Abstract
Nucleosome positioning on the chromatin strand plays a critical role in regulating accessibility of DNA to transcription factors and chromatin modifying enzymes. Hence, detailed information on nucleosome depletion or movement at cis-acting regulatory elements has the potential to identify predicted binding sites for trans-acting factors. Using a novel method based on enrichment of mononucleosomal DNA by bacterial artificial chromosome hybridization, we mapped nucleosome positions by deep sequencing across 250 kb, encompassing the cystic fibrosis transmembrane conductance regulator (CFTR) gene. CFTR shows tight tissue-specific regulation of expression, which is largely determined by cis-regulatory elements that lie outside the gene promoter. Although multiple elements are known, the repertoire of transcription factors that interact with these sites to activate or repress CFTR expression remains incomplete. Here, we show that specific nucleosome depletion corresponds to well-characterized binding sites for known trans-acting factors, including hepatocyte nuclear factor 1, Forkhead box A1 and CCCTC-binding factor. Moreover, the cell-type selective nucleosome positioning is effective in predicting binding sites for novel interacting factors, such as BAF155. Finally, we identify transcription factor binding sites that are overrepresented in regions where nucleosomes are depleted in a cell-specific manner. This approach recognizes the glucocorticoid receptor as a novel trans-acting factor that regulates CFTR expression in vivo.
INTRODUCTION
Recent progress in generating maps of open chromatin genome wide (1,2) has greatly accelerated the discovery of regulatory mechanisms both for individual genes and for coordinated transcriptional networks. However, advances in the technologies that enable de novo characterization of trans-acting factors that interact with these regions of open chromatin are still in development (3–5). Until recently, prediction of transcription factor binding sites in genomic DNA was largely dependent on the use of in silico approaches followed by in vivo verification by chromatin immunoprecipitation (ChIP) using specific antibodies for individual factors. As the regions of open chromatin identified by DNase-seq (DNase I hypersensitivity mapping followed by deep sequencing) and FAIRE (formaldehyde-assisted identification of regulatory elements) can encompass >1 kb of DNA sequence, it remains a challenge to characterize the cis-regulatory elements that are responsible for the chromatin landscape. One approach that can assist in this effort is to fine map the location of nucleosomes across the region of open chromatin, as nucleosome positioning is known to play a critical role in accessibility of the DNA strand to interacting proteins (6,7). The dynamic competition between DNA unwinding, nucleosome positioning and transcription factor binding is not yet fully understood (8,9). However, there are many cases, for example, the androgen receptor (4,10), where loss of a specific nucleosome can be directly correlated with occupancy of a regulatory element by one or more of its cognate protein partners. Here, we combine the techniques of DNase-seq and a novel method to fine map nucleosome positions by bacterial artificial chromosome (BAC) enrichment of mononucleosomal DNA and deep sequencing, to identify trans-acting factors regulating cystic fibrosis transmembrane conductance regulator (CFTR) gene expression. The CFTR gene spans 189 kb of genomic DNA at chromosome 7q31 (11), although regulatory elements for the locus map to regions far upstream and downstream of the gene, in addition to within introns [reviewed in (12)]. Hence, to fully understand the CFTR cis-regulatory sequences and trans-acting factors that control cell-specific expression of the gene, a region of >250 kb must be evaluated. The availability of a CFTR BAC clone that includes the majority of the critical regions (13) greatly facilitated this analysis.
CFTR shows tight tissue-specific regulation of expression, which is largely determined by cis-regulatory elements that lie outside the gene promoter (14). We recently showed that the active locus exists in a looped conformation that brings distal regulatory elements, located within cell-type–specific DNase I hypersensitive sites (DHS) into close proximity with the gene promoter (15). Moreover, we showed that the regulatory mechanisms in airway epithelial cells, which generally express low levels of CFTR in comparison with intestinal epithelial cells, achieve this through different cis-acting elements (15,16). However, in either cell type, the identification of the critical trans-acting factors that activate these elements remains incomplete. Several methods have recently been developed to accurately map transcription factor binding sites including direct methods, such as high-resolution in vivo footprinting by DNase-seq, (3,4) and indirect methods provided by mapping nucleosomes in vivo (17). Here, we use a novel method (Yigit et al., 2012, in review) to search for cell-type–specific depletion of individual nucleosomes at the cores of regions of open chromatin that correspond to well-characterized regulatory elements associated with DHS in the CFTR gene. We find specific loss of nucleosomes that correspond to well-characterized binding sites for known trans-acting factors. Next, we identify predicted binding sites for factors that have not previously been implicated in CFTR regulation, which interact with nucleosome depleted regions in vivo and demonstrate occupancy of these factors in relevant cell types by ChIP. Finally, we use a bioinformatic approach to identify transcription factor binding sites that are overrepresented in regions where a nucleosome is lost in a cell-specific manner in a bronchial epithelial cell line (16HBE14o-) in comparison with skin fibroblasts. This approach predicts sites for several factors including the glucocorticoid receptor (GR), and we show by ChIP that dexamethasone-induced occupancy of multiple GR elements (GREs) causes repression of CFTR transcription.
MATERIALS AND METHODS
Cell culture
The human colon carcinoma cell lines Caco2 (18) and 16HBE14o- bronchial epithelial cells (19), were grown in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% FBS. Primary skin fibroblasts (GM08333) were grown in MEM (Invitrogen) supplemented with 15% FBS. NHBE cells, a mixture of primary human bronchial and tracheal epithelial cells (Lonza, CC-2541), were cultured in BEGM (Lonza) as per the manufacturer’s instructions. Two independent biological samples were analysed, NHBE1 (7F3506) and NHBE2 (8F3407). Primary human fetal epididymis cells were cultured in CMRL1066 medium with 10% FCS (20).
For glucocorticoid receptor binding experiments, 16HBE14o- cells at 100% confluence were treated with 100 nM dexamethasone (Sigma-Aldrich, St. Louis, MO, USA) in ethanol or the same volume of ethanol alone for 3 h and then harvested for ChIP or RNA extraction. CFTR mRNA was assayed by quantitative reverse transcriptase–polymerase chain reaction (RT–qPCR) as described previously (21).
Isolation of nuclei, MNase digestion and DNA purification
Cells were harvested with trypsin or non-enzymatic dissociation solution (NHBE), washed twice in chilled phosphate-buffered saline at 4°C and pelleted by centrifugation at 200 g for 1 min. Cells were then resuspended in ice-cold NP-40 lysis buffer [10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, 0.5% Nonidet P-40, 0.15 mM spermine, 0.5 mM spermidine supplemented with one complete ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor cocktail/50 ml] for 5 min on ice. Nuclei were pelleted at 19 g for 10 min in a pre-cooled swinging bucket rotor at 4°C. Nuclei were gently re-suspended in micrococcal nuclease (MNase) digestion buffer (10 mM Tris, pH 7.4, 60 mM KCl, 15 mM NaCl, 0.15 mM spermine, 0.5 mM spermidine supplemented with one complete EDTA-free protease inhibitor cocktail/ 50 ml) and pelleted again as described earlier in the text. For micrococcal nuclease digestion, nuclei were resuspended in 1 ml MNase buffer and then adjusted to 4 × 107 nuclei/ml in MNase buffer, on ice. MNase digestion was performed at 37°C in the presence of 1 mM CaCl2 until the 147-bp (core nucleosome) and 167-bp (chromatosome) bands were clearly identifiable. The digestion was stopped with a final concentration of 10 mM EDTA and 1% sodium dodecyl sulphate (SDS). After the digestion, the nucleosomes were treated with RNase A at 37°C for 30 min, and proteins were digested by proteinase K treatment. DNA was extracted with phenol/chloroform, precipitated with ethanol and resuspended in 1 × TE buffer.
Gel extraction and purification of mononucleosome DNA
Digested chromatin DNA was separated on 3% agarose gel (NusieveGTG, Lonza) in 1 × TBE buffer until 147- and 167-bp DNA bands were clearly identifiable. The 147-bp DNA band was carefully excised from the agarose gel, and it was eluted in crush and soak buffer (300 mM sodium acetate, 1 mM EDTA) with gentle overnight agitation. The excess agarose was removed by centrifugation, and supernatant containing nucleosome DNA was cleaned up by QIAquick PCR Purification Kit (Qiagen).
Preparation of SOLiD library
About 6–7 µg of DNA was end repaired and ligated to the forward (P1) and reverse (P2) adaptors according to the SOLiD System library preparation protocol using reagents from New England Biolabs (NEB). P1 adaptors had unique 6-bp barcodes at their 3′-ends to distinguish nucleosomes from each of the six cell types analysed. Ligation products were separated by 5% native polyacrylamide gel, and they were extracted by the crush and soak method described earlier in the text. DNA was further purified and concentrated by QIAquick PCR purification kit and then nick translated by DNA polymerase I (NEB) as described in the SOLiD System library preparation protocol. The reaction was stopped with buffer PB and purified by QIAquick PCR purification kit.
BAC DNA isolation and enrichment of targets by BAC hybridization
BAC123s has a 250.3-kb insert containing the whole-coding region of the CFTR gene, also 40.1 kb of DNA 5′ and 25 kb 3′ to the gene (13). BAC DNA was isolated using the PureLink HiPure Midi Kit (Life Technologies) according to the manufacturer’s protocol and verified by digestion with NotI and NruI followed by pulsed-field gel electrophoresis (13). Enrichment of SOLiD nucleosome libraries (2 µg) was done essentially as described previously (22). In summary, ∼150 ng of BAC DNA was labelled with biotin-dUTP (Enzo Life Sciences) and [α-32P] dCTP using a nick translation kit (Roche), according to the manufacturer’s protocol at 15°C for 1 h. [α-32P] was used to indirectly determine the incorporation of biotin-dUTP. The biotinylated products were purified through P-30 columns (Bio-Rad) and lyophilized. Labelled BAC was dissolved in Cot-1 DNA (2 μg/μl) (Invitrogen). The solution was overlaid with mineral oil, denatured by incubation at 95°C for 5 min and incubated at 65°C for 15 min. Five microlitres of 2 × hybridization buffer (22) was added and incubated at 65°C for 6 h.
To capture nucleosome DNA, 1–2 μg of adapter-ligated genomic DNA in 5 μl of water was overlaid with mineral oil, 5 μl of 2 × hybridization buffer was added and then the entire sample was transferred to the tube containing the Cot-1 blocked BAC DNA. The hybridization reaction was incubated at 65°C for a further 72–80 h. In all, 150 μl of streptavidin-coated beads (Invitrogen) were washed twice with 200 μl streptavidin bead-binding buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, pH 8, and 1 M NaCl) and then resuspended in 150 μl of the same buffer. Next, the whole hybridization reaction was added to the streptavidin bead suspension and bound at room temperature for 30 min on a rotator. Streptavidin beads were removed from the binding buffer and washed once, at 25°C for 15 min in 1 ml of 1 × SSC with 0.1% SDS, and then three times, each at 65°C for 15 min in 1 ml 0.1 × SSC with 0.1% SDS. The nucleosome DNA hybridized to the labelled BAC DNA was eluted by addition of 100 μl of 0.1 M NaOH at 25°C for 10 min. Eluted DNA solution was neutralized by addition of 100 μl of 1 M Tris–HCl (pH 7.5) and desalted by using a P-30 column.
After enrichment, DNA was amplified by PCR (18 cycles) using P1 and P2 primers, and PCR products were gel extracted. Enriched paired-end libraries were combined in equal molar amount and sequenced on SOLiD System 4 (ABI, Carlsbad, CA, USA) instrument by paired-end chemistry (35/25 bp) using a quarter of the slide.
Sequence alignments
After sequencing, the colourspace reads were grouped by barcode and aligned to the human reference genome (GRCh37/hg19) using BioScope version 1.3.1 from ABI (Carlsbad, CA, USA). After alignment, the BAM files were processed, and a text file was generated, which contains, for each read, the genomic coordinates, strand, sequence, alignment length and the number of locations the read has aligned to in the genome. Only the uniquely mapped reads were kept for the subsequent analyses. To construct the nucleosome occupancy score, we further filtered out the reads of length between 137 and 157 bp. Let Rj be the number of sequencing reads centred at genomic location j. We defined the centre weighted nucleosome occupancy score at location i as follows:
| (1) |
where G is a Gaussian density with standard deviation 20. This Gaussian weight function covers ±60 bp of the projected nucleosome centre and decays towards the two ends. Compared with uniform weight function, the Gaussian function is effective in avoiding spikes in the reads occupancy curve frequently observed in the linker region because of overlap of reads arising from two neighbouring nucleosomes, and thus provides a sharper boundary of closely positioned adjacent nucleosomes.
Bioinformatic identification of overrepresented transcription factor binding sites in nucleosome depleted regions
The paired-end SOLiD reads that mapped to the CFTR locus on Genome Reference Consortium Human Build 37 (GRCh37) were compared across different cell types by using the edgeR Bioconductor package (23). The edgeR package was initially designed for examining differential expression of replicated count data, but it is equally applicable to analysis of high-throughput sequencing of nucleosome-depleted regions. First, discrete genomic intervals were defined over which to compare the number of overlapping sequence reads. These (∼25 k) intervals consisted of a 100-bp window starting at the beginning of the CFTR locus and sliding forwards 10 bp at a time until reaching the end of the locus (chr7: 117 082 000–117 332 130). Next, the number of sequencing reads that overlapped (at the midpoint of the sequence) with each of these intervals, in each cell type, was computed with a perl program. Finally, edgeR was run on the resultant sequence counts for each of these intervals, comparing each cell type with all other cell types to determine which intervals were consistently nucleosome depleted, or were depleted in one cell type (e.g. 16HBE14o-) compared with another cell type (e.g. skin fibroblasts). The edgeR program output all intervals that were represented at a statistically different level between cell types, and different P-value cut-offs were then used to select the most significant differences between cell types. Next, these regions were analysed with the Clover program for identification of functional sequence motifs in DNA sequences (24). The CFTR locus was used as the background sequence for these analyses.
Chromatin immunoprecipitation
ChIP experiments were carried out by standard protocols. The following antibodies were used: GR (SantaCruz sc1003); FOXA1 (Abcam, ab5089) and BAF155 (sc10756). Rabbit and goat IgG were from Santa Cruz (sc-2027 and sc-2028). All primers used for qPCR are listed in Supplementary Table S1.
RESULTS
Statistics of SOLiD DNA sequence analysis
Data summarizing the sequence analysis for the six cell types (including two biological replicas of the primary NHBE cells) are shown in Table 1. Total unique sequence tags mapping to the hg19 human genome build ranged from 1 412 498 for primary epididymis cells to 11 411 833 for 16HBE14o- cells, with the range likely reflecting absolute amounts of barcoded, purified mononucleosomal DNA in the complex mixture that was sequenced. The unique tags within the CFTR BAC ranged from 1 040 691 to 6 936 596, respectively, in the same samples, demonstrating percentage enrichment efficiencies of 74–61%. The lowest enrichment efficiency was 34% in NHBE2, whereas fibroblasts, NHBE1 and Caco2 all showed enrichments of between 52 and 56%. Sequence coverage by nucleosome was calculated according to the formula (number of mapped reads) × (147)/(BAC size) (7) and ranged from 612 to 4079, thus confirming adequate coverage of nucleosome positions across the BAC for each cell type.
Table 1.
Statistics of SOLiD paired-end DNA sequence analysis
| Cell type/barcode | Fibroblast | NHBE1 | NHBE2 | 16HBE14o- | Caco2 | Epididymis |
|---|---|---|---|---|---|---|
| AGCTTA | GTCATC | GCATGT | AAGTAA | GTGCCT | TAAAGT | |
| Unique tags within CFTR BAC | 5 635 722 | 4 695 178 | 1 054 913 | 6 936 596 | 3 121 914 | 1 040 691 |
| Unique tags at NON-target region | 4 491 093 | 4 282 556 | 2 051 985 | 4 475 238 | 2 832 115 | 371 808 |
| Unique tags in hg19 | 10 126 815 | 8 977 734 | 3 106 898 | 11 411 833 | 5 954 029 | 1 412 498 |
| Non-unique tags in hg19 | 639 090 | 262 297 | 196 299 | 823 494 | 361 968 | 76 100 |
| Total number of tags in hg19 | 10 765 905 | 9 502 327 | 3 303 197 | 12 235 327 | 6 315 997 | 1 488 598 |
| Enrichment efficiency (%) | 56 | 52 | 34 | 61 | 52 | 74 |
| Unique pairs in hg19 (%) | 94 | 94 | 94 | 93 | 94 | 95 |
| Coverage per nucleosome | 3314 | 2761 | 620 | 4079 | 1836 | 612 |
Comparative analysis of nucleosome positions using different methodologies
We previously developed a method of detecting nucleosome positioning by tiled qPCR of purified mononucleosomal DNA (17). This method was applied to analysis of the CFTR promoter and the intron 11 (1811 + 0.1 kb) DHS region (where 1811 is the last coding base in exon 11), which contains a potent enhancer of CFTR promoter activity (15). It was of interest to compare the nucleosome positioning data generated by this method and the BAC hybridization method described here. There was generally good overlap between nucleosome mapping generated by the two methods across the 2-kb region analysed (Supplementary Figure S1), with the exception of the 500-bp region immediately 5′ to the transcription start site. Where we previously noted three well-positioned nucleosomes using the qPCR method, there is an apparent depletion of nucleosomes in the BAC hybridization-generated data. Nucleosome positioning data from the ENCODE consortium (Stanford/BYU) show a comparable depletion of signal in this area, which is likely because of the >70% GC content of this region. The coincidence of nucleosome positioning between the two methods was high in the core of the intron 11 DHS enhancer sequence (Supplementary Figure S2).
Specific loss of nucleosomes at well-characterized binding sites for known trans-acting factors
Enhancer elements
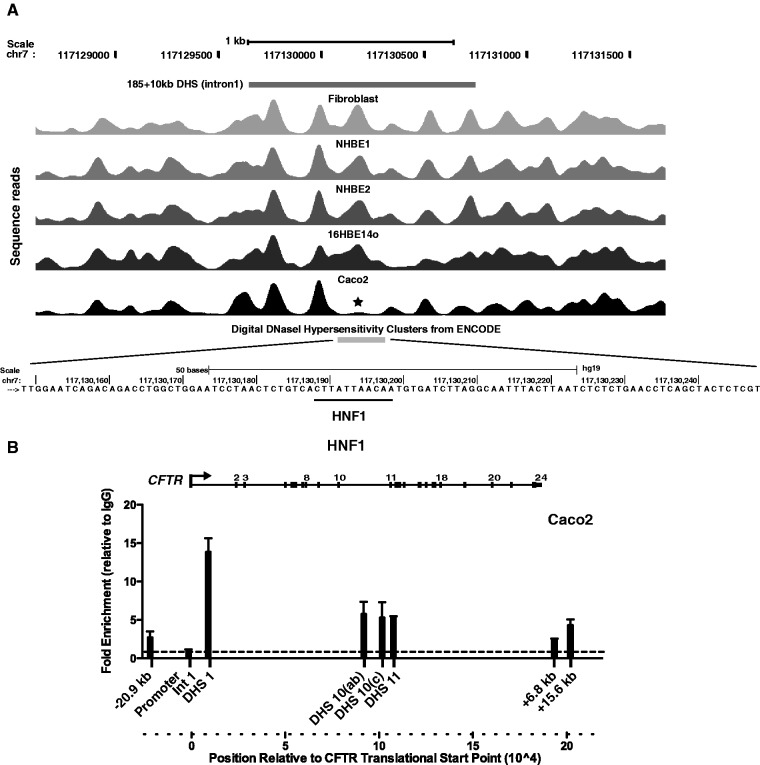
We previously identified several enhancer elements associated with DHS within introns of the CFTR gene that interact with the promoter in a cell-type–specific manner. In some cases, we also characterized critical transcription factors that bind to the elements and contribute to enhancer function. A weak intestinal-specific enhancer in the first intron of the gene at 185 + 10 kb (185 is the last coding base in exon 1) (25) was shown to bind hepatocyte nuclear factor 1 (HNF1) both in vitro and in vivo (15,26). Inspection of the nucleosome positioning data for this region reveals specific loss/repositioning of nucleosomes within the core region of this enhancer element in Caco2 intestinal carcinoma cells (Figure 1A). We next analysed the precise region (chr7: 117 130 188–117 130 198) of the conserved HNF1 binding site that we showed to be critical for the activity of the enhancer (26) and found it to coincide with the nucleosome depleted region. Enrichment of HNF1 binding at this element in Caco2 cells was confirmed by ChIP (Figure 1B). This region was also enriched for the enhancer signature protein p300 in ChIP experiments (15).
Figure 1.
Nucleosome positions across CFTR intron 1 (185 + 10 kb) DHS identify the location of a critical HNF1 binding site. (A) Sequence reads showing nucleosome positions across CFTR intron 1 (185 + 10 kb) DHS (25) in fibroblasts, NHBE, 16HBE14o- and Caco2 cells. The location of the DHS is marked above the reads, and digital DNase hypersensitivity data from ENCODE confirm its location below the reads. Asterisk denotes depletion of a nucleosome at the DHS core that corresponds to the HNF1 binding site in Caco2 cells. Below the reads, single-base resolution of the region marked by asterisk in (A) shows the HNF1 binding site (26). (B) Enrichment of HNF1 assayed by ChIP is greatest at the intron 1 DHS, although it is also seen at other elements across the CFTR locus in Caco2 cells.
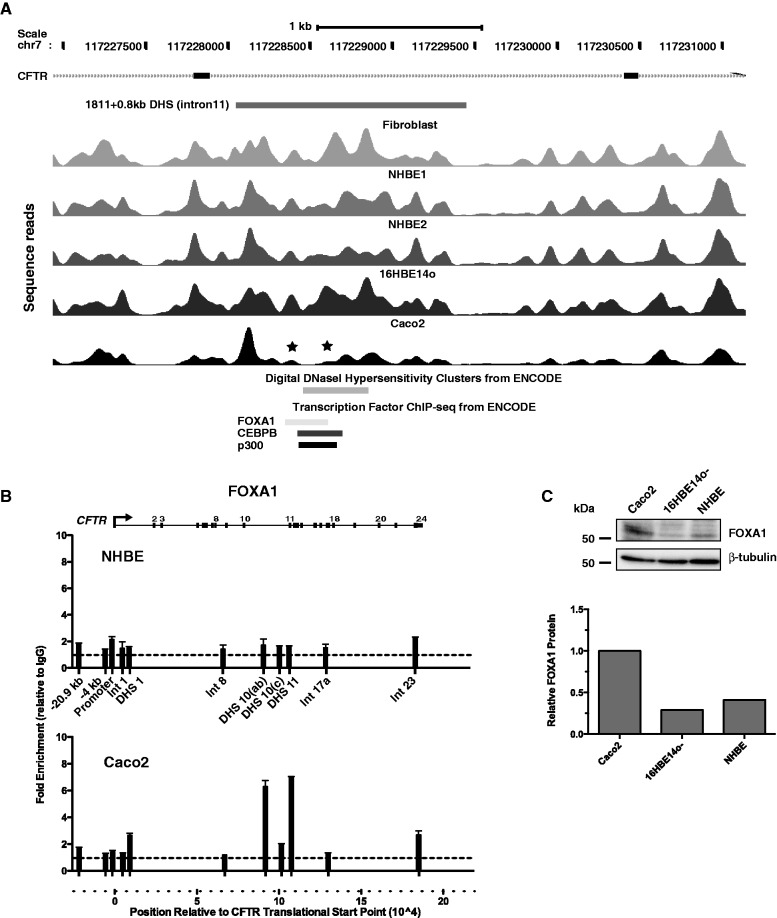
A stronger cell-type–selective enhancer exists in intron 11 of the CFTR gene at 1811 + 0.8 kb, which is also enriched for p300 and was shown by chromosome conformation capture (3 C) to interact directly with the CFTR promoter (15,27). ChIP-seq data from the ENCODE consortium identify binding sites for a number of transcription factors, including FOXA1 and C/EBPβ in this core of this enhancer element (Figure 2A). This is consistent with the depletion/relocation of at least two nucleosomes in this region in Caco2 cells in which the intron 11 element is highly active (15). In contrast, well-positioned nucleosomes are seen at this site in skin fibroblast and primary airway (NHBE) cells in which the element is inactive. Moreover, using ChIP with an antibody specific for FOXA1, we show no enrichment of this factor in the DHS11 region (Figure 2B), despite the expression of FOXA1 protein in these cells (Figure 2C). In contrast, there is strong enrichment of FOXA1 at the DHS 11 enhancer in Caco2 cells (Figure 2B), consistent with our recent data showing binding of both FOXA1 and FOXA2 (which recognize the same DNA sequence motif) to this region (28).
Figure 2.
Nucleosome positions across CFTR intron 11 (1811 + 0.8 kb) DHS show the FOXA binding site that contributes to enhancer activity of this element. (A) Sequence reads showing nucleosome positions across CFTR intron 11 (1811 + 0.8 kb) DHS (15) in fibroblasts, NHBE, 16HBE14o- and Caco2 cells. The location of the DHS is marked above the reads and digital DNase hypersensitivity data and transcription factor ChIP-seq data from ENCODE are shown below the reads. Asterisk denotes depletion of nucleosomes at the DHS core that corresponds to the FOXA binding site in Caco2 cells. (B) ChIP with an antibody specific for FOXA1 shows no enrichment in NHBE cells where the DHS 11 enhancer is inactive, despite the presence of low levels of FOXA1 protein and strong enrichment of DHS 11 and other cis-acting regulatory elements in CFTR in Caco2 cells. (C) Relative expression of FOXA1 protein in Caco2, 16HBE14o- and NHBE cells. Western blot of whole-cell lysates probed with the same antibody used in ChIP experiments or anti β-tubulin and ImageJ quantitation of a scan of the same blot, relative to the β-tubulin signal.
Insulator elements
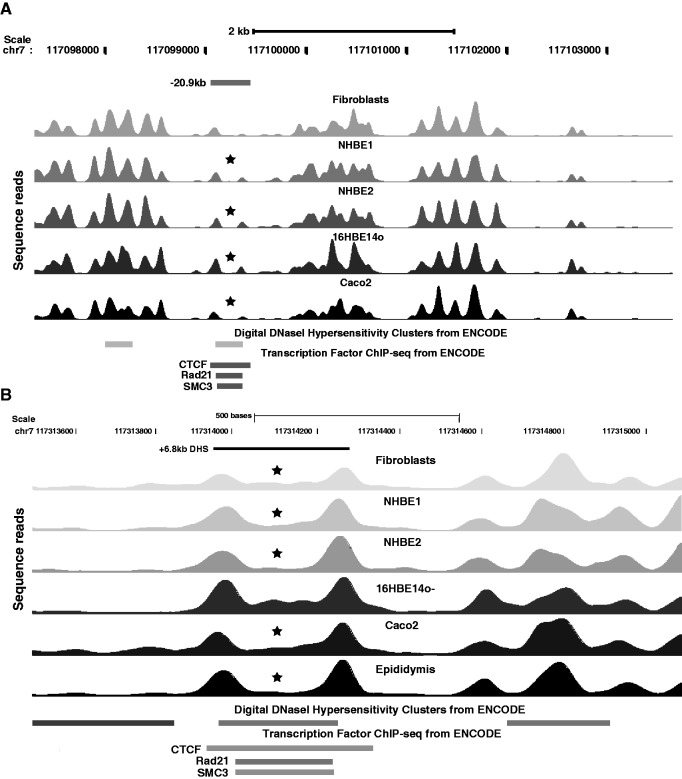
Enhancer-blocking insulator elements are important for establishing chromatin domains and transcriptional hubs genome wide and are often located at the functional boundaries of individual loci. CCCTC binding factor (CTCF) is frequently associated with these insulator elements where it interacts with cohesin complex components (29,30). We previously described CTCF-binding, enhancer-blocking insulator elements associated with DHS at −20.9 kb with respect to the CFTR translational start site and at a DHS at +6.8-kb downstream from the last coding base (31–33). The −20.9-kb DHS is seen in most epithelial cell types examined and also in some other cell types. In contrast, the +6.8-kb DHS is only evident in human lung tissue and epididymis epithelial cells, although low levels of CTCF binding are also seen in Caco2 cells. These observations are markedly consistent with the nucleosome positioning data at these sites (Figure 3A and B). The CTCF binding site at the core of the −20.9-kb DHS is flanked by two well-positioned nucleosomes, with the 5′-peak being more evident in all cases than the 3′-peak (Figure 3A). Moreover, these flanking nucleosomes seem to have been displaced to increase the nucleosome-free region coinciding with the CTCF binding site. The usual periodicity of ∼150 bp between nucleosome peaks is increased to 270 bp for nucleosomes flanking the CTCF binding site. At the +6.8-kb insulator element, the nucleosomes flanking the CTCF binding site are also well positioned, likely with a loss/repositioning of at least one nucleosome, as the distance between the peaks is ∼300 bp (Figure 3B). Also of note is the difference in profile of the sequence peaks between the flanking nucleosomes in different cell types. ChIP data for CTCF binding in the DHS +6.8-kb region showed 15-fold enrichment in primary epididymis cells in which the DHS is evident in comparison with 6-fold in Caco2 cells that lack the DHS (33). Consistent with this observation, complete loss of nucleosomes between the flanking peaks is seen in epididymal cells, whereas there is low occupancy of this region in Caco2 cells (Figure 3B). The +6.8-kb DHS is also evident in adult human lung tissue (34); therefore, it is of interest that one culture of primary human airway epithelial cells (NHBE2) shows a profile similar to the primary epididymis, whereas the other (NHBE1) is more similar to Caco2.
Figure 3.
The −20.9 and +6.8 kb DHS in CFTR that encompass enhancer-blocking insulators show nucleosome positioning by CTCF binding. Sequence reads showing nucleosome positions across (A) CFTR −20.9-kb DHS (15) and (B) +6.8-kb DHS in fibroblasts, NHBE, 16HBE14o-, Caco2 and primary epididymis cells. The location of the DHS is marked above the reads and digital DNase hypersensitivity data, and transcription factor ChIP-seq data from ENCODE are shown below the reads. Asterisk denotes nucleosome depletion at the DHS core that corresponds to the CTCF site.
Other elements
We examined nucleosome positioning across multiple other cell-type–specific DHS in the CFTR locus. One such element is associated with a DHS at 21.5 kb with respect to last coding base of the gene and is only evident in primary airway epithelial cells (15,16). Again, specific depletion of nucleosomes is seen in these cell types; however, in this case, at least three nucleosomes are affected, two lie adjacent to each other, whereas the third is separated from them by two well-positioned nucleosomes (Supplementary Figure S3). The underlying trans-acting factors are currently being investigated.
Identification of predicted binding sites for novel factors that bind in nucleosome depleted regions in vivo
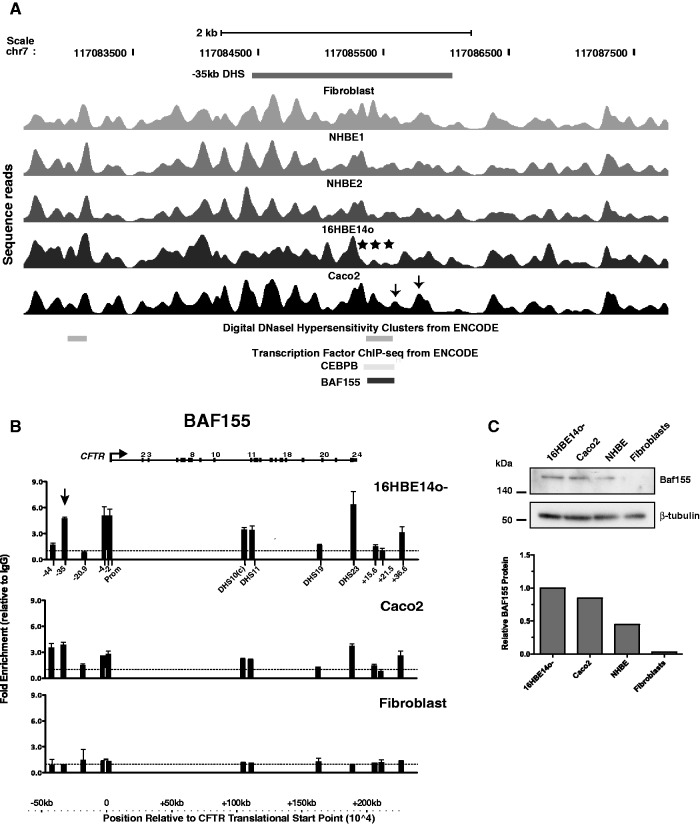
We also evaluated the nucleosome positioning at a cell-type–specific DHS at −35 kb with respect to the CFTR translational start site that is particularly evident in 16HBE14o- cells and primary epididymis cells and is associated with enhancer activity in airway epithelial cells (16). To date, we have not identified the critical transcription factors that interact with this region. However, the availability of nucleosome positioning data across this region identified BAF155 (a component of the SWI/SNF-related, matrix associated, actin-dependent regulator of chromatin structure, SMARC) as a strong candidate for interaction with this element. Selective depletion of at least three nucleosomes is seen in 16HBE14o- cells at the core of the −35-kb DHS (Figure 4A), a region that corresponds to ChIP-seq peaks for C/EBPβ (weak enrichment) and BAF155 in ENCODE data. We were unable to confirm C/EBPβ binding to this site by ChIP in 16HBE14o- cells, although this transcription factor is expressed at a high level in these cells (microarray expression data not shown). However, the predicted BAF155 occupancy at the −35-kb DHS was confirmed by ChIP in 16HBE14o-cells (arrowed in Figure 4B). BAF155 protein is expressed in 16HBE14o- and Caco2 cells but not in fibroblasts (Figure 4C). Consistent with these data, a low level of BAF155 enrichment is seen at multiple DHS across the locus by ChIP in Caco2 cells but not in fibroblasts (Figure 4B). Moreover, inspection of the nucleosome positioning profile at the core of the −35-kb DHS in Caco2 cells reveals some displacement of nucleosomes (marked by arrows in Figure 4A) that may correspond to the activity of BAF155 at this site.
Figure 4.
Nucleosome depletion at the −35-kb DHS in 16HBE14o- cells identifies a role for BAF155 in CFTR regulation in airway epithelial cells. (A) Sequence reads showing nucleosome positions across CFTR −35-kb DHS (15,16) in fibroblasts, NHBE, 16HBE14o- and Caco2 cells. The location of the DHS is marked above the reads, and digital DNase hypersensitivity data and transcription factor ChIP-seq from ENCODE are shown below the reads. Asterisk denotes depletion of several nucleosomes at the DHS core that correspond to the BAF155 binding site in 16HBE14o- cells. (B) ChIP with an antibody specific for BAF155 shows enrichment of −35 kb DHS (arrow) and other cis-acting regulatory elements in CFTR in 16HBE14o- and Caco2 cells but not in fibroblasts. (C) Relative expression of BAF155 protein in Caco2, 16HBE14o-, NHBE cells and fibroblasts. Western blot of whole-cell lysates probed with the same antibody used in ChIP experiments or anti β-tubulin and ImageJ quantitation of a scan of the same blot, relative to the β-tubulin signal.
Identification of transcription factor binding sites that are overrepresented in regions where a specific nucleosome is lost in a cell-specific manner
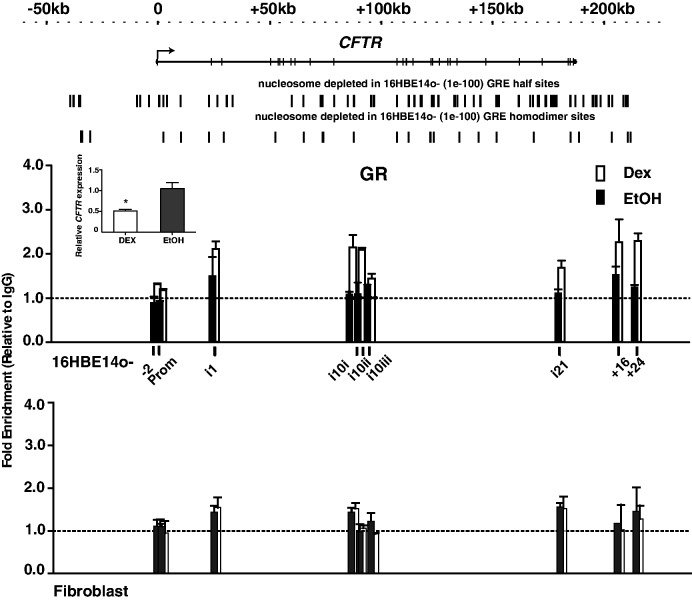
Next, we investigated whether we could identify transcription factors de novo that regulated CFTR expression by binding to nucleosome-depleted regions, either by themselves displacing nucleosome(s) or by occupying regions that were made accessible by the interaction of other factors. To achieve this, nucleosome occupancy across the CFTR locus in skin fibroblasts (where the gene is inactive and shows consistent nucleosome positioning) was compared with that of 16HBE14o- cells (where the gene is highly expressed) to identify nucleosome-depleted regions that were restricted to the latter cell type. The sequence of these regions was then evaluated using the CLOVER algorithm to look for overrepresented transcription factor binding sites (Supplementary Table S2), using the whole CFTR BAC as a background sequence for comparison. This analysis revealed half-sites for multiple nuclear hormone receptors including the glucocorticoid receptor (GR) and for hepatocyte nuclear factor 4 (HNF4). To determine whether these sites were of functional relevance in CFTR expression, we treated 16HBE14o- cells with dexamethasone (100 nM) for 3 h and then performed ChIP with an antibody specific for GR. As the GR can bind to chromatin as a homodimer or in combination with other hormone receptors, changes in GR occupancy at predicted nucleosome-free GRE homodimer and GRE half binding sites in CFTR were then analysed (Figure 5, GRE half-sites and GRE homodimers sites in nucleosome-depleted regions are shown above the ChIP graph, and the motifs used for these predictions are shown in Supplementary Table S3). Multiple sites showed enrichment of GR after dexamethasone induction, including in intron 1 chr7: 117 143 050–117 143 219, intron 10 at chr7: 117 207 782–117 208 258 and chr7: 117 207 992–117 208 126, intron 21 at chr7: 117 297 508–117 297 802 and 3′ to the gene at +16 kb (at chr7: 117 323 222–117 323 391) and +24 kb (at chr7: 117 331 607–117 331 776) with respect to the end of the coding sequence (Figure 5). In contrast, no changes in GR occupancy were seen at any site after dexamethasone induction of skin fibroblasts. Moreover, using a RT–qPCR assay for CFTR mRNA, dexamethasone was shown to decrease CFTR expression under the same experimental conditions (Figure 5 inset panel), suggesting the observed GR occupancy changes were functionally relevant.
Figure 5.
Dexamethasone enhances occupancy of GR binding sites in 16HBE14o- selective nucleosome-free regions and suppresses CFTR expression. ChIP with an antibody specific for GR shows dexamethasone-activated occupancy of GR binding sites in 16HBE14o- cells at CFTR introns 1, 10 and 21 and 3′ to the gene at +16 and + 24 kb. Fibroblasts show no change in GR occupancy. Black bars: EtOH, open bars: 100 nM dexamethasone in EtOH. Clover predictions for GRE half sites and homodimer sites (Supplementary Table S3) in 16HBE14o- selective nucleosome-depleted regions are shown above the graph. Inset: Dexamethasone significantly reduces CFTR mRNA as measured by qRT–PCR. *P < 0.01, t-test.
DISCUSSION
The dynamic competition between nucleosome positioning on the chromatin fibre and the ability of transcription factors to bind their recognition sequences and modulate gene expression patterns is complex (6,7). Although some proteins, such as pioneer factors, have the capability to interact with and disrupt compacted heterochromatin by recruiting chromatin-remodelling complexes (35), others can only bind to open chromatin. Moreover, all transcription factors must overcome the energetically favourable nucleosome core particle by destabilizing its interaction with the DNA strand (8,9). Despite the power of multiple search engines to predict transcription factor binding sites in silico based on DNA sequence alone, these bioinformatic methods have their limitations. Hence, we examined the potential of using experimentally determined nucleosome positioning information across a 250-kb locus to identify cell-type–specific transcription factor binding sites. Applying a novel method to map nucleosomes in primary human epithelial cells and epithelial cell lines, we generated precise nucleosome occupancy data across the CFTR locus. Purified mononucleosomal DNA from each cell type was hybridized to a 250-kb BAC encompassing the locus, purified, amplified and sequenced. Sequences were aligned with hg19, and the nucleosome positions were then compared between the diverse cell types, enabling specific differences in nucleosome occupancy to be correlated with transcription factor interactions. Moreover, bioinformatics approaches to reveal TF binding motifs that were overrepresented in nucleosome-depleted regions identified the involvement of the GR in CFTR regulation, a mechanism that was previously unknown.
The application of this method for nucleosome positioning for defined regions of the human genome has some advantages over the ENCODE genome-wide data that are available on the UCSC genome browser (http://genome.ucsc.edu). Specifically the ability to map nucleosomes in chromatin derived from primary cell types for which limited material is available and the ability to generate greater sequence coverage per nucleosome. Moreover, by combining multiple BACs, it should be possible to generate a high-resolution map of nucleosome positioning over megabase regions of genomic DNA in addition to the 250–300 kb regions cloned in single BACs. Two potential technical limitations of the method should be noted. First, although the use of MNase I to map nucleosome positions is generally well accepted, there is some evidence (36) that MNase I has a sequence-dependent bias. In the context of our experiments, this is not significant, as we are comparing nucleosome occupancy/depletion in different cell types which all have essentially the same genomic sequence. Moreover, recent data showed that chemical mapping produces nucleosome-positioning data that are similar to those derived from MNase I (37). Second, the deep sequencing approach to nucleosome positioning may reveal apparently depleted regions that are artifacts of high GC content. An example is the presence of a gap of ∼500 bp immediately 5′ to the CFTR transcription start site when compared with our previous PCR tiling approach to map nucleosomes. However, this gap is also present in the nucleosome positioning data on the ENCODE browser and corresponds to a region of high GC content, which are difficult to sequence. The high GC content could also potentially influence the kinetics of BAC hybridization.
The identification of novel factors involved in regulation of CFTR expression by analysing cell-type–selective nucleosome-free regions warrants further discussion. First, we show enrichment of BAF155 in vivo at the enhancer element within the −35 kb DHS. BAF155 is a component of the SWI/SNF chromatin-remodelling complex, and it is likely recruited to this element by other transcription factors. Our ChIP data demonstrate enrichment of BAF155 at several DHS across the CFTR locus in 16HBE14o- cells and differential relative occupancy in Caco2 cells, as would be predicted from the divergent regulatory mechanisms in these cell types (16). Moreover, no BAF155 enrichment is seen at the inactive CFTR locus in fibroblasts.
Next, identification of the GR as a suppressor of CFTR expression in 16HBE14o- cells, based on the data showing overrepresentation of its binding motif in nucleosome-depleted regions in this cell type, is novel. Hormonal regulation of CFTR expression has not been studied extensively, although there is some evidence for its upregulation by oestrogen (38). With the exception of an abstract reporting repression of CFTR expression by dexamethasone in rat alveolar epithelial cells (39), the effects of dexamethasone on CFTR gene expression have not been investigated. However, downregulation of gene expression by GR is not uncommon [reviewed in (40)]. Data generated by the Myers Laboratory at the Hudson Alpha Institute for Biotechnology and available on the ENCODE browser show GR enrichment by ChIP after dexamethasone stimulation of another airway cell line A549 (41). This cell line does not express CFTR, and it is of interest that although two of the A549 ChIP-seq peaks for GR (in introns 10 and 21) coincide with sites that were also occupied in 16HBE14o- cells after activation by ligand, only one (in intron 21) corresponds to a nucleosome-depleted heterodimer binding site for the receptor. The possibility that ligand activation of the GR downregulates CFTR expression more generally is of potential therapeutic relevance. For example, blocking ligand:receptor interactions in this well-studied pathway could increase CFTR expression in patients with mutations causing reduced transcript levels.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–3.
FUNDING
National Institutes of Health [R01HL094585, R01HD068901 to A.H; R01GM058617, U54CA143869 to J.W.]; Cystic Fibrosis Foundation (to A.H.); Fellowship from the Cancer Research Institute (to E.Y.). Funding for open access charge: NIH and Institutional Funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to the Northwestern University Genomics Core for all sequencing and sequence alignment completed for this project and also to A. E. Gillen for BAC purification.
REFERENCES
- 1.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle AP, Song L, Lee BK, London D, Keefe D, Birney E, Iyer VR, Crawford GE, Furey TS. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He HH, Meyer CA, Chen MW, Jordan VC, Brown M, Liu XS. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res. 2012;22:1015–1025. doi: 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy TE, Gertz J, Pauli F, Kucera KS, Varley KE, Newberry KM, Marinov GK, Mortazavi A, Williams BA, Song L, et al. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res. 2012;22:860–869. doi: 10.1101/gr.131201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat. Rev. Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moyle-Heyrman G, Tims HS, Widom J. Structural constraints in collaborative competition of transcription factors against the nucleosome. J. Mol. Biol. 2011;412:634–646. doi: 10.1016/j.jmb.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tims HS, Gurunathan K, Levitus M, Widom J. Dynamics of nucleosome invasion by DNA binding proteins. J. Mol. Biol. 2011;411:430–448. doi: 10.1016/j.jmb.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreu-Vieyra C, Lai J, Berman BP, Frenkel B, Jia L, Jones PA, Coetzee GA. Dynamic nucleosome-depleted regions at androgen receptor enhancers in the absence of ligand in prostate cancer cells. Mol. Cell Biol. 2011;31:4648–4662. doi: 10.1128/MCB.05934-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 12.Gillen AE, Harris A. Transcriptional regulation of CFTR gene expression. Front. Biosci. (Elite Ed.) 2012;4:587–592. doi: 10.2741/401. [DOI] [PubMed] [Google Scholar]
- 13.Kotzamanis G, Abdulrazzak H, Gifford-Garner J, Haussecker P, Cheung W, Harris A, Kotsinas A, Gorgoulis V, Huxley C. CFTR expression from a BAC carrying the complete human gene and associated regulatory elements. J. Cell Mol. Med. 2009;13:2938–2948. doi: 10.1111/j.1582-4934.2008.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ott CJ, Blackledge NP, Leir SH, Harris A. Novel regulatory mechanisms for the CFTR gene. Biochem. Soc. Trans. 2009;37:843–848. doi: 10.1042/BST0370843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl Acad. Sci. USA. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Ott CJ, Lewandowska MA, Leir SH, Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J. Cell Mol. Med. 2012;16:1321–1330. doi: 10.1111/j.1582-4934.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ott CJ, Bischof JM, Unti KM, Gillen AE, Leir SH, Harris A. Nucleosome occupancy reveals regulatory elements of the CFTR promoter. Nucleic Acids Res. 2012;40:625–637. doi: 10.1093/nar/gkr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fogh J, Wright WC, Loveless JD. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J. Natl Cancer. Inst. 1977;58:209–214. doi: 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- 19.Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, Finkbeiner WE, Widdicombe JH, Gruenert DC. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1994;10:38–47. doi: 10.1165/ajrcmb.10.1.7507342. [DOI] [PubMed] [Google Scholar]
- 20.Harris A, Coleman L. Ductal epithelial cells cultured from human foetal epididymis and vas deferens: relevance to sterility in cystic fibrosis. J. Cell Sci. 1989;92 doi: 10.1242/jcs.92.4.687. Pt 4, 687–690. [DOI] [PubMed] [Google Scholar]
- 21.Mouchel N, Henstra SA, McCarthy VA, Williams SH, Phylactides M, Harris A. HNF1alpha is involved in regulation of expression of the CFTR gene. Biochem. J. 2004;378:909–918. doi: 10.1042/BJ20031157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bashiardes S, Veile R, Helms C, Mardis ER, Bowcock AM, Lovett M. Direct genomic selection. Nat. Methods. 2005;2:63–69. doi: 10.1038/nmeth0105-63. [DOI] [PubMed] [Google Scholar]
- 23.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith AN, Barth ML, McDowell TL, Moulin DS, Nuthall HN, Hollingsworth MA, Harris A. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J. Biol. Chem. 1996;271:9947–9954. doi: 10.1074/jbc.271.17.9947. [DOI] [PubMed] [Google Scholar]
- 26.Ott CJ, Suszko M, Blackledge NP, Wright JE, Crawford GE, Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J. Cell Mol. Med. 2009;13:680–692. doi: 10.1111/j.1582-4934.2008.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gheldof N, Smith EM, Tabuchi TM, Koch CM, Dunham I, Stamatoyannopoulos JA, Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–4336. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerschner JL, Harris A. Transcriptional networks driving enhancer function in the CFTR gene. Biochem. J. 2012;446:203–212. doi: 10.1042/BJ20120693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 31.Nuthall H, Vassaux G, Huxley C, Harris A. Analysis of a DNAse I hypersensitve site located −20.9 kb upstream of the CFTR gene. Eur. J. Biochem. 1999;266:431–443. doi: 10.1046/j.1432-1327.1999.00872.x. [DOI] [PubMed] [Google Scholar]
- 32.Blackledge NP, Carter EJ, Evans JR, Lawson V, Rowntree RK, Harris A. CTCF mediates insulator function at the CFTR locus. Biochem. J. 2007;408:267–275. doi: 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blackledge NP, Ott CJ, Gillen AE, Harris A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 2009;37:1086–1094. doi: 10.1093/nar/gkn1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nuthall HN, Moulin DS, Huxley C, Harris A. Analysis of DNase I hypersensitive sites at the 3′ end of the cystic fibrosis transmembrane conductance regulator gene. Biochem. J. 1999;341:601–611. [PMC free article] [PubMed] [Google Scholar]
- 35.Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung HR, Dunkel I, Heise F, Linke C, Krobitsch S, Ehrenhofer-Murray AE, Sperling SR, Vingron M. The effect of micrococcal nuclease digestion on nucleosome positioning data. PLoS One. 2010;5:e15754. doi: 10.1371/journal.pone.0015754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rochwerger L, Dho S, Parker L, Foskett JK, Buchwald M. Estrogen-dependent expression of the cystic fibrosis transmembrane regulator gene in a novel uterine epithelial cell line. J. Cell Sci. 1994;107 doi: 10.1242/jcs.107.9.2439. Pt 9, 2439–2448. [DOI] [PubMed] [Google Scholar]
- 39.Laube M, Thome UH. CFTR is negatively regulated by glucocorticoids in alveolar epithelial cells. Pediatr. Res. 2011;70:522–522. doi: 10.1203/PDR.0b013e3182070ec8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos GM, Fairall L, Schwabe JW. Negative regulation by nuclear receptors: a plethora of mechanisms. Trends Endocrinol. Metab. 2011;22:87–93. doi: 10.1016/j.tem.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, Parks WP. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.