Abstract
Nucleophosmin (NPM1) is an abundant nucleolar protein implicated in ribosome maturation and export, centrosome duplication and response to stress stimuli. NPM1 is the most frequently mutated gene in acute myeloid leukemia. Mutations at the C-terminal domain led to variant proteins that aberrantly and stably translocate to the cytoplasm. We have previously shown that NPM1 C-terminal domain binds with high affinity G-quadruplex DNA. Here, we investigate the structural determinants of NPM1 nucleolar localization. We show that NPM1 interacts with several G-quadruplex regions found in ribosomal DNA, both in vitro and in vivo. Furthermore, the most common leukemic NPM1 variant completely loses this activity. This is the consequence of G-quadruplex–binding domain destabilization, as mutations aimed at refolding the leukemic variant also result in rescuing the G-quadruplex–binding activity and nucleolar localization. Finally, we show that treatment of cells with a G-quadruplex selective ligand results in wild-type NPM1 dislocation from nucleoli into nucleoplasm. In conclusion, this work establishes a direct correlation between NPM1 G-quadruplex binding at rDNA and its nucleolar localization, which is impaired in the acute myeloid leukemia-associated protein variants.
INTRODUCTION
Nucleophosmin (hereby NPM1, also known as B23, No38 and numatrin) is an abundant non-ribosomal nucleolar protein playing a key role in ribosome biogenesis. In fact, NPM1 is able to bind RNA and DNA; it has intrinsic RNAse activity that preferentially cleaves pre-rRNA (1); and it directs the nuclear export of both 40S and 60S ribosomal subunits (2). NPM1 also plays several roles outside the nucleolus. It shuttles between the nucleus and the cytoplasm and displays molecular chaperone activity (3,4). Furthermore, a key role of NPM1 in controlling centrosome duplication has been suggested (5). This is confirmed by the analysis of NPM1−/− mice, who show unrestricted centrosome duplication, genomic instability and mid-gestation embryonic lethality (6). NPM1 has also been shown to regulate the function of several tumor suppressors, such as p53 (7), p14arf (8,9) and Fbw7γ (10).
NPM1 is a commonly altered gene in hematological malignancies (11). In particular, it was identified as the most frequently mutated gene in acute myeloid leukemia (AML), accounting for ∼35% of cases (12). More than 50 mutations have been characterized so far; they are always heterozygous, mostly localized at the terminal exon of the gene, and consist of insertions or duplications of short-base sequences (13). At the protein level, all mutations cause similar abnormalities: the reading frame is altered, thus leading to a mutated protein that (i) has acquired four additional residues at the C-terminus, which define a newly formed nuclear export signal, and (ii) is largely destabilized in its C-terminal domain because of the loss of one or both of critical Trp288 and Trp290 residues (13–16). Taken together, these variations account for the aberrant and stable cytoplasmic localization of mutated NPM1 (13,17). Furthermore, as mutant NPM1 oligomerizes with the wild-type through its N-terminal domain (18), the mutated protein is capable of displacing the wild-type counterpart to the cytosol (13). Therefore, in AML patients with NPM1 mutations, NPM1 is largely found in the cytosol, and only a limited portion of the protein is retained in nucleoli (13). This feature characterizes this type of leukemia that has been included as a new provisional entity in the 2008 World Health Organization classification of myeloid neoplasms (4).
The C-terminal domain of NPM1, which is the site of AML-associated mutations, binds both RNA and DNA, with a preference for single-stranded over double-stranded oligonucleotides (19). Recently, we further investigated this issue and established that a domain encompassing the last 70 residues of the protein (NPM1-C70), even though it is able to interact with any DNA oligonucleotide tested, binds with higher affinity two oligonucleotide sequences with G-quadruplex structure found in the superoxide dismutase 2 (SOD2) and c-MYC gene promoters (20).
G-quadruplexes are non-canonical nucleic acid structures resulting from the formation of guanine tetrads, stabilized by Hoogsteen-type hydrogen bonds, that stack onto each other to form extremely stable assemblies (21,22). They can be formed both by DNA and RNA, and they are gaining increasing attention, as they are abundant at telomeric DNA, gene promoters and mRNA 5′-untranslated region (UTRs), and they have been shown to regulate a variety of cellular processes at the translational and post-translational levels (23–25).
Recently, we performed a structural analysis of the interaction of NPM1-C70 with the G-quadruplex region of the c-MYC promoter and showed that this association is mainly electrostatic in nature. A stretch of backbone phosphates, contributing to the formation of each of the three stacked guanine tetrads in the G-quadruplex scaffold, accommodates into a specific groove between helices H1–H2 of the NPM1 C-terminal three-helix bundle (26). This analysis suggested that NPM1-C70 recognizes a conserved feature in the G-quadruplex scaffold; therefore, it may be able to interact with several G-quadruplexes in vivo.
Nucleoli are dynamic nuclear organelles that are mainly formed by tandemly repeated clusters of rDNA and by factors involved in ribosomal RNA transcription and processing, including two of the most abundant protein components, i.e. NPM1 and nucleolin. Interestingly, Drygin et al. (27) have recently shown the presence of several putative G-quadruplex–forming sequences (PQS) in the non-template strand of the rDNA gene, which are bound by nucleolin. As we have recently characterized NPM1 as a G-quadruplex–binding protein, we hypothesized here that, like nucleolin, NPM1 may also bind G-quadruplex regions at rDNA. Here, we show that this is indeed the case, both in vitro and in vivo. Importantly, we also show that the G-quadruplex–binding activity is completely lost by the most common NPM1 AML-associated mutant, which is unfolded in its C-terminal DNA-binding domain, and that mutations aimed at refolding the leukemic NPM1 variant also result in rescuing the G-quadruplex–binding activity. Finally, to further establish a link between NPM1 G-quadruplex binding and its nucleolar localization, we show that treatment of OCI-AML2 cells, bearing wild-type NPM1 at both alleles, with the G-quadruplex selective ligand TmPyP4 is sufficient to completely displace NPM1 from nucleoli to the nucleoplasm.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides used in this study were 5′-GGGTCGGGGGGTGGGGCCCGGGCCGGGG-3′ (2957NT); 5′-AGGGAGGGAGACGGGGGGG-3′ (5701NT); 5′-GGGTGGCGGGGGGGAGAGGGGGG-3′ (6960NT); 5′-GGGGTGGGGGGGAGGG-3′ (13079NT). High-performance liquid chromatography purified oligos were purchased from IDT (Coralville, IA, USA). Oligos for SPR analysis (see later in the text) were also biotynylated at their 5′-end. Lyophilized oligos were dissolved in a buffer containing 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) 20 mM, pH 7.0, and KCl 150 mM before annealing. For annealing, oligos were heated at 95°C for 15 min and were then let to gently cool down overnight at room temperature.
NPM1 and NPM1 variants protein constructs
NPM1-C70 was expressed and purified as previously described (20). The hexa-histidine tag was thrombin-cleaved and removed by Nickel-nitrilotriacetic acid (Ni-NTA) affinity.
The coding sequence for NPM1-Cter-MutA (residues 225–298 of the NPM1 Mutant A protein) was obtained through gene synthesis and cloned into pGEX-6P1 vector (GE Healthcare) within BamHI and EcoRI restriction sites. Plasmid was transformed in Escherichia coli BL21(DE3) cells. Cells were grown in Luria broth supplemented with glucose 20 mM at 37°C to an OD600 = 0.5, induced with 1.0 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) and then transferred to 16°C for 1 h before harvesting. The cell pellet was resuspended in Buffer A (Tris–HCl 20 mM, pH 7.5, and NaCl 150 mM) supplemented with protease inhibitor cocktail (Roche) and sonicated. After separating the debris by centrifugation, the supernatant was loaded on a GSTrap-HP column pre-equilibrated with Buffer A. After extensive washing with Buffer A, the glutathione transferase (GST)-tagged protein was eluted with a linear gradient of reduced glutathione (0–10 mM) in an FLPC system (AktaPrime, GE Healthcare). Fractions containing the GST-tagged protein were pooled and diluted 3-fold with Buffer A and supplemented with dithiothreitol (DTT) 1 mM and ethylenediaminetetraacetic acid (EDTA) 1 mM. At this point, the GST-tagged protein was digested with Prescission protease (GE Healthcare) at 4°C for 4 h. The cleavage mixture obtained was then loaded again onto a GSTrap column to separate NPM1-Cter-MutA from the GST and the uncleaved fusion protein. NPM1-Cter-MutA was recovered from the flow through and then loaded on an SP column (GE Healthcare) and eluted with a linear gradient of NaCl (0.03–1.0 M). Fractions containing the protein were pooled, supplemented with 3.0 M NaCl and then loaded onto a HighTrap Phenyl FF (high sub) column (GE Healthcare) pre-equilibrated with Tris 50 mM, pH 7.5, and 3.0 M NaCl. Elution was performed with a linear reverse gradient of NaCl (3.0–0.0 M). The final protein product was buffer exchanged to Tris 50 mM, pH 7.5, concentrated and stored at −80°C.
The MutA-Ref288, MutA-Ref290 and MutA-Ref288–290 constructs, encompassing the point mutations C288W, A290W or both, on the NPM1-Cter-MutA construct, were obtained by polymerase chain reaction (PCR) amplification using the Quickchange II Lightning Site-Directed Mutagenesis kit (Stratagene) following manufacturer’s instructions. MutA-Ref288, MutA-Ref290 and MutA-Ref288–290 proteins were expressed and purified using the same protocol adopted for NPM1-Cter-MutA.
Circular dichroism
All circular dichroism (CD) experiments were performed using a Jasco J710 instrument (Jasco Inc., Easton, MD, USA) equipped with a Peltier apparatus for temperature control. Static spectra of rDNA oligos were collected at 25°C, using oligos annealed in the appropriate buffer and concentrated to 20 µM. Static spectra of protein samples (NPM1-C70, NPM1-Cter-MutA, MutA-Ref288, MutA-Ref290 and MutA-Ref 288–290) were collected at 25°C. Protein concentration was 20 µM. Spectra were collected using a quartz cell with 1-mm optical path length (Hellma, Plainview, NY, USA) and a scanning speed of 100 nm/min. The reported spectra are the average of five scans. The spectral contribution of buffers was subtracted as appropriate.
Thermal denaturation experiments on protein samples (20 µM) were performed using a quartz cell with 1-mm optical path length and monitoring the variation of CD signal at 222 nm. Temperature was progressively increased, in 1°C/min steps, from 20°C to 90°C. Data were analyzed as previously reported (15). The Kaleidagraph software was used for spectra representation.
Surface plasmon resonance
The interactions between DNA oligonucleotides (ligands) with the purified proteins NPM1-C70, NPM1-Cter-MutA and MutA-Ref288–290 (analytes) as well as with the porphyrin TmPyP4 (analyte) [meso-5,10,15,20-Tetrakis-(N-methyl-4-pyridyl)porphine, Tetratosylate] (Calbiochem), were all measured by surface plasmon resonance (SPR) technique using a Biacore X100 instrument (Biacore, Uppsala, Sweden). Each biotynylated DNA construct was immobilized on a Sensor Chip SA, pre-coated with streptavidin from Biacore AB (Uppsala, Sweden). Capturing procedure on biosensor surface was performed according to manufacturer’s instructions and setting the aim for ligand immobilization to 1000 RU. Running buffer was HEPES-buffered saline-EP (HBS-EP), which contains 10 mM HEPES (pH 7.4), 0.15 M NaCl, 3 mM EDTA and 0.005% (v/v) Surfactant P20 (Biacore AB, Uppsala, Sweden). NPM1-C70, NPM1-Cter-MutA and MutA-Ref288–290 (SA analytes) were dissolved in running buffer, and binding experiments were performed at 25°C with a flow rate of 30 µl/min. The association phase (kon) was followed for 180 s, whereas the dissociation phase (koff) was followed for 300 s. The complete dissociation of active complex formed was achieved by addition of 10 mM HEPES, 2 M NaCl, 3 mM EDTA and 0.005% (v/v) P20, pH 7.4, for 60 s before each new cycle start, as expected if the interaction formed is mainly electrostatic. SA analytes were tested in a wide range of concentrations to reach at least a 2-fold increase from the lower concentration tested.
To test the interaction between NPM1-C70 and the 13079NT oligo, alone and in presence of increasing amounts of TmPyP4, his-tagged NPM1-C70 (NTA ligand) was immobilized on an NTA Sensor Chip, pre-coated with nitrilotriacetate group from Biacore AB (Uppsala, Sweden). Capturing procedure on the biosensor surface was performed according to manufacturer’s instructions and setting the contact time of the his-tagged protein to 60 s at 0.7 µg/ml. Running buffer was HEPES-buffered saline-P (HBS-P), which contains 10 mM HEPES (pH 7.4), 0.15 M NaCl and 0.005% (v/v) Surfactant P20, supplemented with 50 µM EDTA to neutralize contaminating metal ions (Biacore AB, Uppsala, Sweden). The 13079NT oligo and TmPyP4 (NTA analytes) were dissolved in running buffer at varying molar ratios in the different binding experiments, starting from the oligo alone until reaching a 5-fold exess of TmPyP4 versus 13079NT. Binding experiments were performed at 25°C with a flow rate of 30 µl/min. The association phase (kon) was followed for 180 s, whereas the dissociation phase (koff) was followed for 600 s. The complete dissociation of active complex formed was achieved by addition of 10 mM HEPES, 150 mM NaCl, 350 mM EDTA and 0.005% (v/v) P20, pH 8.3, for 30 s before each new cycle start.
When experimental data met quality criteria, kinetic parameters were estimated according to the selected binding model using Biacore X100 Evaluation Software. Two possible models were used to fit the kinetic data, which are the simple 1:1 binding model and the so called heterogeneous ligand one; the latter describes the experimental condition of one ligand with two independent binding sites.
Cell cultures
OCI-AML2 cells were grown in MEM-α-Medium with high glucose (GibcoBRL, Gaithersburg, MD, USA). Cells were grown in suspension at 37°C in a humidified atmosphere of 5% (v/v) CO2 in air. All the media were supplemented with 20% (v/v) fetal bovine serum (GibcoBRL). When indicated, cells were treated with 50 or 100 µM TmPyP4.
Viable count of cells was performed using Trypan Blue solution (Sigma-Aldrich).
Chromatin immunoprecipitation
The chromatin immunoprecipitation assay was performed using the EpiQuik chromatin immunoprecipitation kit (Epigentek). For each immunoprecipitation, 6 × 106 OCI-AML2 cells were cross-linked with 1% formaldehyde at room temperature for 8 min in the MEM-α 10% fetal bovine serum medium. The reaction was stopped by adding glycine to the medium to the final concentration of 0.125 M. Cells were harvested, pelleted and washed once with ice-cold phosphate-buffered saline (PBS) and then re-suspended and incubated in lysis buffer containing protease inhibitors. Samples were sonicated on ice six times for 12 s pulses with 1 min intervals between each pulse to obtain 500- to 200-bp chromatin fragments as confirmed by agarose gel electrophoresis. After centrifugation (14 000g for 10 min), an aliquot of the supernatant was incubated (60 min) with 3 µg of anti-NPM antibody (Abcam) previously cross-linked (90 min) to the 96-well strips. An aliquot of each supernatant was used as the input control. Positive and negative controls were processed using 1 µg of anti-RNA polymerase II IgG and anti-hemagglutin (HA) antibodies, respectively. The immunoprecipitation of the protein–DNA complex was performed as recommended by the manufacturer. The immuno-complexes and input controls were incubated with proteinase K at 65°C, the samples transferred to the column, washed with 70 and 90% ethanol and the purified DNA eluted. To verify the binding of NPM1 to the ribosomal DNA quadruplex regions, PCR was carried out according to standard procedure, using 1 µl of the immunoprecipitated DNA. The PCR primers used to amplify the different rDNA G-quadruplex regions are reported in Supplementary Table S1.
Immunofluorescence
Immunofluorescence was performed as described previously (28). Briefly, cells were spotted on microscope slides with cytospin at 900 g for 3 min and then were fixed in 4% (wt/vol) paraformaldehyde in PBS for 10 min. After fixation, cells were permeabilized with 0.1% Triton X-100 for 10 min and then incubated for 1 h in 10% goat serum. After blocking, they were incubated over night with rabbit anti-NPM antibody (1:50 dilution, Cell Signaling Technology), rabbit anti-fibrillarin antibody (1:400, Cell Signaling Technology) or mouse anti-nucleolin antibody (1:100, Upstate, Lake Placid, NY, USA) in blocking solution. Cells were washed three times and incubated for 1 h with secondary antibodies: goat anti-rabbit-Alexa 488; goat anti-mouse-Alexa 488 (1:2000) (Molecular Probes, Eugene, OR, USA) in blocking solution. Then, cells were washed twice and were fixed with Prolong Gold antifade with Dapi kit (Molecular Probes, Invitrogen).
Confocal imaging was performed using a 480-nm ion argon laser and a 542-nm helium–neon laser connected to a Nikon C1 microscope (Nikon, Tokyo, Japan) with a 60× numerical aperture 1.4 lens and analyzed with EZC1 software from Nikon.
Western blotting
Total cell extracts were resolved on a sodium dodecyl sulfate–polyacrylamide gel and blotted onto a Westran clear signal polyvinylidene fluoride (PVDF) membrane (Whatman, Sigma-Aldrich). Membranes were blocked with phosphate buffered saline with Tween 20 (PBST) 5% non-fat dry milk, incubated with primary antibodies for 2 h at room temperature, washed and hybridized for 1 h at room temperature using the appropriate horseradish peroxidase-conjugated secondary antibody (rabbit and mouse; BioRad, Hercules, CA, USA). Detection was performed with the ECL chemiluminescence kit (Perkin Elmer, Waltham, MA, USA). The antibodies used were anti-nucleophosmin (#3542, Cell Signaling), anti-nucleolin clone 3G4B2 (#05-565, Millipore), anti-fibrillarin (#2639, Cell Signaling) and anti–β-actin (#A5441, Sigma).
RESULTS
NPM1 C-terminal domain interacts in vitro with rDNA G-quadruplexes
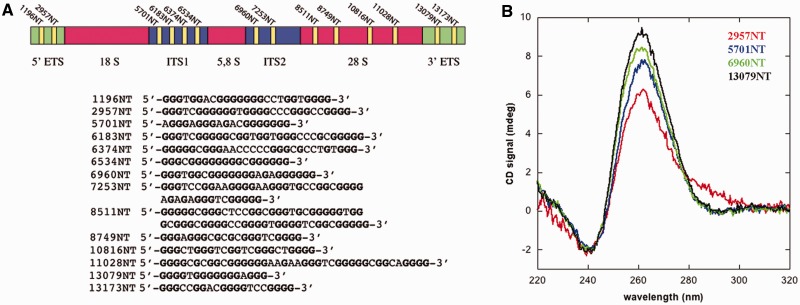
The rDNA gene is transcribed in a 47S precursor that is then processed to generate the 28S, 18S and 5.8S rRNAs (29). Figure 1A shows the organization of the rDNA gene, with two external transcribed regions (ETS) and two internal transcribed regions (ITS) that are present in the 47S pre-rRNA. Several putative quadruplex sequences (PQS) are present in the non-template strand of the rDNA gene (27). These are mainly found in the ETS and ITS regions as well as in the 28S one (yellow bars in Figure 1A). We selected four PQS, each belonging to one of the processed regions, and synthesized the corresponding oligonucleotides (namely, 2957NT, 5701NT, 6960NT and 13079NT). The four oligos property to assume a G-quadruplex assembly in solution was confirmed by circular dichroism. Spectra recorded after annealing the oligos in a buffer containing 150 mM KCl as a stabilizing cation show a positive peak at 262 nm and a through at 240 nm (Figure 1B). Both features are considered typical signatures of a parallel G-quadruplex structure (21).
Figure 1.
G-quadruplex sequences at ribosomal DNA. (A) The rDNA gene is transcribed as a 47S pre-mRNA that is then processed by removal of external and internal transcribed regions (ETS and ITS). The gene contains several putative G-quadruplex sequences in the non-template strand. Their positions are highlighted by yellow bars, and their sequences are reported. Numbering indicates the position of the starting base with respect to the transcription start site. (B) CD spectra of four representative PQS sequences, namely, 2957NT (red), 5701NT (blue), 6960NT (green) and 13079NT (black), annealed in the presence of 150 mM KCl. All spectra present a peak at 262 nm and a through at 240 nm, which are signatures of parallel G-quadruplex assembly.
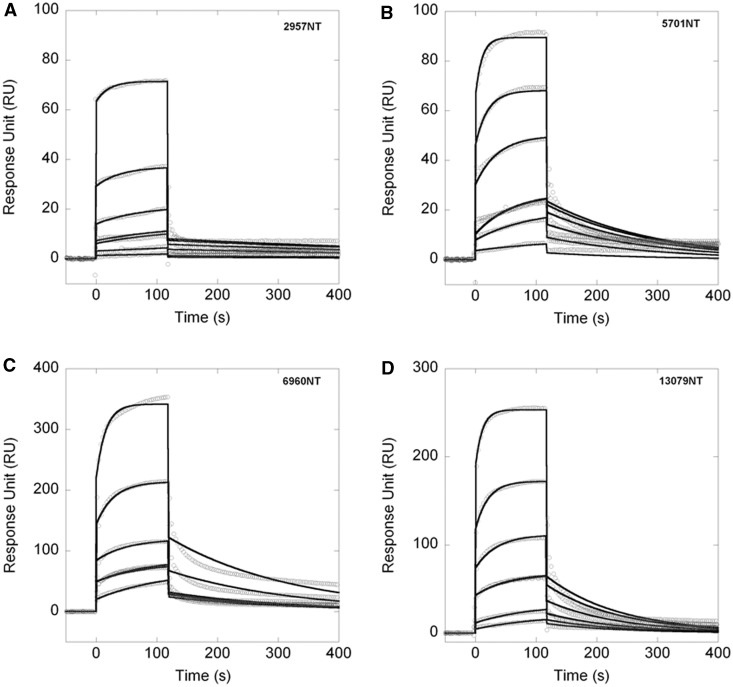
Next, we assessed whether these G-quadruplex regions are recognized by NPM1 in vitro by surface plasmon resonance analysis. As the analyte, we used NPM1-C70, i.e. a construct comprising the last 70 residues of NPM1, namely, residues 225–294, which we have previously shown to be the functional NPM1 domain for G-quadruplex binding (20,26). Figure 2A–D shows the sensorgrams measured for these interactions. In all cases, it was possible to fit the association and dissociation curves with a single-exponential model, indicating that NPM1-C70 binds the four G-quadruplex regions with a 1:1 stoichiometry. Table 1 reports the kinetic and the dissociation constants obtained. The four rDNA G-quadruplex regions are all bound with similar dissociation constants for the 1:1 complexes, ranging from 4.3 to 13.3 µM depending on the oligonucleotides tested (Table 1). These values are similar to those previously obtained with the G-quadruplex regions from the c-MYC and SOD2 promoters (20) and reveal that NPM1-C70 is able to bind several different G-quadruplexes with similar affinities.
Figure 2.
NPM1-C70 binds G-quadruplex sequences at rDNA. (A) Sensorgrams of the interaction of NPM1-C70, used as the analyte, with the immobilized 2957NT oligo. Different traces correspond to increasing NPM1-C70 concentrations. Panels B–D same as panel A using NPM1-C70 as the analyte and the immobilized 5701NT, 6960NT and 13079NT oligos as ligands, respectively. In all cases, a 1:1 kinetic model was used to fit (black lines) the experimental curves (open circles). Kinetic and dissociation constants are reported in Table 1.
Table 1.
SPR kinetic parameters of the interaction between NPM1-C70 and its mutants with different oligos
| Ligand | Analyte | kon (M−1s−1) | koff (s−1) | KD (µM) |
|---|---|---|---|---|
| 2957NT | NPM1-C70 | 360 ± 20 | (1.6 ± 0.2) × 10−3 | 4.3 ± 0.7 |
| 5701NT | NPM1-C70 | 680 ± 40 | (5.6 ± 0.2) × 10−3 | 8.2 ± 0.8 |
| 6960NT | NPM1-C70 | 470 ± 30 | (4.86 ± 0.08) × 10−3 | 10.3 ± 0.7 |
| 13079NT | NPM1-C70 | 630 ± 40 | (8.3 ± 0.1) × 10−3 | 13.3 ± 0.9 |
| 13079NT | NPM1-Cter-MutA | No interaction detected | ||
| 13079NT | MutA-Ref288–290 | 280 ± 10 | (3.02 ± 0.03) × 10−3 | 11.0 ± 0.6 |
Mutant A NPM1 does not interact with G-quadruplex rDNA
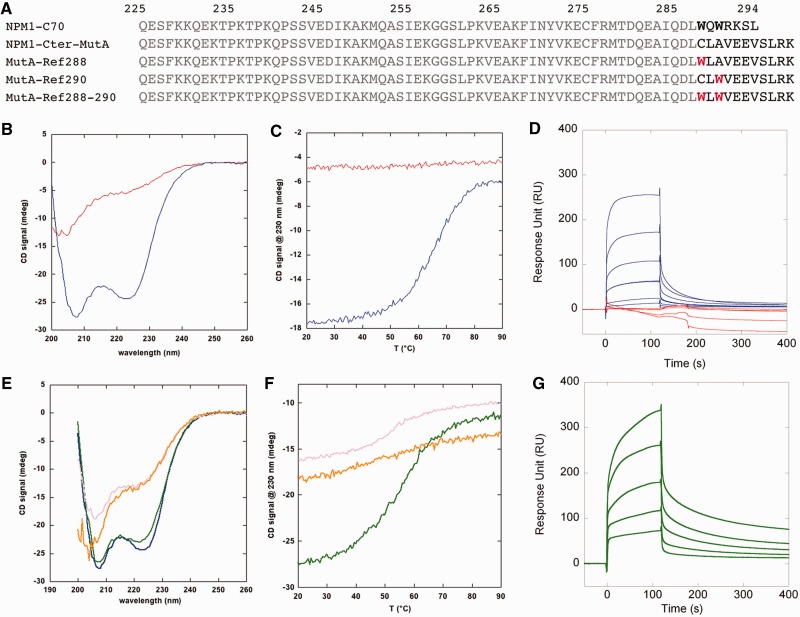
AML-associated NPM1 mutations map to the C-terminal domain of the protein and cause its aberrant cytosolic translocation (12). As we showed that NPM1-C70 is able to interact in vitro with G-quadruplex regions at rDNA, we assessed whether this activity is retained by the mutated protein. To this purpose, we expressed and purified a construct encompassing residues 225–298 of the most common AML-associated mutant NPM1, namely, mutant A, which accounts for nearly 75% of AML cases with NPM1 mutations (4). This construct (NPM1-Cter-MutA) differs from NPM1-C70 because it has four additional residues at the C-terminus, and a different sequence in the last seven, including the replacement of both Trp288 and Trp290 (Figure 3A). Grummit et al. (14) previously reported that a shorter construct for mutant A, encompassing residues 242–298, is natively unstructured. Therefore, we first assessed whether this was also the case for NPM1-Cter-MutA. Figure 3B shows the CD spectra for both NPM1-Cter-MutA and NPM1-C70. Although the latter is the typical spectrum of an α-helical protein, the spectrum for NPM1-Cter-MutA resembles that of a random coil peptide. This is further confirmed by looking at the thermal melting profiles (Figure 3C). A clear transition from the folded to the unfolded species is observed for NPM1-C70. Conversely, no difference in signal at increasing temperatures is observed for NPM1-Cter-MutA.
Figure 3.
AML associated mutant A is unfolded in its DNA binding domain and does not bind rDNA G-quadruplexes. (A) NPM1-Cter-MutA differs from its wild-type counterpart (NPM1-C70) because it is four residues longer, and it has a different sequence in the last seven (residues in black). Refolding mutants were prepared by introducing Trp288 or Trp290, or both (in red), at their wild-type topological position, in the NPM1-Cter-MutA sequence. (B) CD spectra of NPM1-C70 (blue trace) and NPM1-Cter-MutA (red trace). (C) Thermal denaturations of NPM1-C70 (blue trace) and NPM1-Cter-MutA (red trace). (D) Sensorgrams of the interaction between the immobilized oligo 13079NT and NPM1-C70 (blue traces) or NPM1-Cter-MutA (red traces) as the analytes. No binding at all is observed with the leukemic variant. (E) CD spectra of MutA-Ref288 (pink trace), MutA-Ref290 (orange), MutA-Ref288–290 (green) and NPM1-C70 (blue). (F) Thermal denaturation profiles of MutA-Ref288 (pink), MutA-Ref-290 (orange) and MutA-Ref288–290 (green). (G) Sensorgrams of the interaction between the immobilized oligo 13079NT and MutA-Ref288–290 as the analyte. A 1:1 kinetic model was used to fit the association and dissociation curves. Kinetic and dissociation constants are reported in Table 1.
Then, we measured the interaction of NPM1-Cter-MutA with G-quadruplex DNA by surface plasmon resonance. Figure 3D reports the sensorgrams for the interaction between the immobilized 13079NT oligonucleotide and both NPM1-C70 and NPM1-Cter-MutA, as ligands. The association and dissociation curves are clearly detected with NPM1-C70 at all analyte concentrations. Conversely, no binding at all is detected for NPM1-Cter-MutA and, by increasing its concentration, the signal becomes negative, reflecting a larger specific association to the reference cell (30).
Taken together, these data indicate that NPM1-Cter-MutA is completely unfolded and does not bind G-quadruplex regions at rDNA in vitro.
Rescue of Mutant A G-quadruplex binding by stabilizing mutations
NPM1-Cter-MutA has four additional residues and a different sequence in the last seven with respect to its wild-type counterpart (Figure 3A). Therefore, its loss of affinity for G-quadruplexes might in principle be because of the different sequence, rather than to C-terminal domain unfolding. To test this hypothesis, we reintroduced, by site-directed mutagenesis, Trp residues at their topological positions, in the NPM1-Cter-MutA construct, thus obtaining three new variant proteins, namely, MutA-Ref288, MutA-Ref290 and MutA-Ref288–290 (Figure 3A). In both the MutA-Ref288 and MutA-Ref290 constructs, we observed only a partial recovery of folding (Figure 3E and F), which was slightly more pronounced in the MutA-Ref290 construct. Conversely, with the MutA-Ref288–290 construct, where both Trp residues are present in their native topological positions, we observed the typical spectrum of an α-helical protein, almost completely superimposible to that of NPM1-C70 (Figure 3E) and a clear transition from the folded to the unfolded species as a function of increasing temperatures (Figure 3F). These results clearly show that the MutA-Ref288–290 construct is folded similarly to the wild-type, albeit with a reduced stability as indicated by a Tm of 55.6°C versus 66.7°C for the wild-type construct (compare Figure 3F and C).
Next, we tested the ability of the MutA-Ref288–290 construct to interact with the 13079NT rDNA G-quadruplex by SPR. As shown in Figure 3G, MutA-Ref288–290 binds this oligonucleotide similarly to the wild-type domain, with a KD = 11 µM (Table 1).
Thus, the folding of NPM1 C-terminal domain and its ability to bind G-quadruplex regions are strictly correlated.
NPM1 interacts with rDNA G-quadruplex in OCI-AML2 cells
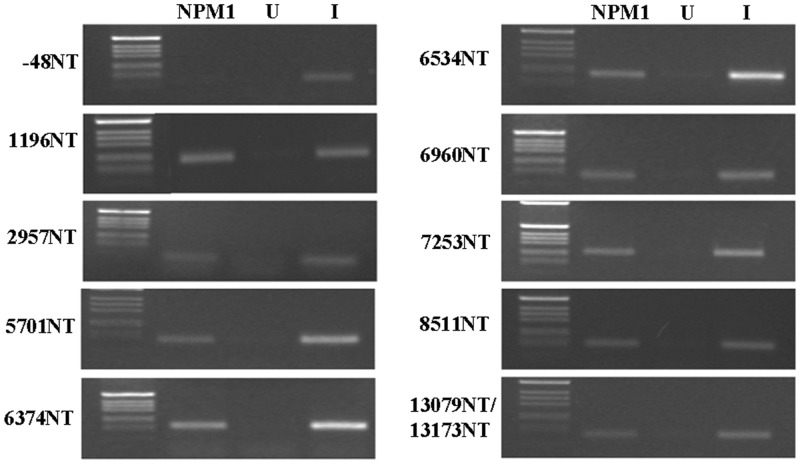
Having shown that NPM1-C70 interacts with G-quadruplex regions in the rDNA gene and that the most common AML-associated mutation impairs this activity in vitro, we wanted to assess whether NPM1 is equally able to interact with G-quadruplex regions within a cellular context. To this end, we performed chromatin immunoprecipitation experiments on OCI-AML2 cell lysates. In these AML cells, bearing wild-type NPM1 at both alleles, the protein is mainly localized in the nucleoli. Cell lysates were immunoprecipitated with either an anti-NPM1 antibody or an unrelated antibody, as negative control, and the immunoprecipitates were PCR amplified with oligos matching PQS belonging to different regions of the rDNA gene. As an internal negative control, we also amplified a region in between the rDNA gene promoter and the 5′-ETS (−48NT), which does not contain PQS (27). As shown in Figure 4, NPM1 binds to all the regions containing PQS but not to the −48NT region. Therefore, these data indicate that NPM1 binds G-quadruplex regions on the rDNA gene also in vivo.
Figure 4.
NPM1 binds G-quadruplex sequence at rDNA in vivo. Chromatin immunoprecipitation experiments on OCI-AML2 cell lysates. Immunoprecipitates with either a NPM1 antibody (NPM1) or the unrelated HA antibody (U) were PCR amplified with oligos matching selected PQS in the rDNA gene. A region in between the end of the gene promoter and the transcription start site that does not contain PQS (−48NT) was also amplified for negative control. Input (I) lanes represent PCR amplifications of the cell lysate, without immunoprecipitation.
TmPyP4 competes with NPM1-C70 for G-quadruplex binding
As NPM1 interacts with rDNA G-quadruplexes both in vitro and in vivo, we asked whether a G-quadruplex selective ligand, like the porphyrin TmPyP4 (31–33), might interfere with this activity.
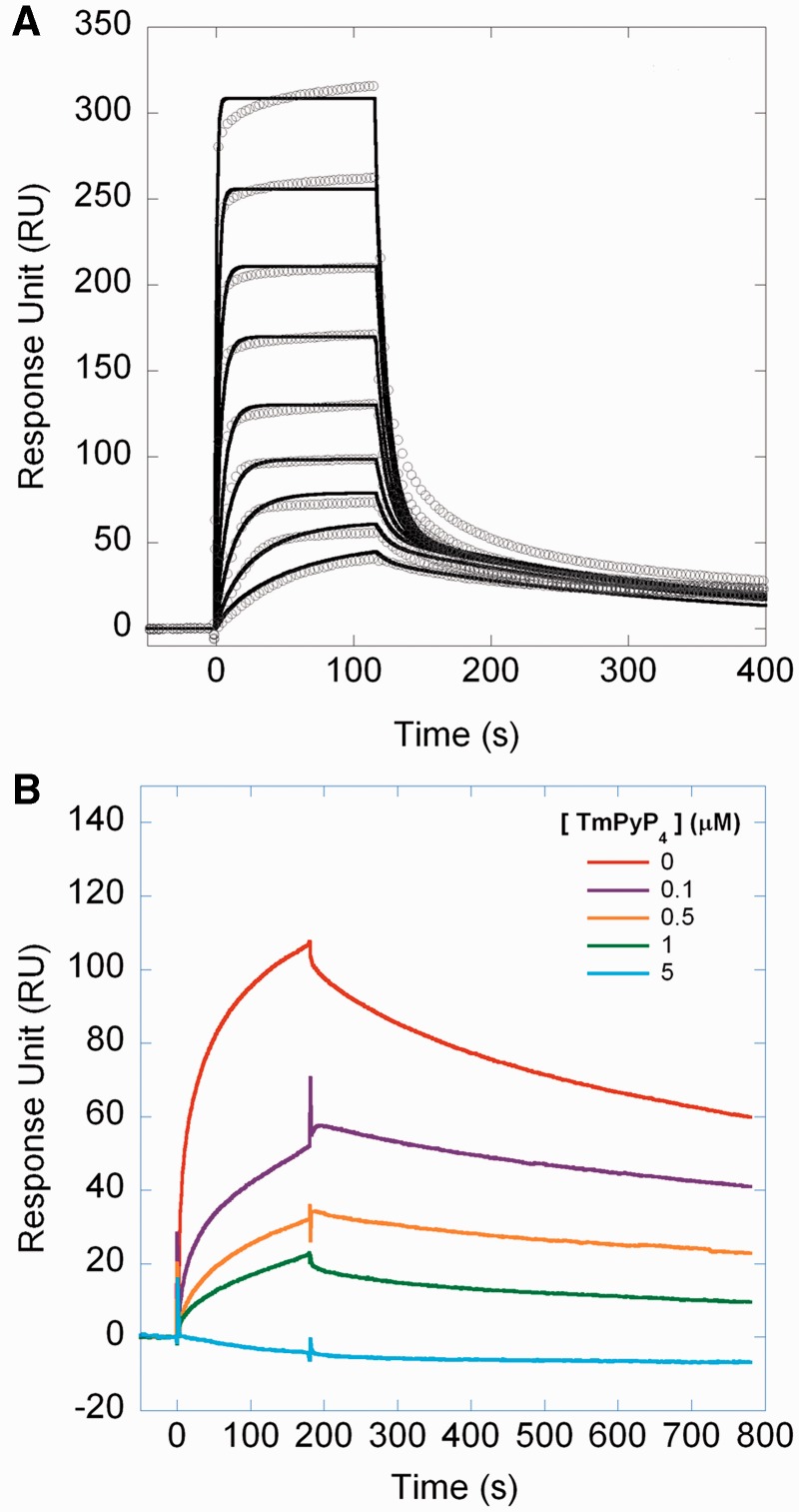
First, we tested the interaction between the four rDNA G-quadruplexes, biotynylated and immobilized on streptavidin chips as ligands and TmPyP4 as the analyte in a SPR assay. As shown in Figure 5A for the 13079NT oligonucleotide and in Supplementary Figure S1 for the other three oligonucleotides, all sensorgrams are fitted with a heterogenous ligand kinetic model, which postulates two binding sites for TmPyP4 on the DNA scaffold, with different affinities. The kinetic and dissociation constants are reported in Table 2, with values ranging from 8 to 16 nM and from 1 to 3 µM, for the first and second KD, respectively. Both the stoichiometry of the interaction and the KD values are those typical for the interaction of TmPyP4 with a G-quadruplex scaffold, whereas the same molecule binds unstructured oligos with 1:1 stoichiometry and reduced affinity (20,31–33).
Figure 5.
TmPyP4 competes with NPM1-C70 for G-quadruplex binding. (A) The 13079NT G-quadruplex oligonucleotide was immobilized onto a streptavidin (SA) chip and TmPyP4 used as the analyte in SPR experiments. All traces (open circles) were fitted (black lines) with a heterogeneous ligand kinetic model. Kinetic ad dissociation constants are reported in Table 2. (B) The histidine-tagged NPM1-C70 protein was immobilized onto a NTA chip. The analyte was the 13079NT G-quadruplex oligonucleotide (1 µM) alone (red) or in combination with 0.1 (magenta), 0.5 (orange), 1.0 (green) and 5 µM (cyan) TmPyP4. Data show that the signal decreases at increasing amounts of TmPyP4.
Table 2.
SPR kinetic parameters of the interaction between TmPyP4 and different oligos
| kon1 (M−1s−1) | koff1 (s−1) | KD1 (nM) | kon2 (M−1s−1) | koff2 (s−1) | KD2 (µM) | |
|---|---|---|---|---|---|---|
| 2957NT | (22.82 ± 0.01) × 105 | (1.91 ± 0.01) × 10−2 | 8.35 ± 0.01 | (1.27 ± 0.01) × 105 | 0.145 ± 0.001 | 1.14 ± 0.01 |
| 5701NT | (8.1 ± 0.3) × 105 | (1.306 ± 0.005) × 10−2 | 16.1 ± 0.7 | (1.78 ± 0.06) × 105 | 0.44 ± 0.01 | 2.5 ± 0.2 |
| 6960NT | (32 ± 1) × 105 | (3.2 ± 0.1) × 10−2 | 10 ± 1 | (3.96 ± 0.09) × 104 | 0.132 ± 0.002 | 3.2 ± 0.1 |
| 13079NT | (4.30 ± 0.08) × 105 | (3.76 ± 0.04) × 10−3 | 8.7 ± 0 .3 | (9.9 ± 0.2) × 104 | 0.115 ± 0.001 | 1.16 ± 0.04 |
Having shown that TmPyP4 binds rDNA G-quadruplexes with much higher affinity than NPM1-C70, we next wanted to assess whether TmPyP4 effectively competes with NPM1-C70 for G-quadruplex binding. To this purpose, we immobilized the histidine-tagged NPM1-C70 protein on an NTA chip and used the 13079NT as the analyte in a SPR assay. Figure 5B shows the sensorgrams obtained when challenging the immobilized protein with 1 µM 13079NT alone or mixed with increasing amounts of TmPyP4. We observe that, in the presence of 0.1 µM TmPyP4, the signal decreases by ∼50%. This is ascribed to the fact that the association time course is heterogeneous and considerably slowed down, preventing equilibrium to be reached within the time of the association phase (180 s). By further increasing the ratio between TmPyP4 and the G-quadruplex, the signal is consistently reduced. An 80% reduction of the signal is observed with an equimolar ratio and, at a 5-fold molar excess of TmPyP4, the binding is completely lost. This experiment demonstrates that the G-quadruplex, on binding TmPyP4, completely loses affinity for NPM1-C70 and that a ternary complex does not form.
Interfering with G-quadruplex binding displaces NPM1 from nucleoli in cells
NPM1 mutations cause loss of nucleolar localization, and because of the appearance of an additional nuclear export signal (Figure 3A), they also cause the stable and aberrant cytosolic localization of the variant protein (12,34). As NPM1-Cter-MutA loses the ability to bind G-quadruplex regions at rDNA, this might correlate with the loss of nucleolar localization of NPM1-MutA. If this is the case, it should be equally possible to displace wild-type NPM1 from nucleoli by competing with its binding to G-quadruplex regions in the rDNA.
Having shown (Figure 5A and Table 2) that TmPyP4 binds rDNA G-quadruplexes with higher affinity than NPM1-C70, and that it effectively competes with NPM1-C70 for G-quadruplex binding (Figure 5B), we next tested whether TmPyP4 treatment impacts on NPM1 localization within OCI-AML2 cells.
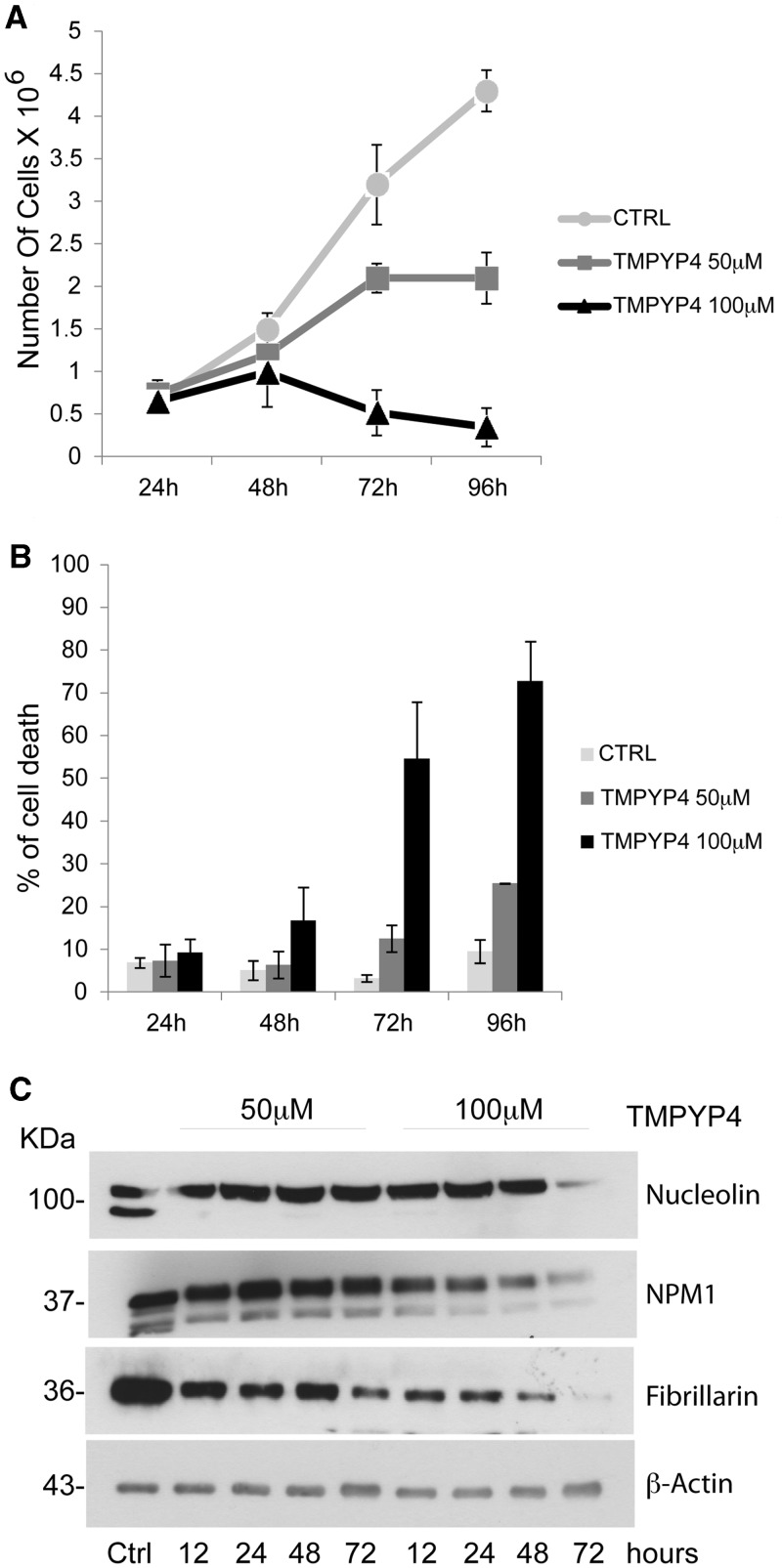
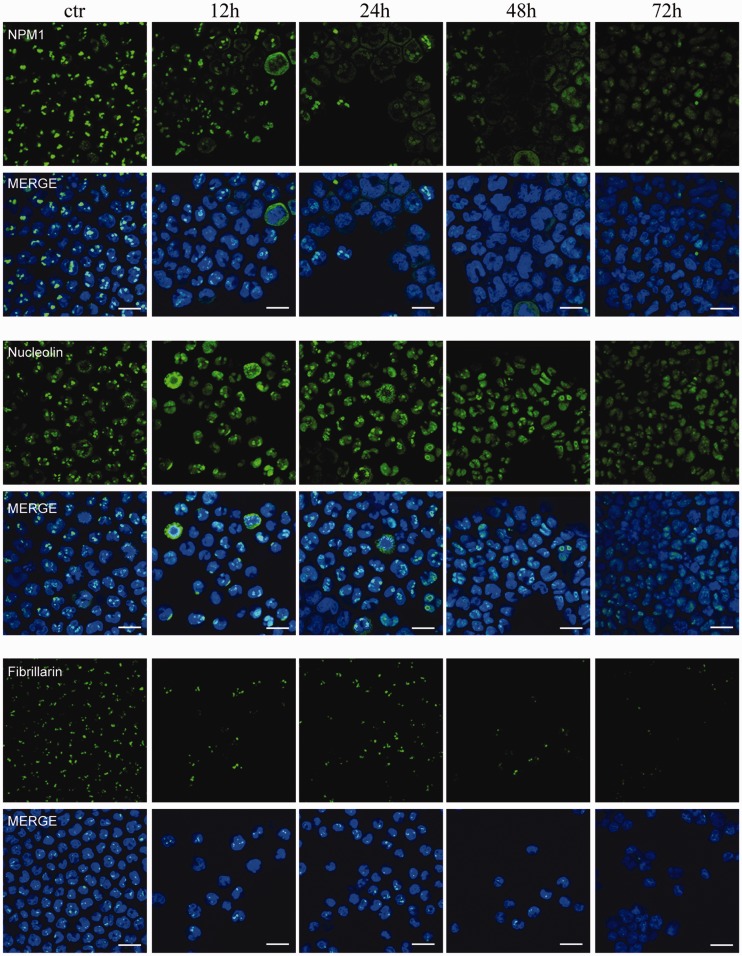
We initially tested several concentrations of TmPyP4 on these cells to assess its potential toxicity. As shown in Figure 6AB, OCI-AML2 cells treated with 100 µM TmPyP4 are still viable after 48 h, but they show a reduced growth rate, whereas at longer time points, cells undergo cell death. At a lower TmPyP4 concentration (50 µM), cells remain viable throughout most of the experiment, showing 30% of cell death only after 72 h of treatment. To investigate the effect of TmPyP4 on the nucleolar localization of NPM1, OCI-AML2 cells were, therefore, treated with the non-toxic concentration of 50 µM, and endogenous NPM1 localization was visualized at various time points by immunofluorescence. Figure 7 shows that already 12 h after treatment, NPM1 starts to localize outside the nucleolus and shows an almost complete diffused nuclear staining 24 h after treatment. Other two nucleolar proteins commonly used as nucleolar markers, nucleolin and fibrillarin also leave the nucleolus and show a more diffused staining. Both these proteins are known to bind NPM1 (35,36), and nucleolin has also been shown to directly bind rDNA G-quadruplexes (27). Interestingly, the staining for fibrillarin after treatment with TmPyP4 seems to be faint, suggesting that, contrary to NPM1 and nucleolin, this protein might be rapidly degraded on delocalization from the nucleolus. Indeed, WB analysis of the three proteins after treatment with 50 µM TmPyP4 shows that although nucleolin and nucleophosmin levels are stable, fibrillarin seems to be degraded (Figure 6C). Treatment with TmPyP4 100 µM shows a more dramatic effect on fibrillarin protein levels, but it still has no or modest effect on NPM1 and nucleolin protein levels at early time points. At longer time points, at this concentration, the reduction of all three protein levels is probably linked to the fact that the cells are undergoing massive cell death (Figure 6C).
Figure 6.
Effect of TmPyP4 treatment on OCI-AML2 cell growth and protein levels. (A) Viability assay of OCI-AML2 cells untreated (ctrl) and treated with 50 µM and 100 µM TmPyP4 for the indicated time points (hours). (B) Percentage of OCI-AML2 cell death after treatment with 50 and 100 µM TmPyP4 for the indicated times (hours). (C) Western blot analysis of OCI-AML2 cells treated as described earlier in the text, using antibodies against NPM1, nucleolin, fibrillarin and β-actin as a loading control. Numbers on the left indicate the corresponding molecular weights. NPM1 and nucleolin levels seem to be stable at both TmPyP4 concentrations at early time points. After 72 h, with 100 µM TmPyP4, both NPM1 and nucleolin levels decrease, probably because of cell death. Conversely, fibrillarin levels start decreasing at both TmPyP4 concentrations as soon as 12 h after treatment, in a dose-dependent manner.
Figure 7.
Immunofluorescence of OCI-AML2 cells treated with 50 µM TmPyP4 for the indicated times and stained with anti-NPM1 antibody (green) anti-nucleolin antibody (green), anti-fibrillarin antibody (green) and with Dapi dye (blue) to highlight the nucleus (scale bar: 20 μm).
DISCUSSION
The nucleolus is a highly dynamic compartment whose functions extend beyond the classical and well established role of ‘ribosome factory’ (37). For instance, a great deal of data suggest an involvement of nucleoli in the responses to several stress signals and in the regulation of cell cycle and cell growth (37,38). The highly regulated trafficking of NPM1 seems to be critical for many of these functions (3). In fact, although the bulk of the protein is mainly found at nucleoli during interphase, NPM1 is also capable of shuttling between nucleoli, nucleus and cytoplasm in response to a variety of post-translational modifications and at different stages of the cell cycle (39). NPM1 trafficking is mediated by specific signals in the protein sequence. In particular, two nuclear export signals (NES) and two nuclear localization signals are present in NPM1 N-terminal core domain and central acidic domain, respectively (1,13,34). Furthermore, a highly specific and strong nucleolar localization signal is found in the C-terminal end of the protein. This signal was originally identified by Nishimura et al. (40) who observed, using green fluorescent protein (GFP)-tagged protein deletions, that the two tryptophan residues at positions 288 and 290 were necessary for nucleolar localization. Both truncation of the protein sequence immediately before Trp288, or mutation to alanine of both Trp residues, was sufficient to dislocate the protein in the nucleoplasm (40).
In NPM1 AML-associated mutations, the insertion or duplication of short-base sequences at the terminal exon of the gene results in the aberrant export of the encoded protein in the cytoplasm (41). This is likely because of the increased probability of exportin1 - chromosome region maintenance 1 (CRM1) to interact with NPM1 mutants. In fact, on the one hand, NPM1 mutants carry an additional NES motif (the sequence specifically recognized by CRM1) as compared with native NPM1, and on the other hand, they roam in the nucleoplasm where CRM1 is also located. Roaming of NPM1 mutants in the nucleoplasm is the result of their absent or markedly reduced capability to bind the nucleolus because of the loss of Trp288 and Trp290, or Trp290 alone. However, the underlying molecular mechanism responsible for the impaired capability of the NPM1 mutants to bind the nucleolus remained unknown.
This work establishes a direct link between NPM1 nucleolar localization and its G-quadruplex–binding properties. We have shown that G-quadruplex sequences found at rDNA genes, which are clustered in tandem arrays in the nucleoli, are specifically recognized by NPM1, both in vitro and in vivo. This activity seems to be correlated with the nucleolar localization of the protein. Consistently, treatment of OCI-AML2 cells with TmPyP4, a G-quadruplex ligand, results in a rapid and complete displacement of NPM1 from nucleoli. Importantly, nucleolar displacement was observed at early time points, and the protein was mostly nucleoplasmic 24 h after treatment, with still viable and dividing cells. This fact argues in favor of a competitive effect of TmPyP4 on NPM1 G-quadruplex binding, rather than on an indirect consequence of ligand mediated nucleolar disruption. However, it has to be noted that TmPyP4 is also able to bind, albeit with reduced affinity, duplex DNA; therefore, it may not be excluded that other NPM1 interactions with nucleic acids, at the nucleolar level, might be affected by TmPyP4. At longer time points, TmPyP4 treatment results in OCI-AML2 cell death in a dose-dependent manner. This behavior may not be necessarily or solely ascribed to NPM1 dislocation from nucleoli. For instance, we have shown that, on TmPyP4 treatment, nucleolin and fibrillarin are also displaced from nucleoli, with a kinetic similar to NPM1, and that, although NPM1 and nucleolin remain stable in nucleoplasm, fibrillarin is rapidly degraded. Nucleolin was previously shown to specifically interact with G-quadruplex regions at rDNA (27), whereas fibrillarin was reported to interact with both NPM1 and nucleolin (35,42), thus explaining the effect of TmPyP4 on both these proteins.
NPM1 and nucleolin are thought to act as hub proteins in the nucleoli (37). They participate to the initial formation of nucleolus-derived foci around clusters of rDNA, and then they contribute to the final nucleolar assembly by segregating in this compartment a wide array of other protein components (37,43). Hence, it is not surprising that TmPyP4 treatment finally results in massive cell death, possibly as a consequence of nucleolar stress induced by NPM1 and nucleolin dislocation.
Having established that NPM1 nucleolar localization is dictated by G-quadruplex binding at rDNA, the following step was to assess whether the loss of nucleolar localization of AML-associated NPM1 variants might be because of impaired G-quadruplex binding at rDNA. To do this, we expressed the C-terminal domain of mutant A NPM1, the most common leukemic variant which is devoid of both Trp288 and Trp290, and we showed that the G-quadruplex–binding activity is completely lost in the mutant. We further investigated this issue by re-introducing the Trp residues in their native topological positions but in the context of the mutant A sequence, and we showed that (i) by introducing Trp288 or Trp290 separately, only a partial rescue of folding is observed; (ii) when both Trp288 and Trp290 are inserted in their topological position, the complete refolding of the domain is observed in the context of the variant sequence; and (iii) the NPM1-Cter-MutA refolded variant binds to G-quadruplex regions at rDNA with an affinity similar to the native construct. Importantly, previous work already demonstrated that such variant re-acquire nucleolar localization (41). Taken together, these data demonstrate that a direct correlation exists between NPM1 G-quadruplex binding at rDNA and its nucleolar localization. In the NPM1 mutated counterpart, the loss of C-terminal domain native structure results in decreased affinity for rDNA and, consequently, in the movement of the protein to the nucleoplasm. The presence of an additional NES in the C-termini reinforces CRM1-mediated nuclear export and finally results in the aberrant and stable cytoplasmic localization of the protein, which is the hallmark of this AML subtype (12,13,34). Furthermore, NPM1-MutA interacts with wild-type NPM1 through the N-terminal oligomerization domain, and this results in cytosolic translocation of the majority of both wild-type and mutated proteins, even though in AML patients mutations are always heterozygous (12,13).
Previous data on NPM1−/− knockout mice have shown early embryonic lethality (6) and suggested that NPM1 heterozygosity may be obligate in this kind of leukemia (44). In fact, in patients’ leukemic blasts, although eminently cytosolic, a small fraction of the wild-type protein always resides in nucleoli, and this probably avoids exessive nucleolar stress that may be a disadvantage for the blasts (44). We have shown here that treatment with TmPyP4 displaces NPM1 and other important nucleolar components, such as nucleolin and fibrillarin, from their native compartment and this, at longer time points, results in cell death. We plan in the future to further investigate this issue and to better characterize the molecular mechanism underlying cell death in these cells and in primary AML cells bearing NPM1 mutations, also extending these studies to other, more specific, G-quadruplex ligands. In fact, it has been previously suggested (44) that one possible strategy to specifically treat this kind of leukemia might be that of targeting residual wild-type NPM1 nucleolar localization. As leukemic blasts do possess low amounts of the protein in nucleoli with respect to healthy cells, it might be possible to selectively induce nucleolar stress only in the former by adjusting the dosage of a putative drug aimed at displacing NPM1 from nucleoli. Alternatively, as we have shown that NPM1 C-terminal domain folding and G-quadruplex binding are correlated, one other strategy might be that of searching for a ligand aimed at refolding the mutated protein, possibly restoring nucleoplasmic and/or nucleolar localization.
Future work in our laboratories will investigate both these possibilities in search for a specific treatment for AML with mutated NPM1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figure 1.
FUNDING
Associazione Italiana Ricerca sul Cancro (AIRC) [IG2011-11712 to L.F. and IG-10111 to B.F.]; Financial support by the Italian Ministry of University and Research [Linea D.1, 2011 Università Cattolica Sacro Cuore, Roma to A.A and RBRN07BMCT_007 to A.D.M.]. Funding for open access charge: Associazione Italiana Ricerca sul Cancro (AIRC) (to L.F.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof. Maurizio Brunori for his precious support and valuable advice.
REFERENCES
- 1.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat. Rev. Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 2.Maggi LB, Jr, Kuchenruether M, Dadey DY, Schwope RM, Grisendi S, Townsend RR, Pandolfi PP, Weber JD. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the mammalian ribosome. Mol. Cell Biol. 2008;28:7050–7065. doi: 10.1128/MCB.01548-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindstrom MS. NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem. Res. Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falini B, Martelli MP, Bolli N, Sportoletti P, Liso A, Tiacci E, Haferlach T. Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood. 2011;117:1109–1120. doi: 10.1182/blood-2010-08-299990. [DOI] [PubMed] [Google Scholar]
- 5.Wang W, Budhu A, Forgues M, Wang X. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat. Cell Biol. 2005;7:823–830. doi: 10.1038/ncb1282. [DOI] [PubMed] [Google Scholar]
- 6.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 7.Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 2002;4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 8.Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol. Cell Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colombo E, Martinelli P, Zamponi R, Shing DC, Bonetti P, Luzi L, Volorio S, Bernard L, Pruneri G, Alcalay M, et al. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res. 2006;66:3044–3050. doi: 10.1158/0008-5472.CAN-05-2378. [DOI] [PubMed] [Google Scholar]
- 10.Bonetti P, Davoli T, Sironi C, Amati B, Pelicci PG, Colombo E. Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7 gamma. J. Cell Biol. 2008;182:19–26. doi: 10.1083/jcb.200711040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falini B, Nicoletti I, Bolli N, Martelli MP, Liso A, Gorello P, Mandelli F, Mecucci C, Martelli MF. Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica. 2007;92:519–532. doi: 10.3324/haematol.11007. [DOI] [PubMed] [Google Scholar]
- 12.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 13.Falini B, Bolli N, Liso A, Martelli MP, Mannucci R, Pileri S, Nicoletti I. Altered nucleophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009;23:1731–1743. doi: 10.1038/leu.2009.124. [DOI] [PubMed] [Google Scholar]
- 14.Grummitt CG, Townsley FM, Johnson CM, Warren AJ, Bycroft M. Structural consequences of nucleophosmin mutations in acute myeloid leukemia. J. Biol. Chem. 2008;283:23326–23332. doi: 10.1074/jbc.M801706200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scaloni F, Gianni S, Federici L, Falini B, Brunori M. Folding mechanism of the C-terminal domain of nucleophosmin: residual structure in the denatured state and its pathophysiological significance. FASEB J. 2009;23:2360–2365. doi: 10.1096/fj.08-128306. [DOI] [PubMed] [Google Scholar]
- 16.Scaloni F, Federici L, Brunori M, Gianni S. Deciphering the folding transition state structure and denatured state properties of nucleophosmin C-terminal domain. Proc. Natl Acad. Sci. USA. 2010;107:5447–5452. doi: 10.1073/pnas.0910516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falini B, Martelli MP, Bolli N, Bonasso R, Ghia E, Pallotta MT, Diverio D, Nicoletti I, Pacini R, Tabarrini A, et al. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006;108:1999–2005. doi: 10.1182/blood-2006-03-007013. [DOI] [PubMed] [Google Scholar]
- 18.Lee HH, Kim HS, Kang JY, Lee BI, Ha JY, Yoon HJ, Lim SO, Jung G, Suh SW. Crystal structure of human nucleophosmin-core reveals plasticity of the pentamer-pentamer interface. Proteins. 2007;69:672–678. doi: 10.1002/prot.21504. [DOI] [PubMed] [Google Scholar]
- 19.Hingorani K, Szebeni A, Olson MO. Mapping the functional domains of nucleolar protein B23. J. Biol. Chem. 2000;275:24451–24457. doi: 10.1074/jbc.M003278200. [DOI] [PubMed] [Google Scholar]
- 20.Federici L, Arcovito A, Scaglione GL, Scaloni F, Lo Sterzo C, Di Matteo A, Falini B, Giardina B, Brunori M. Nucleophosmin C-terminal leukemia-associated domain interacts with G-rich quadruplex forming DNA. J. Biol. Chem. 2010;285:37138–37149. doi: 10.1074/jbc.M110.166736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 22.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Huppert JL, Balasubramanian S. Prevalence of quadruplexes at the human genome. Nucleic Acid Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallo A, Lo Sterzo C, Mori M, Di Matteo A, Bertini I, Banci L, Brunori M, Federici L. Structure of nucleophosmin DNA-binding domain and analysis of its complex with a G- quadruplex sequence from the c-MYC promoter. J. Biol. Chem. 2012;287:26539–26548. doi: 10.1074/jbc.M112.371013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, et al. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 28.Munarriz E, Barcaroli D, Stephanou A, Townsend PA, Maisse C, Terrinoni A, Neale MH, Martin SJ, Latchman DS, Knight RA, et al. PIAS-1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73. Mol. Cell Biol. 2004;24:10593–10610. doi: 10.1128/MCB.24.24.10593-10610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- 30.Leonard P, Hearty S, O’Kennedy R. Measuring protein-protein interactions using Biacore. Methods Mol. Biol. 2011;681:403–418. doi: 10.1007/978-1-60761-913-0_22. [DOI] [PubMed] [Google Scholar]
- 31.Freyer MW, Buscaglia R, Kaplan K, Cashman D, Hurley LH, Lewis EA. Biophysical studies of the c-MYC NHE III1 promoter: model quadruplex interactions with a cationic porphyrin. Biophys. J. 2007;92:2007–2015. doi: 10.1529/biophysj.106.097246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei C, Jia G, Yuan J, Feang Z, Li C. A spectroscopic study on the interactions of porphyrin with G-quadruplex DNAs. Biochemistry. 2006;45:6681–6691. doi: 10.1021/bi052356z. [DOI] [PubMed] [Google Scholar]
- 33.Wei C, Wang J, Zhang M. Spectroscopic study on the binding of porphyrins to (G4T4G4)4 parallel G-quadruplex. Biophys. Chem. 2010;148:51–55. doi: 10.1016/j.bpc.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Bolli N, Nicoletti I, De Marco MF, Bigerna B, Pucciarini A, Mannucci R, Martelli MP, Liso A, Mecucci C, Fabbiano F, et al. Born to be exported: COOH-terminal nuclear export signals of different strength ensure cytoplasmic accumulation of nucleophosmin leukemic mutants. Cancer Res. 2007;67:6230–6237. doi: 10.1158/0008-5472.CAN-07-0273. [DOI] [PubMed] [Google Scholar]
- 35.Amin MA, Matsunaga S, Uchiyama S, Fukui K. Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem. J. 2008;415:345–351. doi: 10.1042/BJ20081411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li YP, Bush RK, Valdez BC, Bush H. C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur. J. Biochem. 1996;237:153–158. doi: 10.1111/j.1432-1033.1996.0153n.x. [DOI] [PubMed] [Google Scholar]
- 37.Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colombo E, Alcalay M, Pelicci PG. Nucleophosmin and its complex network: a possible therapeutic target in hematological diseases. Oncogene. 2011;30:2595–2609. doi: 10.1038/onc.2010.646. [DOI] [PubMed] [Google Scholar]
- 40.Nishimura Y, Ohkubo T, Furuichi Y, Umekawa H. Tryptophans 286 and 288 in the C-terminal region of protein B23.1 are important for its nucleolar localization. Biosci. Biotechnol. Biochem. 2002;66:2239–2242. doi: 10.1271/bbb.66.2239. [DOI] [PubMed] [Google Scholar]
- 41.Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A, Bigerna B, Pasqualucci L, Mannucci R, Rosati R, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107:4514–4523. doi: 10.1182/blood-2005-11-4745. [DOI] [PubMed] [Google Scholar]
- 42.Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K. Nucleolin functions in nucleolus formation and chromosome congression. J. Cell Sci. 2007;120:2091–2105. doi: 10.1242/jcs.008771. [DOI] [PubMed] [Google Scholar]
- 43.Hernandez-Verdun D, Roussel P, Gébrane-Younès J. Emerging concepts of nucleolar assembly. J. Cell Sci. 2002;115:2265–2270. doi: 10.1242/jcs.115.11.2265. [DOI] [PubMed] [Google Scholar]
- 44.Falini B, Gionfriddo I, Cecchetti F, Ballanti S, Pettirossi V, Martelli MP. Acute Myeloid Leukemia with mutated nucleophosmin (NPM1): any hope for a targeted therapy? Blood Rev. 2011;25:247–254. doi: 10.1016/j.blre.2011.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.