Abstract
The use of alternative poly(A) sites is common and affects the post-transcriptional fate of mRNA, including its stability, subcellular localization and translation. Here, we present a method to identify poly(A) sites in a genome-wide and strand-specific manner. This method, termed 3′T-fill, initially fills in the poly(A) stretch with unlabeled dTTPs, allowing sequencing to start directly after the poly(A) tail into the 3′-untranslated regions (UTR). Our comparative analysis demonstrates that it outperforms existing protocols in quality and throughput and accurately quantifies RNA levels as only one read is produced from each transcript. We use this method to characterize the diversity of polyadenylation in Saccharomyces cerevisiae, showing that alternative RNA molecules are present even in a genetically identical cell population. Finally, we observe that overlap of convergent 3′-UTRs is frequent but sharply limited by coding regions, suggesting factors that restrict compression of the yeast genome.
INTRODUCTION
The 3′-untranslated regions (UTRs) of mRNAs, located directly after the stop codon, harbor signals for transcript stability, localization and translational control (reviewed in (1) and (2)). The control of mRNA expression by 3′-UTRs is mediated by trans-acting factors, including RNA-binding proteins and microRNAs (miRNAs), which interact with cis-regulatory elements within the 3′-UTRs (3). More than half of mammalian genes produce transcripts with 3′-UTRs of different lengths (4). Shorter 3′-UTRs have been associated with proliferation and transformation, which is partially explained by the exclusion of miRNA-binding sites in the 3′-UTRs of proto-oncogenes (5,6). Shortening of 3′-UTRs is also observed at early stages of development in mice (7), flies (8) and worms (9) as well as during reprogramming of stem cells (10). Longer 3′-UTRs, on the other hand, tend to be more frequent in differentiated cells (11,12). Unicellular eukaryotes such as yeast have also been shown to use different poly(A) sites in response to stress conditions (13). To understand the mechanisms underlying alternative poly(A) site usage and its functional significance, there is a need for tools that accurately and efficiently map these sites on a genome-wide scale.
With the emergence of next-generation sequencing technologies, it has become possible to sequence entire transcriptomes within days. However, standard mRNA sequencing (RNA-Seq) is inefficient for poly(A) site mapping: only a small fraction of sequencing reads contain poly(A) tails, making it difficult to distinguish transcript end isoforms. Therefore, protocols have been developed to enrich for transcript ends of poly(A) RNA prior to RNA-Seq and/or to identify poly(A) sites (13–20). However, efficient poly(A) site identification remains technologically challenging because in many protocols, a substantial fraction of reads either does not reach the poly(A) site or is of low quality and must be discarded. In protocols where sequencing proceeds from within the transcript toward the poly(A) tail (13,14,16,20), the readout strongly depends on a stringent size selection to ensure that each read contains enough of the 3′-UTR to map to the genome while still reaching the poly(A) tail. Other Illumina-based protocols (17,19), in which sequencing starts at the 3′-end and reads through the poly(A) site, suffer from low sequence quality (Supplementary Figure S1). This is likely due to desynchronization of base incorporation caused by polymerase slippage during clustering and sequencing or mispriming of the sequencing oligo. To overcome this, the poly(A) tail can be shortened (18). However, even a few remaining nucleotides of the poly(A) tail can compromise the delineation of clusters by the sequencing software (18). Furthermore, performing the poly(A) shortening directly on the RNA could be damaging.
Therefore, we have developed a poly(A) site mapping protocol that circumvents reading through the poly(A) tail, termed ‘3′T-fill’. In this protocol, the poly(A) stretch is filled in with dTTPs before the sequencing reaction starts (Figure 1). This enables sequencing to start immediately after the poly(A) tail. After assessing the accuracy of this approach and experimentally comparing it with alternative methods, we analyzed poly(A) sites in the yeast transcriptome and discovered that simultaneous presence of alternative terminal mRNA isoforms is the rule rather than the exception. In addition, we provide new insight into the genome-wide organization of poly(A) sites in Saccharomyces cerevisiae, showing that antisense poly(A) sites are sharply limited by the presence of the coding region, but not by the length of 3′-UTRs.
Figure 1.
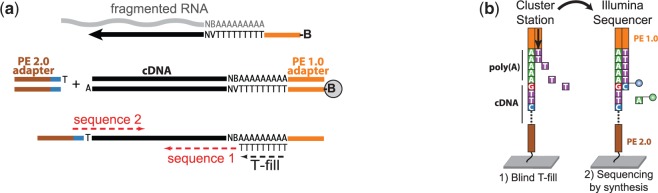
3′T-fill method overview. (a) Schematic of the 3′T-fill protocol. Fragmented RNA is reverse transcribed with an oligo(dT) primer coupled to adapter PE 1.0 (orange) and biotin (B). After second strand synthesis, fragments are captured on beads and the barcoded adapter PE 2.0 (brown) is ligated to the bound fragment. (b) On the cluster station, the poly(A) tail is filled in with complementary, unlabeled dTTPs. Sequencing starts directly at the end of the 3′-UTR (‘sequence 1’ in panel (a)). For multiplexing, the barcode is read by paired-end sequencing (‘sequence 2’).
MATERIALS AND METHODS
Yeast RNA isolation
We grew S. cerevisiae strain SLS045 (S288c background) (21) to mid-log phase (OD ∼1) using either YPD (1% yeast extract, 2% peptone and 1% glucose) or YPGal (1% yeast extract, 2% peptone and 1% galactose). Total RNA was isolated by a standard hot phenol method and treated with RNase-free DNaseI using Turbo DNA-free kit (Ambion). To each 600 µg of DNaseI-treated total RNA, 1.36 ng pGIBS-LYS, 3.6 ng pGIBS-PHE and 10.7 ng pGIBS-THR polyadenylated in vitro transcripts (IVTs) were added as external controls (ATCC 87482, 87483 and 87484, respectively).
Library preparation
For the results presented, 10 µg of total RNA was used as starting material. This amount could be reduced to 500 ng without a significant loss in quality (results not shown). The RNA was fragmented by incubating the samples at 80°C for 5 min in the presence of RNA fragmentation buffer (40 mM Tris-acetate, pH 8.1, 100 mM KOAc and 30 mM MgOAc). The fragmented RNA was purified using 1.5× Ampure XP Beads (Beckman Coulter Genomics) and eluted in 12.8 µl elution buffer (EB) (10 mM Tris–HCl, pH 8). For retrotranscription, 11.2 µl of the eluted RNA was mixed with 1 µl of biotinylated oligo P5_dT16VN (1 µM; Supplementary Table S3) and 1 µl of 10 mM dNTPs. The samples were incubated at 65°C for 5 min and transferred to ice. Four microliters of 5× first-strand buffer (Invitrogen), 2 µl DTT 0.1 M, 0.32 µl actinomycin D (1.25 mg/µl) and 0.5 µl RNasin plus RNase inhibitor (Promega) were added to each sample, and samples were incubated at 42°C for 2 min to minimize possible mispriming. Following this, 0.5 µl Superscript II reverse transcriptase (200 U/µl; Invitrogen) was used for retrotranscription (Figure 1a). The reaction was performed at 42°C for 50 min and inactivated at 70°C for 15 min. The samples were purified using 1.5× of Ampure XP beads and eluted in 40 µl EB. For producing the second cDNA strand, 40 µl of sample was mixed with 5 µl of 10× DNA polymerase buffer (Fermentas), 2.5 µl of dNTPs (10 mM), 0.5 µl of RNaseH (5 U/µl; NEB) and 2 µl of DNA polymerase I (10 U/µl; Fermentas). The samples were incubated at 16°C for 2.5 h, purified with 0.8× Ampure XP beads and eluted in 20 µl EB. Twenty microliters of Dynabeads M-280 Streptavidin (Invitrogen) were washed two times with 200 µl 1× B&W buffer (5 mM Tris–HCl, pH 7.5, 0.5 mM ethylenediaminetetraacetic acid (EDTA) and 1 M NaCl) and resuspended in 20 µl of 2× B&W buffer. Twenty microliters of the double-stranded cDNA sample was bound to the 20 µl of Dynabeads by mixing them for 15 min at 25°C. The beads were washed twice with 200 µl of 1× B&W buffer, once with 200 µl EB and resuspended in 21.25 µl EB. 2.5 µl end repair buffer and 1.25 µl end repair enzyme mix (NEBNext DNA Sample Prep Master Mix Set 1, NEB) were added and the samples incubated at 20°C for 30 min. The beads were washed twice with 200 µl of 1× B&W buffer, once with 200 µl EB and resuspended in 21 µl EB. 2.5 µl dA tailing buffer (10× NEBuffer 2 from NEB and 0.2 mM dATP) and 1.5 µl Klenow Fragment (3′→5′ exo–) 5 U/µl (NEB) were added and the samples incubated at 37°C for 30 min. The beads were washed twice with 200 µl 1× B&W buffer, once with 200 µl EB and resuspended in 8 µl EB. 12.5 µl 2× Quick ligation buffer (NEB), 2 µl P7_T1_Mpx linker (2.5 µM; Supplementary Table S3) and 2 µl T4 DNA ligase were added (2000 U/µl; NEB). The samples were incubated while shaking for 15 min at 20°C to ligate the adapters. The beads were washed four times with 200 µl 1× B&W buffer, once with 200 µl EB and resuspended in 50 µl EB. Enrichment polymerase chain reaction (PCR) was performed using 24 µl of beads, 25 µl Phusion Master Mix 2× (NEB) and 0.5 µl each of oligos PE1.0 and PE2.0 (10 µM; Illumina). The PCR program was 30 s at 96°C, 18 cycles of (10 s at 96°C, 10 s at 65°C and 10 s at 72°C) and 5 min at 72°C. The PCR product was purified with 1.8× Ampure XP Beads. 300 bp libraries were size selected using e-Gel 2% SizeSelect (Invitrogen).
The 3′T-fill reaction
The final libraries were loaded into the cluster station (cBot, Illumina) and the priming buffer was exchanged for the T-fill solution: 101 µl water, 20 µl Phusion buffer HF (5×) (NEB), 3 µl dTTPs (10 mM), 0.8 µl genomic DNA Sequencing primer V2 (100 µM) (Illumina), 3 µl non-hot start Phusion polymerase (2 U/µl, NEB) and 2 µl Taq polymerase E (5 U/µl; Genaxxon). The addition of the latter Taq polymerase was crucial for a complete T-fill. During the 5 min incubation at 60°C (standard cBot step for primer annealing), the primer aligns and the poly(A) stretch is filled in with dTTPs (Figure 1b). After clustering, the samples were sequenced on a HiSeq 2000 (Illumina).
Read pre-processing and alignment
The python package HTSeq (Anders,S. http://www-huber.embl.de/users/anders/HTSeq/) was used for processing of the sequencing data. De-multiplexed samples were trimmed to remove the poly(A) tail and adapter sequences before alignment. Read alignment was performed using the GSNAP aligner (22). As a reference genome, we used the S. cerevisiae S288c genome (SGD R63) combined with the three IVTs from Bacillus subtilis used as spike-in controls in this study. Reads with low mapping quality (<30) or which were composed of >80% As or Ts were filtered out. We also filtered out potential internal priming events by discarding reads mapping to a genomic region with eight or more As downstream, or high A/T content (27 out of 30 preceding bases).
Assessing signal enrichment at transcript ends
The S. cerevisiae transcriptome annotation of Xu et al. (21) was updated to reflect the genome sequence version used. Aligned reads from all tested protocols were mapped to the 7272 full-length annotated transcript features of this annotation, with either a +200 bp extension downstream of the transcriptional termination site (TTS) to account for poly(A) sites extending beyond current annotation or a ±200 bp extension from TTS. Reads with ambiguous assignment (0.8% of assigned reads for 3′T-fill) were assigned to the transcript with the closest proximal 3′-end.
Estimating poly(A) site calling accuracy using IVTs
Poly(A) site calls were defined as the read base closest to the poly(A) tail (e.g. the first base in the 3′T-fill protocol). The accuracy of poly(A) site calling was calculated as the fraction of reads supporting the known poly(A) site, of total non-filtered reads mapping to each IVT.
Distribution of poly(A) sites within transcripts
Poly(A) sites were uniquely mapped to the updated transcriptome annotation from Xu et al. (21) with a +200 bp extension downstream of the TTS. Within these features, poly(A) site counts were grouped to those assigning to UTRs, coding regions and extension regions for genes. Only verified, monocistronic genes were considered for this analysis.
Definition and analysis of high-confidence poly(A) sites
Poly(A) site calls from all the samples were merged to create a common poly(A) site map, such that non-zero counts on consecutive bases were merged. High-confidence poly(A) sites were defined as sites detected in all three replicates of a given condition and with a minimum of 10 supporting reads in each. For estimating the number of poly(A) sites per gene, we further selected only poly(A) sites accounting for at least 10% of the total expression of that gene. The alternative presence of 14 RNA-binding protein target sequences, obtained from Supplementary Data S1 from Riordan et al. (23), was determined for this set of poly(A) sites. The fraction of reads accounted for by the major 3′-isoform for each gene was calculated for all genes with multiple high-confidence poly(A) sites, and the presence of antisense expression was computed using all high-confidence poly(A) sites.
Expression quantification
3′T-fill and RNA-Seq reads were assigned to the full-length transcript annotation of Xu et al. (21) as described above. Differential expression calling was performed using the R/Bioconductor package DESeq (24). As our goal was to compare the technical suitability of 3′T-fill and RNA-Seq for expression quantification, only technical replicates were used. Expression levels of poly(A) isoforms detected by 3′T-fill were computed by comparing counts supporting poly(A) sites in the merged poly(A) site map by different replicates. Changes in poly(A) site influenced by the growth medium (glucose versus galactose) were assessed by extension of a statistical method developed for differential exon usage (25).
RESULTS
Comparison to alternative methods
To assess the performance of our 3′T-fill approach, we conducted an extensive comparison with alternative methods for poly(A) mapping (Supplementary Figures S2 and S3 and Supplementary Table S1). In addition to a strand-specific RNA-Seq protocol (26), with very few reads that map poly(A) sites (Figure 2), we evaluated four poly(A)-mapping methods, including one RNA-Seq following enrichment of poly(A)-containing RNA fragments (as in (13)), one which sequences into the poly(A) tail called ‘3′Internal’ (27), similar to (14,16), one which cleaves off the poly(A) tail called ‘3′BpmI’ (similar principle to Jan et al. (18), but cleavage is performed on cDNA rather than RNA) and the 3′T-fill method which fills up the poly(A) stretch before sequencing. Details of the compared methods can be found in Supplementary Methods and Supplementary Figures S2 and S3. Our 3′T-fill protocol (Figure 1) avoids delicate RNA processing steps by reverse transcribing the RNA directly after fragmentation. Binding the fragments to magnetic beads early in the protocol allows for automation and easy scaling up to 96-well plates. Compared with protocols where sequencing proceeds from the 5′-direction into the poly(A) tail, this protocol offers several advantages: size selection is not critical, no trimming is necessary and both forward and reverse reads are informative. Importantly, the 3′T-fill method is the most easily performed of all protocols developed thus far. The 3′T-fill protocol is also the most efficient of all the protocols we tested, with all mapped reads identifying poly(A) sites with high confidence (74% of raw reads, Supplementary Table S1).
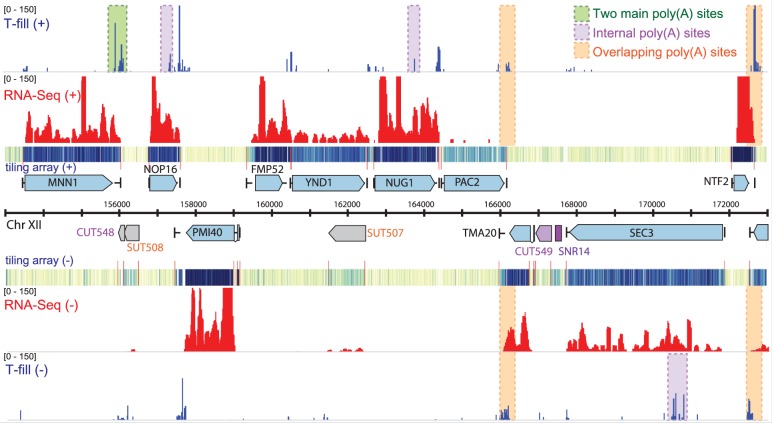
Figure 2.
Example reads from 3′T-fill, RNA-Seq and tiling array (21) in yeast grown in rich glucose media. Only a small fraction of the tiling array and total RNA-Seq signals arises from 3′-ends of transcripts. With the 3′T-fill protocol many alternative (green), internal (purple) and overlapping poly(A) sites (orange) were detected. Visualization was performed with the Integrative Genomics Viewer (IGV) (28).
Accuracy of poly(A) site mapping
To estimate the accuracy of the 3′T-fill protocol for mapping the precise poly(A) sites, three IVTs (1–2 kb each) from B. subtilis with a DNA-encoded poly(A) tail were spiked into the starting RNA material. On average, 83% of 3′T-fill reads mapping to these transcripts identified the exact poly(A) site and 97% within a window of ±2 nt. For the Helicos technology, which applies a similar approach to fill the poly(A) tail (15), but used much shorter IVTs (40 nt), 65.6% of the IVT reads were reported to map the exact poly(A) site and 99.7% within ±2 nt (Supplementary Table S1). Compared with other methods, the proportion of 3′T-fill reads that uniquely mapped to the yeast 3′-UTRs was highest in the T-fill method, which confirms its specificity for identifying the correct poly(A) sites.
RNA quantification
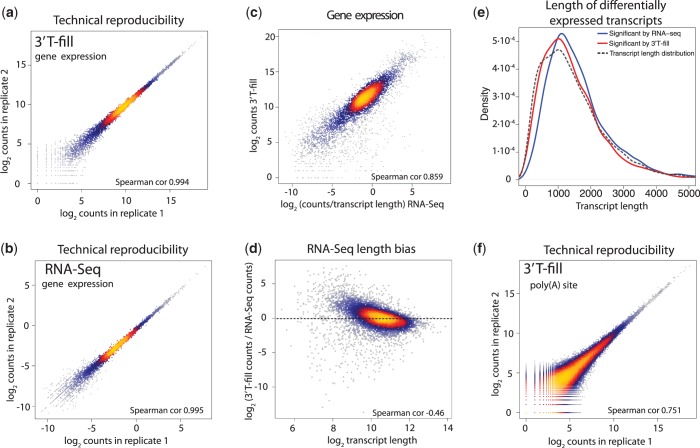
To benchmark the RNA quantification efficiency of our approach, we compared the results obtained with our 3′T-fill method to those from standard, strand-specific RNA-Seq. The technical reproducibility of the 3′T-fill method is high and at similar levels to RNA-Seq (Figure 3a and b). Importantly, 3′T-fill and RNA-Seq measurements of gene expression highly correlate (Figure 3c). Thus, the 3′T-fill approach can also be used to identify genes that are differentially expressed between conditions (Supplementary Figure S4). However, in contrast to RNA-Seq experiments, in which more reads are produced from long transcripts, 3′T-fill only produces one read per transcript, resulting in a size-independent representation of transcript abundance (Figure 3d and e). This represents a major advantage over RNA-Seq, in which short transcripts might be underrepresented when assessing differential expression. Most importantly, a poly(A) mapping approach allows for discrimination of transcript end isoforms, which are mostly overlooked in RNA-Seq. The number of reads with the same poly(A) position also correlated well between two technical replicates of the T-fill method (Figure 3f).
Figure 3.
Comparison between 3′T-fill and RNA-Seq. Reproducibility of transcript quantification was high between technical replicates of 3′T-fill (a) and RNA-Seq (b), as well as between the two methods (c). (d) Since 3′T-fill does not display the length bias of RNA-Seq, 3′T-fill captures more shorter transcripts, providing a size-independent estimate of their abundance. (e) The length distribution of differentially expressed genes (in glucose versus galactose media) in 3′T-fill (red) is closer to the global transcript length (dashed line) than transcript length in RNA-Seq (blue). 2441 and 3401 genes were differentially expressed for 3′T-fill and RNA-Seq, respectively (adjusted P < 0.1). (f) A high reproducibility in mapping individual poly(A) sites was observed between technical replicates of 3′T-fill samples. Data from panels a–c and f correspond to cells grown in galactose.
Poly(A) site organization in the yeast transcriptome
Having established the accuracy of our method, we collectively analyzed RNA quantification and alternative 3′-ends to investigate the nature of poly(A) site organization in the yeast transcriptome (excerpt in Figure 2). We detected 22 512 different transcript end isoforms with high confidence (see ‘Materials and Methods’ section for definition) present in glucose medium alone. Approximately 43% of all transcripts have multiple poly(A) sites, and ∼700 genes express at least two transcript isoforms where the poly(A) sites are at least 100 bp apart. Overall, the major 3′-isoform for each gene only accounts for 67.5% of the reads, indicating that alternative mRNA molecules are present even in a genetically identical cell population. In fact, we observe that 13% of genes have differential isoforms with alternative presence of RNA-binding protein target sequences (23) and thus potentially varying posttranscriptional fate. Although glucose and galactose conditions had remarkably similar global poly(A) site profiles, we detected 187 transcripts with differences in poly(A) site usage between these conditions (Supplementary Table S2). Furthermore, 3.3% of the reads were detected within coding sequences, indicating internal polyadenylation that could affect the length of the protein product.
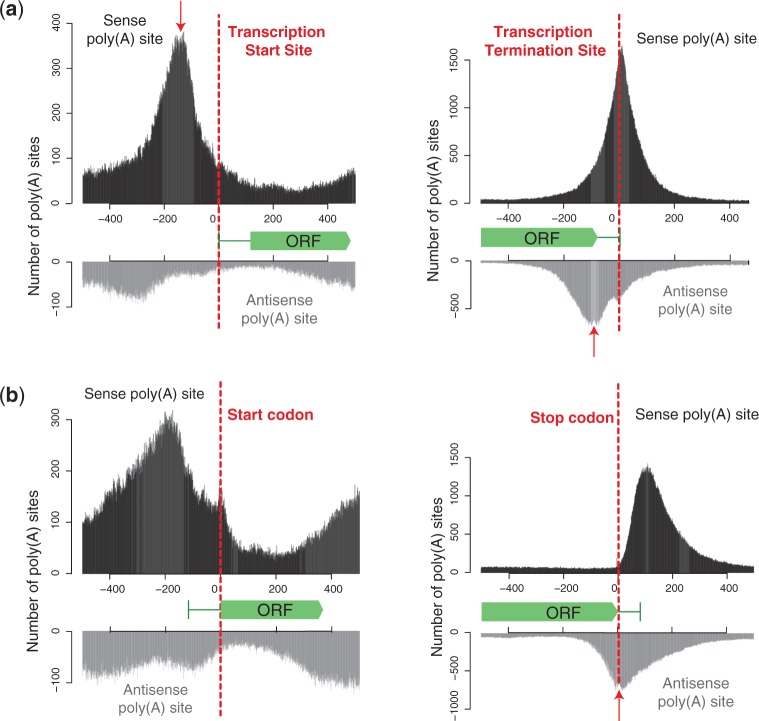
The detected poly(A) sites for coding transcripts (Figure 4a) yielded well-defined 3′-ends that are in good agreement with the existing transcriptome annotation (21). A similar pattern was observed for stable unannotated transcripts (SUTs) (Supplementary Figure S5a), whereas poly(A) sites for cryptic unstable transcripts (21) were much less defined (Supplementary Figure S5b), which is in accordance with their alternative termination pathway (29). Forty percent of transcripts have antisense transcripts that terminate within their annotated 3′-UTRs. This is more than the 12% (30) and 14% (15) previously reported and likely due to the high coverage in our dataset. We found that the antisense poly(A) site signal peaks ∼100 bp upstream from the annotated transcription termination site for coding transcripts (Figure 4a) and SUTs (Supplementary Figure S5a). Interestingly, the signal of these antisense peaks declines sharply after the sense open reading frame (ORF) stop codon (Figure 4b). This decline, together with the observation that poly(A) sites accumulate directly upstream of the transcription start site (Figure 4a), suggests that the promoter as well as the coding sequence limit further compaction of the yeast genome.
Figure 4.
Accumulation of poly(A) sites relative to annotated gene features. Poly(A) site positions on sense (upper panels in black) and antisense strands (lower panels in gray) relative to annotated (a) transcription start sites (left), transcription termination sites (right), (b) ORF starts (left) and ORF ends (right). Arrows indicate the following: sharp drop-off in poly(A) site abundance of upstream genes just before ORF promoters/transcription start sites (a, left); peak of convergent 3′-UTR overlap at 100 bp upstream of transcription termination sites (a, right) and sharp drop in overlap of antisense 3′-UTRs beyond ORF stop codons (b, right).
DISCUSSION
As only a very small fraction of reads obtained from a strand-specific RNA-Seq run contain the poly(A) site (Figure 2), several methods that aim to identify the poly(A) sites have been developed, yet to date no comparative analysis of these protocols has been performed. In this study, we tested and compared four methods for poly(A) site mapping. Being among the most accurate methods in calling the exact poly(A) site and having the highest number of reads in annotated 3′-UTRs, the 3′T-fill outperforms currently available protocols in both simplicity and efficiency. It avoids delicate RNA processing and is flexible in library size and read length. In addition to its capacity to distinguish and quantify alternative poly(A) site usage, it enables quantification of RNA expression in a length-independent manner as only one read is produced per transcript. This method can easily be extended to non-polyadenylated RNAs (e.g. bacterial RNA, eukaryotic non-polyadenylated RNAs) by artificial RNA polyadenylation. The T-fill step has the potential to improve poly(A) sequencing on other sequencing platforms, as slippage during poly(A) amplification is likely to occur on any platform that requires an amplification step prior to sequencing (e.g. 454, Solid, Ion Torrent). Also, single cell poly(A) sequencing (31) where efficiency is crucial for the outcome would greatly profit from our protocol. One limitation of 3′T-fill compared with ‘direct RNA sequencing’ (Helicos) (15) (instrument no longer commercially available) is that it requires amplification of cDNA prior to sequencing. To circumvent this step, it would in principle be possible to combine our method with ‘FRT-seq’ (32) and synthesize the cDNA directly on the flow cell.
In addition to presenting a new poly(A) sequencing method, our analysis of poly(A) site usage in S. cerevisiae expands the current view of yeast transcriptome architecture and indicates factors that limit the compression of the yeast genome. This method constitutes a powerful tool for future studies of fundamental biological processes, such as comparisons of different developmental or disease stages in higher eukaryotes, in which it can be used to clarify the role of differential poly(A) site usage and expression level in regulating cellular function.
ACCESSION NUMBERS
Raw and processed data are available from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/, accession number GSE40110) and at our website, http://steinmetzlab.embl.de/polyAmapping/, browsable (IGV) files are provided.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–5 and Supplementary Methods.
FUNDING
University of Luxembourg—Institute for Systems Biology Program and the Deutsche Forschungsgemeinschaft (to L.M.S.) and an EMBO fellowship (to V.P.). Funding for open access charge: Deutsche Forschungsgemeinschaft (to S.W.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Raeka Aiyar, Julien Gagneur, Nicolas Delhomme, Louise-Amelie Schmitt and Wu Wei for helpful discussions and EMBL Genomics Core Facility for technical support.
REFERENCES
- 1.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:22–31. doi: 10.1002/wrna.47. [DOI] [PubMed] [Google Scholar]
- 3.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 4.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl Acad. Sci. USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomsen S, Azzam G, Kaschula R, Williams LS, Alonso CR. Developmental RNA processing of 3′UTRs in Hox mRNAs as a context-dependent mechanism modulating visibility to microRNAs. Development. 2010;137:2951–2960. doi: 10.1242/dev.047324. [DOI] [PubMed] [Google Scholar]
- 9.Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. The landscape of C. elegans 3′UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon OK, Brem RB. Noncanonical transcript forms in yeast and their regulation during environmental stress. RNA. 2010;16:1256–1267. doi: 10.1261/rna.2038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox-Walsh K, Davis-Turak J, Zhou Y, Li H, Fu XD. A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3′ end formation. Genomics. 2011;98:266–271. doi: 10.1016/j.ygeno.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck AH, Weng Z, Witten DM, Zhu S, Foley JW, Lacroute P, Smith CL, Tibshirani R, van de Rijn M, Sidow A, et al. 3′-end sequencing for expression quantification (3SEQ) from archival tumor samples. PLoS One. 2010;5:e8768. doi: 10.1371/journal.pone.0008768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, Vrielink JA, Agami R. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol. 2012;13:R59. doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riordan DP, Herschlag D, Brown PO. Identification of RNA recognition elements in the Saccharomyces cerevisiae transcriptome. Nucleic Acids Res. 2011;39:1501–1509. doi: 10.1093/nar/gkq920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22: 2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelechano V, Wilkening S, Järvelin AI, Tekkedil MM, Steinmetz LM. Genome wide Analyses of Chromatin and Transcripts. In: Carl Wu CDA, editor. Nucleosomes, Histones & Chromatin, Part B. Vol. 513. San Diego, CA: Academic Press; 2012. pp. 271–294. [Google Scholar]
- 28.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 30.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Mamanova L, Turner DJ. Low-bias, strand-specific transcriptome Illumina sequencing by on-flowcell reverse transcription (FRT-seq) Nat. Protoc. 2011;6:1736–1747. doi: 10.1038/nprot.2011.399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.