Abstract
Ribosome biogenesis GTPase A protein (RbgA) is an essential GTPase required for the biogenesis of the 50S subunit in Bacillus subtilis. Homologs of RbgA are widely distributed in bacteria and eukaryotes and are implicated in ribosome assembly in the mitochondria, chloroplast and cytoplasm. Cells depleted of RbgA accumulate an immature large subunit that is missing key ribosomal proteins. RbgA, unlike many members of the Ras superfamily of GTPases, lacks a defined catalytic residue for carrying out guanosine triphosphate (GTP) hydrolysis. To probe RbgA function in ribosome assembly, we used a combined bioinformatics, genetic and biochemical approach. We identified a RNA-binding domain within the C-terminus of RbgA that is structurally similar to AmiR–NasR Transcription Anti-termination Regulator (ANTAR) domains, which are known to bind structured RNA. Mutation of key residues in the ANTAR domain altered RbgA association with the ribosome. We identified a putative catalytic residue within a highly conserved region of RbgA, His9, which is contained within a similar PGH motif found in elongation factor Tu (EF-Tu) that is required for GTP hydrolysis on interaction with the ribosome. Finally, our results support a model in which the GTPase activity of RbgA directly participates in the maturation of the large subunit rather than solely promoting dissociation of RbgA from the 50S subunit.
INTRODUCTION
Ribosomes are complex molecular machines that, in bacteria, contain three RNA molecules and >50 proteins (1–4). The accurate and timely assembly of ribosomes is critical for organisms to achieve the fast growth rates observed (4). Although active ribosomes can be generated in vitro without the need for additional assembly factors, non-physiological temperatures and salt conditions are required (1,2,5). In light of this observation, Nomura and co-workers suggested that ribosome assembly factors would be required in vivo at key steps that required these non-physiological conditions in vitro. However, these assembly factors have remained elusive in bacteria (4,6). Over the past decade, studies have shown that several guanosine triphosphatases (GTPases) likely play a critical role in ribosome assembly (RA-GTPases) in both bacteria and eukaryotes (6–8). However, the precise molecular function carried out by RA-GTPases during in vivo ribosome assembly remains unknown.
Ribosome biogenesis GTPase A (RbgA) is an essential widely conserved GTPase that is required for the assembly of the 50S subunit in Bacillus subtilis (9–12). RbgA homologs are widely distributed evolutionarily, and eukaryotic homologs of RbgA such as Mtg1 (mitochondria), Lsg1, Nug1 and Nog2 have been implicated in ribosome assembly (13–16). RbgA proteins have several unique features that distinguish them from conventional Ras-like GTPases. First, RbgA belongs to a family of circularly permuted GTPases (cpGTPases) in which the order of the highly conserved G motifs (G1–G4) is altered such that the G4 motif precedes the G1 motif in the sequence of the protein (17). Although the reason for this circular permutation of the G domain is unclear, in almost every case, cpGTPases contain a second protein domain directly downstream of the Switch II (G3) region. This would allow guanine nucleotide–dependent movement of this second protein domain, which is likely involved in binding to RNA or interacting with ribonucleoprotein structures (17). Second, RbgA and its homologs are also classified as hydrophobic amino acid–substituted GTPases (HAS-GTPases) (18). HAS-GTPases are involved in a wide variety of cellular functions such as ribosome biogenesis, tRNA modification and cell cycle regulation (18). In this family of GTPases, the catalytic glutamine that is usually present in Ras-like GTPases is replaced by a hydrophobic amino acid. Several prokaryotic GTPases such as FeoB, MnmE, YphC YqeH, Obg and Era fall into this family (19–25). These GTPases use alternative mechanisms for guanosine triphosphate (GTP) hydrolysis that have been deduced with the help of crystal structures in two cases, MnmE and FeoB, but not for RbgA (26,27).
Cells depleted of RbgA do not form mature 50S subunits but rather accumulate a novel 45S complex lacking three ribosomal proteins—L16, L27 and L36 (9,11). In yeast, the RbgA homolog Lsg1 is proposed to participate in the association of Rpl10p (L16 homolog) with the 60S subunit at a late stage during large-subunit biogenesis (14). Incorporation of L16 is also predicted to occur at a late stage of 50S assembly and is accompanied by a large conformational change in vitro (28), suggesting that RbgA proteins regulate an evolutionarily conserved step in ribosome assembly. The GTPase activity of RbgA is stimulated ∼60-fold by 50S subunits, and physical association with 50S subunits is maximal in the presence of a non-hydrolyzable analog of GTP (12), indicating that GTP hydrolysis is responsible for dissociating RbgA from the ribosome. Interaction with the 45S yields only a mild increase in GTPase activity, and the association of RbgA with 45S subunits is equal in the presence of GTP or a non-hydrolyzable analog of GTP (12). These and other observations have led to a model in which RbgA functions by interacting with the large ribosomal subunit before the incorporation of L16 and either directly recruits L16 to the ribosome complex or indirectly facilitates L16 binding by remodeling the ribosome (12). The molecular basis of RbgA-mediated ribosome assembly, in particular the role of specific amino acid residues, is poorly understood.
To elucidate how RbgA participates in ribosome assembly, we used a site-directed mutagenesis approach combined with in vivo and biochemical characterization of RbgA mutants. We have discovered a RNA-binding motif that mediates, in part, RbgA interaction with the 50S subunit. We also identified a potential catalytic residue that mediates GTP hydrolysis. We discuss our findings in relation to other ribosome-associated GTPases and suggest that future studies of RbgA in bacteria will also provide additional insight into mitochondrial and eukaryotic ribosome assembly.
MATERIALS AND METHODS
Growth conditions
All strains were grown at 37°C in LB (lysogeny broth) medium. Antibiotics were added at the following concentrations, when required: chloramphenicol (5 µg/ml), kanamycin (25 µg/ml), spectinomycin (100 µg/ml) and ampicillin (100 µg/ml). In addition, IPTG was added at a concentration of 1 mM, unless mentioned otherwise, and xylose was added at a concentration of 2%, when needed.
Construction of strains
All strains used in this study were derived from wild-type B. subtilis strain JH642 (RB247). Strain RB301 was constructed as described (9). Strain RB562 was constructed by switching the antibiotic resistance cassette linked to Pspank and rbgA in RB301 from chloramphenicol to spectinomycin. RB611 was constructed by transforming RB562 with plasmid pMAP65 (29) that overexpresses lacI repressor to control the expression of wild-type rbgA gene under the control of Pspank promoter. Strain RB613 was constructed by cloning wild-type rbgA into the pSWEET plasmid (30) with a chloramphenicol resistance cassette and rbgA under the control of the PxylA promoter. This construct was linearized and inserted at the amyE locus in RB611, creating strain RB613. Chloramphenicol-resistant colonies were confirmed to have lost the ability to degrade starch, indicating disruption of the amyE gene. The desired mutations were introduced into the pSWEET plasmid using the QuikChange IIXL kit (Stratagene) by following the manufacturer’s instructions. The resultant plasmids with mutated rbgA gene were linearized and transformed in RB611 strain. Resultant strains are dependent on isopropyl β-D-1-thiogalactopyranoside (IPTG) for growth and are listed in Supplementary Table S1. Escherichia coli BL21(DE3) cells transformed with plasmid pET21b containing full-length rbgA placed under the control of the T7 promoter were used to overexpress RbgA proteins (9). Desired mutations were constructed using the QuikChange IIXL kit (Stratagene) by following the manufacturer’s instructions.
Characterization of association between RbgA or mutants and 45S or 50S complexes
Wild-type and mutant proteins were purified as described (12). 50S subunits and 45S intermediates were prepared by sucrose density gradient (12), and 50S and 45S complexes were isolated from lysates of RB418 and RB301 cells, respectively. The cells were grown to OD600 of 0.5 at 37°C in LB medium with or without IPTG. RB301 and RB418 cells were grown without IPTG for several generations to deplete the cells of RbgA and IF-2, respectively, until a doubling time of ∼150 min was reached. Chloramphenicol (Sigma) was added to a final concentration of 100 μg/ml 5 min before harvesting; 50S subunits and 45S complexes were isolated as described (12). The association assay was performed with varying levels of RbgA protein to 45S or 50S to determine the linear range of the assay. At each ratio, the nucleotide concentration was 400 µM, which is well above the Km of RbgA to ensure maximal RbgA protein was bound to the nucleotide. All assays reported here were performed at 37°C with 50 nM RbgA or mutant protein, 10 nM 45S or 50S and 400 µM nucleotide [guanosine diphosphate (GDP), GTP or Guanosine 5′-(β,γ-imido) triphosphate GMP-PNP]. The binding protocol was used as described (12). Briefly, 60 pmol of RbgA was pre-incubated with 1.5 mM of different guanine nucleotides for 15 min followed by addition of 10 pmol purified subunits. The binding was allowed to proceed for 15 min at 37°C, and then centrifugation was performed once through Microcon 100 filters (Millipore) with a cutoff of 100 kDa. The RbgA–subunit complexes were washed once with buffer A and twice with buffer C before elution. Four controls were performed for every assay—45S/50S with no RbgA/mutant protein and RbgA/mutant protein with each of the three nucleotides but no subunit. This was to ensure that no RbgA was bound to the subunit while preparation of subunits and RbgA/mutant protein was not retained on the filter in the absence of subunit. The subunit-bound RbgA was detected by separating the complex on sodium dodecyl sulphate–polyacrylamide gel electrophoresis gels (12% Bis-Tris NuPAGE gel, Invitrogen) followed by western blot analysis using polyclonal rabbit anti-RbgA and horseradish peroxidase-conjugated goat anti-rabbit polyclonal antibodies (PerkinElmer Life Sciences). The bands were visualized using Western Lightening chemiluminescent detection system (PerkinElmer Life Sciences). A series of incremental exposures was obtained to determine that chemiluminescent signal was in the linear range of detection. A single time point from the linear range was chosen, and mutant and wild-type protein, present on the same blot membrane, was quantified using FujiFilm Multi Gauge V3.0 software. The binding level of RbgA/mutant protein was determined, and the data are reported as a percentage of wild-type RbgA association with either the 45S or the 50S subunit in the presence of GMP-PNP, which displays maximal binding.
Characterization of GTPase activity of RbgA/mutants
The assay was performed as described (12). Briefly, for measuring GTPase activity in the presence of 50S subunits, 100 nM RbgA/mutant protein was incubated with 100 nM 50S subunit and 200 µM GTP at 37°C for 30 min, and for measuring intrinsic GTPase activity, 2 µM RbgA/mutant protein was incubated with 200 µM GTP at 37°C for 30 min. We predetermined that under these conditions, the values were in the linear range of the assay. The phosphate released was measured by malachite green colorimetric system (BioAssay systems). The mutant proteins were assayed on the same 96-well plate (Corning) as the wild-type protein.
Bioinformatics analysis and superimposition
Coordinates from the C-terminus of RbgA were used to search for similar structures using the DALI server (http://ekhidna.biocenter.helsinki.fi/dali_server/) (31), and resulting structures were superimposed using PyMol (32). Superimposition of free EF-Tu (pdb id: 1EFT) and ribosome-bound EF-Tu (pdb id:3FIC) was superimposed on RbgA (pdb id:1PUJ) using ligalign script (33) in PyMol. All the figures were made in Chimera (34).
RESULTS
Multiple sequence alignment of RbgA homologs revealed four regions of high conservation
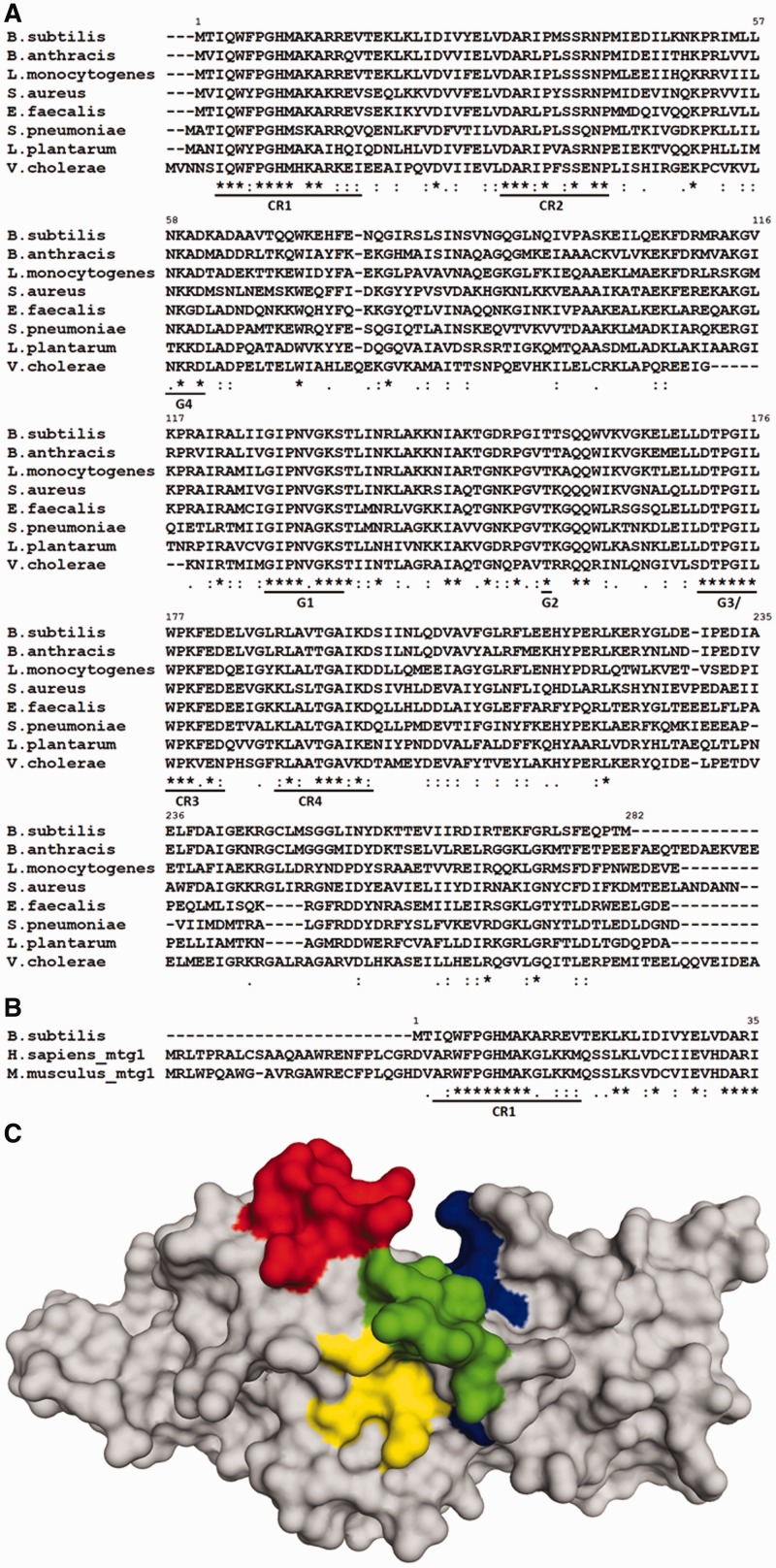
RbgA contains two structural domains, an N-terminal circularly permuted G-domain and a C-terminal domain that contains no sequence similarity to any known functional protein domain. To identify key residues within conserved regions in RbgA, we generated a multiple sequence alignment of bacterial RbgA homologs using CLUSTAL-W (Figure 1A). In addition to the universally conserved G-domain motifs G1–G4, we identified four regions of high conservation designated CR1–CR4 (Figure 1A). Although these conserved regions are dispersed throughout the protein sequence, they lie in close vicinity in the crystal structure of the protein (Figure 1C). The first two regions CR1 and CR2 are contained in the N-terminal domain of RbgA and precede the circularly permuted G-domain. Of these two, CR1 was of particular interest, as this stretch of 15 amino acids found at the N-terminus of RbgA is largely conserved among all bacterial RbgA homologs as well as eukaryotic Mtg1 proteins (Figure 1B). Most of CR1 is unstructured in all available RbgA crystal structures deposited to date, indicating its dynamic nature and thus its role in RbgA function cannot be assessed by its structure. CR3 is a highly conserved loop that links the N-terminal domain to the C-terminal domain and contains the Switch II motif. Thus it is likely to participate in communication between the N and C terminal domains when bound to either GTP or GDP. The C-terminal domain following the CR3 showed low sequence similarity, and only one moderately conserved region (CR4) was identified in this domain.
Figure 1.
(A) Multiple sequence alignment of selected bacterial RbgA homologs. Highly conserved regions are underlined. G-domain motifs are indicated as G4 (NKXD), G1 (GXXXXGKS/T), G2 (T) and G3 (DXXG) in addition to four conserved regions (CR1-CR4). (B) Multiple sequence alignment of RbgA with eukaryotic homologs Mtg1 from humans and mouse highlighting conserved region CR1. Alignments were constructed with ClustalW with default parameters and species indicated to the left. ‘Asterisk’ indicates positions that have a single fully conserved residue; ‘colon’ indicates conservation between groups of strongly similar physicochemical properties; ‘dot’ indicates conservation between groups of weakly similar physicochemical properties. (C) Structure of RbgA depicting CR1, CR2, CR3 and CR4. Crystal structure of RbgA (1PUJ) depicting the three conserved regions. First nine amino acid residues are not structured in the crystal. CR1 (9–17) is shown in red, CR2 (30–42) is shown in yellow, CR3 (175–182) is shown in green and CR4 (188–197) is shown in blue. All four conserved regions are dispersed across the protein sequence and lie in close vicinity in the structure of the protein. The amino acid numbering of RbgA from B. subtilis is indicated in each alignment.
The C-terminal domain of RbgA contains an ANTAR RNA-binding domain
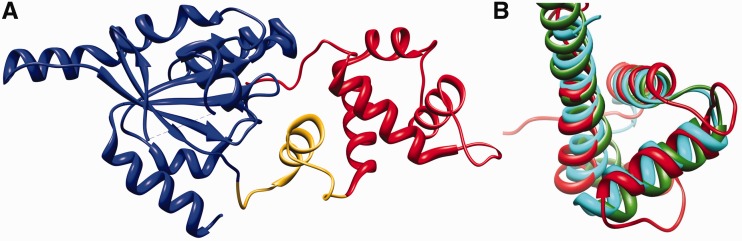
Because the C-terminal domain of RbgA contained low sequence similarity to proteins of known function in the NCBI database (other than RbgA homologs), we asked whether the C-terminal domain is structurally similar to any previously characterized domains. To identify proteins that contain a structurally similar domain, we used the structure of only the C-terminal domain (177–269 residues) of RbgA as a query to search the DALI database (31). The first four hits (Supplementary Table S2) were from crystal structures of either RbgA or its homologs, as expected (DALI Z-score = 7.3–14.8, r.m.s.d = 0–2.8 Å). The DALI score provides a measure of structural similarity between a given pair of protein domains, and structures that have significant similarities have a Z-score >2. The first non-RbgA hit was the C-terminal domain of the NasR, a protein that mediates transcription anti-termination (Z-score = 4.1, r.m.s.d. = 3.6 Å) (35). The next few hits were with incomplete domains of exodeoxyribonuclease and Cdc4, followed by other potential transcription anti-terminators, including AmiR (36). These results showed that the C-terminal domain of RbgA had significant structural similarity to the C-terminal domains of two proteins that are involved with promoting transcription anti-termination, AmiR and NasR (Supplementary Table S2). This C-terminal domain present in both AmiR and NasR interacts with structured RNA and encompasses a RNA-binding domain denoted the AmiR–NasR Transcription Anti-termination Regulator (ANTAR) domain (37). The ANTAR domain, characterized by a three-helix structure, is structurally similar to the C-terminal domain of RbgA (Figure 2) (37). Superimposition of the RbgA C-terminal structure on the C-terminal regions of NasR and AmiR shows a topologically similar three-helix arrangement, typical of ANTAR domains (r.m.s.d. = 3.6 and 3.4 Å, respectively) (Figure 2). It is noteworthy that the structural similarity notwithstanding, the sequence similarity of these proteins with the RbgA C-terminal domain is remarkably low (13% for NasR and15% for AmiR).
Figure 2.
C-terminal domain of RbgA contains RNA-binding domain ANTAR. (A) The crystal structure of RbgA (PDB: 1PUJ). The N-terminal G-domain is depicted in blue (9–176), the C-terminal domain is depicted in red (201–282) and a linker helix connecting the two domains is depicted in yellow (177–200). (B) The superposition of the ANTAR domains of RbgA (red), AmiR (green) and NasR (cyan).
Site-directed mutagenesis and phenotypic analysis of RbgA mutants
As rbgA is an essential gene, null mutations will not survive and cannot be studied. Therefore, we developed a strain that allowed mutations that negatively affect RbgA function to be generated and characterized. A conditional rbgA mutant strain was created by placing a wild-type copy of the rbgA at its native locus under the control of the Pspank promoter, yielding a strain that is dependent on IPTG for growth. Mutant versions of a second copy of the rbgA gene were placed under the control of the Pxyl promoter and introduced at the amyE locus. The expression of the mutant rbgA gene is therefore dependent on the presence of xylose in the growth media. To study mutations in rbgA, we constructed strains in the presence of IPTG and then interrogated their phenotype by growing cells in the presence of xylose and absence of IPTG. As a control, we created a strain (RB613) that contains a wild-type copy of the rbgA gene under the control of the Pxyl promoter at the amyE locus. As expected, RB613 grows equally well in the presence of 1 mM IPTG or 2% xylose (data not shown).
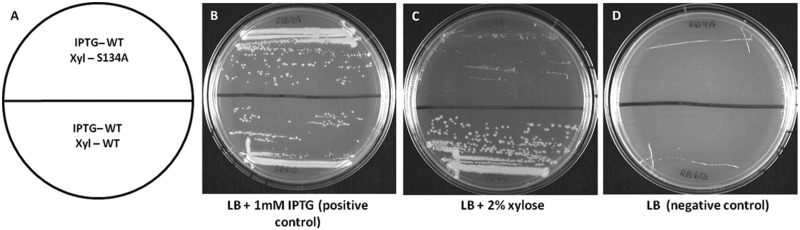
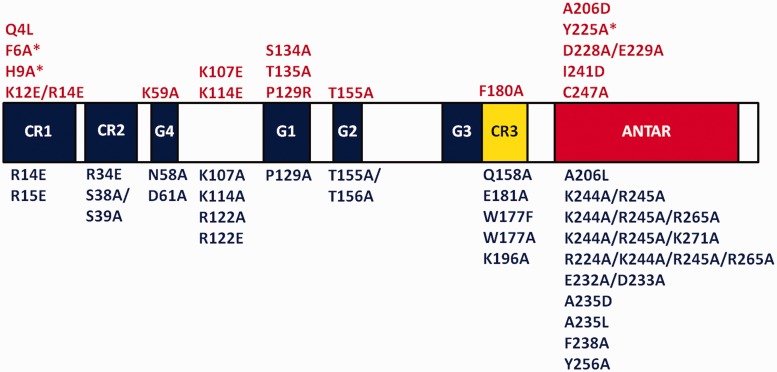
Using this system, we screened 44 mutations in rbgA to assess their growth phenotypes by streaking each mutant rbgA strain on plates containing either 1 mM IPTG or 2% xylose. A representative result is shown in Figure 3. Previous work on GTPases has shown that mutating the S/T residue in the P-loop (motif G1 -GXXXXGKS/T) abolishes GTP hydrolysis (38). We constructed the corresponding RbgA mutant strain (S134A) and found that this mutation was lethal and no colonies were observed on plates containing 2% xylose, in contrast to the control strain RB613 (Figure 3C). Both strains displayed wild-type growth when streaked on plates containing 1 mM IPTG (Figure 3B), whereas no growth was observed in the absence of either inducer (Figure 3D). Using this system, we identified 17 mutations that negatively affected cell growth (Figure 4). We further quantified the effect of each mutation by measuring growth rates in liquid media. Mutations that reduced the growth rate 3-fold or less compared with wild-type growth were classified as causing a mild growth effect, whereas mutations that reduced cell growth >3-fold were classified as severely affecting cell growth. We found that 12 mutations cause a severe growth defect, whereas five mutations have a milder effect on cell growth (Supplementary Table S3 and Figure S1). To verify that defective RbgA proteins that did not support growth were able to accumulate inside the cell, we confirmed that the expression of all mutant RbgA proteins was similar to wild-type expression levels, with the exception of one case (D228A/E229A) (data not shown). Mutations that alter the CR1, CR3 and ANTAR residues and that caused a severe growth defect were characterized further and are discussed later in the text.
Figure 3.
Testing the growth phenotype of RbgA mutants. (A) All strains were constructed with the wild-type rbgA gene under the control of an IPTG-inducible promoter and a mutated rbgA gene (S134A is shown as an example here) under the control of a xylose-inducible promoter. In contrast, the control strain had the wild-type rbgA gene under the control of both promoters (B). The two strains were streaked on IPTG-containing plates. Wild-type growth is seen in both strains due to the expression of rbgA. (C) The two strains were streaked on plates containing xylose. No colonies were observed for S134A, and hence the mutation was characterized as lethal. The RB613 control strain showed wild-type growth as rbgA is expressed from xylose-inducible promoter (D). The two strains were streaked on plates without IPTG or xylose. No growth is seen due to the absence of protein induction in either case.
Figure 4.
Consolidated results of mutational analysis of RbgA. Functional motifs are labeled by color. G domain (blue), linker region (yellow), ANTAR domain (red). CR1-3: conserved region; G1–G4: conserved GTP-binding domains found in GTPase superfamily; ANTAR: AmiR–NasR transcription anti-termination domain. The mutations shown in red above the protein are either sick or unable to support growth, whereas the mutations shown in blue below the protein do not affect growth. Asterisk indicates partial dominant negative effect.
In constructing our strains, we did not find any indication of a strong dominant negative effect of any mutation in RbgA. Previously it was reported that a deletion of the first 10 amino acids of RbgA (referred to as ΔN10) resulted in a dominant negative phenotype (39). We tested all of our mutants and the ΔN10 RbgA mutant for dominant negative effects using 1 mM IPTG and 2% xylose. Under these conditions, none of the mutations tested displayed any phenotype, including ΔN10, as all cells grew at a wild-type growth rate. Reduction of IPTG levels to 100 µM, which reduces expression of the wild-type copy of rbgA, still allows cells to proliferate at wild-type growth levels. Growing cells with 100 µM IPTG and 2% xylose uncovered the dominant negative phenotype of the ΔN10 mutant. Under these conditions, we identified three additional mutations in our collection that also had dominant negative effects, F6A and H9A in CR1 and Y225A in the ANTAR domain (Supplementary Table S4). We note that when cells are grown under 100 µM IPTG and 2% xylose, the protein expression level of mutant copy of RbgA is at least 10-fold higher than the wild-type copy, indicating that these RbgA mutations have a weak dominant negative phenotype.
Biochemical characterization of RbgA mutants
To fully characterize the effects of mutations that were found to be severely deleterious to growth, we (i) monitored the ability of RbgA mutants to associate with both 50S subunits and 45S complexes, and (ii) assessed their ability to catalyze GTP hydrolysis in the presence and absence of 50S subunits. The results of these two experiments are discussed in the following two sections. Seven mutants were selected for these studies based on their location in the protein and their phenotype of severely reduced growth rates relative to wild-type RbgA; three are from CR1 (F6A, H9A, K12E/R14E), one from CR3 (F180A) and three (A206D, Y225A, I241D) from the ANTAR domain. These RbgA mutants were purified as C-terminal His-tag fusions; previous work has shown that wild-type RbgA with a C-terminal His-tag is fully functional (9).
Mutations in the ANTAR domain alter RbgA:ribosome association
We have previously shown that RbgA stably binds to the 45S complex and 50S subunit in a nucleotide-dependent manner (12). RbgA binds to the 50S subunit in the presence of a non-hydrolyzable GTP analog, GMP-PNP, and binding in the presence of GTP is reduced, likely due to GTP hydrolysis on interaction with the 50S subunit (12). In contrast, both GTP and GMP-PNP enhance binding of RbgA to the 45S complex equally (12). We hypothesized that the ANTAR domain of RbgA is essential for association with the ribosome and targeted five key structural residues in the ANTAR domain for mutation, as described later in the text.
The ANTAR domain comprises three alpha-helices and is stabilized by coiled-coil interactions (Figure 2). ANTAR domains display low sequence conservation among homologs and contain only five strictly conserved residues that have their side-chains exposed into the cavity formed between the three helices and hence stabilize the structural fold (37). RbgA contains four of these conserved residues at the corresponding position in the domain: A206, A235, F238 and I241. Our results show that substitutions in two of these residues (A206D and I241D) caused a severe growth defect, whereas substitutions in A235 or F238 had no effect on growth (Figure 4 and Supplementary Table S3). We also generated a fifth mutant, Y225A, which likely disrupts non-covalent interactions between the helices and may alter the structure of the domain. Y225A caused a severe growth defect and displayed a weak dominant negative phenotype (Figure 4 and Supplementary Table S4). We proceeded to test association of mutated RbgA proteins with the 45S intermediate and the 50S subunit under different nucleotide conditions using a quantitative in vitro binding assay.
Purified RbgA proteins were incubated with either purified 50S subunits or 45S intermediates for 15 min at 37°C. These complexes were then centrifuged through a 100-kDa Millipore filter to remove unbound RbgA from the sample. The remaining RbgA that is associated with the ribosomal subunit was then analyzed and quantified by western blot analysis. A representative example of the results is depicted in Figure 5. Wild-type RbgA binds most efficiently to the 50S subunit in the presence of GMP-PNP, with a ∼5-fold reduction in association in the presence of GTP and ∼30-fold reduction in the presence of GDP. A206D, a strictly conserved residue in the ANTAR domain, renders RbgA unable to associate with the 50S subunit (Figure 5) or 45S intermediate (Table 1) under any nucleotide condition. Because A206D retains some GTPase activity (see later in the text), we do not expect that the lack of binding observed is solely due to a complete loss of the tertiary structure of the entire protein.
Figure 5.
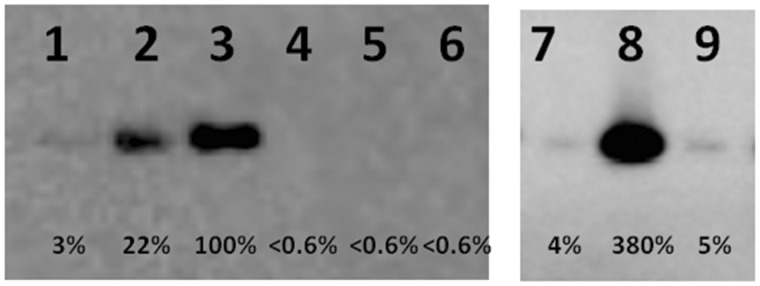
In vitro binding assay for RbgA mutants to 50S subunit under different nucleotide conditions. Purified RbgA-his6 or RbgA mutant-his6 (50 nM) was incubated with purified 50S (10 nM) subunit and GDP, GTP or GMP-PNP (400 µM) at 37°C. The presence of RbgA was tested by western blot with custom RbgA-antibody and quantified. Percentages were generated by assigning RbgA + 50S + GMP-PNP as 100%. Lane 1: RbgA + 50S + GDP; lane 2: RbgA + 50S + GTP; lane 3: RbgA + 50S + GMP-PNP; lane 4: A206D + 50S + GDP; lane 5: A206D + 50S + GTP; lane 6: A206D + 50S + GMP-PNP; lane 7: Y225A + 50S + GDP; lane 8: Y225A + 50S + GTP; lane 9: Y225A + 50S + GMP-PNP.
Table 1.
Results for in vitro binding of RbgA mutants to the 45S and 50S subunits under different nucleotide conditions
| Domain/region | 50S |
45S |
|||||
|---|---|---|---|---|---|---|---|
| GDP | GTP | GMP-PNP | GDP | GTP | GMP-PNP | ||
| RbgA wt | 4 ± 1%a | 18 ± 4% | 100% | 5 ± 2% | 85 ± 5% | 100% | |
| ANTAR | A206D | <0.6% | <0.6% | <0.6% | <0.6% | <0.6% | <0.6% |
| Y225A | 4 ± 1% | 313 ± 61% | 7 ± 2% | 6 ± 2% | 200 ± 20% | 9 ± 2% | |
| I241D | 2 ± 1% | 4 ± 2% | 51 ± 6% | 3 ± 1% | 6 ± 1% | 62 ± 5% | |
| CR1 | F6A | 2 ± 1% | 82 ± 13% | 83 ± 14% | 2 ± 2% | 80 ± 9% | 81 ± 8% |
| H9A | 1 ± 1% | 83 ± 16% | 80 ± 10% | 3 ± 1% | 81 ± 10% | 82 ± 9% | |
| K12E/R14E | 3 ± 1% | 78 ± 13% | 82 ± 14% | 3 ± 1% | 80 ± 7% | 84 ± 9% | |
| CR3 | F180A | 5 ± 1% | 16 ± 4% | 84 ± 5% | 4 ± 1% | 88 ± 2% | 93 ± 2% |
aThe values represent the average of three independent experiments ± S.D.
The binding of each RbgA mutant protein was measured in a similar manner, and the results are reported as a percentage of wild-type RbgA association with either the 45S or the 50S subunit in the presence of GMP-PNP, which displays maximal binding. Notably, of the eight mutant proteins tested, the most striking results were seen with substitutions in the ANTAR domain that changed the binding of RbgA to the 50S or 45S subunits. Mutations in CR1 were only altered in GTP-bound form, a result of the altered GTPase activity of these mutants (discussed later in the text). A mutation in CR3 displayed near wild-type levels of binding and retained the same nucleotide dependence as wild-type RbgA. In contrast, all three mutants targeting the ANTAR domain displayed altered binding and surprisingly had different phenotypes. A206D completely lost all ability to stably associate with the ribosomal subunits, whereas I241D displayed reduced association (∼10 fold) with the 50S or 45S subunits in the presence of GTP. The most surprising result was observed with the Y225A mutant RbgA protein. This RbgA derivative was greatly reduced in its association with the subunits in the presence of GMP-PNP, displaying binding levels comparable with those observed with GDP (Figure 5). However, Y225A associated with both the 50S and 45S subunits in the presence of GTP at higher levels than observed with the wild-type RbgA protein, indicating that this mutation may stabilize interaction in state of partial GTP hydrolysis (Figure 5). Thus all three ANTAR domain mutants display altered association with both the 45S and 50S subunits, highlighting the importance of this domain in association with the ribosomal subunit.
Mutations in CR1 severely reduce GTPase activity of RbgA
RbgA has a low intrinsic GTPase activity, which is stimulated ∼60-fold in the presence of mature 50S subunits (12). We therefore assessed the ability of the 50S subunit to stimulate the GTPase activity of RbgA mutants. As a negative control, we used a mutant protein containing the S134A substitution in the P-loop, which shows no detectable intrinsic GTPase activity and is inert to stimulation in the presence of the 50S subunit. Our results show that mutations in CR1 and the ANTAR domain impact the GTPase activity of RbgA, whereas the mutation in CR3 had no effect (Table 2). However, mutations in the ANTAR domain also disrupted interaction of RbgA with both the 45S as well as the 50S subunit, and therefore, the attenuated GTPase activity seen in A206D and I241D is likely due, in part, to an indirect effect of poor association with the 50S subunit (Table 1, and aforementioned discussion). It is also possible that A206D has a reduced affinity for guanine nucleotides.
Table 2.
Measurement of GTPase activity of RbgA mutants in the presence and absence of the 50S subunit
| Domain/Region | Mutation | Intrinsic GTPase activity |
Stimulation of GTPase activity with 50S |
Fold stimulation in the presence of 50S |
|---|---|---|---|---|
| V = pmol phosphate/min/mg | ||||
| RbgA wt | 0.207 ± 0.008a | 12.570 ± 0.180 | 61 ± 3.0 | |
| CR1 | F6A | 0.018 ± 0.002 | 0.283 ± 0.050 | 15 ± 4.0 |
| H9A | 0.031 ± 0.006 | <0.002 | ||
| K12E/R14E | 0.121 ± 0.004 | 0.226 ± 0.070 | 2 ± 0.5 | |
| G-domains | S134A | <0.002 | <0.002 | |
| ANTAR | A206D | 0.040 ± 0.001 | 0.978 ± 0.060 | 24 ± 2.0 |
| Y225A | 0.139 ± 0.005 | 3.970 ± 0.300 | 29 ± 3.0 | |
| I241D | 0.104 ± 0.007 | 0.597 ± 0.110 | 6 ± 0.5 | |
| CR3 | F180A | 0.097 ± 0.012 | 8.080 ± 0.190 | 84 ± 11.0 |
aThe values represent the average of three independent experiments ± S.D.
Mutations in CR1 had a strong impact on the GTPase activity of RbgA. Mutation F6A resulted in a ∼11-fold reduction in intrinsic GTPase activity and was stimulated ∼15-fold in the presence of the mature 50S subunit, demonstrating that this mutant is reduced but still capable of GTP hydrolysis. Mutation K12E/R14E had a moderate reduction in intrinsic GTPase activity but was stimulated only 2-fold in the presence of the 50S subunit. These results suggest that the flexible highly conserved N-terminal region of RbgA encompassing CR1 is required for GTPase activation in the presence of the ribosomal subunit.
A recent crystal structure of EF-Tu in complex with the ribosome demonstrated that interaction between the ribosome and His84 of EF-Tu was required for efficient hydrolysis of GTP (40). Voorhees et al. noted that His84 is preceded by a proline and glycine (denoted the PGH motif), which were proposed to be critical for the correct positioning of His84 on interaction with nucleotide A2662 of the ribosome during activation. We observed that the CR1 region of RbgA also contains a PGH motif (amino acids 7–9) that is highly conserved in bacterial and eukaryotic RbgA homologs (Figure 1A and B). An alignment of crystal structures of RbgA and free EF-Tu indicates that His9 from PGH motif of RbgA is in a similar position as the catalytic His84 of EF-Tu (Figure 6 and Supplementary Figure S3). To test the role of this histidine residue (His9) in the GTPase activity of RbgA, we generated a H9A mutant and characterized its GTPase activity in the absence and presence of the 50S subunit. Our results show that mutation H9A reduced intrinsic GTPase activity of RbgA by ∼6 fold relative to the wild-type protein. Most significantly, this mutant displayed no detectable GTPase activity in the presence of the 50S subunit (Table 2). Together this result, in conjunction with the phenotype of F6A and K12E/R14E, shows that mutations in CR1 reduce GTPase activity of RbgA and that His9 is required for GTPase activity in the presence of the 50S subunit. The absence of GTPase activity in the mutant is unlikely to be due to reduced association with the 50S subunit, as this mutant, in particular, and other CR1 mutants associated with the 50S subunit at similar levels compared with wild-type RbgA (Table 1).
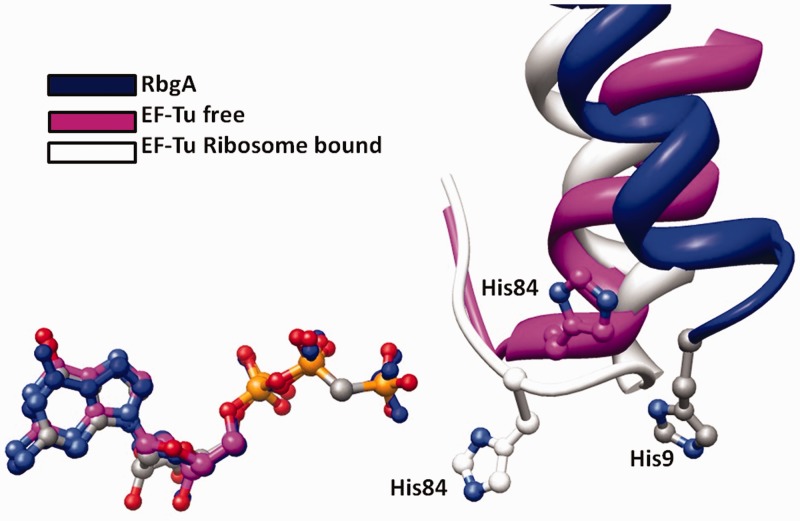
Figure 6.
A ligand-based structural superimposition of EF-Tu (free-magenta and ribosome bound-light gray) over the free RbgA (blue). For clarity, only the helices containing the catalytic histidine residues are shown in complex with their respective GTP analogs.
Linker region is required for RbgA function
All cpGTPases have a C-terminal domain linked to the G3 motif (Switch II) (17). In RbgA, a highly conserved loop (CR3) links the C-terminal ANTAR domain to the G3 motif (Switch II). We hypothesized that the CR3 could be crucial for communicating conformational changes during GTP hydrolysis between the G3 motif and the ANTAR domain. We identified one mutation in CR3, F180A, that was lethal. We tested this mutation using the in vitro binding assay described previously to determine whether this residue was required for association of RbgA to the ribosomal subunits, and found that this mutation did not alter association of RbgA to the 45S complex or the 50S subunit (Table 1). Next, we characterized the GTPase activity of this mutant and found that mutation F180A has a mild effect on GTPase activity of RbgA with and without the 50S subunit (Table 2). Thus, even though this mutation is defective neither in ribosome association nor in ribosome-stimulated GTPase activity, it is unable to support growth and yields ribosome assembly defects in vivo.
Strains expressing mutated RbgA proteins do not accumulate a novel intermediate
A previously proposed model suggested that GTP-bound RbgA binds to the 45S intermediate and either directly or indirectly recruits the ribosomal protein L16 to the intermediate (12). Stable integration of L16 and formation of a mature 50S subunit likely triggers GTP hydrolysis and dissociation of RbgA from the mature 50S subunit. We hypothesized that strains that show a growth defect phenotype due to the expression of a compromised RbgA protein may accumulate a trapped intermediate that differs from the 45S intermediate observed on depletion of the wild-type protein. We analyzed ribosome profiles of strains expressing RbgA mutants that negatively affected cell growth and isolated the 45S intermediate from each mutant (a representative set is shown in Supplementary Figure S2A). Our results show that strains expressing mutated RbgA proteins do not trap a new intermediate. These strains also accumulated the 45S intermediate similar to RbgA depleted cells, and this intermediate also lacked the ribosomal protein L16 (Supplementary Figure S2B). Thus, no novel ribosomal intermediate could be detected for the RbgA mutants that were analyzed.
DISCUSSION
RA-GTPases are critical for the efficient and accurate biogenesis of cellular, mitochondrial and chloroplast ribosomes; however, their roles in the assembly process are poorly understood (4,6,8). RbgA and its closest eukaryotic homolog Mtg1 are involved in the assembly of the large ribosomal subunit in bacteria and mitochondria in eukaryotes, respectively (9,11,13). RA-GTPases use a variety of RNA-binding domains to interact with RNA, including OB-folds (Obg) and KH-domains (Era) (24,41). The crystal structure of RbgA showed that the C-terminal domain (201–269) is connected to the N-terminal G-domain (1–176) through a linker helix (177–200) (Figure 2). We have identified a RNA-binding domain within the C-terminus of RbgA, the ANTAR domain, which was discovered in proteins that bind structured mRNA leader regions and promote transcription anti-termination (37). ANTAR domain–containing proteins such as NasR and EutV promote anti-termination by interacting with mRNA and promoting the formation of stem-loop structures that preclude the formation of intrinsic transcription terminators (35,42,43). It is attractive to speculate that RbgA may influence ribosome assembly by interacting with rRNA through its ANTAR domain and stabilizing the formation of a critical stem-loop in the 23S rRNA that does not form spontaneously and prevents the formation of other non-productive loops that may hamper further assembly. It is possible that in in vitro experiments, these non-productive loops are circumvented by high-temperature or high-salt conditions, thereby facilitating proper assembly. Efforts to identify the RbgA-binding site on the ribosome are currently underway. Mutations in the ANTAR domain reduced association of RbgA to the 45S or 50S to varying degrees. Residue A206 is likely important for the structural stability of the ANTAR domain. This residue is one of four highly conserved alanine residues seen in most ANTAR structures (37). In addition, mutants such as Y225A could prove to be good candidates for structural and CryoEM studies, as they show a unique binding profile compared with wild-type RbgA, and could shed light on the interaction of RbgA with the 50S subunit.
Most ANTAR domain proteins are two-domain proteins in which the ANTAR domain is precluded from interacting with RNA until a signal or ligand is sensed by the receiver domain, which opens up ANTAR for RNA binding (42,43). One example of this arrangement is seen in the recently published NasR structure where the signal receiver domain (NIT) and the ANTAR domain are joined by a linker (35). Thus, a similar arrangement in RbgA suggests that the guanine nucleotide–dependent conformational change of the N-terminal G-domain could be communicated to the ANTAR domain through the conserved linker region. In principle, the mechanistic reverse wherein binding of the ANTAR domain to rRNA could be coupled to a conformational change in the N-terminal G-domain that facilitates GTP hydrolysis could also occur. In both scenarios, communication between the N- and C- terminal domains is important for RbgA function and would likely rely on the linker CR3, similar to the role of the linker in NasR protein connecting the signal receiver domain and the ANTAR domain. Our results indicate that this mutation in CR3 is deleterious to growth, although neither the association with the ribosome nor the GTPase activity of this mutant protein is affected. This suggests that although the function of either domain is not affected, the mutant protein cannot function in vivo, and may indicate the linker is critical for communication between the two structural domains.
The GTPase activity of RbgA is essential to its function, and mutations in conserved G-domains that abolish GTP hydrolysis are lethal. However, due to the absence of a crystal structure of RbgA bound to 50S, the exact mechanism of GTP hydrolysis remains elusive. Recent structural evidence demonstrating how EF-Tu carries out GTP hydrolysis on interaction with the ribosome during codon recognition has shown that positioning of His84 through interaction with 23S rRNA nucleotide A2662 is critical for GTP hydrolysis (40). Several previous studies on EF-Tu have also demonstrated the essential role of His84 for catalysis of GTP (44,45). His84 is part of a PGH motif that is conserved in nearly all EF-Tu homologs identified to date, and a similar PGH motif is found in bacterial and eukaryotic homologs of RbgA. A key difference between the conserved PGH motifs in EF-Tu and RbgA is the fact that whereas His84 is found in the Switch II (G3) region (His84 substitutes for the catalytic Gln normally found in Ras superfamily GTPases), His9 in RbgA is found in the highly flexible CR1 region and is not ordered in the available crystal structures (1PUJ) (46). In RbgA, the Switch II region is instead linked to the C-terminal ANTAR domain. Mutation of His9 of RbgA resulted in a severely attenuated intrinsic rate of GTP hydrolysis, and no stimulation of GTPase activity in the presence of the 50S subunit could be detected. Importantly, this mutation had no significant effect on association with the 50S subunit. Mutations in other parts of CR1 also greatly impacted GTP hydrolysis; however, all other mutations showed at least some stimulation in the presence of the 50S subunit. Our data are consistent with a model in which the highly flexible CR1 region, on interaction with the ribosome, adopts a conformation that suitably positions His9 for GTP hydrolysis.
This assertion is also supported by analysis of the EF-Tu crystal structure (40). Vorhees et al. suggested that the EF-Tu GTP hydrolysis mechanism could be a general mechanism used by translation GTPases to carry out GTP hydrolysis. To this end, we superimposed the G-domain of RbgA and EF-Tu (Figure 6 and Supplementary Figure S3) by using the GDPNP bound to each of these proteins in the crystal structure as a point of reference. In the superimposed structure, His9 from RbgA is in close proximity to His84 in the structure of free EF-Tu, consistent with our aforementioned assertion (Figure 6 and Supplementary Figure S3). It should be noted that the position of catalytic His84 in EF-Tu is different in the free EF-Tu structure compared with the ribosome-bound structure (Figure 6 and Supplementary Figure S3). As only the structure of free RbgA is available, it is difficult to predict whether His9 would move to the exact position as EF-Tu His84 on interaction with the ribosome and whether it would interact with the same 23S rRNA nucleotide A2662. It is also noteworthy that due to relocation of the putative catalytic residue His9 in RbgA from Switch II to the N-terminal helix, a direct interaction between histidine and the activated water positioned to attack the γ-phosphate requires a small translocation of Switch II and potentially the ANTAR domain as well.
Based on previously published mechanisms of GTP hydrolysis, this can be accomplished by one of the following ways. First, binding of the ANTAR domain to the 50S subunit could result in a conformational change that is transmitted through the linker CR3 to Switch II. This would likely be coupled with the rearrangement of the N-terminal helix with concomitant positioning of His9 that would facilitate catalysis. The presence of a dynamic and unstructured N-terminal CR1, as evidenced by relatively high B-factors of this region in the RbgA crystal structure, as well as a flexible linker domain connecting Switch II to the ANTAR domain, further strengthens this possibility. A nucleotide-dependent large domain rearrangement has also been shown to occur in other ribosomal GTPases such as YphC (21). A second possibility is that the histidine residue might take part in catalysis through additional intermediate water molecule(s), as has previously been shown for other HAS-GTPases (MnmE and FeoB) (26,27). In these GTPases, the catalytic residue interacts with a secondary (in the case of MnmE, or tertiary in the case of FeoB) water molecule that in turn activates the catalytic water molecule and results in GTP hydrolysis (26,27). Further, it is also possible that the mechanism in RbgA might be a combination of both these mechanisms. Additional structural and biochemical studies are required to test either of these mechanisms and to further ascertain whether the nucleotide equivalent to A2662 in B. subtilis is required for hydrolysis. This will require the development of tools to isolate mutant ribosomes from this organism, similar to the tools that have been developed for E. coli (47,48). The GTPase activity of RbgA was unaffected by E. coli 50S ribosomes, which was not unexpected, given that E. coli lacks a RbgA homolog, and thus we could not test the interaction of RbgA with A2662 from the E. coli 50S subunit.
Although we have uncovered important insights into the biochemistry of RbgA and how RbgA interacts with the ribosome, several important questions regarding RbgA (and RA-GTPases in general) remain. First, does the GTPase activity of RbgA play a direct role in the assembly process? If the GTPase activity was solely involved in displacing RbgA from the ribosome once assembly had been completed, then we expected to identify RbgA mutations that completed assembly but did not dissociate RbgA from the ribosome. This was not the case, as all mutations reported in this study that were deleterious to growth yielded a 45S complex that lacked L16, which supports a direct role for GTP hydrolysis in the assembly process. Second, what part of the 23S rRNA does RbgA interact with during the assembly process? At this point there are limited chemical footprinting data that show RbgA interacting with the central protuberance (11). We are currently mapping the structure of the 23S rRNA in the 50S subunit and 45S complex to identify key sites that are altered in the 45S complex. In addition, as RbgA homologs are not found in E. coli, we are also targeting unique stem-loops within the B. subtilis 23S rRNA as possible sites where RbgA may act. Third, does RbgA serve as a checkpoint to monitor large-subunit assembly and correct incorporation of L16? Recent work in yeast has uncovered a novel type of translational checkpoint used by ribosome assembly factors (49). Finally, why do most organisms, ranging from bacteria to humans, require RbgA to form ribosomes, whereas many proteobacteria have lost this requirement? Future work will uncover the precise role of RbgA in the assembly of ribosomes in all three kingdoms of life.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–3.
FUNDING
National Science Foundation CAREER Award [0643565 to R.A.B.]. Funding for open access charge: National Science Foundation CAREER Award [0643565].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank David Achila and Brian Shy for technical assistance.
REFERENCES
- 1.Nomura M, Erdmann VA. Reconstitution of 50S ribosomal subunits from dissociated molecular components. Nature. 1970;228:744–748. doi: 10.1038/228744a0. [DOI] [PubMed] [Google Scholar]
- 2.Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 3.Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 5.Fahnestock S, Erdmann V, Nomura M. Reconstitution of 50S ribosomal subunits from protein-free ribonucleic acid. Biochemistry. 1973;12:220–224. doi: 10.1021/bi00726a007. [DOI] [PubMed] [Google Scholar]
- 6.Britton RA. Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 2009;63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- 7.Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell. Res. 2004;296:43–50. doi: 10.1016/j.yexcr.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Karbstein K. Role of GTPases in ribosome assembly. Biopolymers. 2007;87:1–11. doi: 10.1002/bip.20762. [DOI] [PubMed] [Google Scholar]
- 9.Uicker WC, Schaefer L, Britton RA. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 2006;59:528–540. doi: 10.1111/j.1365-2958.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer L, Uicker WC, Wicker-Planquart C, Foucher AE, Jault JM, Britton RA. Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J. Bacteriol. 2006;188:8252–8258. doi: 10.1128/JB.01213-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuo Y, Morimoto T, Kuwano M, Loh PC, Oshima T, Ogasawara N. The GTP-binding protein YlqF participates in the late step of 50 S ribosomal subunit assembly in Bacillus subtilis. J. Biol. Chem. 2006;281:8110–8117. doi: 10.1074/jbc.M512556200. [DOI] [PubMed] [Google Scholar]
- 12.Achila D, Gulati M, Jain N, Britton RA. Biochemical characterization of ribosome assembly GTPase RbgA in Bacillus subtilis. J. Biol. Chem. 2012;287:8417–8423. doi: 10.1074/jbc.M111.331322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrientos A, Korr D, Barwell KJ, Sjulsen C, Gajewski CD, Manfredi G, Ackerman S, Tzagoloff A. MTG1 codes for a conserved protein required for mitochondrial translation. Mol. Biol. Cell. 2003;14:2292–2302. doi: 10.1091/mbc.E02-10-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kallstrom G, Hedges J, Johnson A. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell Biol. 2003;23:4344–4355. doi: 10.1128/MCB.23.12.4344-4355.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassler J, Kallas M, Hurt E. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60 S pre-ribosomal particles. J. Biol. Chem. 2006;281:24737–24744. doi: 10.1074/jbc.M604261200. [DOI] [PubMed] [Google Scholar]
- 16.Im CH, Hwang SM, Son YS, Heo JB, Bang WY, Suwastika IN, Shiina T, Bahk JD. Nuclear/nucleolar GTPase 2 proteins as a subfamily of YlqF/YawG GTPases function in pre-60S ribosomal subunit maturation of mono- and dicotyledonous plants. J. Biol. Chem. 2011;286:8620–8632. doi: 10.1074/jbc.M110.200816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anand B, Verma SK, Prakash B. Structural stabilization of GTP-binding domains in circularly permuted GTPases: implications for RNA binding. Nucleic Acids Res. 2006;34:2196–2205. doi: 10.1093/nar/gkl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra R, Gara SK, Mishra S, Prakash B. Analysis of GTPases carrying hydrophobic amino acid substitutions in lieu of the catalytic glutamine: implications for GTP hydrolysis. Proteins. 2005;59:332–338. doi: 10.1002/prot.20413. [DOI] [PubMed] [Google Scholar]
- 19.Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. USA. 2002;99:16243–16248. doi: 10.1073/pnas.242338299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monleon D, Martinez-Vicente M, Esteve V, Yim L, Prado S, Armengod ME, Celda B. Structural insights into the GTPase domain of Escherichia coli MnmE protein. Proteins. 2007;66:726–739. doi: 10.1002/prot.21186. [DOI] [PubMed] [Google Scholar]
- 21.Muench SP, Xu L, Sedelnikova SE, Rice DW. The essential GTPase YphC displays a major domain rearrangement associated with nucleotide binding. Proc. Natl. Acad. Sci. USA. 2006;103:12359–12364. doi: 10.1073/pnas.0602585103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uicker WC, Schaefer L, Koenigsknecht M, Britton RA. The essential GTPase YqeH is required for proper ribosome assembly in Bacillus subtilis. J. Bacteriol. 2007;189:2926–2929. doi: 10.1128/JB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan SM, Mishra R, Neubig RR, Maddock JR. Analysis of guanine nucleotide binding and exchange kinetics of the Escherichia coli GTPase Era. J. Bacteriol. 2000;182:3460–3466. doi: 10.1128/jb.182.12.3460-3466.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, Andrews PC, Maddock JR. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 2006;188:6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wout P, Pu K, Sullivan SM, Reese V, Zhou S, Lin B, Maddock JR. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 2004;186:5249–5257. doi: 10.1128/JB.186.16.5249-5257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scrima A, Wittinghofer A. Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J. 2006;25:2940–2951. doi: 10.1038/sj.emboj.7601171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ash MR, Maher MJ, Guss JM, Jormakka M. The initiation of GTP hydrolysis by the G-domain of FeoB: insights from a transition-state complex structure. PLoS One. 2011;6:e23355. doi: 10.1371/journal.pone.0023355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teraoka H, Nierhaus KH. Protein L16 induces a conformational change when incorporated into a L16-deficient core derived from Escherichia coli ribosomes. FEBS Lett. 1978;88:223–226. doi: 10.1016/0014-5793(78)80179-3. [DOI] [PubMed] [Google Scholar]
- 29.Petit M-A, Dervyn E, Rose M, Entian K-D, McGovern S, Ehrlich SD, Bruand C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol. 1998;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- 30.Bhavsar AP, Zhao X, Brown ED. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl. Environ. Microbiol. 2001;67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. The PyMOL Molecular Graphics System [computer program], Version 1.2r3pre. Schrödinger, LLC; http://pymol.sourceforge.net/faq.html#CITE.
- 33.Heifets A, Lilien RH. LigAlign: flexible ligand-based active site alignment and analysis. J. Mol. Graph. Model. 2010;29:93–101. doi: 10.1016/j.jmgm.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Boudes M, Lazar N, Graille M, Durand D, Gaidenko TA, Stewart V, van Tilbeurgh H. The structure of the NasR transcription antiterminator reveals a one-component system with a NIT nitrate receptor coupled to an ANTAR RNA-binding effector. Mol. Microbiol. 2012;85:431–444. doi: 10.1111/j.1365-2958.2012.08111.x. [DOI] [PubMed] [Google Scholar]
- 36.O'Hara BP, Norman RA, Wan PT, Roe SM, Barrett TE, Drew RE, Pearl LH. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J. 1999;18:5175–5186. doi: 10.1093/emboj/18.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shu CJ, Zhulin IB. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 2002;27:3–5. doi: 10.1016/s0968-0004(01)02036-9. [DOI] [PubMed] [Google Scholar]
- 38.Sprang SR. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 39.Matsuo Y, Oshima T, Loh PC, Morimoto T, Ogasawara N. Isolation and characterization of a dominant negative mutant of Bacillus subtilis GTP-binding protein, YlqF, essential for biogenesis and maintenance of the 50 S ribosomal subunit. J. Biol. Chem. 2007;282:25270–25277. doi: 10.1074/jbc.M703894200. [DOI] [PubMed] [Google Scholar]
- 40.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma MR, Barat C, Wilson DN, Booth TM, Kawazoe M, Hori-Takemoto C, Shirouzu M, Yokoyama S, Fucini P, Agrawal RK. Interaction of Era with the 30S ribosomal subunit implications for 30S subunit assembly. Mol. Cell. 2005;18:319–329. doi: 10.1016/j.molcel.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 42.Ramesh A, Debroy S, Goodson JR, Fox KA, Faz H, Garsin DA, Winkler WC. The mechanism for RNA recognition by ANTAR regulators of gene expression. PLoS Genet. 2012;8:e1002666. doi: 10.1371/journal.pgen.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart V, van Tilbeurgh H. Found: the elusive ANTAR transcription antiterminator. PLoS Genet. 2012;8:e1002773. doi: 10.1371/journal.pgen.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daviter T, Wieden HJ, Rodnina MV. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J. Mol. Biol. 2003;332:689–699. doi: 10.1016/s0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- 45.Scarano G, Krab IM, Bocchini V, Parmeggiani A. Relevance of histidine-84 in the elongation factor Tu GTPase activity and in poly(Phe) synthesis: its substitution by glutamine and alanine. FEBS Lett. 1995;365:214–218. doi: 10.1016/0014-5793(95)00469-p. [DOI] [PubMed] [Google Scholar]
- 46.Kim do J, Jang JY, Yoon HJ, Suh SW. Crystal structure of YlqF, a circularly permuted GTPase: implications for its GTPase activation in 50S ribosomal subunit assembly. Proteins. 2008;72:1363–1370. doi: 10.1002/prot.22112. [DOI] [PubMed] [Google Scholar]
- 47.Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires CL. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 1999;181:3803–3809. doi: 10.1128/jb.181.12.3803-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asai T, Zaporojets D, Squires C, Squires CL. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. USA. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.