Abstract
The array of genome editing strategies based on targeted double-stranded DNA break formation have recently been enriched through the introduction of transcription activator-like type III effector (TALE) nucleases (TALENs). To advance the testing of TALE-based approaches, it will be crucial to deliver these custom-designed proteins not only into transformed cell types but also into more relevant, chromosomally stable, primary cells. Viral vectors are among the most effective gene transfer vehicles. Here, we investigated the capacity of human immunodeficiency virus type 1- and adenovirus-based vectors to package and deliver functional TALEN genes into various human cell types. To this end, we attempted to assemble particles of these two vector classes, each encoding a monomer of a TALEN pair targeted to a bipartite sequence within the AAVS1 ‘safe harbor’ locus. Vector DNA analyses revealed that adenoviral vectors transferred intact TALEN genes, whereas lentiviral vectors failed to do so, as shown by their heterogeneously sized proviruses in target cells. Importantly, adenoviral vector-mediated TALEN gene delivery resulted in site-specific double-stranded DNA break formation at the intended AAVS1 target site at similarly high levels in both transformed and non-transformed cells. In conclusion, we demonstrate that adenoviral, but not lentiviral, vectors constitute a valuable TALEN gene delivery platform.
INTRODUCTION
The ability to design DNA-binding motifs that recognize pre-defined sequences of choice within complex eukaryotic genomes is opening a wide range of basic and applied research possibilities that involves the modulation of endogenous gene expression and chromosomal DNA editing. The latter aim has been mostly pursued by designing zinc-finger nucleases (ZFNs) comprising a sequence-specific array of synthetic zinc-finger motifs fused to the nuclease domain of the type IIS restriction endonuclease FokI (1,2). In addition, the recoding of natural homing endonucleases for heritable genome modification purposes has also been undergoing intense investigation (3,4). These artificial enzymes are tailored to generate double-stranded DNA breaks (DSBs) at pre-defined genomic sequences. As a consequence of their activity, targeted gene knockout or chromosomal insertion of exogenous DNA can ensue following the activation and engagement of the error-prone non-homologous end-joining (NHEJ) or the error-free homologous recombination (HR) DNA repair pathways, respectively.
The recent discovery of the rules governing the recognition of specific DNA sequences by bacterial transcription activator-like type III effectors (TALEs) (5,6) has translated in a novel and straightforward methodology to construct proteins with customized DNA binding specificities. TALEs are proteins found in phytopathogenic bacteria of the genus Xanthomonas, whose main well-established function is to induce expression of specific host plant genes that enhance virulence. TALEs comprise an N-terminal translocation domain, a central region of tandem direct repeats that confers DNA binding specificity and a nuclear localization motif preceding a C-terminal transcriptional activation domain (7). The DNA binding domain of TALEs displays a unique architecture composed of a tandem array of typically 15.5–19.5 repeat units with circa 34 residues each (7). Often, the only distinguishing feature among the different repeats is a 2-amino acid hypervariable polymorphism at positions 12 and 13 dubbed ‘repeat-variable di-residue’ (RVD). Crucially, it has been discovered that each individual RVD dictates the binding of the repeat in which it is embedded to a single nucleotide (5,6). Therefore, this one repeat to one nucleotide code directly establishes a simple rule governing TALE–DNA interactions. This insight combined with the modular nature of the DNA binding domain, is permitting researchers to harness TALE-derived repeats as custom-made scaffolds on which to assemble functional heterologous domains. Hitherto, designer TALEs tested in vitro and in cellula have included those with transcriptional activation or FokI nuclease domains (8,9). The latter, called TALE nucleases (TALENs) operate similarly to ZFNs in that a pair is assembled at a given DNA sequence consisting of two half-target sites separated by a spacer sequence (10). The directional binding of each TALEN monomer to its respective half-target site induces dimerization of the FokI portions resulting in site-specific DNA cleavage. Importantly, in contrast to zinc-finger modules (11), there are as of yet no indications that binding of an individual TALE repeat to its cognate base pair is altered by neighboring sequences. This context-independent feature combined with the simple DNA binding code suggests that, when compared with ZFNs, TALENs can bind a wider range of DNA sequences and be programmed in an easier and more predictable manner. Moreover, the ability to construct functionally viable TALEN pairs with relatively large arrays of repeats has the potential to render them more specific than ZFNs.
Thus far, nucleic acids encoding TALENs have been introduced into cells by nucleic acid microinjection or by transfection methods based on chemical agents or electroporation (8,9,12–14). Unfortunately, these methods suffer from low throughput or are too cytotoxic or too inefficient in populations of non-transformed cells. Notably, the therapeutic application and the thorough assessment of the genome-wide TALEN specificity, for instance, will be dependent on highly efficient TALEN delivery systems into relevant karyotypically stable cells. In the realm of biomedical research, this will include tissue-specific stem and progenitor cells. From the broad variety of methods to introduce foreign nucleic acids into cells, viral vectors have proven to be the most effective thus far. In particular, vectors derived from the double-stranded linear DNA human adenovirus serotype 5 and from the single-stranded positive-sense RNA lentivirus human immunodeficiency type 1 (HIV-1) have become broadly used (15,16). Owing to the infection mechanisms evolved by their parental viruses (17,18), adenoviral and lentiviral vectors are particularly fit for the in vitro and in vivo transduction of dividing and non-dividing cells. In the current study, we investigated the suitability of these two major vector classes as delivery vehicles of TALEN sequences. We report that, in contrast to HIV-1-based lentiviral vectors, adenoviral vectors can package and deliver functional TALEN-encoding expression units into transformed, immortalized and primary human cells.
MATERIALS AND METHODS
Cells
The 293T and HeLa cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen). The medium used for the culturing of the former and latter cell types was supplemented with 10 and 5% fetal bovine serum (Invitrogen), respectively. The adenoviral vector packaging cell lines PER.C6 (19) and PER.E2A (20) were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 10 mM MgCl2 (Sigma-Aldrich) without and with 250 µg/ml of Geneticin (Invitrogen), respectively. These cells were kept in an atmosphere of humidified air with 10% CO2. The human myoblasts were obtained from a Duchenne muscular dystrophy patient and were immortalized through constitutive expression of Bmi-1 and TERT. The generation of and culture conditions for these immortalized human myoblasts have been specified elsewhere (21). The human bone marrow-derived primary fetal mesenchymal stem cells (hMSCs) were cultured as previously specified (22). The human myoblasts and hMSCs were kept in an atmosphere of humidified air with 5% CO2.
Recombinant DNA
A TALEN pair (Supplementary Figure S1) targeting intron 1 of the PPP1R12C locus (AAVS1 target site: 5′-TCTGTCCCCTCCACCCCAC-spacer-GACAGGATTGGTGACAGAA-3′) were generated by Golden Gate cloning, as previously described (9). To generate pAdShu.TALEN-LS1.IRES.RFP and pAdShu.TALEN-RS1.IRES.RFP, plasmids 1383.pVAX.AAVS1.TALEN.L-94 and 1384.pVAX.AAVS1.TALEN.R-95 were digested with Eco32I (Fermentas) and PmeI (New England Biolabs), and the resulting 3.0-kb TALEN open reading frame (ORF) fragments were isolated and ligated into the Eco32I-linearized 8.8-kb plasmid vector backbone pShuttle.IRES.DsRedEx2.1. Cells transduced with vectors based on this construct can be traced and quantified via the encoded red fluorescent protein (RFP) reporter DsRedEx2.1. The first-generation (i.e. early region 1 [E1]-deleted) fiber-modified adenoviral vector molecular clones pAd.ΔE1.TALEN-LS1.F50 and pAd.ΔE1.TALEN-RS1.F50 (Figure 2A) were assembled by HR following transformation of Escherichia coli cells BJ5183pAdEasy-1.50 (23) with shuttle plasmids pAdShu.TALEN-LS1.IRES.RFP and pAdShu.TALEN-RS1.IRES.RFP, respectively, whereas the second-generation adenoviral vector molecular clones pAd.ΔE1ΔE2A.TALEN-LS1.F50 and pAd.ΔE1ΔE2A.TALEN-RS1.F50 (Figure 2A) were assembled by HR following transformation of E. coli cells BJ5183pAdEasy-2.50 (23) with shuttle plasmids pAdShu.TALEN-LS1.IRES.RFP and pAdShu.TALEN-RS1.IRES.RFP, respectively. Before transformation, all shuttle plasmids were digested to completion with MssI. The structure and composition of lentiviral vector constructs pLV.TALEN-LS1.i.RFP, pLV.TALEN-RS1.i.RFP, pLV.TALEN-LS1 and pLV.TALEN-RS1 are described in Figure 1A. The enhanced green fluorescent protein (eGFP)-encoding plasmid CMVPRES [(24); herein referred to as pLV.CMV.eGFP] served as a negative control in the polymerase chain reaction (PCR) amplifications of TALEN sequences.
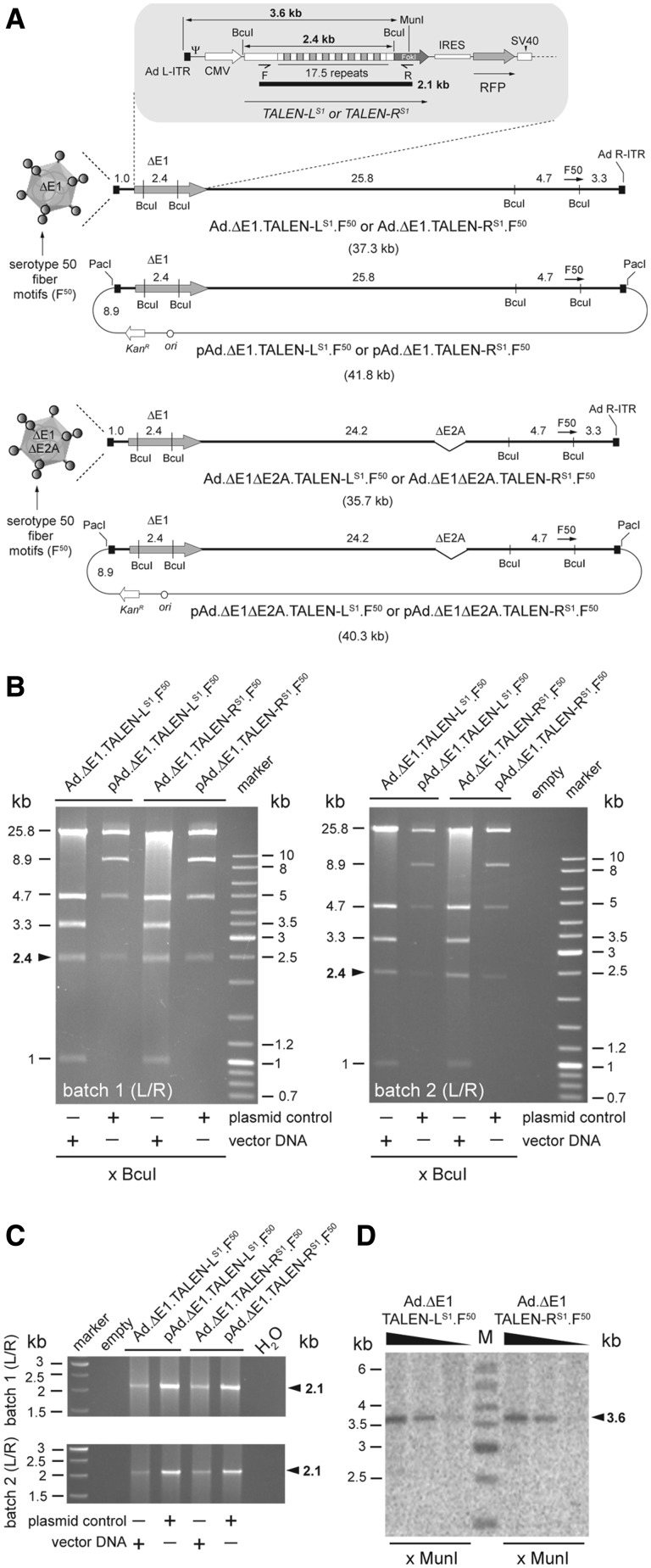
Figure 2.
Structural analysis of TALEN-encoding adenoviral vector genomes. (A) DNA structures of the first-generation adenoviral vectors Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 and of the second-generation adenoviral vectors Ad.ΔE1ΔE2A.TALEN-LS1.F50 and Ad.ΔE1ΔE2A.TALEN-RS1.F50 drawn in relation to those of their respective plasmid clones. The adenoviral vector and shuttle plasmid DNA templates are depicted as BcuI physical maps with numerals specifying the different restriction fragment sizes in kilobases (kb). These DNA molecules harbor a bi-cistronic expression unit coding for the AAVS1-specific artificial nucleases TALEN-LS1 or TALEN-RS1 and, through an IRES sequence, an RFP reporter. The ORFs are under the transcriptional control of the cytomegalovirus immediate-early gene promoter (CMV) and the simian virus 40 (SV40) polyadenylation signal. The central regions of TALEN-LS1 and TALEN-RS1 encode a tandem of 17.5 TALE repeats that define specificity to the AAVS1 target sequence at the human chromosome 19 (19q13.42-qter). C-terminally both TALENs contain the non-specific cleavage domain of FokI, whose catalytic activity is contingent on DNA binding-dependent dimerization. Each of the TALEN proteins are tagged by an HA antigen located close to their N-termini (not drawn). The thick and thin lines represent adenoviral vector and plasmid backbone sequences, respectively. Ψ, human adenovirus serotype 5 packaging signal; Ad L-ITR and Ad R-ITR, ‘left’ and ‘right’, respectively, adenoviral inverted terminal repeat. F50, ORF encoding chimeric fiber composed of basal shaft domains from adenovirus serotype 5 fused to apical shaft and knob motifs from adenovirus serotype 50. The kanamycin-resistance gene (KanR) and the prokaryotic origin of replication (ori) are also indicated. (B) Restriction fragment length analysis of Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 genomes treated with BcuI. Plasmids pAd.ΔE1.TALEN-LS1.F50 and pAd.ΔE1.TALEN-RS1.F50 served as a reference. The sizes (in kb) of the DNA restriction fragments resulting from the BcuI treatment are indicated on the left. The arrowhead points to the 2.4-kb DNA fragments diagnostic for intact, non-rearranged, TALE-derived DNA sequences within TALEN-encoding genes. Marker, molecular-weight marker Gene Ruler DNA Ladder Mix. (C) PCR analysis on DNA isolated from Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 particles using primers TALEN-Seq-F and TALEN-Seq-R (F and R half arrows, respectively). These primers, drawn in relation to their respective target sequences, amplify most of the TALEN-LS1 and TALEN-RS1 DNA including the 17 102-bp tandem DNA repeats. The position (arrowheads) and size (in kb) of the 2.1-kb amplicon diagnostic for intact TALEN transgenes are indicated at the right. Marker, Gene Ruler DNA Ladder Mix. H20, PCR performed with nuclease-free water instead of vector DNA template. (D) Autoradiogram of MunI-treated extrachromosomal DNA isolated from HeLa cells transduced with Ad.ΔE1.TALEN-LS1.F50 or with Ad.ΔE1.TALEN-RS1.F50 at MOIs of 10, 5 and 1 HTU/cell. The DNA, extracted at 3 days post-transduction, was subjected to Southern blot analysis using a 397-bp probe hybridizing to a sequence encoding the NH2 terminus of the TALEN-LS1 and TALEN-RS1 proteins.
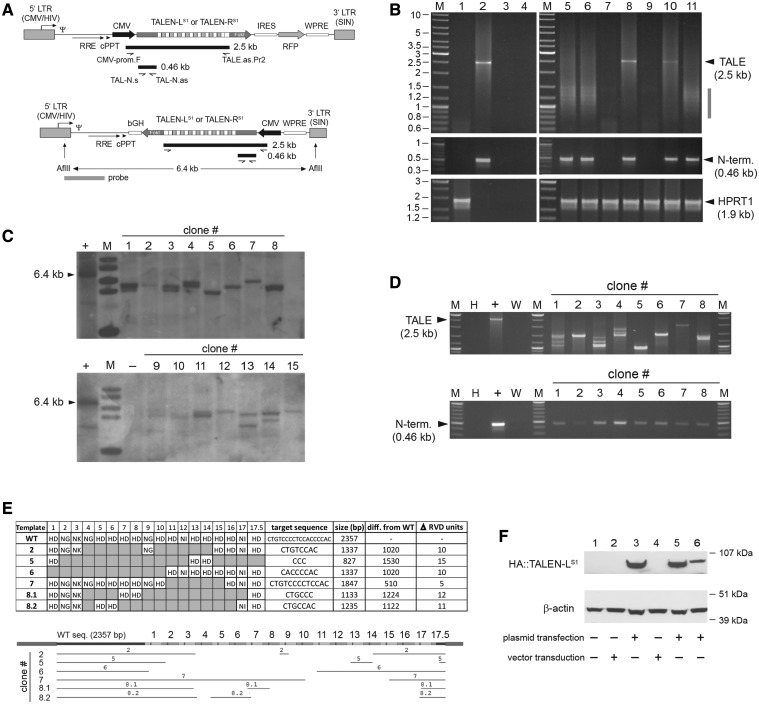
Figure 1.
Structural analyses of HIV-1-based lentiviral vectors harboring TALEN sequences in transduced cells. (A) Genetic organization of lentiviral vector constructs. Diagram of bicistronic pLV.TALEN-LS1.i.RFP and pLV.TALEN-RS1.i.RFP and of monocistronic pLV.TALEN-LS1 and pLV.TALEN-RS1 lentiviral vector transfer plasmids. The bicistronic constructs code for the AAVS1-specific custom-made nucleases TALEN-LS1 or TALEN-RS1 and, through an IRES sequence, an RFP reporter, whereas the monocistronic plasmids encode exclusively the AAVS1-specific designer nucleases. Each of the TALEN proteins are tagged by an HA antigen located close to their N-termini (not drawn). Gray boxes with broken arrow, hybrid 5′ LTR containing CMV and HIV-1 sequences; gray boxes without broken arrow, self-inactivating (SIN) 3′ LTR; Ψ, HIV-1 packaging signal; RRE, Rev-responsive element; cPPT, central polypurine tract; WPRE, Woodchuck hepatitis virus posttranscriptional regulatory element. In pLV.TALEN-LS1.i.RFP and pLV.TALEN-RS1.i.RFP, the ORFs are under the transcriptional control of the CMV promoter and the polyadenylation signal present within the SIN 3′ LTR, whereas the ORFs in pLV.TALEN-LS1 and pLV.TALEN-RS1 are under the control of the CMV promoter and the bovine growth hormone polyadenylation signal (bGH). Primers, PCR products and Southern blot probe used are depicted as half-arrows, black bars and gray bar, respectively, and are drawn in relation to their respective vector DNA sequences. For the sake of simplicity, plasmid backbone sequences are not shown. The sizes of the PCR products expected from the amplification of full-length lentiviral vector genomes are indicated. The 6.4-kb AflII restriction fragment corresponding to intact DNA from LV.TALEN-LS1 or LV.TALEN-RS1 is also indicated. (B) PCR analyses of TALEN-encoding HIV-1-based lentiviral vector genomes following transduction of HeLa cells. PCR amplifications with the aid of the primers shown in Figure 1A were carried out using, as template, total cellular DNA from mock-transduced HeLa cells, plasmid pAd.ΔE1.TALEN-LS1.F50, plasmid pLV.CMV.eGFP or nuclease-free H20 (lanes 1, 2, 3 and 4, respectively). PCR amplifications using the same primer pairs were performed in parallel on DNA extracted from HeLa cells transduced with integration-competent LV.TALEN-RS1 (lane 5), integrase-defective IDLV.TALEN-RS1 (lane 6), Ad.ΔE1.TALEN-RS1.F50 (lane 10) or integration-competent LV.TALEN-RS1.i.RFP (lane 11). Extra PCR controls supplementing those corresponding to lanes 1 through 4 were provided by using DNA isolated from HeLa cells exposed to an LV.TALEN-RS1 preparation made in parallel but deploying pUC19 instead of lentiviral vector packaging construct psPAX2 (lane 7) or incubated with pLV.TALEN-RS1 mixed or not mixed with the DNA transfection agent ExGen500 (lanes 8 and 9, respectively). Lanes M, Gene Ruler DNA Ladder Mix. A primer pair targeting the human HPRT1 served to control the integrity of the cellular DNA. (C) Clonal analysis. Southern blot analysis of AflII-digested genomic DNA from HeLa cell clones 1 through 15 stably transduced with LV.TALEN-LS1. The vector-specific probe that was used is shown in Figure 1A (horizontal gray bar). Lane M, 1 kb molecular-weight marker. The parental transfer plasmid pLV.TALEN-LS1 treated with AflII served as an internal control for intact vector DNA. (D) PCR screening of genomic DNA from HeLa cell clones stably transduced with LV.TALEN-LS1. Upper and lower panel, PCR products resulting from the use of the primer pairs CMV-prom.F/TALE.as.Pr2 and TAL-N.s/TAL-N.as, respectively, on HeLa cell clones 1 through 8 (note: clone 8 turned out to yield two closely sized and vector-specific amplicons, thus the nomenclature 8.1 and 8.2). PCR amplifications on plasmid pLV.TALEN-LS1 served as a positive control (+), whereas those on nuclease-free water (W) and HeLa genomic DNA (H) provided for negative controls. Lanes M, Gene Ruler DNA Ladder Mix. (E) Structural analyses of proviral vector DNA. DNA sequencing of amplicons retrieved from HeLa cell clones 2, 5, 6, 7 and 8 after PCR amplifications with primers CMV-prom.F and TALE.as.Pr2 (Figure 1D, upper panel). Upper panel, summary of the TALE repeat arrays present in the indicated LV.TALEN-LS1-transduced HeLa cell clones. The respective predicted target sites, lengths, size reduction in relation to the wild-type sequence and the number of 102-bp long TALE repeat units found to be deleted, are also indicated. Lower panel, alignment of the TALEN-LS1 DNA sequences retained in the various proviruses (horizontal thin lines) drawn in relation to the reference wild-type sequence (horizontal thick line). (F) TALEN protein detection. HA tag-directed western blot analysis of full-length TALEN-LS1 expression in 293T producer cells (lane 1), IDLV.TALEN-LS1- and LV.TALEN-LS1-transduced HeLa cells (lane 2 and lane 4, respectively) and in pLV.TALEN-LS1-transfected 293T cells used to generate these IDLV.TALEN-LS1 and LV.TALEN-LS1 vector preparations (lanes 3 and 5, respectively). Lane 6, HeLa cells transfected with pLV.TALEN-LS1. The β-actin served as loading control.
Viral vectors
The production of adenoviral vectors Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 took place in PER.C6 cells, whereas that of adenoviral vectors Ad.ΔE1ΔE2A.TALEN-LS1.F50 and Ad.ΔE1ΔE2A.TALEN-RS1.F50 occurred in PER.E2A cells. The rescue of vector particles was initiated by transfecting each of the PacI-linearized adenoviral vector molecular clones into their respective packaging cells by using a polyethyleneimine-based protocol (23). The procedures to propagate and purify the resulting viral vectors have been described elsewhere (23). Vector titers were determined through limiting dilutions on HeLa indicator cells seeded at a density of 0.8 × 105 cells per well of 24-well plates (Greiner). At 3 days post-transduction, frequencies of RFP-positive cells were determined through flow cytometry by using a BD LSR II FACS (Becton Dickinson). As a result, the titers are expressed in terms of HeLa cell-transducing units (HTUs) per milliliter. The HIV-1-based and vesicular stomatitis virus glycoprotein-G (VSV-G)-pseudotyped lentiviral vectors were generated by transfecting 293T cells essentially as specified elsewhere (25) except for the use of 7.5 × 106 cells in T75-cm2 flasks instead of 1.75 × 107 cells in T175-cm2 flasks. A total amount of 12.85 µg of DNA was transfected corresponding to 2:1:1 mixtures of bi-cistronic lentiviral vector transfer plasmid, packaging construct psPAX2 (Addgene plasmid 12260) or psPAX2D116N (25) and VSV-G pseudotyping construct pLP/VSVG (Invitrogen). The psPAX2- and psPAX2D116N-based production systems yielded second-generation integration competent and integrase-defective lentiviral vectors (IDLVs), respectively. The mono-cistronic lentiviral vectors with an integration-competent and integrase-defective phenotype were made instead with a third-generation packaging system based on the pMDLg/pRRE (Addgene plasmid 12251) and pMDLg/pRRED64V packaging plasmids, respectively. Vector titers were determined by using HIV-1 p24gag immunocapture (Perkin Elmer).
Immunofluorescence microscopy
Fifty thousand HeLa cells were seeded in wells of 24-well plates and, after an overnight incubation period, they were either mock-transduced or were transduced with Ad.ΔE1.TALEN-LS1.F50 or with Ad.ΔE1.TALEN-RS1.F50 at 3 HTU/cell. Three days post-transduction, the cells were subjected to immunostaining essentially as described elsewhere (26) except for the use of an influenza hemagglutinin (HA)-specific mouse monoclonal IgG1 antibody (HA.11 clone 16B12; Covance) together with a goat anti-mouse IgG (H+L) Alexa 488-conjugated secondary antibody (A-11001; Molecular Probes).
Western blot analysis
HeLa cells seeded at a density of 1 × 105 cells per well of 24-well plates (Greiner) were transduced with different amounts of Ad.ΔE1.TALEN-LS1.F50 or Ad.ΔE1.TALEN-RS1.F50. At 72 h post-transduction, the cell monolayers were rinsed with ice-cold phosphate buffered saline (pH 7.4) and lysed in 100 µl of ice-cold RIPA buffer composed of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% sodium dodecyl sulphate (SDS), 0.5% sodium deoxycholate and 1% Nonidet P-40 plus a cocktail of protease inhibitors (Complete Mini, Roche Applied Science). Next, the cell lysates were spun at 4°C for 8 min at 16 400 g. The proteins were subjected to electrophoresis through an SDS 7.5% polyacrylamide gel after which they were transferred onto an Immobilon-P membrane (Millipore) by electroblotting overnight. After incubation with Tris-buffered saline and tween (TBST) blocking solution consisting of 10 mM Tris-HCl (pH 8.0), 150 mM NaCl and 0.2% Tween-20 supplemented with 10% non-fat dry milk powder (Elk, Campina), the membranes were incubated overnight at 4°C with the HA-specific monoclonal antibody F-7 (SC-7392, Santa Cruz Biotechnology) diluted 1:1000. Subsequently, the membranes were washed once with TBST at room temperature and subjected to a 3-h incubation period with a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (SC-2005; Santa Cruz Biotechnology) diluted 1:5000 in blocking solution. Finally, the membranes were rinsed and processed for protein detection by chemiluminescence as described elsewhere (27).
The western blot analysis to detect TALEN expression following lentiviral vector transduction was performed as follows. Hundred-thousand HeLa cells were seeded in wells of 6-well plates and were transduced with 200 ng of p24gag of LV.TALEN-LS1 or 400 ng of p24gag of IDLV.TALEN-LS1 in the presence of polybrene (8 μg/ml). Forty-eight hours after transduction, the cells were harvested, and total protein lysates were prepared as aforementioned. Fifty micrograms of protein were subjected to electrophoresis through a NuPAGE 4–12% Bis-Tris Gel (Invitrogen) and were transferred onto a nitrocellulose membrane (Hybond-ECL, Amersham) that was subsequently exposed overnight at 4°C to a mouse monoclonal antibody against the HA-tag (clone 12CA5 a gift from Valeria Marigo). A horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Amersham) was diluted 1:2000 in blocking solution. The proteins were detected by using a chemiluminescence labelling detection reagent (ECL; GE Healthcare Life Sciences). For protein loading normalization, the same membrane was stripped and incubated with a 2000-fold diluted mouse monoclonal antibody directed against β-actin (sc-81178, Santa Cruz Biotechnology).
Isolation and structural analysis of adenoviral vector DNA
DNA was isolated from purified adenoviral vector particles as follows. To a 50-µl vector preparation aliquot it was added 12 µl of DNaseI buffer [130 mM Tris-HCl (pH 7.5), 1.2 mM CaCl2 and 50 mM MgCl2], 8 µl of DNaseI (10 mg/ml; Fermentas) and 50 µl of Milli-Q water (Millipore). This mixture was incubated for 30 min at 37°C, after which, the DNaseI was inactivated for 1 h at 55°C after the addition of 2.4 µl of 0.5 M ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 6 µl of 10% SDS and 1.5 µl of a Proteinase K (21.4 mg/ml; Fermentas). Next, the vector DNA was purified by using the QIAEX II Gel Extraction Kit (Qiagen) and was eluted in 30 µl of 10 mM Tris-HCL (pH 7.8). After measuring its concentration with the aid of a Nanodrop (Thermo Scientific), the recovered DNA was subjected to BcuI (Fermentas) digestion and agarose gel electrophoreses (200 ng) and to PCR analyses (1 ng). The PCR amplifications were carried out with primers TALEN-Seq-F (5′-CCAGCTGCTGAAGATCGCCAAGC-3′) and TALEN-Seq-R (5′-CTATCCTGAGTGGAATTTCTGGC-3′) whose target sequences flank the TALEN DNA-binding motifs. The DNA samples were subjected to PCR with 0.4 µM of these primers (Eurofins MWG Operon), 0.1 mM of each dNTP (New England Biolabs), 1× Colorless GoTaq Flexi buffer, 1 mM MgCl2 and 2.5 units of GoTaq DNA polymerase (all from Promega). The PCR cycles were performed in a DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad) using the following cycling parameters. After a 3-min denaturation step at 95°C, the samples were subjected to 30 cycles consisting of 30 s at 95°C, 30 s at 60°C and 4 min at 72°C. The reactions were terminated by a final elongation period of 3 min at 72°C. One-fifth of these reactions were subjected to electrophoresis through 1% agarose gels in 1× Tris-acetate-EDTA (TAE) buffer. Moreover, to establish the integrity of TALEN sequences in adenoviral vector particles at the nucleotide level, the 2.4-kb BcuI fragments from Ad.ΔE1.TALEN-LS1.F50 or from Ad.ΔE1.TALEN-RS1.F50 DNA were inserted into a XbaI-linearized pUC19 backbone and were sequenced by using M13-F (5′-GACGTTGTAAAACGACGGCCAGT-3′) and M13-R (5′-CAGGAAACAGCTATGACCATGA-3′) primers.
Surveyor nuclease S mutation detection assay
Genomic DNA was extracted from mock-transduced or adenoviral vector-transduced target cells as described before (28). Briefly, at 72 h post-infection, the cells were collected by trypsinization and incubated overnight at 55°C in 500 µl of lysis buffer [100 mM Tris-HCl (pH 8.5), 5 mM EDTA, 0.2% SDS, 200 mM NaCl] supplemented with freshly added Proteinase K at a final concentration of 100 ng/ml. Next, the cell lysates were extracted twice with a buffer-saturated phenol:chloroform:isoamyl alcohol mixture (25:24:1) and once with chloroform. The DNA in the aqueous phase was precipitated by the addition of 2.5 volumes of absolute ethanol and 0.5 volumes of 7.5 M ammonium acetate (pH 5.5). After washing with 70% ethanol, the DNA pellet was mildly dried and dissolved in 100 µl of Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA) supplemented with RNase A (Fermentas) at a final concentration of 100 µg/ml.
The recovered DNA was used for PCR analyses. The extent of AAVS1 processing by the error-prone NHEJ DNA repair pathway was evaluated by using the Surveyor assay (SURVEYOR Mutation Detection Kit, Transgenomics) according to the manufacturer’s instructions. In brief, genomic DNA purified from mock-transduced or adenoviral vector-transduced cells was used as a template to PCR amplify a 469-bp AAVS1 sequence flanking the TALEN pair target site. PCR mixtures contained 0.4 µM of primers AAVS1-CEL1-F (5′-TTCGGGTCACCTCTCACTCC-3′) and AAVS1-CEL1-R (5′-GGCTCCATCGTAAGCAAACC-3′), 0.1 mM of each dNTP, 1× Colorless GoTaq Flexi buffer, 1 mM MgCl2 and 2.5 units of GoTaq DNA polymerase. The PCR cycles were performed using the following cycling conditions. An initial denaturation step at 95°C for 5 min was followed by 40 cycles of 30 s at 95°C, 30 s at 61°C and 30 s at 72°C. The reactions were terminated by a final elongation period of 5 min at 72°C. The PCR products, after being heat denaturated and re-annealed according to the manufacturer’s specifications, were treated with the nucleotide mismatch-sensitive Surveyor nuclease S (SNS) enzyme to detect small insertions and deletions (indels) caused by NHEJ-dependent DNA repair. Samples were loaded on a 1.7% agarose gel, and electrophoresis was performed in 1× TAE buffer. The DNA was visualized by in-gel staining with 2 µg/ml of ethidium bromide (Sigma-Aldrich) in 1× TAE buffer followed by stain removal in Milli-Q water. The frequencies of target DNA cleavage were determined by using the ImageJ software (National Institutes of Health, USA). Genomic DNA derived from hMSC cells co-transduced with a 1:1 mixture of Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 at a total multiplicity of infection (MOI) of 24 HTU/cell, served as template to PCR amplify the AAVS1 target site region. The pGEM-T Easy TA cloning vector (Promega) was ligated to the resulting 469-bp amplicons and was transformed into E. coli strain DH5α. Plasmid DNA from 20 randomly selected colonies was extracted, and the respective inserts were subjected to DNA sequencing by using the M13-F primer.
PCR analyses of TALEN sequences following lentiviral vector-mediated gene transfer
Total cellular DNA from HeLa cells incubated with integration-competent or IDLVs was isolated as specified elsewhere (28). The recovered DNA was subjected to three different PCR protocols. To determine the integrity of the cellular DNA, a PCR targeting the human housekeeping gene hypoxanthine phosphoribosyltransferase 1 (HPRT1) was performed. To this end, reactions containing 0.4 µM of primers hHPRT.1 and hHPRT.2 (29), 0.1 mM of each dNTP, 1× Colorless GoTaq Flexi buffer, 1 mM MgCl2 and 2.5 units of GoTaq DNA polymerase were assembled. The PCR cycles were performed using the following cycling conditions. An initial denaturating step at 95°C for 5 min was followed by 30 cycles of 30 s at 95°C, 30 s at 60°C and 120 s at 72°C.
A second set of PCR mixtures were assembled to amplify a 458-bp TALEN sequence corresponding to the N-terminal, non-repetitive region, of each nuclease. These consisted of 0.4 µM of primers TAL-N.s (5′-CCTTACGACGTGCCTGACTACG-3′) and TAL-N.as (5′-CGCTTGGCGATCTTCAGC-3′), 0.1 mM of each dNTP, 1× Colorless GoTaq Flexi buffer, 1 mM MgCl2 and 2.5 units of GoTaq DNA polymerase. The PCR cycles were performed using the following cycling conditions. An initial denaturating step at 95°C for 5 min was followed by 30 cycles of 30 s at 95°C, 30 s at 62°C and 30 s at 72°C.
Finally, the third PCR set was designed to amplify a 2.5-kb TALEN sequence encompassing the region encoding the repetitive DNA-binding domain of each nuclease. To this end, reactions containing 0.4 µM of primers CMV-prom.F (5′-AATGGGCGGTAGGCGTGTA-3′) and TALE.as.Pr2 (5′-GTCGGGTCTGCTCAGCTG-3′), 0.1 mM of each dNTP, 1× Colorless GoTaq Flexi buffer, 1 mM MgCl2 and 2.5 units of GoTaq DNA polymerase were assembled. The PCR cycling parameters consisted of an initial denaturating step at 95°C for 5 min followed by 40 cycles of 30 s at 95°C, 30 s at 60°C and 180 s at 72°C. In all three protocols, the PCR amplifications were terminated by a final elongation period of 5 min at 72°C. Control samples subjected to the same PCR conditions were carried out in parallel. These controls consisted of using, in place of DNA from lentiviral vector-transduced HeLa cells, DNA from mock-transduced HeLa cells, DNA from Ad.ΔE1.TALEN-RS1.F50-transduced HeLa cells, pAd.ΔE1.TALEN-LS1.F50, pLV.CMV.eGFP or MilliQ water. Additional controls were provided by PCR amplifications on DNA isolated from HeLa cells exposed to an LV.TALEN-RS1 preparation made with the aid of pUC19 instead of the lentiviral vector packaging construct psPAX2 (Addgene plasmid 12260) or on DNA extracted from cells transfected with pLV.TALEN-RS1 transfer plasmid mixed or not mixed with ExGen500 (Fermentas).
DNA sequencing of proviral vector DNA in LV.TALEN-LS1-transduced HeLa cell clones was performed as follows. The PCR conditions and cycling parameters specified earlier in the text were applied on chromosomal DNA isolated from HeLa cell clones 2, 5, 6, 7 and 8. Next, the resulting PCR products were inserted into the pGEM-T Easy TA cloning vector (Promega) and were transformed into GT115 E. coli cells (InvivoGen). Finally, the vector inserts were subjected to DNA sequencing by using the oligodeoxyribonucleotides M13-F, M13-R and TALEN-Seq-F.
Southern blot analysis
Total cellular DNA from 4 × 105 hMSCs exposed to three different MOIs of Ad.ΔE1.TALEN-LS1.F50 or of Ad.ΔE1.TALEN-RS1.F50 was extracted 72 h post-transduction and purified as described before (28). After overnight digestion with BamHI (Fermentas), 10 µg of DNA per sample was resolved in a 1% agarose gel in 1× TAE buffer. Next, the DNA was transferred by capillary action onto an Amersham Hybond-XL membrane (GE Healthcare Life Sciences) using a standard Southern blot technique. A 397-bp probe specific for TALEN DNA corresponding to the N-terminal region of the nucleases was isolated by digestion of 1383.pVAX.AAVS1.TALEN.L-94 with XbaI and PstI (both from Fermentas) and was purified with the aid of the QIAEX II gel extraction kit (Qiagen). This DNA probe was radiolabelled with EasyTide (α-32P) dCTP (3000 Ci/mmol; Perkin Elmer) by using the DecaLabel DNA labelling system (Fermentas). Before its addition onto the membrane, the radiolabelled fragment was separated from unincorporated dNTPs through size-exclusion chromatography with the aid of Sephadex-50 columns (GE Healthcare Life Sciences). A Storm 820 PhosphorImager (Amersham) as used for the detection of the probe-hybridizing DNA. Finally, images were acquired and processed by using the Storm scanner control 5.03 and the ImageQuant Tools 3.0 software, respectively (both from Amersham).
HeLa cell clones were derived from an LV.TALEN-LS1-transduced population by limiting dilution using a seeding density of 0.3 cells per well in 96-well plates. Southern blot analysis of genomic DNA extracted from various HeLa cell clones was performed as follows. Genomic DNA was extracted from 5 × 106 cells from each clone using a QIAamp DNA Mini Kit according to the manufacturer’s instructions (Qiagen). Ten micrograms of genomic DNA was digested overnight with AflII (New England Biolabs), subjected to electrophoresis through a 0.8% agarose gel and was transferred onto a Hybond-XL nylon membrane. Before DNA detection, this membrane was incubated with a (α-32P)-labelled 814-bp probe corresponding to the lentiviral vector long-terminal repeat (LTR) elements R and U5, the packaging signal and part of the Rev-responsive element.
RESULTS
HIV-1-based lentiviral vectors introduce rearranged TALEN genes into human cells
We started by investigating the feasibility of generating HIV-1-based lentiviral vectors to deliver TALEN-encoding expression units into human cells. Although the long-term presence of artificial nucleases in cells is, clearly, undesirable in most experimental settings, lentiviral vectors can be made with in-built stringent drug-controllable transgene expression systems (30) or class I integrase mutant moieties (31,32). These integrase mutations are non-pleiotropic, as they confer an integration-defective phenotype to the vectors by specifically disabling the chromosomal insertion step of their DNA. IDLVs are, therefore, being increasingly exploited to deliver transgenes whose expression is only required during a short timeframe in populations of cycling cells. Examples include IDLV-mediated delivery of transposases, homing endonucleases and ZFNs (33–36).
Therefore, we constructed the transfer plasmids pLV.TALEN-LS1 and pLV.TALEN-RS1 to package the lentiviral vector pairs LV.TALEN-LS1/IDLV.TALEN-LS1 and LV.TALEN-RS1/IDLV.TALEN-RS1, respectively. These constructs encode a TALEN enzyme (i.e. TALEN-LS1 or TALEN-RS1) specific for the AAVS1 locus at the human chromosome 19 (19q13.42-qter). Moreover, we also constructed pLV.TALEN-LS1.i.RFP and pLV.TALEN-RS1.i.RFP to make lentiviral vector particles carrying bicistronic cassettes encoding TALEN-LS1 and TALEN-RS1, respectively, and through an IRES, the RFP reporter.
To investigate the integrity of vector genomes in target cells, we carried out transduction experiments on HeLa cells deploying the HIV-1-based lentiviral vector set harboring the TALEN-RS1 ORF. Following DNA extraction from vector-transduced cells, we set up a PCR assay (Figure 1A) based on the use of primer pairs framing the repeats of the TALEN ORF and its upstream region encoding the nuclease’s N-terminus (CMV-prom.F/TALE.as.Pr2) or targeting exclusively the latter segment (TAL-N.s/TAL-N.as). PCR amplifications with primers CMV-prom.F and TALE.as.Pr2 on DNA extracted from HeLa cells transduced with LV.TALEN-RS1, IDLV.TALEN-RS1 or LV.TALEN-RS1.i.RFP did not yield detectable levels of 2.5-kb amplicons diagnostic for full-length TALEN-RS1 sequences (Figure 1B, upper panels, lanes 5, 6 and 11, respectively). In fact, in these samples, the only PCR products that could be discerned were those consistent with heterogeneously sized sub-genomic templates whose molecular-weight range spanned from about 0.8 to 1.5 kb. Significantly, PCR amplifications carried out on the same DNA templates with primers TAL-N.s and TAL-N.as, gave rise to properly sized 0.46-kb PCR products indicating that the non-repetitive upstream portion of the TALEN-RS1 ORF did not suffer major rearrangements in transduced cells (Figure 1B, middle panels). Of note, equivalent results were obtained in complementary transduction experiments that made use of the HIV-1-based lentiviral vector set carrying the TALEN-LS1 ORF (not shown). To validate these data, PCR amplifications were done in parallel on control samples. These negative and positive controls consisted of water (Figure 1B, lane 4) or DNA extracted from mock-transduced HeLa cells (Figure 1B, lane 1), Ad.ΔE1.TALEN-RS1.F50-transduced HeLa cells (Figure 1B, lane 10), plasmid pAd.ΔE1.TALEN-LS1.F50 (Figure 1B, lane 2) or plasmid pLV.CMV.eGFP (Figure 1B, lane 3). Additional controls consisted of PCR amplifications on DNA isolated from HeLa cells exposed to an LV.TALEN-RS1 preparation made with pUC19 instead of the lentiviral vector packaging construct psPAX2 (Figure 1B, lane 7) or incubated with plasmid pLV.TALEN-RS1 mixed or not mixed with the DNA transfection agent ExGen500 (Figure 1B, lanes 8 and 9, respectively). PCR amplifications targeting HPRT1 served as a control for DNA template integrity (Figure 1B, lower panels).
To complement these PCR results on whole target cell populations, we carried out Southern blot analysis on genomic DNA from individual HeLa cell clones derived from a bulk population transduced with LV.TALEN-LS1. To this end, the chromosomal DNA from 15 clones was digested with AflII, Southern blotted and incubated with the probe depicted in Figure 1A. To serve as a molecular-weight reference, the parental transfer plasmid pLV.TALEN-LS1 was subjected to the same procedures. Results depicted in Figure 1C demonstrate that none of the tested clones yielded the AflII 6.4-kb fragment diagnostic for full-length LV.TALEN-LS1 proviruses. The PCR screening of chromosomal DNA from HeLa cell clones 1 through 8 with primers pairs CMV-prom.F/TALE.as.Pr2 and TAL-N.s/TAL-N.as (Figure 1A) established that the proviral LV.TALEN-LS1 deletions located specifically within the TALE repeat region (Figure 1D). To obtain a more detailed knowledge about these rearrangements, we cloned and sequenced the 2.5-kb amplicons obtained after PCR on genomic DNA from HeLa cell clones 2, 5, 6, 7 and 8 (Figure 1D, upper panel). Interestingly, this data revealed that the various clones comprised TALE unit-length deletions leading, as a result, their sizes to differ by multiples of 102 bp (Figure 1E, upper panel). In these clones, the number of deleted RVD-containing repeat units (ΔRVD units) varied between a minimum of 5 (clone 7) and a maximum of 15 (clone 5). Moreover, the alignment of the retrieved DNA sequences with that of the original wild-type DNA shows the involvement of the majority of the 17.5 TALE repeats in the vector rearrangements (Figure 1E, lower panel and Supplementary Figure S2). Thus, these data provide no evidence for any particular ‘hotspot’ within the TALE array at which template recombination events preferentially take place.
Finally, we investigated through HA tag-directed western blot analysis, TALEN-LS1 expression following addition of 400 ng of p24gag of LV.TALEN-LS1 or IDLV.TALEN-LS1 onto HeLa cultures. Consistent with the vector DNA analyses, these experiments failed to yield detectable HA-specific signals at a molecular-weight expected from full-length TALEN-LS1 proteins in protein lysates corresponding to vector-transduced cells. In contrast, these HA-specific signals could be readily detected in control lysates derived from pLV.TALEN-LS1-transfected HeLa cells or 293T producer cells (Figure 1F). To serve as controls for assembly of LV.TALEN-LS1 or IDLV.TALEN-LS1 particles, we produced in parallel LV and IDLV vector preparations carrying a human PGK1 promoter-driven eGFP reporter (LV.PGK.eGFP and IDLV.PGK.eGFP, respectively). Exposure of HeLa cell cultures to 400 ng of p24gag of LV.PGK.eGFP or IDLV.PGK.eGFP led to the transduction of 98% and 91% of the target cells, respectively (not shown).
Taken together, these data indicate that TALEN sequences introduced into human cells through HIV-1-based lentiviral vectors are prone to extensive rearrangements resulting presumably from deletions involving the TALE repeats.
Adenoviral vectors deliver full-length TALEN genes into human cells inducing high-level DSB formation at an endogenous chromosomal target locus
In light of the previous results, we sought to investigate the feasibility of using adenoviral vectors as an alternative delivery platform for the introduction of intact TALEN genes into human cells. To this end, the full-length adenoviral vector molecular clones pAd.ΔE1.TALEN-LS1.F50 and pAd.ΔE1.TALEN-RS1.F50 were assembled through HR in E. coli to produce the E1-deleted adenoviral vectors Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50, respectively (Figure 2A). These clones contain, in place of E1 from the prototypic adenovirus serotype 5, a bicistronic expression unit encoding a monomer of the AAVS1-specific TALEN pair and, for tracing and quantification purposes, the RFP reporter (Figure 2A). In addition, their adenoviral genomic sequences harbor an ORF coding for a vector retargeting chimeric fiber composed of serotype 5 basal shaft domains fused to apical shaft and knob domains derived from the species B adenovirus serotype 50 (F50). The latter CD46-interacting motif allows for entry of vectors displaying species B fiber domains into human cells lacking on their surface the Coxsackie B virus and adenovirus receptor, CAR. Of note, among these cells, there are various therapeutically relevant tissue-specific normal and malignant stem and progenitor cell types (37–41). In addition, plasmids pAd.ΔE1ΔE2A.TALEN-LS1.F50 and pAd.ΔE1ΔE2A.TALEN-RS1.F50 were also constructed to assemble fiber-modified E1- plus E2A-deleted particles Ad.ΔE1ΔE2A.TALEN-LS1.F50 and Ad.ΔE1ΔE2A.TALEN-RS1.F50, respectively (Figure 2A). By virtue of their lack of E2A, these so-called second-generation adenoviral vectors have been shown to display a much-reduced de novo replication and ‘leaky’ viral gene expression profile in target cells when compared with their E1-deleted, first-generation, counterparts [see, for instance, (20)].
We started by testing whether TALEN sequences are stably maintained in first- and second-generation adenoviral vectors following their rescue and subsequent propagation on their respective packaging cell lines. Thus, these producer cells were transfected with PacI-treated plasmids pAd.ΔE1.TALEN-LS1.F50, pAd.ΔE1.TALEN-RS1.F50, pAd.ΔE1ΔE2A.TALEN-LS1.F50 or pAd.ΔE1ΔE2A.TALEN-RS1.F50 to assemble the corresponding adenoviral vector particles (Figure 2A). The rescued particles were subsequently amplified following serial propagation on increasing amounts of packaging cells. The frequency of RFP-positive producer cells, as monitored by direct fluorescence microscopy, revealed a clear propagation round-dependent increase in vector yields (not shown). The Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 vectors, corresponding to two independent batches, were purified through CsCl buoyant density gradient ultracentrifugation, after which, the integrity of their genomes was probed by BcuI restriction fragment length analysis. To serve as an additional molecular-weight reference, the parental plasmids pAd.ΔE1.TALEN-LS1.F50 and pAd.ΔE1.TALEN-RS1.F50 were taken along in this analysis. Results depicted in Figure 2B show that the overall DNA restriction pattern is consistent with that expected from linear full-length adenoviral vector genomes (Figure 2A). Thus, importantly, the internal TALE-encompassing restriction fragments derived from Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 DNA did co-migrate with those corresponding to their respective 2.4-kb parental plasmid counterparts (Figure 2B, arrowheads). Results equivalent to these were obtained following the structural analyses of DNA isolated from Ad.ΔE1ΔE2A.TALEN-LS1.F50 and Ad.ΔE1ΔE2A.TALEN-RS1.F50 (Supplementary Figure S3). Next, we performed a more sensitive PCR-based assay using primers bracketing the 17.5 repeats of TALEN-LS1 and TALEN-RS1 to search for rearranged TALEN sequences in the purified vector preparations. Again, parental constructs pAd.ΔE1.TALEN-LS1.F50 and pAd.ΔE1.TALEN-RS1.F50 served as control DNA templates. This assay led to the detection of 2.1-kb amplicons whose size is diagnostic for vector DNA genomes harboring intact TALEN-encoding genes (Figure 2C). Moreover, Southern blot analysis of extrachromosomal DNA isolated from hMSCs exposed to different amounts of Ad.ΔE1.TALEN-LS1.F50 or Ad.ΔE1.TALEN-RS1.F50 also led to the sole detection of full-length TALEN sequences in the transduced cells (Figure 2D). Finally, we cloned into pUC19 the 2.4-kb DNA species from BcuI-treated Ad.ΔE1.TALEN-LS1.F50 or BcuI-treated Ad.ΔE1.TALEN-RS1.F50 and sequenced the inserts from six individual clones. DNA sequence information corresponding to the 7 most downstream and to the 3–4 most upstream TALEN repeats could be retrieved (Supplementary Figure S4). The nucleotide sequences analysed revealed to be isogenic to that in the parental constructs with no evidence for point mutations, local rearrangements and/or deletions.
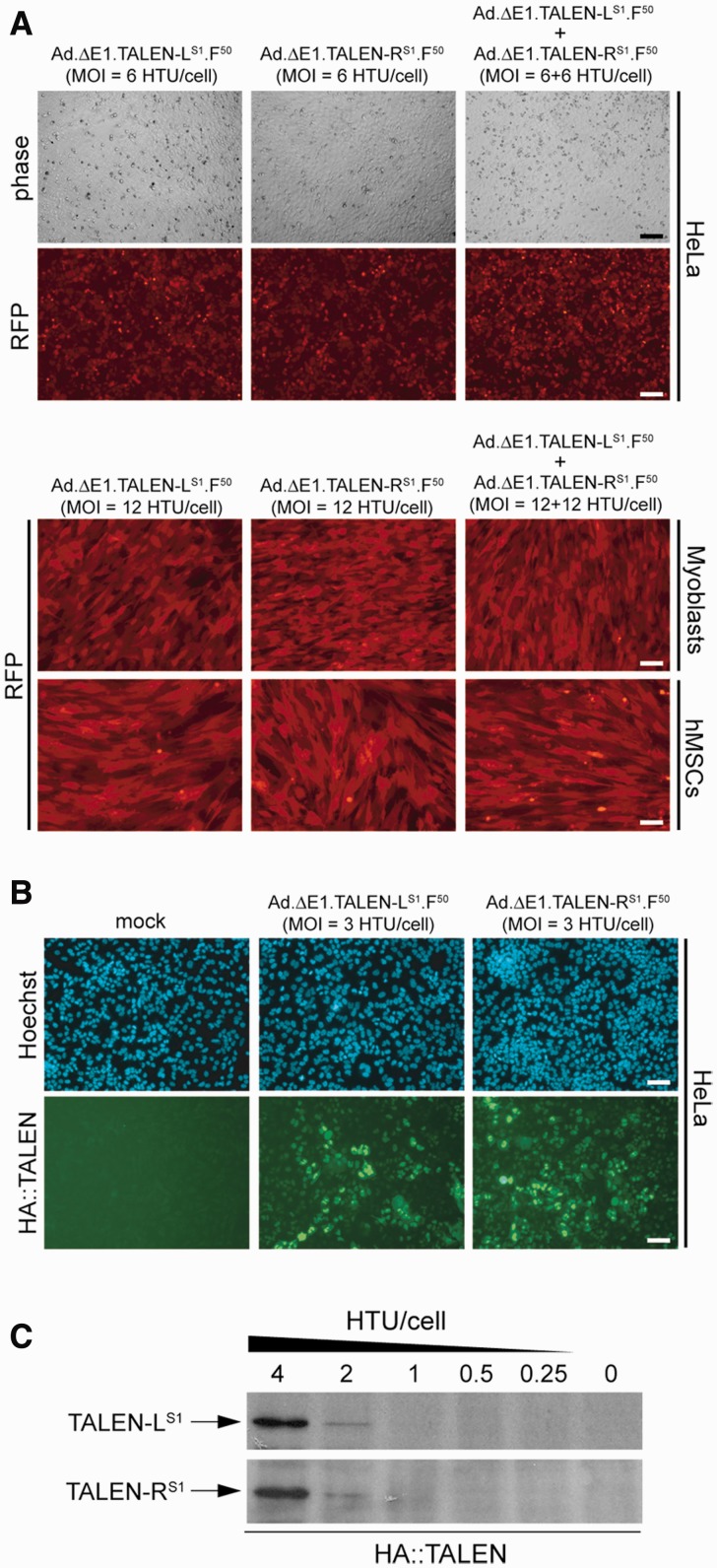
Transduction experiments deploying Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 alone or mixed together were carried out on CAR-positive human cervix carcinoma HeLa cells (42) and on two CAR-negative cell types, namely, immortalized myoblasts from a Duchenne muscular dystrophy (DMD) patient and hMSCs (42,43). The use of relatively modest MOIs was sufficient to transduce most target cells in the various cultures (Figure 3A). Importantly, HA tag-specific immunofluorescence microscopy and western blot analyses (Figure 3B and 3C, respectively) demonstrated the expression of TALEN-LS1 and TALEN-RS1 genes in target HeLa cells following their transduction with Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50, respectively. Taken together, we conclude that first-generation adenoviral vectors can deliver expression units encoding full-length TALENs into human cells.
Figure 3.
Adenoviral vector-mediated delivery of TALEN genes into transformed and non-transformed human cells. (A) Gene transfer efficiency. Upper panel, phase-contrast and live-cell RFP direct fluorescence microscopy on HeLa cells transduced with Ad.ΔE1.TALEN-LS1.F50 or with Ad.ΔE1.TALEN-RS1.F50 at an MOI of 6 HTU/cell or with a mixture of the two adenoviral vectors at an MOI of 6 HTU/cell each. Lower panel, RFP direct fluorescence microscopy on immortalized DMD myoblasts and on primary hMSCs transduced with Ad.ΔE1.TALEN-LS1.F50 or with Ad.ΔE1.TALEN-RS1.F50 at an MOI of 12 HTU/cell or co-transduced with a 1:1 mixture of Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 at a total MOI of 24 HTU/cell. The fluorescence microscopy images were acquired at 48 h post-infection. (B) HA tag-specific immunofluorescence microscopy on HeLa cells mock-transduced or transduced with Ad.ΔE1.TALEN-LS1.F50 or with Ad.ΔE1.TALEN-RS1.F50 at an MOI of 3 HTU/cell. The direct fluorescence microscopy and the immunofluorescence microscopy were carried out at 48 and 72 h post-infection, respectively. (C) HA tag-directed western blot analysis of protein lysates derived from HeLa cell cultures incubated for 72 h with Ad.ΔE1.TALEN-LS1.F50 or with Ad.ΔE1.TALEN-RS1.F50. The MOIs deployed are indicated.
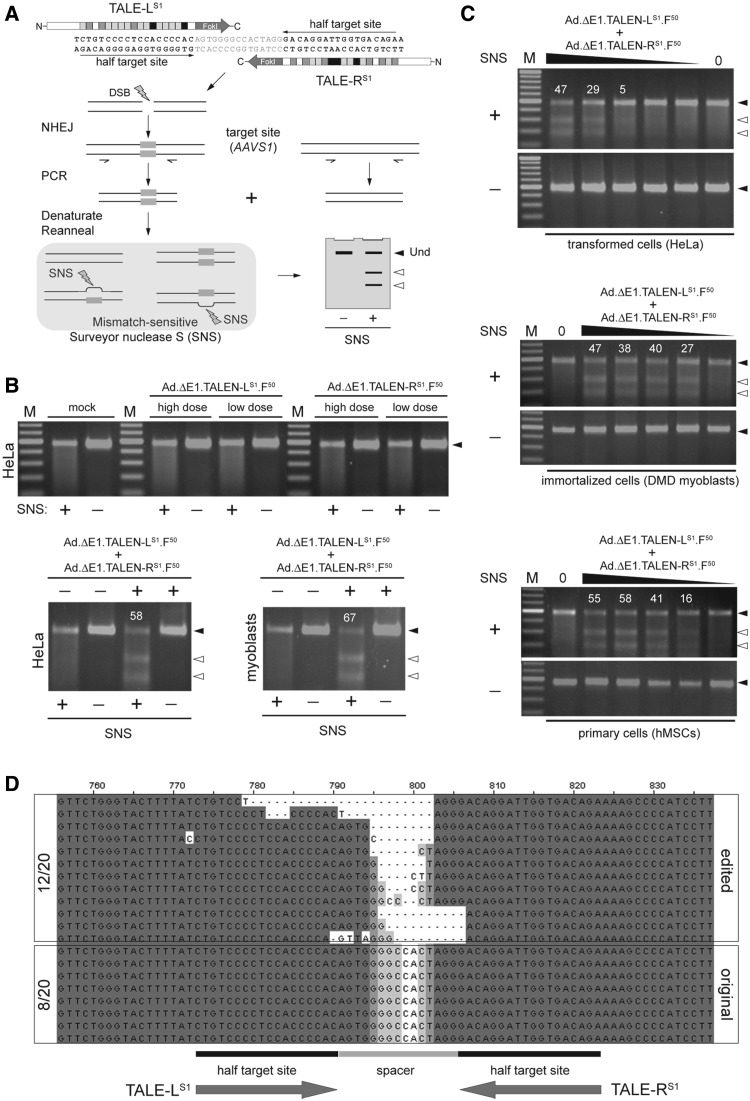
Finally, to investigate the functionality of TALENs introduced into human cells through adenoviral vector-mediated gene transfer, we deployed the Surveyor mutation detection assay (Figure 4A). PCR amplification of AAVS1 target DNA from mock-transduced cells or from cells transduced with Ad.ΔE1.TALEN-LS1.F50 or with Ad.ΔE1.TALEN-RS1.F50 yielded amplicons that, on denaturation and reannealing, were not susceptible to site-specific cleavage by the mismatch-sensitive SNS enzyme (Figure 4B, solid arrowhead in the upper panel). These data validated the read-out system by demonstrating the absence of indels at the AAVS1 locus under these experimental control conditions. However, co-transduction of HeLa cells and DMD myoblasts with Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 resulted in efficient cleavage of the endogenous chromosomal target sequence, as evidenced by the detection of large amounts of SNS-digested products (Figure 4B, open arrowheads in the lower panels). This conclusion was substantiated through vector dose-response experiments on HeLa cells, human myoblasts and hMSCs (Figure 4C). Additional transduction experiments on hMSCs that made use of the second-generation adenoviral vectors Ad.ΔE1ΔE2A.TALEN-LS1.F50 and Ad.ΔE1ΔE2A.TALEN-RS1.F50 produced results similar to those shown in Figure 4C (Supplementary Figure S5). Finally, we sought to confirm the involvement of the NHEJ pathway in the repair of the TALEN-induced DSBs. To this end, we cloned the AAVS1-specific PCR products resulting from genomic DNA of hMSCs exposed to Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 at an MOI of 12 HTU/cell each. DNA sequencing of 20 randomly selected clones revealed that 12 of these harbored indels characteristic of NHEJ-mediated repair of DSBs (Figure 4D).
Figure 4.
Targeted disruption of the chromosomal AAVS1 locus. (A) Schematic representation of the Surveyor mutation detection assay used to probe TALEN activity in human cells. Assembly of the TALEN pair at the bipartite target sequence results in local DSB formation. The error-prone NHEJ DNA repair pathway links the broken chromosomal ends introducing in the process small insertions and deletions (indels) at the junction (gray boxes). PCR on chromosomal DNA with primers framing the target site yields a mixture of amplicons representing the heterogeneous population of mutated sequences and non-modified alleles. Denaturation and re-annealing of this mixture results in DNA species with internal base pairs that are mismatched. The SNS enzyme, by preferentially recognizing and cleaving at DNA sequences with such mismatched base pairs, yields low-molecular-weight DNA fragments (open arrowheads) that can be resolved from undigested DNA (solid arrowheads) through conventional agarose gel electrophoreses. Parallel samples not treated with SNS (−) served as a negative control. (B) TALEN activity at AAVS1. Upper panel, Surveyor assay on chromosomal DNA isolated from HeLa cells not exposed (mock) or exposed to Ad.ΔE1.TALEN-LS1.F50 or to Ad.ΔE1.TALEN-RS1.F50 at a high and a low dose (i.e. 6 and 2 HTU/cell, respectively). Lower panels, Surveyor assay on chromosomal DNA extracted from HeLa cells and DMD myoblasts (left and right panels, respectively). HeLa cells were mock-transduced (−) or were co-transduced with 6 HTU/cell of Ad.ΔE1.TALEN-LS1.F50 and 6 HTU/cell of Ad.ΔE1.TALEN-RS1.F50 (+). Human myoblasts were mock-transduced (−) or were co-transduced with 12 HTU/cell of Ad.ΔE1.TALEN-LS1.F50 and 12 HTU/cell of Ad.ΔE1.TALEN-RS1.F50 (+). Solid and open arrowheads indicate the positions of undigested and SNS-digested DNA molecules, respectively. White numerals correspond to the percentage of target site cleavage. (C) Dose-response of TALEN activity. Surveyor assay on cellular DNA from HeLa cells, DMD myoblasts and hMSCs untreated or treated with different doses of 1:1 mixtures of Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50. In these dose-response co-transduction experiments, the total amounts of vector applied on HeLa cells were 12, 4, 1.3, 0.4 and 0.15 HTU/cell, whereas on DMD myoblasts and hMSCs, HTU/cell were 24, 8, 2.7, 1.0 and 0.3. Solid and open arrowheads indicate the positions of undigested and SNS-digested DNA molecules, respectively. White numerals correspond to the percentage of target site cleavage. (D) Sequence analysis. DNA sequencing of cloned AAVS1-specific amplicons resulting from PCR amplifications on genomic DNA isolated from hMSCs co-transduced with Ad.ΔE1.TALEN-LS1.F50 and Ad.ΔE1.TALEN-RS1.F50 at 12 HTU/cell each. The presence of indels in 12 of 20 clones, ‘footprint’ for NHEJ processing of DSBs, demonstrates at the nucleotide level the TALEN-mediated cleavage of the chromosomal AAVS1 target site.
We conclude that delivery of TALENs by first- and second-generation adenoviral vectors is a highly effective approach to generate in a cell type-independent manner site-specific DSBs at an endogenous human target locus.
DISCUSSION
The approaches aiming at editing or modulating the expression of specific endogenous genes and targeting chromosomal insertion of exogenous DNA have recently been enriched with the introduction of sequence-specific designer TALEs. To broaden the use of TALE-based proteins and to most optimally screen and evaluate their performance in many settings, it will be crucial to expand the range of cellular delivery tools for these molecules. Despite their promise and favorable set of attributes when compared with other custom-made sequence-specific proteins, TALEs are relatively large proteins whose DNA binding specificity is governed by an extensive tract of repetitive sequences. Of note, DNA repeats are known to be particularly unstable motifs. Indeed, examples of both physiological and function-impairing processes associated with instability of tandem repeats are copious and cover virtually all biological systems from viruses and prokaryotes to eukaryotes (44–46). Also in the context of gene delivery vehicles, such as those based on adenoviral and lentiviral vectors, examples exist pointing to certain repetitive sequence arrangements as the culprits behind DNA instability (47–50). Bearing this in mind, in the present study, we sought to investigate the suitability of adenoviral and HIV-1-based lentiviral vectors, two of the most commonly used viral vector systems, to serve as TALEN gene delivery platforms.
We report that HIV-1-based lentiviral vector genomes bearing TALEN sequences are prone to rearrangements in target cells. In this regard, the structural analyses of these genomes in HeLa cell populations and in individual HeLa cell clones by PCR, Southern blot and DNA sequencing indicate that most of the rearrangements occurred through recombination events involving the TALE repeat array, ultimately leading to deletions with various sizes. The DNA sequence analyses in particular revealed that these rearrangements of the TALE array, albeit variable in number, consisted of precise as opposed to randomly truncated deletions of individual TALE repeats.
Conversely, we found that TALEN sequences are stably maintained in first- and second-generation adenoviral vectors following their serial propagation in producer cells. Indeed, the TALEN transgenes in the adenoviral vector genomes presented neither large rearrangements nor small-scale mutations as revealed by restriction fragment length analysis and DNA sequencing, respectively. Importantly, transduction experiments with the TALEN-encoding adenoviral vectors led to DSB formation at the intended target chromosomal locus in transformed and non-transformed human cells at similarly high frequencies.
Clearly, viruses from the Adenoviridae and Retroviridae families display diametrically different structural features, life cycles and replication strategies (15,51,52). The latter aspect in particular is likely to contribute to the disparate stability of TALEN ORFs in adenoviral versus HIV-1-based lentiviral vectors. Adenoviruses replicate via a DNA strand-displacement mechanism using a virus-encoded high-fidelity DNA polymerase and a single-stranded DNA-binding protein that, by cooperatively coating the displaced single-stranded replicative intermediates, presumably inhibits spurious intramolecular hybridization (52,53). Retroviruses, on the other hand, encode error-prone reverse transcriptases (RTs), which convert their double-copy RNA genomes into linear double-stranded DNA templates that serve as substrates for proviral chromosomal insertion (51). The RTs of simple and complex retroviruses also display a RNase H activity responsible for the digestion of the RNA component of hybrid RNA:DNA duplex intermediates. Moreover, they share a low template affinity and processivity. These features seem to have evolved to allow RT to undergo at least two obligate template switching events (i.e. minus-strand and plus-strand DNA transfer) to complete full-length viral DNA synthesis and, as ‘byproduct’, generate genotypic diversity through recombination. It is known, however, that the presence of direct repeats in retroviral genomes increases the likelihood for additional inter- and intra-molecular template switching events, often resulting in the deletion of one of the repeats plus intervening sequences (47,48,54,55). In fact, the insertion of direct repeats in retroviral genomes provides for an elegant model system to study the RT template switching process in vivo or to induce precise rearrangements of cis- and trans-acting elements in retroviral vectors (54,56). Of note, the frequency of template switching increases with the size of and the distance between the direct repeat units. The latter finding suggests that spacing favors template ‘looping’, which, in turn, aids in the intra-strand alignment of direct repeats, RT template switching and ensuing intra-molecular deletions (55). The models for these direct repeat-induced deletions (48,54,55) postulate that during reverse transcription, RT and the nascent DNA strand dissociates from the RNA template owing to the aforementioned RNaseH activity and intrinsic low template affinity. Segments and, eventually, all nascent DNA strand hybridizes to the RNA template at the distal ‘acceptor’ repeat serving as a novel ‘ectopic’ initiation point for RNA-dependent DNA synthesis. The net result of these processes is often homogenously sized deletions of the retroviral genome. In contrast to simple direct repeat pairs, TALE sequences present a tract of numerous and contiguous repeats. Thus, each of these arrayed repeat units provide for multiple potential positions at which RT template switching may occur. This corollary is consistent with the heterogeneous and subgenomic-sized population of TALEN-containing HIV-1-based lentiviral vector proviruses that were detected in stably transduced clones, as it is with the proviral vector DNA sequences found in their cells. In fact, by showing precise as opposed to truncated deletions of TALE repeats, the DNA sequencing data do indicate the involvement of a mechanism based on hybridization of similar sequences, which underlies RT template switching. This is strengthen by the observation that, of the six independent molecular clones analysed, four contain sequence ‘footprints’ consistent with the occurrence of more than one RT template switch event (Figure 1E, clones 2, 5, 8.1 and 8.2). These data have implications for the delivery of TALE-containing transgenes through lentiviral vectors in that proteins with specificities for unintended and multiple chromosomal DNA positions can be assembled, potentially leading to deleterious effects in transduced cells. Therefore, cells exposed to TALE protein derivatives that operate as monomers, such as artificial transcriptional activators or repressors, might be particularly vulnerable. Finally, in this regard, HA-directed immunofluorescence microscopy on HeLa cells transduced with TALEN-encoding HIV-1-based lentiviral vector particles did reveal the presence of HA tag-positive cells. These data suggest that despite their variable number of deleted repeats, TALE-containing transgenes transferred by HIV-1-based lentiviral vectors can yield enough protein to be detected through this assay (not shown). Notwithstanding these findings, one can envision strategies that might minimize or overcome the instability of TALEN sequences in these vectors. For instance, by exploiting the degeneracy of the genetic code, the individual TALE motifs within the array could be re-engineered by the introduction of judiciously chosen silent substitutions to increase sequence divergence among repeats. Regarding this issue, it has been shown via a lentiviral vector direct repeat deletion assay that deletion frequencies can decrease steeply with incremental repeat sequence divergence (57). Finally, it is worth pointing out that future experiments will be required to establish whether the assembly of rearranged TALE repeats following HIV-1-based lentiviral vector transductions also takes place while deploying other classes of retroviral vector systems.
In conclusion, TALEN sequences are prone to rearrangements following their introduction into human cells by HIV-1-based lentiviral vectors. As a result, vector-mediated TALEN gene transfer using these gene carriers will be dependent on new optimization strategies. Conversely, we demonstrate that first- and second-generation adenoviral vectors are amenable to the transfer of full-length TALEN genes into human cells. Importantly, the resulting TALEN protein yields suffice to induce high-level DSB formation at an endogenous chromosomal target locus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Prinses Beatrix Spierfonds [W.OR11-18 to M.A.F.V.G.]; the European Community’s 7th Framework Programme for Research and Technological Development [PERSIST grant agreement number 222878 to A.R., T.C. and M.A.F.V.G.]. Funding for open access charge: Prinses Beatrix Spierfonds [W.OR11-18].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Zeinab Neshati, Antoine A.F. de Vries (both from the Department of Cardiology, Leiden University Medical Center, The Netherlands) and Jaroslav Skokan for generating the constructs precursors of pShuttle.IRES.DsRedEx2.1. They also thank Harald Mikkers and Marie-José Goumans for providing us with the hMSCs and Laetitia Pelascini and Rob Hoeben for critically reading the manuscript (all from the Department of Molecular Cell Biology, Leiden University Medical Center, The Netherlands). Didier Trono (École Polytechnique Fédérale de Lausanne, Switzerland) is acknowledged for kindly making available the immortalized DMD myoblasts.
REFERENCES
- 1.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klug A. The discovery of Zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 3.Redondo P, Prieto J, Muñoz IG, Alibés A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 4.Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, Pâques F. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 6.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:150. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 7.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 8.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 9.Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mussolino C, Cathomen T. TALE nucleases: tailored genome engineering made easy. Curr. Opin. Biotech. 2012;23:1–7. doi: 10.1016/j.copbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Cathomen T, Joung KJ. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 12.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong C, Huang G, Ashton C, Wu H, Yan H, Ying QL. Rapid and cost-effective gene targeting in rat embryonic stem cells by TALENs. J. Genet. Genomics. 2012;39:275–280. doi: 10.1016/j.jgg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol. Biosyst. 2012;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 15.Gonçalves MAFV, de Vries AAF. Adenovirus: from foe to friend. Rev. Med. Virol. 2006;16:167–186. doi: 10.1002/rmv.494. [DOI] [PubMed] [Google Scholar]
- 16.Dropulić B. Lentiviral vectors: their molecular design, safety, and use in laboratory and preclinical research. Hum. Gene Ther. 2011;22:649–657. doi: 10.1089/hum.2011.058. [DOI] [PubMed] [Google Scholar]
- 17.Yamashita M, Emerman M. Retroviral infection of non-dividing cells: old and new perspectives. Virology. 2006;344:88–93. doi: 10.1016/j.virol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Henaff D, Salinas S, Kremer EJ. An adenovirus traffic update: from receptor engagement to the nuclear pore. Future Microbiol. 2011;6:179–192. doi: 10.2217/fmb.10.162. [DOI] [PubMed] [Google Scholar]
- 19.Fallaux FJ, Bout A, van der Velde I, van den Wollenberg DJ, Hehir KM, Keegan J, Auger C, Cramer SJ, van Ormondt H, et al. New helper cells and matched early region 1–deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther. 1998;9:1909–1917. doi: 10.1089/hum.1998.9.13-1909. [DOI] [PubMed] [Google Scholar]
- 20.Havenga MJ, Holterman L, Melis I, Smits S, Kaspers J, Heemskerk E, van der Vlugt R, Koldijk M, Schouten GJ, Hateboer G, et al. Serum-free transient protein production system based on adenoviral vector and PER.C6 technology: high yield and preserved bioactivity. Biotechnol. Bioeng. 2008;100:273–283. doi: 10.1002/bit.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cudré-Mauroux C, Occhiodoro T, König S, Salmon P, Bernheim L, Trono D. Lentivector-mediated transfer of Bmi-1 and telomerase in muscle satellite cells yields a duchenne myoblast cell line with long-term genotypic and phenotypic stability. Hum. Gene Ther. 2003;14:1525–1533. doi: 10.1089/104303403322495034. [DOI] [PubMed] [Google Scholar]
- 22.Gonçalves MAFV, Janssen JM, Nguyen QG, Athanasopoulos T, Hauschka SD, Dickson G, de Vries AAF. Transcription factor rational design improves directed differentiation of human mesenchymal stem cells into skeletal myocytes. Mol. Ther. 2011;19:1331–1341. doi: 10.1038/mt.2010.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen JM, Liu J, Skokan J, Gonçalves MAFG, de Vries AAF. Development of an AdEasy-based system to produce first- and second-generation adenoviral vectors with tropism for CAR- or CD46-positive cells. J. Gene Med. 2012;15:1–11. doi: 10.1002/jgm.2687. [DOI] [PubMed] [Google Scholar]
- 24.Seppen J, Rijnberg M, Cooreman MP, Oude Elferink RP. Lentiviral vectors for efficient transduction of isolated primary quiescent hepatocytes. J. Hepatol. 2002;36:459–465. doi: 10.1016/s0168-8278(01)00308-7. [DOI] [PubMed] [Google Scholar]
- 25.Pelascini PL, Janssen JM, Gonçalves MAFV. Histone deacetylase inhibition activates transgene expression from integration-defective lentiviral vectors in dividing and non-dividing cells. Hum. Gene Ther. 2012;24:78–96. doi: 10.1089/hum.2012.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonçalves MAFV, Swildens J, Holkers M, Narain A, van Nierop GP, van de Watering MJ, Knaän-Shanzer S, de Vries AAF. Genetic complementation of human muscle cells via directed stem cell fusion. Mol. Ther. 2008;16:741–748. doi: 10.1038/mt.2008.16. [DOI] [PubMed] [Google Scholar]
- 27.Gonçalves MAFG, Holkers M, van Nierop GP, Wieringa R, Pau MG, de Vries AAF. Targeted chromosomal insertion of large DNA into the human genome by a fiber-modified high-capacity adenovirus-based vector system. PLoS One. 2008;29:e3084. doi: 10.1371/journal.pone.0003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Nierop GP, de Vries AAF, Holkers M, Vrijsen KR, Gonçalves MAFV. Stimulation of homology-directed gene targeting at an endogenous human locus by a nicking endonuclease. Nucleic Acids Res. 2009;37:5725–5736. doi: 10.1093/nar/gkp643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonçalves MAFV, van der Velde I, Knaän-Shanzer S, Valerio D, de Vries AAF. Stable transduction of large DNA by high-capacity adeno-associated virus/adenovirus hybrid vectors. Virology. 2004;321:287–296. doi: 10.1016/j.virol.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat. Methods. 2006;3:109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- 31.Philpott NJ, Thrasher AJ. Use of nonintegrating lentiviral vectors for gene therapy. Hum. Gene Ther. 2007;18:483–489. doi: 10.1089/hum.2007.013. [DOI] [PubMed] [Google Scholar]
- 32.Wanisch K, Yáñez-Muñoz RJ. Integration-deficient lentiviral vectors: a slow coming of age. Mol. Ther. 2009;17:1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staunstrup NH, Mikkelsen JG. Integrase-defective lentiviral vectors-a stage for nonviral integration machineries. Curr. Gene Ther. 2011;11:350–362. doi: 10.2174/156652311797415881. [DOI] [PubMed] [Google Scholar]
- 34.Cornu TI, Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- 35.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 36.Izmiryan A, Basmaciogullari S, Henry A, Paques F, Danos O. Efficient gene targeting mediated by a lentiviral vector-associated meganuclease. Nucleic Acids Res. 2011;39:7610–7619. doi: 10.1093/nar/gkr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shayakhmetov DM, Papayannopoulou T, Stamatoyannopoulos G, Lieber A. Efficient gene transfer into human CD34(+) cells by a retargeted adenovirus vector. J. Virol. 2000;74:2567–2583. doi: 10.1128/jvi.74.6.2567-2583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knaän-Shanzer S, van der Velde I, Havenga MJ, Lemckert AA, de Vries AAF, Valerio D. Highly efficient targeted transduction of undifferentiated human hematopoietic cells by adenoviral vectors displaying fiber knobs of subgroup B. Hum. Gene Ther. 2001;12:1989–2005. doi: 10.1089/104303401753204562. Erratum in: Hum. Gene Ther. 2003 Aug 10;14. [DOI] [PubMed] [Google Scholar]
- 39.Knaän-Shanzer S, van de Watering MJ, van der Velde I, Gonçalves MAFV, Valerio D, de Vries AAF. Endowing human adenovirus serotype 5 vectors with fiber domains of species B greatly enhances gene transfer into human mesenchymal stem cells. Stem Cells. 2005;23:1598–1607. doi: 10.1634/stemcells.2005-0016. [DOI] [PubMed] [Google Scholar]
- 40.Tuve S, Wang H, Ware C, Liu Y, Gaggar A, Bernt K, Shayakhmetov D, Li Z, Strauss R, Stone D, et al. A new group B adenovirus receptor is expressed at high levels on human stem and tumor cells. J. Virol. 2006;80:12109–12120. doi: 10.1128/JVI.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann D, Meyer B, Wildner O. Improved glioblastoma treatment with Ad5/35 fiber chimeric conditionally replicating adenoviruses. J. Gene Med. 2007;9:764–778. doi: 10.1002/jgm.1076. [DOI] [PubMed] [Google Scholar]
- 42.Gonçalves MAFV, Holkers M, Cudré-Mauroux C, van Nierop GP, Knaän-Shanzer S, van der Velde I, Valerio D, de Vries AAF. Transduction of myogenic cells by retargeted dual high-capacity hybrid viral vectors: robust dystrophin synthesis in duchenne muscular dystrophy muscle cells. Mol. Ther. 2006;13:976–986. doi: 10.1016/j.ymthe.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 43.Gonçalves MAFV, de Vries AAF, Holkers M, van de Watering MJ, van der Velde I, van Nierop GP, Valerio D, Knaän-Shanzer S. Human mesenchymal stem cells ectopically expressing full-length dystrophin can complement Duchenne muscular dystrophy myotubes by cell fusion. Hum. Mol. Genet. 2006;15:213–221. doi: 10.1093/hmg/ddi438. [DOI] [PubMed] [Google Scholar]
- 44.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 45.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holkers M, de Vries AAF, Gonçalves MAFV. Nonspaced inverted DNA repeats are preferential targets for homology-directed gene repair in mammalian cells. Nucleic Acids Res. 2012;40:1984–1999. doi: 10.1093/nar/gkr976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhode BW, Emerman M, Temin HM. Instability of large direct repeats in retrovirus vectors. J. Virol. 1987;61:925–927. doi: 10.1128/jvi.61.3.925-927.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pathak VK, Temin HM. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc. Natl Acad. Sci. USA. 1990;87:6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinwaerder DS, Carlson CA, Lieber A. Generation of adenovirus vectors devoid of all viral genes by recombination between inverted repeats. J. Virol. 1999;73:9303–9313. doi: 10.1128/jvi.73.11.9303-9313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belousova N, Harris R, Zinn K, Rhodes-Selser MA, Kotov A, Kotova O, Wang M, Aurigemma R, Zhu ZB, Curiel DT, et al. Circumventing recombination events encountered with production of a clinical-grade adenoviral vector with a double-expression cassette. Mol. Pharmacol. 2006;70:1488–1493. doi: 10.1124/mol.106.025619. [DOI] [PubMed] [Google Scholar]
- 51.Coffin JM, Hughes SH, Varmus HE. Retroviruses. NY: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1997. [PubMed] [Google Scholar]
- 52.de Jong RN, van der Vliet PC, Brenkman AB. Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP. Curr. Top. Microbiol. Immunol. 2003;272:187–211. doi: 10.1007/978-3-662-05597-7_7. [DOI] [PubMed] [Google Scholar]
- 53.Zijderveld DC, Stuiver MH, van der Vliet PC. The adenovirus DNA binding protein enhances intermolecular DNA renaturation but inhibits intramolecular DNA renaturation. Nucleic Acids Res. 1993;21:2591–2598. doi: 10.1093/nar/21.11.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Julias JG, Hash D, Pathak VK. E- vectors: development of novel self-inactivating and self-activating retroviral vectors for safer gene therapy. J. Virol. 1995;69:6839–6846. doi: 10.1128/jvi.69.11.6839-6846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delviks KA, Pathak VK. Effect of distance between homologous sequences and 3' homology on the frequency of retroviral reverse transcriptase template switching. J. Virol. 1999;73:7923–7932. doi: 10.1128/jvi.73.10.7923-7932.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delviks KA, Hu WS, Pathak VK. Psi- vectors: murine leukemia virus-based self-inactivating and self-activating retroviral vectors. J. Virol. 1997;71:6218–6224. doi: 10.1128/jvi.71.8.6218-6224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.An W, Telesnitsky A. Effects of varying sequence similarity on the frequency of repeat deletion during reverse transcription of a human immunodeficiency virus type 1 vector. J. Virol. 2002;76:7897–7902. doi: 10.1128/JVI.76.15.7897-7902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.