Abstract
Monocyte chemoattractant protein 1-induced protein 1 (MCPIP1), belonging to the MCPIP family with highly conserved CCCH-type zinc finger and Nedd4-BP1, YacP Nuclease domains, has been implicated in negative regulation of the cellular inflammatory responses. In this report, we demonstrate for the first time that this RNA-binding nuclease also targets viral RNA and possesses potent antiviral activities. Overexpression of the human MCPIP1, but not MCPIP2, MCPIP3 or MCPIP4, inhibited Japanese encephalitis virus (JEV) and dengue virus (DEN) replication. The functional analysis of MCPIP1 revealed that the activities of RNase, RNA binding and oligomerization, but not deubiqutinase, are required for its antiviral potential. Furthermore, infection of other positive-sense RNA viruses, such as sindbis virus and encephalomyocarditis virus, and negative-sense RNA virus, such as influenza virus, as well as DNA virus, such as adenovirus, can also be blocked by MCPIP1. Moreover, the endogenous MCPIP1 gene expression was induced by JEV and DEN infection, and knockdown of MCPIP1 expression enhanced the replication of JEV and DEN in human cells. Thus, MCPIP1 can act as a host innate defense via RNase activity for targeting and degrading viral RNA.
INTRODUCTION
Control of RNA processing such as accumulation and degradation can have an important role in host defense against viral infection. Zinc finger antiviral protein (ZAP), a CCCH-type zinc finger protein, prevents the accumulation of viral mRNA by directly binding and recruiting the exosome to degrade the target RNA (1–3). The CCCH-type zinc finger proteins are characterized by three cysteine residues and one histidine residue that coordinate zinc ion binding (4,5). Tristetraprolin (TTP), the first identified CCCH-type zinc finger protein, binds to Adenylate-uridylate-rich (AU-rich) elements (AREs) in mRNA, removes the poly(A) tail and increases RNA turnover (6–8). A genome-wide survey revealed 58 and 55 CCCH-type zinc finger genes in the human and mouse genome, respectively (9). Gene expression profiling suggested that these CCCH-type zinc finger genes are associated with macrophage activation (9). A novel CCCH-type zinc finger protein found to be induced by monocyte chemoattractant protein 1 (MCP-1, also known as CCL-2) was named MCP-1-induced protein 1 (MCPIP1) (10).
MCPIP1 (also known as zinc finger CCCH-type containing 12A; ZC3H12A) belongs to MCPIP protein family, which contains other three members, MCPIP2, MCPIP3 and MCPIP4 (also known as ZC3H12B, ZC3H12C and ZC3H12D) (9,11). MCPIP proteins all contain a single CCCH-type zinc finger domain with RNA-binding potential at the middle region and a highly conserved Nedd4-BP1, YacP Nuclease (NYN) domain with RNase activity at the N-terminus (12–14). The CCCH-type zinc finger domain of MCPIP1 is characterized by three Cys (C306, C312 and C318) and one His (H322), which coordinate zinc ion binding for RNA-binding capacity (11). MCPIP1 is an Mg2+- or Mn2+-dependent RNase, and the crystal structure of the N-terminal MCPIP1 RNase domain revealed a catalytic pocket composed of several conserved acidic residues such as D141, D225, D226 and D244 involved in Mg2+ binding (14). MCPIP1 acts as an RNase to degrade certain mRNA of inflammatory cytokines such as interleukin 6 (IL-6), IL-12p40 and IL-1β (12,15). MCPIP1 also functions as a deubiquitinase (DUB) to inhibit lipopolysaccharide (LPS)-, IL-1β- and tumour necrosis factor alpha (TNF-α)-mediated NF-κB and c-Jun N-terminal kinase (JNK) signalling pathways by removing the ubiquitin moieties of TNF receptor-associated factors (TRAFs), including TRAF2, TRAF3 and TRAF6 (16). MCPIP4, originally identified as a potential tumour suppressor gene (17,18), has recently been shown to inhibit TLR signalling and macrophage activation, mainly through its deubiquitination activity (19). MCPIP1, but not MCPIP2, MCPIP3 and MCPIP4, could suppress miRNA biosynthesis and activity via cleavage of the terminal loops of precursor miRNAs (pre-miRNAs) (13). Thus, MCPIP family negatively regulates cellular inflammatory responses and maintains cellular immune homeostasis by distinct functions and diverse molecular mechanisms.
Japanese encephalitis virus (JEV) and dengue virus (DEN), members of the flavivirus genus of the Flaviviridae family, are important mosquito-borne human pathogens causing hemorrhagic, febrile and severe encephalitic illnesses. DEN infection causes an estimated 50–100 million cases of dengue fever and several hundred thousand cases of dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) annually around the world (20). JEV infection causes human epidemic encephalitis, with an estimated 10 000–15 000 deaths annually in South and Southeast Asia (21). JEV and DEN are enveloped and contain a single-stranded, positive-sense RNA genome, which encodes a long polyprotein that is processed into three structural proteins [core (C), precursor membrane (prM), envelope (E)] and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5). Flavivirus genome replication takes place by viral RNA replicase complex through RNA-dependent RNA polymerization (22,23). The positive-sense genomic RNA is transcribed into a replication-intermediate negative-sense RNA, which is then used as a template to synthesize genomic RNAs for translation and assembly of virion progeny.
MCPIP1 is rapidly induced by proinflammatory molecules such as TNF-α, MCP-1, IL-1β and LPS (10–12,24,25). Cytokines and chemokines such as TNF-α, MCP-1, IL-1β and IL-6 have been implicated in the development of dengue fever and DHF/DSS (26). High levels of TNF-α have been found in the serum and cerebrospinal fluid samples of JE patients with higher mortality rates (27). Thus, MCPIP1 is likely induced with JEV and DEN infection in humans; however, its role in viral replication has not been addressed. In this study, we examined the antiviral potential of human MCPIP family members and found that overexpression of MCPIP1, but not the related MCPIP2, MCPIP3 or MCPIP4 exhibited potent antiviral activity against JEV and DEN infection. We also examined the molecular mechanism of antiviral activity of MCPIP1 by using various mutants with defects on its RNase, RNA binding, oligomerization and DUB activity. We then tested the antiviral spectrum of MCPIP1 against various RNA and DNA viruses and found a broad antiviral activity of MCPIP1. Finally, we addressed the antiviral potential of endogenous MCPIP1 by knockdown of the expression of MCPIP1 gene in human cells. Thus, for the first time, MCPIP1 is identified as a host antiviral factor that is able to bind and degrade viral RNA.
MATERIALS AND METHODS
Cell lines, viruses, chemicals and antibodies
Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma) containing 10% fetal bovine serum (FBS). The tetracycline (Tet)-regulated expression HEK 293 cell line T-REx-293 (Invitrogen) was cultured in DMEM containing 10% FBS and 5 µg/ml of blasticidin. Baby hamster kidney BHK-21 cells were grown in RPMI 1640 medium containing 5% FBS. The human lung epithelial carcinoma cell line A549 was maintained in F-12 medium (Invitrogen) supplemented with 10% FBS. JEV strain RP-9 (28) and DEN-2 strain PL046 (29) were propagated in the C6/36 mosquito cell line grown in RPMI 1640 medium containing 5% FBS. A recombinant sindbis virus expressing enhanced green fluorescent protein (eGFP) was prepared, and the titer was determined as previously described (30). Vesicular stomatitis virus (VSV) and encephalomyocarditis virus (EMCV) were propagated in Vero cells with minimum essential medium (Eagle) containing 10% FBS. The adenovirus expressing a GFP (ZsGreen1) was generated and titrated by using the Adeno-X ViraTrak ZsGreen1-Express Expression System 2 (Clontech). Vaccinia virus (VV) growth and viral titration were done in BHK-21 cells. Hygromycin and blasticidin were from InvivoGen. Doxycycline (Dox) and puromycin were from Clontech and Sigma, respectively. Mouse monoclonal antibodies against HA-tag (Covance), GFP (Roche), influenza A nucleoprotein (NP) (Abcam) and enterovirus 71 (EV71) capsid protein VP1 (Chemicon) were used. Rabbit polyclonal antibody against ZC3H12A (GeneTex) was used.
Plasmid constructs and establishment of stable cell lines
The cDNAs encoding human MCPIP1 and MCPIP3 were amplified from RNA of LPS-treated K562 cells with the primer pairs for MCPIP1, 5′-ATGAGTGGCCCCTGTGGAG-3′ and 5′-TTACTCACTGGGGTGCTGGG-3′; and MCPIP3, 5′-ATGCCGGGTGGCGGC-3′ and 5′-TCAATAACCCAGCTGGGATTTCTCCACTAAAATGGCTG-3′. The cDNAs encoding human MCPIP2 and MCPIP4 were amplified from RNA of K562 cells with primer pairs for MCPIP2, 5′-ATGGAGAAGAGTGCCTCCAAGG-3′ and 5′-TCAACGTGCAGCCCTAAGCTT AGC-3′; and MCPIP4, 5′-ATGGAGCACCCCAGCAAGATG-3′ and 5′-TTAGGGCTTGCCCAGGGGCGCCC-3′. The cDNA was cloned to HA-tagged pcDNA3 vector to create an in-frame-fused HA-tag at the N terminus. The sequences were checked and were as reported in GenBank NM_025079, NM_001010888, NM_033390 and NM_207360 for MCPIP1, MCPIP2, MCPIP3 and MCPIP4, respectively.
The HA-tagged mutant forms of MCPIP1 (D141N, C157A and D225/226A) were generated by single-primer mutagenesis as described (31) by use of the following primers annealing to MCPIP1 cDNA with the mutated codons underlined: 5′-CGACCTGAGACCAGTGGTCATCAACGGGAGCAACGTGGCCAT-3′, 5′-GGAACAAGGAGGTCTTCTCCGCCCGGGGCATCCTGCTGGCAGT-3′ and 5′-AAGCGGGTGGTGTGCTATGCCGCCAGATTCATTGTGAAGCTGGC-3′, respectively. HA-tagged truncated constructs of MCPIP1, Δ305–325 and Δ458–536, were generated by single-primer mutagenesis. The truncated MCPIP1 constructs were generated by using the single-primer method (31) by designing the primer annealing to the flanking sequences of the deleted region. The primer used to generate Δ305–325 construct: 5′-CTTTGGAGCACAGGAAGCAGCCAAGCTGCCCCCAGCG-3′ anneals to nt. 893–912 (underlined with thin line) and nt 976–992 (underlined with thick line) of MCPIP1 mRNA. For MCPIP1 Δ458–536 construct, the primer (5′-GGGGGTTCGAGGAGGAGGCGGGGCTGGCAGGAGCC-3′) anneals to nt 1353–1371 and nt 1609–1624 of MCPIP1 mRNA underlined with thin and thick lines, respectively. The cDNA fragments with an N-terminal HA-tag were subcloned into a pcDNA5/TO vector (Invitrogen) for inducible expression.
To generate stable Tet-regulated expressing cells, T-REx-293 cells were transfected with various pcDNA5/TO plasmids and selected with hygromycin (250 µg/ml) and blasticidin (5 µg/ml) for 8 days. Individual colony was picked and expanded in DMEM containing 10% FBS, hygromycin and blasticidin.
Establishment of short-hairpin RNA (shRNA)-expressing stable cell lines
To generate human MCPIP1- and control-knockdown cells, we used the shMCPIP lentiviral particles (Santa Cruz Biotechnology, sc-78944 -V) that express three shRNAs targeting human MCPIP1 mRNA (5′-CTTCGTCAATGACAAGTTT-3′; 5′-GTGTCCCTATGGAAGGAAA-3′; 5′-CGATCACCCTGTTGATACA-3′) and the control shRNA lentiviral particles (sc-108080), respectively. Briefly, A549 cells were transduced with lentivirus for 24 h and selected with puromycin (2.5 µg/ml) for 72 h.
Western immunoblotting
Cell lysates were prepared and analysed as previously described (32). Briefly, equivalent amounts of proteins determined by Bio-Rad DC Protein Assay Kit were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. Membranes were blocked with 5% skim milk and incubated with primary antibodies. The blots were then incubated with a horseradish peroxidase-conjugated secondary antibody and developed with an enhanced chemiluminescence system (PerkinElmer). For re-blotting, the membrane was washed with 1× ReBlot plus strong antibody stripping solution (Chemicon) for 15 min at room temperature and re-probed with the primary antibody.
Immunofluorescence assay
Cells were fixed with 4% formaldehyde and permeabilized with phosphate buffered saline containing 0.5% Triton X-100, then incubated in blocking solution (3% skim milk in phosphate buffered saline) for 1 h. Viral protein expression was detected with a primary antibody for the indicated viral protein and Alexa-fluor 488 goat anti-mouse secondary antibody (Molecular Probes). The nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI; Molecular Probes). Cells were observed under an Olympus fluorescence microscope.
Reverse transcriptase polymerase chain reaction
Total cellular RNA was extracted with the RNeasy Total RNA kit (Qiagen), and then cDNA was synthesized with the ThermoScript reverse transcriptase polymerase chain reaction (RT-PCR) system (Invitrogen) by using oligo(dT)20 as a primer. Quantitative real-time PCR was performed by TaqMan Master Mix with TaqMan probes for MCPIP1 (Hs00962356_m1) and GAPDH (Hs02758991_g1) on an ABI-Prism 7500 real-time PCR system (Applied Biosystems). The relative mRNA levels were assessed by the comparative threshold cycle (ΔCt) method and normalized with GAPDH internal control. For viral RNA detection, RT-PCR was carried out by using random hexamer as RT primer and the specific primers for PCR (5′-ACCATGACTAAAAAACCAGGAGG-3′ and 5′-TCAGATGACCCTGTCTTCCTG-3′ for JEV; 5′-TGATGAATAACCAACGAA-3′ and 5′-CTACCACAGGACTCCTGCCTCTTCC-3′ for DEN-2; and 5′-TCCTGT GGCATCCACGAAACT-3′ and 5′-GAAGCATTTGCGGTGGACGAT-3′ for β-actin). The amplified PCR products were analysed by 0.8% agarose gel electrophoresis.
RNA binding assay
We used coimmunoprecipitation and RT-PCR to detect the association of MCPIP1 with viral RNA in virus-infected cells. Briefly, T-REx-293/MCPIP1 cells were infected with DEN-2 or JEV (multiplicity of infection [MOI] of 1) for 18 h and then cultured in medium with or without Dox (1 µg/ml) for 18 h. The cell extracts (containing protease inhibitor and RNasin) were mixed with prewashed HA-beads (Sigma) and incubated at 4°C for overnight. The complex of HA-tagged MCPIP1 protein and viral RNA was washed three times with HA lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA and 1% Triton X-100), and viral RNA was extracted by use of the RNeasy Total RNA kit (Qiagen). RT-PCR was performed by using the primers for DEN-2 3′-untranslated region (UTR) (5′-TAGAAGGCAAAACTAACATGAAACAAGG-3′, and 5′-AGAACCTGTTGATTCAACAGC-3′) or JEV 3′-UTR (5′-TAGTGTGATTTAAGGTAGAAAAGTAGACT-3′ and 5′-AGATCCTGTGTTCTTCCTCAC-3′).
In vitro RNA cleavage assay
The recombinant HA-tagged MCPIP1 and its D141N mutant were pulled down by use of HA-beads from T-REx-293 cells treated with Dox (1 µg/ml). The full-length viral RNA was in vitro synthesized from SP6-driven DEN-2 and JEV infectious clones (33–35) by use of MEGAscript high-yield transcription kit (Applied Biosystems). Viral RNA (1.5 µg) and purified HA-tagged MCPIP1 proteins (wild-type or D141N) (∼100 ng) were incubated in RNA cleavage buffer (25 mM HEPES, 50 mM potassium acetate, 5 mM DTT and 0.5 unit RNasin) with or without 5 mM Mg2+ at 30°C for 1 h. RNA integrity was analysed by 0.8% agarose gel electrophoresis and detected by ethidium bromide staining.
Oligomerization assay of MCPIP1
T-REx-293/MCPIP1 cells were cultured in medium with Dox (1 µg/ml) for 24 h, and then cell lysates were harvested in conjugation buffer (0.5% Triton X-100, 20 mM HEPES, 150 mM NaCl) containing protease inhibitor (Roche). Cell extracts were incubated with a final concentration of 1 mM disuccinimidyl suberate (Thermo Scientific) at room temperature for 30 min. The cross-linking reaction was stopped by adding quenching buffer Tris–HCl (pH 7.5) to a final concentration of 50 mM for 30 min at room temperature. The cell extracts were mixed with pre-washed HA-beads (Sigma) and incubated at 4°C for overnight. HA-tagged MCPIP1 proteins were washed three times with conjugation buffer (0.5% Triton X-100, 20 mM HEPES, 150 mM NaCl) containing protease inhibitor and eluted by HA peptides (Sigma). Samples were separated by SDS-PAGE and immunoblotted with anti-HA antibody.
RESULTS
Human MCPIP1, but not the other three MCPIP proteins, blocks JEV and DEN-2 infection
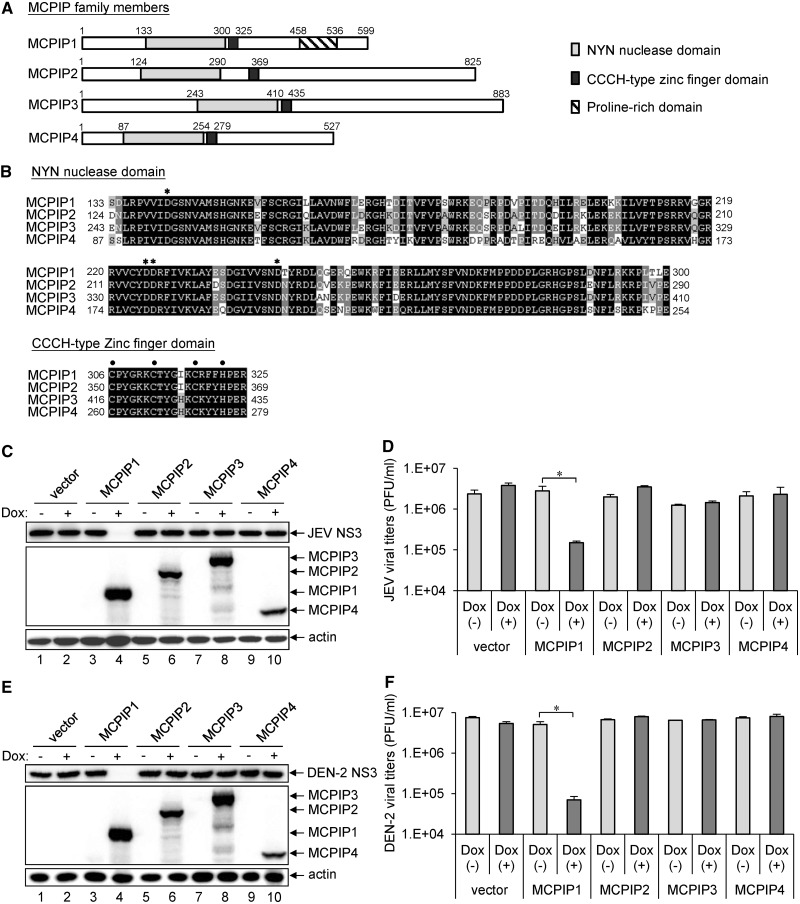
The human MCPIP protein family consists of four members: MCPIP1, MCPIP2, MCPIP3 and MCPIP4 (9,11); all contain homologous NYN and CCCH-type zinc finger domains (Figure 1A). The NYN domain with four conserved negative-charged Asp residues (D141, D225, D226 and D244) for Mg2+ binding and the CCCH-type zinc finger domain characterized by three Cys (C306, C312 and C318) and one His (H322) for Zn2+ binding, function in RNase and RNA-binding activities, respectively (Figure 1B) (9,14). We cloned the human cDNAs encoding these MCPIP proteins and established inducible cell lines expressing the MCPIP proteins with an N-terminal HA-tag in HEK T-REx-293 cells. With Dox induction, these cells expressed the corresponding MCPIP proteins with the expected molecular sizes recognized by antibody against the HA-tag (Figure 1C and E). To assess the antiviral potential of these human MCPIP proteins, we infected cells with JEV or DEN serotype 2 (DEN-2) and measured viral NS3 protein expression by western blotting and viral production by plaque-forming assays. The replication of JEV and DEN-2 was greatly reduced in cells with MCPIP1, but not with the other three MCPIP proteins (Figure 1C–F), which indicates a unique antiviral potential of MCPIP1. As overexpression of MCPIP1 induced apoptosis in monocytes and cardiac myocytes (10), we determined whether the antiviral effect of MCPIP1 was owing to cellular toxicity. In our T-REx-293 cells cultured in high density, the cell survival measured by trypan-blue exclusion and cytotoxicity measured by Lactate dehydrogenase release showed no significant difference between cells with or without MCPIP1 expression (Supplementary Figure S1). However, we noticed that MCPIP1 overexpression caused growth arrest when the cells were cultured in low density. Thus, the human MCPIP1 exhibits potent antiviral effects without causing cytotoxicity by itself.
Figure 1.
The human MCPIP1 exhibits potent antiviral activity against JEV and DEN-2 infection. (A) Schematic representation of human MCPIP family proteins containing NYN and CCCH-type zinc finger domains. (B) Amino acid sequence alignment of NYN and CCCH-type zinc finger domains in human MCPIP family proteins. Identical amino acids are in bold and similar amino acids are in grey. Four conserved negative-charged Asp (D) residues in the NYN domain are indicated by asterisks. The three Cys and one His residues in the CCCH-type zinc finger domain are indicated by dots. The human T-REx-293 cells stably transfected with pcDNA5/TO vector or pcDNA5/TO with MCPIP1/2/3/4 were cultured in medium without (−) or with (+) Dox (1 µg/ml) for 12 h, and then cells were infected with JEV or DEN-2 (MOI 5) for 24 h. (C, E) Western blot analysis of protein expression of HA for MCPIP, JEV NS3, DEN-2 NS3 and actin. (D, F) Viral titration was done by plaque-forming assay on BHK-21 cells. Virus titers are shown as means and standard deviations of two independent experiments. The data of the indicated groups were compared by two-tailed Student’s t-tests. *P ≤ 0.05.
RNase activity of MCPIP1 is required for its antiviral potential
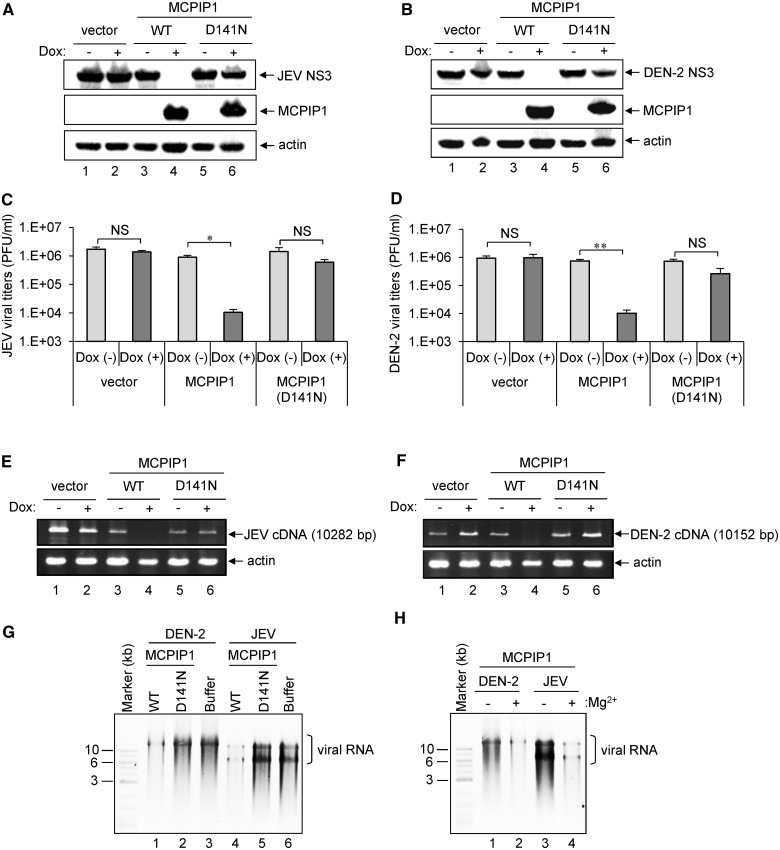
The NYN domain of MCPIP1 shows RNase activity, but the D141N mutation in the NYN domain abolishes its RNase activity (12). To determine whether the RNase activity of MCPIP1 is involved in its antiviral effects, we established T-REx-293 cells inducibly expressing the D141N nuclease-dead mutant of MCPIP1. As compared with the wild-type MCPIP1, the D141N mutant showed no anti-JEV or anti-DEN-2 effects as measured by western blot analysis of viral NS3 protein (Figure 2A and B), by immunofluorescence assay of viral NS1 protein (Supplementary Figure S2) and viral titration determined by plaque-forming assay (Figure 2C and D and Supplementary Figure S2). The viral RNAs determined by RT-PCR were also greatly reduced in cells expressing wild-type, but not the D141N-mutated MCPIP1 (Figure 2E and F). Thus, the RNase activity of MCPIP1 appears to be essential for its antiviral activities.
Figure 2.
RNase activity of human MCPIP1 is essential for its antiviral effect against JEV and DEN-2 infection. (A, B) Human T-REx-293 cells with inducible expression of vector, MCPIP1 or MCPIP1-D141N mutant were cultured in medium without (−) or with (+) Dox (1 µg/ml) for 12 h, then infected with JEV or DEN-2 (MOI 5). At 24 h post-infection, cell lysates were harvested for western blot analysis of viral protein NS3 expression for JEV (A) and DEN-2 (B). Virus titers of JEV (C) and DEN-2 (D) in the culture supernatants were determined by plaque-forming assays on BHK-21 cells. Results are means and standard deviations of two independent experiments. Data were compared by two-tailed Student’s t-tests. *P ≤ 0.05; **P ≤ 0.01; NS: not significant. The viral RNA levels of JEV (E) and DEN-2 (F) were analysed by RT-PCR as described in ‘Materials and Methods’ section. The expected sizes of JEV and DEN-2 PCR products are shown on the right. (G) HA-tagged wild-type (WT) or MCPIP1-D141N mutant proteins were incubated with in vitro transcribed full-length DEN-2 or JEV RNA in reaction buffer with 5 mM Mg2+ at 30°C for 1 h. RNA was separated by 0.8% agarose gel electrophoresis and stained with ethidium bromide. The photos are shown as inverse images. (H) In vitro cleavage of human MCPIP1 degrading viral RNA was analysed without (−) or with (+) Mg2+.
We examined whether MCPIP1 could directly degrade viral RNA by in vitro assay. Immunoprecipitated wild-type and MCPIP1-D141N mutant were incubated with in vitro-transcribed full-length JEV or DEN-2 viral RNA with or without Mg2+ for 1 h, and then viral RNA integrity was analysed by agarose gel electrophoresis and ethidium bromide staining. JEV and DEN-2 viral RNA level was reduced after incubation with wild-type MCPIP1, but not the D141N-mutant or buffer alone (Figure 2G), through an Mg2+-dependent mechanism (Figure 2H). We further used replicon system to address whether MCPIP1 also degrades viral RNA in vivo. We measured the replicon reporter expression, which depend on the replicon RNA levels, in cells with wild-type or MCPIP1-D141N expression. As shown in Supplementary Figure S3, the luciferase activities derived from JEV and DEN-2 replicons were much lower in cells expressing wild-type MCPIP1, but not in cells with MCPIP1-D141N mutant. Thus, human MCPIP1 may function as an RNase to target viral RNA during JEV and DEN-2 infection.
CCCH-type zinc finger domain is essential for viral RNA binding and antiviral activities of MCPIP1
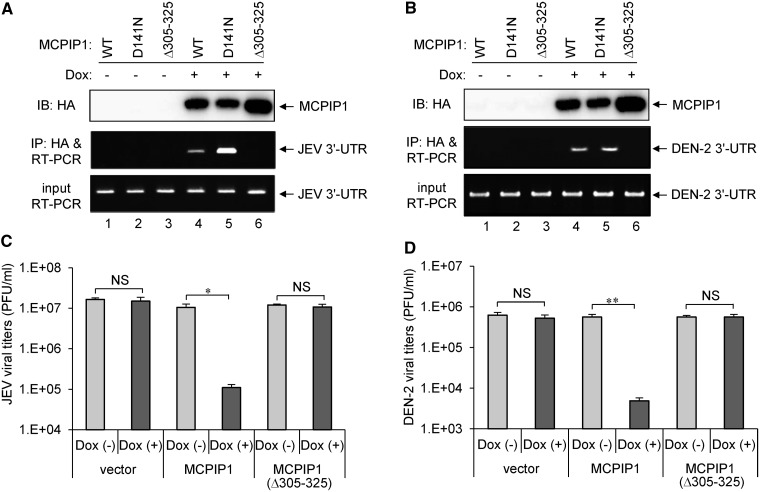
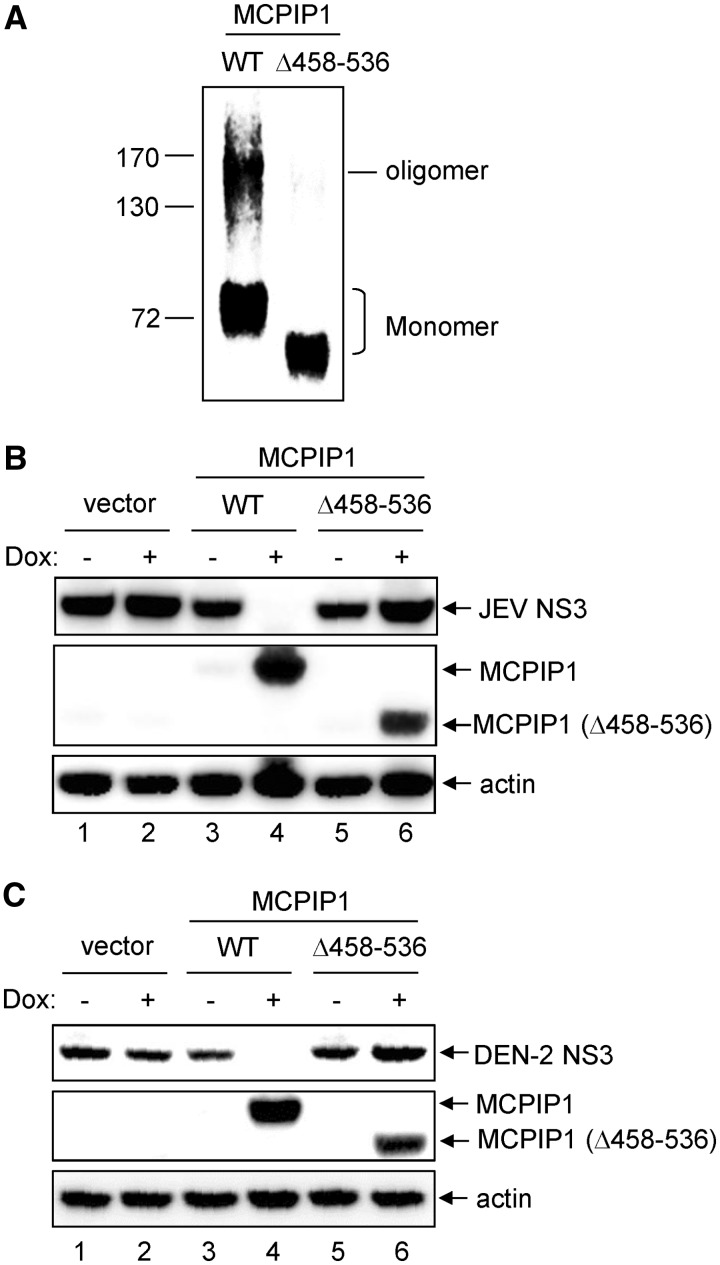
To verify whether the CCCH-type zinc finger domain of MCPIP1 is involved in viral RNA binding and is essential for its antiviral activity, we established T-REx-293 cells inducibly expressing an HA-tagged CCCH-type zinc finger-deleted mutant of MCPIP1 (Δ305–325). The viral RNA-binding ability of MCPIP1 was analysed by immunoprecipitation with antibody against HA-tag, then RT-PCR of JEV and DEN-2 RNA. Both the wild-type and MCPIP1-D141N mutant pulled down JEV and DEN-2 viral RNA, but the MCPIP1-Δ305–325 mutant lost its viral RNA-binding activity (Figure 3A and B). Furthermore, cells expressing MCP1P1-Δ305–325 mutant no longer showed anti-JEV and anti-DEN-2 activities of MCPIP1 (Figure 3C and D), indicating that CCCH-type zinc finger domain of human MCPIP1 is essential for viral RNA-binding and antiviral activity against JEV and DEN-2 infection.
Figure 3.
The CCCH-type zinc finger domain of human MCPIP1 is required for viral RNA binding and antiviral activity against JEV and DEN-2 infection. (A, B) Human T-REx-293 cells with WT or MCPIP1-D141N or −Δ305 to 325 mutant were infected with JEV or DEN-2 (MOI 5) for 18 h, then cultured in medium without (−) or with (+) Dox (1 µg/ml) for 18 h. The viral RNA bound with MCPIP1 protein was pulled down with HA-beads and amplified by RT-PCR with JEV or DEN-2 3′-UTR specific primers (middle panels). RT-PCR of mRNA levels of JEV or DEN-2 3′-UTR in virus-infected cells (bottom panels). Western blot analysis of HA-tagged MCPIP1 protein expression in cell lysates (top panels). (C, D) Human T-REx-293 cells with vector, MCPIP1 or MCPIP1-Δ305–325 mutant were cultured without (−) or with (+) Dox (1 µg/ml) for 12 h and then infected with JEV or DEN-2 (MOI 5) for 24 h. Virus titers of JEV (C) and DEN-2 (D) were determined by plaque-forming assays on BHK-21 cells. Results are means and standard deviations of two independent experiments. The titers of the indicated groups were compared by two-tailed Student’s t-tests. *P ≤ 0.05; **P ≤ 0.01; NS: not significant.
DUB activity is not required for the antiviral effect of MCPIP1
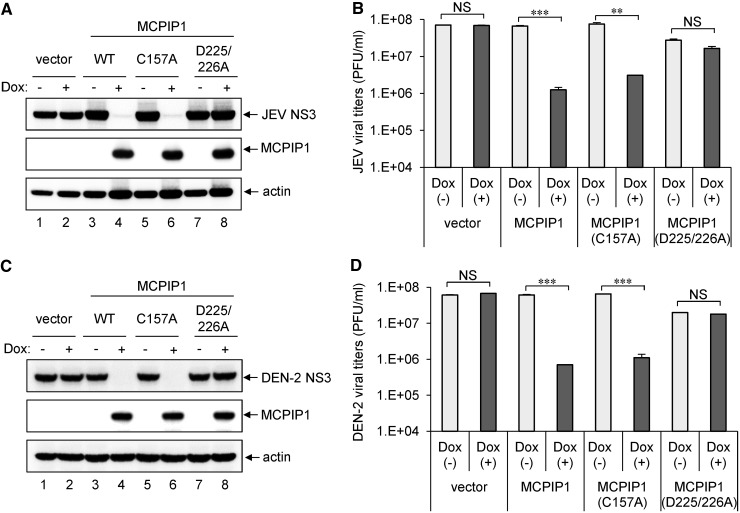
MCPIP1 can function as a DUB to remove the ubiquitin moieties of TRAFs (16). As cellular ubiquitinating/deubiquitinating enzymes have been implicated in virus replication (36,37), we further assessed whether the DUB activity of human MCPIP1 is involved in the inhibition of JEV and DEN-2 replication. The MCPIP1-C157A mutation abolishes DUB, but not RNase activity, whereas the MCPIP1-D225/226A mutant retains DUB, but not RNase activity (16,38). We thus established T-REx-293 cells inducibly expressing MCPIP1-C157A and -D225/226A mutants. The replication of JEV and DEN-2 detected by viral NS3 protein expression (Figure 4A and C) and viral production (Figure 4B and D) were reduced with the RNase-positive MCPIP1-C157A mutant, but not the DUB-positive MCPIP1-D225/226A mutant, to levels similar to that of wild-type, suggesting that RNase, but not DUB activity of MCPIP1, is required for its antiviral effect.
Figure 4.
Deubiqutinase activity of human MCPIP1 is not involved in its antiviral potential against JEV and DEN-2 infection. Human T-REx-293 cells with vector, WT or MCPIP1-C157A or -D225/226A were cultured without (−) or with (+) Dox (1 µg/ml) for 12 h, then infected with JEV or DEN-2 (MOI 5) for 24 h. Western blot analysis of levels of indicated proteins (A, C). Virus titers of JEV (B) and DEN-2 (D) are means and standard deviations of two independent experiments. The titers of the indicated groups were compared by two-tailed Student’s t-tests. **P ≤ 0.01; ***P ≤ 0.001; NS: not significant.
Oligomerization of MCPIP1 is required for anti-JEV and anti-DEN-2 activities
A unique C-terminal proline-rich domain noted in MCPIP1, but not in other MCPIP proteins, can trigger oligomerization and is involved in miRNA silencing activity of MCPIP1 (13). To examine whether the oligomerization of MCPIP1 is involved in its antiviral activity, we established T-REx-293 cells inducibly expressing an HA-tagged proline-rich domain-deleted mutant of MCPIP1 (Δ458–536). The wild-type and MCPIP1-Δ458–536 mutant were subjected to oligomerization analysis by chemical cross-linking, then immunoprecipitation and immunoblotting with anti-HA-tag antibody. High molecular-weight oligomers noted in wild-type were greatly lost in MCPIP1-Δ458–536 mutant (Figure 5A). The antiviral effect of MCPIP1 was lost in MCPIP1-Δ458–536 mutant as measured by immunoblotting for the viral NS3 protein (Figure 5B and C) and viral titration with plaque-forming assays (Supplementary Figure S4), indicating that oligomerization of MCPIP1 is required for its antiviral activity.
Figure 5.
The proline-rich domain of human MCPIP1 is essential for its oligomerization and antiviral activity. (A) Human T-REx-293 cells with WT or MCPIP1-Δ458–536 mutant were cultured with Dox (1 µg/ml) for 24 h. The cell proteins were cross-linked with disuccinimidyl suberate, and HA-tagged MCPIP1 proteins were pulled down with HA-beads. The oligomerization of MCPIP1 was analysed by SDS-PAGE and immunoblotted with anti-HA antibody. (B, C) Human T-REx-293 cells with vector, WT or MCPIP1-Δ458–536 mutant were cultured without (−) or with (+) Dox (1 µg/ml) for 12 h and then infected with JEV or DEN-2 (MOI 5) for 24 h. The cell lysates were harvested for western blotting with the indicated antibodies.
The antiviral potential of MCPIP1 against a variety of RNA and DNA viruses
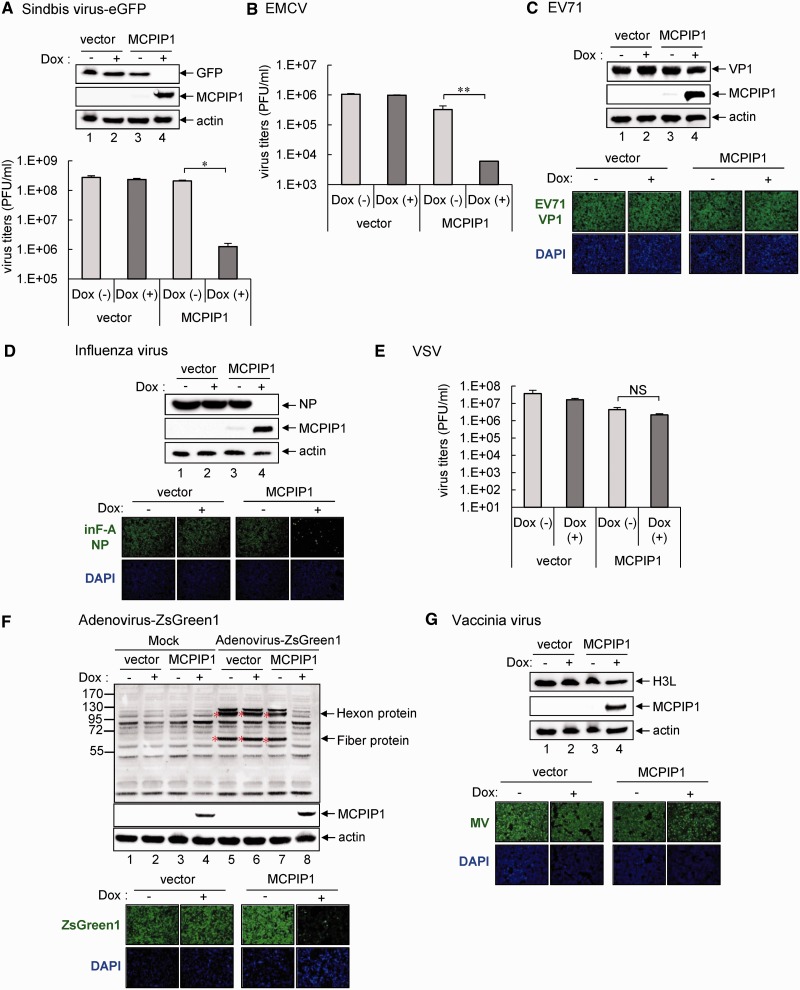
To assess whether the antiviral potential of MCPIP1 is unique to JEV and DEN-2, which are flaviviruses with positive-sense single-stranded RNA genome, we tested the effect of MCPIP1 on a variety of viruses ranging from RNA viruses with positive- or negative-sense genome, as well as DNA viruses. Among the other three RNA viruses with a positive-sense RNA genome, sindbis virus, an alphavirus, showed hampered replication by MCPIP1 as determined by western blot analysis of the eGFP reporter and progeny production by plaque-forming assay (Figure 6A). However, EMCV and EV71, two members of Picornaviridae, responded differently to MCPIP1. EMCV replication was blocked in cells with MCPIP1 expression (Figure 6B), whereas EV71 replication was not affected (Figure 6C). MCPIP1 expression had distinct antiviral potential against members of the negative-sense RNA viruses: it blocked the replication of influenza A virus (Figure 6D), an orthomyxovirus, but not that of VSV (Figure 6E), a rhabdovirus.
Figure 6.
The antiviral potential of human MCPIP1 against several viruses. Human T-REx-293 cells with MCPIP1 or vector control were cultured without (−) or with (+) Dox (1 µg/ml) for 12 h and then infected with the indicated viruses for 24 h. (A) Western blot analysis of the indicated proteins in cells with sindbis-eGFP infection (MOI 20) (upper panel). Virus titers of sindbis-eGFP were determined by plaque-forming assays on BHK-21 cells (lower panel). (B) For EMCV infection (MOI 1), virus titers were determined by plaque-forming assays on Vero cells. (C) Western blot analysis of levels of indicated proteins with EV71 infection (MOI 5) (upper panels). Immunofluorescence assay of EV71 viral protein VP1 (green) and DAPI (blue) was photographed by fluorescent microscopy (lower panels). (D) Western blot analysis of levels of indicated proteins to detect influenza virus infection (MOI 1) (upper panels). Immunofluorescence assay of influenza viral protein NP (green) and DAPI (blue) (lower panels). (E) For VSV infection (MOI 0.01), virus titers were determined by plaque-forming assays on Vero cells. (F) Western blot analysis of the protein expression of adenovirus hexon and fiber proteins to detect adenovirus-ZsGreen1 infection (MOI 20) (upper panels). Immunofluorescence assay of adenovirus expression of ZsGreen1 (green) and DAPI (blue) (lower panels). (G) Western blot analysis of level of vaccinia viral protein H3L to detect VV infection (MOI 5) (upper panels). Immunofluorescence assay of mature vaccinia virus (MV) (green) and DAPI (blue) (lower panels). The results are means and standard deviations of two independent experiments. The viral titers were compared by two-tailed Student’s t-tests. *P ≤ 0.05; **P ≤ 0.01. NS: not significant.
Interestingly, the antiviral potential of MCPIP1 was not limited to RNA viruses, as replication of a recombinant adenovirus carrying ZsGreen1, a bright GFP, was blocked in cells with MCPIP1 by western blot analysis of adenoviral hexon and fiber proteins and immunofluorescence assay of ZsGreen1 reporter expression (Figure 6F). However, replication of another DNA virus, VV, was not hindered by MCPIP1 expression (Figure 6G). Thus, human MCPIP1 shows a broad-spectrum, but not promiscuous, antiviral activity against various RNA and DNA viruses.
The antiviral potential of endogenous MCPIP1 in human cells
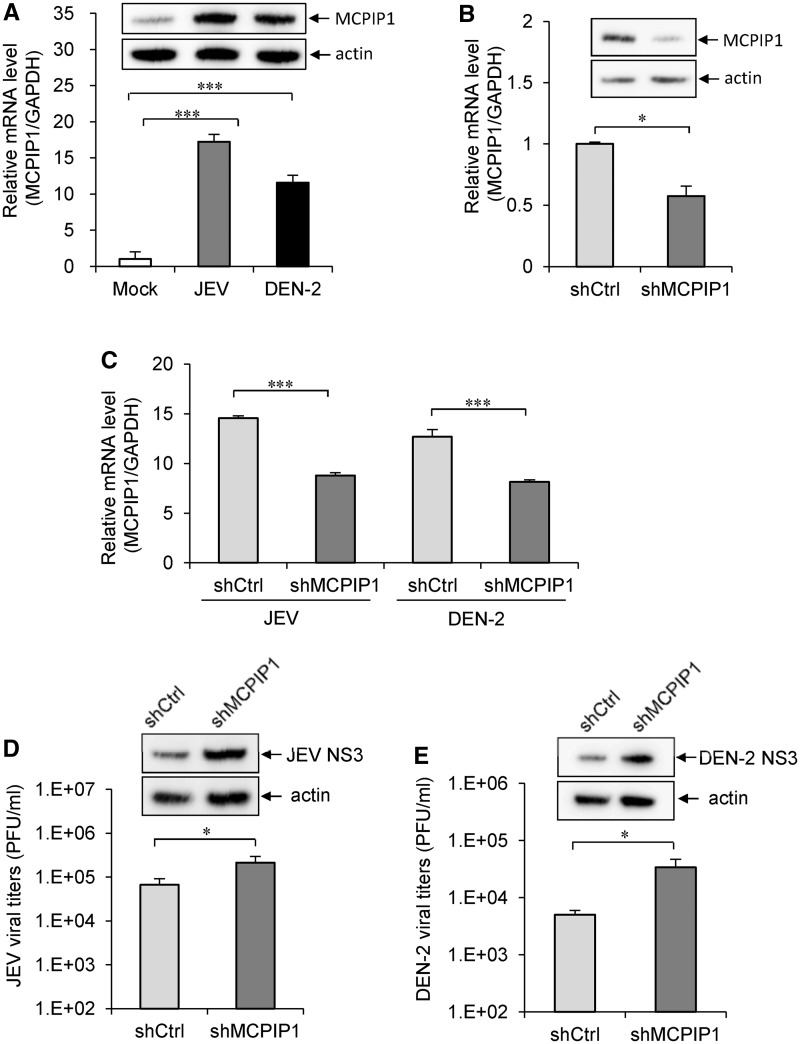
To address the role of endogenous MCPIP1, we examined whether MCPIP1 gene expression is induced by viral infection in human cells. By measuring the RNA and protein expression levels with real-time RT-PCR and western blotting, MCPIP1 was readily induced by JEV and DEN-2 infection (Figure 7A). To reduce the endogenous MCPIP1 expression, we transduced A549 cells with lentivirus expressing shRNA targeting three different regions of MCPIP1 gene transcript (shMCPIP1) and evaluated the knockdown effect at RNA and protein levels (Figure 7B). Even though the knockdown effect was <50% (Figure 7C), higher viral replication of JEV and DEN-2 was noted in cells with reduced MCPIP1 expression (Figure 7D and E). These results thus suggest that MCPIP1 can be induced by viral infection and may play a role in host antiviral immunity.
Figure 7.
The antiviral potential of endogenous MCPIP1. (A) The human A549 cells were mock infected or infected with JEV (MOI 5 for 24 h) or DEN-2 (MOI 5 for 48 h). The relative MCPIP1 mRNA levels were analysed by quantitative real-time RT-PCR and normalized to that of GAPDH mRNA. The MCPIP1 protein levels were assessed by western blotting with MCPIP1-specific antibody. (B) A549 cells were transduced with MCPIP shRNA lentiviral particles (shMCPIP1) or control shRNA lentiviral particles (shCtrl). The knockdown effect was evaluated by real-time RT-PCR and western blotting as described in panel A. (C) The MCPIP1 expression levels in A549-shCtrl and A549-shMCPIP1 cells infected with JEV (MOI 5 for 24 h) or DEN-2 (MOI 5 for 48 h) were analysed by real-time RT-PCR. A549-shCtrl and A549-shMCPIP1 cells infected with (D) JEV (MOI 5 for 24 h) or (E) DEN-2 (MOI 5 for 48 h) were analysed for viral production by plaque-forming assay and viral protein expression by western blotting. Data are shown as means and standard deviations of three independent experiments. The data of the indicated groups were compared by two-tailed Student’s t-tests. *P ≤ 0.05; ***P ≤ 0.001.
DISCUSSION
MCPIP1 is a multifunctional protein involved in various biological and physiological functions such as negative regulation of cellular inflammatory response (12,15,16), glial differentiation of neuroprogenitor cells (39), cell death of cardiomyocytes (10), adipogenesis and angiogenesis (40), as well as inhibition of miRNA biogenesis (13). Here, we extend the function of MCPIP1 to host antiviral defense. The potent antiviral activity of wild-type MCPIP1 was diminished in four mutants, D141N, Δ305–325, D225/226A and Δ458-536, but not in C157A-mutant. Three of the mutants involve RNase or DUB activity of MCPIP1: D141N is RNase (−)/DUB (−), D225/226A is RNase (−)/DUB (+) and C157A is RNase (+)/DUB (−) (38). As MCPIP1’s antiviral activity was noted in C157A, but not in D141N and D225/226A, the activity of RNase, but not DUB, is required in the antiviral action of MCPIP1. Different from the other members of the CCCH-type zinc finger family, such as TTP (6–8,41,42) and ZAP (43), which recruit cellular mRNA decay machinery exosome to degrade RNA molecules, MCPIP1 appears to function as an antiviral RNase by itself. The Δ305–325 mutant lacking the RNA binding CCCH-type zinc finger (12) failed to block viral replication; therefore, MCPIP1 likely binds to and degrades viral RNA directly.
The CCCH-type zinc finger domain of human MCPIP1 located within amino acid residue 305–325 is characterized by three Cys (C306, C312 and C318) and one His (H322), which coordinate zinc ion binding for RNA-binding capacity. Both MCPIP1-Δ305–325 and MCPIP1-C306R mutants have been used to demonstrate the importance of this CCCH domain in previous studies on IL-6 mRNA and pre-miRNA (12,13). We also have constructed a single-point mutant MCPIP1-C306R and established stable T-REx-293 cells with inducible expression of C306R mutant. To our surprise, MCPIP1-C306R was still able to bind with viral RNA and showed antiviral activities, although to a lesser extent when compared with the wild-type MCPIP1 (Supplementary Figure S5). The different binding properties of MCPIP1 with cellular and viral RNAs remain elusive.
Replication of several viruses, including positive-sense RNA viruses (JEV, DEN-2, sindbis virus, and EMCV), negative-sense RNA virus (influenza A virus) and DNA virus (adenovirus), was reduced in cells with MCPIP1 overexpression. However, not all viruses tested are sensitive to the antiviral activity of MCPIP1: replication of EV71, VSV and VV was not suppressed by MCPIP1 overexpression. Similarly, MCPIP1 can destabilize the mRNA of IL-6, IL-12p40 and IL-1β, but not TNF-α or CXCL1 (12,15), and MCPIP1 appears to target the 3′-UTR of IL-6 and IL-1β mRNA (12,13,15). The RNA sequences recognized by CCCH-type zinc finger proteins are critical in determining their targets. TTP has a preferential RNA target sequence, a 5′-UUAUUUAUU-3′ nonamer, located in AREs (44), but ZAP does not recognize any of the three types of AREs (45). The viral sequences sensitive to ZAP have been mapped to the 3′ long terminal repeat of Moloney murine leukemia virus and to multiple fragments in the sindbis virus genome (45). We used an in vitro cleavage assay to determine the viral RNA recognized by MCPIP1. Similar to the result for the full-length JEV RNA (Figure 2E), four different JEV RNA subfragments with deletions of nucleotides 2520–7116, 290–5863, 6965–10910 and 2811–10044 could still be degraded by the wild-type, but not by the D141N nuclease-dead MCPIP1 (Supplementary Figure S6). These results suggest that MCPIP1 may target multiple sites of the JEV RNA, or it may target the 5′ and 3′ sequences present in all of these RNA subfragments. MCPIP1 preferentially cleaves the unpaired regions around the terminal loops of pre-miRNA; therefore, the structure of terminal loop in a stem-loop might act as a platform for MCPIP1 recognition (13). Thus, whether specific viral RNA sequences and/or structures are required for recognition and cleavage of MCPIP1 remains unclear.
Four members of the human MCPIP family share a highly conserved NYN nuclease and CCCH-type zinc finger domains. However, only MCPIP1 exhibits antiviral activity. Similarly, a recent study showed that MCPIP1, but not the other MCPIP 2/3/4 proteins, cleaves pre-miRNA and suppresses miRNA biosynthesis (13). A unique proline-rich domain at the C terminus of MCPIP1, showing little similarity to the C termini of MCPIP2/3/4, contributes to MCPIP1 oligomerization and efficient interaction with pre-miRNA. We found that the Δ458–536 mutant, lacking the proline-rich domain of MCPIP1, lost its oligomeric potential and antiviral activity, suggesting that oligomerization of MCPIP1 is also involved in its antiviral action. Because MCPIP1 is a broad suppressor of the miRNA pathway, the potential involvement of miRNA in the antiviral activity of MCPIP1 cannot be excluded. However, our in vitro cleavage data suggests that MCPIP1 per se can cleave viral RNA, regardless of miRNA machinery, in an Mg2+-dependent manner, as was previously reported for cellular mRNA and pre-miRNA (12,13).
MCPIP1 is rapidly induced in macrophages by proinflammatory molecules such as TNF-α, MCP-1, IL-1β and LPS (11,12,24,25). Here, we find that MCPIP1 could also be induced by viral infection (Figure 7). As high levels of TNF-α, and to a lesser for IL-1β and MCP-1, could be detected in cells with JEV and DEN-2 infection (Supplementary Figure S7), the virus-triggered MCPIP1 induction might result from the action of proinflammatory cytokines. However, the induction of MCPIP1 by IL-1β has been reported to be mediated via NF-κB and extracellular signal-regulated kinases (ERK) pathways (15,24). As JEV and DEN-2 infection could also activate NF-κB and ERK pathways (46–48), the possibility that MCPIP1 is induced by virus-triggered NF-κB and ERK activation cannot be excluded. Furthermore, different from IL-1β, interferon (IFN)-α readily induced IFN-stimulated genes such as Stat1 and IRF-9, but failed to induce MCPIP1 (Supplementary Figure S8), indicating that human MCPIP1 is not induced by type-I IFN.
Induction of MCPIP1 functions in cellular modulation and helps to control the inflammatory response and immune homeostasis (12,16). MCPIP1 is a negative regulator controlling the stability of a set of inflammatory gene transcripts; Zc3h12a/MCPIP1-deficient mice showed severe immune disorders and spontaneously died within 12 weeks of birth (12). Elevated proinflammatory cytokines such as TNF-α, IL-1β and MCP-1 have been implicated in the development of DHF/DSS in severe dengue patients and viral encephalitis in JE patients (26,27). Although MCPIP1 expression has not been documented in patients with DEN or JEV infection, MCPIP1 induction may benefit the host in two ways: providing host defense against viral infection by degrading the viral RNA and controlling excess immune activation by negatively regulating the inflammatory response. Thus, assessing the role of MCPIP1 in viral pathogenesis and its therapeutic potential for viral diseases will be of interest.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Text and Supplementary Figures 1–8.
FUNDING
National Science Council (NSC), Taiwan [NSC 101-2321-B-001-028-MY3 to R.J.L. and Y.L.L., NSC 100-2320-B-038-033 to R.J.L., and NSC 100-2923-B-001-002-MY3 to Y.L.L.]; Academia Sinica, Taiwan. Funding for open access charge: Academia Sinica, Taiwan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Michael Lai (Institute of Molecular Biology, Academia Sinica, Taiwan) for providing the Influenza A WSN H1N1; Dr. Mei-Shang Ho (Institute of Biomedical Sciences, Academia Sinica, Taiwan) for enterovirus 71 New strain 5; Dr. Song-Kun Shyue (Institute of Biomedical Sciences, Academia Sinica, Taiwan) for antiserum against adenovirus; and Dr. Wen Chang (Institute of Molecular Biology, Academia Sinica, Taiwan) for antibodies against vaccinia virus intracellular mature virus and H3L.
REFERENCES
- 1.Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J. Virol. 2003;77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 3.Muller S, Moller P, Bick MJ, Wurr S, Becker S, Gunther S, Kummerer BM. Inhibition of filovirus replication by the zinc finger antiviral protein. J. Virol. 2007;81:2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown RS. Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 7.Carrick DM, Lai WS, Blackshear PJ. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res. Ther. 2004;6:248–264. doi: 10.1186/ar1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One. 2008;3:e2880. doi: 10.1371/journal.pone.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou L, Azfer A, Niu J, Graham S, Choudhury M, Adamski FM, Younce C, Binkley PF, Kolattukudy PE. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ. Res. 2006;98:1177–1185. doi: 10.1161/01.RES.0000220106.64661.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE, Fu M. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J. Biol. Chem. 2008;283:6337–6346. doi: 10.1074/jbc.M707861200. [DOI] [PubMed] [Google Scholar]
- 12.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, Sugimoto K, Miyazono K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol. Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Xu J, Peng W, Sun Y, Wang X, Xu Y, Li X, Gao G, Rao Z. Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase. Nucleic Acids Res. 2012;40:6957–6965. doi: 10.1093/nar/gks359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizgalska D, Wegrzyn P, Murzyn K, Kasza A, Koj A, Jura J, Jarzab B. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. FEBS J. 2009;276:7386–7399. doi: 10.1111/j.1742-4658.2009.07452.x. [DOI] [PubMed] [Google Scholar]
- 16.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, Kolattukudy PE, Fu M. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J. Exp. Med. 2010;207:2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minagawa K, Yamamoto K, Nishikawa S, Ito M, Sada A, Yakushijin K, Okamura A, Shimoyama M, Katayama Y, Matsui T. Deregulation of a possible tumour suppressor gene, ZC3H12D, by translocation of IGK@ in transformed follicular lymphoma with t(2;6)(p12;q25) Br. J. Haematol. 2007;139:161–163. doi: 10.1111/j.1365-2141.2007.06752.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Vikis HG, Wang Y, Jia D, Wang D, Bierut LJ, Bailey-Wilson JE, Amos CI, Pinney SM, Petersen GM, et al. Identification of a novel tumor suppressor gene p34 on human chromosome 6q25.1. Cancer Res. 2007;67:93–99. doi: 10.1158/0008-5472.CAN-06-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang S, Qi D, Liang J, Miao R, Minagawa K, Quinn T, Matsui T, Fan D, Liu J, Fu M. The putative tumor suppressor Zc3h12d modulates toll-like receptor signaling in macrophages. Cell. Signal. 2012;24:569–576. doi: 10.1016/j.cellsig.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10:100–103. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 21.Solomon T. Recent advances in Japanese encephalitis. J. Neurovirol. 2003;9:274–283. doi: 10.1080/13550280390194037. [DOI] [PubMed] [Google Scholar]
- 22.Uchil PD, Satchidanandam V. Characterization of RNA synthesis, replication mechanism, and in vitro RNA-dependent RNA polymerase activity of Japanese encephalitis virus. Virology. 2003;307:358–371. doi: 10.1016/s0042-6822(02)00130-7. [DOI] [PubMed] [Google Scholar]
- 23.Westaway EG, Mackenzie JM, Khromykh AA. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 2003;59:99–140. doi: 10.1016/s0065-3527(03)59004-2. [DOI] [PubMed] [Google Scholar]
- 24.Kasza A, Wyrzykowska P, Horwacik I, Tymoszuk P, Mizgalska D, Palmer K, Rokita H, Sharrocks AD, Jura J. Transcription factors Elk-1 and SRF are engaged in IL1-dependent regulation of ZC3H12A expression. BMC Mol. Biol. 2010;11:14. doi: 10.1186/1471-2199-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skalniak L, Mizgalska D, Zarebski A, Wyrzykowska P, Koj A, Jura J. Regulatory feedback loop between NF-kappaB and MCP-1-induced protein 1 RNase. FEBS J. 2009;276:5892–5905. doi: 10.1111/j.1742-4658.2009.07273.x. [DOI] [PubMed] [Google Scholar]
- 26.Martina BE, Koraka P, Osterhaus AD. Dengue virus pathogenesis: an integrated view. Clin. Microbiol. Rev. 2009;22:564–581. doi: 10.1128/CMR.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravi V, Parida S, Desai A, Chandramuki A, Gourie-Devi M, Grau GE. Correlation of tumor necrosis factor levels in the serum and cerebrospinal fluid with clinical outcome in Japanese encephalitis patients. J. Med. Virol. 1997;51:132–136. [PubMed] [Google Scholar]
- 28.Chen LK, Lin YL, Liao CL, Lin CG, Huang YL, Yeh CT, Lai SC, Jan JT, Chin C. Generation and characterization of organ-tropism mutants of Japanese encephalitis virus in vivo and in vitro. Virology. 1996;223:79–88. doi: 10.1006/viro.1996.0457. [DOI] [PubMed] [Google Scholar]
- 29.Lin YL, Liao CL, Chen LK, Yeh CT, Liu CI, Ma SH, Huang YY, Huang YL, Kao CL, King CC. Study of Dengue virus infection in SCID mice engrafted with human K562 cells. J. Virol. 1998;72:9729–9737. doi: 10.1128/jvi.72.12.9729-9737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin RJ, Chang BL, Yu HP, Liao CL, Lin YL. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarova O, Kamberov E, Margolis B. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques. 2000;29:970–972. doi: 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- 32.Lin RJ, Yu HP, Chang BL, Tang WC, Liao CL, Lin YL. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J. Immunol. 2009;183:8035–8043. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 33.Chien HL, Liao CL, Lin YL. The FUSE binding protein 1 interacts with untranslated regions of Japanese encephalitis virus RNA and negatively regulates viral replication. J. Virol. 2011;85:4698–4706. doi: 10.1128/JVI.01950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee TC, Lin YL, Liao JT, Su CM, Lin CC, Lin WP, Liao CL. Utilizing liver-specific microRNA-122 to modulate replication of dengue virus replicon. Biochem. Biophys. Res. Commun. 2010;396:596–601. doi: 10.1016/j.bbrc.2010.04.080. [DOI] [PubMed] [Google Scholar]
- 35.Liang JJ, Liao CL, Liao JT, Lee YL, Lin YL. A Japanese encephalitis virus vaccine candidate strain is attenuated by decreasing its interferon antagonistic ability. Vaccine. 2009;27:2746–2754. doi: 10.1016/j.vaccine.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Liao TL, Wu CY, Su WC, Jeng KS, Lai MM. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J. 2010;29:3879–3890. doi: 10.1038/emboj.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Si X, Gao G, Wong J, Wang Y, Zhang J, Luo H. Ubiquitination is required for effective replication of coxsackievirus B3. PLoS One. 2008;3:e2585. doi: 10.1371/journal.pone.0002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi D, Huang S, Miao R, She ZG, Quinn T, Chang Y, Liu J, Fan D, Chen YE, Fu M. Monocyte chemotactic protein-induced protein 1 (MCPIP1) suppresses stress granule formation and determines apoptosis under stress. J. Biol. Chem. 2011;286:41692–41700. doi: 10.1074/jbc.M111.276006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vrotsos EG, Kolattukudy PE, Sugaya K. MCP-1 involvement in glial differentiation of neuroprogenitor cells through APP signaling. Brain Res. Bull. 2009;79:97–103. doi: 10.1016/j.brainresbull.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu J, Azfer A, Zhelyabovska O, Fatma S, Kolattukudy PE. Monocyte chemotactic protein (MCP)-1 promotes angiogenesis via a novel transcription factor, MCP-1-induced protein (MCPIP) J. Biol. Chem. 2008;283:14542–14551. doi: 10.1074/jbc.M802139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 42.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 43.Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl Acad. Sci. USA. 2007;104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J. Biol. Chem. 2002;277:48558–48564. doi: 10.1074/jbc.M206505200. [DOI] [PubMed] [Google Scholar]
- 45.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 2004;78:12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang TH, Liao CL, Lin YL. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-kappaB activation. Microbes Infect. 2006;8:157–171. doi: 10.1016/j.micinf.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 47.Gupta N, Bhaskar AS, Lakshmana Rao PV. Transcriptional regulation and activation of the mitogen-activated protein kinase pathway after Japanese encephalitis virus infection in neuroblastoma cells. FEMS Immunol Med. Microbiol. 2011;62:110–121. doi: 10.1111/j.1574-695X.2011.00792.x. [DOI] [PubMed] [Google Scholar]
- 48.Huerta-Zepeda A, Cabello-Gutierrez C, Cime-Castillo J, Monroy-Martinez V, Manjarrez-Zavala ME, Gutierrez-Rodriguez M, Izaguirre R, Ruiz-Ordaz BH. Crosstalk between coagulation and inflammation during Dengue virus infection. Thromb. Haemost. 2008;99:936–943. doi: 10.1160/TH07-08-0438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.