two adjacent elements. There are three CRE-like elements at the promoter region of human GLUT3. Other transcriptional factors are ETS (E-26), Sp1 (Specificity Protein 1) and EGR (early growth response protein 1). (C) Activation of PKA enhanced the GLUT3 expression. SH-SY5Y cells were differentiated with ATRA for 7 days and treated with various concentration of forskolin for 10 h. The total RNA was extracted and amplified to measure mRNA expressions of GLUT3, BDNF and GAPDH by qPCR. (D, E) Overexpression of CREB increased the expression of GLUT3 mRNA. SH-SY5Y cells under differentiation condition were transfected with pCI/CREB and empty plasmid (Con) for 3 days and the mRNA levels of GLUT3 and GAPDH were measured by qPCR (D) and regular PCR products of GLUT3 mRNA with same primers were subjected to agarose electrophoresis (E). The level of GLUT3 mRNA was presented as mean ± SD (n = 3); **P < 0.01.
To understand the molecular mechanism of GLUT3 expression regulation, we first analyzed the promoter of the human GLUT3 gene by MatInspector software analysis (40,41), a Genomatix internationally renowned program for the identification of transcription factor-binding sites. The bioinformatic analysis revealed an array of putative nuclear factor-binding sites including three potential CRE-like elements, namely CRE1, CRE2 and CRE3 (Figure 1B), which may be the binding sites of CREB.
Activation of CREB is dependent on its phosphorylation at Ser133 by activated PKA (37). So, we determined whether PKA/CREB regulates human GLUT3 expression. We used SH-SY5Y human neuroblastoma cells and differentiated them to neuronal-like cells with 10 µM ATRA for 7 days. Then, the cells were treated with various concentrations of forskolin for 10 h to activate PKA. The total RNA was extracted, reverse-transcripted to cDNA and subjected to qPCR to measure the level of GLUT3 mRNA. Because BDNF expression is regulated by CREB, we included it in this study as a positive control. The results showed that forskolin treatment promoted the expressions of BDNF and GLUT3 dose dependently (Figure 1C) but with a stronger effect on GLUT3 expression. The highest expression of GLUT3 was induced by 5 µM forskolin treatment (Figure 1C). This result supports our hypothesis that PKA/CREB may promote GLUT3 expression.
To explore the role of CREB on human GLUT3 expression, we overexpressed CREB in SH-SY5Y cells for 3 days under the differentiation condition. The level of GLUT3 mRNA was measured by qPCR (Figure 1D). Similarly, the cDNA was amplified by PCR with the same primers as that used for qPCR, and the products were also subjected to agarose electrophoresis (Figure 1E). We observed that an increased level of GLUT3 mRNA was induced by overexpression of CREB (Figure 1D and E), suggesting that CREB enhances the expression of GLUT3.
CREB binds to CRE1, CRE2 and CRE3 and promotes the expression of luciferase driven by GLUT3 promoter
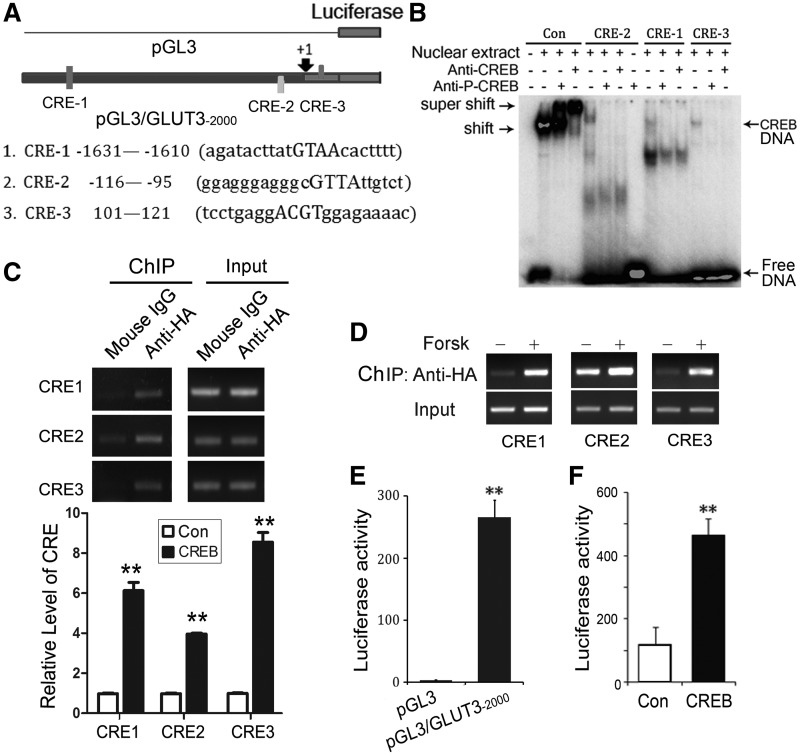
There are three CREs in human GLUT3 promoter. We first measured the binding of CREB to CRE1 (−1631 to −1610 bp), CRE2 (−116 to −95 bp) and CRE3 (+101 bp to +121 bp) (Figure 2A) by Electrophoretic Mobility Shift Assay (EMSA). We incubated the nuclear extracts of CREB-expressing HEK-293T cells with 32P-labeled DNA probes of CRE1, CRE2 or CRE3. The reaction mixtures were applied for native gel electrophoresis. We observed a mobility shift up of CRE-1, CRE-2 or CRE-3 induced by adding CREB containing nuclear extract, suggesting the formation of protein–CRE (1, 2 or 3) complex (Figure 2B). However, this mobility shift was detected when the total protein of nuclear extract reached to 10 µg, whereas only 5 µg nuclear extract was needed to completely shift the positive control consensus CRE (Figure 2B), suggesting that protein(s) in the extract could bind to the CREs but are weaker than the positive-control CRE.
Figure 2.
CREB binds to the CRE-like elements in human GLUT3 promoter and promotes the luciferase expression driven by this promoter. (A) Schematic diagram of three CRE-like elements in GLUT3 promoter and its luciferase reporter. (B) CREB bound to three CRE-like elements detected by EMSA. 32P-labeled consensus CRE (Con) as a positive control and three CRE-like elements, CRE1, CRE2 and CRE3, of human GLUT3 promoter were incubated with the nuclear extracts (5 µg for consensus CRE and 10 µg for three CRE-like elements) in the absence or presence of anti-CREB or anti-P-CREB and subjected to native PAGE. The gel was dried and autoradiographed with a PhosphorImager (BAS-1500, Fijifilm, Japan). (C) CRE-like elements in human GLUT3 promoter were co-immunoprecipitated with CREB by ChIP assay. SH-SY5Y cells under differentiation condition were transfected with pCI/CREB and then CREB was immunoprecipitated by anti-HA. Three CRE-like elements in the immune-complexes were amplified by PCR using the primers specific to CRE1, CRE2 or CRE3. The PCR products were quantified by densitometry and presented as mean ± SD (n = 3); **P < 0.01. (D) Forskolin treatment enhanced the binding of CREB with the CRE of GLUT3 promoter. SH-SY5Y cells under differentiation condition were transfected with pCI/CREB, treated with forskolin for 10 h and then CREB was immunoprecipitated by anti-HA. The three CREs were amplified by PCR with their specific primers. (E) Promoter of human GLUT3 promoted luciferase expression. Human GLUT3 promoter (−2000 to +241) was inserted into pGL3-basic vector to generate pGL3/GLUT3−2000. pGL3/GLUT3−2000 or its control pGL3 showed in panel A was co-transfected with pRL-TK into HEK-293T cells for 48 h. The firefly luciferase activity and Renilla luciferase activity were measured subsequently and the firefly luciferase activity was normalized with Renilla luciferase activity and presented as mean ± SD (n = 3); **P < 0.01. (F) CREB enhanced GLUT3 promoter-driving luciferase expression. pGL3/GLUT3−2000 and pRL-TK was co-transfected with pCI (Con) or pCI/CREB into HEK-293T cells for 48 h. The luciferase activity was measured and presented as mean ± SD (n = 3); **P < 0.01.
To validate the binding of CREB to the CREs, we added anti-CREB or anti-pS133-CREB into the incubation mixtures. We observed that addition of anti-CREB or anti-pS133-CREB antibody into the mixture containing control CRE induced a super-shift (Figure 2B). However, addition of these two antibodies into the mixture containing CRE1, CRE2 or CRE3 abolished the shift (CREB-CRE) induced by the nuclear extracts (Figure 2B), suggesting that CREB binds weakly but specifically to the three CREs, which are located in the promoter region of human GLUT3 gene.
To confirming the binding of CREB onto the CREs of human GLUT3 promoter, we performed ChIP in SH-SY5Y cells. CREB tagged with HA at its N-terminus was expressed in SH-SY5Y cells and immunoprecipitated by anti-HA. DNA in the immunocomplex was amplified by PCR using three sets of primers covering CRE1, CRE2 and CRE3, respectively. We observed that significant amount of CRE1, CRE2 or CRE3 was detected in the immunocomplex of anti-HA (Figure 2C), indicating the binding of CREB with these three CREs.
We further determined the effect of PKA activation by forskolin on the binding of CREB with CREs. We treated CREB-expressing-SH-SY5Y cells with 10 mM forskolin for 10 h and then carried out the ChIP with anti-HA. We found that forskolin treatment enhanced the binding of CREB with the CREs in human GLUT3 promoter (Figure 2D). Thus, these data confirm that CREB acts on the three CREs in the promoter region of the human GLUT3 gene and PKA activation enhances the binding.
Our above results have shown that CREB specifically bound to the CREs in the promoter of human GLUT3 gene in vitro and in intact cells. It is possible that CREB acts on the promoter to regulate the transcription of human GLUT3 gene. To test this possibility, we inserted the promoter region of human GLUT3, −2000 to +241, into pGL3 basic vector to generate reporter plasmid of pGL3/GLUT3−2000 (Figure 2A). We transfected pGL3/GLUT3−2000 and pRL-Tk into HEK-293T cells and then measured luciferase activity by the dual luciferase assay. We found that promoter of human GLUT3 increased luciferase activity by ∼100-fold (Figure 2E), indicating that the promoter of human GLUT3 drives the luciferase expression.
By using this luciferase reporter, hence, we test the promotion of GLUT3 expression by CREB. We co-transfected pCI/CREB or pCI with pGL3/GLUT3−2000 and pRL-Tk into HEK-293T cells and measured the luciferase activity 48 h after transfection. We observed an ∼3-fold elevation in luciferase activity in pCI/CREB-transfected HEK-293T cells when compared with the control transfected cells (Figure 2F), suggesting that CREB enhances the expression of luciferase driven by GLUT3 promoter. Thus, CREB may act onto the CRE elements of GLUT3 promoter to promote the expression of human GLUT3.
CRE2 and CRE3 are responsible for the promotion of GLUT3 expression
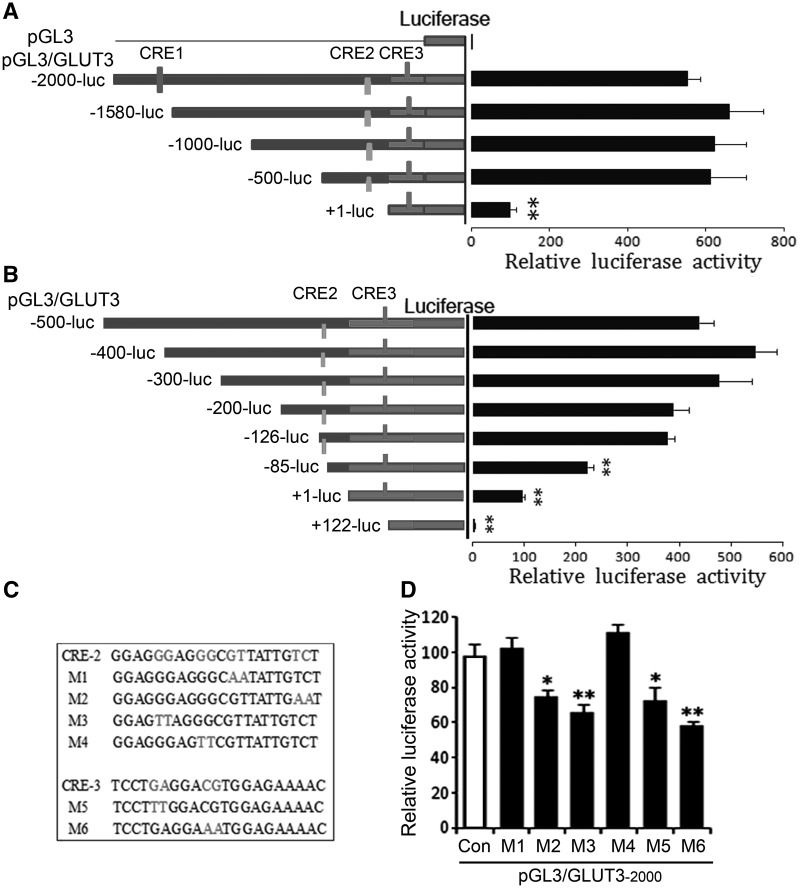
To verify the regulation of GLUT3 expression by the three CREs in the promoter region of human GLUT3, we constructed luciferase reporter plasmids, pGL3/GLUT3, with sequential deletion mutants of human GLUT3 promoter (Figure 3A, left). The generated plasmids were transfected into HEK-293T cells. The luciferase activity was determined and used to present the expression level of GLUT3. We observed that the luciferase activity in the cells transfected with pGL3/GLUT3−2000, pGL3/GLUT3−1580, pGL3/GLUT3−1000 or pGL3/GLUT3−500 was almost at the same level (Figure 3A, right), indicating that CRE1 is not required for the promotion of GLUT3 expression. The luciferase activity in cells transfected with pGL3/GLUT3+1, in which another 500-bp DNA, including CRE2, was deleted, was reduced by ∼6-fold (Figure 3A, right), suggesting that CRE2 plays a significant role in promoting GLUT3 expression. The luciferase activity in the cells transfected with pGL3/GLUT3+1, which contained CRE3, was much higher than that of pGL3 (Figure 3A, right), suggesting that CRE3 may also act as a transcription enhancer of GLUT3.
Figure 3.
CRE2 and CRE3 act as transcription enhancer of human GLUT3. (A, B) From −126 to +122 of human GLUT3 promoter was required for its promotion activity. Sequential deletions of the promoter region of GLUT3 were cloned into pGL3-basic plasmid containing the luciferase reporter gene and the plasmids were co-transfected with pRL-TK into HEK-293T cells. The luciferase activity was measured, normalized with Renilla luciferase and presented as mean ± SD (n = 3); **P < 0.01. (C) Mutations of CRE2 and CRE3 used in this study. (D) CRE2 and CRE3 of human GLUT3 promoter were required for its promotion activity. pGL3/GLUT3−2000 carrying different mutations described in panel C were co-transfected with pRL-TK into HEK-293T cells for 48 h, and then the luciferase activity was measured and presented as mean ± SD (n = 3); **P < 0.01.
To confirm the role of CRE2 and CRE3 in promoting GLUT3 gene expression, we created more detailed deletions of GLUT3 promoter from −500 to +122 (Figure 3B, left). We found that the luciferase activity decreased significantly when the region from −126 to −85 containing CRE2 was deleted (Figure 3B). Further deletion of −85 to +1 decreased more luciferase activity (Figure 3B), suggesting the involvement of other enhancers at this region in the regulation of GLUT3 expression. Deletion of +1 to +122 containing CRE3 almost abolished the luciferase activity (Figure 3B), indicating a role of CRE3 in promoting GLUT3 expression.
Following, we specified the role of CRE2 and CRE3 in the promotion of GLUT3 expression. We mutated CRE2 and CRE3 (Figure 3C) and measured their effect on the luciferase activity. We found that M1 and M4 mutations of CRE2 did not affect the activity of luciferase, but M2 and M3 mutations of CRE2 caused reduction of luciferase activity (Figure 3D). Similarly, mutation of CRE3 significantly decreased luciferase activity (Figure 3D). These results confirmed the role of CRE2 and CRE3 in the promotion of GLUT3 expression.
PKA/CREB promotes the expression of luciferase driven by human GLUT3 promoter
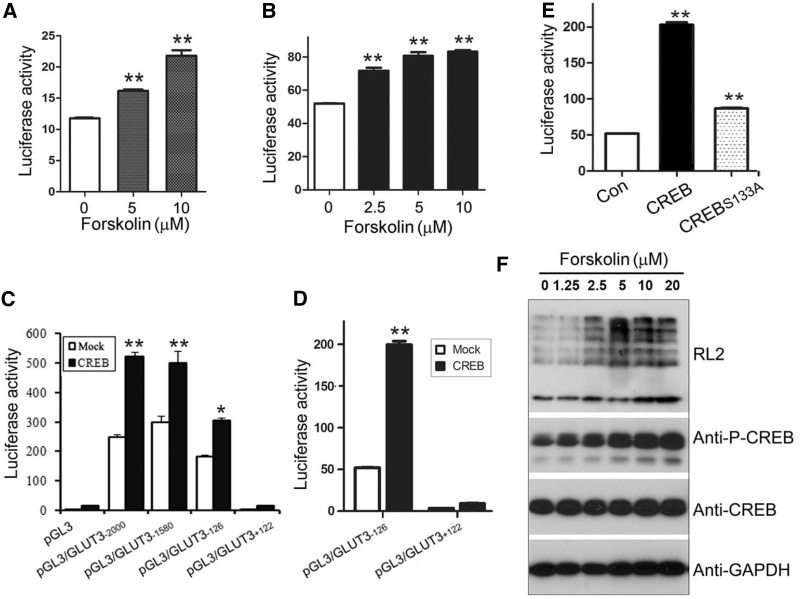
We have shown that PKA/CREB regulates endogenous GLUT3 expression. So, we investigated the regulation of PKA/CREB on luciferase expression. We transfected pGL3/GLUT3−2000 (Figure 4A) or pGL3/GLUT3−126 (Figure 4B) into SH-SY5Y cells under differentiation and treated the cells with forskolin for 10 h. The luciferase activity was measured at total 48 h after transfection. We observed that forskolin treatment enhanced luciferase activity in both pGL3/GLUT3−2000 (Figure 4A) and pGL3/GLUT3−126 (Figure 4B) transfected cells, supporting that PKA promotes GLUT3 expression.
Figure 4.
PKA/CREB enhances GLUT3 promoter driving-luciferase expression. (A, B) Activation of PKA increased luciferase expression. pGL3/GLUT3−2000 (A) or pGL3/GLUT3−126 (B) was co-transfected with pRL-TK into SH-SY5Y cells and treated with various concentration of forskolin for 10 h. The luciferase activity was measured and presented as mean ± SD (n = 3); **P < 0.01. (C, D) CREB enhanced luciferase expression. HEK-293FT (C) or SH-SY5Y cells (D) were co-transfected with pGL3/GLUT3s and pCI/CREB and pRL-TK. Luciferase activity was measured and presented as mean ± SD (n = 3); **P < 0.01. (E) CREBS133A had much less activity in the promotion of GLUT3 expression. pCI/CREB or pCI/CREBS133A was co-transfected with pGL3/GLUT3−126 into SH-SY5Y cells, and the luciferase activity was measured 48 h after transfection. The relative activity of luciferase was presented as mean ± SD (n = 3); **P < 0.01. (F) Activation of PKA increased O-GlcNAcyaltion. Differentiated SH-SY5Y cells were treated with various concentrations of forskolin for 10 h. Total proteins were analyzed by western blots developed with antibodies as indicated. GAPDH was included as a loading control.
We next determined the role of CREB on luciferase expression driven by varied lengths of the GLUT3-promoter. We co-expressed pGL3/GLUT3s with CREB in HEK-293T cells and then measured the luciferase activity 48 h after transfection. We found that CREB enhanced the expression of pGL3/GLUT3−2000, pGL3/GLUT3−1580 and pGL3/GLUT3−126 similarly but not pGL3/GLUT3+122 (Figure 4C). We observed similar results in neuronal SH-SY5Y cells (Figure 4D). These data further confirm that PKA/CREB works on the promoter of GLUT3 to promote its expression.
Ser133 phosphorylation is required for CREB transcriptional activity. Thus, we mutated serine 133 to alanine (CREBS133A) and then co-transfected with the pGL3/GLUT3−126 into SH-SY5Y cells under differentiation condition for 48 h and measured the luciferase activity. We found that CREBS133A elevated luciferase activity but much weaker than wild-type CREB (Figure 4E), suggesting that phosphorylation of CREB at Ser133 enhances the promotion of GLUT3 expression.
Increased intracellular glucose generally leads to upregulation of protein O-GlcNAcylation by providing donor substrate UDP-GlcNAc derived from glucose (42–44). To determine whether the increased expression of GLUT3 by PKA/CREB has physiological activity to transport more glucose into cells, we measured the O-GlcNAcylation of global protein to present intracellular glucose level. We found that forskolin treatment significantly increased O-GlcNAcylation of global proteins determined by anti-RL2 (Figure 4F), coinciding with increased CREB phosphorylation (Figure 4F) and GLUT3 expression (Figure 1C). Thus, GLUT3 expression induced by PKA/CREB facilitates to transport glucose and sequentially leads to increased global protein O-GlcNAcylation.
Full-length CREB is decreased and truncated CREB is increased in AD brain
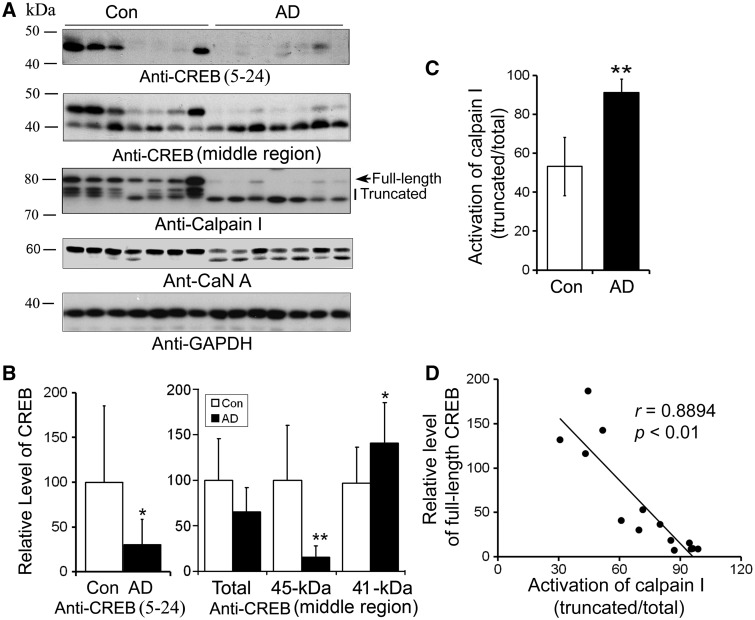
Above results indicate that CREB involves in the regulation of GLUT3 expression, and decreased GLUT3 in AD brain may result from altered CREB signaling in AD. Hence, we measured CREB levels in the homogenates of the frontal cortices from seven AD and seven age- and post-mortem interval-matched control brains by western blots. We found that CREB level detected by anti-CREB against 5–24 amino acids was dramatically decreased in AD brain (Figure 5A and B). To further confirm this phenomenon, we employed another antibody against middle region of CREB to perform the western blot. We observed a similar decrease of full-length CREB at 45-kDa and an increased 41-kDa band in AD brain, without a change total level of CREB (Figure 5A). These results suggest that the truncation of CREB is increased in AD brain (Figure 5A and B).
Figure 5.
The level of full-length CREB is decreased in AD brain and is correlated to activation of calpain I in human brain. (A) Western blots of CREB, calpain I and CaN A. Frontal cortical homogenates from seven AD and seven control cases were subjected to western blots developed with antibodies against CREB, calpain I, CaN A or GAPDH. (B) The level of full-length CREB was decreased and truncation of CREB was increased in AD brain significantly. Blots as shown in (A) developed with anti-CREB (upper panel) were quantitated by densitometry, and the levels of CREB was normalized with GAPDH and presented as mean ± SD; *P < 0.05; **P < 0.01. (C) Truncation and activation of calpain I were significantly increased in AD brain. Arrow indicates full length of calpain I and vertical line indicates truncated and active form of calpain I. Blots as shown in (A) developed with anti-calpain I were quantitated by densitometry, the truncation and activation of calpain I were presented by the ratio of the truncated (76–78 kDa) over the total calpain I. (D) CREB level was correlated with the activation of calpain I in human brain. The relative activation of calpain I [truncated/(full length + truncated)] (x-axis) is plotted against the relative CREB level (y-axis).
Calpain I is activated by calcium-dependent autoproteolysis from an 80-kDa inactive form into a 76- to 78-kDa active form (45). This process is increased in AD (46,47). So, we determined calpain I activation (ratio of truncated/total calpain I) in these brain samples and performed Pearson correlation analysis. In addition, calcinurine A (CaN), a well-known substrate of calpain I (44), was included in this study (Figure 5A). We observed a marked increase in calpain I activation in AD brain (Figure 5A and C), and this activation was highly and negatively correlated with full-length CREB with a correlation coefficient r= 0.8894 (P < 0.001) in human brain (Figure 5D), suggesting that calpain I activation might underlie the increased truncation of CREB in AD brain.
Calpain I is responsible for the truncation of CREB
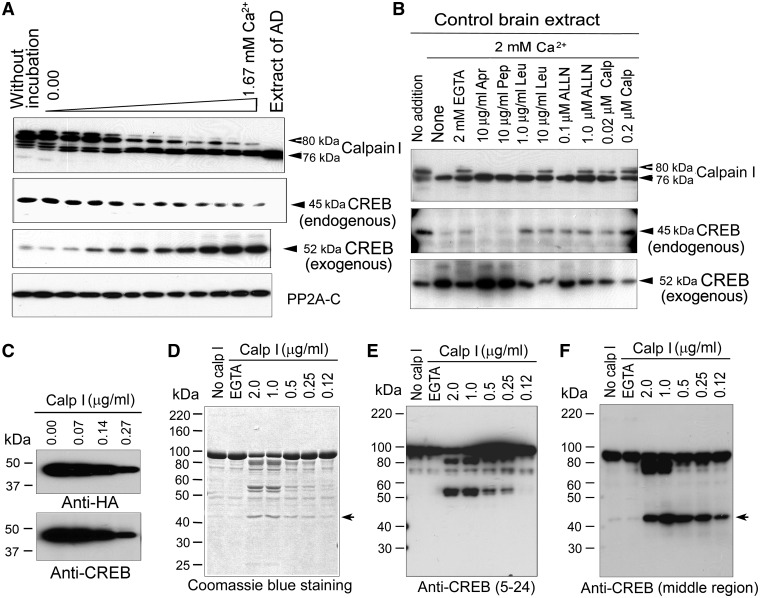
Calpain I is a calcium-dependent neutral protease. To determine whether CREB truncation in human brain is indeed caused by activated calpain I, we incubated normal human brain extracts in the presence of various concentration of calcium for 10 min at 30°C and detected the levels of calpain I and CREB by western blots. We found that calpain I was proteolysed from 80 to 78 and 76 kDa in a Ca2+ dose-dependent manner (Figure 6A, upper panel), suggesting that calcium initiates the autoproteolysis and activation of calpain I. Concurrently, the 45-kDa CREB detected by anti-N-terminal CREB was significantly decreased in a calcium concentration-dependent manner (Figure 6A, middle panel). As an alternative, we added recombinant MBP–CREB fusion protein into the proteolysis reaction and performed in vitro proteolysis, followed by western blots to detect the proteolysed products. We observed a calcium dose-dependent increase in the 52-kDa band detected by anti-N-terminal CREB (Figure 6A, middle panel). However, PP2A catalytic subunit (PP2A–C) in human brain extract was not proteolysed (Figure 6A, lower panel). Because MBP was fused at N-terminus of CREB, this 52-kDa band represented the fusion protein of MBP (∼43 kDa) plus the N-terminal fragment of CREB. Thus, this result further confirms that CREB is proteolysed by a calcium-dependent protease(s) in human extract and the truncation site might be located at its N-terminus.
Figure 6.
Proteolysis of CREB is catalyzed by calcium-mediated truncation/activation of calpain I. (A) In vitro proteolyses of calpain I and CREB in human brain extracts were activated by calcium. A normal human brain extract was incubated for 10 min at 30°C in the absence (upper two panels) or presence (3th panel) of recombinant MBP–CREB and analyzed by western blots. PP2A catalytic subunit was used as negative control. (B) Calcium-activated proteolysis of calpain I and CREB was inhibited by calpain inhibitors selectively. Recombinant MBP–CREB was incubated with normal human brain extract in the presence of EGTA or CaCl2 plus various selective protease inhibitors for 10 min at 30°C, followed by western blots to detect the proteolysis. Abbreviations: Apr, aprotinin; Pep, pepstatin; Leu, leupeptin; Calp, calpastatin peptide. (C) Immunopurified CREB from mammalian cells was proteolysed by calpain I. HA-CREB was immunopurified from HEK-293FT cells and incubated with various concentrations of calpain I for 10 min at 30°C. The reaction products were analyzed with western blots. (D–F) Recombinant MBP–CREB was proteolysed by calpain I. Recombinant MBP–CREB was incubated with various concentrations of calpain I for 10 min at 30°C. The proteolytic products were analyzed by Coomassie blue staining (D) and western blots developed with anti-CREB (against amino acids 5–24) (E) or anti-CREB (against middle region) (F).
Because the above incubation was carried out in the presence of 2.0 mM EDTA that chelated all endogenous Ca2+ when no additional Ca2+ was added and most of the added Ca2+ that allowed only free Ca2+ > 2.0 mM chelating capacity of EDTA, only micromolar levels of free Ca2+ were present in the reaction mixture during incubation of the brain extracts. Hence, these results suggest that the CREB proteolysis in the human brain extracts resulted from activation of calpain I rather than calpain II, which requires millimolar concentrations of free calcium for activation.
We next verified above proteolysis of CREB in human brain extract is caused by calpain. Calpain I is a cysteine protease. So, we performed proteolysis of MBP–CREB in vitro in the presence of various protease inhibitors. When aprotinin, a serine protease inhibitor, and pepstatin, an aspartic protease inhibitor, were included in the normal human brain extracts during incubation, no significant inhibition of the proteolysis of either calpain I or CREB was observed (Figure 5B). These results excluded the involvement of serine proteases and aspartic proteases in the proteolysis of calpain I and CREB. In contrast, when leupeptin, a selective inhibitor of cysteine and serine proteases, and N-acetyl-Leu-Leu-Nle-CHO (ALLN), a calpain and cysteine protease inhibitor, were included in the incubation mixtures, a marked inhibition of calpain I proteolysis and an almost complete blockage of CREB proteolysis were observed (Figure 5B). A specific calpain inhibitor, calpepstatin peptide, also inhibited the autoproteolysis of calpain I and prevented CREB proteolysis. These results are consistent with the above studies that CREB is proteolysed by calcium-dependent cleavage/activation of calpain I.
To confirm CREB is proteolysed by calpain I, we overexpressed HA-tagged CREB at N-terminus in HEK-293FT cells and immunopurified HA-CREB with anti-HA. The immunopurified HA-CREB was incubated with various concentration of calpain I and subjected to western blots using anti-HA and anti-CREB. We observed a decrease of CREB in dose-dependent manner (Figure 6C), suggesting that CREB is cleaved by calpain I in vitro.
No specific amino acid sequence is uniquely recognized by calpain (45). To address the cleavage of CREB by calpain I, we incubated purified calpain I with recombinant MBP–CREB and detected the proteolysed products with coomassie blue staining (Figure 6D) and western blots by using anti-N-terminal CREB (Figure 6E) or anti-middle region of CREB (Figure 6F). We observed that, similar to the proteolysis by proteases in human brain extract, purified calpain I cleaved MBP–CREB in a calcium- and dose-dependent manner (Figure 6D–F). MBP–CREB (93-kDa) was cleaved by lower concentration calpain I into 52-kDa and 41-kDa fragments mainly (Figure 6D). The 52-kDa could not be recognized by anti-middle region of CREB but by anti-N-terminal CREB (Figure 6E and F), whereas 41-kDa band was recognized by anti-middle region of CREB but not by anti-N-terminal CREB (Figure 6E and F). However, high concentration calpain I also cleaved MBP-CREB to 85-kDa big fragments, which was recognized by both anti-N-terminus and anti-middle region of CREB (Figure 6E and F). Thus, we speculated that 93-kDa MBP–CREB was cut by calpain I at N-terminal CREB and generated 41-kDa truncated CREB, which contained C-terminal CREB and 52-kDa fragment which consists of MBP and N-terminal CREB. So, we dissected the 41-kDa fragment and subjected to N-terminal sequence analysis for five circles. The sequence of 41-kDa from N-terminus was AQPQI. Thus, we conclude that CREB was cleaved by calpain I at Gln28–Ala29 (Figure 7A).
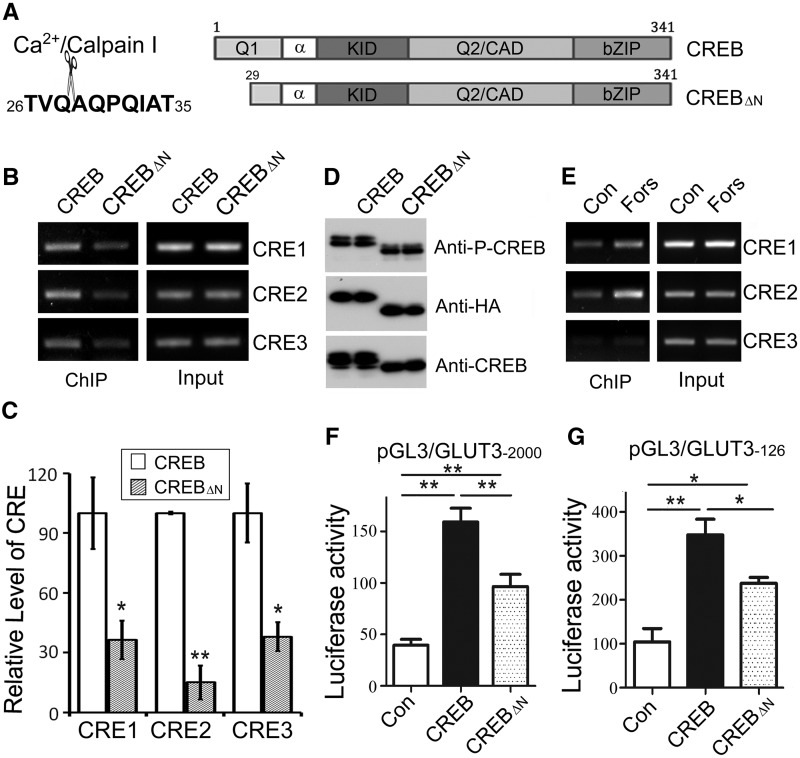
Figure 7.
Truncated CREB has less activity to promote GLUT3 expression. (A) Schematic diagram of CREB and truncated CREB (CREBΔN). (B, C) CREBΔN bound less than CREB to the CREs in GLUT3 promoter. pCI/CREB or pCI/CREBΔN was transfected into SH-SY5Y cells under differentiation condition and then immunoprecipitated by anti-HA. Three CRE-like elements in the immune-complexes were amplified by PCR using the primers specific to CRE1, CRE2 or CRE3. PCR products were quantified by densitometry and presented as mean ± SD (n = 3); *P < 0.05; **P < 0.01. (D) Truncation of CREB did not affect its phosphorylation at Ser133. CREB or CREBΔN was expressed in HEK-293T cells and analyzed by western blots developed with anti-HA, anti-CREB1 and phosphorylated CREB at Ser133. (E) Forskolin treatment enhanced the binding of CREBΔN with the CRE of GLUT3 promoter. SH-SY5Y cells under differentiation condition were transfected with pCI/CREBΔN and treated with forskolin for 10 h, and then CREBΔN was immunoprecipitated by anti-HA. The three CREs were amplified by PCR with their specific primers. (F, G) CREBΔN had less ability to promote the luciferase expression. Luciferase activity was measured in the SH-SY5Y cells cotransfected with pGL3/GLUT3−2000 (F) or pGL3/GLUT3−126 (G) and pCI vector, pCI/CREB or pCI/CREBΔN and presented as mean ± SD (n = 3); *P < 0.05; **P < 0.01.
Truncated CREB has less ability to promote GLUT3 expression
Calpain I cleaves CREB at Gln28–Ala29 to generated 41-kDa truncated CREB (CREBΔN). Hence, we investigated the effect of the truncation on CREB-regulated GLUT3 expression. We constructed truncated pCI/CREBΔN (Figure 7A), transfected it into SH-SY5Y cells and performed ChIP with anti-HA as described previously. We observed that CREs in the promoter of GLUT3 precipitated by CREBΔN were much less than that by CREB (Figure 7B and C), suggesting weaker binding of CREBΔN than CREB. Transcriptional activity of CREB requires its phosphorylation at Ser133 (37). Thus, we expressed both CREB and CREBΔN in HEK-293FT cells and detected the phosphorylation level by western blots. Similar Ser133 phosphorylation of CREB was detected in CREB and CREBΔN (Figure7D). Moreover, forskolin treatment also increased binding of CREBΔN to CREs of GLUT3 promoter (Figure 7E), suggesting that truncation did not suppressed Ser133 phosphorylation.
To investigate the role of CREB truncation on its transcriptional activity to promote GLUT3 expression, we co-expressed CREBΔN or CREB in HEK-293T with pGL3/GLUT3−2000 (Figure 7F) and pGL3/GLUT3−126 (Figure 7G). The luciferase activity was assessed with dual luciferase assay. The luciferase activity in the HEK-293T cells transfected with CREBΔN was significantly lower than that in the cells transfected with CREB, which was independent of the various promoter regions of GLUT3 gene (Figure 7F and G). These results suggest that the N-terminal truncation of CREB may reduce its ability to promote GLUT3 expression.
Level of GLUT3 is decreased in AD brain and correlates to CREB level positively
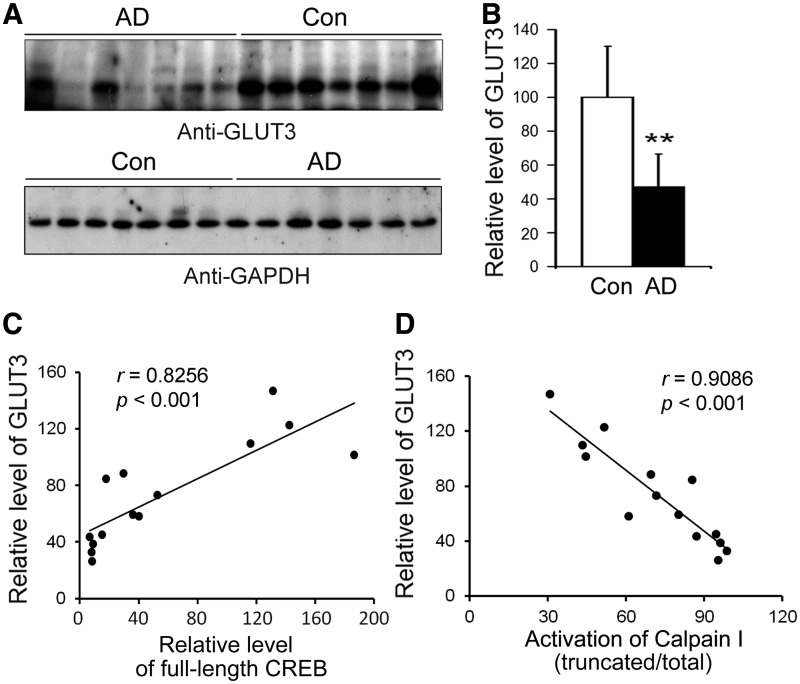
To determine whether decreased GLUT3 results from decreased CREB in AD brain, we analyzed GLUT3 level in frontal cortices from seven AD and seven control brains using western blots. GLUT3 level was decreased markedly in the frontal cortices of AD brains (Figure 8A and B), which is consistent with previous studies (29,30).
Figure 8.
GLUT3 level is decreased in AD brain and related with CREB and calpain I activation in human. (A, B) Protein level of GLUT3 in AD was decreased. Western blots of frontal cortical homogenates from seven AD and seven control cases developed with anti-GLUT3. GAPDH blot was included as a loading control (A). Western blots were quantitated by densitometry and the relative level of GLUT3 is presented as mean ± SD; **P < 0.01. (C) The level of GLUT3 correlated to CREB in human brain. The relative level of GLUT3 (y-axis) in the frontal cortical homogenates from seven AD and seven control cases is plotted against the level of CREB (x-axis). (D) Activation of calpain I correlated to the level of GLUT3 in human brain. The relative activation of calpain I [truncated/(full length + truncated)] (x-axis) is plotted against the level of GLUT3 (y-axis).
Furthermore, we found that GLUT3 level was positively correlated with the full-length CREB (Figure 8C). Because we have found above that overactivation of calpain I in AD brain was responsible for the decreased CREB, we analyzed the relationship between GLUT3 level and calpain I activation by using Pearson correlation analysis. We found that activation of calpain I was negatively correlated with GLUT3 level in the human brain (Figure 8D).
DISCUSSION
Previous studies have demonstrated a decrease of GLUT3 protein level in AD brain (30), which might be a cause of the impaired brain glucose uptake/metabolism and associated neurodegeneration in this disease. In this study, we proved that PKA/CREB promotes the expression of GLUT3 by acting on the CRE-like elements, mainly CRE2 and CRE3, of the promoter region of human GLUT3. We further found that in AD brain, full-length CREB is decreased, truncation of CREB is increased and the level of full-length CREB negatively correlates with the activation of calpain I in human brain. In vitro, calpain I cleaves CREB at Gln28–Ala29 and generates a 41-kDa truncated CREB, which has less ability to promote GLUT3 expression. Moreover, the level of GLUT3 is correlated with the level of full-length CREB positively and with activation of calpain I negatively in human brain. It appears that in AD brain, the dysregulated calcium homeostasis probably leads to overactivation of calpain I, which, in turn, cleaves CREB, resulting in a weaker promotion in the expression of GLUT3 and other genes, consequently impairment of brain glucose uptake and metabolism. Thus, these findings provide a novel possible mechanism explaining the regulation of GLUT3 by CREB, the negative impact of the proteolysis of CREB by calpain I on the expression of GLUT3 and the involvement of this mechanism in impairment of brain glucose uptake and AD pathogenesis.
Calpain is a family of calcium-dependent intracellular cysteine proteases that catalyzes limited proteolytic cleavage of a variety of cellular proteins in all eukaryotes (45,48). Calpain I, which is the major calpain isoform in the neuron, is fully activated by low micromolar concentrations of calcium (hence also called µ-calpain), whereas calpain II requires low millimolar levels of calcium for optimal activity (hence also called m-calpain). Calpain I is activated by removal of the N-terminal 14 amino acids of the 80-kDa subunit first to produce the 78-kDa intermediate product followed by removal of an additional 12 amino acids to produce the 76-kDa autolytic fragment (49). Many putative etiologic factors of AD, including excitotoxicity, β-amyloid neurotoxicity and free-radical injury, have in common the potential for disrupting intracellular calcium homeostasis (50–52). Though there is a lack of direct evidence of altered calcium homeostasis in AD brain, dysregulation of calcium is one of the major hypotheses that may explain the pathogenic mechanism of the disease (53,54). Calpain is thought to play a critical role in activation of neuronal cdk5 (55–57), MAPK pathway (58), PKA (59) and protein phosphatase 2B (46,60), as well as the truncation of tau, which in turn causes its abnormal hyperphosphorylation and leads to neuronal death (61,62). Elevated truncation and activation of calpain have been previously reported in early-stage AD (46,47). This study demonstrates that calpain I activation may also play a role in the impairment of glucose uptake via down-regulation of GLUT3 expression due to truncation of CREB in AD brain.
CREB is the principal transcriptional factor exhibiting the most significant relevance to molecular networks of AD related genes (63). In the incipient of AD, studies on transcriptional network have shown that CREB is the most significant transcriptional regulator, suggesting that functional impairment of CREB in AD could begin at the early stage of the disease (63). The β-amyloid (Aβ) peptide, which plays an important role in the pathogenesis of AD, alters hippocampal-dependent synaptic plasticity and memory and mediates synapse loss through the CREB-signaling pathway (64). The CREB-target genes play key roles in neuronal development, synaptic plasticity and neuroprotection in the central nervous system. Among those, BDNF is a well-studied CREB-target gene and has important roles during development and adult life (65,66) in neuronal growth, survival, differentiation and synaptic function (67,68). Numerous reports have indicated that the relative levels of BDNF mRNA and proteins are decreased in severe AD when compared with aged-matched controls (69–74).
CREB binds onto CRE of promoters as a dimer and activates gene expression. CRE typically appears as either palindrome (TGACGTCA) or half-site (CGTCA) sequences (75,76). Half-site is less active than the full-CRE palindrome for CREB binding and AMP responsiveness (37,77,78). The human GLUT3 gene does not have the typical CRE element but has three potential CRE-like sequences. These three CRE-like elements have much weaker CREB-binding activity than the typical ‘TGACGTCA’ CRE consensus sequence. These three CRE-like elements are located from −1631 to −1610 bp for CRE1, from −116 to −95 bp for CRE2 and from +101 to +121 bp for CRE3. In this study, deletion of CRE1 did not affect GLUT3 promoter-driven luciferase expression, suggesting that CRE1 does not act as a significant enhancer in the expression of GLUT3, even though it could bind to CREB. Deletion of the region containing either CRE2 or CRE3 significantly reduced the expression of luciferase, suggesting that CRE2 and CRE3 serve as enhancers of GLUT3. Mutations of CRE2 or CRE3 significantly decreased the promotion of GLUT3 expression, further supporting their requirement for promotion of GLUT3 expression.
CREB contains an N-terminal transactivation domain (TAD) and a C-terminal basic Leu zipper DNA-binding and dimerization domain. In the TAD, a central kinase-inducible domain (KID), containing a PKA phosphorylation site, Ser133, is flanked by hydrophobic glutamine-rich domains, Q1 and Q2. The Q domains enhance transcription by interacting with other associated factors (79). In this study, we found that CREB was proteolysed in vitro by calpain I at Gln28–Ala29 and generated truncated CREB, which lacks a part of Q1. Truncated CREB bound less to CREs in the promoter of GLUT3 and had less activity to promote GLUT3 expression but did not affect its Ser133 phosphorylation. These results further verify that Q1 domain enhances transcription (79) and that removal of a part of N-terminal Q1 by calpain I may reduce its transcription activity.
The transcription activity of CREB is mainly regulated by its phosphorylation at Ser133 by PKA (80). We have shown in this study that CREB phosphorylation is also required for promoting GLUT3 expression, because the Ser133 to Ala mutation eliminated CREB’s ability to promote GLUT3-droved luciferase expression. In AD brain, PKA activity is decreased due to overactivation of calpain I (59). Therefore, calpain I over-activation may result in decreased GLUT3 expression by two pathways, PKA down-regulation and CREB truncation, synergistically.
Most glucose in the brain is metabolized to produce ATP to maintain neuronal activity. Approximately 2–5% of total glucose feeds into the hexosamine biosynthesis pathway to produce glucosamine-6-phosphate, and, ultimately, UDP-Nacetylglucosamine (UDP-GlcNAc) (42). UDP-GlcNAc is the donor substrate for O-linked-β-Nacetylglucosamine (O-GlcNAc) transferase (OGT), which catalyzes protein O-GlcNAcylation, a process transferring GlcNAc from UDP-GlcNAc to Ser/Thr residues of proteins. O-GlcNAcylation is a recently recognized post-translational modification of numerous cytoplasmic and nuclear proteins. The activity of OGT is sensitive to relatively small changes in UDP-GlcNAc availability over a wide range of concentrations (43). Therefore, decreased GLUT3 may cause impairment of glucose up-take, leading to down-regulation of O-GlcNAcylation of proteins.
Microtubule-associated protein tau is modified by O-GlcNAcylation. In AD brain, tau is abnormally hyperphosphorylated and accumulated in the form of NFTs (81,82). Many studies have demonstrated that abnormal hyperphosphorylation of tau is critical to neurodegeneration and promotes its aggregation into tangles (83–89). We recently found that tau phosphorylation is inversely regulated by O-GlcNAcylation and that decreased O-GlcNAcylation induces hyperphosphorylation of tau (44). In AD brain, protein O-GlcNAcylation is markedly decreased and correlated negatively with phosphorylation at most phosphorylation sites of tau protein (90). Therefore, decreased GLUT3 may down-regulate O-GlcNAcylation via reduction of neuronal glucose uptake, leading to abnormal hyperphosphorylation of tau and neurofibrillary degeneration in AD.
In summary, overactivation of calpain I due to overload of calcium in AD brain proteolyses CREB to truncated CREB, which has less ability to promote GLUT3 expression and might cause a reduction of glucose uptake and metabolism in the brain and consequently neurodegeneration and dementia.
FUNDING
The National Natural Science Foundation of China [Grants 30973143 and 81030059 to F.L.]; a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institution (PAPD); Basic Research Program of Jiangsu Education Department [Grants 10KJA310040 to F.L. and 11KJD310002 to W.Q.]; the Brooklyn Home for Aged Men; U.S. Alzheimer’s Association [Grant IIRG-10-173154 to F.L.]; National Basic Research Program of China [973 Program, 2013CB531005]; Brain Donation Program is partially supported by a National Institute on Aging grant [P30 AG19610, Arizona Alzheimer’s Disease Core Center]. Funding for open access charge: the National Natural Science Foundation of China [Grant 81030059].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Ms M. Marlow for editorial suggestions and Ms J. Murphy for secretarial assistance.
REFERENCES
- 1.Hoyer S. Causes and consequences of disturbances of cerebral glucose metabolism in sporadic Alzheimer disease: therapeutic implications. Adv. Exp. Med. Biol. 2004;541:135–152. doi: 10.1007/978-1-4419-8969-7_8. [DOI] [PubMed] [Google Scholar]
- 2.Hoyer S. Brain glucose and energy metabolism abnormalities in sporadic Alzheimer disease. Causes and consequences: an update. Exp. Gerontol. 2000;35:1363–1372. doi: 10.1016/s0531-5565(00)00156-x. [DOI] [PubMed] [Google Scholar]
- 3.Jagust WJ, Seab JP, Huesman RH, Valk PE, Mathis CA, Reed BR, Coxson PG, Budinger TF. Diminished glucose transport in Alzheimer's disease: dynamic PET studies. J. Cereb. Blood Flow Metab. 1991;11:323–330. doi: 10.1038/jcbfm.1991.65. [DOI] [PubMed] [Google Scholar]
- 4.Minoshima S, Frey KA, Foster NL, Kuhl DE. Preserved pontine glucose metabolism in Alzheimer disease: a reference region for functional brain image (PET) analysis. J. Comput. Assist. Tomogr. 1995;19:541–547. doi: 10.1097/00004728-199507000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Friedland RP, Budinger TF, Ganz E, Yano Y, Mathis CA, Koss B, Ober BA, Huesman RH, Derenzo SE. Regional cerebral metabolic alterations in dementia of the Alzheimer type: positron emission tomography with [18F]fluorodeoxyglucose. J. Comput. Assist. Tomogr. 1983;7:590–598. doi: 10.1097/00004728-198308000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Minoshima S, Giordani B, Berent S, Frey KA, Foster NL, Kuhl DE. Metabolic reduction in the posterior cingulate cortex in very early Alzheimer's disease. Ann. Neurol. 1997;42:85–94. doi: 10.1002/ana.410420114. [DOI] [PubMed] [Google Scholar]
- 7.Ferris SH, Crook T, Clark E, McCarthy M, Rae D. Facial recognition memory deficits in normal aging and senile dementia. J. Gerontol. 1980;35:707–714. doi: 10.1093/geronj/35.5.707. [DOI] [PubMed] [Google Scholar]
- 8.Foster NL, Chase TN, Mansi L, Brooks R, Fedio P, Patronas NJ, Di Chiro G. Cortical abnormalities in Alzheimer's disease. Ann. Neurol. 1984;16:649–654. doi: 10.1002/ana.410160605. [DOI] [PubMed] [Google Scholar]
- 9.Grady CL, Haxby JV, Schlageter NL, Berg G, Rapoport SI. Stability of metabolic and neuropsychological asymmetries in dementia of the Alzheimer type. Neurology. 1986;36:1390–1392. doi: 10.1212/wnl.36.10.1390. [DOI] [PubMed] [Google Scholar]
- 10.Haxby JV, Grady CL, Koss E, Horwitz B, Heston L, Schapiro M, Friedland RP, Rapoport SI. Longitudinal study of cerebral metabolic asymmetries and associated neuropsychological patterns in early dementia of the Alzheimer type. Arch. Neurol. 1990;47:753–760. doi: 10.1001/archneur.1990.00530070043010. [DOI] [PubMed] [Google Scholar]
- 11.Desgranges B, Baron JC, de la Sayette V, Petit-Taboue MC, Benali K, Landeau B, Lechevalier B, Eustache F. The neural substrates of memory systems impairment in Alzheimer's disease. A PET study of resting brain glucose utilization. Brain. 1998;121(Pt 4):611–631. doi: 10.1093/brain/121.4.611. [DOI] [PubMed] [Google Scholar]
- 12.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch. Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy AM, Frackowiak RS, Newman SK, Bloomfield PM, Seaward J, Roques P, Lewington G, Cunningham VJ, Rossor MN. Deficits in cerebral glucose metabolism demonstrated by positron emission tomography in individuals at risk of familial Alzheimer's disease. Neurosci. Lett. 1995;186:17–20. doi: 10.1016/0304-3940(95)11270-7. [DOI] [PubMed] [Google Scholar]
- 14.Kennedy AM, Newman SK, Frackowiak RS, Cunningham VJ, Roques P, Stevens J, Neary D, Bruton CJ, Warrington EK, Rossor MN. Chromosome 14 linked familial Alzheimer's disease. A clinico-pathological study of a single pedigree. Brain. 1995;118(Pt 1):185–205. doi: 10.1093/brain/118.1.185. [DOI] [PubMed] [Google Scholar]
- 15.Mosconi L, Sorbi S, de Leon MJ, Li Y, Nacmias B, Myoung PS, Tsui W, Ginestroni A, Bessi V, Fayyazz M, et al. Hypometabolism exceeds atrophy in presymptomatic early-onset familial Alzheimer's disease. J. Nucl. Med. 2006;47:1778–1786. [PubMed] [Google Scholar]
- 16.Small GW, Mazziotta JC, Collins MT, Baxter LR, Phelps ME, Mandelkern MA, Kaplan A, La Rue A, Adamson CF, Chang L, et al. Apolipoprotein E type 4 allele and cerebral glucose metabolism in relatives at risk for familial Alzheimer disease. JAMA. 1995;273:942–947. [PubMed] [Google Scholar]
- 17.Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, Thibodeau SN, Osborne D. Preclinical evidence of Alzheimer's disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N. Engl. J. Med. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- 18.Reiman EM, Caselli RJ, Chen K, Alexander GE, Bandy D, Frost J. Declining brain activity in cognitively normal apolipoprotein E epsilon 4 heterozygotes: a foundation for using positron emission tomography to efficiently test treatments to prevent Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2001;98:3334–3339. doi: 10.1073/pnas.061509598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc. Natl Acad. Sci. USA. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Small GW, Ercoli LM, Silverman DH, Huang SC, Komo S, Bookheimer SY, Lavretsky H, Miller K, Siddarth P, Rasgon NL, et al. Cerebral metabolic and cognitive decline in persons at genetic risk for Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2000;97:6037–6042. doi: 10.1073/pnas.090106797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosconi L, Brys M, Switalski R, Mistur R, Glodzik L, Pirraglia E, Tsui W, De Santi S, de Leon MJ. Maternal family history of Alzheimer's disease predisposes to reduced brain glucose metabolism. Proc. Natl Acad. Sci. USA. 2007;104:19067–19072. doi: 10.1073/pnas.0705036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosconi L, Mistur R, Switalski R, Brys M, Glodzik L, Rich K, Pirraglia E, Tsui W, De Santi S, de Leon MJ. Declining brain glucose metabolism in normal individuals with a maternal history of Alzheimer disease. Neurology. 2009;72:513–520. doi: 10.1212/01.wnl.0000333247.51383.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheepers A, Joost HG, Schurmann A. The glucose transporter families SGLT and GLUT: molecular basis of normal and aberrant function. JPEN. J. Parenter. Enteral. Nutr. 2004;28:364–371. doi: 10.1177/0148607104028005364. [DOI] [PubMed] [Google Scholar]
- 24.Nagamatsu S, Kornhauser JM, Burant CF, Seino S, Mayo KE, Bell GI. Glucose transporter expression in brain. cDNA sequence of mouse GLUT3, the brain facilitative glucose transporter isoform, and identification of sites of expression by in situ hybridization. J. Biol. Chem. 1992;267:467–472. [PubMed] [Google Scholar]
- 25.Duelli R, Maurer MH, Staudt R, Sokoloff L, Kuschinsky W. Correlation between local glucose transporter densities and local 3-O-methylglucose transport in rat brain. Neurosci. Lett. 2001;310:101–104. doi: 10.1016/s0304-3940(01)02060-2. [DOI] [PubMed] [Google Scholar]
- 26.Khan JY, Rajakumar RA, McKnight RA, Devaskar UP, Devaskar SU. Developmental regulation of genes mediating murine brain glucose uptake. Am. J. Physiol. 1999;276:R892–R900. doi: 10.1152/ajpregu.1999.276.3.R892. [DOI] [PubMed] [Google Scholar]
- 27.Zeller K, Duelli R, Vogel J, Schrock H, Kuschinsky W. Autoradiographic analysis of the regional distribution of Glut3 glucose transporters in the rat brain. Brain Res. 1995;698:175–179. doi: 10.1016/0006-8993(95)00888-w. [DOI] [PubMed] [Google Scholar]
- 28.Manolescu AR, Witkowska K, Kinnaird A, Cessford T, Cheeseman C. Facilitated hexose transporters: new perspectives on form and function. Physiology. 2007;22:234–240. doi: 10.1152/physiol.00011.2007. [DOI] [PubMed] [Google Scholar]
- 29.Simpson IA, Davies P. Reduced glucose transporter concentrations in brains of patients with Alzheimer's disease. Ann. Neurol. 1994;36:800–801. doi: 10.1002/ana.410360522. [DOI] [PubMed] [Google Scholar]
- 30.Simpson IA, Chundu KR, Davies-Hill T, Honer WG, Davies P. Decreased concentrations of GLUT1 and GLUT3 glucose transporters in the brains of patients with Alzheimer's disease. Ann. Neurol. 1994;35:546–551. doi: 10.1002/ana.410350507. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Liu F, Iqbal K, Grundke-Iqbal I, Gong CX. Decreased glucose transporters correlate to abnormal hyperphosphorylation of tau in Alzheimer disease. FEBS Lett. 2008;582:359–364. doi: 10.1016/j.febslet.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mobasheri A, Richardson S, Mobasheri R, Shakibaei M, Hoyland JA. Hypoxia inducible factor-1 and facilitative glucose transporters GLUT1 and GLUT3: putative molecular components of the oxygen and glucose sensing apparatus in articular chondrocytes. Histol. Histopathol. 2005;20:1327–1338. doi: 10.14670/HH-20.1327. [DOI] [PubMed] [Google Scholar]
- 33.Rajakumar RA, Thamotharan S, Menon RK, Devaskar SU. Sp1 and Sp3 regulate transcriptional activity of the facilitative glucose transporter isoform-3 gene in mammalian neuroblasts and trophoblasts. J. Biol. Chem. 1998;273:27474–27483. doi: 10.1074/jbc.273.42.27474. [DOI] [PubMed] [Google Scholar]
- 34.Rajakumar A, Thamotharan S, Raychaudhuri N, Menon RK, Devaskar SU. Trans-activators regulating neuronal glucose transporter isoform-3 gene expression in mammalian neurons. J. Biol. Chem. 2004;279:26768–26779. doi: 10.1074/jbc.M402735200. [DOI] [PubMed] [Google Scholar]
- 35.Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc. Natl Acad. Sci. USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker WH, Sanborn BM, Habener JF. An isoform of transcription factor CREM expressed during spermatogenesis lacks the phosphorylation domain and represses cAMP-induced transcription. Proc. Natl Acad. Sci. USA. 1994;91:12423–12427. doi: 10.1073/pnas.91.26.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto KK, Gonzalez GA, Biggs WH, 3rd, Montminy MR. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334:494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 38.Braak H, Braak E. Staging of Alzheimer's disease-related neurofibrillary changes. Neurobiol. Aging. 1995;16:271–278. doi: 10.1016/0197-4580(95)00021-6. discussion 278–284. [DOI] [PubMed] [Google Scholar]
- 39.Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, Vogel FS, Hughes JP, van Belle G, Berg L. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 40.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 42.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci. STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 43.Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 44.Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong CX. O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2004;101:10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 46.Liu F, Grundke-Iqbal I, Iqbal K, Oda Y, Tomizawa K, Gong CX. Truncation and activation of calcineurin A by calpain I in Alzheimer disease brain. J. Biol. Chem. 2005;280:37755–37762. doi: 10.1074/jbc.M507475200. [DOI] [PubMed] [Google Scholar]
- 47.Saito K, Elce JS, Hamos JE, Nixon RA. Widespread activation of calcium-activated neutral proteinase (calpain) in the brain in Alzheimer disease: a potential molecular basis for neuronal degeneration. Proc. Natl Acad. Sci. USA. 1993;90:2628–2632. doi: 10.1073/pnas.90.7.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mattson MP, Chan SL. Neuronal and glial calcium signaling in Alzheimer's disease. Cell. Calcium. 2003;34:385–397. doi: 10.1016/s0143-4160(03)00128-3. [DOI] [PubMed] [Google Scholar]
- 49.Zimmerman UJ, Schlaepfer WW. Two-stage autolysis of the catalytic subunit initiates activation of calpain I. Biochim. Biophys. Acta. 1991;1078:192–198. doi: 10.1016/0167-4838(91)99009-h. [DOI] [PubMed] [Google Scholar]
- 50.Choi DW, Koh JY. Zinc and brain injury. Annu. Rev. Neurosci. 1998;21:347–375. doi: 10.1146/annurev.neuro.21.1.347. [DOI] [PubMed] [Google Scholar]
- 51.Khachaturian ZS. Calcium, membranes, aging, and Alzheimer's disease. Introduction and overview. Ann. N Y Acad. Sci. 1989;568:1–4. doi: 10.1111/j.1749-6632.1989.tb12485.x. [DOI] [PubMed] [Google Scholar]
- 52.Mattson MP, Cheng B, Davis D, Bryant K, Lieberburg I, Rydel RE. Beta-amyloid peptides destabilize calcium homeostasis and render human cortical neurons vulnerable to excitotoxicity. J. Neurosci. 1992;12:376–389. doi: 10.1523/JNEUROSCI.12-02-00376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bezprozvanny I, Mattson MP. Neuronal calcium mishandling and the pathogenesis of Alzheimer's disease. Trends Neurosci. 2008;31:454–463. doi: 10.1016/j.tins.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LaFerla FM. Calcium dyshomeostasis and intracellular signalling in Alzheimer's disease. Nat. Rev. Neurosci. 2002;3:862–872. doi: 10.1038/nrn960. [DOI] [PubMed] [Google Scholar]
- 55.Kusakawa G, Saito T, Onuki R, Ishiguro K, Kishimoto T, Hisanaga S. Calpain-dependent proteolytic cleavage of the p35 cyclin-dependent kinase 5 activator to p25. J. Biol. Chem. 2000;275:17166–17172. doi: 10.1074/jbc.M907757199. [DOI] [PubMed] [Google Scholar]
- 56.Lee MS, Kwon YT, Li M, Peng J, Friedlander RM, Tsai LH. Neurotoxicity induces cleavage of p35 to p25 by calpain. Nature. 2000;405:360–364. doi: 10.1038/35012636. [DOI] [PubMed] [Google Scholar]
- 57.Nath R, Davis M, Probert AW, Kupina NC, Ren X, Schielke GP, Wang KK. Processing of cdk5 activator p35 to its truncated form (p25) by calpain in acutely injured neuronal cells. Biochem. Biophys. Res. Commun. 2000;274:16–21. doi: 10.1006/bbrc.2000.3070. [DOI] [PubMed] [Google Scholar]
- 58.Veeranna, Kaji T, Boland B, Odrljin T, Mohan P, Basavarajappa BS, Peterhoff C, Cataldo A, Rudnicki A, Amin N, et al. Calpain mediates calcium-induced activation of the erk1,2 MAPK pathway and cytoskeletal phosphorylation in neurons: relevance to Alzheimer's disease. Am. J. Pathol. 2004;165:795–805. doi: 10.1016/S0002-9440(10)63342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang Z, Liu F, Grundke-Iqbal I, Iqbal K, Gong CX. Down-regulation of cAMP-dependent protein kinase by over-activated calpain in Alzheimer disease brain. J. Neurochem. 2007;103:2462–2470. doi: 10.1111/j.1471-4159.2007.04942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang KK, Roufogalis BD, Villalobo A. Characterization of the fragmented forms of calcineurin produced by calpain I. Biochem. Cell. Biol. 1989;67:703–711. doi: 10.1139/o89-105. [DOI] [PubMed] [Google Scholar]
- 61.Litersky JM, Johnson GV. Phosphorylation by cAMP-dependent protein kinase inhibits the degradation of tau by calpain. J. Biol. Chem. 1992;267:1563–1568. [PubMed] [Google Scholar]
- 62.Yang LS, Ksiezak-Reding H. Calpain-induced proteolysis of normal human tau and tau associated with paired helical filaments. Eur. J. Biochem. 1995;233:9–17. doi: 10.1111/j.1432-1033.1995.009_1.x. [DOI] [PubMed] [Google Scholar]
- 63.Satoh J, Tabunoki H, Arima K. Molecular network analysis suggests aberrant CREB-mediated gene regulation in the Alzheimer disease hippocampus. Dis. Markers. 2009;27:239–252. doi: 10.3233/DMA-2009-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saura CA, Valero J. The role of CREB signaling in Alzheimer's disease and other cognitive disorders. Rev Neurosci. 2011;22:153–169. doi: 10.1515/RNS.2011.018. [DOI] [PubMed] [Google Scholar]
- 65.Hyman C, Hofer M, Barde YA, Juhasz M, Yancopoulos GD, Squinto SP, Lindsay RM. BDNF is a neurotrophic factor for dopaminergic neurons of the substantia nigra. Nature. 1991;350:230–232. doi: 10.1038/350230a0. [DOI] [PubMed] [Google Scholar]
- 66.Marini AM, Jiang X, Wu X, Tian F, Zhu D, Okagaki P, Lipsky RH. Role of brain-derived neurotrophic factor and NF-kappaB in neuronal plasticity and survival: from genes to phenotype. Restor. Neurol. Neurosci. 2004;22:121–130. [PubMed] [Google Scholar]
- 67.Lowenstein DH, Arsenault L. Dentate granule cell layer collagen explant cultures: spontaneous axonal growth and induction by brain-derived neurotrophic factor or basic fibroblast growth factor. Neuroscience. 1996;74:1197–1208. doi: 10.1016/0306-4522(96)00226-6. [DOI] [PubMed] [Google Scholar]
- 68.Ando S, Kobayashi S, Waki H, Kon K, Fukui F, Tadenuma T, Iwamoto M, Takeda Y, Izumiyama N, Watanabe K, et al. Animal model of dementia induced by entorhinal synaptic damage and partial restoration of cognitive deficits by BDNF and carnitine. J. Neurosci. Res. 2002;70:519–527. doi: 10.1002/jnr.10443. [DOI] [PubMed] [Google Scholar]
- 69.Connor B, Young D, Yan Q, Faull RL, Synek B, Dragunow M. Brain-derived neurotrophic factor is reduced in Alzheimer's disease. Brain Res. Mol. Brain Res. 1997;49:71–81. doi: 10.1016/s0169-328x(97)00125-3. [DOI] [PubMed] [Google Scholar]
- 70.Phillips HS, Hains JM, Armanini M, Laramee GR, Johnson SA, Winslow JW. BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer's disease. Neuron. 1991;7:695–702. doi: 10.1016/0896-6273(91)90273-3. [DOI] [PubMed] [Google Scholar]
- 71.Ferrer I, Marin C, Rey MJ, Ribalta T, Goutan E, Blanco R, Tolosa E, Marti E. BDNF and full-length and truncated TrkB expression in Alzheimer disease. Implications in therapeutic strategies. J. Neuropathol. Exp. Neurol. 1999;58:729–739. doi: 10.1097/00005072-199907000-00007. [DOI] [PubMed] [Google Scholar]
- 72.Hock C, Heese K, Hulette C, Rosenberg C, Otten U. Region-specific neurotrophin imbalances in Alzheimer disease: decreased levels of brain-derived neurotrophic factor and increased levels of nerve growth factor in hippocampus and cortical areas. Arch. Neurol. 2000;57:846–851. doi: 10.1001/archneur.57.6.846. [DOI] [PubMed] [Google Scholar]
- 73.Holsinger RM, Schnarr J, Henry P, Castelo VT, Fahnestock M. Quantitation of BDNF mRNA in human parietal cortex by competitive reverse transcription-polymerase chain reaction: decreased levels in Alzheimer's disease. Brain Res. Mol. Brain Res. 2000;76:347–354. doi: 10.1016/s0169-328x(00)00023-1. [DOI] [PubMed] [Google Scholar]
- 74.Michalski B, Fahnestock M. Pro-brain-derived neurotrophic factor is decreased in parietal cortex in Alzheimer's disease. Brain Res. Mol. Brain Res. 2003;111:148–154. doi: 10.1016/s0169-328x(03)00003-2. [DOI] [PubMed] [Google Scholar]
- 75.Comb M, Birnberg NC, Seasholtz A, Herbert E, Goodman HM. A cyclic AMP- and phorbol ester-inducible DNA element. Nature. 1986;323:353–356. doi: 10.1038/323353a0. [DOI] [PubMed] [Google Scholar]
- 76.Short JM, Wynshaw-Boris A, Short HP, Hanson RW. Characterization of the phosphoenolpyruvate carboxykinase (GTP) promoter-regulatory region. II. Identification of cAMP and glucocorticoid regulatory domains. J. Biol. Chem. 1986;261:9721–9726. [PubMed] [Google Scholar]
- 77.Fink JS, Verhave M, Kasper S, Tsukada T, Mandel G, Goodman RH. The CGTCA sequence motif is essential for biological activity of the vasoactive intestinal peptide gene cAMP-regulated enhancer. Proc. Natl Acad. Sci. USA. 1988;85:6662–6666. doi: 10.1073/pnas.85.18.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Craig JC, Schumacher MA, Mansoor SE, Farrens DL, Brennan RG, Goodman RH. Consensus and variant cAMP-regulated enhancers have distinct CREB-binding properties. J. Biol. Chem. 2001;276:11719–11728. doi: 10.1074/jbc.M010263200. [DOI] [PubMed] [Google Scholar]
- 79.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell. Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 81.Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl Acad. Sci. USA. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grundke-Iqbal I, Iqbal K, Quinlan M, Tung YC, Zaidi MS, Wisniewski HM. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J. Biol. Chem. 1986;261:6084–6089. [PubMed] [Google Scholar]
- 83.Iqbal K, Grundke-Iqbal I, Zaidi T, Merz PA, Wen GY, Shaikh SS, Wisniewski HM, Alafuzoff I, Winblad B. Defective brain microtubule assembly in Alzheimer's disease. Lancet. 1986;2:421–426. doi: 10.1016/s0140-6736(86)92134-3. [DOI] [PubMed] [Google Scholar]
- 84.Alonso AC, Zaidi T, Novak M, Grundke-Iqbal I, Iqbal K. Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proc. Natl Acad. Sci. USA. 2001;98:6923–6928. doi: 10.1073/pnas.121119298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alonso AC, Grundke-Iqbal I, Iqbal K. Alzheimer's disease hyperphosphorylated tau sequesters normal tau into tangles of filaments and disassembles microtubules. Nat. Med. 1996;2:783–787. doi: 10.1038/nm0796-783. [DOI] [PubMed] [Google Scholar]
- 86.Alonso AC, Zaidi T, Grundke-Iqbal I, Iqbal K. Role of abnormally phosphorylated tau in the breakdown of microtubules in Alzheimer disease. Proc. Natl Acad. Sci. USA. 1994;91:5562–5566. doi: 10.1073/pnas.91.12.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lucas JJ, Hernandez F, Gomez-Ramos P, Moran MA, Hen R, Avila J. Decreased nuclear beta-catenin, tau hyperphosphorylation and neurodegeneration in GSK-3beta conditional transgenic mice. EMBO J. 2001;20:27–39. doi: 10.1093/emboj/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fath T, Eidenmuller J, Brandt R. Tau-mediated cytotoxicity in a pseudohyperphosphorylation model of Alzheimer's disease. J. Neurosci. 2002;22:9733–9741. doi: 10.1523/JNEUROSCI.22-22-09733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jackson GR, Wiedau-Pazos M, Sang TK, Wagle N, Brown CA, Massachi S, Geschwind DH. Human wild-type tau interacts with wingless pathway components and produces neurofibrillary pathology in Drosophila. Neuron. 2002;34:509–519. doi: 10.1016/s0896-6273(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 90.Liu F, Shi J, Tanimukai H, Gu J, Gu J, Grundke-Iqbal I, Iqbal K, Gong CX. Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer's disease. Brain. 2009;132:1820–1832. doi: 10.1093/brain/awp099. [DOI] [PMC free article] [PubMed] [Google Scholar]