Abstract
CENP-B is a widely conserved centromeric satellite DNA-binding protein, which specifically binds to a 17-bp DNA sequence known as the CENP-B box. CENP-B functions positively in the de novo assembly of centromeric nucleosomes, containing the centromere-specific histone H3 variant, CENP-A. At the same time, CENP-B also prevents undesired assembly of the CENP-A nucleosome through heterochromatin formation on satellite DNA integrated into ectopic sites. Therefore, improper CENP-B binding to chromosomes could be harmful. However, no CENP-B eviction mechanism has yet been reported. In the present study, we found that human Nap1, an acidic histone chaperone, inhibited the non-specific binding of CENP-B to nucleosomes and apparently stimulated CENP-B binding to its cognate CENP-B box DNA in nucleosomes. In human cells, the CENP-B eviction activity of Nap1 was confirmed in model experiments, in which the CENP-B binding to a human artificial chromosome or an ectopic chromosome locus bearing CENP-B boxes was significantly decreased when Nap1 was tethered near the CENP-B box sequence. In contrast, another acidic histone chaperone, sNASP, did not promote CENP-B eviction in vitro and in vivo and did not stimulate specific CENP-B binding to CENP-A nucleosomes in vitro. We therefore propose a novel mechanism of CENP-B regulation by Nap1.
INTRODUCTION
Kinetochores assemble on the centromeric region of each chromosome, where they form sites for microtubule attachment. Kinetochores are highly complex, with >100 proteins (1–7), but centromeric chromatin is slightly simpler, and its main constituents are CENPs A-C (8–11), CENP-S (3), CENP-T (3), CENP-X (12) and CENP-W (5). CENP-T, -W, -S and -X are assembled into a novel nucleosome-like particle (13). CENP-A is a centromere-specific histone H3 variant that is widely conserved among eukaryotes. CENP-A is a key essential component for the formation of functional kinetochores, and significant chromosome missegregation has been observed in CENP-A-depleted cells (14–19).
Structural studies revealed that human CENP-A and its yeast Saccharomyces cerevisiae homolog, Cse4, form nucleosomes containing a histone octamer with two each of histones H2A, H2B, H4 and CENP-A (or Cse4) (20–22). The DNA winds left-handedly around the histone octamer containing CENP-A (or Cse4) (20–24). Unusual centromeric nucleosomes such as hemisomes and hexasomes, which may be cell-cycle dependent intermediates, have also been proposed (25–33). In the CENP-A nucleosome, the DNA at the entry/exit sites is detached from the histone octamer surface, and is therefore more accessible than its counterpart in canonical nucleosomes (21–24,34,35). Indeed, CENP-A nucleosomes are slightly less stable than canonical H3 nucleosomes (36).
CENP-A marks the centromeric chromatin as the site of kinetochore assembly, and CENP-C assembly requires the C-terminal six residues of CENP-A (37). The targeting of CENP-A to its particular assembly sites on the DNA is complex and depends on epigenetic factors, rather than the specific DNA sequence (38,39). This is a topic that is currently being intensively studied in a number of laboratories. In contrast, the binding of CENP-B to α-satellite DNA appears to be more straightforward—the protein binds specifically to a 17-bp DNA sequence known as the CENP-B box (40).
The CENP-B box sequence appears in centromeric satellite repeats in human (α-satellite DNA) and mouse (minor satellite DNA) (10,40) and may be located at the entry/exit regions of the CENP-A nucleosome (21,41–43). CENP-B is required for stable de novo assembly of CENP-A and the functional centromere on transfected α-satellite DNA, during the formation of human artificial chromosomes (HACs) (44). However, CENP-B function is complex. When CENP-B binds to an ectopic chromosomal locus, CENP-A nucleosome assembly is suppressed by the induction of heterochromatin formation around the CENP-B binding site (44). The CENP-B mediated heterochromatin formation is also observed in the fission yeast Schizosaccharomyces pombe (45,46). These findings suggested that CENP-B not only functions positively during de novo centromere formation on the proper chromosome locus but may also suppress centromere formation at non-specific chromosome loci. This in turn implied that non-specific CENP-B binding to chromosomes could potentially be harmful, as it might induce heterochromatin formation on inappropriate chromosome loci. However, the mechanism by which CENP-B is evicted from non-specific binding sites is not known.
Nap1 and sNASP are acidic histone chaperones that promote nucleosome assembly with the core histones H2A, H2B, H3 and H4 in vitro (47–55). Nap1 and sNASP also stimulate linker histone H1 binding to chromatin in vitro (56–58). Thus, Nap1 and sNASP appear to promote proper DNA binding of basic proteins, such as core and linker histones, which have a propensity to interact randomly with the phosphate backbone of DNA.
In the present study, we found that human Nap1 stimulates the formation in vitro of specific complexes between the CENP-B DNA-binding domain (CENP-B DBD) and CENP-A or H3 nucleosomes containing CENP-B box sequences. Nap1 also potently reverses the non-specific binding of CENP-B DBD to CENP-A or H3 nucleosomes lacking the CENP-B box. In model experiments with human cells, CENP-B eviction activity was observed in vivo when Nap1 was tethered near bona fide CENP-B binding sites, either in the centromere of a HAC or at an ectopic chromosome locus. In contrast, another acidic histone chaperone, sNASP, did not affect CENP-B binding to the CENP-A nucleosome in vitro and did not exhibit CENP-B eviction activity in vivo. We suggest that Nap1 may function to eliminate non-specific CENP-B binding to chromosomes and may promote its proper loading onto correct chromosome loci.
MATERIALS AND METHODS
Purification of human histones, CENP-A, Nap1 and sNASP
Three human histones (H2A, H2B and H4) and CENP-A were produced in Escherichia coli cells and purified by the same methods described previously (21). CENP-B DBD (1–129 amino acid residues) was overexpressed in E. coli JM109(DE3) cells, as an N-terminally hexahistidine-tagged protein. The DNA fragment encoding CENP-B DBD was ligated into the NdeI and BamHI sites of the pET15b vector (Novagen). Freshly transformed JM109(DE3) cells were grown on an LB plate containing ampicillin (100 µg/ml) at 37°C. After a 16 h incubation, five colonies were inoculated into LB medium (2 l) containing ampicillin (100 µg/ml). The cells were cultured at 37°C. When the cell density reached an A600 = 0.6, isopropyl-β-d-thiogalactopyranoside (0.5 mM) was added to induce CENP-B DBD expression, and the cells were further cultured at 37°C for 12 h. The CENP-B DBD was extracted from the cells and purified by the same method as that for recombinant histones, including the removal of the hexahistidine tag by thrombin protease treatment (21). Human Nap1 and sNASP were produced as N-terminally hexahistidine-tagged proteins and were purified as described previously (52,55). The hexahistidine tag was removed by protease treatment, and therefore, the Nap1 and sNASP proteins purified by these methods lacked the hexahistidine tag. These samples were dialyzed against 10 mM Tris–HCl buffer (pH 7.5), containing 0.5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM DTT and 10% glycerol.
Reconstitution and purification of CENP-A or H3 nucleosomes
The histone octamer containing histones H2A, H2B, H4 and CENP-A or H3 was reconstituted as described previously (21,59). The 192-bp DNA fragments, with or without the CENP-B box, were amplified by polymerase chain reaction (PCR), as described previously (42). The CENP-A nucleosomes were reconstituted with the histone octamer and the 192-bp DNA fragment by the salt dialysis method, and the reconstituted CENP-A nucleosomes were purified by the method described previously (21).
Assay for nucleosome formation with CENP-B DBD
For the specific complex formation assay, CENP-A or H3 nucleosomes containing the CENP-B box sequence (Cb+) were mixed with the CENP-B DBD in the presence or absence of Nap1 or sNASP in 20 mM Tris–HCl (pH 7.5) buffer, containing 140 mM NaCl and 1 mM DTT. After incubation for 20 min at 37°C, the samples were analyzed by 5% polyacrylamide gel electrophoresis in 0.2 × TBE buffer (18 mM Tris base, 18 mM boric acid and 0.4 mM EDTA) at 16 V/cm for 55 min, and the DNA bands were visualized by ethidium bromide staining.
The inhibition assay for non-specific CENP-B DBD binding to CENP-A or H3 nucleosomes was performed using CENP-A nucleosomes lacking CENP-B boxes (Cb−). These CENP-A or H3 nucleosomes (Cb−) were mixed with the CENP-B DBD in the presence of Nap1 or sNASP in 20 mM Tris–HCl (pH 7.5) buffer, containing 140 mM NaCl and 1 mM DTT. For the control experiment without the histone chaperone, CENP-A or H3 nucleosomes (Cb−) were mixed with the CENP-B DBD in 20 mM Tris–HCl (pH 7.5) buffer, containing 140 mM NaCl and 1 mM DTT. After incubation for 20 min at 37°C, the samples were analyzed by 5% polyacrylamide gel electrophoresis in 0.2 × TBE buffer (18 mM Tris base, 18 mM boric acid and 0.4 mM EDTA) at 16 V/cm for 55 min, and the DNA bands were visualized by ethidium bromide staining.
Assay for Nap1-CENP-B DBD binding
Hexahistidine-tagged Nap1 (His6-Nap1) was prepared by the method described previously (52), without the removal of the hexahistidine tag. The His6-Nap1 (14 µg, 600 nM) was mixed with the CENP-B DBD (1.5 µg, 200 nM) in 500 µl of 20 mM Tris–HCl (pH 7.5) buffer, containing 100 mM NaCl, 30 mM imidazole and 10 µg/ml bovine serum albumin (BSA). Ni-NTA-agarose beads (3 µl, 50% slurry) were added to the reaction mixtures, and the samples were incubated for 60 min at 4°C. After the incubation, the beads were pelleted and washed three times with 500 µl of 20 mM Tris–HCl (pH 7.5) buffer, containing 100 mM NaCl, 50 mM imidazole and 0.2% Tween 20. The proteins bound to the beads were analyzed by sodium dodecyl sulphate (SDS)-15% polyacrylamide gel electrophoresis with Coomassie brilliant blue staining.
Cell lines
The HeLa-HAC-2-4 cell line was established by the transfection-based delivery (60) of an alphoidtetO HAC from the HeLa-HAC-R5 cell line (61) to the common HeLa cell line.
Constuction of tetR-EYFP-fusion protein expressing plasmids
PCR products of Nap1 and sNASP were cloned into the PacI and NotI sites of pJETY3-tetR-EYFP plasmid (61), which expresses tetR-EYFP-alone.
Cell culture and transfection
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (Nacalai Tesque) supplemented with 10% tet-approved FBS (Clontech) at 37°C in a 5% CO2 atmosphere. FuGENE HD (Roche) was used for transfection.
Indirect immunofluorescent staining
Cells grown on coverslips were washed once with phosphate buffered saline (PBS), fixed with 2.6% formaldehyde in PBS for 5 min at RT and then quenched with PBS containing 0.5% Triton X-100 and 125 mM glycine for 5 min at RT. The cells were blocked with PBS containing 2% BSA and 0.1% Triton X-100 for 30 min at RT, incubated with primary antibodies for 60 min, washed twice with PBS containing 0.1% Triton X-100 and incubated with secondary antibodies for 60 min. The cells were stained with Qnuclear™ Deep Red Stain (Invitrogen), at a 1:30 000 dilution in PBS containing 0.1% Triton X-100, for 20 min at RT. After a final set of washes, the cells were mounted with VectaShield for Fluorescence (Vector Labs).
The anti-CENP-B N-ter polyclonal antibody (BN1) was used at 0.8 µg/ml. The anti-CENP-A monoclonal antibody (A1) was used at 1 µg/ml. Secondary antibodies [DyLight 405-conjugated Goat Anti-Mouse IgG (Thermo), Alexa Fluor® 594-conjugated Goat Anti-mouse IgG (Invitrogen) and Alexa Fluor® 594-conjugated Goat Anti-mouse IgG (Invitrogen)] were used at 1 µg/ml. PBS containing 0.2% BSA and 0.1% Triton X-100 was used for antibody dilution.
Microscopy
Z-stack images with a spacing of 0.5 µm were acquired on an LSM700 microscope (Zeiss) equipped with an Objective Plan-Apochromat 40 × /1.3 oil lens (Zeiss). For quantification analysis, Z-stack images covering an entire single nuclear signal were used for maximum intensity projections.
Quantification of CENP-B and CENP-A assembly level
ImageJ (National Institutes of Health) was used for quantification. The maximum intensity of each CENP-B and CENP-A channel was normalized to 255. The amount of the CENP-B or CENP-A signal on alphoidtetO HAC was measured, after the ‘Subtract Background’ function was performed to eliminate non-specific signals.
The total CENP-B (or CENP-A) signals on the alphoidtetO HAC and endogenous centromeres in the same single nucleus were measured, after subtracting the residual noise below the threshold (=20). The CENP-B (or CENP-A) assembly level on the alphoidtetO HAC was standardized by dividing the CENP-B (or CENP-A) signals on the HAC by the total of the CENP-B (or the average of the CENP-A) signals from endogenous centromeres in the same single nucleus.
Fluorescent recovery after photobleaching
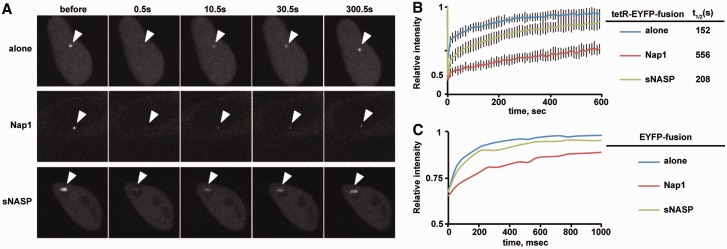
HeLa-Int-03 cells were transfected with a set of tetR-EYFP-fusion expressing plasmids. A focus of the tetR-EYFP-fusion proteins on the tetO array was bleached, and the recovery rate was measured. The Fluorescent recovery after photobleaching (FRAP) assay was performed 1 day after transfection, using a confocal microscope (FV-1000; Olympus) with a PlanSApo 60 × (NA = 1.35) oil-immersion lens. For Figure 6A and B, images (0.5 s/frame) were collected at 10 s intervals, and a 3 µm diameter spot was bleached (100% 488-nm laser transmission; two iterations) after three images. For Figure 6C, 10 images were collected (65 ms/frame) before bleaching, and 90 images were further collected. ImageJ (National Institutes of Health) was used for intensity measurements and fitting analysis. As a control experiment, YFP-H2B expressing cells were also measured by bleaching the same diameter in the nucleus. The YFP-H2B signals did not show a significant recovery within the same time scale (data not shown).
Figure 6.
Turnover of tetR-EYFP-fusion proteins on the tetO site. HeLa-Int-03 cells were transfected with a set of tetR-EYFP-fusion expressing plasmids. A focus of the tetR-EYFP-fusion proteins on the tetO array was bleached, and the recovery rate was measured. (A) FRAP examples. A 3-µm diameter area containing the tetO array was bleached, and the fluorescence intensity in the bleached area was measured. Arrowheads indicate tetO arrays. (B) FRAP results. The relative intensity of the bleached area for the indicated tetR-EYFP-fusion protein was plotted (averages of 10–15 cells with the standard deviations), and each halftime of recovery t1/2, obtained by fitting the curve to single exponential association kinetics, is shown. (C) FRAP results for EYFP-alone, EYFP-NAP1 and EYFP-sNASP. The same diameter in the nucleus was bleached, and the recovery was measured (averages of 15–20 cells). Note that the x-axis unit is ms. The FRAP assay was performed 1 day after transfection, using a confocal microscope (FV-1000; Olympus) with a PlanSApo 60 × (NA = 1.35) oil-immersion lens. For panel (A) and (B), images (0.5 s/frame) were collected at 10 s intervals, and a 3-µm diameter spot was bleached (100% 488-nm laser transmission; two iterations) after three images. For panel (C), 10 images were collected (65 ms/frame) before bleaching, and 90 images were further collected. ImageJ (National Institutes of Health) was used for intensity measurements and fitting analysis.
Chromatin immunoprecipitation assay
Cells were collected 2 days after transfection and fixed with 0.5% formaldehyde (Sigma: F8775) at 22°C for 10 min. The cells were then sonicated with a Bioruptor (Cosmobio) to generate an average DNA size of 0.5–1 kb, in sonication buffer (10 mM HEPES, 1 mM EDTA, 1.5 μM aprotinin, 10 μM leupeptin, 1 mM DTT, 0.05% SDS and 40 μM MG132). The soluble chromatin (input) was recovered by centrifugation and immunoprecipitated with chromatin immunoprecipitation (ChIP) buffer (55 mM HEPES, 150 mM NaCl, 1 mM EGTA, 2 mM MgCl2, 2 mM ATP, 1.5 μM aprotinin, 10 μM leupeptin, 1 mM DTT, 0.01% SDS and 1% NP-40), containing 2 µg of anti-CENP-B C-terminus antibody (5E6C1) and Protein G Sepharose 4 Fast Flow (GE Healthcare). The DNA was purified from the immunoprecipitates and quantified by real-time PCR, using the following primer sets: tetOF (5′-CTCTTTTTGTGGAATCTGCAAGTG) and tetOR (5′-TCTATCACTGATAGGGAGAGCTCT) for alphoidtetO, and 11-10 R and mCbox-4 for the 11-mer of chromosome 21 alphoid DNA (21-I alphoid).
RESULTS
Reconstitution of non-specific CENP-B binding to CENP-A and H3 nucleosomes
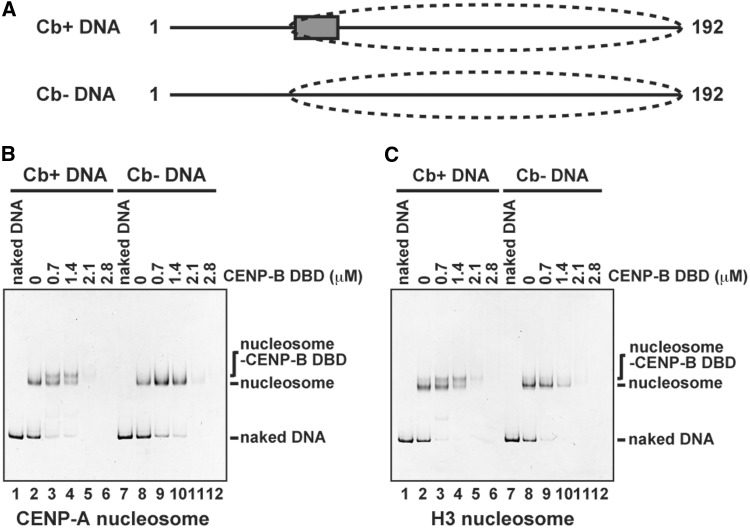
We reconstituted CENP-A and H3 nucleosomes with a 192-bp DNA (Supplementary Figure S1), in which a CENP-B box sequence was located at the entry/exit regions of the nucleosome (Figure 1A). This nucleosome positioning was previously confirmed by micrococcal nuclease mapping with the same DNA fragment (42). We then tested CENP-B binding to the CENP-A and H3 nucleosomes by an electrophoretic mobility shift assay, using polyacrylamide gels. The CENP-B DBD, which contains amino acid residues 1–129 of human CENP-B (Supplementary Figure S1B) (41,62,63), was used in this electrophoretic mobility shift assay. We previously reported that the CENP-B DBD forms specific complexes with these CENP-A and H3 nucleosomes by the salt dialysis method (42). However, when the CENP-B DBD was incubated with the reconstituted CENP-A and H3 nucleosomes under physiological salt conditions, we detected only a small amount of the specific complex with the CENP-B DBD bound to the CENP-B box sequence of these nucleosomes (Figure 1B and C, lanes 3 and 4). Thus, the salt-dialysis method may suppress non-specific CENP-B binding to nucleosomes. Furthermore, when the CENP-B DBD concentration was increased, the bands corresponding to the specific complex and the CENP-A and H3 nucleosomes disappeared (Figure 1B and C, lanes 5 and 6), possibly owing to the formation of aggregates resulting from non-specific binding of the CENP-B DBD to nucleosomes.
Figure 1.
Specific and non-specific CENP-B DBD binding to the CENP-A and H3 nucleosomes. (A) Schematic representations of 192-bp α-satellite DNAs. The box and the dashed circle indicate the location of the CENP-B box and the nucleosome positioning, respectively, as revealed by the previous MNase mapping (42). (B) CENP-B DBD binding to the CENP-A nucleosomes with or without the CENP-B box DNA. The CENP-A nucleosomes (140 nM) with the CENP-B box sequence (Cb+) (lanes 2–6) or without the CENP-B box sequence (Cb−) (lanes 8–12) were incubated with the CENP-B DBD for 20 min at 37°C. CENP-B DBD concentrations were 0 µM (lanes 2 and 8), 0.7 µM (lanes 3 and 9), 1.4 µM (lanes 4 and 10), 2.1 µM (lanes 5 and 11) and 2.8 µM (lanes 6 and 12). The samples were analyzed by non-denaturing 5% polyacrylamide gel electrophoresis, followed by ethidium bromide staining. Lanes 1 and 7 indicate the 192-bp naked DNA. (C) CENP-B DBD binding to the H3 nucleosomes with or without the CENP-B box DNA. The H3 nucleosomes (140 nM) with the CENP-B box sequence (Cb+) (lanes 2–6) or without the CENP-B box sequence (Cb−) (lanes 8–12) were incubated with the CENP-B DBD for 20 min at 37°C. CENP-B DBD concentrations were 0 µM (lanes 2 and 8), 0.7 µM (lanes 3 and 9), 1.4 µM (lanes 4 and 10), 2.1 µM (lanes 5 and 11) and 2.8 µM (lanes 6 and 12). The samples were analyzed by non-denaturing 5% polyacrylamide gel electrophoresis, followed by ethidium bromide staining. Lanes 1 and 7 indicate the 192-bp naked DNA.
To test the non-specific binding of the CENP-B DBD to nucleosomes, we reconstituted the CENP-A and H3 nucleosomes with DNA lacking the CENP-B box sequence (Supplementary Figure S1). As shown in Figure 1B and C (lanes 7–12), the CENP-B DBD did not form specific complexes with either the CENP-A or H3 nucleosomes in the absence of the CENP-B box sequence. The bands corresponding to the nucleosomes disappeared in the presence of excess CENP-B DBD, probably by the formation of non-specific aggregates that did not enter the gel (Figure 1B and C, lanes 11 and 12). Therefore, excess CENP-B DBD apparently forms aggregates by binding non-specifically to CENP-A and H3 nucleosomes under physiological salt conditions.
Nap1 promotes specific CENP-B binding to nucleosomes by eliminating non-specific CENP-B binding
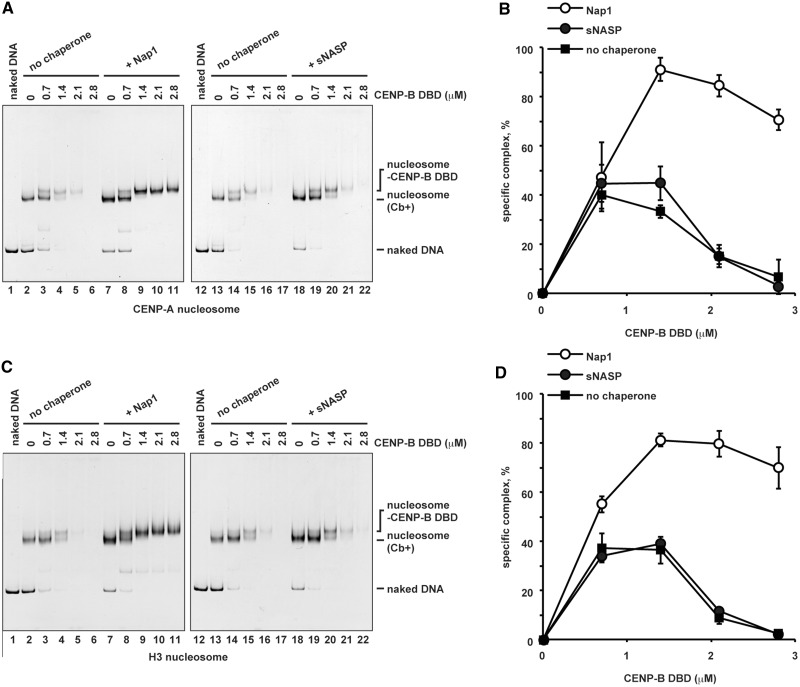
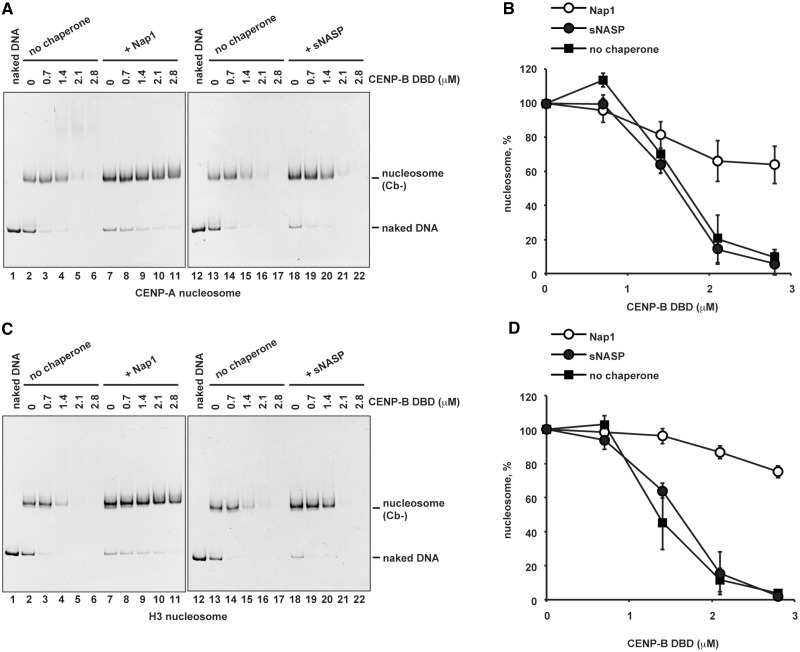
Nap1, an acidic histone chaperone, reportedly inhibits non-specific histone binding to DNA and promotes correct nucleosome assembly (64). We therefore tested the effects of purified recombinant Nap1 (Supplementary Figure S2) on CENP-B binding to the CENP-A nucleosome. Remarkably, we found that Nap1 significantly stimulated specific CENP-B binding to the CENP-A nucleosome (Figure 2A, lanes 7–11, and B). The Nap1-mediated stimulation of CENP-B DBD binding was also observed with the H3 nucleosome containing the CENP-B box (Figure 2C, lanes 7–11, and D). We postulated that this might be caused by Nap1-mediated inhibition of non-specific CENP-B binding to the CENP-A and H3 nucleosomes. Consistent with this idea, Nap1 dramatically inhibited the formation of non-specific aggregates of the CENP-A and H3 nucleosomes without the CENP-B box, driven by the CENP-B DBD (Figure 3A and C, lanes 7–11, and B and D). In contrast, a second acidic histone chaperone, sNASP (Supplementary Figure S2), did not significantly stimulate specific complex formation (Figure 2A and C, lanes 18–22, and B and D) and did not inhibit the formation of non-specific aggregates in the presence of excess CENP-B DBD (Figure 3A and C, lanes 18–22, and B and D).
Figure 2.
Nap1 stimulates specific binding of the CENP-B DBD to the CENP-A and H3 nucleosomes. (A) CENP-B DBD binding to the CENP-A nucleosomes with the CENP-B box DNA, in the presence of Nap1 or sNASP. The CENP-A nucleosomes (Cb+) (140 nM) were incubated with the CENP-B DBD in the presence of Nap1 (2.8 µM, lanes 7–11) or sNASP (2.8 µM, lanes 18–22) for 20 min at 37°C. Lanes 2–6 and 13–17 indicate control experiments in the absence of Nap1 or sNASP. CENP-B DBD concentrations were 0 µM (lanes 2, 7, 13 and 18), 0.7 µM (lanes 3, 8, 14 and 19), 1.4 µM (lanes 4, 9, 15 and 20), 2.1 µM (lanes 5, 10, 16 and 21)s and 2.8 µM (lanes 6, 11,17 and 22). The samples were analyzed by non-denaturing 5% polyacrylamide gel electrophoresis, followed by ethidium bromide staining. Lanes 1 and 12 indicate the 192-bp naked DNA. (B) Graphic representation of the specific complex formation. The relative band intensities of the specific CENP-B DBD-CENP-A nucleosome complexes were plotted with the standard deviations (n = 3). (C) CENP-B DBD binding to the H3 nucleosomes with the CENP-B box DNA, in the presence of Nap1 or sNASP. The H3 nucleosomes (Cb+) (140 nM) were incubated with the CENP-B DBD in the presence of Nap1 (2.8 µM, lanes 7–11) or sNASP (2.8 µM, lanes 18–22) for 20 min at 37°C. Lanes 2–6 and 13–17 indicate control experiments in the absence of Nap1 or sNASP. CENP-B DBD concentrations were 0 µM (lanes 2, 7, 13 and 18), 0.7 µM (lanes 3, 8, 14 and 19), 1.4 µM (lanes 4, 9, 15 and 20), 2.1 µM (lanes 5, 10, 16 and 21) and 2.8 µM (lanes 6, 11,17 and 22). The samples were analyzed by non-denaturing 5% polyacrylamide gel electrophoresis, followed by ethidium bromide staining. Lanes 1 and 12 indicate the 192-bp naked DNA. (D) Graphic representation of the specific complex formation. The relative band intensities of the specific CENP-B DBD-H3 nucleosome complexes were plotted with the standard deviations (n = 3).
Figure 3.
Nap1 dissociates non-specifically bound CENP-B DBD from the CENP-A nucleosome. (A) CENP-B DBD binding to the CENP-A nucleosomes without the CENP-B box DNA, in the presence of Nap1 or sNASP. The CENP-A nucleosomes (Cb−) (140 nM) were incubated with the CENP-B DBD in the presence of Nap1 (2.8 µM, lanes 7–11) or sNASP (2.8 µM, lanes 18–22) for 20 min at 37°C. Lanes 2–6 and 13–17 indicate control experiments in the absence of Nap1 or sNASP. CENP-B DBD concentrations were 0 µM (lanes 2, 7, 13 and 18), 0.7 µM (lanes 3, 8, 14 and 19), 1.4 µM (lanes 4, 9, 15 and 20), 2.1 µM (lanes 5, 10, 16 and 21) and 2.8 µM (lanes 6, 11,17 and 22). The samples were analyzed by non-denaturing 5% polyacrylamide gel electrophoresis, followed by ethidium bromide staining. Lanes 1 and 12 indicate the 192-bp naked DNA. (B) Graphic representation of the experiments shown in panel A. The relative band intensities of the CENP-A nucleosomes were plotted with the standard deviations (n = 3). (C) CENP-B DBD binding to the H3 nucleosomes without the CENP-B box DNA, in the presence of Nap1 or sNASP. The H3 nucleosomes (Cb−) (140 nM) were incubated with the CENP-B DBD in the presence of Nap1 (2.8 µM, lanes 7–11) or sNASP (2.8 µM, lanes 18–22) for 20 min at 37°C. Lanes 2–6 and 13–17 indicate control experiments in the absence of Nap1 or sNASP. CENP-B DBD concentrations were 0 µM (lanes 2, 7, 13 and 18), 0.7 µM (lanes 3, 8, 14 and 19), 1.4 µM (lanes 4, 9, 15 and 20), 2.1 µM (lanes 5, 10, 16 and 21) and 2.8 µM (lanes 6, 11,17 and 22). The samples were analyzed by non-denaturing 5% polyacrylamide gel electrophoresis, followed by ethidium bromide staining. Lanes 1 and 12 indicate the 192-bp naked DNA. (D) Graphic representation of the experiments shown in panel C. The relative band intensities of the H3 nucleosomes were plotted with the standard deviations (n = 3).
We conclude that Nap1, but not sNASP, promotes the specific binding of CENP-B to the CENP-B box in a nucleosomal context by suppressing non-specific aggregate formation. As sNASP is a highly acidic protein (pI = 4.35), the CENP-B regulating activity of Nap1 does not appear to be a consequence of non-specific interactions between the acidic chaperone and the basic CENP-B DBD.
Nap1 can remove CENP-B from chromosomes in vivo
The results of the aforementioned experiments strongly suggested that Nap1 regulates CENP-B binding to nucleosomes in vitro. We therefore developed a protocol to test whether Nap1 can regulate the association of CENP-B with chromosomes in vivo.
Incubation with excess Nap1 in vitro dissociates the CENP-B DBD from specific complexes with the CENP-A nucleosomes containing the CENP-B box (Supplementary Figure S3). Eviction of the CENP-B DBD from specific complexes was never observed in the presence of excess amounts of sNASP (Supplementary Figure S3). Thus, we postulated that if Nap1 regulates CENP-B binding in vivo, as it does in vitro, then Nap1 should be able to remove CENP-B from chromatin containing CENP-B boxes.
To detect CENP-B eviction from chromatin by Nap1 in vivo, we tethered Nap1 adjacent to bona fide CENP-B binding sites in a functional centromere. For this purpose, we fused Nap1 to tetR-EYFP and expressed the chimeric protein in HeLa cells bearing the alphoidtetO HAC (Figure 4A). This HAC is based on a synthetic alphoid DNA dimer, in which one monomer contains a CENP-B box, and the adjacent monomer contains a tetracycline operator sequence, instead of the CENP-B box (61,65).
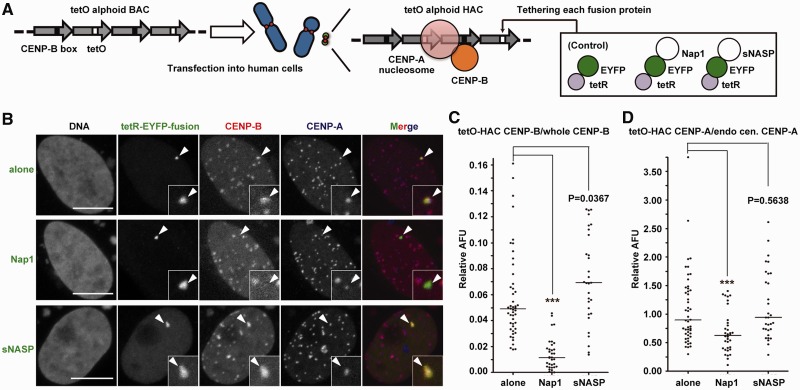
Figure 4.
The CENP-B and CENP-A assembly levels are reduced by the tethering of Nap1, but not sNASP, to the centromere. (A) A schematic drawing of the tetR-fusion/alphoidtetO array tethering system on the HAC. A HeLa cell line containing a stable alphoidtetO HAC (61, HeLa-HAC-2-4) was transfected with the tetR-EYFP-fusion protein expressing plasmids (tetR-EYFP alone, -Nap1 or -sNASP). Immunofluorescence analysis was performed 2 days after transfection. (B) Cells were co-stained with antibodies against CENP-B and CENP-A. The HAC centromere signals were determined by the EYFP signals (arrowhead), and DNA was visualized with Qnuclear™ Deep Red Stain (Invitrogen). The scale bars represent 10 µm (light gray). (C and D) Immunofluorescence signals of CENP-B (C) and CENP-A (D) on the HAC centromere, against those of all centromeres on host chromosomes within the same single nucleus, were quantified and plotted as relative arbitrary fluorescence units (AFU). Solid lines indicate the median. Asterisks indicate significant differences, with P < 0.001 (Mann–Whitney test).
The tethering of tetR-EYFP alone did not affect the levels of CENP-B and CENP-A at the HAC kinetochore (Figure 4B) (61,65). In contrast, tethering of tetR-EYFP-Nap1 dramatically decreased CENP-B levels at the alphoidtetO HAC centromere. In controls, tethering of tetR-EYFP-sNASP actually caused a small increase in the CENP-B levels (Figure 4B and C). Similar results were obtained when these proteins were tethered to an ectopic insertion of alphoidtetO sequences on a chromosomal arm (Figure 5). This ectopic insertion lacks CENP-A but contains bound CENP-B. As the protein levels of tetR-EYFP-Nap1 and tetR-EYFP-sNASP were similar (Supplementary Figure S4), the difference between Nap1 and sNASP is unlikely to be explained by for protein production and stability.
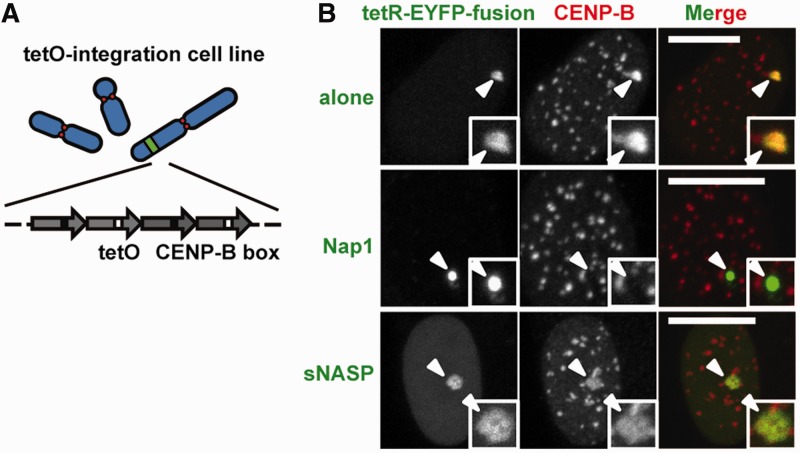
Figure 5.
Nap1 tethering reduces the CENP-B assembly level on non-centromeric alphoid chromatin in vivo. (A) A schematic drawing of the alphoidtetO ectopic integration site. (B) An alphoidtetO ectopic integration cell line (61, HeLa-Int-03) was transfected with a set of tetR-EYFP-fusion expressing plasmids. The CENP-B signals on the ectopic site were apparently reduced by Nap1 tethering, but not by sNASP tethering. Arrowheads indicate alphoidtetO DNA integration sites. The scale bars represent 10 µm.
It is also possible that the binding residence time of tetR-EYFP-Nap1 and tetR-EYFP-sNASP to the alphoidtetO differs significantly and affects the CENP-B removal activity. For example, if tetR-EYFP-sNASP binds stably to the alphoidtetO, then it may not be able to interact with CENP-B associated with the different sequences elsewhere in the repeats. To test this possibility, the turnover of each tetR-EYFP-fusion protein on the alphoidtetO was monitored by the FRAP technique (Figure 6). Both tetR-EYFP-Nap1 and tetR-EYFP-sNASP recovered over a time scale of minutes (t1/2 ∼ 9.3 and ∼3.5 min, respectively). These turnover rates are relatively rapid compared with the analytical time scale of CENP-B localization (1 day after the induction). Thus, both fusion proteins should be able to diffuse around the binding site to allow the interaction with nearby CENP-B. Indeed, the FRAP analyses of EYFP-Nap1 and EYFP-sNASP revealed that these proteins diffuse in the nucleus within seconds (Figure 6). The decreased mobility of the Nap1 fusions than sNASPs might be owing to the oligomerization of Nap1. These experiments clearly indicated that Nap1, but not sNASP, can regulate the association of CENP-B with chromatin in vivo.
Nap1 inhibits non-specific CENP-B binding in cells
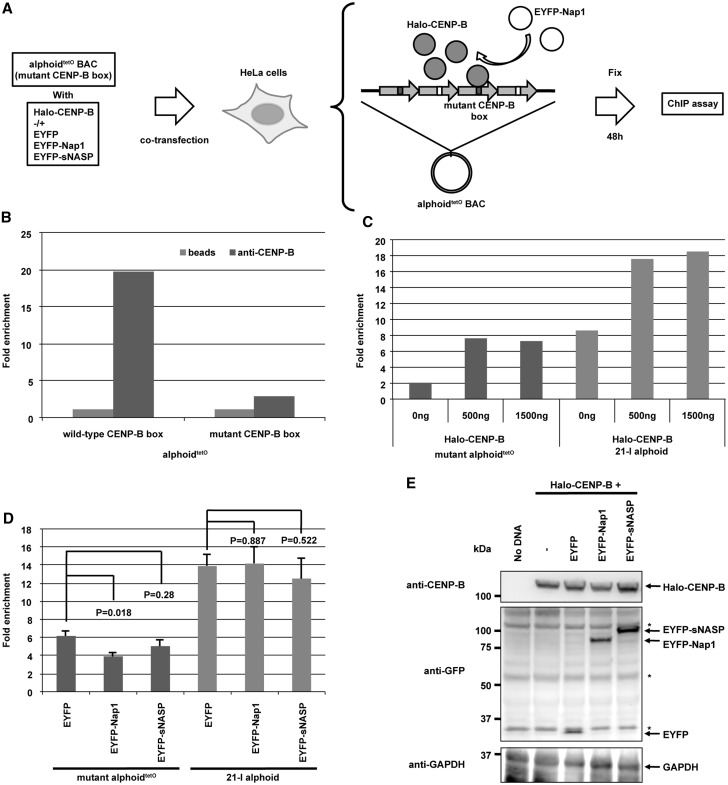
We next tested whether the Nap1 actually inhibits non-specific CENP-B binding in cells by ChIP (Figure 7A). Two point mutations within the canonical CENP-B box sequence in alphoid DNA abolish the specific CENP-B binding (Figure 7B). Consistently, these CENP-B box mutations cause significant deficiencies in de novo CENP-A assembly and de novo HAC formation in human and mouse cells (44,66,67). However, the non-specific binding of CENP-B to the mutant CENP-B box was increased under conditions where Halo-CENP-B was overexpressed (Figure 7C). We then tested whether Nap1 co-expression with Halo-CENP-B reduces non-specific CENP-B binding to the mutant CENP-B box. As shown in Figure 7D, Nap1 overexpression significantly reduced the non-specific CENP-B binding to the mutant CENP-B box selectively, whereas no significant reduction of the specific CENP-B binding to the host centromere with canonical CENP-B boxes was observed under the same conditions. In controls, the CENP-B expression level was not substantially decreased on Nap1 overexpression (Figure 7E). In addition, an interaction between CENP-B and Nap1 was detected by in vitro and in vivo pull-down assays (Supplementary Figure S5). These results suggested that the selective inhibition of non-specific CENP-B-DNA binding by Nap1 may regulate proper CENP-B binding to chromosomes in vivo.
Figure 7.
Nap1 decreases non-specific CENP-B binding in cells. (A) Schematic diagram of the ChIP analysis. HeLa cells were transfected with alphoidtetO DNA plus or minus each expression plasmid. The ChIP analysis was performed using an anti-CENP-B C-terminus antibody [5E6C1 (66)]. (B) CENP-B specifically binds to wild-type canonical CENP-B box on the alphoidtetO DNA. HeLa cells were transfected with alphoidtetO containing wild-type or mutant CENP-B boxes (66). The ChIP analysis was performed on the introduced alphoidtetO DNAs. The rate of recovery of immunoprecipitates using the anti-CENP-B antibody was normalized to the control IP without the antibody (beads). (C) CENP-B overexpression increased its non-specific binding to the introduced alphoidtetO-containing mutant CENP-B boxes and its specific binding to endogenous alphoid DNA containing canonical CENP-B boxes on the chromosome 21 centromere (21-I alphoid). The ChIP analysis was performed by co-transfecting the alphoidtetO DNA-containing mutant CENP-B boxes and the Halo-CENP-B expression plasmid (0, 500 and 1500 ng). Real-time PCR analysis was performed on the introduced alphoidtetO DNA and 21-I alphoid DNA. (D) Non-specific binding of CENP-B to the introduced alphoidtetO-containing mutant CENP-B boxes was decreased by Nap1 overexpression. The ChIP analysis was performed by co-transfecting the alphoidtetO DNA-containing mutant CENP-B boxes and the Halo-CENP-B expression plasmid (500 ng) with the EYFP, EYFP-Nap1 or EYFP-sNASP expression plasmid. Real-time PCR analysis was performed as in panel C. Error bars, s.d. (n = 3). P-values (t-test) are indicated in the figure. (E) Expression of Halo-CENP-B and EYFP-fusion proteins in HeLa cells. HeLa cells co-transfected with each plasmid set were analyzed by western blotting, using antibodies against CENP-B, GAPDH and GFP. Asterisks indicate non-specific signals with the anti-GFP antibody.
The CENP-A assembly level positively correlates with the CENP-B assembly level in vivo
Interestingly, Nap1-mediated dissociation of CENP-B from centromeric chromatin was accompanied by a significant decrease in the levels of CENP-A at the centromere on the alphoidtetO HAC (Figure 4B and D). No such correlation was observed when tetR-EYFP-alone or tetR-EYFP-sNASP was tethered (Figure 4B and D and Supplementary Figure S6). These results suggested that CENP-B may be able to modulate CENP-A levels at kinetochores, as suggested by previous findings in which CENP-B promoted the de novo assembly of CENP-A chromatin and stable HAC formation on input alphoid DNA (44,66,67).
DISCUSSION
CENP-B positively functions to promote de novo CENP-A assembly on transfected alphoid DNA (44,66–68). Paradoxically, CENP-B also acts as an inhibitor of CENP-A loading onto alphoid DNA, through heterochromatin formation (44). This suggests that if CENP-B were improperly loaded on a chromosome arm, then this might induce inappropriate heterochromatin formation. Therefore, the non-specific binding of CENP-B may need to be kept to a minimum.
In the present study, we found that human Nap1 significantly inhibits non-specific CENP-B binding to nucleosomes in vitro. Furthermore, Nap1 also markedly stimulates the specific binding of CENP-B to nucleosomes. These in vitro data strongly suggested that Nap1 functions to promote specific CENP-B binding to the CENP-B box within CENP-A and H3 nucleosomes, by inhibiting its non-specific binding. Consistent with this, Nap1 interacted with the CENP-B DBD in vitro and full-length CENP-B in vivo and markedly inhibited non-specific CENP-B binding to DNA in vivo.
Intriguingly, similar Nap1-mediated inhibition of non-specific DNA binding had been previously found with histones (64). Andrews et al. (64) reported that Nap1 eliminates non-nucleosomal histone–DNA interactions and promotes correct nucleosome assembly in vitro. Full-length CENP-B is an acidic protein (10,69), but its DBD (1–129 amino acid residues) is highly basic (pI = 10.49). Therefore, one role of Nap1 might be to mediate specific DNA binding by basic proteins, such as histones and the CENP-B DBD, which have a propensity for non-specific electrostatic interactions with the negatively charged DNA phosphate backbone.
Nap1 is an acidic histone-binding protein that could in theory simply bind electrostatically to the basic CENP-B DBD and non-specifically inhibit the CENP-B–DNA interaction. However, we do not think this is the case because another acidic histone-binding protein, sNASP, affected neither specific nor non-specific CENP-B-binding to nucleosomes in vitro and chromosomes in vivo. Furthermore, in vivo model experiments, in which Nap1 or sNASP was tethered near CENP-B-binding sites, revealed that the Nap1 tethering, but not the sNASP tethering, significantly reduced CENP-B levels on kinetochore chromatin of a synthetic HAC and also on an ectopic array of alphoid DNA containing CENP-B-binding sites in a chromosome arm. The ectopic array lacked bound CENP-A. Therefore, the promotion of specific CENP-B binding to both H3 and CENP-A nucleosomes may be a novel unidentified function of Nap1.
Our Nap1 tethering experiments also revealed that CENP-A levels at the HAC kinetochore were correlated with CENP-B levels. This suggests that either CENP-B assembly on the HAC promotes CENP-A loading or CENP-B stripping by Nap1 somehow destabilizes CENP-A nucleosomes. The former is consistent with previous observations that CENP-B promotes the de novo formation of stable CENP-A-containing chromatin during HAC formation from transfected alphoid DNA (44). Our data therefore suggested that Nap1 may indirectly regulate the specificity of CENP-A assembly, by regulating CENP-B binding. It is also possible that Nap1 may directly promote the assembly/disassembly of CENP-A nucleosomes on HACs, independently of CENP-B. We prefer the former explanation, as sNASP tethering does not affect the levels of CENP-A on the HAC in vivo, although both Nap1 and sNASP promote the assembly of CENP-A nucleosomes with almost the same efficiency in vitro (21,55).
These studies have led us to propose a novel Nap1 function: the regulation of specific CENP-B loading at centromeric chromatin by the inhibition of non-specific CENP-B binding to other chromosome loci. Thus, Nap1 may indirectly promote the specific assembly of CENP-A nucleosomes, key structural elements of active centromeres.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
Grants-in-Aid from the Japanese Society for the Promotion of Science (JSPS); Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (in part); Waseda Research Institute for Science and Engineering, the Sagawa Foundation for Promotion of Cancer Research, and NOVARTIS Foundation (Japan) for the Promotion of Science (to H.K.); Kazusa DNA Research Institute Foundation (to H.M.); Intramural Research Program of the NIH, NCI, CCR (to V.L.). Work in the WCE lab is funded by The Wellcome Trust, of which WCE is a Principal Research Fellow [073915]; Wellcome Trust Centre for Cell Biology [077707 and 092076]. Funding for open access charge: Waseda University.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- 2.Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, III, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, III, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 4.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, III, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 5.Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Dorn JF, Maddox PS. Kinetochore dynamics: how protein dynamics affect chromosome segregation. Curr. Opin. Cell Biol. 2012;24:57–63. doi: 10.1016/j.ceb.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Takeuchi K, Fukagawa T. Molecular architecture of vertebrate kinetochores. Exp. Cell Res. 2012;18:1367–1374. doi: 10.1016/j.yexcr.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 8.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 9.Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saitoh H, Tomkiel J, Cooke CA, Ratrie H, III, Maurer M, Rothfield NF, Earnshaw WC. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 12.Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, Fukagawa T. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J. Cell Biol. 2009;186:173–182. doi: 10.1083/jcb.200903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 15.Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 17.Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Régnier V, Vagnarelli P, Fukagawa T, Zerjal T, Burns E, Trouche D, Earnshaw W, Brown W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 22.Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat. Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingston IJ, Yung JSY, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J. Biol. Chem. 2011;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 27.Talbert PB, Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 28.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc. Natl Acad. Sci. USA. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddox PS, Corbett KD, Desai A. Structure, assembly and reading of centromeric chromatin. Curr. Opin. Genet. Dev. 2011;22:1–9. doi: 10.1016/j.gde.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tachiwana H, Kurumizaka H. Structure of the CENP-A nucleosome and its implications for centromeric chromatin architecture. Genes Genet. Syst. 2011;86:357–364. doi: 10.1266/ggs.86.357. [DOI] [PubMed] [Google Scholar]
- 32.Shivaraju M, Unruh JR, Slaughter BD, Mattingly M, Berman J, Gerton JL. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell. 2012;150:304–316. doi: 10.1016/j.cell.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bui M, Dimitriadis EK, Hoischen C, An E, Quénet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell. 2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 35.Panchenko T, Sorensen TC, Woodcock CL, Kan ZY, Wood S, Resch MG, Luger K, Englander SW, Hansen JC, Black BE. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc. Natl Acad. Sci. USA. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, Colmenares SU, Karpen GH. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol. Cell. 2012;45:263–269. doi: 10.1016/j.molcel.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- 39.Saffery R, Irvine DV, Griffiths B, Kalitsis P, Wordeman L, Choo KH. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum. Mol. Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- 40.Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pluta AF, Saitoh N, Goldberg I, Earnshaw WC. Identification of a subdomain of CENP-B that is necessary and sufficient for localization to the human centromere. J. Cell Biol. 1992;116:1081–1093. doi: 10.1083/jcb.116.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka Y, Tachiwana H, Yoda K, Masumoto H, Okazaki T, Kurumizaka H, Yokoyama S. Human centromere protein B induces translational positioning of nucleosomes on α-satellite sequences. J. Biol. Chem. 2005;280:41609–41618. doi: 10.1074/jbc.M509666200. [DOI] [PubMed] [Google Scholar]
- 43.Tachiwana H, Kagawa W, Kurumizaka H. Comparison between the CENP-A and histone H3 structures in nucleosomes. Nucleus. 2012;3:1–6. doi: 10.4161/nucl.18372. [DOI] [PubMed] [Google Scholar]
- 44.Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 45.Cam HP, Noma K-I, Ebina H, Levin HL, Grewal SIS. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2007;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 46.Lorenz DR, Mikheyeva IV, Johansen P, Meyer L, Berg A, Grewal SIS, Cam HP. CENP-B cooperates with Set1 in bidirectional transcriptional silencing and genome organization of retrotransposons. Mol. Cell Biol. 2012;32:4215–4225. doi: 10.1128/MCB.00395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishimi Y, Hirosumi J, Sato W, Sugasawa K, Yokota S, Hanaoka F, Yamada M. Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur. J. Biochem. 1984;142:431–439. doi: 10.1111/j.1432-1033.1984.tb08305.x. [DOI] [PubMed] [Google Scholar]
- 48.Ito T, Bulger M, Kobayashi R, Kadonaga JT. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McBryant SJ, Park YJ, Abernathy SM, Laybourn PJ, Nyborg JK, Luger K. Preferential binding of the histone (H3-H4)2 tetramer by NAP1 is mediated by the amino-terminal histone tails. J. Biol. Chem. 2003;278:44574–44583. doi: 10.1074/jbc.M305636200. [DOI] [PubMed] [Google Scholar]
- 50.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 2005;280:1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 51.Mazurkiewicz J, Kepert JF, Rippe K. On the mechanism of nucleosome assembly by histone chaperone NAP1. J. Biol. Chem. 2006;281:16462–16472. doi: 10.1074/jbc.M511619200. [DOI] [PubMed] [Google Scholar]
- 52.Tachiwana H, Osakabe A, Kimura H, Kurumizaka H. Nucleosome formation with the testis-specific histone H3 variant, H3t, by human nucleosome assembly proteins in vitro. Nucleic Acids Res. 2008;36:2208–2218. doi: 10.1093/nar/gkn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, Walsh STR, Parthun MR. Expanded binding specificity of the human histone chaperone NASP. Nucleic Acids Res. 2008;36:5763–5772. doi: 10.1093/nar/gkn574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okuwaki M, Kato K, Nagata K. Functional chracterization of human nucleosome assembly protein 1-like proteins as histone chaperones. Genes Cells. 2010;15:13–27. doi: 10.1111/j.1365-2443.2009.01361.x. [DOI] [PubMed] [Google Scholar]
- 55.Osakabe A, Tachiwana H, Matsunaga T, Shiga T, Nozawa RS, Obuse C, Kurumizaka H. Nucleosome formation activity of human somatic nuclear autoantigenic sperm protein (sNASP) J. Biol. Chem. 2010;285:11913–11921. doi: 10.1074/jbc.M109.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richardson RT, Batova IN, Widgren EE, Zheng LX, Whitfield M, Marzluff WF, O'Rand MG. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J. Biol. Chem. 2000;275:30378–30386. doi: 10.1074/jbc.M003781200. [DOI] [PubMed] [Google Scholar]
- 57.Finn RM, Browne K, Hodgson KC, Ausió J. sNASP, a histone H1-specific eukaryotic chaperone dimer that facilitates chromatin assembly. Biophys. J. 2008;95:1314–1325. doi: 10.1529/biophysj.108.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saeki H, Ohsumi K, Aihara H, Ito T, Hirose S, Ura K, Kaneda Y. Linker histone variants control chromatin dynamics during early embryogenesis. Proc. Natl Acad. Sci. USA. 2005;102:5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tachiwana H, Kagawa W, Osakabe A, Kawaguchi K, Shiga T, Hayashi-Takanaka Y, Kimura H, et al. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc. Natl Acad. Sci. USA. 2010;107:10454–10459. doi: 10.1073/pnas.1003064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suzuki N, Itou T, Hasegawa Y, Okazaki T, Ikeno M. Cell to cell transfer of the chromatin-packaged human β-globin gene cluster. Nucleic Acids Res. 2010;38:e33. doi: 10.1093/nar/gkp1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohzeki J-I, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H. Breaking the HAC barrier: Histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012;31:2391–2402. doi: 10.1038/emboj.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoda K, Kitagawa K, Masumoto H, Muro Y, Okazaki T. A human centromere protein, CENP-B, has a DNA binding domain containing four potential alpha helices at the NH2 terminus, which is separable from dimerizing activity. J. Cell Biol. 1992;119:1413–1427. doi: 10.1083/jcb.119.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanaka Y, Nureki O, Kurumizaka H, Fukai S, Kawaguchi S, Ikuta M, Iwahara J, Okazaki T, Yokoyama S. Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 2001;20:6612–6618. doi: 10.1093/emboj/20.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrew AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohzeki J-I, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okamoto Y, Nakano M, Ohzeki J-I, Larionov V, Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J. 2007;26:1279–1291. doi: 10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat. Biotech. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 69.Earnshaw WC. Anionic regions in nuclear proteins. J. Cell Biol. 1987;105:1479–1482. doi: 10.1083/jcb.105.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.