Abstract
ParB proteins are one of the three essential components of partition systems that actively segregate bacterial chromosomes and plasmids. In binding to centromere sequences, ParB assembles as nucleoprotein structures called partition complexes. These assemblies are the substrates for the partitioning process that ensures DNA molecules are segregated to both sides of the cell. We recently identified the sopC centromere nucleotides required for binding to the ParB homologue of plasmid F, SopB. This analysis also suggested a role in sopC binding for an arginine residue, R219, located outside the helix-turn-helix (HTH) DNA-binding motif previously shown to be the only determinant for sopC-specific binding. Here, we demonstrated that the R219 residue is critical for SopB binding to sopC during partition. Mutating R219 to alanine or lysine abolished partition by preventing partition complex assembly. Thus, specificity of SopB binding relies on two distinct motifs, an HTH and an arginine residue, which define a split DNA-binding domain larger than previously thought. Bioinformatic analysis over a broad range of chromosomal ParBs generalized our findings with the identification of a non-HTH positively charged residue essential for partition and centromere binding, present in a newly identified highly conserved motif. We propose that ParB proteins possess two DNA-binding motifs that form an extended centromere-binding domain, providing high specificity.
INTRODUCTION
Low-copy number plasmids and most bacterial chromosomes have acquired partitioning (Par) mechanisms to ensure faithful segregation in dividing cells. This stable inheritance depends on active partition systems [reviewed in (1)]. Virtually, all Par systems are composed of three essential elements: a centromere, a centromere-binding protein (CBP) and an NTPase. Interactions between these partners are important for the partition process, and of these, the specific recognition of centromeres by CBPs to assemble partition complexes is crucial. Partition complexes are substrates for partition machineries, as they are responsible for the proper intracellular localization of the replicon onto which these are assembled. CBPs are functional equivalents of eukaryotic kinetochores, in which they act as the link between centromeres and NTPases.
Bacterial partition systems have been classified in three types according to the nature of the NTPase involved (2). The partition system we study here is of type I, representative of the most widespread system, present on most plasmids and on all chromosomes carrying a Par system. Although, the molecular mechanisms underlying partitioning of type I systems are still to be unraveled, evidence is now accumulating that in vivo type I adenosine triphosphatases (ATPases) generally show dynamic behaviors, either oscillating over the nucleoid (3–5) or forming metastable filaments (6,7). Most of the ParA proteins have also been found to polymerize in vitro in response to adenosine triphosphate (ATP) binding (7–10). These dynamic behaviors depend on the presence of the three Par elements. Notably, ParB stimulates ParA activities, such as polymerization (8,9) and ATP hydrolysis (11,12). This latter activity is further stimulated with centromere-bound ParB (13). Partition complexes assemblies on centromere DNA are thus important for the regulation of the partition process.
Bacterial centromeres, generically called parS sites, are diverse in composition and organization. Chromosomal parS sites typically consist of 16-bp inverted repeats clustered around the replication origin or spread throughout the Ori domain (14). Plasmid centromeres consist of inverted and/or direct repeats [reviewed in (15)]. The repeats can be extensive, for example, sopC of the F plasmid consists of 12 tandem repeats of a 43-bp motif (16), and parS of pTAR contains 13 heptameric repeats that are separated by integral helical turns (17). By contrast, the partition site OB3 of plasmid RK2/RP4 is a simpler palindromic core sequence of 13 bp, although related sites dispersed throughout this plasmid might also contribute to segregation (18). Centromeres of more complex organization also exist, such as that of plasmid P1 (19). This ∼80-bp parS consists of two arms that harbor non-symmetrical heptamer (A-box) and hexamer (B-box) DNA motifs, separated by a central binding site for the host-encoded DNA-bending protein, integration host factor. Despite this high diversity, ParBs generally bind to centromere sites through helix-turn-helix (HTH) or in some cases ribbon-helix-helix (RHH) motifs [for review (15)].
In F plasmid, the partitioning locus sop specifies the ATPase, SopA, the CBP, SopB and the centromere, sopC. Each of the 12 tandem 43-bp repeat contains a 16-bp inverted repeat that constitutes the SopB-binding site (20). SopB is composed of several functional domains (Figure 1A). Owing to its high flexibility, the structure of the N-terminal part of SopB is unknown, which is also the case with others ParBs (21,22). This part contains the SopA-interaction domain (23), the arginine-finger and the putative arginine-loop motifs that stimulate SopA ATPase activity (13) and possibly a SopB dimer–dimer interaction region (24). SopB not only forms a SopB–sopC partition complex that recruits SopA but also extends the complex by spreading along the DNA on either side of sopC (25), and also probably in trans to different DNA molecules (26). By coating DNA, SopB forms extended partition complexes whose role in partitioning is not understood. In all cases, specific assembly of SopB on sopC to nucleate the formation of extended partition complexes is essential for F partitioning.
Figure 1.
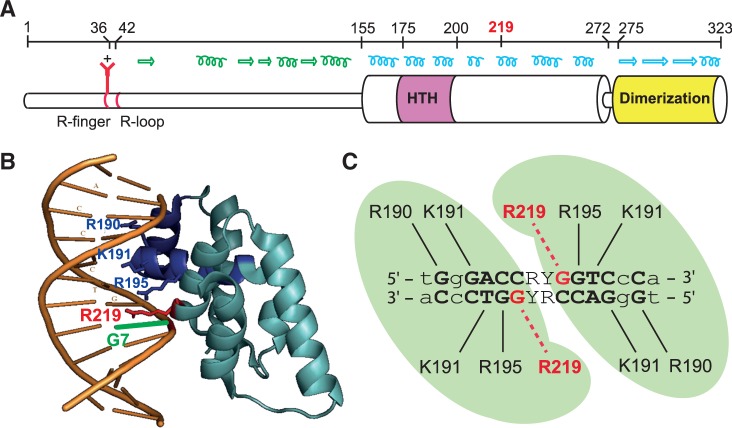
Model of SopB–sopC interaction. (A) Schematic representation of SopB. Top; amino acid numbering of SopB to scale. Middle; secondary structure of SopB. Helices and arrows represent α-helix and β-sheet, respectively, as determined by crystal analysis in blue or predicted (predator, JPRED) in green. Bottom; functional domains and motifs of SopB. Thick cylinders (from amino acids 155–272 and 275–323) represent the parts of SopB structure that have been solved, whereas thin cylinders (from amino acids 1–155 and 272–275) have not (26). (B) Representation of SopB–sopC structure drawn from accession number 3EZF (26), with R219 colored in red, R190, K191 and R195 in blue and the guanine 7 in green. Helices 2 and 3 of the HTH motif are colored in dark blue. (C) Predicted interactions between amino acids of SopB with sopC nucleotides, adapted from the study by Pillet et al. (27). R190, R219, R195 and K191 correspond to arginines 190, 195, 219 and lysine 191. Straight and dotted lines represent suggested amino acid–nucleotide interactions. R and Y stand for purine and pyrimidine nucleotides, respectively. Green zones represent the subunits of an SopB dimer.
Our present knowledge of the protein–centromere contacts that define binding specificity is based on structural studies of only three ParB–parS complexes: KorB–OB3 (RP4), ParB–parS (P1) and SopB–sopC (F). KorB binds OB3 through a classical HTH motif, and the recognition specificity is provided by two amino acid side chains external to the HTH motif (22). The P1 partition complex is more complicated owing to the unusual organization of its centromere site (see earlier in the text). Each HTH motif of ParB dimer contacts an A-box on each parS arm, and an additional DNA-binding module contacting the B-box is provided by the dimerization domain (21). The strong specificity of P1 ParB–parS interaction is shared between these two distinct DNA-binding motifs. Specificity determinants of SopB–sopC partition complex have been recently investigated at the nucleotide level by high-throughput analysis (27). This revealed that all 16 bp of sopC are required, to varying degrees, for maximal SopB binding strength. In addition, the co-crystal structure of a SopB–sopC complex was solved at ∼3.5 A (26), enabling prediction of the likely contacts between sopC and residues of the HTH motif (Figure 1C). Because previous results had suggested binding specificity was entirely contained within the HTH (23), it was interesting that an arginine residue (SopB-R219) outside the HTH was close to the sopC guanine at position 7 (sopC-G7) in the co-crystal.
To better understand the underlying mechanisms involved in plasmid partitioning and centromere binding, we have now investigated the predicted interaction between SopB-R219 and sopC-G7. Strikingly, we found that this interaction is essential for SopB–sopC binding in vitro and is necessary for partition in vivo. These findings reveal that the specific contacts between SopB and sopC occur both inside and outside the HTH motif of SopB. Our results refine the basis of the high-binding specificity of F partition complex assembly. Based on sequence alignments and the known structure of a chromosomal ParB, we identified and showed that an arginine residue provides a specific contact with the chromosomal centromere. Extensive sequence analysis revealed that this positively charged residue is part of a newly defined and highly conserved motif present in chromosomal ParBs over the whole bacterial domain of life.
MATERIALS AND METHODS
Bacterial strains and plasmids
Strains are derivatives of Escherichia coli K-12 or E. coli B (C2833) and are listed, together with plasmids, in Supplementary Tables S1 and S2, respectively. Construction of plasmids is detailed in Supplementary Figure S4.
Media and growth conditions
Cultures were essentially grown at 37°C with aeration in LB (Miller, 1972) containing thymine (10 µg ml−1) and antibiotics (Sigma) as appropriate (µg ml−1): kanamycin (Km, 50), chloramphenicol (Cm, 10), streptomycin (Sm, 200) and spectinomycin (Sp, 20 in liquid medium, 30 in solid). Luria Broth (LB) was supplemented with 1.5% agar (Difco) for solid medium. For stability assay, cultures were grown at the indicated temperature with aeration in LB or in M9 minimal medium supplemented with glucose (0.4%), casamino acids (0.2%), MgSO4 (1 mM), CaCl2 (0.1 mM), thiamine (1 µg ml−1), leucine (20 µg ml−1) and thymine (40 µg ml−1).
DNA manipulation and related procedures
Enzyme reactions, DNA preparation, agarose gel electrophoresis and transformation with plasmid DNA were carried out using standard procedures, according to suppliers’ recommendations where applicable (restriction endonuclease, DNA polymerase and ligase, New England BioLabs; polymerase chain reaction with Phusion, Fermentas; plasmid and gel-fractionated fragment DNAs, Qiagen or Promega). SopB mutations were introduced in pYAS6 and pYAS47 plasmids by quick-change mutagenesis and verified by DNA sequencing (Supplementary Figure S4).
Protein purification
Intein-tagged SopB-G324, SopB-R219 mutants, ParBPput-G291 and ParBPput-G291-R194A proteins were purified from strain C2833 essentially as described previously (13), except that fractions were directly collected and frozen at −80°C after elution from the chitin column. ParBBcen-G306 does not bind to the chitin column. ParBBcen-G306 and ParBBcen-G306-R192A were prepared as described earlier in the text, except that FrI was incubated directly with dithiothreitol to induce full intein cleavage (see Supplementary Figure S3) and was stored in aliquots at −80°C. ParB proteins from Bcen were assayed directly with crude extracts.
Plasmids stability assay
Experiments were started from colonies of cells freshly transformed with the plasmids under test. SopB alleles, expressed from a pSC101 derivative under plac, were assayed in strain MC1061. Overnight cultures in M9–glucose–casamino acid medium containing selective antibiotics were diluted 400-fold into the same medium and grown to A600 = 0.25. Samples were then diluted serially into fresh M9 medium without the antibiotic selective for the plasmid under test and were processed as described previously (28). To determine the fraction of cells that retained the plasmid, samples were taken at the beginning and after 20 and 40 generations. The calculated loss rate per generation is given by the slope A of the equation (y = B.e−Ax) from the fitted curves (using Graph Pad Prism 4 software) on graph plotting the per cent of cell mini-F+ as a function of the generation time, which provides a good approximation of the loss frequency (f) when <10% [f = 1 − (cell mini-F+/total cell)1/n, where n is the generation number]. ParB alleles of Bcen or Pput were assayed for the stabilization of pDAG504 in same conditions as previously published (29,30). Briefly, for Bcen alleles, plasmids under test were introduced in strain DH10B, and growth cultures were performed in M9–glucose–casamino acid medium at 37°C. For Pput alleles, plasmids under test were introduced into strain DH5α, and growth cultures were performed in LB at 37°C.
Electromobility shift assay
Experiments were performed as previously described (9), using 1 nM radiolabeled DNA probes in the presence of sonicated salmon sperm DNA as competitor (100 µg ml−1). Binding reaction products were resolved at 4°C by electrophoresis on 6% polyacrylamide gels in TGE (25 mM Tris base, 190 mM Glycine, 1 mM Ethylenediaminetetraacetic acid) for 2–4 h at 180 V. Polyacrylamide gel electrophoresis-purified complementary oligonucleotides (Sigma-Aldrich) were used to assemble duplex DNA probes by annealing (only top strands are shown; 16-bp SopB/ParB binding sites are indicated by upper case letters; bases corresponding to positions 7 and 10 are indicated in bold): sopC2: 5′-attagtcTGGGACCACGGTCCCActcgtat-3′; sopC63: 5′-attagtcTGGGACAACTGTCCCActcgtat-3′; sopC64: 5′-attagtcTGGGACTACAGTCCCActcgtat-3′; parS66: 5′-attagtcTGTTCCACGTGGAACActcgtat-3′; and parS67: 5′-attagtcTGTTTCACGTGAAACActcgtat.
RESULTS
We have previously investigated the bases involved in the specific recognition of the F plasmid centromere by SopB [(27); summarized in Figure 1C]. In particular, the guanine at position 7 (sopC-G7) in the left arm (and its counterpart at position 10 in the right arm) was found to play a role in SopB–sopC interaction. In addition, the co-crystal structure of SopB–sopC complexes, determined at a medium resolution [∼3 A; (26)], suggests that G7 is close to an arginine residue (R219), in the major groove (Figure 1B). We estimated the distance between the O group of G7 and the NH group of R219 at 2.6 A (Supplementary Figure S1), compatible with hydrogen-bonding between them (31). This R219 residue does not belong to the HTH motif responsible for the primary binding to DNA (Figure 1A), which contains three positively charged residues (R190, K191 and R195; Figure 1B) involved in specificity of binding to sopC (13,26). SopB-R219 is part of an α-helix separated from the HTH by an other α-helix (Figure 1A). Its potential role in centromere binding and thus in the partition process was further investigated.
SopB-R219 mutants are unable to stabilize mini-F
To determine whether the R219 residue plays a direct role in the partition of the mini-F plasmid, we constructed two SopB-R219 mutants by converting R219 to alanine (R219A) or lysine (R219K). These mutations were introduced into a pSC101 derivative, pYAS47, which carries sopAB under the control of the lac promoter, yielding pJYB161 (sopB-R219A) and pJYB162 (sopB-R219K). These plasmids were introduced into strain MC1061 carrying pDAG209, an unstable mini-F sopC+ but ΔsopAB, and loss of the mini-F during growth without selection was measured (Supplementary Figure S2A and Table 1). When wild-type SopB was provided in trans from pYAS47, no cell without pDAG209 was observed after 40 generations, indicating that the mini-F sopC+ was completely stable (loss rate < 0.01%). Without SopB (pAM238), the mini-F was lost at a frequency of 4.0% per generation as expected from the copy number (∼5–6 at division) for segregation at random. The mutant alleles SopB-R219A (pJYB161) and -R219K (pJYB162) failed to stabilize pDAG209, which was lost at 3.4 and 3.1% per generation, respectively. Western blot analysis showed that in the stability assays, the mutant SopBs were produced at the same level as wild-type (wt) (Supplementary Figure S2B). These results indicate that the R219 residue of SopB is essential for the maintenance of mini-F during generation.
Table 1.
Stability of mini-F plasmids carrying F sopC or chromosomal parS centromeres
| F plasmids | Multicopy plasmids | Relevant characteristics | Loss/generation (%) |
|---|---|---|---|
| pDAG209 | pAM238 | 4.0 (±0.5) | |
| (sopC) | pYAS47 | sopA+B+ | <0.01 |
| pJYB161 | sopA+B-R219A | 3.4 (±0.3) | |
| pJYB162 | sopA+B-R219K | 3.1 (±0.1) | |
| pDAG551 | pBBR1MCS5 | 2.6 (±0.1) | |
| (parS) | pDAG562 | Bcen parA+B+ | 0.15 (±0.1) |
| pJYB167 | Bcen parA+B-R192A | 2.7 (±0.4) | |
| pDAG504 | pAM238 | 3.6 (±0.7) | |
| (parSPput) | pDAG503 | Pput parA+B+ | 0.25 (±0.1) |
| pJYB168 | Pput parA+B-R194A | 2.1 (±0.5) |
Derivatives of strain MC1061 carrying the mini-F pDAG209 and the indicated plasmids were grown at 30°C. Strains carrying the mini-Fs pDAG551 (consensus parS) or pDAG504 (parSPput) were grown at 37°C. The sop and par operons are under the control of the lac promoter on pAM238 or pBBR1MCS5. Samples of the cultures were plated at intervals for estimation of the proportion of mini-F-positive cells. Loss rates per generation, given by the slopes (A) of the fitted curves as presented in Supplementary Figure S2A, are average of at least three experiments, and standard deviations are indicated in brackets.
SopB-R219 variants do not bind sopC centromere
To test whether the inability of sopB-R219 alleles in stabilizing the mini-F is primarily because of a default in centromere-binding activity, we performed electromobility shift assays (EMSA). Wild-type SopB and R219 variants were expressed as intein-tagged proteins and purified as native proteins after cleavage of the intein (see ‘Materials and Methods’ section and Supplementary Figure S3). Typical EMSA experiments are shown in Figure 2A. The 32P-labeled substrate, a 30-bp DNA duplex containing the 16-bp consensus SopB-binding site, was specifically bound by SopB as previously observed (27). By contrast, SopB-R219A showed almost no sopC-binding activity: at 1 µM, the amount of protein–DNA complex formed is much less than with 10 nM of wild-type SopB, indicating that the loss of sopC-binding activity in SopB-R219A is >200-fold. The arginine 219 of SopB is, therefore, essential for specific SopB binding to the centromeric site. Similarly, SopB-R219K showed a significant reduction in sopC-binding activity, although slightly less than SopB-R219A. The ∼100-fold loss in sopC binding indicates that the lysine present in this variant is not sufficient to compensate for the positive charge of the arginine residue in wild-type SopB.
Figure 2.
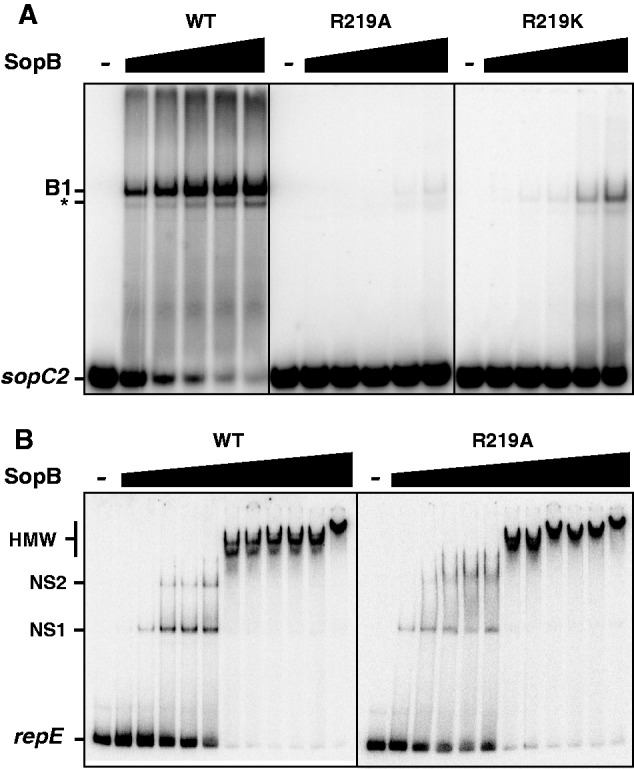
Interaction of SopB wild-type and R219 variants with DNA. The ability of wild-type SopB, SopB-R219A and -R219K to interact with an sopC DNA duplex or with non-specific DNA was tested in a gel mobility shift assay. Reaction mixtures, assembled on ice and incubated at 30°C, were analyzed by electrophoresis on polyacrylamide gels. (A) SopBs binding to sopC centromere site. 32P-labeled 30-bp sopC2 probe was incubated alone (−) or with increasing concentrations (from left to right; 10, 30, 100, 300 and 1000 nM) of wild-type SopB (WT), SopB-R219A or SopB-R219K as indicated on top, in the presence of 100 µg ml−1 competitor DNA. Positions of DNA probes and protein complexes are indicated on the left: B1 denotes the specific complex, whereas asterisk corresponds to a specific shift from a secondary form of the duplex DNA. (B) SopBs binding to non-specific DNA probe. 32P-labeled 114-bp repE probe was incubated alone (−) or with increasing concentrations (from left to right; 100, 200, 300, 400, 500, 600, 700, 800, 900, 1000 and 2000 nM) of wild-type SopB (WT) or SopB-R219A as indicated on top, in the absence of competitor DNA. NS1 and NS2 denote discrete complexes, and HMW corresponds to high molecular weight complexes.
In addition to its site-specific DNA-binding activity, SopB also binds non-specific DNA (9), for spreading along the DNA outside of sopC (25). To determine the extent to which SopB-R219A variant is deficient in DNA-binding activity, we also tested its ability to still bind DNA non-specifically. For this, we performed EMSA in the absence of competitor DNA, using a 114-bp duplex DNA that does not carry the 16-bp sopC site (Figure 2B). In these conditions, SopB-wt and SopB-R219A bound similarly the non-specific DNA probe. For both alleles, one discrete complex (NS1) was observed between 100 and 200 nM, and a second discrete complex (NS2) was detected between 200 and 500 nM. At ∼500 nM SopB, ∼50% of the probe was shifted, and above this concentration, high molecular weight complexes were formed. Thus, both proteins displayed about the same affinity for non-specific DNA, indicating that SopB-R219 is deficient in sopC binding only. This result strengthens the finding that the R219 residue of SopB is strongly involved in site-specific DNA binding.
In vivo SopB-R219A does not form foci
SopB, like all ParB proteins, forms bright foci in cells carrying its cognate centromere (32). These foci correspond to the assembly of partition complexes on centromeres and are usually located near the cell center or the quarter positions (33). To determine whether SopB binding to sopC was also affected in vivo by the alanine mutation at position 219, we constructed mini-F derivatives, pJYB214 and pJYB223, with Venus-Yfp fused to SopB-wt and SopB-R219A, respectively. pJYB214 was fully stable in strain DLT1215 (data not shown), indicating that the fusion is functional. In this strain, DLT2950, fluorescence microscopy showed that all cells harbor two to four bright regularly positioned foci (Figure 3, left panel). By contrast, cells transformed with pJYB223, which carries the sopB-R219A allele, did not form foci, and fluorescence was spread throughout the cytoplasm (Figure 3, right panels). In addition, some cells (∼15%) did not display fluorescence, suggesting that they had lost the mini-F, as expected for an unstable mini-F derivative. The absence of focus formation implies that SopB-R219 does not support centromere binding in vivo.
Figure 3.
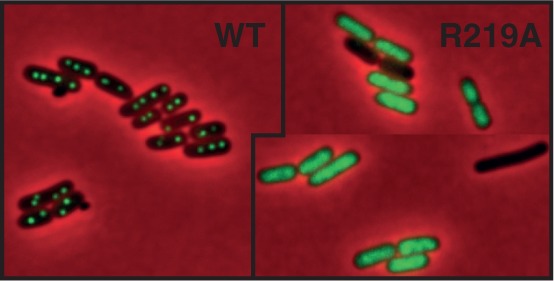
SopB-R129A allele does not form foci cells. Left; localization of SopB–Venus fusion. Fluorescent foci from the mini-F expressing SopB–Venus display the expected pattern for partition complexes faithfully partitioned. Right; localization of SopB-R219A–Venus fusion. All cells displayed diffuse fluorescence throughout the cytoplasm. Cells without fluorescence are presumed to lack the mF carrying SopB-R219A–Venus as expected from the loss rate (see Supplementary Figure S2A and main text).
Guanines 7 and 10 in sopC site are critical for SopB binding
The SopB–sopC co-crystal structure (26) predicts that the R219 residue of an SopB monomer is near the guanine in position 7 of the left arm of sopC, and thus that R219 of the second SopB monomer should be close to the guanine 10 on the right arm (Figure 1B and C). We previously observed by SPRi that mutations of these bases affect SopB binding to sopC (27). However, because of the low sensitivity of this technique, quantification of the loss in DNA-binding activity was not possible. Because SopB-R219 variants are deficient in sopC binding, we expected that if R219 directly interacts with guanines 7 and 10, we should observe a comparable loss in SopB binding if these bases are mutated.
We, therefore, performed EMSA in same conditions as described earlier in the text with wild-type SopB but with 30-bp sopC DNA probes carrying either transversion (C to A; probe sopC63) or transition (C to T; probe sopC64) at positions 7 and 10 (Figure 4). Both probes were barely shifted even at the highest SopB concentration (1 µM), indicating that these bases are strictly required for the formation of SopB-sopC complexes. This result is consistent with a direct interaction between R219 residues and guanine at positions 7 and 10.
Figure 4.
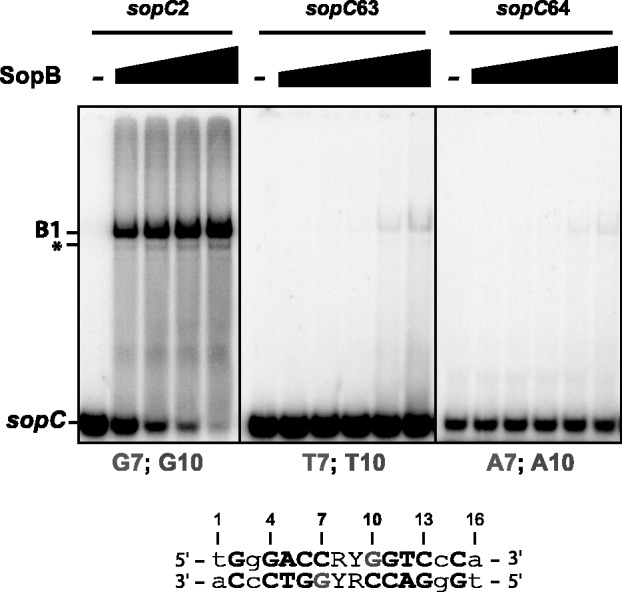
Interaction of wild-type SopB with mutated sopC centromeric DNA. Top; the ability of wild-type SopB to interact with different sopC DNA duplexes (indicated on top) was assayed as in Figure 2. 32P-labeled 30-bp sopC probes were incubated alone (−) or with increasing concentrations of wild-type SopB (from left to right; 10, 30, 100, 300 and 1000 nM), except for sopC2 where the 1000 nM SopB reaction was omitted. Legend is as in Figure 2. Bottom; bases at positions 7 (bottom strand) and 10 (top strand) are indicated in grey. Numbers indicate positions of the bases in the palindromic SopB-binding site according to the study by Pillet et al. (27).
Chromosomal CBPs harbors extended DNA-binding domains essential for partition activity
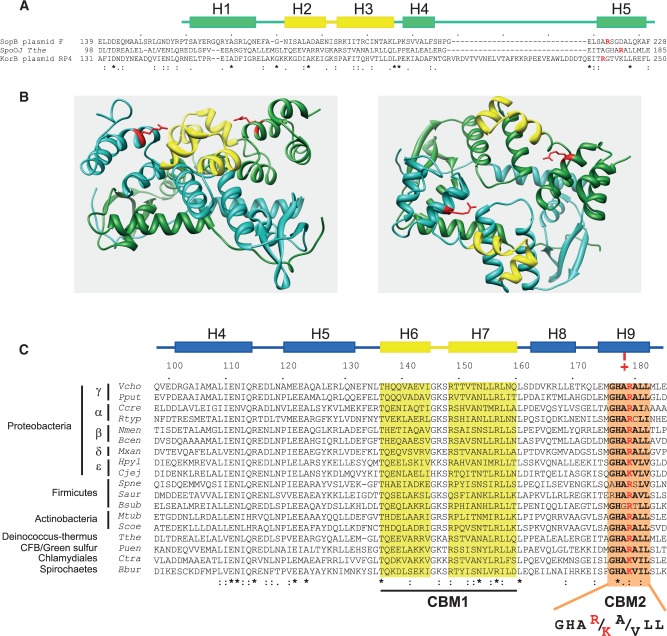
The structure of a chromosomal homologue of SopB, Spo0J of Thermus thermophilus, solved in the absence of the parS centromeric DNA, also displays a typical HTH proposed to be the parS-binding motif (34). Interestingly, a helix (H9) located two helices away from the HTH recognition helix (H7) was shown to correspond to helix H8 of KorB (35), and it also aligns with helix H5 of SopB (Figure 6A). H5SopB and H8KorB carry the R219 and the R240 residues, respectively, involved in specific recognition of their cognate centromere [this study, (22)]. Helix H9Spo0J also harbors an arginine residue (R179) in the corresponding helix (Figure 6A). In the 3D structure of the Spo0J dimer, the orientation of R179 relative to the HTH supports the possibility that it could provide a direct interaction with a specific base of parS (Figure 6B). Indeed, each arginine residue on both monomers has its side chain directed towards where the parS DNA would be bound in the HTH motif alignment.
Figure 6.
ParB conserved motifs and residues involved in centromere binding. (A) Amino acid alignment of ParB homologues. Sequence alignments of SopB (F) with KorB (RP4) and Spo0J (T. thermophilus) are based on the study by Leonard et al. (34). Identical residues and highly conserved residues are indicated by asterisks and dots, respectively. Helices H1 to H5 of SopB, based on the crystal structure analysis (26), are schematically represented on top; Helices H2 and H3, corresponding to the HTH motif, are drawn in yellow. Arginine residues R219 (SopB), R240 (KorB) and R179 (Spo0J) are colored in red. (B) Ribbon diagrams of a ParB dimer. Structure is from T. thermophilus Spo0J (34). One monomer is colored in cyan and the other in green. The right diagram is a 90° horizontal rotation of the left diagram. HTH motifs and R179 residues of each monomer are colored in yellow and red, respectively. Each arm of the 16-bp parS DNA is expected to bind one HTH motifs of the Spo0J dimer. Orientations of R179 residues are compatible for providing additional specificity contacts in parS DNA. (C) Amino acid alignment of chromosomal ParBs. ParBs from a large spectrum of bacterial species were aligned with ClustalW; in the case of multichromosomal bacteria, ParBs are from the main chromosome. These ParBs are expected to bind the parS consensus sequence 5′-tGTTNCACGTGNAACa-3′ (30,48). Sequences highlighted in yellow and orange correspond to the HTH motif and the CBM2 (centromere-binding motif 2) box, respectively. Helices are represented as in (A) based on the crystal structure of Spo0J of T. thermophilus (34); amino acid sequence are numbered according to Tthe Spo0J. The red plus sign (+) schematically represents the conserved R or K positively charged residues. ParBs are from Borrelia burgdorferi B31(Bbur), B. cenocepacia (Bcen), B. subtilis (Bsub), Campylobacter jejeuni (Cjej), Caulobacter crescentus (Ccre), Chlamydia trachomatis (Ctra), Helicobacter pylori (Hpyl), Mycobacterium tuberculosis (Mtub), Myxococcus xanthus (Mxan), Neisseria meningitidis (Nme), P. putida (Pput), Porphyromonas uenonis (Puen), Rickettsia typhi (Rtyp), Staphylococcus aureus (Saur), Streptomyces coelicolor (Scoe), Streptococcus pneumonia (Spne), T. thermophilus (Tthe) and Vibrio cholera (Vcho).
Sop0JTthe binds the pseudopalindromic parS sequence 5′-TGTTCCACGTGAAACA-3′. This sequence corresponds to the highly conserved parS, characteristic of centromeres present on bacterial chromosomes, whose consensus is 5′-tGTTNCACGTGNAACa-3′. If the Spo0J-R179 residue is involved in specific binding, we expect that it should be conserved at least in a subset of chromosomal ParBs. To test this hypothesis, we aligned ParBs from a large range of bacteria (Figure 6C). We found that R179 is highly conserved and present in most phyla. In the ε-Proteobacteria, the Chlamydiales and the Spirochaete groups, the arginine is replaced by a lysine, suggesting that the positively charged nature of the residue is the important feature, compatible with a direct interaction with a base of the parS site. If the H9 helices of ParB are functional equivalents to the H5 helix of SopB, these R/K residues should also be involved in chromosome segregation.
ParABS partition activity on bacterial chromosome is difficult to be assessed directly because of pleiotropic effects. However, partition function could be examined by the ability of parAB expressed from plac on a multicopy vector to stabilize a mini-F (Δsop) plasmid carrying a single parS in E. coli in trans. Indeed, this assay has been used previously to demonstrate the partition capacity of the Bacillus subtilis soj-spo0J-parS and the Pseudomonas putida and Burkholderia cenocepacia parABS systems (29,30,36). To determine whether these positively charged residues are involved in partition, we tested the ParABSPput and ParABSBcen systems in stability assays by introducing mutations at positions equivalent to Spo0J-R179, parB-R194A and parB-R192A, respectively. We observed, as previously reported using the same experimental conditions (29,30), that both ParAB systems were able to efficiently stabilize their cognate parS carrying mF, pDAG504 or pDAG551 (Table 1; loss rates = 0.15–0.25%). We found, however, that both parB-R194A and parB-R192A alleles were impaired in their ability to support stabilization of pDAG504; Bcen ParB-R192A was completely deficient (loss rate = 2.7%), whereas Pput ParB-R194A was highly but not completely deficient (loss rate = 2.1%). These results thus clearly demonstrate that these positively charged residues in H9 helices are essential, as is SopB-R219, for the partition function of chromosomal ParBs.
By performing EMSA, we then tested whether these arginine residues are, as for their SopB counterparts, directly involved in parS-specific binding (Figure5). ParB proteins from P. putida were purified as for SopBs, whereas ParB proteins from B. cenocepacia were directly assayed from crude extracts expressing intein-tagged ParBs (see ‘Material and Methods’ section and Supplementary Figure S3). In contrast to wild-type, mutant ParBs from Pput and Bcen were not able to bind to their respective parS sites. In both cases, the DNA binding deficiency is so high that no specific retardation was detected. These results thus indicate that R194 and R192 residues from Pput and Bcen, respectively, are directly involved in centromere binding.
Figure 5.
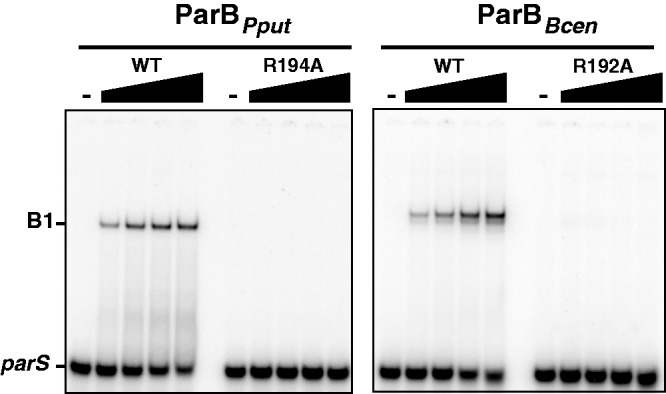
Chromosomal ParBs binding to parS sites. DNA binding interaction between Pput or Bcen parS, left and right panel, respectively, and wild-type ParB from Pput (ParBPput), Bcen (ParBBcen) or variant ParB-R192ABcen and ParB-R194APput proteins were assayed by EMSA as in Figure 2. 32P-labeled 30-bp parS probes were incubated alone (−) or with increasing concentrations of purified ParBPput proteins (left panel; from left to right: 0, 10, 30, 100 and 300 nM) or ParBBcen proteins in crude extracts (right panel; from left to right: 0, 2.8, 8.4, 28 and 84 mg ml−1). Legend is as in Figure 2.
Finally, we found that the R/K residues of H9 helices are part of a highly conserved motif, GHAR/KA/VLL, that covered most of the H9 helix (Figure 6C). Secondary structure analysis on these motifs predicted that they all harbor an α-helical conformation (data not shown).We named this conserved motif box ‘CBM2’ for centromere-binding motif 2; the CBM1 being the typical HTH motif.
DISCUSSION
SopB is known to bind DNA specifically on the sopC centromere to function in the active partitioning of the F plasmid. We have shown here that the specificity of SopB–sopC interaction is more complex than anticipated, by identifying a new specific DNA-binding determinant, an arginine at position 219, located outside of the HTH motif. We demonstrated that SopB-R219A/K variants are completely deficient in sopC binding in vitro, whereas both mutants are still able to bind DNA non-specifically (Figure 2). Fluorescence microscopy revealed that, in contrast to wt SopB, neither mutant allele was able to form intracellular foci (Figure 4), supporting the idea that in vivo they are also deficient in sopC binding. In addition, these single mutations totally abolished active segregation (Table 1), demonstrating that the R219 residue is a crucial DNA-binding determinant of the SopB–sopC interaction. Extension of this result to chromosomal partition systems reveals that most ParBs also harbor a secondary CBM that was shown to be essential for partitioning functions (Figure 6 and Table 1).
Direct interaction between R219 and guanine 7 defines an extended DNA-binding domain in F SopB
HTH motifs are composed of two helices that contact the DNA in the major groove. The second helix, called the recognition helix, usually provides specificity by contacting specific bases of the target binding sequence. In the case of SopB, the recognition helix makes three specific base contacts through positively charged residues (K190, K191 and K195) and possibly two others through residues S189 and I192 (26). These numerous contacts were thought to be sufficient for providing a high specificity for centromere binding. The proximity evident in the SopB–sopC structure of the positively charged residue, R219, located two helices away from the HTH, to guanine at position 7 (G7) was thus intriguing, especially because G7 was found be involved in SopB binding (27). We found the same strong deficiency of binding, as judged by gel retardation, between SopB-R219A binding to sopC and wild-type SopB binding to sopC carrying either transitions or transversions at positions 7 and 10 (compare Figures 2A and 4). In addition, replacing the arginine by a lysine at position 219 does not restore binding to sopC (Figure 2A). A positive charge at this position is thus not sufficient for DNA binding, arguing for specific contact between SopB-R219 and sopC-G7. In agreement with this finding, bioinformatic analysis (27) showed that (i) the arginine at position 219 is conserved among all closely related SopB homologues; and (ii) all sopC centromere sequences present in the databases harbor a guanine at position 7. Along with these data, our results thus clearly support a direct interaction between R219 residues of each SopB monomer with each G7 base of the palindromic sopC sequence.
If the R219-G7 interaction was anticipated from the structure, the finding that it is a critical one was not. Single mutations at position R219 completely abolish sopC recognition, revealing that the HTH is not sufficient to provide the specificity of the SopB–sopC interaction. However, specific contacts made by the recognition helix are also crucial for binding specificity, as mutations of two of the positively charged residues, K191 and K195, also abolish specific, but not non-specific, sopC binding (13). The centromere-binding specificity of plasmid F is thus shared by two motifs, the HTH and the R219 containing helix, which together define an extended DNA-binding domain.
Centromere-binding proteins harbor extended DNA-binding domains for highly specific interactions
Is this extended centromere-binding domain present in SopB a common characteristic among CBPs? Some co-crystal structures of ParB–parS are available to address such a question. Similarly to SopB, KorB of RK2/RP4 harbors two DNA-binding motifs (22). However, in sharp contrast to SopB, residues from the helices forming the HTH motif of KorB do not make specific contacts with bases in the OB site (13-bp parS centromere). The KorB–OB recognition specificity is mainly based on two side chain interactions outside the HTH (T211 and R240) that form direct hydrogen bonds with two bases next to the central base pair binding sequence. Thus, although the specificity determinants are not distributed as for F SopB, KorB also has an extended centromere-binding domain.
In the case of the P1 ParB, the specificity recognition is shared by two separate DNA-binding motifs, each contacting its specific binding sequence, the A- and B-boxes; the P1 centromere site being composed of a complex arrangement of these two DNA-binding boxes (37,38). The A-boxes are specifically bound by the HTH motifs, and the B-boxes are bound by the dimerized dimer motifs (21). These two motifs participate to the efficient binding of ParB to parS; therefore, they could be regarded as an extended DNA-binding domain. Although P1 ParB and F SopB are closely related to each other, SopB does not carry this second DNA-binding motif because of the lack of two small extra loops residues providing ‘wings’ that are specifically present in the ParB dimerization motif (26). This difference is in relation with the centromere structures where, in the particular case of P1, ParB recognizes two different binding sequences. Thus, as in KorB and SopB, P1 ParB has an extended DNA-binding domain composed of two centromere-binding motifs.
In the course of this study, we have identified the SopB-R219 counterpart present on chromosomal ParBs in most bacterial phyla (Figure 6C). As for SopB, this residue present in the two chromosomal Par systems tested here is essential for the partition function (Table 1) and is absolutely required for binding to its cognate centromere sequence (Figure 5). In addition, it belongs to a newly identified and well-conserved α-helical motif, GHAR/KA/VLL. This motif aligns with helices H5 and H8 of SopB and KorB, respectively, which are also essential for centromere binding. We, therefore, named this new motif CBM2 for centromere-binding motif 2. We proposed that chromosomal ParBs also harbor an extended centromere-binding domain composed of two DNA-binding motifs, the classical HTH (CBM1) and the newly identified CBM2.
We were unable to identify the CBM2 box and the positively charged residue on annotated chromosomal ParBs in only a few phyla, e.g. in cyanobacteria. The CBM2 motif, GHAR/KA/VLL, is also not conserved on ParBs from secondary chromosomes in the case of bacteria carrying two or three chromosomes (data not shown). This could be because of divergent parS sites present on these chromosomes. Indeed, in the case of secondary chromosomes, parS sequences are different from the consensus one present on the main chromosome (14,39). It will be of interest to investigate whether a second DNA-binding motif is present on these Par systems.
In the case of type Ib ParBs that carry RHH DNA-binding motifs in place of an HTH, no co-crystal structure is available to determine whether specificity determinants are also present outside the primary binding motif. However, further DNA contacts providing additional binding specificity have been suggested by comparison of ParG, the ParB analogue of TP228, with the Arc and Mnt repressors harboring RHH motifs (40), supporting the possibility that type Ib ParBs could also carry extended centromere-binding domains.
Initially, the HTH motif of SopB was identified as the only determinant for centromere-specific binding (23). Our results along with these examples indicate that ParB–parS interactions are not as simple as initially proposed. Rather they suggest that bacterial CBPs have evolved, using different strategies, to possess extended DNA-binding domains. We thus propose that the requirement for at least two independent DNA-binding motifs is imposed by the need for a high centromere-binding specificity. Mutational analysis and co-crystal structure determination of other ParB–parS complexes, especially from chromosomal Par systems but also from ParBs with RHH, will be of particular interest to determine the extent of the different strategies developed for providing such high specificity for centromere binding.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–4 and Supplementary References [13,29,30,41–47].
FUNDING
Agence National pour la Recherche [2010 BLAN 1316 01]; Fonds Europeen de Developpement Regional (for microscope equipments); Association pour le Recherche sur le Cancer (for microscope equipments). Funding for open access charge: Agence National pour la Recherche [2010 BLAN 1316 01].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank S. Depaul for technical assistance with stability assays and media preparation, N. Seddiki for help with PyMol software, Y. Ah-Seng for providing pYAS47 before publication, V. Morales for her expertise in protein purification and all members of the ‘segregation’ team for discussions. They are grateful to C. Johnson and reviewers for improvements in the manuscript.
REFERENCES
- 1.Salje J. Plasmid segregation: how to survive as an extra piece of DNA. Crit. Rev. Biochem. Mol. Biol. 2010;45:296–317. doi: 10.3109/10409238.2010.494657. [DOI] [PubMed] [Google Scholar]
- 2.Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 3.Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebersbach G, Gerdes K. The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc. Natl Acad. Sci. USA. 2001;98:15078–15083. doi: 10.1073/pnas.261569598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marston AL, Errington J. Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol. Cell. 1999;4:673–682. doi: 10.1016/s1097-2765(00)80378-0. [DOI] [PubMed] [Google Scholar]
- 6.Jakimowicz D, Zydek P, Kois A, Zakrzewska-Czerwinska J, Chater KF. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 2007;65:625–641. doi: 10.1111/j.1365-2958.2007.05815.x. [DOI] [PubMed] [Google Scholar]
- 7.Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L. A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell Biol. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barilla D, Rosenberg MF, Nobbmann U, Hayes F. Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. EMBO J. 2005;24:1453–1464. doi: 10.1038/sj.emboj.7600619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouet JY, Ah-Seng Y, Benmeradi N, Lane D. Polymerization of SopA partition ATPase: regulation by DNA binding and SopB. Mol. Microbiol. 2007;63:468–481. doi: 10.1111/j.1365-2958.2006.05537.x. [DOI] [PubMed] [Google Scholar]
- 10.Hui MP, Galkin VE, Yu X, Stasiak AZ, Stasiak A, Waldor MK, Egelman EH. ParA2, a Vibrio cholerae chromosome partitioning protein, forms left-handed helical filaments on DNA. Proc. Natl Acad. Sci. USA. 2010;107:4590–4595. doi: 10.1073/pnas.0913060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis MA, Martin KA, Austin SJ. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol. Microbiol. 1992;6:1141–1147. doi: 10.1111/j.1365-2958.1992.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe E, Wachi M, Yamasaki M, Nagai K. ATPase activity of SopA, a protein essential for active partitioning of F-plasmid. Mol. Gen. Genet. 1992;234:346–352. doi: 10.1007/BF00538693. [DOI] [PubMed] [Google Scholar]
- 13.Ah-Seng Y, Lopez F, Pasta F, Lane D, Bouet JY. Dual role of DNA in regulating ATP hydrolysis by the SopA partition protein. J. Biol. Chem. 2009;284:30067–30075. doi: 10.1074/jbc.M109.044800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J. Bacteriol. 2007;189:8693–8703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes F, Barilla D. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- 16.Helsberg M, Eichenlaub R. Twelve 43-base-pair repeats map in a cis-acting region essential for partition of plasmid mini-F. J. Bacteriol. 1986;165:1043–1045. doi: 10.1128/jb.165.3.1043-1045.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalnin K, Stegalkina S, Yarmolinsky M. pTAR-encoded proteins in plasmid partitioning. J. Bacteriol. 2000;182:1889–1894. doi: 10.1128/jb.182.7.1889-1894.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams DR, Macartney DP, Thomas CM. The partitioning activity of the RK2 central control region requires only incC, korB and KorB-binding site O(B)3 but other KorB-binding sites form destabilizing complexes in the absence of O(B)3. Microbiology. 1998;144:3369–3378. doi: 10.1099/00221287-144-12-3369. [DOI] [PubMed] [Google Scholar]
- 19.Martin KA, Davis MA, Austin S. Fine-structure analysis of the P1 plasmid partition site. J. Bacteriol. 1991;173:3630–3634. doi: 10.1128/jb.173.12.3630-3634.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori H, Mori Y, Ichinose C, Niki H, Ogura T, Kato A, Hiraga S. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J. Biol Chem. 1989;264:15535–15541. [PubMed] [Google Scholar]
- 21.Schumacher MA, Funnell BE. Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature. 2005;438:516–519. doi: 10.1038/nature04149. [DOI] [PubMed] [Google Scholar]
- 22.Khare D, Ziegelin G, Lanka E, Heinemann U. Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat. Struct. Mol. Biol. 2004;11:656–663. doi: 10.1038/nsmb773. [DOI] [PubMed] [Google Scholar]
- 23.Ravin NV, Rech J, Lane D. Mapping of functional domains in F plasmid partition proteins reveals a bipartite SopB-recognition domain in SopA. J. Mol. Biol. 2003;329:875–889. doi: 10.1016/s0022-2836(03)00525-4. [DOI] [PubMed] [Google Scholar]
- 24.Hanai R, Liu RP, Benedetti P, Caron PR, Lynch AS, Wang JC. Molecular dissection of a protein SopB essential for Escherichia coli F plasmid partition. J. Biol. Chem. 1996;271:17469–17475. doi: 10.1074/jbc.271.29.17469. [DOI] [PubMed] [Google Scholar]
- 25.Lynch AS, Wang JC. SopB protein-meditated silencing of genes linked to the sopC locus of Escherichia coli F plasmid. Proc. Natl Acad. Sci. USA. 1995;92:1896–1900. doi: 10.1073/pnas.92.6.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumacher MA, Piro KM, Xu W. Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes. Nucleic Acids Res. 2010;38:4514–4526. doi: 10.1093/nar/gkq161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pillet F, Sanchez A, Lane D, Anton Leberre V, Bouet JY. Centromere binding specificity in assembly of the F plasmid partition complex. Nucleic Acids Res. 2011;39:7477–7486. doi: 10.1093/nar/gkr457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemonnier M, Bouet JY, Libante V, Lane D. Disruption of the F plasmid partition complex in vivo by partition protein SopA. Mol. Microbiol. 2000;38:493–505. doi: 10.1046/j.1365-2958.2000.02101.x. [DOI] [PubMed] [Google Scholar]
- 29.Godfrin-Estevenon AM, Pasta F, Lane D. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol. Microbiol. 2002;43:39–49. doi: 10.1046/j.1365-2958.2002.02735.x. [DOI] [PubMed] [Google Scholar]
- 30.Dubarry N, Pasta F, Lane D. ParABS systems of the four replicons of Burkholderia cenocepacia: new chromosome centromeres confer partition specificity. J. Bacteriol. 2006;188:1489–1496. doi: 10.1128/JB.188.4.1489-1496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon GS, Sitnikov D, Webb CD, Teleman A, Straight A, Losick R, Murray AW, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 33.Niki H, Hiraga S. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 34.Leonard TA, Butler PJ, Lowe J. Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol. Microbiol. 2004;53:419–432. doi: 10.1111/j.1365-2958.2004.04133.x. [DOI] [PubMed] [Google Scholar]
- 35.Leonard TA, Butler PJ, Lowe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer-a conserved biological switch. EMBO J. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaichi Y, Niki H. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:14656–14661. doi: 10.1073/pnas.97.26.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Funnell BE, Gagnier L. The P1 plasmid partition complex at parS: II. Analysis of ParB protein binding activity and specificity. J. Biol. Chem. 1993;268:3616–3624. [PubMed] [Google Scholar]
- 38.Bouet JY, Surtees JA, Funnell BE. Stoichiometry of P1 plasmid partition complexes. J. Biol. Chem. 2000;275:8213–8219. doi: 10.1074/jbc.275.11.8213. [DOI] [PubMed] [Google Scholar]
- 39.Passot FM, Calderon V, Fichant G, Lane D, Pasta F. Centromere binding and evolution of chromosomal partition systems in the burkholderiales. J. Bacteriol. 2012;194:3426–3436. doi: 10.1128/JB.00041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golovanov AP, Barilla D, Golovanova M, Hayes F, Lian LY. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol. Microbiol. 2003;50:1141–1153. doi: 10.1046/j.1365-2958.2003.03750.x. [DOI] [PubMed] [Google Scholar]
- 41.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl Acad. Sci. USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouet JY, Bouvier M, Lane D. Concerted action of plasmid maintenance functions: partition complexes create a requirement for dimer resolution. Mol. Microbiol. 2006;62:1447–1459. doi: 10.1111/j.1365-2958.2006.05454.x. [DOI] [PubMed] [Google Scholar]
- 44.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 45.Bouet JY, Campo NJ, Krisch HM, Louarn JM. The effects on Escherichia coli of expression of the cloned bacteriophage T4 nucleoid disruption (ndd) gene. Mol. Microbiol. 1996;20:519–528. doi: 10.1046/j.1365-2958.1996.5411067.x. [DOI] [PubMed] [Google Scholar]
- 46.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 47.Ravin N, Lane D. Partition of the linear plasmid N15: interactions of N15 partition functions with the sop locus of the F plasmid. J. Bacteriol. 1999;181:6898–6906. doi: 10.1128/jb.181.22.6898-6906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin DCH, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.