Abstract
Histone H3 of nucleosomes positioned on active genes is trimethylated at Lys36 (H3K36me3) by the SETD2 (also termed KMT3A/SET2 or HYPB) methyltransferase. Previous studies in yeast indicated that H3K36me3 prevents spurious intragenic transcription initiation through recruitment of a histone deacetylase complex, a mechanism that is not conserved in mammals. Here, we report that downregulation of SETD2 in human cells leads to intragenic transcription initiation in at least 11% of active genes. Reduction of SETD2 prevents normal loading of the FACT (FAcilitates Chromatin Transcription) complex subunits SPT16 and SSRP1, and decreases nucleosome occupancy in active genes. Moreover, co-immunoprecipitation experiments suggest that SPT16 is recruited to active chromatin templates, which contain H3K36me3-modified nucleosomes. Our results further show that within minutes after transcriptional activation, there is a SETD2-dependent reduction in gene body occupancy of histone H2B, but not of histone H3, suggesting that SETD2 coordinates FACT-mediated exchange of histone H2B during transcription-coupled nucleosome displacement. After inhibition of transcription, we observe a SETD2-dependent recruitment of FACT and increased histone H2B occupancy. These data suggest that SETD2 activity modulates FACT recruitment and nucleosome dynamics, thereby repressing cryptic transcription initiation.
INTRODUCTION
Post-translational modifications of histone N-terminal tails play a crucial role in the regulation of transcription by RNA polymerase II (RNAPII) (1,2). Histone H3 trimethylated on lysine 36 (H3K36me3) is found in actively transcribed genes and is more abundant in exons than in introns (3,4). Misteli and colleagues recently found that H3K36me3 may influence splicing decisions by acting as a docking site for the chromatin-binding adaptor protein MRG15, which in turn recruits the splicing factor PTB, which inhibits the inclusion of PTB-dependent alternative exons (5). However, the level of H3K36me3 detected along the body of an actively transcribed gene can also be modulated by splicing (6,7), suggesting that splicing and chromatin are bi-directionally interwoven (8). The methyltransferase responsible for H3K36 trimethylation is termed Set2 in yeast (9) and SETD2, KMT3A/SET2 or HYPB in mammals (10). Previous studies have shown that depletion of Set2 in yeast (11–14) leads to increased levels of spurious transcripts initiated from cryptic promoter-like sequences within genes. These cryptic alternative start sites are widespread in eukaryotic genomes and can be found on virtually every single gene (15). Thus, H3K36me3 is thought to be required for repression of cryptic transcriptional initiation, but the mechanisms involved are not yet completely understood. In yeast, Set2-dependent histone H3 methylation is required for the activation of Rpd3S after its recruitment to the RNAPII C-terminal domain (11–13,16,17). Rpd3S, a histone deacetylase complex that erases histone acetylation created to facilitate passage of the elongating polymerase, then restores the original structure of chromatin, preventing cryptic transcription initiation from within coding regions (11–13,16,17). However, a recent study failed to detect changes in global levels of various H3 and H4 acetyl modifications after knocking down SETD2 in mouse cells (18), arguing that H3K36 trimethylation may inhibit intragenic transcription in mammalian cells by a mechanism that does not involve histone acetylation. Another study using mouse embryonic stem cells found that H3K36me3 recruits histone H3K4me2/3 demethylase KDM5B via an Rpd3S-like complex and proposed that KDM5B represses cryptic transcription by removing intragenic H3K4me3 (19). In addition to Set2, factors that restore normal chromatin structure during transcriptional elongation such as the histone chaperone FACT (a complex termed after its ability to ‘FAcilitate Chromatin Transcription’) (20,21) and Spt6 are crucial to repress transcription from cryptic promoters located within protein-coding genes (11,12,22–24). Here we demonstrate that human SETD2 and FACT act on transcription through a common mechanism. We show that RNA interference (RNAi)-mediated downregulation of SETD2 in HeLa cells reduces recruitment of the FACT complex to active genes and affects nucleosome organization. Notably, we found that depletion of SPT16 alone or depletion of SPT16 and SETD2 results equally in reduced nucleosome occupancy on gene bodies, suggesting that FACT and H3K36me3 combine to maintain chromatin integrity on transcribed genes. We further show that transcriptional activation leads to a SETD2-dependent reduction of histone H2B occupancy and enhanced reformation of nucleosomes after passage of RNAPII. Altogether, these results suggest that H3K36 trimethylation by SETD2 associated with transcribed genes enhances the recruitment of FACT, which promotes the exchange of H2A–H2B dimers. Moreover, SETD2-dependent FACT recruitment is crucial to reassemble nucleosomes in the wake of RNAPII elongation, thereby repressing cryptic intragenic transcription initiation.
MATERIALS AND METHODS
Cells and drugs
HeLa and human embryonic kidney (HEK) 293T cells were grown as monolayers in DMEM, supplemented with 10% (v/v) FBS, 2 mM l-glutamine and 100 U/ml penicillin–streptomycin. U2OS cells stably expressing tetracycline-inducible β-globin transgenes were cultured in DMEM, supplemented with 10% (v/v) FBS, 2 mM l-glutamine, 100 U/ml penicillin–streptomycin, 200 μg/ml G418 and 1 μg/ml puromycin. All cell culture reagents were from Invitrogen. When indicated, the histone deacetylase inhibitor trichostatin A (TSA; Sigma) was added to HeLa cells for 12 h at a final concentration of 0.5 μM. Unfolded protein response (UPR) was induced on HeLa cells by incubation with 2 mM dithiothreitol (DTT; Sigma) for 1 h. To inhibit transcription, HeLa cells were treated with 75 μM 5,6-dichloro-1-β-d-ribobenzimidazole (DRB; Sigma) for 30 min.
RNA interference
SETD2 RNAi was performed with three different siRNA duplexes. On each transfection, two duplexes were used. As unspecific siRNA control, a sequence targeting the firefly luciferase gene (GL2), was used. All oligonucleotides were synthesized by Eurogentec. SPT16 RNAi was performed with a pool of three specific siRNAs from Santa Cruz (sc-37875), transfected onto HeLa or HEK 293T cells on two consecutive rounds. HeLa or HEK 293T cells were plated such that they were 50–60% confluent at the time of transfection. siRNA duplexes were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Cells were retransfected 24 h after the first transfection and harvested on the following day. The siRNA sequences targeting SETD2 are listed in Supplementary Table S1.
RNA isolation and quantitative RT-PCR
To quantify nascent RNA transcripts, we used a modified cell fractionation protocol that was initially described by Wuarin and Schibler (25,26). Chromatin-associated nascent RNA was extracted with TRIzol (Invitrogen). cDNA was made using Superscript II Reverse Transcriptase (Invitrogen). RT-qPCR was performed in the 7000 Real-Time PCR System (Applied Biosystems), using SYBR Green PCR master mix (Applied Biosystems). The relative RNA expression was estimated as follows: 2(Ct reference − Ct sample), where Ct reference and Ct sample are mean threshold cycles of RT-qPCR done in duplicate on cDNA samples from U6 snRNA (reference) and the cDNA from our genes of interest (sample), respectively. All primer sequences are presented in Supplementary Table S2.
Western blot
Whole-cell lysates were prepared, resolved and transferred as described (26). The primary antibodies used were anti-histone H3 (ab1791; Abcam); anti-H3K36me3 (ab9050; Abcam) and anti-H3Ac (ab47915; Abcam). Detection was performed with the appropriate secondary antibodies (BioRad) and enhanced luminescence substrate (Amersham).
Co-immunoprecipitation
Immunoprecipitation of endogenous H3K36me3 and SPT16 from HeLa cells was performed using the Nuclear Complex Co-IP kit from Active Motif according to the manufacturer’s protocol. Anti-H3K36me3 (ab9050; Abcam) and anti-SPT16 (sc-28734; Santa Cruz) antibodies were used. The immune complexes were pulled down using protein A-Sepharose beads from Sigma. The pulled-down complexes were then subjected to western blot using the antibody against SPT16.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed on HeLa or HEK 293T cells as described (6). The relative occupancy of the immunoprecipitated protein at each DNA site was estimated by RT-qPCR as follows: 2(Ct Input − Ct IP), where Ct Input and Ct IP are mean threshold cycles of RT-qPCR done in duplicate on DNA samples from input and specific immunoprecipitations, respectively. Gene-specific and intergenic-region primer pairs are presented in Supplementary Table S2. The following antibodies were used: anti-RNAPII (N20 sc-899; Santa Cruz), anti-histone H3 (ab1791; Abcam), anti-H3K36me3 (ab9050; Abcam), anti-H2BUb (ab10812; Abcam), anti-SPT6 (ab49066; Abcam), anti-SPT16 (sc-28734; Santa Cruz) and anti-SSRP1 (609702; Biolegend).
Fluorescence recovery after photobleaching
Live cells were imaged using a 63×/1.4 oil immersion objective on a Zeiss LSM 510 META (Carl Zeiss) confocal microscope equipped with an incubator (Pecon) maintained at 37°C. Fluorescence recovery after photobleaching (FRAP) experiments were performed as previously described (27). The t80% value was determined for each FRAP curve as the first time point for which there was no statistically significant difference between the mean of the distribution of normalized fluorescence values and the value 0.8 (P > 0.05, one-sample t-test).
RNA-seq data analysis
We analyzed previously reported poly(A)+ RNA-seq data from human mesenchymal stem cells transfected with control and targeting siRNAs directed against SETD2 (5). High-throughput sequencing reads were aligned to the reference human Genome hg19 using TopHat (28), removing reads mapping to multiple locations. Our analysis was restricted to transcriptionally active genes defined as the genes with at least 50 reads/kb in both samples. Gene annotations were obtained from UCSC knownGene table (29). To identify genes with intragenic transcription initiation, we compared read counts for expressed exons (minimum of 5 reads/100 bp required) in depleted and control cells. First, we selected all transcripts with significantly higher read counts for at least one exon in SETD2-depleted cells. Second, we discarded transcripts with significantly higher read counts for the first annotated and expressed exon (likely corresponding to upregulated full-length mRNAs), Third, we defined the new cryptic TSS as the first significantly higher exon and filtered out all transcripts that showed significantly higher read counts for <60% of the downstream exons (possibly corresponding to isolated alternative splicing events). Significantly higher exons were defined as those having exonic read counts (normalized reads per million) fold change higher than 1.2 between SETD2-depleted and control cells, and Fisher’s proportion test P < 0.05 (alternative hypothesis: read count proportions relative to the total library size in siSETD2 greater than siControl). To withdraw restrictions by previous gene annotation, we performed transcriptome assembly using Cufflinks (30).
Micrococcal nuclease assay
The assay was performed as described (31) using HeLa cells transfected with the indicated siRNA duplexes. DNA was digested with 10 U of micrococcal nuclease (MNase; Fermentas) for 1, 2.5, 5, 10 or 20 min before addition of stop buffer. Mononucleosome-sized DNA obtained after 20-min digestion and non-digested (total) DNA were used as templates for RT-qPCR (32). The amount of MNase-resistant DNA at each gene region analyzed was estimated as follows: 2(Ct t0 − Ct t20), where Ct t0 and Ct t20 are mean threshold cycles of RT-qPCR done in duplicate on DNA samples from non-digested (t0) and 20 min MNase-digested (t20) samples, respectively. Results were further normalized to the amount of MNase-resistant DNA measured with primers for an intergenic region (estimated using the same 2(Ct t0 − Ct t20) formula). The sequences of gene-specific and intergenic primers are shown in Supplementary Table S2.
RESULTS
Downregulation of SETD2 leads to higher RNAPII turnover at the transcription site
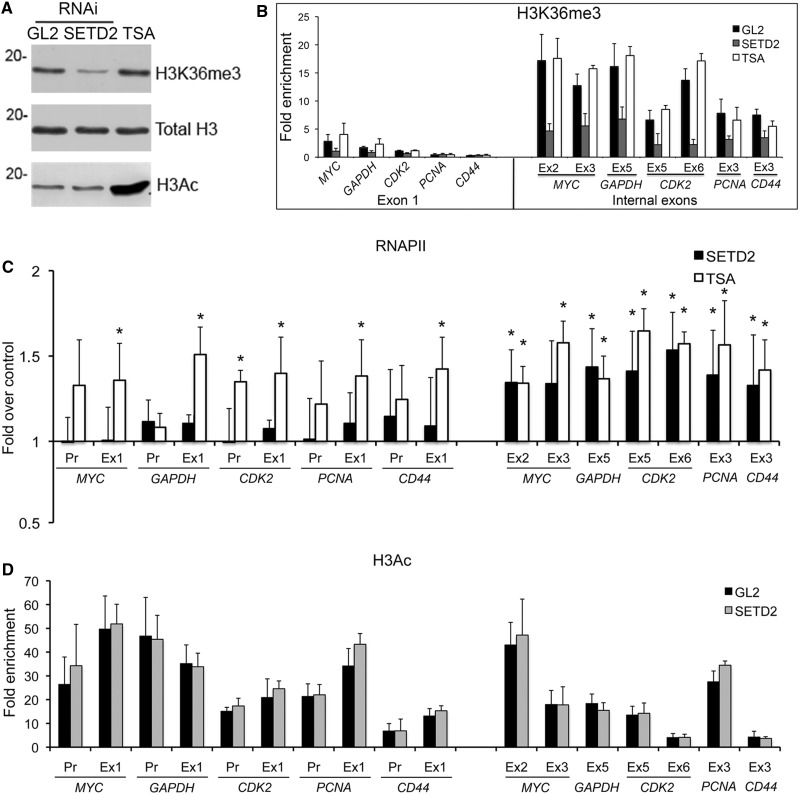
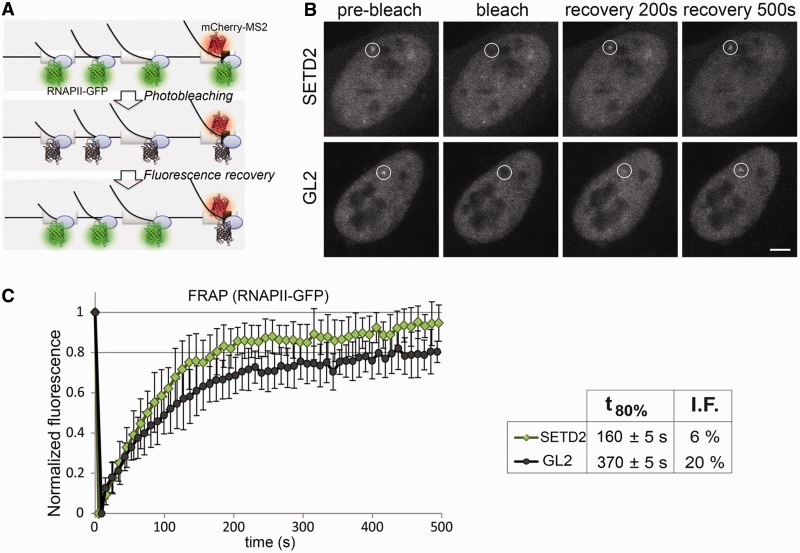
To study the impact of H3K36me3 on transcription, we reduced the levels of SETD2 methyltransferase in HeLa cells by RNAi (Figure 1A and Supplementary Figure S1A, B and G). Three different synthetic small interfering RNA duplexes targeting SETD2 were designed and tested, and two of them were alternatively used throughout this study (Supplementary Figure S1A). As a negative control, we used the GL2 duplex, which targets firefly luciferase (33). ChIP was carried out using antibodies directed against histone H3 or H3K36me3, followed by real-time RT-qPCR with primers targeting the indicated regions of five randomly selected genes (Figure 1B). The histone H3 ChIP signal was used to normalize H3K36me3 levels, thus correcting for differences in nucleosome density. We have previously reported that the level of H3K36me3 in the first exon is low and increases significantly in the second and subsequent internal exons (6). Accordingly, when compared with control GL2-treated cells, downregulation of SETD2 causes marginal reductions in H3K36me3 signal associated with first exons (Figure 1B); in contrast, the level of H3K36me3 on internal exons is drastically reduced in SETD2-depleted cells (Figure 1B). Next, we carried out ChIP analysis with an antibody (N-20) that binds to RNAPII largest subunit independently of the phosphorylation status of its C-terminal domain (Figure 1C). The results, depicted as fold changes relative to control GL2-treated cells, reveal that SETD2 depletion does not significantly alter RNAPII occupancy on the promoter region and on the first exon, but leads to higher density of RNAPII on internal exons (Figure 1C). Similar data were obtained when this experiment was repeated in HEK 293T cells (Supplementary Figures S1C, D and S2A). Because in yeast, methylated H3K36 activates the Rpd3S histone deacetylase complex (11–13,16,17,34), we sought to compare the effect of SETD2 depletion with inhibition of histone deacetylases. As expected, treatment of cells with the histone deacetylase inhibitor TSA induces an increase in global histone H3 acetylation without affecting the global level of H3K36me3 (Figure 1A). Inversely, downregulation of SETD2 causes a significant reduction in H3K36me3 (Figure 1B), but does not affect histone H3 acetylation (Figure 1A and D), as previously reported (18). ChIP analysis shows that TSA does not alter the distribution of H3K36me3 (Figure 1B), but significantly increases RNAPII occupancy (Figure 1C). Unlike SETD2 depletion, TSA equally affects RNAPII occupancy on the promoter region, first exon and internal exons, most likely because it increases the DNA accessibility (35). Altogether, these findings suggest that the increased RNAPII levels observed at the internal exons of SETD2-depleted cells are not the result of higher levels of transcription initiation at the canonical promoter. Instead, these data are consistent with transcription initiation from cryptic promoter-like sequences within gene bodies, as previously reported in yeast cells depleted of Set2 (11–14). To test this view, we analyzed the effect of SETD2 depletion on transcriptional dynamics of RNAPII in living cells using FRAP. Human osteosarcoma–derived cells (U2OS) were engineered to stably express tetracycline-inducible β-globin transgenes integrated tandemly in the genome, as previously reported (27). On transcription activation, the site of transcription is detected by a red fluorescent protein (mCherry) fused to the MS2 protein that binds to an array of six MS2-binding sequences inserted in the third exon of β-globin (Figure 2A). Simultaneously, the cells express α-amanitin–resistant RNAPII fused to green fluorescent protein (RNAPII–GFP) (36). Incubation with α-amanitin causes degradation of the endogenous RNAPII, whereas the resistant RNAPII–GFP remains active (37) (Supplementary Figure S1H). Transcriptional activation in the presence of α-amanitin leads to accumulation of RNAPII–GFP at the sites of β-globin transcription. The FRAP analysis consists of bleaching the RNAPII–GFP fluorescence at the transcription site of β-globin, and subsequently monitoring the fluorescence recovery by time-lapse microscopy (Figure 2B). While control (GL2) cells take 370 ± 5 s to recover 80% of the initial RNAPII–GFP fluorescence intensity, in SETD2-depleted cells, RNAPII–GFP fluorescence recovers significantly faster (160 ± 5 s; Figure 2C). Recovery of RNAPII–GFP fluorescence at the transcription site of β-globin results from recruitment of unbleached fluorescent polymerase molecules through transcription initiation. Taken together with the increased intragenic occupancy of RNAPII on SETD2 depletion (Figure 1C), the faster fluorescence recovery observed in the FRAP experiments (Figure 2C) is consistent with the view that downregulation of SETD2 facilitates intragenic transcription initiation. Moreover, the fraction of RNAPII molecules immobilized at the transcription site decreases from ∼20% in control cells to 6% in SETD2-depleted cells (Figure 2C). We have previously shown that ∼20% of the total population of RNAPII–GFP in control cells remains transiently immobilized at the β-globin transcription site owing to transcriptional pausing associated with termination (27). Possibly, spurious transcripts initiated at intragenic sites are not normally terminated, allowing faster dissociation of the polymerase from the gene template.
Figure 1.
SETD2 depletion impacts on transcription by RNAPII. HeLa cells were either transfected with siRNAs targeting GL2 or SETD2 or treated with 0.5 μM TSA for 12 h. (A) Total protein extracts were analyzed by western blot with anti-histone antibodies, as indicated on the right. Molecular weight markers are shown on the left. (B, C and D) ChIP analysis with antibodies that recognize H3K36me3 (B), RNAPII (C) and H3Ac (D). Histone modifications ChIP data are normalized against total histone H3 ChIP. RNAPII ChIP signals are shown as fold change over the signal obtained on the control cells. Means and standard deviations from at least three independent experiments are shown. Two SETD2 siRNA duplexes were used randomly on each individual RNAi experiment. Statistically significant changes between SETD2-depleted cells and control cells (GL2 RNAi) and between TSA-treated cells and control cells are indicated (*P < 0.05).
Figure 2.
SETD2 depletion alters the dynamics of RNAPII transcription. (A) Schematic representation of the MS2 system. Dimers of MS2 coat protein fused to mCherry bind to MS2 stem–loops inserted in the third exon of the β-globin gene, while transcription is carried out by α-amanitin–resistant RNAPII–GFP. (B) FRAP experiments were performed on U2OS cells transfected with siRNAs targeting GL2 and SETD2. A circular region corresponding to the site of transcription of the β-globin gene is bleached, and fluorescence recovery is subsequently monitored. Scale bar: 5 µm. (C) Normalized fluorescence recovery curves measured at the transcription site for SETD2-depleted (n = 19) and GL2-depleted (n = 18) cells from four individual experiments. t80% denotes the time required to recover 80% of the initial fluorescence. I.F. indicates the percentage of the immobile fraction of bleached RNAPII–GFP molecules at the site of transcription. Error bars represent standard deviations.
Downregulation of SETD2 leads to intragenic transcription initiation in human cells
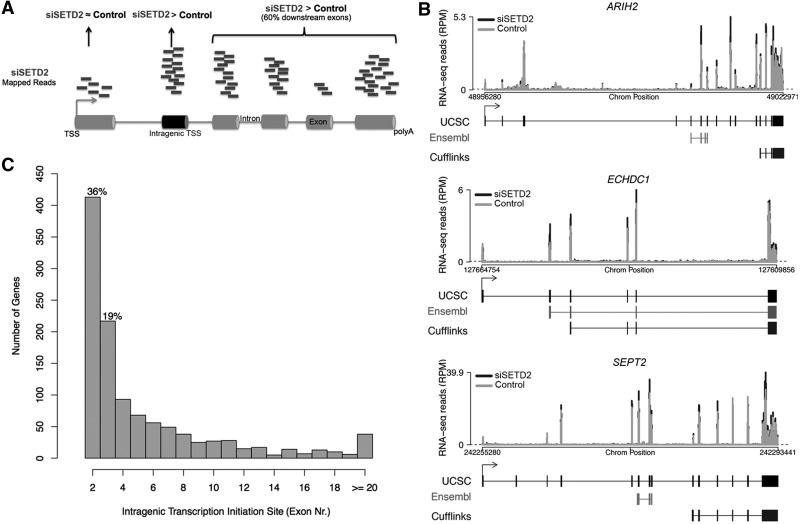
To obtain an estimate of how many transcripts are initiated at cryptic intragenic promoters in SETD2-depleted cells, we analyzed high-throughput cDNA sequencing data from human mesenchymal stem cells transfected with control and targeting siRNAs directed against SETD2 (5). To identify genes with intragenic initiation, we compared exonic read counts in depleted and control cells (Figure 3A). First, we selected all transcripts with higher read counts for at least one exon in SETD2-depleted cells; next, we discarded transcripts with higher read counts for the first annotated exon (likely corresponding to upregulated full-length mRNAs), and transcripts with higher read counts for isolated internal exons (possibly corresponding to alternative splicing events). Poly(A)+ spliced isoforms were reconstructed using algorithms (TopHat and Cufflinks), which do not depend on gene annotation (28,30). Reconstructed isoforms are depicted in Figure 3B. Our analysis shows that downregulation of SETD2 by RNAi leads to increased levels of transcripts that are normally spliced and polyadenylated but start downstream of the first annotated exon (Figure 3B). We found evidence for intragenic transcription initiation in 1139 genes among 10 772 active protein-coding loci (11%). A list of these genes is shown on Supplementary Table S3. Our analysis further revealed that most (55%) intragenic transcriptions in SETD2-depleted cells are initiated at the second or third expressed exons (Figure 3C), and at least 13% start at annotated alternative promoters (Supplementary Table S3). Taking into account that alternative transcription is emerging as a major contributor to transcriptome diversity (38), this observation raises the possibility that H3K36me3 may play a regulatory role in modulating alternative promoter usage.
Figure 3.
Genome-wide analysis of intragenic transcription initiation induced by SETD2 depletion. (A) Schematic representation describing the constraints for identification of genes with intragenic transcription initiation in SETD2-depleted cells. Genes showing significantly higher read counts for exons downstream of the first annotated exon in SETD2-depleted cells were selected. The intragenic transcription initiation site is defined as the first exon with significantly higher read counts for which at least 60% of the downstream exons also show significantly higher read counts. (B) RNA-seq profiles for the genes ARIH2, ECHDC1 and SEPT2 showing intragenic transcription initiation on different internal exons after SETD2 knockdown (black) relative to control (gray). Dashed line represents the minimum threshold 5 reads/100 bp (normalized reads per million). For each gene, the UCSC transcript annotations compared with annotated alternative promoter from Ensembl and Cufflinks transcript reconstruction are shown below the graph. Boxes represent exons, separated by introns shown as solid lines (C) Histogram of intragenic transcription initiation site location (exon number).
SETD2-dependent recruitment of FACT and nucleosome organization
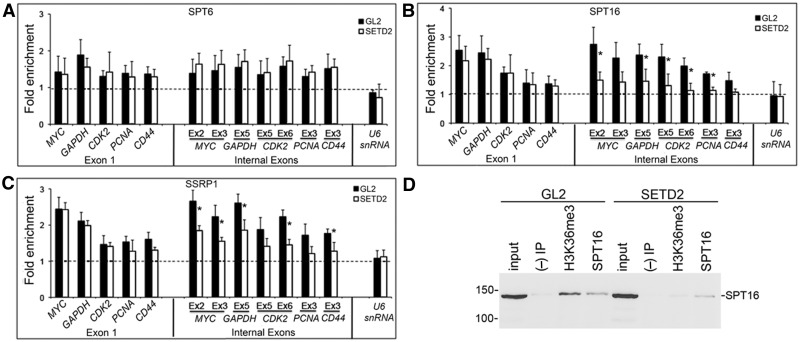
Previous studies in yeast have shown that, in addition to Set2 histone methyltransferase, factors important to restore normal chromatin structure on transcriptional elongation such as FACT and Spt6 are crucial to repress transcription from cryptic promoters located within protein-coding genes (11,12,22–24). To determine whether these different factors act through a common repression mechanism in human cells, we reduced the levels of SETD2 in HeLa cells by RNAi, and analyzed by ChIP the occupancy of SPT6 and FACT subunits (SPT16 and SSRP1) along five randomly selected genes (Figure 4). Relative to control (GL2-treated) cells, the density of SPT6 across the analyzed genes is not affected by SETD2 downregulation (Figure 4A). In contrast, significantly lower levels of FACT subunits SPT16 and SSRP1 are detected over internal exons in SETD2-depleted cells (Figure 4B and C). No significant differences occur over first exons, consistent with our previous observation that H3K36me3 levels are low in this region (6). As a control, we analyzed the human U6 snRNA gene set, which is transcribed by RNA polymerase III (39) and lacks enrichment for H3K36me3 (40). Lack of H3K36me3 marking in the U6 snRNA gene is consistent with the finding that in yeast, the H3K36 methyltransferase Set2 binds directly to the phosphorylated C-terminal domain of RNAPII (14,41,42), a structure that is absent from RNA polymerase III. As expected, the density of FACT subunits SPT16 and SSRP1 on the U6 snRNA gene was not altered by SETD2 downregulation (Figure 4B and C). A similar reduction in the recruitment of FACT to the gene body after SETD2 depletion was observed in HEK 293T cells (Supplementary Figure S2B and C).
Figure 4.
SETD2 is required for FACT recruitment. HeLa cells were transfected with siRNAs targeting GL2 or SETD2. (A–C) ChIP analysis was carried out using antibodies against SPT6 (A), SPT16 (B) and SSRP1 (C). For each antibody, the ChIP signals were normalized against the ChIP signal of a non-transcribed intergenic region. Graphs depict mean and standard deviation from at least four independent experiments. Two SETD2 siRNA duplexes were used randomly on each individual RNAi experiment. Statistically significant changes between GL2- and SETD2-depleted cells are indicated (*P < 0.05). (D) Co-IP of endogenous H3K36me3 and SPT16 on control (GL2) or SETD2-depleted HeLa cells. H3K36me3 and SPT16 were immunoprecipitated from HeLa cells with specific antibodies. ‘Input’ represents the total cell lysate, and the ‘(−) IP’ is a negative control obtained from a beads-only immunoprecipitation. The pulled-down complexes were subjected to western blot with an antibody against SPT16. Molecular weight markers are shown on the left. Three experiments with similar results were performed.
The impaired FACT recruitment to transcribed chromatin templates in SETD2-depleted cells is most likely due to the absence of H3K36me3. To address this possibility, we performed co-immunoprecipitation (co-IP) of endogenous SPT16 and H3K36me3 in HeLa cells (Figure 4D). This experiment revealed that SPT16 is promptly detected on immune complexes pulled down with an antibody against H3K36me3 (Figure 4D). To confirm the specificity of this interaction, we repeated the experiment in SETD2-depleted cells, which display only residual levels of H3K36me3 despite having high RNAPII occupancy (Figure 1 and Supplementary Figure S1). As expected, the co-IP of SPT16 and H3K36me3 was lost on SETD2 depletion (Figure 4D). These data provide additional support to the hypothesis that H3K36me3 acts as a signal that directs FACT to the nucleosomes positioned on transcribed chromatin templates.
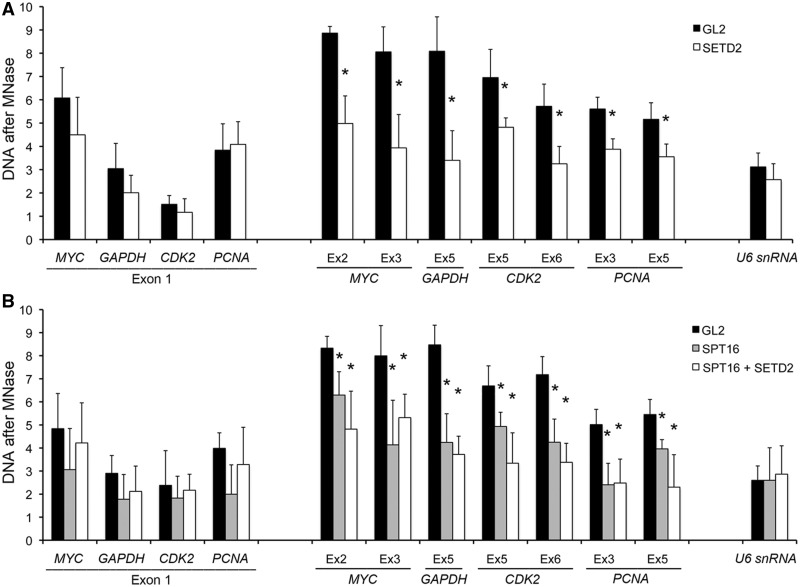
Given that FACT is critical for nucleosome reorganization after transcriptional elongation in actively transcribed genes (43), we predicted that the reduced levels of SPT16 and SSRP1 observed in active genes after SETD2 downregulation should affect nucleosome density. To test this view, chromatin was digested with MNase and mononucleosome-sized DNA was isolated (Supplementary Figure S3A). Nucleosome density at the selected loci was then estimated by RT-qPCR analysis (32). As expected, SETD2-depleted cells have a significantly lower density of nucleosomes across internal exons, whereas no significant changes are detected in first exons or in the U6 snRNA gene (Figure 5A). Taken together, these results suggest that the high levels of H3K36 trimethylation characteristic of actively transcribed genes enhance the recruitment of the FACT complex, thereby maintaining a high density of nucleosomes on these genes. To further address this possibility, we examined the nucleosome occupancy of chromatin extracted from HeLa cells depleted of SPT16 alone or SPT16 and SETD2 together (Figure 5B). The high efficiency (>90%) of the SPT16 and SETD2 knockdown was confirmed by western blot and RT-qPCR (Supplementary Figures S1E, F and 3B). When compared with control (GL2 RNAi) cells, the MNase sensitivity pattern of the SPT16-depleted cells revealed lower nucleosome occupancy on the body of the selected genes (Figure 5B). These data are consistent with the previous finding that mutation of FACT results in histone depletion over coding regions of yeast genes (44). Our results further showed that nucleosomes positioned over first exons or over the U6 snRNA gene are less sensitive to SPT16 depletion (Figure 5B). One possibility is that additional histone chaperones replace FACT-mediated nucleosome assembly on the 5′ regions of transcribed genes. Importantly, the increased sensitivity of coding regions to MNase digestion on SPT16 depletion was not further enhanced by the additional loss of SETD2 (Figure 5B). This suggests that FACT and H3K36me3 cooperate to maintain chromatin structure at transcriptionally active genes.
Figure 5.
SETD2 regulates nucleosome organization. Nucleosome occupancy on GL2- and SETD2-depleted (A) or GL2-, SPT16- and SPT16+SETD2-depleted (B) cells was estimated by the amount of MNase-resistant DNA after 20-min digestion. The graphs show the amount of DNA recovered after 20-min digestion with MNase relative to the amount of DNA present in undigested samples. The data were normalized against the nucleosome occupancy of a non-transcribed intergenic region. Means and standard deviations from at least three independent experiments are shown. Statistically significant changes between GL2- and SETD2-, SPT16-depleted or SETD2+SPT16-depleted cells are indicated (*P < 0.05).
SETD2-dependent nucleosome reorganization during transcription
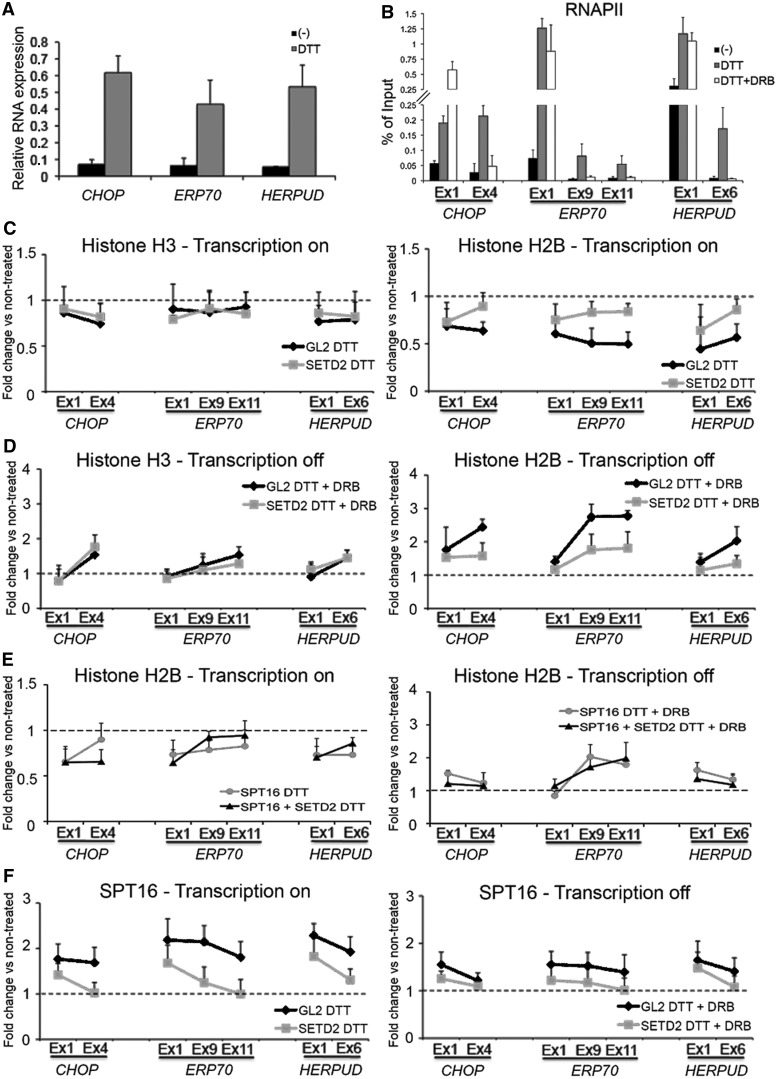
The finding that actively transcribed genes are densely packed with nucleosomes appears counter-intuitive. Yet, a recent study reported compelling evidence suggesting that transcription elongation is facilitated by higher chromatin compaction (45). To further address this issue, we monitored nucleosome organization after transcriptional activation. We took advantage of the gene expression program activated during the UPR, a cellular reaction triggered by misfolded proteins in the endoplasmic reticulum (46). During the UPR, which can be induced by incubating cells with the reducing agent DTT, the transcription of several genes, including CHOP, ERP70 and HERPUD, is rapidly stimulated (46,47). Accordingly, after incubation of HeLa cells with DTT for 1 h, we detect higher mRNA levels for CHOP, ERP70 and HERPUD nascent transcripts (Figure 6A). Increased RNAPII occupancy and H3K36me3 levels along these genes after DTT treatment are also consistent with transcriptional activation (Figure 6B and Supplementary Figure S4A). Transcriptional activation induced by DTT did not significantly alter the distribution of histone H3 along UPR-responsive genes (Figure 6C); similarly, downregulation of SETD2 had no significant effect on histone H3 occupancy (Figure 6C). In contrast, the levels of histone H2B were significantly reduced in DTT-treated cells compared with non-treated cells (Figure 6C). Notably, after SETD2 downregulation, histone H2B levels on DTT treatment did not decrease as sharply as in control cells (Figure 6C). This difference was particularly noticeable on the internal exons, where the decrease in H2B occupancy was statistically significant (P < 0.05) in control cells only. Taking into account that SETD2 downregulation reduces FACT loading onto the gene body (Figure 4B and C), and that FACT is involved in the exchange of histone H2A–H2B dimers (48), our results suggest that the high levels of H3K36me3 characteristic of actively transcribed genes play a role in facilitating transcription elongation by recruiting FACT. FACT recruitment would displace H2A–H2B dimers in the first instance to allow passage of RNAPII through nucleosomes. Later, in the wake of RNAPII elongation, FACT promotes the reassembly of the displaced H2A–H2B dimers, thereby restoring the original chromatin structure, as revealed by the MNase sensitivity assays (Figure 5). To further test this view, we treated cells with the transcription elongation inhibitor DRB to investigate the reassembly of the nucleosomal histones after passage of RNAPII. Within 30 min after addition of DRB to HeLa cells that had been incubated for 1 h in the presence of DTT, RNAPII occupancy along the body of UPR-responsive genes decreased. The reduction was most evident toward the 3′ end of the genes, whereas at the 5′ end, RNAPII occupancy remained unaltered or even higher (Figure 6B). This is consistent with the inability of RNAPII to escape the promoter-proximal pause and resume elongation in the presence of DRB (49,50). ChIP analysis of histones H3 and H2B along the UPR-responsive genes in cells that were incubated with DTT for 1 h and then treated with DRB for 30 min reveals an accumulation of both histones in the analyzed genes (Figure 6D). This accumulation was significant (P < 0.05) for histone H2B positioned on internal exons, and was impaired by SETD2 depletion (Figure 6D). In contrast, downregulation of SETD2 had negligible effects on the accumulation of histone H3 after DRB treatment (Figure 6D). These data support the view that SETD2 activity is crucial to destabilize histone H2A–H2B dimers when transcription is activated, and to reassemble nucleosomes when transcription is inhibited. To investigate whether this dual role involves the histone chaperone FACT, we measured the levels of histone H2B exchange during transcription in SPT16-depleted cells and in cells depleted of both SPT16 and SETD2 (Figure 6E). On transcription activation with DTT, the reduction of histone H2B levels in SPT16-depleted cells (Figure 6E) was comparable with the reduction observed in SETD2-depleted cells (Figure 6C). Next and more importantly, the combined depletion of SPT16 and SETD2 did not alter the displacement level of histone H2B on transcription activation (Figure 6E). Similarly, the reassembly of histone H2B in SPT16-depleted cells on DRB addition did not differ from the level observed in the absence of SETD2 (compare Figure 6E and D). Moreover, histone H2B reassembly in SPT16-depleted cells was not affected by the additional knockdown of SETD2 (Figure 6E). These data suggest that H3K36me3 and FACT combine to regulate nucleosome dynamics during transcription. In this sense, we then investigated the transcription-dependent recruitment of SPT16 to the chromatin of SETD2-depleted cells (Figure 6F). In the UPR-responsive genes, SPT16 was recruited to chromatin in a transcription-dependent manner: its levels increased significantly (P < 0.05) on transcription activation with DTT and decreased after DRB addition (Figure 6F). The recruitment of SPT16 after DTT addition is also H3K36me3-dependent because in SETD2-depleted cells, the levels of this FACT subunit in the internal exons did not increase significantly (P > 0.05) relatively to control (no DTT treatment) cells (Figure 6F and Supplementary Figure S4B). We conclude that FACT is recruited to the chromatin templates in a SETD2-dependent manner most likely through recognition of H3K36me3-tagged nucleosomes. This mechanism is crucial to regulate nucleosome dynamics during transcription by promoting the exchange of histone H2A–H2B dimers and, more importantly, to restore the chromatin structure in the wake of RNAPII elongation.
Figure 6.
Transcription affects nucleosome organization in a SETD2-dependent manner. Transcription of CHOP, ERP70 and HERPUD genes was stimulated by treating HeLa cells with 2 mM DTT for 1 h; as a control, cells were left untreated (−). To inhibit transcription, cells were first treated with 2 mM DTT for 1 h and then incubated with 75 μM DRB for 30 min (DTT + DRB). Cells were transfected with siRNAs targeting GL2, SETD2, SPT16 or SETD2 + SPT16 as indicated. Means and standard deviations from at least four independent experiments are shown. (A) Cells were fractionated into cytoplasmic, nucleoplasmic and chromatin fractions; RNA associated with chromatin was isolated; and RT-qPCR was carried out with primers to detect nascent RNA from the indicated genes. The amount of PCR product estimated by RT-qPCR was normalized to the levels of U6 snRNA. (B) ChIP analysis with antibodies to RNAPII. Data are shown as percentage of the input chromatin. (C and D) ChIP analysis with antibodies to total histone H3 and H2B when transcription is either active or inhibited (+DRB). The decrease in H2B occupancy on DTT treatment (C) and its accumulation after DRB addition (D) was statistically significant (P < 0.05) in control (GL2) cells, on the internal exons of the three genes only. (E) ChIP analysis of histone H2B when transcription is either active or inhibited (+DRB) in cells depleted of SPT16 or SPT16 + SETD2. (F) ChIP analysis with antibodies to SPT16. Recruitment of SPT16 to the chromatin templates of the three genes after transcription activation with DTT was significant (P < 0.05) in control cells. In SETD2-depleted cells, the recruitment of SPT16 to the internal exons of the three genes on addition of DTT was significantly impaired. Data shown in (C), (D), (E) and (F) are represented as fold change over the corresponding ChIP signal of cells that were not treated with DTT.
DISCUSSION
In this study, we show that trimethylation of H3K36 by SETD2, a hallmark of active genes, plays a critical role in restoring normal chromatin structure after disruption on passage of RNAPII. As shown previously in yeast, restoring chromatin structure behind RNAPII is important for the fidelity of transcription initiation. In mutant strains defective for Spt6, Spt16 and Set2, aberrant transcripts are initiated at cryptic promoter-like sequences within the body of several genes (23,34,51–53). Cryptic transcription was reported to be widespread, occurring in ∼1000 genes (17% of all genes) across the Saccharomyces cerevisiae genome (22). A large number of factors that control chromatin structure and transcription have been identified as required to repress cryptic intragenic transcription in yeast (22). Here, we have performed a comprehensive genome-wide analysis of cryptic transcription induced by downregulation of SETD2 in human cells and we found evidence for poly(A)+ transcripts initiated from intragenic sites in at least 1139 genes, corresponding to 11% of all active genes. We also observed increased RNAPII occupancy at internal coding regions of active genes, consistent with internal initiation induced by SETD2 depletion at sites downstream the canonical promoter (Figure 1D). Live-cell imaging further showed that the turnover of RNAPII at the transcription site is faster in SETD2-depleted cells (Figure 2), suggesting that at any given time, there are more RNAPII molecules engaging on transcription (owing to intragenic initiation), and that on average, RNAPII molecules spend less time associated with the template, probably reflecting aberrant transcription.
Whether nucleosomes are restored after passage of RNAPII by depositing new histones or transferring the original histones remains unclear. Our observation that on transcriptional activation the density of H2B is reduced while H3 occupancy remains unaltered is consistent with the model that nucleosomes do not completely disassemble during transcription but only H2A–H2B dimers are removed (54,55). In particular, FACT and SPT6 are thought to serve as histone chaperones facilitating passage of the elongation machinery through nucleosomes. Unexpectedly, we found that histone exchange induced by transcriptional activation is SETD2 dependent. Downregulation of SETD2 leads to reduced H3K36me3 levels and results in lower density of FACT subunits SPT16 and SSRP1, but not SPT6, on active genes. Together with our finding that SPT16 co-precipitates with H3K36me3, these data raise the possibility that H3K36me3 serves as a docking site for nucleosome binding of FACT. In support of this model, a number of previous studies showed that FACT activity is influenced by several post-translational modifications of histone tails such as ubiquitylation, which plays a role in nucleosome dynamics during transcription (56), or methylation (21). Very recently, the Workman laboratory showed that in yeast, H3K36 methylation impacts on the interaction of histone H3 with histone chaperones suppressing histone exchange over coding regions, thereby preventing spurious cryptic transcripts from initiating within coding regions (57). The same group reported that H3K36me2 binds Spt16 more tightly than H3K36me3; yet, Spt16 was amongst the histone chaperones found to associate with H3K36me3-containing mononucleosomes (58). Importantly, yeast Setd2 adds both di- and tri-methyl marks to H3K36 (59–61), while human SETD2 is responsible for trimethylation of H3K36 only (18). Therefore, it is possible that Setd2 regulates Spt16 recruitment in yeast by adjusting the relative levels of H3K36me2 and H3K36me3 on transcribed genes. In contrast, SETD2 activity in human cells results in augmented levels of H3K36me3 and enhanced recruitment of FACT.
The biological implications of the findings reported here are vast. For instance, as a mechanism that impacts on gene expression, intragenic transcription initiation has the potential to generate transcriptome diversity, which might potentiate diseases such as cancer. Indeed, current genome-wide research tools have revealed the presence of SETD2 mutations in an increasing number of cancers, leading to the classification of SETD2 as a novel tumor suppressor (62–65). Although the tumor suppressor mechanism on which SETD2 impinges is not yet known, its role as a guardian of the genome integrity to prevent intragenic transcription initiation warrants further investigation in the context of cancer.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–4.
FUNDING
Fundação para a Ciência e Tecnologia (FCT), Portugal [PTDC-BIA-BCM-111451-2009 to S.F.d.A.] and [PTDC-SAU-GMG-113440-2009 to M.C.-F.]; Fellowships from FCT [SFRH-BPD-34679-2007 and SFRH-BPD-62911-2009 to S.F.d.A. and A.R.G.]; RNPnet, a Marie Curie Network for Initial Training [PITN-GA-2011-289007 to S.C.S.]. Funding for open access charge: Fundação para a Ciência e Tecnologia (FCT), Portugal [PTDC-BIA-BCM-111451-2009 to S.F.d.A.].
Conflict of interest statement: None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Nuno Barbosa-Morais for critical comments on the bioinformatics analysis.
REFERENCES
- 1.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc. Natl Acad. Sci. USA. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Almeida SF, Carmo-Fonseca M. Design principles of interconnections between chromatin and pre-mRNA splicing. Trends Biochem. Sci. 2012;37:248–253. doi: 10.1016/j.tibs.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, Zhang QH, Chen SJ, Huang QH, Chen Z. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J. Biol. Chem. 2005;280:35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 11.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 12.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 13.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 14.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lickwar CR, Rao B, Shabalin AA, Nobel AB, Strahl BD, Lieb JD. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One. 2009;4:e4886. doi: 10.1371/journal.pone.0004886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 21.Formosa T. The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta. 2012;1819:247–255. doi: 10.1016/j.bbagrm.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 24.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 25.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Almeida SF, Garcia-Sacristan A, Custodio N, Carmo-Fonseca M. A link between nuclear RNA surveillance, the human exosome and RNA polymerase II transcriptional termination. Nucleic Acids Res. 2010;38:8015–8026. doi: 10.1093/nar/gkq703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins SB, Rino J, Carvalho T, Carvalho C, Yoshida M, Klose JM, de Almeida SF, Carmo-Fonseca M. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat. Struct. Mol. Biol. 2011;18:1115–1123. doi: 10.1038/nsmb.2124. [DOI] [PubMed] [Google Scholar]
- 28.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carey M, Smale ST. Micrococcal Nuclease-Southern Blot Assay: I. MNase and Restriction Digestions. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4890. pdb.prot4890. [DOI] [PubMed] [Google Scholar]
- 32.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 34.Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 36.Chapman RD, Conrad M, Eick D. Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol. Cell Biol. 2005;25:7665–7674. doi: 10.1128/MCB.25.17.7665-7674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen VT, Giannoni F, Dubois MF, Seo SJ, Vigneron M, Kedinger C, Bensaude O. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 1996;24:2924–2929. doi: 10.1093/nar/24.15.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, Dahmane N, Davuluri RV. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 2011;21:1260–1272. doi: 10.1101/gr.120535.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egloff S, O'Reilly D, Murphy S. Expression of human snRNA genes from beginning to end. Biochem. Soc. Trans. 2008;36:590–594. doi: 10.1042/BST0360590. [DOI] [PubMed] [Google Scholar]
- 40.Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat. Struct. Mol. Biol. 2010;17:629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc. Natl Acad. Sci. USA. 2005;102:17636–17641. doi: 10.1073/pnas.0506350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J. Biol. Chem. 2006;281:13–15. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- 43.Winkler DD, Luger K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011;286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamai A, Puglisi A, Strubin M. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell. 2009;35:377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Vaillant C, Palmeira L, Chevereau G, Audit B, d'Aubenton-Carafa Y, Thermes C, Arneodo A. A novel strategy of transcription regulation by intragenic nucleosome ordering. Genome Res. 2010;20:59–67. doi: 10.1101/gr.096644.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 47.Kokame K, Agarwala KL, Kato H, Miyata T. Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. J. Biol. Chem. 2000;275:32846–32853. doi: 10.1074/jbc.M002063200. [DOI] [PubMed] [Google Scholar]
- 48.Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell. 2009;35:365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 50.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 52.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, Reinberg D, Lis JT. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- 54.Campos EI, Reinberg D. Histones: annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 55.Avvakumov N, Nourani A, Cote J. Histone chaperones: modulators of chromatin marks. Mol. Cell. 2011;41:502–514. doi: 10.1016/j.molcel.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 56.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 57.Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 58.Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 60.Li B, Howe L, Anderson S, Yates JR, III, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 61.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, Kok K. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 64.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Al Sarakbi W, Sasi W, Jiang WG, Roberts T, Newbold RF, Mokbel K. The mRNA expression of SETD2 in human breast cancer: correlation with clinico-pathological parameters. BMC Cancer. 2009;9:290. doi: 10.1186/1471-2407-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.