Abstract
Previously, we published a method for creating a novel DNA substrate, the double Holliday junction substrate. This substrate contains two Holliday junctions that are mobile, topologically constrained and separated by a distance comparable with conversion tract lengths. Although useful for studying late stage homologous recombination in vitro, construction of the substrate requires significant effort. In particular, there are three bottlenecks: (i) production of large quantities of single-stranded DNA; (ii) the loss of a significant portion of the DNA following the recombination step; and (iii) the loss of DNA owing to inefficient gel extraction. To address these limitations, we have made the following changes to the protocol: (i) use of a helper plasmid, rather than exogenous helper phage, to produce single-stranded DNA; (ii) use of the unidirectional ϕC31 integrase system in place of the bidirectional Cre recombinase reaction; and (iii) gel extraction by DNA diffusion. Here, we describe the changes made to the materials and methods and characterize the substrates that can be produced, including migratable single Holliday junctions, hemicatenanes and a quadruple Holliday junction substrate.
INTRODUCTION
Homologous recombination is an important process in the cell for replication fork recovery, double-strand break repair and meiotic crossovers (1). The Szostak model of homologous recombination proposes a double Holliday junction intermediate following strand invasion (2), and such junction structures have been observed in yeast (3). Several studies have shown that the complete absence of Holliday junction (HJ) processing enzymes results in lethality (4,5).
The recent discovery of Gen1 (6), a human HJ resolvase similar to bacterial RuvC, has brought renewed interest in HJ processing enzymes. Many studies use oligonucleotide-based HJ structures, which are easy to create but may not accurately reflect the structures formed in vivo. First, it is difficult to create mobile HJs from oligonucleotides, as the base pairing may shift during annealing. Second, although oligonucleotide-based HJs typically form folded, antiparallel structures (7), the junctions formed by neighboring homologous regions are necessarily parallel following invasion (2) to allow for replication and migration. Third, oligonucleotides have free ends, and therefore do not retain topological constraints that will occur between larger molecules.
We previously published a method for creating a novel DNA substrate, the double Holliday junction substrate (DHJS) (8). This substrate contains two HJs that are mobile, topologically constrained and separated by a distance comparable with conversion tract lengths (9,10). The DHJS has been used to characterize the yeast and Drosophila dissolution complexes (11–13). However, the difficulty of producing the substrate has hindered its use in the field. In this work, we describe new methodology for creating a variety of HJ substrates. A helper plasmid that allows constitutive expression of the desired single-stranded DNA (ssDNA) circles streamlines production of this essential starting material, which can be used to make single, double or quadruple HJ substrates. In addition to previously described substrates (containing single and double HJs), replacing the Cre/lox recombination system with the unidirectional ϕC31 integrase system greatly increases DNA yield and creates a quadruple HJ substrate that is dissolved in the same manner as the original DHJS.
MATERIALS AND METHODS
Creation of the pM13g plasmid
The construct M13cp was kindly provided by Dr. Andrew Bradbury (14). This construct maintains the M13 coding genome as a plasmid with a p15a origin of replication and a chloramphenicol resistance marker. To use this construct in Escherichia coli XL2-Blue MRF′, the chloramphenicol cassette of the M13cp construct was replaced with a gentamicin resistance gene amplified from the pFastBac1 plasmid by standard cloning methods. Clones were functionally tested by transformation into E. coli XL2-Blue MRF′/pDHJS AN+ and assayed for phagemid expression. A successful clone was transformed into DH5α and made chemically competent. Although pM13g demonstrated successful phagemid expression in all stains tested, DH5α proved to have the most consistent, stable expression and was therefore used for large-scale phagemid expression.
Insertion of the attB/attP sites
Oligonucleotides containing the attB and attP sites plus appropriate sticky ends were obtained (Integrated DNA Technologies) Table 1 and annealed using a thermocycler. The attB and attP sites were sequentially inserted into the pDHJS plasmids using the BglII and EcoRI sites and the SphI/NheI and NdeI sites, respectively. For ease of screening, the BglII and NdeI sites were eliminated in the final plasmid owing to the oligonucleotide sequence.
Table 1.
Primers for inserting attB and attP into the pDHJS plasmids
| Name | Sequence |
|---|---|
| attB F | GATCGCGCGGTGCGGGTGCCAGGGCGTGCCCTTGGGCTCCCCGGGCGCGTACTG |
| attB R | AATTCAGTACGCGCCCGGGGAGCCCAAGGGCACGCCCTGGCACCCGCACCGCGA |
| attP F (SphI) | CGCACACAATTGATGACTTGATCTAGAGTAGTGCCCCAACTGGGGTAACCTTTGAGTTCTCTCA GTTGGGGGCGTAGG |
| attP R (SphI) | ATATCCTACGCCCCCAACTGAGAGAACTCAAAGGTTACCCCAGTTGGGGCACTAC TCTAGA TCAAGTCATCAATTGTGTGCGCATG |
| attP F (NheI) | CTAGCGCACACAATTGATGACTTGATCTAGAGTAGTGCCCCAACTGGGGTAACCTTTGAGTTCTCTCA GTTGGGGGCGTAGG |
| attP R (NheI) | ATATCCTACGCCCCCAACTGAGAGAACTCAAAGGTTACCCCAGTTGGGGCACTACTCTAGATCAAGTCA TCAATTGTGTGCC |
Purification of ϕC31 integrase
The construct pHS62, containing the full coding sequence of ϕC31 integrase, was kindly provided by Dr. Margaret Smith (15). A decahistidine tag was added to the N-terminus of the integrase by cloning a small, double-stranded DNA fragment created from complementary oligonucleotides into an NdeI site at the 5′ end of the gene using standard methods. The orientation of the insert was confirmed by restriction digestion. The resulting plasmid was named pHPhiC31Int. This plasmid was transformed into chemically competent BL21(DE3) pLysS cells.
An overnight culture of BL21(DE3) pLysS pHPhiC31Int cells was diluted to OD600 = 0.1. Cells were grown at 25°C until OD600 = 0.6, then grown at 18°C until OD600 = 0.8. Cells were then induced with 0.1 mM isopropyl β-1-thiogalactopyranoside (IPTG) and grown overnight. Cells were pelleted and frozen. Cell pellet was resuspended in lysis buffer (150 mM HEPES pH 7.0, 100 mM NaCl, 10% glycerol) with 1 mg/ml lysozyme and incubated on ice for 1 h. Cell lysate was then sonicated. One volume of 2 M NaCl was slowly added, followed by one volume of 1 M NaCl, 18% PEG 8000. The solution was slowly stirred for 30 min on ice, followed by centrifugation at 20 000g for 20 min. The supernatant was applied to a nickel metal affinity column (Qiagen), which was subsequently washed with ten column volumes each of Wash I (50 mM Hepes pH 7.0, 1 M NaCl, 10% glycerol) and Wash II (50 mM HEPES pH 7.0, 100 mM NaCl, 10% glycerol, 50 mM imidazole pH 7.0). Protein was eluted in 1 ml fractions with Elution buffer (50 mM HEPES pH 7.0, 500 mM NaCl, 10% glycerol, 400 mM imidazole pH 7.0). Bradford reagent (BioRad) was used to find the peak fractions, which were then pooled and dialyzed into Storage Buffer (50 mM HEPES pH 7.0, 500 mM NaCl, 50% glycerol). All buffers were at 4°C and contained 2.5 mM 2-mercaptoethanol and 0.1 mM phenyl methanesulfonylfluoride (PMSF).
Creation and dissolution of the quadruple HJ substrate
The quadruple HJ substrate was constructed like the DHJS (8), with the exceptions outlined in this article. The pDHJS plasmids containing attB/attP were transformed into DH5α cells containing pM13g and plated onto LB agar containing carbenicillin (50 μg/ml) and gentamicin (3.5 μg/ml). Single colonies were selected and inoculated into 10 ml of Terrific Broth with carbenicillin and gentamicin. These small cultures were incubated at 37°C at 200 rpm until culture growth was apparent, usually ∼12 h. For each pDHJS plasmid, multiple colonies were grown in small cultures, and the fastest growing was inoculated into 2.8 l flasks containing 0.5 l of Terrific Broth with carbenicillin and gentamicin, which were incubated at 37°C at 170 rpm overnight to provide sufficient aeration while avoiding foaming of the culture.
Cells were spun down, and the phagemids were precipitated from the cell media (supernatant) by mixing with one-fourth volume of 5× Phage Precipitation Buffer (2.5 M NaCl, 25% PEG 6000) and stirring the mixture slowly at 4°C overnight. Samples of the media were run on a gel to confirm the presence of DNA before the preparation procedure. Precipitated phagemids were pelleted by centrifugation at 10 000g for 15 min, and DNA was extracted according to the protocol for the Qiagen Maxiprep kit. After the centrifugation step, the supernatant was filtered and applied to a Qiagen tip-500 column that was pre-equilibrated with Buffer QBT. The column was washed with 30 ml Qiagen Buffer QC and eluted with 20 ml Buffer QM (50 mM MOPS pH 7.0, 4 M urea, 0.1 M EDTA NaCl, 30% ethanol). To precipitate the DNA, 20 ml of 100% ethanol was added to the eluate, which was then mixed and incubated on ice for 20 min, followed by centrifugation at 5000g for 1 h. The pellet was allowed to dry and dissolved in 0.5 ml TE [10 mM Tris pH 7.9, 0.1 mM EDTA ethylenediaminetetraacetic acid (EDTA)]. The concentration of the DNA was determined spectrophotometrically, and the DNA was ethanol precipitated and re-suspended in TE (10 mM Tris pH 7.9, 0.1 mM EDTA) to make a final solution of 1 mg/ml.
The ssDNA circles were appropriately annealed and linked using Archaeoglobus fulgidus Reverse Gyrase as previously described (8). The unique re-annealing activity of Reverse Gyrase allows for efficient production of a plasmid (retaining two bubble regions) from two ssDNA circles of opposing strands (resulting from plus and minus origins, respectively). After the reactions were cooled to 37°C, the large heterodimers were linearized by adding XhoI (New England Biolabs) directly to the reactions at 1.5 units/μg of DNA, and incubating at 37°C for 1 h. The linearized DNA was diluted out to 13 μg/ml in three-fourths the reaction volume of ϕC31 integrase buffer (10 mM Tris pH 7.0, 1 mM EDTA, 100 mM NaCl, 5% glycerol, 10 mM spermidine, 5 mM dithiothreitol (DTT), 0.5 mg/ml bovine serum albumin); the ϕC31 integrase was diluted into the remaining one-fourth reaction volume to 20 ng/μl, and mixed into the DNA solution. The reaction was incubated at 30°C for 30 min, and stopped by the addition of sodium dodecyl sulphate to 0.5% followed by incubation at 70°C for 10 min. Potassium chloride was then added to 600 mM, and the solution incubated on ice for 30 min followed by centrifugation at 20 000g for 20 min. Reaction products were purified over a diethylaminoethanol (DEAE) column (Qiagen). The small heterodimers were gel purified from the linear reaction products and recovered by incubation with rotation in Buffer MEN (50 mM MOPS pH 7.0, 10 mM EDTA, 750 mM NaCl) overnight followed by concentration over a DEAE column (Qiagen). Following quantitation, the small heterodimers were annealed and linked with Reverse Gyrase as described earlier.
Dissolution of the quadruple HJ substrate was performed as in (12).
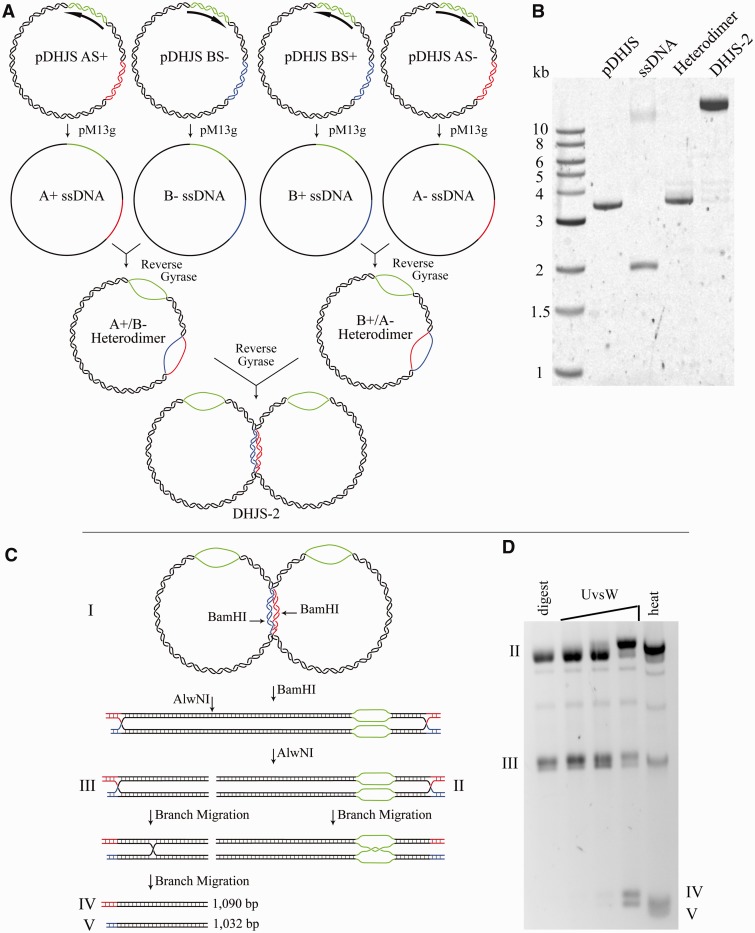
Creation and migration of the DHJS-2
DHJS-2 was produced as in (16) with the exceptions noted in this article. Specifically, the starting plasmids contained the attB/attP sites, and the ssDNA was produced using the pM13g system, as aforementioned. The UvsW migration of DHJS-2 was performed as in (16).
Electron microscopy
DNA samples (∼100 ng) were incubated with or without T4 gp32 as appropriate. The protein–DNA complexes were fixed with 0.6% glutaraldehyde (v/v) for 10 min at room temperature followed by gel filtration using a 1 ml Bio-Gel A-5 m (BioRad) column to remove free protein and fixatives.
The samples were prepared for electron microscopy as described previously (17). Briefly, the samples were adsorbed to thin carbon foils supported by 400-mesh copper grids in the presence of spermidine, then washed with a water/ethanol series, air dried and rotary shadowcast with tungsten. The grids were visualized in a Tecnai 12 TEM. Images for publication were captured using a Gatan Orius CCD camera.
RESULTS
Production of ssDNA using pM13g
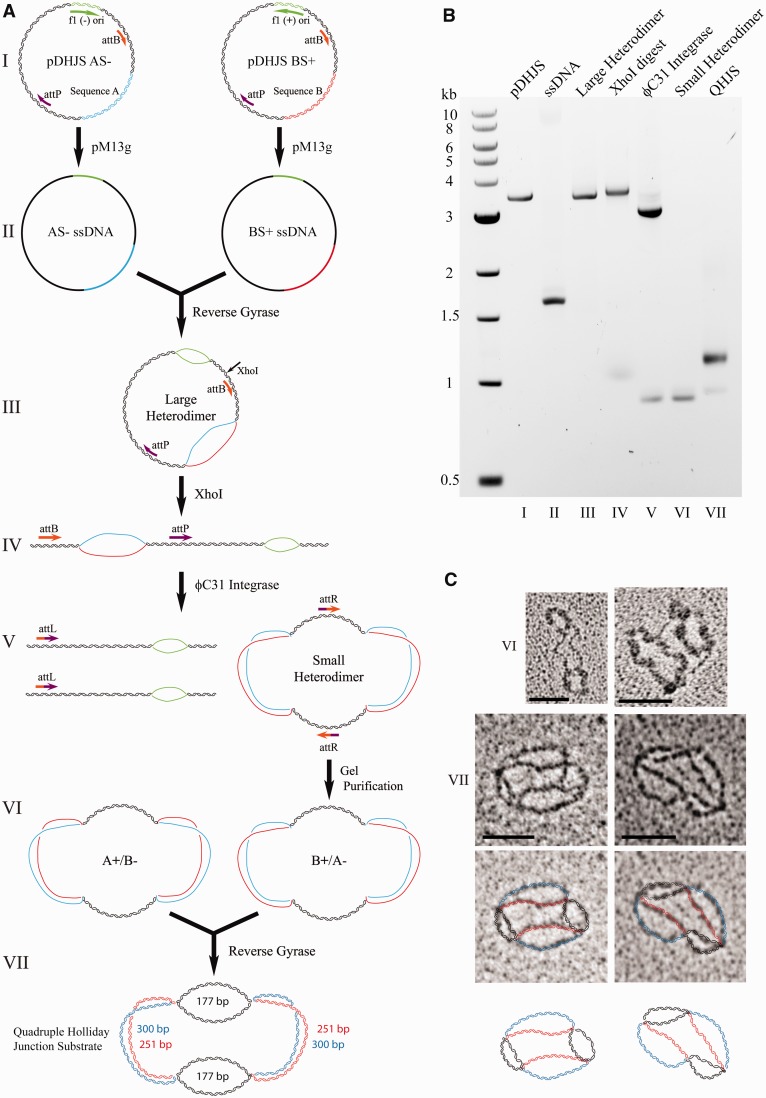
Each of the described HJ substrates is constructed from the same series of ssDNA circles. Therefore, one of the keys to the preparation of useful amounts of HJ substrates is the expression and purification of large quantities of the component ssDNA circles (Figures 1A and 3A, I and II). Previously, the M13K07 helper phage was used, requiring that large preparations of helper phage must first be produced and titered. The pM13g helper plasmid expresses the components of the viral coat proteins that are normally expressed by the helper phage. Cells containing pM13g and a plasmid with the f1 origin will constitutively produce phagemid-containing ssDNA from the origin-containing plasmid.
Figure 1.
Method for creating the quadruple HJ substrate. (A) Outline for creation of the quadruple HJ substrate, using the ϕC31 integrase system. Although both AS−/BA+ and AS+/BS− reactions are performed, only one is shown for simplicity. DNA molecules are not drawn to scale. (B) Products from (A) on an agarose gel. (C) Electron microscopic images of the VI and VII molecules. For VII, strands are outlined in color to clarify the conformation. Scale bar = 50 nm.
Figure 3.
Creation of the DHJS-2 substrate. (A) Outline of the steps for producing the DHJS-2 substrate. This method is as published, with the exception of the use of pM13g to create the ssDNA and the presence of the attB/attP sites in the plasmids (which are not used). (B) Gel electrophoresis of DNA in (A). (C) Outline of products created by digestion of the DHJS-2. Digestion with BamHI and AlwNI creates two substrates containing single HJs (II and III). The smaller substrate (III) will migrate into two smaller duplex pieces, whereas the larger HJ substrate (II) will migrate and be trapped in the bubble region. (Because the red and blue regions are non-homologous, branch migration cannot occur in this direction.). (D) Products shown on a gel. HJs can be migrated by heat or the phage T4 enzyme UvsW (16).
To produce ssDNA for the substrates, DH5α cells were transformed with pM13g and made chemically competent [using a standard calcium chloride protocol (18)]. These competent cells were then transformed with each of the desired pDHJS plasmids (modified as described later in the text) immediately before large growths for ssDNA production. Single colonies were inoculated into 10 ml of Terrific Broth (containing carbenicillin and gentamicin) and grown for 10–12 hours so that the cultures were visibly growing but remained in log phase (OD600 of ∼0.1–0.2). Cultures grown past log phase had a tendency to stop producing phagemid. These cultures were then used to inoculate 2–3 l of Terrific Broth for the large-scale overnight growth. Half the normal culture volume was grown per flask to ensure sufficient aeration, which was observed to be important for the robust production of phagemid. Because not all colonies grew at the same rate, multiple small growths were prepared, and the fastest growing culture was then used to inoculate the large growths.
Although a strand bias was observed, in that less DNA was produced from the (+) plasmids, sufficient DNA could be obtained from large-scale overnight growths. From 1 l of culture, an average of 250 μg of the (−) strands and 150 μg of the (+) strands were regularly obtained. These values are comparable with expression with the M13K07 helper phage, which was ∼200 μg/l for all strands.
Replacing the DHJS recombination system
Previously, the recombination step of DHJS creation presented a major bottleneck. Recombination between sites adjacent to the inserted ‘A’ and ‘B’ regions of the plasmid (blue and red, respectively) removes the origin bubble (green) and creates a migratable region (black) (Figure 1, IV and V). The previous recombination reaction, which used Cre recombinase and a set of loxP sites, was bidirectional, meaning that the products of the reaction are also substrates. Although small-scale reactions can produce 70% product, the large-scale reactions required for DHJS creation are rarely at or >50% product. Transition of the system to the unidirectional ϕC31 integrase theoretically allows complete recombination, meaning that the vast majority of the DNA molecules put into the reaction will contribute useable product for the downstream reactions.
To convert the DHJS to the ϕC31 integrase system, the two loxP sites in the starting plasmids were replaced with sequential attB and attP sites. These unique sites become attL and attR following recombination, effectively eliminating the reverse reaction (Figure 1, IV and V). Oligonucleotides containing the sequences were obtained and annealed to make DNA duplex with sticky ends, which were then ligated into each of the digested plasmids (see Materials and Methods). Using this recombination system, we regularly observe >90% product formation.
Surprisingly, despite using a dilute DNA solution (that favored intramolecular reactions when using Cre recombinase), ϕC31 integrase produced almost exclusively dimer products (Figue 1A, IV and V, and B, lane 6). Although a mix of monomers and dimers can be produced from the starting plasmids, using the large heterodimer as a substrate produces largely dimers and occasionally tetramers (data not shown). Visualization by electron microscopy (Figure 1C, VI) and reaction with Top2 (data not shown) confirmed that the dimers were produced via recombination rather than catenation.
The ‘small heterodimer’ thus contains two double-stranded arms of equal length (177 bp) connected by two larger bubbles (one arm is 300 nt, the other 251 nt, for a total bubble of 551 nt), which were coated with T4 gp32 for visualization by electron microscopy (Figure 1C, VI). The final product is made from the annealing and linking of a pair of these small heterodimers (Figure 1A, VI and VII). Visualization confirms that the final product contains four HJs (Figure 1C, VII).
Using the ϕC31 recombination system increases the overall yield of product molecules. Approximately five times as much substrate is created, normalized to the amount of starting DNA, when producing the quadruple HJ substrate versus the DHJS.
Gel extraction by diffusion
Following the recombination reaction, the small heterodimer is separated away from the linear byproduct by gel electrophoresis. The ensuing gel extraction step to recover the small heterodimer is the largest source of DNA loss during the procedure (Figure 1, V and VI). Although we have tried a variety of methods, including electroelution, DEAE paper, silica-based gel purification kits and beta-agarase, the most consistently high yields came from diffusion overnight. The gel slices were chopped into pieces ∼2 mm3 and incubated in 10 volumes of buffer (50 mM MOPS pH 7.0, 10 mM EDTA, 750 mM NaCl) with rotation overnight to allow the DNA to diffuse out of the gel. The buffer was then separated from the gel by filtration through Kimwipes, and the DNA was recovered and concentrated using a DEAE column (Qiagen). Using this method, 30% of the DNA was regularly recovered.
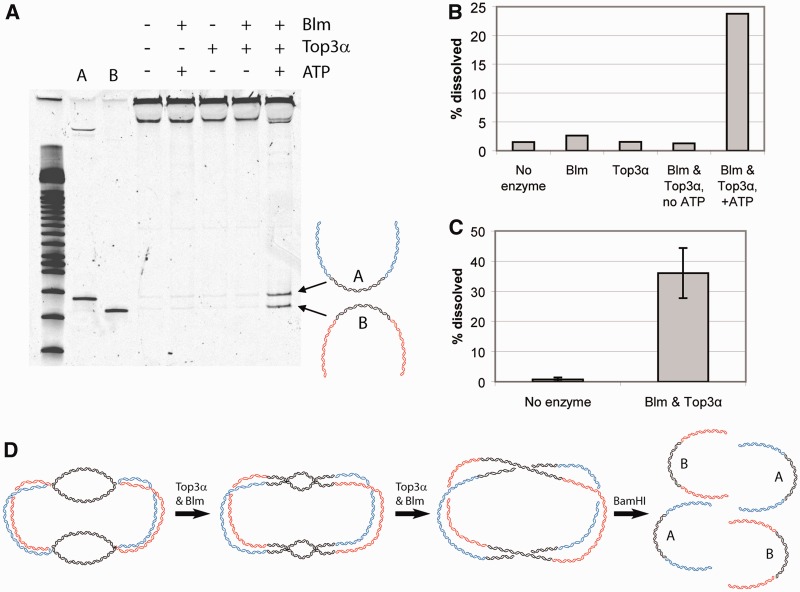
The quadruple HJ substrate can be dissolved by the Blm-Top3α complex
A hallmark of the DHJS is its ability to be dissolved by Blm and Top3α (11,12). To ensure that the new version of the substrate maintains functionality, we performed the dissolution reaction using the Drosophila Blm-Top3α complex. As with the original dHJ substrate, Blm, Top3α and adenosine triphosphate were all required for substrate dissolution (Figure 2). We observed levels of dissolution comparable with that of the previous substrate (∼30–40%).
Figure 2.
Dissolution of the quadruple HJ substrate. (A) Dissolution of the quadruple HJ substrate requires Blm, Top3α and ATP, as with the DHJS (11). Substrates were digested with BamHI after the reaction, before being run on an acrylamide gel. (B) Quantification of (A). (C) Quantification of the amount of dissolution achievable with the quadruple HJ substrate. (n = 5). (D) Schematic of the process of dissolution with Blm and Top3α, followed by digestion with BamHI. Only the black regions are homologous and therefore allow branch migration. The final products are separated by digestion.
Single migratable HJs can be produced
The previously described ‘DHJS-2’ substrate (16), which produces two kinds of migratable single HJs on restriction digestion, can also be produced from the same starting plasmids as the DHJS (Figure 3A). This method uses the same ssDNA and does not require the recombination system. Following annealing and linking with Reverse Gyrase, the substrate can be stored as a stable, large heterodimer. We produced this substrate using the pM13g system and the attB/attP-containing plasmids (Figure 3B).
Before use in a reaction, digestion with BamHI and AlwNI produces two substrates each containing a single migratable HJ (Figure 3C). To ensure that the substrate we produced from the pM13g system maintains the expected functionality, DHJS-2 was tested with UvsW as in previous work (16). As expected, UvsW is able to migrate the junctions into their product molecules (Figure 3D). Incubating the substrate at 37°C for 1 h (heat) in a low magnesium buffer is also sufficient to allow spontaneous migration. To prevent this, digestion and enzymatic migration are performed at 30°C, and reactions are stopped with buffer containing ethidium bromide and magnesium chloride.
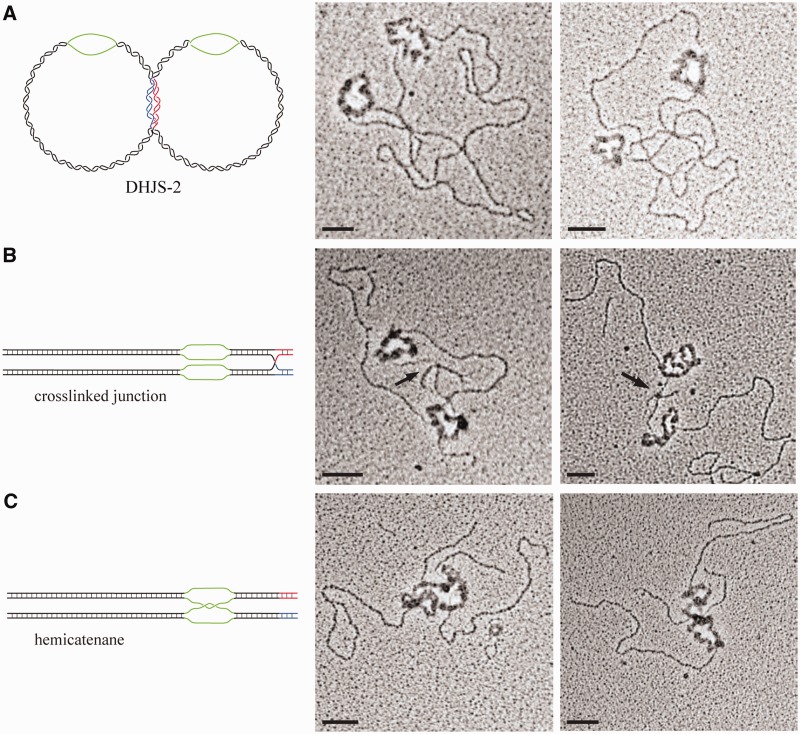
To directly observe this substrate and its derivatives, we used electron microscopy. The substrates were pre-incubated with the phage T4 ssDNA-binding protein gp32 to highlight the ssDNA bubbles. The substrates were then mounted onto carbon-coated grids and shadowed with tungsten.
When visualizing the DHJS-2, a molecule containing two origin bubbles is evident (Figure 4A). Because of the relative size of the plasmids, the location of the HJs is unclear in the starting molecule. To capture the HJs following digestion but before migration, the DHJS-2 was cross-linked with psoralen before digestion. On these molecules, a four-way junction is clearly evident on the small arms extending from the origin bubble (Figure 4B). Molecules that have been digested, but not cross-linked, allow spontaneous branch migration of the HJ in the homologous direction (shown in black), which will terminate in the single-stranded origin bubble (green); these molecules show a hemicatenane connection at their bubble regions (Figure 4C). These images verify our expectations of the substrate conformation.
Figure 4.
Electron microscopic images of DHJS-2 and its derivatives. DHJS-2 and its derivatives were incubated with T4 gp32 to highlight and thicken the single-stranded regions before visualization by electron microscopy. (A) Images of undigested DHJS-2. The thickened, circular regions are the origin bubbles (green). (B) DHJS-2 cross-linked with psoralen before digestion to trap the HJ. Arrow indicates location of junction. (C) Digested and migrated DHJS-2, which creates a hemicatenane in the bubble region. Scale bar = 50 nm.
DISCUSSION
In updating the method for DHJS creation, we sought to make the process less labor intensive and more efficient. The three points we focused on were production of ssDNA, the recombination reaction and gel extraction.
Previously, production of the ssDNA required preparing large quantities of the M13K07 phage and titrating the resulting phage. In addition, the pDHJS plasmids must be freshly transformed into phage competent (F′) cells, and colonies tested for ssDNA expression of the phagemid vector following addition of the helper phage. In the updated system, the pDHJS plasmids are transformed into cells containing the pM13g plasmid, and phagemid is constitutively expressed. The best expression results were obtained from freshly transformed cells, but there was little variation among colonies, alleviating the need to test colonies before scaling up. Once the competent cells are produced, production of the ssDNA requires less work than methods using helper phage. This aspect of the method can be applied to produce ssDNA from the previously published pDHJS plasmids, containing the loxP sites, to create the DHJS.
Gel extraction is always the largest source of DNA loss during the procedure. Although we have tried a variety of methods, the most consistently large yield was when the gel slices were finely chopped, incubated in buffer overnight and recovered through a DEAE column, similar to the method commonly used to recover PAGE-purified oligonucleotides. Although other methods can produce similar or higher yields, they were much less consistent.
Another significant source of DNA loss is the Cre recombination step. The loxP sites that are the substrates for Cre remain intact following recombination; therefore, the products can also become substrates. Although an optimized small-scale reaction can yield up to 70% products, the large-scale reactions needed to produce the DHJS regularly yield only 20–40%, meaning that up to 80% of the initial DNA is not used for further substrate preparation. In contrast, the distinct attB and attP sites, which are the substrates for ϕC31 integrase, recombine to form attL and attR, making the system unidirectional. Product formation, even in large-scale reactions, was regularly >90%, reducing the loss of DNA by 5-fold.
Although the original DHJS was extensively characterized to show the existence of HJs, an additional method to verify the conformation is to directly visualize the DNA using electron microscopy. When observed by electron microscopy, the new version of the DHJS, using the ϕC31 recombination system, unexpectedly revealed four HJs with the appropriate size arms connecting them (Figure 1C). Subsequent ϕC31 recombination assays revealed that linearized starting plasmids form a combination of monomer and dimer products. In contrast, the heterodimers used to produce substrate (which contain a large single-stranded bubble region adjacent to the recombination sites) were unable to form monomer products and produced only dimers (data not shown). Therefore, it appears that the character of the local DNA (single-stranded or double-stranded) affects the ϕC31 recombination reaction and leads to the obligate formation of intermolecular dimer products at the small heterodimer stage (Figure 1A, V). To produce dimers, the attB site of one molecule pairs with the attP site on an adjacent molecule, while simultaneously, the attP site of the first molecule pairs with the attB site of the second molecule, creating complete dimer products with two attR sites. This conformation appears to be preferred, as no molecules with just one recombination event were observed (i.e. where attB on one molecule underwent recombination, but attP of the same molecule did not). This quadruple HJ product can be produced more efficiently than the original DHJS and is equally capable of modeling dHJ dissolution in vitro (Figure 2).
The components of the DHJS can also be used to create the DHJS-2, which can be digested to make two migratable single HJs. Owing to the ability of the junctions to spontaneously migrate, it is important to use the highest concentration of magnesium and the lowest temperature and incubation time possible for the enzyme in question. Reactions should also be stopped with magnesium and ethidium bromide to prevent further migration. Alternatively, thermal branch migration of the substrate will create a hemicatenane structure that can be used for decatenation studies (Figure 3C, II). Indeed, Top3α is able to decatenate this structure (data not shown).
The expected conformations of DHJS-2 and its derivatives were clearly visualized using electron microscopy (Figure 4). Molecules containing two origin regions that produce HJs or hemicatenanes, depending on the conditions, were evident. This substrate is produced efficiently, as no recombination step is necessary, and should be useful for future studies of HJ migration or decatenation. The catenated bubbles in the context of a duplex provide a physiological substrate to study these reactions.
FUNDING
National Institutes of Health [GM029006 to T.H. and GM031819 to J.D.G.]. Funding for open access charge: Academia Sinica.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Andrew Bradbury and Margaret Smith for the kind donation of constructs (pM13cp and pHS62, respectively). They would also like to thank Kenneth Kreuzer for the gift of UvsW protein to test the DHJS-2 substrate.
REFERENCES
- 1.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 2.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 3.Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen SL, Kuo HK, Savukoski D, Brodsky MH, Sekelsky J. Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genet. 2011;7:e1002315. doi: 10.1371/journal.pgen.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wechsler T, Newman S, West SC. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature. 2011;471:642–646. doi: 10.1038/nature09790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 7.Murchie AI, Clegg RM, von Kitzing E, Duckett DR, Diekmann S, Lilley DM. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature. 1989;341:763–766. doi: 10.1038/341763a0. [DOI] [PubMed] [Google Scholar]
- 8.Plank JL, Hsieh T-s. A Novel, Topologically Constrained DNA Molecule containing a double holliday junction: design, synthesis, and initial biochemical characterization. J. Biol. Chem. 2006;281:17510–17516. doi: 10.1074/jbc.M602933200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanton HL, Radford SJ, McMahan S, Kearney HM, Ibrahim JG, Sekelsky J. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 2005;1:e40. doi: 10.1371/journal.pgen.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larocque JR, Jasin M. Mechanisms of recombination between diverged sequences in wild-type and BLM-deficient mouse and human cells. Mol. Cell Biol. 2010;30:1887–1897. doi: 10.1128/MCB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plank JL, Wu J, Hsieh T-s. Topoisomerase III{alpha} and Bloom's helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. PNAS. 2006;103:11118–11123. doi: 10.1073/pnas.0604873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen SH, Wu CH, Plank JL, Hsieh TS. Essential functions of the C-terminus of Drosophila topoisomerase IIIalpha in double Holliday junction dissolution. J. Biol. Chem. 2012;287:19346–19353. doi: 10.1074/jbc.M112.363044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chasteen L, Ayriss J, Pavlik P, Bradbury AR. Eliminating helper phage from phage display. Nucleic Acids Res. 2006;34:e145. doi: 10.1093/nar/gkl772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl Acad. Sci. USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb MR, Plank JL, Long DT, Hsieh TS, Kreuzer KN. The phage T4 protein UvsW drives Holliday junction branch migration. J. Biol. Chem. 2007;282:34401–34411. doi: 10.1074/jbc.M705913200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffith JD, Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu. Rev. Biophys. Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]