Abstract
In all organisms, transfer RNAs (tRNAs) undergo extensive post-transcriptional modifications. Although base modifications in the anticodon are known to alter decoding specificity or improve decoding accuracy, much less is known about the functional relevance of modifications in other positions of tRNAs. Here, we report the identification of an A-to-I tRNA editing enzyme that modifies the tRNA-Ala(AGC) in the model plant Arabidopsis thaliana. The enzyme is homologous to Tad1p, a yeast tRNA-specific adenosine deaminase, and it selectively deaminates the adenosine in the position 3′-adjacent to the anticodon (A37) to inosine. We show that the AtTAD1 protein is exclusively localized in the nucleus. The tad1 loss-of-function mutants isolated in Arabidopsis show normal accumulation of the tRNA-Ala(AGC), suggesting that the loss of the I37 modification does not affect tRNA stability. The tad1 knockout mutants display no discernible phenotype under standard growth conditions, but produce less biomass under environmental stress conditions. Our results provide the first evidence in support of a physiological relevance of the A37-to-I modification in eukaryotes.
INTRODUCTION
Transfer RNAs (tRNAs) are highly modified in all organisms and genetic compartments that have been investigated. For example, as many as 16.4% of the residues in cytosolic tRNAs from the yeast Saccharomyces cerevisiae carry chemical modifications at distinct sites. The number of modifications per tRNA species is in the range between 7 and 17 (1). Most of these modifications are well conserved in other eukaryotes. Modifications in tRNAs are generally introduced at the post-transcriptional level, and, in recent years, a considerable number of tRNA-modifying enzymes have been identified (2–6).
The roles of many tRNA modifications in both prokaryotes and eukaryotes are not fully understood. Several modifications in and near the anticodon have been found to exert crucial functions in translation, for example, by facilitating wobbling (7–10), enhancing decoding (11) or influencing the propensity to ribosomal frameshifting (4,12). tRNA modifications remote from the anticodon loop can also directly influence the translation process and/or tRNA recognition (2) or can have roles in tRNA folding and stability (1). However, to date, the precise functions of many naturally occurring tRNA modifications have remained unknown. Often, loss of a modification does not negatively influence cellular viability and does not even impair growth under laboratory conditions (1,13).
A common base modification in tRNAs is the hydrolytic deamination of adenosine to inosine. This modification is often referred to as tRNA editing because (i) A-to-I modifications also occur in mRNAs of various eukaryotes, and (ii) inosine differs in its base pairing potential from adenosine; therefore, A-to-I modifications alter the coding properties of the affected RNA molecule (14–17). Inosine in the wobble position of the anticodon is often an essential tRNA modification, in which loss of the modifying enzyme is incompatible with cellular viability. For example, the single A-to-I editing event in tRNA-Arg(ACG) of Escherichia coli is performed by a dedicated deaminase (dubbed tadA for tRNA-specific adenosine deaminase) that is encoded by an essential gene (18). Essentiality of A-to-I editing in tRNA-Arg(ACG) is most likely because of the requirement for inosine in the wobble position to decode the three arginine codons CGU, CGC and CGA (7). Eukaryotic wobble position-specific A-to-I tRNA deaminases have been termed ADATs (adenosine deaminases acting on tRNA). Similar to tadA, their deaminase domain comprises three zinc-binding amino acid residues (Supplementary Figure S1) and a proton-shuttling glutamate residue (19–21). Closely related to ADATs are the ADARs (adenosine deaminases acting on RNA), the family of enzymes catalyzing mRNA editing by A-to-I conversions in double-stranded transcript regions (17,22–25).
Plant cells have three genetic compartments: the nucleocytosolic compartment, the mitochondrion and the plastid (chloroplast). Each of the three compartments has its own specific set of tRNAs. Although tRNAs in the chloroplast and, to a lesser extent, also tRNAs in the mitochondrion are relatively well studied (26–32), less is known about cytosolic tRNAs and their modifications (33). This is mainly because of the difficulty of purifying individual cytosolic tRNA species in sufficient quantities. In contrast, isolation of cytoplasmic organelles represents a straightforward procedure that, because of the low complexity of organellar transcriptomes, results in a strong enrichment of tRNAs. To shed some light on tRNA modification in the nucleocytosolic compartment of plant cells, we have begun to identify modifying enzymes based on structural and sequence motifs that are potentially highly conserved in all eukaryotes. Here, we report the identification and functional analysis of a Tad1/ADAT1-like protein from the model plant Arabidopsis thaliana that specifically deaminates the adenosine in the position 3′-adjacent to the anticodon of the cytosolic tRNA-Ala(AGC).
MATERIALS AND METHODS
Plant material, growth conditions and phenotypic assays
All A. thaliana lines used in this study are in the Columbia ecotype background. Surface-sterilized seeds were stratified for 3 days, then sown onto half-strength Murashige and Skoog (MS) medium (34) with 1% sucrose and grown in controlled environment chambers. For phenotypic assays, plants were transferred to soil 7 days after germination on synthetic medium and grown under long-day conditions (16 h light/8 h dark regime) or short-day conditions (8 h light/16 h dark regime) at a light intensity of 120 µE m−2 s−1 at 22°C. For cold stress treatments, plants were transferred, after 20 days of growth under standard conditions, to 4°C for 20 days (at 100 μE m−2 s−1). For heat stress treatments, plants were transferred after 16 days of growth under standard conditions to 30°C for 8 days (at 150 μE m−2 s−1). To exclude possible confounding effects from unequal illumination or temperature, plants from the various lines were randomly mixed.
Homozygous T-DNA insertion lines from the SALK (35) and GABI-Kat (36) collections were identified by polymerase chain reaction (PCR) using genomic DNA and T-DNA–specific primers (LB-SALK-b1.3 and LB-GABI, respectively) in combination with gene-specific primers (Supplementary Table S1). T-DNA insertion sites were determined by sequencing of PCR products. Sequencing revealed that the T-DNA was inserted in the first intron (175 bp downstream of the ATG) in GABI_826F11. PCR conducted using an RB (right border) or LB (left border)-derived primer in combination with a gene-specific primer failed to amplify a product from genomic DNA in SALK_021164, possibly because of border sequence deletion during T-DNA insertion. Failure to obtain a PCR product with the gene-specific primer pair F1 and R1 (Figure 1), but ready amplification of a portion of the TAD1 coding region with the gene-specific primer pair F6 and R6 (Supplementary Table S1) indicated insertion of T-DNA in the promoter region close to the start codon. PCR with gene-specific primers (F1 or R1) in combination with a primer specific for the kanamycin marker gene of the T-DNA (Pkan; Supplementary Table S1) resulted in amplification of specific PCR products with both the forward (F1) and the reverse (R1) gene-specific primers, indicating that the At1g01760 locus in SALK_021164 harbors a tandem T-DNA insertion in inverted orientation. Further sequencing analysis revealed that the T-DNA was inserted 16 bp upstream of the start codon of the At1g01760 reading frame, and the T-DNA insertion was accompanied by the deletion of 52 bp at the insertion site.
Figure 1.
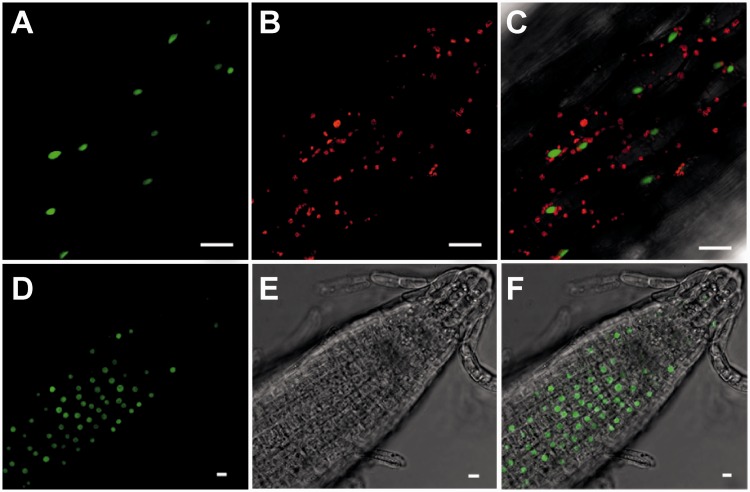
Subcellular localization of a TAD1–GFP fusion protein expressed from the CaMV 35S promoter in stably transformed Arabidopsis plants. Images were obtained by confocal laser-scanning microscopy. Scale bars = 10 µm. (A–C) Localization of the TAD1–GFP fusion protein in hypocotyl cells. (A) GFP fluorescence. (B) Chlorophyll fluorescence. (C) Overlay of GFP and chlorophyll fluorescence. (D–F) Localization of the TAD1–GFP fusion protein in root tips. (D) GFP fluorescence. (E) Bright field image. (C) Overlay of GFP fluorescence and bright field image.
Isolation of mitochondria
Seven-day-old seedlings raised on synthetic medium were transferred to hydroponic growth conditions (liquid 0.5 × MS medium supplemented with 1% sucrose) for 2 weeks. Mitochondria were isolated from whole plant tissue according to Michalecka et al. (37). The purified mitochondria were frozen in liquid nitrogen and stored at −80°C until used for RNA isolation (see later in the text).
Isolation of nucleic acids, hybridization procedures and cDNA synthesis
Total plant DNA was extracted from fresh leaf tissue by a cetyltrimethylammoniumbromide-based method (38). RNA was isolated using a guanidine isothiocyanate/phenol-based method (peqGOLD TriFast; Peqlab GmbH) according to the manufacturer’s instructions. For northern blot analysis, RNA samples (total cellular RNA or purified mitochondrial RNA) were electrophoresed in formaldehyde-containing 2% agarose gels and blotted onto Hybond XL nylon membranes (GE Healthcare). To detect the tRNA-Ala(AGC) in northern blot experiments, an antisense oligonucleotide corresponding to position 1–33 of the tRNA (oligo antiAlaAGC; Supplementary Table S1) was 5′-end–labeled using T4 polynucleotide kinase (New England Biolabs). To this end, 20 pmol of the oligonucleotide was incubated with 10 U T4 polynucleotide kinase and 30 µCi of [γ-32P] adenosine triphosphate for 30 min at 37°C. To detect the mitochondrial tRNA-Cys(GCA) and the mitochondrial 5S rRNA, PCR products generated by amplification with specific primers (Supplementary Table S1) were used as probe. The probe was labeled with [α-32P] dCTP using the Multiprime DNA labeling system (GE Healthcare). Hybridizations were performed at 65°C in Rapid-Hyb buffer, and signals were analyzed using a Typhoon Trio+ variable mode imager (GE Healthcare). For cDNA synthesis, RNA samples treated with TURBO DNase (Ambion) were reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) and oligo(dT)18 primer or random primer following the manufacturer’s protocol.
DNA sequencing and analysis of tRNA editing
For DNA sequencing, amplification products were separated by electrophoresis on agarose gels and purified from excised gel slices using a NucleoSpin Extract II kit (Macherey-Nagel). tRNA purification and tRNA editing analysis were performed as described previously (39). Cytosolic tRNA-Ala(AGC), tRNA-Val(AAC) and tRNA-Thr(AGU) were amplified by reverse transcriptase (RT)–PCR with specific primers (Supplementary Table S1), and purified PCR products were directly sequenced.
Mutant complementation and analysis of subcellular localization
To complement the tad1 mutant and to determine the subcellular localization of the TAD1 protein, the entire TAD1 coding sequence (without the stop codon) was amplified using gene-specific primers (Supplementary Table S1) and cloned into vector pENTR™/SD/D-TOPO (Invitrogen). After sequence verification, the fragment was transferred into the Gateway binary vector pGWB5 by an LR recombination reaction, generating a TAD1–green fluorescence protein (GFP) fusion. The construct was transformed into Agrobacterium tumefaciens strain GV3101 and introduced into A. thaliana wild-type plants or tad1 mutant plants by the floral dip method (40). Transgenic T1 plants were identified by their kanamycin resistance. More than 20 independent transgenic lines expressing the TAD1–GFP fusion protein were identified. Fluorescence was analyzed in 8-day-old seedlings germinated on half-strength MS medium using a confocal laser-scanning microscope (TCS SP5; Leica Microsystems). Fluorescence was excited with the 488-nm line of an argon ion laser. GFP fluorescence was detected via a 505- to 530-nm band-pass filter and chlorophyll fluorescence with a 650- to 710-nm filter.
Quantitative RT–PCR
cDNAs were used as templates for quantitative real-time PCR with gene-specific primers (Supplementary Table S1). Real-time PCR was performed using the StepOnePlus real-time PCR system (Applied Biosystems) using Absolute SYBR Green ROX mix (Thermo Scientific) for quantitation. Three biological and three technical replicates were analyzed. The 2−ΔΔCT method was used to determine the relative transcript levels (41). Reactions for each tested gene in each cDNA sample were independently repeated at least three times. EF1alpha (At5g60390) was used as a reference for cDNA quality.
Aminoacylation assays
Aminoacylation of tRNA-Ala(AGC) was analyzed according to published protocols (42) with minor modifications. Briefly, samples of frozen leaf tissue were ground in liquid nitrogen and extracted with ice-cold RNA elution buffer (0.3 M sodium acetate and 10 mM ethylenediaminetetraacetic acid, pH 4.5). After addition of phenol/chloroform (pH 4.7), the cells were disrupted by vortexing, and, after centrifugation, the aqueous layer was collected. This extraction step was repeated, and the RNA was subsequently precipitated with ethanol. The RNA pellet was then resuspended in 0.3 M sodium acetate (pH 4.5), precipitated again with ethanol, and the pellet was air-dried on ice. After re-suspension in 10 mM sodium acetate (pH 4.5) and spectrophotometric quantitation, samples of 8 µg of RNA were separated by electrophoresis on a 14% polyacrylamide gel with 0.3 M sodium acetate and 7 M urea as gel buffer and 0.3 M sodium acetate (pH 5.0) as running buffer. Gels were run at 60 V for 25 h at 4°C, and the electrophoresis buffer was changed every 6 h to maintain the acidic pH. The RNAs were then electroblotted onto Hybond XL nylon membranes (GE Healthcare) in blotting buffer (10 mM Tris–acetate, pH 7.8, 5 mM sodium acetate and 0.5 mM ethylenediaminetetraacetic acid) at 40 V for 2 h at 4°C and subsequently UV–cross-linked to the membrane. Deacylation of tRNA samples was performed by incubation for 30 min at 70°C after addition of an equal volume of 0.1 M Tris–HCl (pH 9.5) and 0.1 M NaCl.
Analysis of total protein content
To determine total protein content in leaves, 2-week-old plants grown under short-day conditions were transferred to 4°C and grown under cold stress for 3 weeks. For measurements of total protein contents in roots, 1-week-old seedlings grown under long-day conditions on half-strength MS medium were transferred to hydroponic growth conditions for 1 week at 22°C. Subsequently, the seedlings were transferred to cold stress conditions at 4°C for 2 weeks. Total soluble protein was extracted from 100 mg of plant tissue ground in liquid nitrogen and suspended in 100 μl extraction buffer (4 mM Tris–HCl, pH 7.5, 5 mM NaCl, 6.25 µM MgCl2, 10 µM EGTA, 10 µM DTT, 1% TritonX-100 and protease inhibitor cocktail). Cell debris were removed by centrifugation at 12 000 r.p.m. for 20 min at 4°C. The protein concentration of the extract was measured with a protein assay kit (Bio-Rad) using bovine serum albumin as a protein concentration standard.
Measurement of respiration activity
Oxygen consumption was measured using a Clark-type oxygen electrode (Hansatech Instruments). To determine respiration activity of mutant and wild-type plants, leaves were cut into small pieces with a sharp blade, and leaf pieces representing between 15 and 25 mg of fresh weight were placed in the measuring chamber containing 1 ml of incubation medium (10 mM MES–KOH, pH 6.5). Oxygen consumption measurements were performed in the dark at 25°C.
Bioinformatic analyses
TAD1 homologues were identified in GenBank searches using the basic local alignment search tool for proteins (BLASTp; http://www.ncbi.nlm.nih.gov/BLAST/). Phylogenetic analyses were performed on the Phylogeny.fr platform (http://www.phylogeny.fr) using the default settings. Amino acid sequences were aligned with MUSCLE (43) or ClustalW (44).
RESULTS AND DISCUSSION
Identification of an Arabidopsis protein with sequence similarity to Tad1p
We have recently described a genome-wide search for candidate editing deaminases in the model plant A. thaliana (39). This Patmatch search (http://www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl) was based on the consensus sequence motifs in the catalytic domain of all editing deaminases (21) and revealed several candidate proteins that were further analyzed by BLAST searches to identify potential homologues from other species. No putative genes for ADAR-like proteins could be identified in the Arabidopsis genome, consistent with the probable absence of A-to-I conversional editing from plant messenger RNAs. However, we identified a candidate deaminase (encoded by A. thaliana locus At1g01760) that displayed some sequence similarity with tRNA editing deaminases of the Tad1/ADAT family (13,20,21). The putative At1g01760 protein shows 26% sequence identity to Tad1p from the yeast S. cerevisiae, 25% to hADAT1 from humans and 30% to dADAT1 from Drosophila melanogaster (Supplementary Figure S1). Strikingly, the sequence similarity is particularly high in the catalytic domain, including the three zinc-coordinating residues and the proton-shuttling glutamate (Supplementary Figure S1). Searches of other sequenced plant genomes revealed putative homologues in all species analyzed (Supplementary Figure S2), suggesting that the candidate TAD1 protein is encoded by a conserved single-copy gene in most, if not all, plants.
The putative AtTAD1 protein localizes to the nucleus
The Tad1/ADAT1 proteins in yeast and animals specifically deaminate the adenosine residue in the position 3′-adjacent to the anticodon of the cytosolic tRNA-Ala(AGC) (13,19). To test whether At1g01760 encodes a plant tRNA editing deaminase orthologous to Tad1/ADAT1, we first wanted to determine the subcellular localization of the protein. To this end, we translationally fused the full-length reading frame of At1g01760 to the gene for the GFP. The At1g01760–GFP fusion gene was inserted into a plant expression vector and stably introduced into Arabidopsis plants by A. tumefaciens-mediated transformation. Analysis of GFP fluorescence in transgenic plants by confocal laser-scanning microscopy revealed strong green fluorescence that localized exclusively to the nucleus in both hypocotyl cells and root cells (Figure 1). The observed nuclear localization would be compatible with a function of the At1g01760 protein as tRNA editing enzyme and warranted further investigation of At1g01760 by reverse genetics.
Isolation of T-DNA–tagged mutants for At1g01760
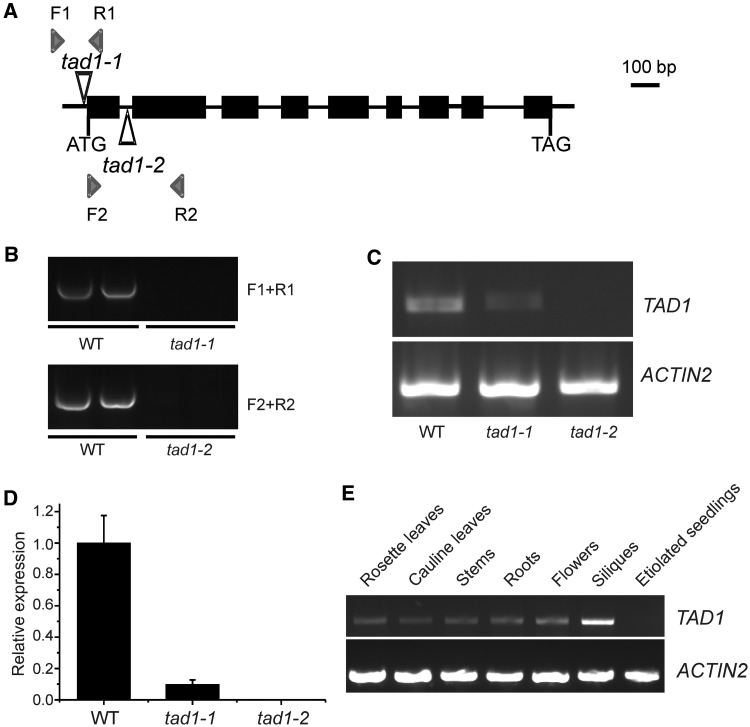
To test whether the protein encoded by locus At1g01760 is indeed an editing deaminase acting on tRNA-Ala(AGC), we sought to address the gene function by isolating mutants defective in At1g01760. To this end, we searched the collection of Arabidopsis T-DNA insertion lines available at the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/) and the GABI-Kat database (http://www.gabi-kat.de/). Two independent T-DNA insertion lines associated with locus At1g01760 were identified (Figure 2A and B). In one of the lines, henceforth referred as the tad1-1 allele, the T-DNA resided 16 bp upstream of the start codon of the At1g01760 reading frame. In the second line, referred as tad1-2, the insertion site was 175 bp downstream of the start codon, disrupting the first intron in the At1g01760 locus. Therefore, in contrast to the tad1-1 allele, the tad1-2 allele can reasonably be expected to represent a loss-of-function allele. However, as T-DNA insertions immediately upstream of the coding region often inhibit gene expression by disrupting important promoter or 5′ untranslated region (5′-UTR) elements, we also included the tad1-1 mutant in all subsequent analyses of At1g01760 gene function. To generate homozygous tad1-1 and tad1-2 mutants, the T-DNA lines were selfed, and plants homozygous for the T-DNA insertion were identified by PCR (Figure 2B).
Figure 2.
Identification and characterization of tad1 mutants in Arabidopsis. (A) Exon–intron structure of the putative TAD1 locus and location of the T-DNA insertions in the tad1-1 and tad1-2 mutants (indicated by open triangles). The arrowheads denote location and orientation of the primers used in (B). (B) Identification of homozygous T-DNA insertions in the tad1 mutants by PCR using genomic DNA as template. Primer combinations are indicated at the right. Two independent samples were analyzed for each plant line. Failure to obtain PCR products with gene-specific primer pairs in the mutants (primers F1 and R1 for tad1-1 and primers F2 and R2 for tad1-2) confirmed homozygosity of the T-DNA insertions in the TAD1 locus. (C) Detection of TAD1 mRNA in the wild-type (WT) and the two tad1 mutants by semiquantitative RT–PCR. The mRNA for an actin isoform (ACTIN2) served as internal RT–PCR control. Note residual TAD1 expression in the tad1-1 mutant, but virtual absence of transcripts from the tad1-2 mutant. (D) Quantitation of TAD1 transcript levels in T-DNA mutants by qRT–PCR. mRNA accumulation in the tad1-1 and tad1-2 mutants is shown relative to the wild-type level (set to 100%). (E) Detection of TAD1 transcripts in various tissues of Arabidopsis wild-type plants by semiquantitative RT–PCR.
Semiquantitative (Figure 2C) and quantitative RT–PCR (Figure 2D) were used to examine expression of At1g01760 in homozygous tad1-1 and tad1-2 mutants. These analyses showed that, as suspected from the T-DNA insertion site disrupting intron 1, the tad1-2 mutant represents a null allele, with no detectable residual expression of At1g01760. In contrast, the T-DNA insertion in the tad1-1 mutant turned out to not completely abolish At1g01760 expression (Figure 2C and D). The residual expression level of ∼10% of the wild-type level (as determined by qRT–PCR; Figure 2D) made the tad1-1 mutant a useful mutant to include in our functional analyses because it can be expected to present an intermediate phenotype between the wild-type and the tad1-2 null mutant.
Semiquantitative RT–PCR assays were used to comparatively analyze expression of the candidate TAD1 gene in different tissues and developmental stages (Figure 2E). Although relatively high expression was detected in siliques, expression levels were significantly lower in all other tissues investigated, including leaves, stems, roots and flowers. Expression levels in etiolated seedlings were below the detection limit of our RT–PCR assays (Figure 2E).
Analysis of tRNA editing in tad1 mutants
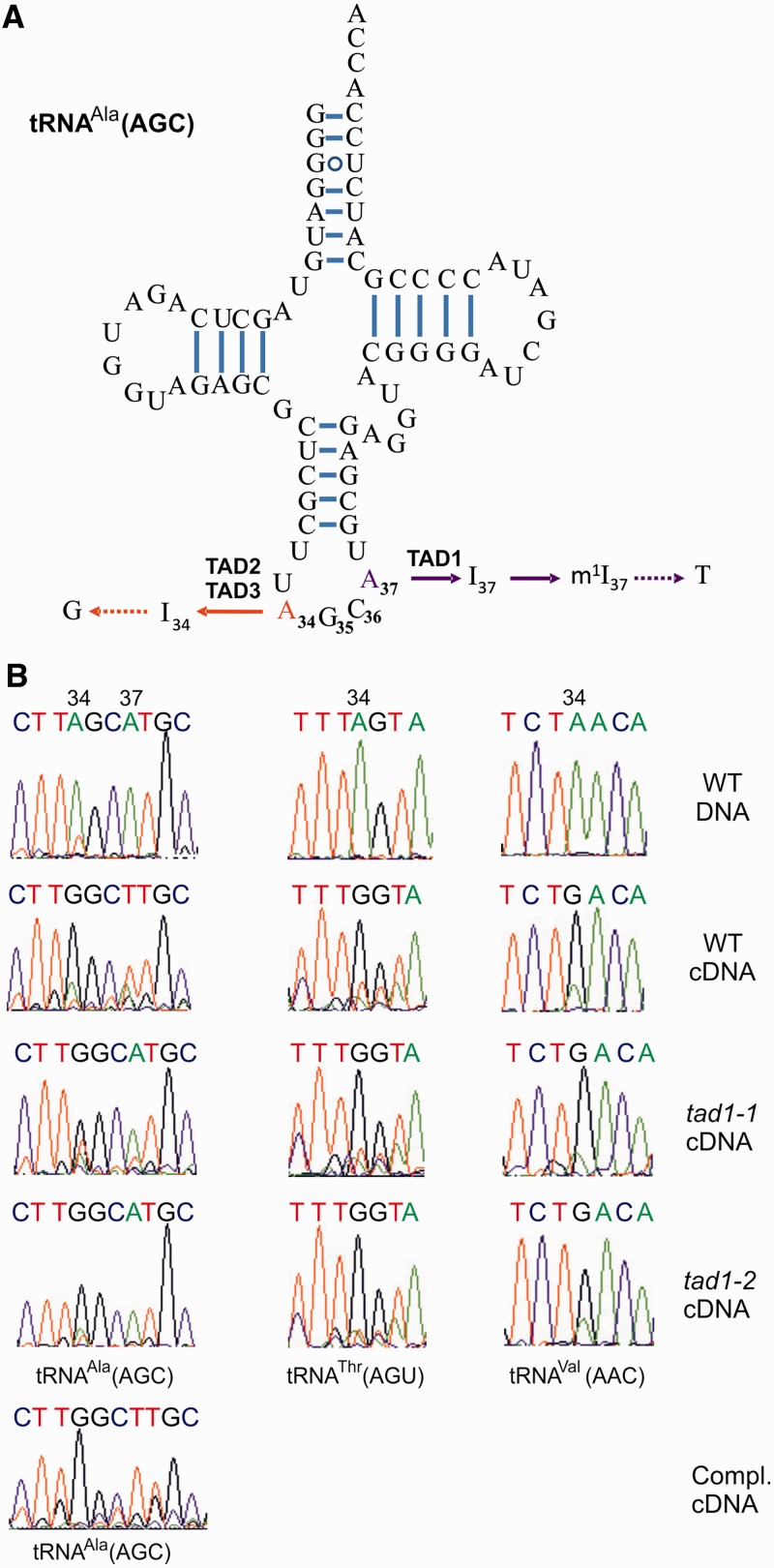
The only substrate of the Tad1 enzyme in yeast and animal cells is adenosine 37 (A37) in the tRNA-Ala(AGC) of the cytosol (13,19; Figure 3A). A37 is oxidatively deaminated to inosine by Tad1 and subsequently methylated in an S-adenosylmethionine–dependent reaction to N1-methylinosine (m1I; Figure 3A; 45). Interestingly, m1I occurs exclusively in this position and has not been found in any other tRNA in eukaryotic organisms (but is present in some tRNAs of extremophile archaea; 45). In addition to the A-to-I editing site in position 37, tRNA-Ala(AGC) also undergoes A-to-I editing in position 34, the wobble position of the anticodon (Figure 3A). The I34 modification is not unique to tRNA-Ala(AGC), but it occurs in altogether eight cytosolic tRNAs in higher eukaryotes (seven in yeast) and in the tRNA-Ala(AGC) of bacteria and chloroplasts (18,39,46,47). In yeast, the A-to-I editing at position 34 is conducted by a complex of two proteins (TAD2 and TAD3), each of which harbors a canonical deaminase motif (20,46; Figure 3A). To test whether the At1g01760 protein is a tRNA editing deaminase and acts specifically on A37 of the cytosolic tRNA-Ala(AGC) in Arabidopsis, we used the homozygous tad1 mutant plants to test for tRNA editing efficiency in the cytosolic tRNA-Ala(AGC). As inosine is read as G and m1I is read as T by reverse transcriptases, the editing events in position 34 and 37 of tRNA-Ala(AGC) can be directly detected by sequencing of amplified cDNA obtained by reverse transcription of the tRNA. Sequencing of reverse transcribed tRNA from wild-type plants confirmed the presence of the I34 and m1I37 modifications known from yeast and animals in the Arabidopsis tRNA-Ala(AGC) (Figure 3B). Interestingly, A37 remained largely unmodified in both the tad1-1 and the tad1-2 mutants. Presence of an A in the amplified cDNA population (and not a G or T) indicates that the modification is blocked at the deamination step and not at the subsequently occurring methylation step (Figure 3A and B). This finding represents strong evidence that the protein encoded by the At1g01760 locus is indeed a functional homologue of the yeast Tad1p editing deaminase. Although A37 editing in the tad1-2 mutant was virtually completely absent, a small proportion of edited tRNA-Ala(AGC) was reproducibly detectable in the tad1-1 mutant. This is consistent with the T-DNA insertion in tad1-1 representing a leaky allele that gives rise to ∼10% residual expression (Figure 2C and D).
Figure 3.
Analysis of tRNA-Ala(AGC) editing in tad1 mutants. (A) Cloverleaf structure of the tRNA-Ala(AGC) from Arabidopsis. Watson–Crick base pairing and UG base pairing are represented by bars and open circles, respectively. TAD1 deaminates A37 in the anticodon loop of the tRNA. I37 undergoes further modification by S-adenosylmethionine–dependent methylation to N1-methylinosine (m1I37; 45). m1I37 is read as T by reverse transcriptases (indicated by the dotted arrow). In yeast, two other deaminase proteins, TAD2 and TAD3, form a heterodimer and specifically deaminate A34 (46). Reverse transcriptases read I as G. (B) Analysis of A-to-I editing at positions 34 and 37 of tRNA-Ala(AGC) in wild-type Arabidopsis plants (WT), the tad1 mutants, tad1-1 and tad1-2, and the complemented tad1-2 line (Compl.). The tRNAs were reverse transcribed, the cDNA sequences amplified by PCR and directly sequenced. Note loss of A-to-I editing at position 37 in tad1 mutants (as evidenced by presence of the genomically encoded A instead of the T originating from reverse transcription of m1I-37 in the wild-type), but unaffected editing at position 34 (G indicating presence of inosine in both the wild-type and the mutants). As a control, two other inosine-containing cytosolic tRNAs (a threonine and a valine tRNA) were also investigated and, likewise, turned out to be unaffected in the Arabidopsis tad1 mutants. DNA indicates genomic DNA; cDNA denotes complementary DNA.
The presence of a clear A peak at position 37 in the cDNA sequence of tRNA-Ala(AGC) from the tad1-2 mutant (Figure 3B) also suggests that, in the absence of A-to-I editing, nucleoside methylation (to m1A instead of m1I; Figure 3A) cannot occur. Previous research on reverse transcription of modified tRNAs demonstrated that m1A base pairs with all four nucleotides (48) during cDNA synthesis. Presence of an unambiguous A peak in the tad1-2 knockout mutant, therefore, tentatively indicates lack of adenosine methylation at position 37; thus, it may suggest that the I37-methylating enzyme is selective for inosine.
In contrast to A37 editing, A-to-I editing at the wobble position A34 was unaffected in both T-DNA mutants (Figure 3B), well in line with the specificity of the Tad1p/ADAT1 enzyme for position A37 of tRNA-Ala(AGC) in yeast and animals. This indicates that I34 is generated by another deaminase that remains to be identified in plants and may or may not be homologous to the TAD2/TAD3 editing enzyme conducting this deamination reaction in yeast (46).
To provide ultimate proof for At1g01760 encoding the plant Tad1p/ADAT1 enzyme, we complemented the tad1-2 mutant with the At1g01760–GFP fusion gene using A. tumefaciens-mediated transformation. Analysis of the resulting transgenic Arabidopsis plants revealed full restoration of A37 editing in tRNA-Ala(AGC) (Figure 3B).
Taken together, our mutant and complementation analyses strongly suggest that the At1g01760 gene encodes a functional homologue of the Tad1/ADAT1 protein that specifically deaminates position A37 in the cytosolic tRNA-Ala(AGC) of A. thaliana. We, therefore, suggest to rename the At1g01760 gene AtTAD1 or, alternatively, AtADAT1.
Accumulation and aminoacylation of tRNA-Ala(AGC) in tad1 mutants
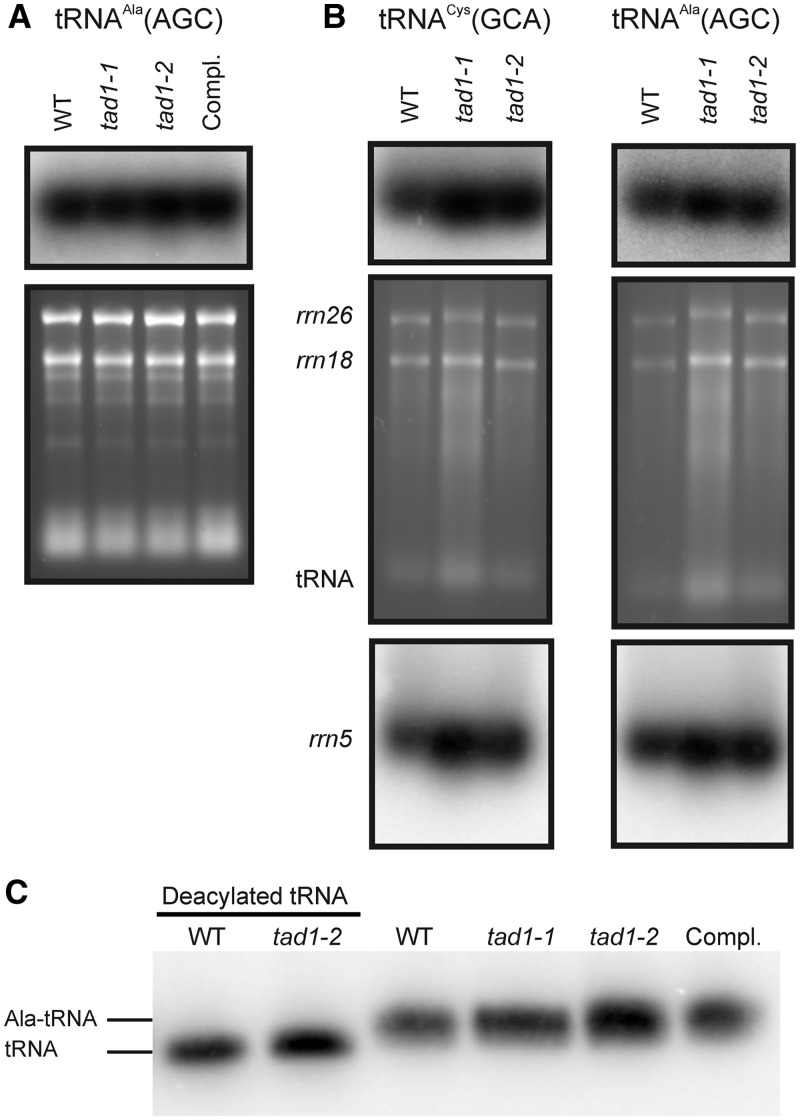
Previous work has established that many nucleoside modifications have a role in stabilizing tRNA molecules in vivo, either by stabilizing their structure and/or by promoting proper folding (reviewed in 1). To test whether the A37 modification in tRNA-Ala(AGC) of Arabidopsis plants also has a beneficial effect on tRNA stability, we investigated tRNA accumulation in the wild-type and the tad1 mutants by northern blot hybridizations. These analyses revealed that the unmodified tRNA-Ala(AGC) in the mutants accumulates to similarly high levels as the modified tRNA in the wild-type (Figure 4A). This indicates that, unlike many other nucleoside modifications, the A37 modification in tRNA-Ala(AGC) has no positive impact on tRNA stability in Arabidopsis.
Figure 4.
Accumulation and aminoacylation of tRNA-Ala(AGC) in wild-type plants (WT), tad1 mutant plants and the complemented tad1-2 line (Compl.). Northern blot analyses were carried out with total cellular RNA (A) or purified mitochondrial RNA (B). To control for loading, the ethidium bromide-stained agarose gels before blotting are also shown. Accumulation of the mitochondrial encoded tRNA-Cys(GCA) was analyzed as a control in (B). The 26S and 18S rRNAs of the mitochondrial ribosome and the tRNA band are indicated in the ethidium bromide-stained agarose gels. To additionally control for loading differences visible in the mitochondrial RNA gel blots, the blots were stripped and re-hybridized to a probe specific to the mitochondrial 5S rRNA. (C) Analysis of aminoacylation of tRNA-Ala(AGC) in the wild-type, the tad1 mutant and the complemented tad1-2 line. In the gel system used here (42), the aminoacylated tRNA migrates slower than its corresponding deacylated species. To visualize the difference in electrophoretic mobility, aliquots of the wild-type sample and the tad1-2 sample were deacylated in vitro. Samples of 8 µg of RNA were separated by electrophoresis, blotted and hybridized to a tRNA-Ala(AGC)-specific probe.
In flowering plants, several nucleus-encoded tRNA species are not only involved in translation in the cytosol but are also imported into the mitochondrial compartment, where they participate in mitochondrial protein biosynthesis (49). In solanaceous plants, the tRNA-Ala(AGC) is among the tRNA species imported into mitochondria (50). To distinguish between tRNA-Ala(AGC) accumulation in the cytosol and the mitochondrion, mitochondria were purified from wild-type and mutant plants, and the effect of the loss of A-to-I editing at position A37 on the mitochondrial tRNA-Ala(AGC) pool was analyzed. Unaltered tRNA accumulation in the mitochondrial compartment (Figure 4B) indicates that loss of A37 editing does not appreciably impair import of tRNA-Ala(AGC) into the mitochondrion and nor does it reduce the stability of the tRNA inside mitochondria.
To exclude the possibility that the loss of A-to-I editing at position A37 affects charging of tRNA-Ala(AGC), the level of aminoacylation of the tRNA was determined in the wild-type, the tad1-1 and tad1-2 mutants, and the complemented line. No difference was seen in that the tRNA pool seems to be fully aminoacylated in vivo in all plant lines analyzed (Figure 4C).
Phenotype of tad1 mutant plants
Having established that AtTAD1 inactivation in the homozygous T-DNA insertion lines prevents A-to-I editing at position A37 of tRNA-Ala(AGC), we next wanted to determine the phenotypic consequences of the loss of this modification. In E. coli, it has been demonstrated that tRNA modifications adjacent to the anticodon can influence translational efficiency and/or accuracy (51). If this were the case for the modification of A37 in the Arabidopsis tRNA-Ala(AGC), the tad1 mutants should display a mutant phenotype under conditions where translational efficiency limits plant growth or fitness. We, therefore, compared tad1 mutant plants with wild-type plants under a range of different growth conditions (Figure 5).
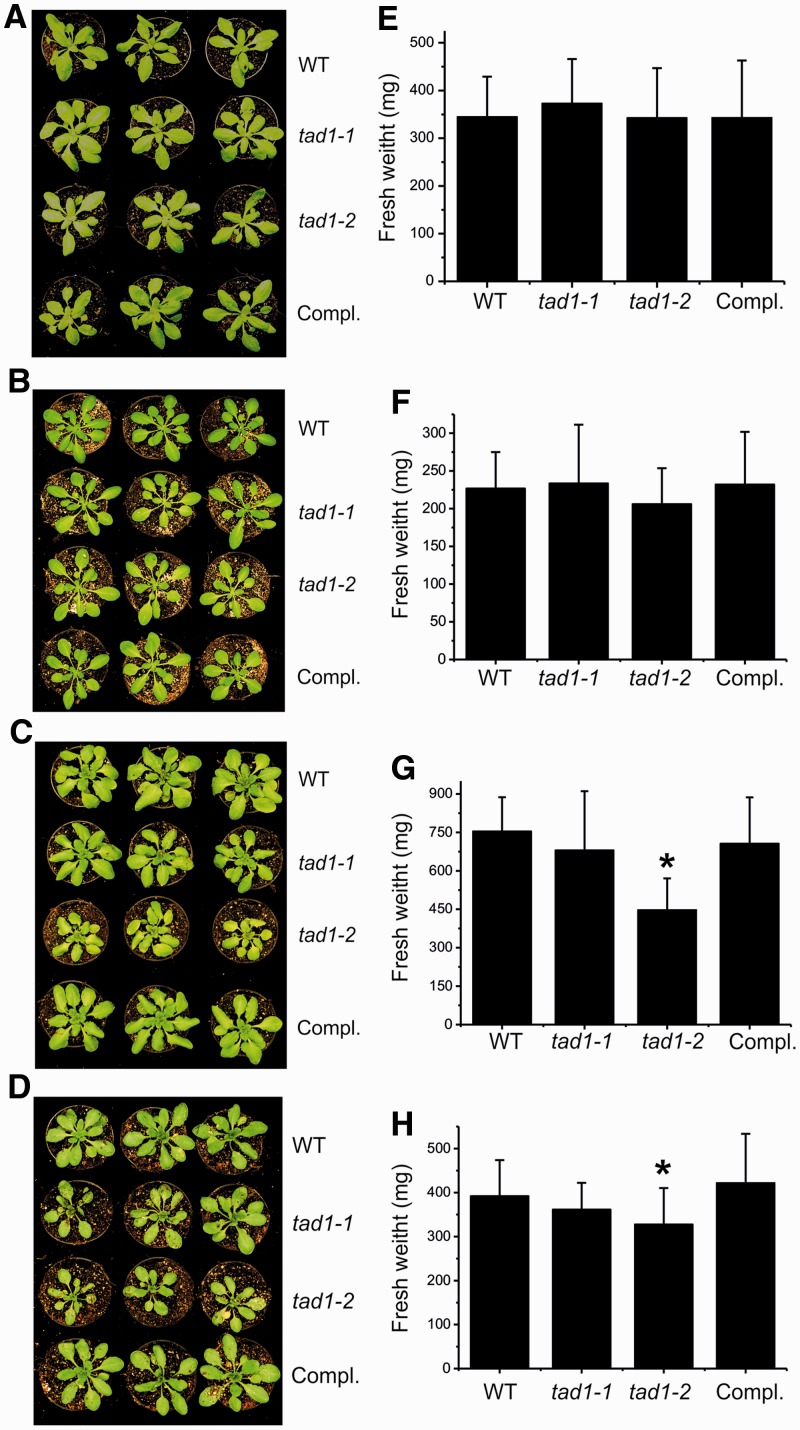
Figure 5.
Phenotype of tad1 mutant plants in comparison with the wild-type (WT) and the complemented tad1-2 line (Compl.). Seven-day-old seedlings raised on synthetic medium were transferred to soil and grown under different environmental conditions in the greenhouse. (A) Phenotype after growth for 15 days under long-day conditions. (B) Phenotype after 21 days under short-day conditions. (C) Twenty-day-old plants grown on soil under short-day conditions were cold stressed at 4°C for 20 days. (D) Sixteen-day-old plants grown on soil under short-day conditions were exposed to heat stress at 30°C for 8 days. (E) Fresh weight of rosettes from plants grown as in (A). (F) Fresh weight of rosettes from plants grown as in (B). (G) Fresh weight of rosettes from plants grown as in (C). (H) Fresh weight of rosettes from plants grown as in (D). Asterisks indicate statistically significant differences (P < 0.05) in Student’s t-test. Error bars represent the standard deviation (n = 16 or 17).
The tad1 mutants showed no discernible phenotype when grown under standard growth conditions (Figure 5A and B). Also, when their biomass was compared with wild-type plants, no significant difference was found (Figure 5E and F), suggesting that the A37 modification is not essential for translation, and fairly high rates of protein biosynthesis can be sustained even with tRNA-Ala(AGC) that is fully unmodified in position 37. However, when the tad1 mutant plants were challenged with stressful environmental conditions, their growth was significantly reduced compared with wild-type plants. Exposure to both heat stress and cold stress resulted in slower growth and lower biomass accumulation. Consistent with tad1-1 representing a leaky allele and tad1-2 representing a complete knockout, this growth phenotype under stress was more pronounced in tad1-2 plants than in tad1-1 plants (and was not statistically significant in tad1-1; Figure 5C, D, G and H). Importantly, wild-type–like growth was fully restored in the complemented line. These growth phenotypes in the tad1 mutants provide evidence for a functional relevance of the A37 modification in tRNA-Ala(AGC). Thus, although the modification is clearly not essential, it seems to be necessary for efficient translation, at least under certain environmental conditions. Having observed growth phenotypes in our tad1 mutants under temperature stress, it seems worth mentioning that the occurrence of A → I → m1I modifications in non-eukaryotic systems is restricted to extremophile archaea. This may lend further support to a role of this modification in translational fidelity under stressful environmental conditions.
As the tRNA-Ala(AGC) is also imported into mitochondria (50), the effect of the loss of A-to-I editing at position A37 on mitochondrial function was characterized. To this end, we measured respiration activity in the dark. The data revealed that respiration rates are reduced in the tad1 mutants, especially under cold stress, and, as expected, more strongly in the tad1-2 plants than in the tad1-1 plants (Figure 6A and B). This finding indicates that not only cytosolic protein biosynthesis but also mitochondrial translation may be affected by the absence of A37 editing.
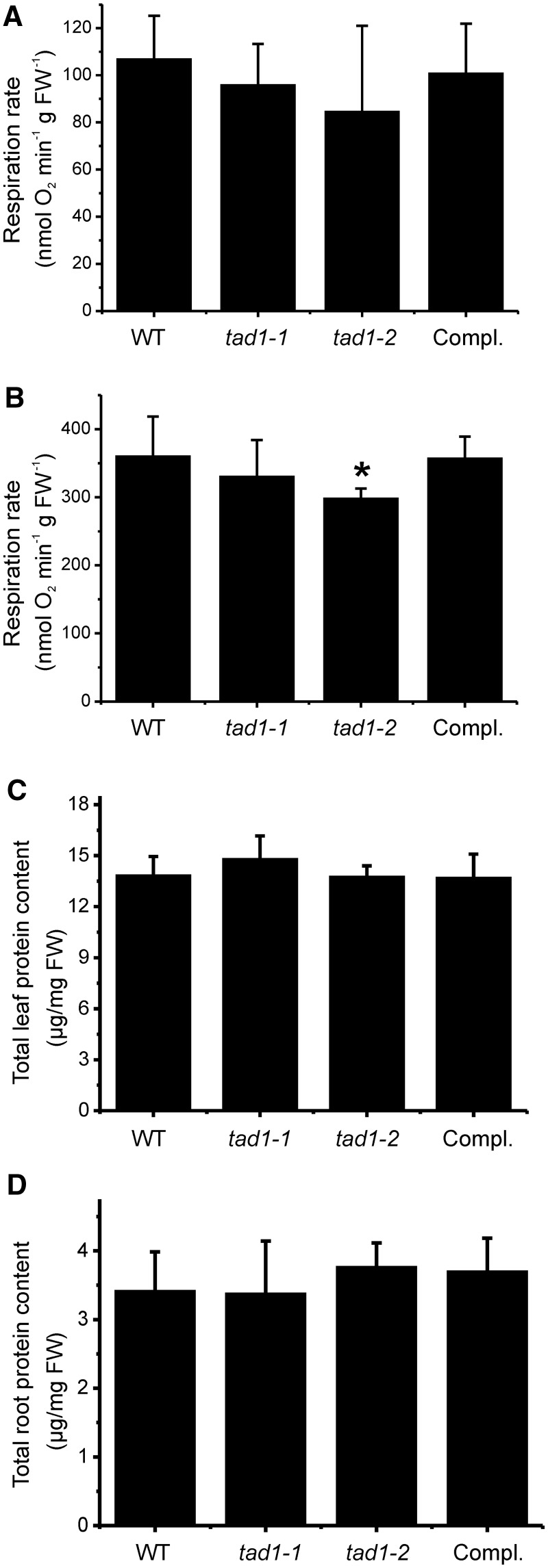
Figure 6.
Measurement of respiration activity and total protein content in tad1 mutant plants, wild-type plants (WT) and the complemented tad1-2 line (Compl.). (A) Leaf respiration rates of 30-day-old plants grown under short-day conditions. (B) Leaf respiration rates of cold stressed plants. Twenty-day-old plants grown in soil under short-day conditions were cold stressed at 4°C for 20 days. Respiration was measured in the dark and is given in nanomoles consumed oxygen per minute and gram fresh weight (FW). Error bars represent the standard deviation (n = 8). Asterisks indicate statistically significant differences (P < 0.05) in Student’s t-test. (C) Analysis of total soluble proteins amounts in leaves (n = 6) from cold-stressed plants. (D) Analysis of total soluble proteins amounts in roots (n = 6) from cold-stressed plants.
Finally, we determined total cellular protein contents under cold stress to test whether the reduced translational efficiency under stress results in lower protein accumulation in the tad1 mutants. In leaves, most of the cellular protein is chloroplast protein; therefore, effects on cytosolic translation could be masked. To exclude this possibility, we additionally measured protein contents in roots, where only low amounts of plastid proteins accumulate (52,53). However, protein contents were not significantly different between the wild-type, the mutants and the complemented line (Figure 6C and D). This suggests that, although editing at position 37 of tRNA-Ala(AGC) is important for translational efficiency in the cold, it does not appreciably affect the cellular protein content, presumably because the tad1 mutants adjust their growth rate to the reduced translation rate (Figure 5).
Although the functional relevance of the A-to-I deamination at position 37 of tRNA-Ala(AGC) in other eukaryotes is currently unknown, an interesting observation was made in connection with a human autoimmune disease. Sera from patients suffering from the chronic inflammatory muscle disorder myositis often contain autoantibodies directed against the anticodon loop of tRNA-Ala(AGC), and the A modifications to m1I at position 37 and I at position 34 seem to play a crucial role in the recognition of tRNA-Ala(AGC) by the autoantibodies (54).
In summary, our work reported here has identified the gene (AtTAD1) that encodes the enzyme responsible for site-specific adenosine-to-inosine deamination at position 37 of the cytosolic tRNA-Ala(AGC) of the model plant A. thaliana. Our data show that the modification (affecting the position 3′-adjacent to the anticodon) is functionally important in that its loss results in reduced plant growth under unfavorable environmental conditions. To our knowledge, this is the first demonstration of the functional significance of a tRNA modification outside the anticodon in plants.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1 and 2.
FUNDING
Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to two anonymous reviewers for helpful suggestions. They thank Eugenia Maximova (MPI-MP) for help with confocal laser-scanning microscopy, Carola Paepke (MPI-MP) for help with respiration measurements and the MPI-MP Green Team for plant cultivation and care. They are grateful to the ABRC at Ohio State University for providing the SALK_021164 (tad1-1) line and to the GABI-Kat consortium for making available the GABI_826F11 (tad1-2) line.
REFERENCES
- 1.Phizicky EM, Alfonzo JD. Do all modifications benefit all tRNAs? FEBS Lett. 2010;584:265–271. doi: 10.1016/j.febslet.2009.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astroem SU, Bystroem AS. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell. 1994;79:535–546. doi: 10.1016/0092-8674(94)90262-3. [DOI] [PubMed] [Google Scholar]
- 3.Slany RK, Kersten H. Genes, enzymes and coenzymes of queuosine biosynthesis in procaryotes. Biochimie. 1994;76:1178–1182. doi: 10.1016/0300-9084(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 4.Brégeon D, Colot V, Radman M, Taddei F. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 2001;15:2295–2306. doi: 10.1101/gad.207701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeuchi Y, Kitahara K, Suzuki T. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J. 2008;27:2194–2203. doi: 10.1038/emboj.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crick FHC. Codon-anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 8.Näsvall SJ, Chen P, Björk GR. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–1673. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agris PF, Vendeix FAP, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 10.Allnér O, Nilsson L. Nucleotide modifications and tRNA anticodon-mRNA codon interactions on the ribosome. RNA. 2011;17:2177–2188. doi: 10.1261/rna.029231.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alfonzo JD, Blanc V, Estévez AM, Rubio MAT, Simpson L. C to U editing of the anticodon of imported mitochondrial tRNATrp allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 1999;18:7056–7062. doi: 10.1093/emboj/18.24.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lecointe F, Namy O, Hatin I, Simos G, Rousset J-P, Grosjean H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 2002;277:30445–30453. doi: 10.1074/jbc.M203456200. [DOI] [PubMed] [Google Scholar]
- 13.Gerber A, Grosjean H, Melcher T, Keller W. Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. EMBO J. 1998;17:4780–4789. doi: 10.1093/emboj/17.16.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 15.Petschek JP, Mermer MJ, Scheckelhoff MR, Simone AA, Vaughn JC. RNA editing in Drosophila 4f-rnp gene nuclear transcripts by multiple A-to-G conversions. J. Mol. Biol. 1996;259:885–890. doi: 10.1006/jmbi.1996.0365. [DOI] [PubMed] [Google Scholar]
- 16.Hurst SR, Hough RF, Aruscavage PJ, Bass BL. Deamination of mammalian glutamate receptor RNA by Xenopus dsRNA adenosine deaminase: Similarities to in vivo RNA editing. RNA. 1995;1:1051–1060. [PMC free article] [PubMed] [Google Scholar]
- 17.Keegan LP, Gallo A, O’Connell MA. The many roles of an RNA editor. Nat. Rev. Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- 18.Wolf J, Gerber AP, Keller W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 2002;21:3841–3851. doi: 10.1093/emboj/cdf362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maas S, Gerber AP, Rich A. Identification and characterization of a human tRNA-specific adenosine deaminase related to the ADAR family of pre-mRNA editing enzymes. Proc. Natl Acad. Sci. USA. 1999;96:8895–8900. doi: 10.1073/pnas.96.16.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerber AP, Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001;26:376–377. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- 21.Rubio MAT, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GAM, Papavasiliou FN, Alfonzo JD. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc. Natl Acad. Sci. USA. 2007;104:7821–7826. doi: 10.1073/pnas.0702394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, George CX, Patterson JB, Samuel CE. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J. Biol. Chem. 1997;272:4419–4428. doi: 10.1074/jbc.272.7.4419. [DOI] [PubMed] [Google Scholar]
- 23.Hough RF, Bass BL. Analysis of Xenopus dsRNA adenosine deaminase cDNA reveals similarities to DNA methyltransferases. RNA. 1997;3:356–370. [PMC free article] [PubMed] [Google Scholar]
- 24.Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA. 2000;6:1004–1018. doi: 10.1017/s1355838200000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer LM, Zhang D, Rogozin IB, Aravind L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 2011;39:9473–9497. doi: 10.1093/nar/gkr691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis MA, Dudock BS. Nucleotide sequence of a spinach chloroplast isoleucine tRNA. J. Biol. Chem. 1982;257:11195–11198. [PubMed] [Google Scholar]
- 27.Greenberg BM, Gruissem W, Hallick RB. Accurate processing and pseudouridylation of chloroplast transfer RNA in a chloroplast transcription system. Plant Mol. Biol. 1984;3:97–109. doi: 10.1007/BF00040034. [DOI] [PubMed] [Google Scholar]
- 28.Francis MA, Suh ER, Dudock BS. The nucleotide sequence and characterization of four chloroplast tRNAs from the Alga Codium fragile. J. Biol. Chem. 1989;264:17243–17249. [PubMed] [Google Scholar]
- 29.Rogalski M, Karcher D, Bock R. Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 2008;15:192–198. doi: 10.1038/nsmb.1370. [DOI] [PubMed] [Google Scholar]
- 30.Alkatib S, Fleischmann TT, Scharff LB, Bock R. Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acids Res. 2012;40:6713–6724. doi: 10.1093/nar/gks350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fey J, Tomita K, Bergdoll M, Maréchal-Drouard L. Evolutionary and functional aspects of C-to-U editing at position 28 of tRNACys(GCA) in plant mitochondria. RNA. 2000;6:470–474. doi: 10.1017/s1355838200992380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fey J, Weil JH, Tomita K, Cosset A, Dietrich A, Small I, Maréchal-Drouard L. Role of editing in plant mitochondrial transfer RNAs. Gene. 2002;286:21–24. doi: 10.1016/s0378-1119(01)00817-4. [DOI] [PubMed] [Google Scholar]
- 33.Chen P, Jäger G, Zheng B. Transfer RNA modifications and genes for modifying enzymes in Arabidopsis thaliana. BMC Plant Biol. 2010;10:201. doi: 10.1186/1471-2229-10-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 35.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 36.Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 37.Michalecka AM, Agius SC, Møller IM, Rasmusson AG. Identification of a mitochondrial external NADPH dehydrogenase by overexpression in transgenic Nicotiana sylvestris. Plant J. 2004;37:415–425. doi: 10.1046/j.1365-313x.2003.01970.x. [DOI] [PubMed] [Google Scholar]
- 38.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 39.Karcher D, Bock R. Identification of the chloroplast adenosine-to-inosine tRNA editing enzyme. RNA. 2009;15:1251–1257. doi: 10.1261/rna.1600609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Jester BC, Levengood JD, Roy H, Ibba M, Devine KM. Nonorthologous replacement of lysyl-tRNA synthetase prevents addition of lysine analogues to the genetic code. Proc. Natl Acad. Sci. USA. 2003;100:14351–14356. doi: 10.1073/pnas.2036253100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grosjean H, Auxilien S, Constantinesco F, Simon C, Corda Y, Becker HF, Foiret D, Morin A, Jin YX, Fournier M, et al. Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review. Biochimie. 1996;78:488–501. doi: 10.1016/0300-9084(96)84755-9. [DOI] [PubMed] [Google Scholar]
- 46.Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999;286:1146–1149. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- 47.Delannoy E, Le Ret M, Faivre-Nitschke E, Estavillo GM, Bergdoll M, Taylor NL, Pogson BJ, Small I, Imbault P, Gualberto JM. Arabidopsis tRNA adenosine deaminase arginine edits the wobble nucleotide of chloroplast tRNAArg(ACG) and is essential for efficient chloroplast translation. Plant Cell. 2009;21:2058–2071. doi: 10.1105/tpc.109.066654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wittig B, Wittig S. Reverse transcription of tRNA. Nucleic Acids Res. 1978;5:1165–1178. doi: 10.1093/nar/5.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salinas T, Duchêne A-M, Maréchal-Drouard L. Recent advances in tRNA mitochondrial import. Trends Biochem. Sci. 2008;33:320–329. doi: 10.1016/j.tibs.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 50.Delage L, Dietrich A, Cosset A, Maréchal-Drouard L. In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum. Mol. Cell. Biol. 2003;23:4000–4012. doi: 10.1128/MCB.23.11.4000-4012.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phelps SS, Malkiewicz A, Agris PF, Joseph S. Modified nucleotides in tRNALys and tRNAVal are important for translocation. J. Mol. Biol. 2004;338:439–444. doi: 10.1016/j.jmb.2004.02.070. [DOI] [PubMed] [Google Scholar]
- 52.Valkov VT, Scotti N, Kahlau S, MacLean D, Grillo S, Gray JC, Bock R, Cardi T. Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: transcriptional and posttranscriptional control. Plant Physiol. 2009;150:2030–2044. doi: 10.1104/pp.109.140483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Ruf S, Hasse C, Childs L, Scharff LB, Bock R. Identification of cis-elements conferring high levels of gene expression in non-green plastids. Plant J. 2012;72:115–128. doi: 10.1111/j.1365-313X.2012.05065.x. [DOI] [PubMed] [Google Scholar]
- 54.Becker HF, Corda Y, Mathews MB, Fourrey J-L, Grosjean H. Inosine and N1-methylinosine within a synthetic oligomer mimicking the anticodon loop of human tRNAAla are major epitopes for anti-PL-12 myositis autoantibodies. RNA. 1999;5:865–875. doi: 10.1017/s1355838299990118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.