Abstract
Previously, Lrp-like transcriptional regulator LysM from the hyperthermoacidophilic crenarchaeon Sulfolobus solfataricus was proposed to have a single target, the lysWXJK operon of lysine biosynthesis, and a single effector molecule, l-lysine. Here we identify ∼70 novel binding sites for LysM in the S. solfataricus genome with a LysM-specific nanobody-based chromatin immunoprecipitation assay coupled to microarray hybridization (ChIP-chip) and in silico target site prediction using an energy-based position weight matrix, and validate these findings with in vitro binding. LysM binds to intergenic and coding regions, including promoters of various amino acid biosynthesis and transport genes. We confirm that l-lysine is the most potent effector molecule that reduces, but does not completely abolish, LysM binding, and show that several other amino acids and derivatives, including d-lysine, l-arginine, l-homoarginine, l-glutamine and l-methionine and branched-chain amino acids l-leucine, l-isoleucine and l-valine, significantly affect DNA-binding properties of LysM. Therefore, it appears from this study that LysM is a much more versatile regulator than previously thought, and that it uses a variety of amino acids to sense nutritional quality of the environment and to modulate expression of the metabolic machinery of Sulfolobus accordingly.
INTRODUCTION
The Leucine-responsive regulatory protein (Lrp) family of transcriptional regulators is one of the largest families of bacterial/archaeal regulators (1–3). Lrp-like regulators exhibit a similar architecture (4–9). The N-terminal DNA-binding domain bears a winged helix-turn-helix motif; the C-terminal Regulation of Amino acid Metabolism (RAM) domain shows a characteristic αβ sandwich fold (βαββαβ) (5) and is involved in effector-binding and oligomerization. Intriguingly, despite these structural similarities, bacterial and archaeal Lrp-like regulators modulate activity of two different transcription machineries. Bacterial Lrp-like regulators modulate the initiation frequency of a unique RNA polymerase that directly binds core promoter elements (10). In contrast, archaeal members regulate the activity of a eukaryotic-like transcription apparatus that consists of a TATA box, transcription factor B responsive element (BRE) and initiator (Inr); three general transcription factors [TATA binding protein (TBP), transcription factor B and transcription factor E] and a complex RNA polymerase that is most homologous to eukaryotic RNA polymerase II (11–13). As in eukarya, the unique archaeal RNA polymerase is recruited by protein–protein interactions with transcription factors. The fundamental differences between the bacterial and archaeal transcriptional machinery appeal for a different mode of action of bacterial and archaeal Lrp-like regulators, in particular concerning transcriptional activation.
Escherichia coli Lrp, archetype of the Lrp-like family, is a global regulator that directly or indirectly affects a plenitude of metabolic pathways (14–16) and was recently shown to use several amino acids as cofactors (17). This regulator is proposed to play an important role in coordination of the metabolic switch of microorganisms on transitions between regimes of feast and famine (18,19). In contrast, most other bacterial Lrp-like regulators act more specific, with one or a few targets only, and are generally involved in control of amino acid metabolism. Much less is known about archaeal members. Frequently, their cofactor has not yet been identified, targets apart from the control region of their own gene (autoregulation) are rarely known and molecular mechanisms of regulatory process have not been unravelled [for a review see (2)]. Nevertheless, a few case studies, in particular with regulators from model organisms Sulfolobus solfataricus (crenarchaeote) and Methanocaldococcus jannaschii (euryarchaeote), indicate that archaeal Lrp-like regulators appear to be more versatile than their bacterial counterparts and that they are also involved in regulation of central metabolism (20,21).
LysM is an Lrp-like regulator from hyperthermoacidopilic crenarchaeon S. solfataricus that was previously shown to bind the lysWXJK control region and was proposed to act as a lysine-sensitive co-activator (22). Both LysM and its target sequence in the lysW operator are highly conserved among Sulfolobus genomes. However, a high-resolution contact map of LysM–operator DNA contacts has not been established, DNA sequence specificity of LysM has not been thoroughly studied and, besides lysW, no additional target genes have been identified to date.
To gain further insights into LysM action and its physiological role, we established a high-resolution contact map of LysM–lysW promoter/operator interactions and performed a saturation mutagenesis of the symmetrical LysM consensus box. Furthermore, we identified additional LysM-binding sites in the genome of S. solfataricus and validated these data with in vitro DNA-binding assays for a subset of selected targets. The data presented here clearly demonstrate that LysM binds with high affinity to several additional binding sites in the Sulfolobus genome both in vivo and in vitro. Furthermore, we show that LysM has a rather broad ligand-binding specificity and that several amino acids besides l-lysine significantly affect its DNA-binding capacity.
MATERIALS AND METHODS
Strains and growth conditions
E. coli strain DH5α was used for all cloning and plasmid propagation purposes. E. coli BL21(DE3) was used as a host for overexpression of recombinant proteins. S. solfataricus P2 (DSM1617) and Sulfolobus acidocaldarius (DSM639) were grown aerobically at 80°C and 75°C, respectively, in Brock basic medium (23) supplemented with 0.2% sucrose, with or without l-lysine (5 mM), as indicated.
DNA and RNA extractions
Genomic DNA (gDNA) from S. solfataricus P2 and S. acidocaldarius was extracted from 4 ml of a culture with a QuickPick SML gDNA kit (BioNobile). Plasmid DNA was extracted from transformed E. coli DH5α with a Miniprep kit (Qiagen). For RNA extraction, 1 ml of an exponentially grown S. solfataricus P2 culture [optical density (OD600 nm) of 0.3] was mixed with 2 ml RNAprotect Bacteria Reagent (Qiagen) and centrifuged. Pelleted cells were subsequently lysed with proteinase K (Qiagen), and RNA was extracted with an RNeasy mini kit (Qiagen). RNA samples were mixed with 10 U of DNase I (Roche) and incubated for 20 min at 37°C to remove any contaminating gDNA. DNase I was removed with the RNeasy mini kit according to the clean-up procedure. All samples were analysed by end-point polymerase chain reaction (PCR) with primers DC1115f and DC1116r (Supplementary Table S1) to confirm absence of gDNA.
Plasmid constructions and DNA manipulations
To construct plasmid pET24lysMSa-6xhis for overexpression of C-terminal 6xHis-tagged S. acidocaldarius LysM (LysMSa), the open reading frame (ORF) region of Saci_0752 was PCR-amplified using gDNA as template and primers DC689f and DC690r. The amplicon was digested with NdeI and XhoI and ligated into kanamycin-resistant expression vector pET24a (Novagen) digested with the same enzymes. Vector pBendLYSM for the circular permutation assay was obtained by ligating the annealed oligonucleotides DC826f and DC827r bearing sticky XbaI sites into pBend2 (24) digested with the same enzyme. All constructs were verified by DNA sequencing. All oligonucleotides used in this work are listed in the Supplementary Data (Supplementary Table S1).
Production and purification of recombinant LysM from S. solfataricus and S. acidocaldarius
Untagged recombinant S. solfataricus LysM (LysMSs) was produced in E. coli BL21(DE3) cells transformed with plasmid pLUW632 (22). Induction was with 1 mM isopropyl-β-d-thiogalactopyranoside at a cell density of 9 × 108 ml−1, followed by overnight growth at 25°C. LysMSs was purified as described (25) with two modifications: after harvesting by centrifugation, cells were sonicated for 15 min at 20% of maximal amplitude in a VibraCell® sonicator equipped with a continuously cooled cell; purified protein was dialyzed overnight at 4°C against LysM storage buffer composed of 20 mM Tris (pH 8.0) and 20% glycerol.
We purified recombinant C-terminal 6xHis-tagged LysM from S. acidocaldarius (LysMSa) from E. coli BL21(DE3) cells containing plasmid pET24-lysMSa-6xhis. LysMSa overexpression was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside at a cell density of 9 × 108 ml−1, followed by overnight growth at 30°C. Cells were collected by centrifugation, resuspended in 6 ml of binding buffer (20 mM phosphate buffer, 0.5 M NaCl and 40 mM imidazole, pH 7.4) and followed by sonication and centrifugation (10 min at 7000 rpm in a Jouan centrifuge with AB50.10 A rotor). Soluble extract was heated at 80°C during 10 min and subsequently centrifuged to remove denatured proteins. Harvested supernatant was loaded on a HisTrapTM FF 1 ml column (GE Healthcare) operated by an AKTATM fast protein liquid chromatography system (GE Healthcare). The column was extensively equilibrated with binding buffer before application of a linear gradient with elution buffer (binding buffer with 500 mM instead of 40 mM imidazole). Peak fractions were analysed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and electrophoretic mobility shift assay (EMSA), and then collected and dialyzed against LysM storage buffer before storage at −80°C. In contrast to N-terminally His-tagged LysMSs (22), purified C-terminally His-tagged LysMSa did not precipitate and was correctly folded, as indicated by DNA-binding activity and cofactor response. All LysMSs and LysMSa protein concentrations are expressed in monomer equivalents.
In vitro DNA binding: EMSAs, in-gel OP–Cu footprinting and pre-modification binding interference analyses
EMSAs were performed as described (26), either with gel-purified single 5′-end 32P-labelled PCR fragments or with purified 47-bp duplexes generated by annealing of complementary oligonucleotides (Supplementary Table S1), of which one was 5′-end labelled with [γ-32P]-ATP by T4 polynucleotide kinase. Unless otherwise stated, separation of free DNA from DNA–protein complexes was performed on 6 and 8% polyacrylamide gels for the PCR fragments and 47-bp duplexes, respectively. LysM-binding reactions were performed in LrpB binding buffer as described previously (27) with 25 µg ml−1 sonicated herring sperm DNA as non-specific competitor.
EMSA autoradiographs were scanned with a Microtek Bio-5000 scanner, and binding equilibrium association constants (KAs) were determined with the Densitometric Image Analysis Software, for which a description will be published elsewhere. To enable comparison of KAs measured at different times, relative KAs were determined by normalization with the KA for binding to the consensus sequence fragment, measured each time in parallel. In-gel footprinting of separated LysM–DNA complexes with the 1,10-phenanthroline-copper [(OP)2-Cu+] ion (Cu–OP) was performed as described (28). Missing contact and pre-methylation binding interference experiments were performed as described previously (29), using LrpB binding buffer to perform binding reactions. Reference ladders were generated by chemical sequencing methods (30).
DNA bending test
Circular permutation assay (24,31) was performed with a set of six fragments of identical length (156 bp) bearing the 15-bp consensus LysM binding site at various distances from the extremities. Fragments were generated by PCR amplification with pBendLYSM plasmid DNA as template and oligonucleotide pairs DC826f-EP31r (fragment I), EP15-EP16r (fragment II), EP17-EP18r (fragment III), EP9-EP10r (fragment IV), EP19-EP20r (fragment V) and EP21-EP22r (fragment VI) as primers. The LysM binding site is located closest to an extremity on fragments I and VI, and approximately in the middle in fragment IV. The apparent bending angle was calculated from relative mobilities of complexes on 8% polyacrylamide, as described (31).
Real-time quantitative PCR
First-strand cDNA was synthesized from 30 ng RNA with SuperScript® III First-Strand Synthesis SuperMix kit (Invitrogen), according to the manufacturer’s instructions. Quantitative real-time PCR (qPCR) was carried out in a Bio-Rad iCycler with iQ™ SYBR® Green Supermix (Bio-Rad) using following amplification protocol: initial denaturation at 95°C for 3 min followed by 40 cycles of 95°C for 10 s and 55°C for 30 s, and one cycle of 95°C for 1 min and 55°C for 1 min. Reactions were performed with 12.5 µl SYBR Green supermix and 20-fold diluted cDNA in a total volume of 25 µl. Amplification reactions were performed in technical duplicates on biological quadruplicates, with a reaction without template as negative control. Specificity was verified by melt curve analysis. Sso0951, encoding TBP, was used as a reference gene for normalization. Of four genes that were tested (TBP, mini chromosome maintenance, 23 S rRNA and RNAP subunit B), TBP proved to have the most stable expression in tested conditions. Quantification cycles (Cqs) were determined with Bio-Rad iQ5 software. Efficiencies of gene-specific primer pairs were calculated by determining the slope of a linear regression curve resulting from Cq values for a 10-fold dilution series with gDNA as a template. Relative expression ratios were calculated by integrating knowledge of primer pair efficiencies (32).
Generation of LysM-specific nanobodies
LysM-specific nanobodies were generated by immunizing an alpaca (Vicugna pacos) with purified full-length 6xHis-tagged LysMSa. A total of six injections at weekly intervals were given, each with 200 µg protein. Plasma obtained 4 days after the last injection showed an end titre of ∼104. Subsequently, using peripheral blood lymphocytes, a variable domain of heavy chain antibodies library was constructed as described (33). The LysMSa-directed nanobodies were generated according to the previously described bio-panning procedure (33). A chromatin immunoprecipitation (ChIP)-grade nanobody that does not disrupt LysMSa- and LysMSs-containing protein–DNA complexes was selected by enzyme-linked immunosorbent assay and EMSA (data not shown).The specificity of the nanobody for LysM was further tested in pull-down assays (Supplementary Figure S1).
ChIP-chip
ChIP was performed according to (34). S. solfataricus P2 (DSM1617) cells were harvested at mid-exponential growth phase (OD600nm ∼0.6). Two biological replicates were used for each growth condition (with 5 mM lysine and without lysine). For enrichment analysis, qPCR was performed, as described previously, with 5 ng DNA as template. The 2-[Delta][Delta]Cq method was applied to calculate ChIP enrichment from the ChIP DNA as compared with input DNA (35). The lysW operator region was amplified with primers DC1102f and DC1103r and for normalization, a reference sequence in E. coli DNA that was spiked into all samples was amplified with the primers DC821 and DC822 (Supplementary Table S1). The DNA tiling microarray was designed and manufactured by NimbleGen (Roche) and sample labelling, hybridization and array processing were executed at NimbleGen, with ChIP input and output samples labelled with Cy3 and Cy5, respectively.
Micro-array data analysis was performed using an extended version of the programme described by (36), which uses the Ringo package of R-Bioconductor (37).The source code of the extended programme is made available through http://micr.vub.ac.be. It includes importing data, data quality assessment, pre-processing of data, identifying ChIP-enriched regions (chers) and determining significant differences between experiments with and without lysine. To avoid detection of peaks in the log2 ratios due to reduced signal-to-noise ratios at low-intensity probes, as part of the pre-processing step, average log2 intensities over red and green channels are calculated for each probe, and probes with resulting values smaller than the average minus twice the standard deviation are replaced by missing values. Next, a standard normalization step is performed in which Tukey’s biweight mean across each sample’s log2 ratios is subtracted from individual log2 ratios, and resulting values are averaged over replicates and smoothed over a 100-bp window to reduce stochastic noise and systematic noise due to differences in hybridization efficiency of different probes.
A region was considered as ChIP-enriched if smoothed log2 ratios of all reporters mapped to this region exceed a threshold of 2 and if the region contains at least five probe-matched positions, each of these positions being <500 bp apart from another matched position within this region. A constant threshold of 2 was chosen rather than the more statistically inspired version used by (36) to maintain an almost constant number of detected chers across biological replicates, rather than ensuring that the number of detected false-positives is smaller than a certain amount. The resulting chers were extended at both sides with 150 bp, and overlapping chers were then combined into one cher. This additional step was necessary to make sure that a cher is only detected once: sometimes a cher could be detected more than once due to, for example, a single reporter level that is under the threshold, whereas a series of neighbouring reporter levels are above the threshold.
Only chers that were present in both biological replicates for at least one of the experiments with and without lysine were taken into account for further analysis. To determine which chers detected in presence of lysine were significantly enhanced compared with the situation without lysine and vice versa, the same procedure to detect chers was repeated with a threshold of 1, and significantly differentially enhanced chers were defined as those that exceeded the threshold of 2 for one set of biological replicates and did not exceed the threshold of 1 for the other set.
In silico binding site prediction
A binding energy-weighted sequence logo for LysM DNA-binding sequence specificity has been calculated according to (38) by equating the frequency of each base to the relative KAfor binding to a consensus variant fragment having the corresponding substitution divided by the sum of the four KAvalues for each of four bases at this position.
For each cher sequence, the best potential binding site was predicted based on the energy-based position weight matrix, and the corresponding theoretical KA was calculated. This is shown in Table 1 and Supplementary Dataset S1.
Table 1.
Predicted binding affinities for potential/confirmed LysM-binding motifs in selected targets that were further studied by in vitro DNA-binding assays
| Predicted KD (nM) | Closest ORF | Predicted binding motif | Observed in vitro binding |
|---|---|---|---|
| 0.3 | Sso2043 | GTACGATTAGCGTAC | ++ |
| 1.7 | Sso0977 | GTTCGTATGTCGAAC | ++ |
| 6 | Sso1906 | GTACGATTACAGTAT | + |
| 9 | Sso0157 | GTTCTAAAATCGTAC | ++ |
| 11 | Sso0684 | GTTCGGAAATCAAAC | ++ |
| 42 | Sso2336 | ATACGCTAGGCTTAC | + |
| 54 | Sso0572 | CTTCGATACACGAAT | + |
| 78 | Sso0155 | ATACGGGCGGAGAAT | − |
| 103 | Sso2824 | GCACGCTATTAGAGT | − |
| 158 | Sso2497 | GTGCGATTTCAGCGT | + |
| 172 | Sso0340 | CGGCGGGATTCGAAC | − |
Binding motifs are ranked according to their predicted KDs, and it is indicated to what extent these targets are bound in vitro: (−) no binding; (+) low-affinity binding [either unstable binding (smearing) or not all free DNA is complexed at high LysM concentrations]; (++) high-affinity binding (all free DNA is complexed at intermediate LysM concentrations). Predicted motifs are given with indication of conservation of specificity-determining residues (bold). We refer to Figures 1 and 6 for the corresponding EMSA images.
RESULTS
The LysM–lysW operator interaction in S. solfataricus and S. acidocaldarius
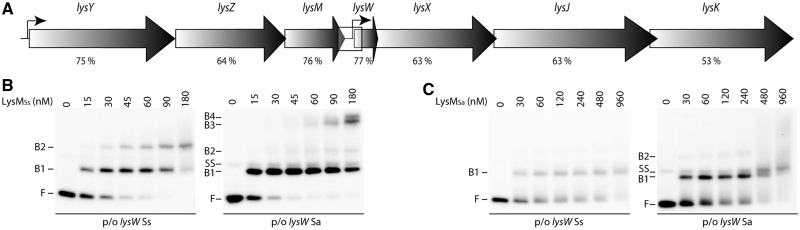
LysM is highly conserved among all sequenced Sulfolobus species, and their lysine biosynthesis genes are invariably organized in two consecutive operons with the same polarity, lysYZM and lysWXJK (Figure 1A). In S. solfataricus, only the latter bears a target site for LysM (22). To analyse functional conservation of LysM and its target site(s), we performed EMSAs of LysMSs and its orthologue LysMSa from S. acidocaldarius binding to the lysW operator of both organisms (Figure 1B and C). The results indicate that each regulator binds the two operators with a similar binding affinity. However, binding and migration patterns showed remarkable differences. LysMSs formed two distinct complexes (B1 and B2) with the cognate lysW operator in a concentration-dependent manner, and some supershifted smear at the highest concentration used. In contrast, binding of LysMSs to the heterologous lysW operator from S. acidocaldarius resulted in the formation of essentially one major complex (B1) and of a small amount of a second, slower migrating complex (B2). At the highest protein concentrations, two additional complexes (B3 and B4) were formed that exhibit an even lower relative mobility. Binding of LysMSa to both lysW operators appeared very similar and produced essentially one major complex (B1), a tiny amount of a slower migrating complex (B2) and, at the highest protein concentrations, supershifted smear.
Figure 1.
Functional conservation of the LysM–lysW operator interaction in Sulfolobus. (A) Schematic overview of genomic organization of the lys locus. ORFs are depicted by arrows, with names of corresponding genes mentioned above. Transcription start sites are indicated with small black arrows. The genomic organization shown here is identical for all sequenced Sulfolobus species, and amino acid sequence identity between S. solfataricus and S. acidocaldarius orthologues is mentioned below each lys gene. The lysW promoter/operator region that is subject of the interaction analysis is indicated by a rectangle. (B) EMSAs of binding of LysMSs to the lysW control regions of S. solfataricus (on a 203-bp fragment) and S. acidocaldarius (on a 188 bp-fragment), as indicated. Protein concentrations are mentioned on top of the autoradiograph. Positions of free DNA (F), free single-stranded DNA (SS) and protein–DNA complexes (B1–B4) are pointed out. (C) EMSAs of binding of LysMSa to the lysW control regions of S. solfataricus and S. acidocaldarius, as indicated. Notations are the same as in subpanel (B).
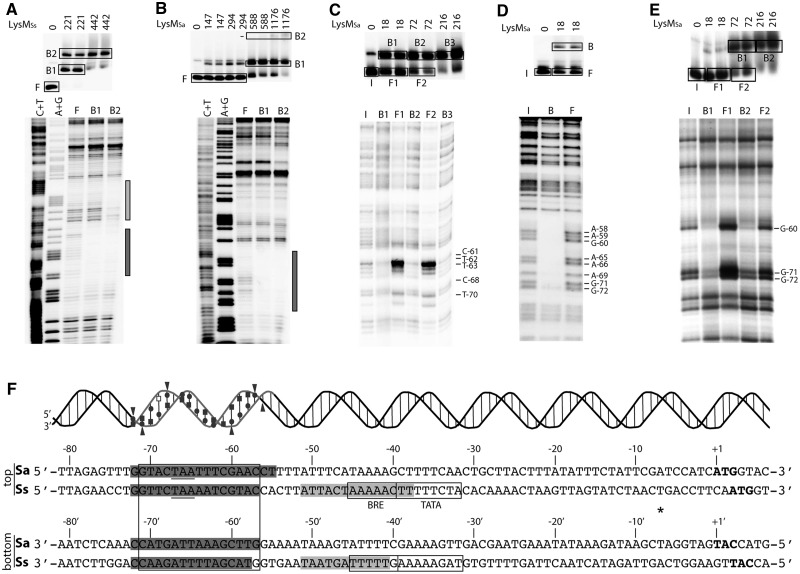
Previously, a single high-affinity binding site for LysMSs was identified by DNase I footprinting at low protein concentrations, whereas higher concentrations resulted in protection of a large, but undefined, zone and in the appearance of hyper-reactive bands (22). In-gel footprinting with the Cu–OP ion (Figure 2A and Supplementary Figure S2) demonstrated that in complex B1, LysMSs protected a stretch of 16 and 15 nucleotides (nt) on the top and bottom strands, respectively (Figure 2F). This zone extends from position −59 to −74 upstream of the initiation codon (−50 to −65 upstream of the transcription start site) and corresponds to the upstream binding site in Figure 2F. The slower migrating complex B2 showed two distinct zones of protection, one identical to that observed in complex B1, and an additional slightly shorter stretch of 14 nt on the top strand and of 11 nt on the bottom strand (Figure 2A and Supplementary Figure S2). This second zone of LysM binding (−31 to −44) is located slightly downstream of the major LysM binding site and covers the BRE and part of the TATA box of the lysW promoter (Figure 2F).The centres of the principal high-affinity site and of the more degenerated accessory site are 21 bp apart; consequently, equivalent positions in both sites are aligned on the same face of the helix.
Figure 2.
High-resolution contact probing of the LysM-–ysW operator interaction. (A) In-gel Cu–OP footprinting experiment of binding of LysMSs to the lysW operator region of S. solfataricus (203-bp fragment, top strand labelled). On top, the EMSA is shown with indication of used protein concentrations (in nM) and populations of free (F) and bound (B1–B2) DNA that were further analysed by denaturing acrylamide gel electrophoresis, of which the autoradiograph is shown below. C + T and A + G indicate Maxam–Gilbert sequencing ladders. The region that is protected in both nucleoprotein populations B1 and B2 is indicated with a black bar; the region that is only protected in complex B2 is indicated with a grey bar. (B) In-gel Cu–OP footprinting experiment of binding of LysMSa to the lysW operator region of S. acidocaldarius (188-bp fragment, top strand labelled). Notations are the same as in subpanel (A). (C) Depyrimidation binding interference experiment of binding of LysMSa to the lysW operator region of S. acidocaldarius. Populations of input (I), free (F1 and F2) and bound DNA (B1, B2 and B3) are indicated. Observed effects are pointed out with horizontal lines with the corresponding nucleotides mentioned. (D) Depurination binding interference experiment of binding of LysMSa to the lysW operator region of S. acidocaldarius. (E) Pre-methylation binding interference experiment of binding of LysMSa to the lysW operator region of S. acidocaldarius. (F) Alignment of the nucleotide sequence of the lysW operator region of S. acidocaldarius and S. solfataricus with a summary of all results of footprinting and binding interference experiments. The lysM translational stop codon is underlined, the lysW translational initiation codon is in bold and the semi-palindromic LysM binding motif is boxed, as are the BRE and TATA box promoter elements [predicted based on transcription start site determination in (22)]. An asterisk indicates the transcription initiation site. Positions are numbered with respect to the lysW initiation codon of S. acidocaldarius. Protection against chemical cleavage in Cu–OP footprinting is indicated with a grey shaded region. On top of the sequence, a helical representation demonstrates minor and major groove orientation of LysMSa binding. Strand regions that were protected against cleavage are grey coloured, while binding interference effects are depicted by following symbols: circles = depurination binding interference; squares = depyrimidation binding interference; triangles = pre-methylation binding interference. Open symbols represent weak effects; filled symbols strong effects. Subpanels (A–E) show representative experiments that were performed with DNA having the top strand labelled; experiments were similarly performed with DNA having the bottom strand labelled, as summarized here and shown in Supplementary Figure S2.
The major complex B1 formed on binding of LysMSa to the lysW operator of S. acidocaldarius showed a continuous stretch of protection of 18 nt on the top strand and 16 nt on the bottom strand (Figure 2B and Supplementary Figure S2). This zone extends from position −55 to −72 upstream of the initiation codon and aligns perfectly with the principal LysM binding site in the lysW operator of S. solfataricus (Figure 2F). Complex B2 did not show an additional clear zone of protection. Formation of this minor complex might therefore be due to binding of a higher oligomeric state of LysMSa (without further contact with the DNA), to establishment of additional non-specific interactions, or to formation of ‘sandwich-type’ structures.
High-resolution contact mapping of the LysM–lysW operator interaction in S. acidocaldarius
Missing contact probing assays (39) performed at different protein concentrations (Figure 2C and F and Supplementary Figure S2) indicated that removal of five pyrimidines of the top strand (T-70, C-68, T-63, T-62 and C-61) and seven of the bottom strand (C-72′, C-71′, T-66′, T-65′, C-60′, T-59′ and T-58′) strongly inhibits complex formation with LysMSa. Similarly, removal of eight purines of the top strand (G-72, G-71, A-69, A-66, A-65, G-60, A-59 and A-58) and six of the bottom strand (A-70′, G-68′, A-64′, A-63′, A-62′ and G-57′) strongly interferes with complex formation (Figure 2D and F and Supplementary Figure S2). Pre-methylation binding interference experiments demonstrated that methylation of three guanine residues of the top strand (G-72, G-71 and G-60) and three of the bottom strand (G-68′, G-57′ and G-56′) strongly reduces binding of LysMSa (Figure 2E and F and Supplementary Figure S2). Removal of any of these six guanine residues also strongly inhibited LysMSa binding, except G-56′. As this position was not protected in the Cu–OP footprinting assay, we may conclude that G-56′ is not directly contacted and that the negative effect of pre-methylation of this base most likely results from steric hindrance exerted by the methyl group on contact of LysMSa with an adjacent base-specific or backbone group. These results confirm recognition of a semi-palindromic sequence centred around position −64, which is highly conserved between S. solfataricus and S. acidocaldarius (22) (Figure 2F). The binding site overlaps part of the lysM ORF and its translational stop codon. Furthermore, it is located 11 bp upstream of the predicted lysW BRE and TATA box. Dimethyl sulphate methylates guanine residues at position N7 in the major groove. Therefore, we may conclude that LysM interacts specifically with two consecutive major groove segments and intervening minor groove of the operator, all aligned on one face of the DNA helix (Figure 2F). It is interesting to note that in this helical orientation, TBP and LysM are expected to bind to the same face of the DNA.
Sequence specificity of LysM binding
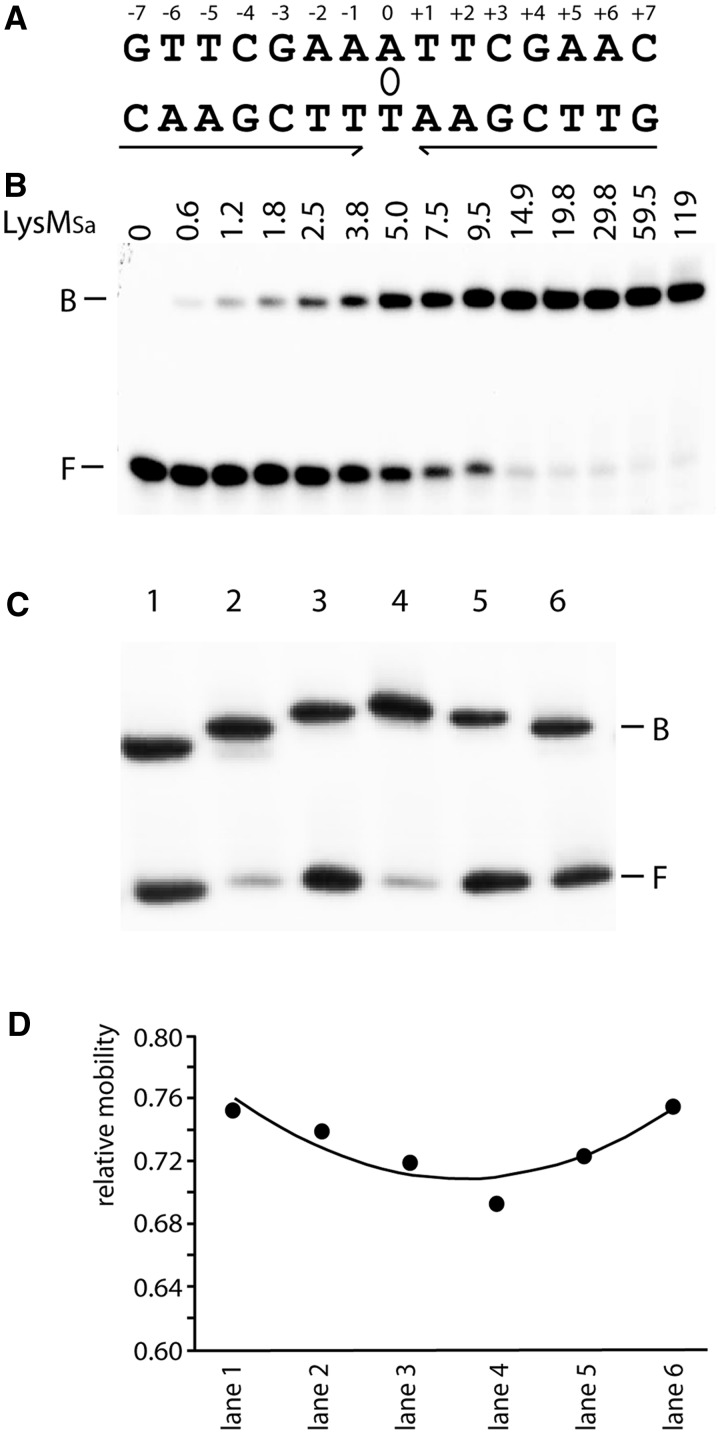
Based on phylogenetic footprinting of the lysW operator region, a degenerate consensus sequence was derived that allowed the definition of a strictly palindromic 15-bp consensus sequence for LysM binding (Figure 3A). LysMSa specifically binds a 47-bp DNA fragment harbouring this consensus sequence and forms a single complex in EMSA (Figure 3B). The average equilibrium association constant (KA) for this interaction was calculated to be 196 μM−1 (corresponding dissociation constant KD = 5 nM).
Figure 3.
Binding and LysMSa-induced DNA bending of the consensus binding site. (A) Cartoon representing the LysM consensus sequence. Inverted repeat elements are indicated with arrows; the axis of 2-fold symmetry is depicted by an ellipse. (B) EMSA of binding of LysMSa to a DNA fragment encompassing the consensus sequence. This fragment was generated by hybridization of 47-nt-long complementary oligonucleotides containing the 15-nt LysM box flanked on either side by the 16-nt stretches that surround the LysMSa binding site of the lysW operator in the S. acidocaldarius genome. Positions of bound (B) and free (F) DNA are indicated, as are applied LysMSa concentrations (in nM). (C) EMSA with permuted DNA fragments bearing the consensus binding site. In all binding reactions, we added 30 nM LysMSa. Characteristics of fragments are further described in Materials and Methods, and lane numbers in the EMSA correspond to fragment numbers. Positions of bound (B) and free (F) DNA are indicated. (D) Graphical representation of the relative mobility (μ) of different complexes as a function of the position of the insert sequence within the DNA fragment. The apparent bending angle (α) was calculated as follows: μM/μE = cos (α/2) (M = insert in the centre of the fragment, resulting in the lowest value of μ, and E = insert at the end of the fragment).
Six fragments of identical length but with the LysM consensus site permuted were used to analyse DNA bending (Figure 3C and D). All six fragments migrated with a similar mobility when being unbound, indicating the absence of a measurable intrinsic curvature (Figure 3C). Therefore, the role of structure specificity in LysM binding is limited. In contrast, LysMSa–DNA complexes displayed clear differences in relative mobilities, which allowed calculation of an average apparent bending angle of 36° (Figure 3C and D). This result contrasts the lack of measurable DNA bending reported for LysMSs using a similar assay (22).
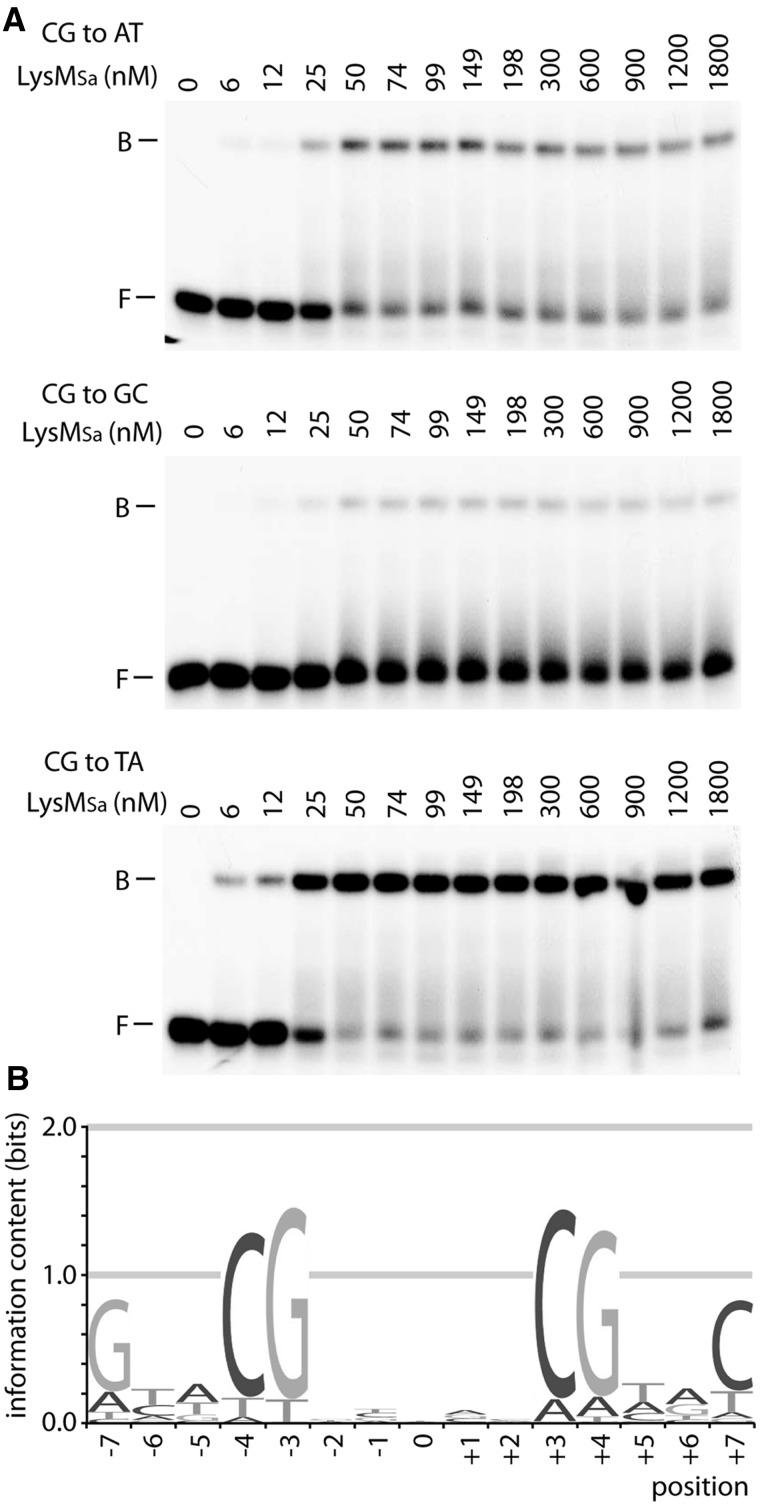
To determine the sequence specificity of LysM binding, EMSAs with LysMSa binding to a set of 22 mutated variants (all possible single-bp substitutions at all positions of one half-site) were performed (Figure 4A), and relative KAs were calculated (Supplementary Table S2). Due to full symmetry of the consensus site, only one substitution was analysed at position 0. This analysis resulted in a quantitative model of binding specificity, representing a significant part of the complete binding energy landscape. This model is graphically represented by an energy-normalized sequence logo (Figure 4B). The total information content of the LysM binding specificity is 8.46, and therefore, LysM binds DNA with a relative high sequence specificity. This specificity is restricted to the half sites, particularly to positions −3 and −4, which are highly discriminative for a G-C and a C-G bp, respectively. The remainder of the sequence specificity in major groove recognition is largely contributed by position −7, displaying a preference for a G-C bp. As information content of individual positions is higher in half sites than in the central region, our analysis confirms recognition of two major groove segments and the intervening minor groove (Figure 4B). These results of the high-resolution contact mapping of the lysW operator are fully compatible with these data (Figure 2F). Unexpectedly, there is no pronounced preference for weak bps at central positions. Generally, the contribution of the minor groove-recognized part of a site to sequence preference is low and mostly arising from a preference for A-T or T-A bps, which facilitate minor groove compression. Most likely, this situation reflects the relative modest LysM-induced DNA bending. This is further confirmed by the observation that LysMSa binds mutant operators bearing an I-C at positions −1 and +1 with only a 1.3-fold higher affinity than mutant operators with G-C substitutions at both positions (Supplementary Figure S3). Inosine is identical to guanine in the major groove but lacks the exocyclic amino group that causes a steric hindrance for minor groove compression.
Figure 4.
Systematic bp substitution of the LysM consensus sequence to analyse sequence specificity of binding. In this figure, a representative example of such a binding analysis is shown for position −4, but analogous EMSAs have been performed and analysed for the other positions (Supplementary Table S2). (A) EMSAs of LysM binding to variants harbouring a single-bp substitution at position −4. The bp change is mentioned on top of each autoradiograph, as are LysMSa concentrations (in nM). DNA fragments were prepared by hybridizing complementary oligonucleotides. (B) Graphical representation of the systematic bp substitution experiment in an energy-normalized sequence logo. The height of the stack of letters corresponds to the information content (bits).
Genome-wide in vivo mapping of LysMSs binding sites in S. solfataricus
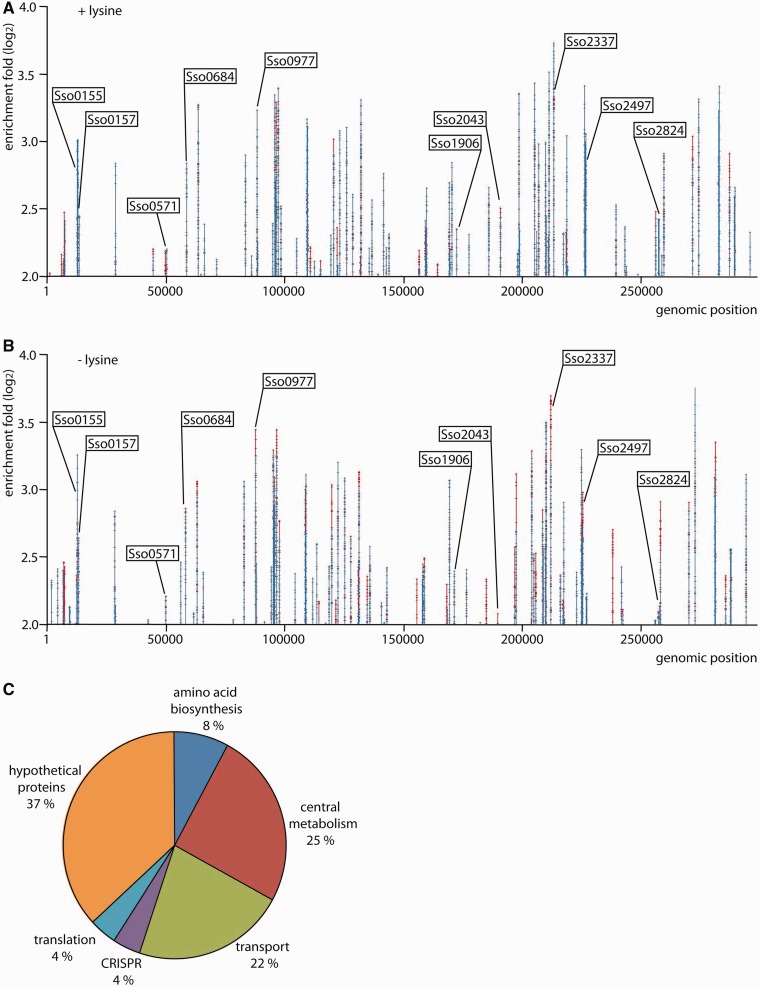
LysM binding sites were identified on a genome-wide scale in vivo, using ChIP-chip assays with S. solfataricus cells grown in either presence or absence of exogenous lysine (5 mM) (Figure 5). Utilization of a LysM-specific nanobody for immunoprecipitation avoided the need of overexpressing or epitope-tagging LysMSs.In total, 73 genomic regions distributed throughout the genome exhibited an enrichment of more than 4-fold (normalized log2 value of 2.0) in either both or one of the growth conditions. For several peaks, this enrichment even exceeded 8-fold. Resulting comprehensive dataset, obtained by setting the threshold at a log2 value of 2.0, is given in Supplementary Dataset S1.
Figure 5.
Genome-wide distribution of LysMSs binding sites that are bound in vivo. (A) LysMSs binding profile across the S. solfataricus chromosome in cells in exponential growth phase grown in medium with supplementation of 5 mM lysine. The enrichment fold-ratio corresponds to the log2 of the signal ratio of ChIP-enriched DNA versus input DNA. Signals arising from biological duplicates are depicted in different colours (red/blue). Selected targets that are called in this growth condition (signals exceeding a log2 value of 2.0 in both biological duplicates), and for which binding is further analysed in this work, are labelled. (B) LysMSs binding profile across the S. solfataricus chromosome in cells grown in medium without additional supplementation of lysine. Notations are the same as in subpanel (A). (C) Pie chart showing the percentage of LysM binding sites associated with genes divided in functional categories.
These ChIP-chip assays provide first direct evidence of LysMSs being bound in vivo to the lysW operator (Supplementary Figure S4). Peak maxima were centred around the characterized binding motif. By applying qPCR to ChIP samples, we quantified enrichment of the lysW operator/promoter region as being ∼586- and 83-fold in absence and presence of lysine, respectively (Supplementary Figure S5). Significant binding was also detected at the lysYZM promoter region (Supplementary Figure S4), which was previously shown not to be bound by LysM in vitro. This observation necessitates revision of the statement that LysMSs is not involved in autoregulation and in regulation of lysY and lysZ (22). However, it remains unclear whether LysMSs is associated at the lysY promoter because of direct recognition of a binding motif, or rather indirectly through protein–protein interactions (see later in the text).
Besides lysW and lysY, 71 previously unknown LysMSs targets were identified. Sequences of these ChIP-enriched regions were scanned with previously developed binding energy-based position weight matrix to predict the most probable binding motif (Supplementary Dataset S1). A comparison with the annotated genome information of S. solfataricus (40) indicates that 76% of all predicted binding motifs are located in ORFs. Given a genome coding density of 84%, this percentage indicates only a slight over-representation of binding motifs being present in intergenic regions, which is unexpected for a specific transcription factor.
Positions of predicted motifs located in intergenic regions with respect to the closest translational start varied significantly, but many are located between 40 and 70 bp upstream of an initiation codon (Supplementary Dataset S1). There are also several cases in which, as for the lysW operator, the predicted motif is located at the 3′-end of an ORF and close to the promoter of a downstream gene or operon (see later, Figure 6A and F). Transcription units, of which expression is potentially influenced by LysMSs, encode proteins with various functions, which can be classified in following categories: amino acid metabolism, central metabolism, transport, clustered regularly interspaced short palindromic repeats (CRISPR) immunity system, translation and hypothetical proteins For a distribution over the different functional categories, see Figure 5 and Supplementary Dataset S1. Curiously, LysMSs is associated with several tRNA genes and CRISPR loci. Furthermore, of particular interest are potential target genes that function in biosynthesis and transport of amino acids other than lysine, some of which are studied in more detail later in the text.
Figure 6.
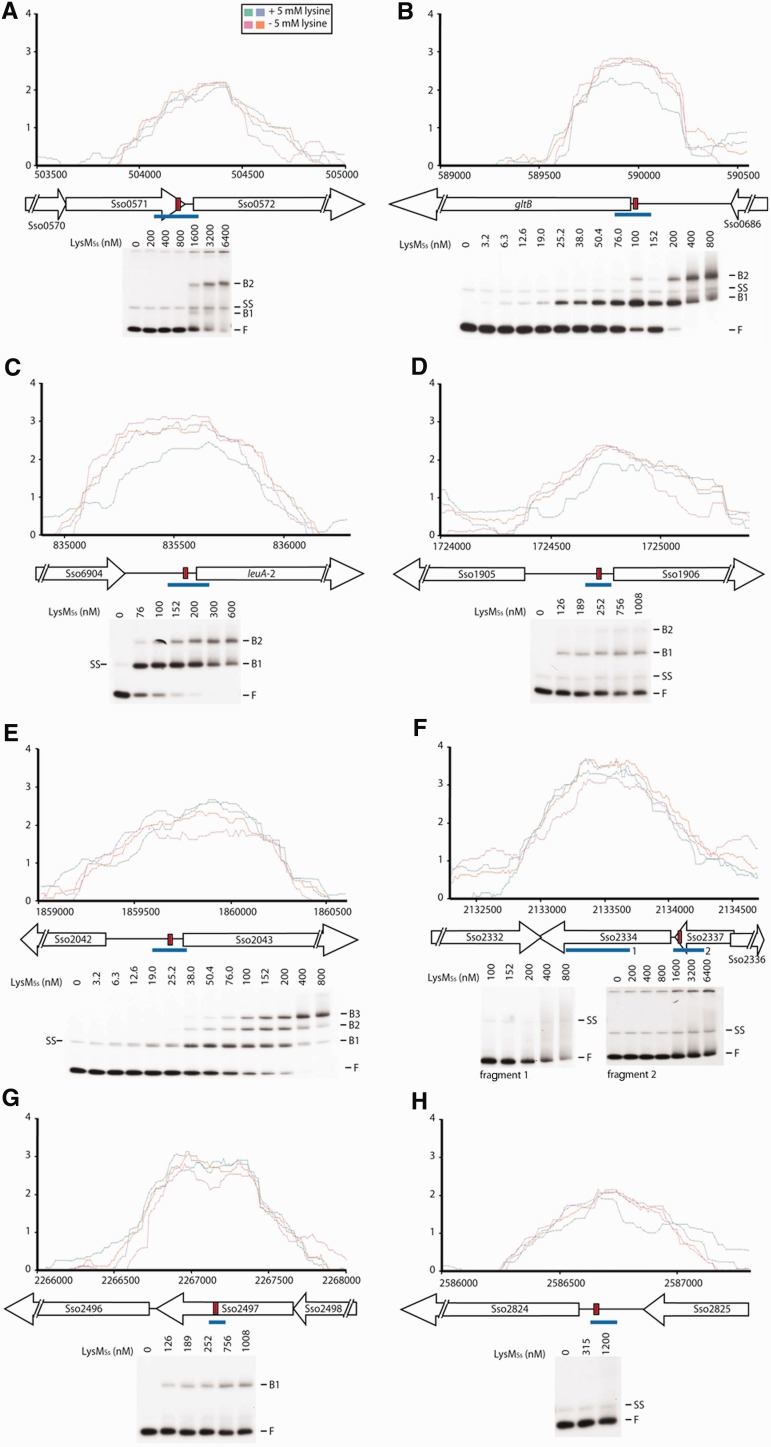
In vivo and in vitro LysMSs binding for a selection of called ChIP-enriched regions: Sso0571 (A), gltB (B), leuA-2 (C), Sso1906 (D), Sso2043 (E), Sso2336 (F), Sso2497 (G) and Sso2824 (H). Zoomed profiles are plotted as the log2 of enrichment fold (y-axis) versus genomic position (x-axis). Signals arising from biological replicates are depicted in different colours, of which the code is provided in the figure inset in subpanel (A). Aligned with genomic positions, a schematic overview is given of intergenic and ORF regions. ORFs are depicted by open arrows and labelled. Intergenic regions are represented by a horizontal line. In each ChIP-enriched region, a red rectangle indicates the 15-bp sequence that is predicted to show the highest similarity to an LysM-binding motif. Below each binding profile, an EMSA is shown probing a fragment spanning the part of the ChIP-enriched region containing the predicted motif, which is indicated by a blue line [in subpanel (F), two fragments are tested called 1 and 2]. LysMSs concentrations are mentioned on top of the autoradiograph. Positions of free DNA (F), free single-stranded DNA (SS) and protein–DNA complexes (B1–B3) are indicated.
In vitro DNA-binding affinity of LysMSs for the lysW operator decreases on addition of lysine (22). However, the in vivo LysMSs DNA-binding profiles appeared similar in either presence or absence of l-lysine (Figure 5).Based on the main criteria used for peak calling, namely, that peaks exceed a log2 value of 2.0 in both biological duplicates, not all peaks were called in both growth conditions. Nevertheless, for all peaks called in one condition, signals exceeded a log2 threshold of 1.0 in the other condition, indicating that there is no significant difference in binding profile in cells grown with and without exogenous lysine (Supplementary Dataset S1). However, it should be taken into account that ChIP-chip profiles do not provide a quantitative measure of DNA binding, as demonstrated for lysW: whereas ChIP profiles in both conditions appear similar (Supplementary Figure S4), qPCR quantification, which is more accurate because of a higher dynamic range, demonstrates a difference in enrichment of >4-fold (Supplementary Figure S5). Consequently, despite the observation that ChIP binding profiles are similar on growth in presence and absence of lysine, DNA-binding characteristics such as affinity or stoichiometry might nevertheless be different.
Validation of ChIP-chip data with in vitro DNA-binding assays
In vitro DNA binding was analysed with EMSA for a selected subset of eight potential targets with varying predicted binding affinities and for which predicted binding motifs are located either in intergenic control regions or in ORFs (Figure 6 and Table 1). In this selection, we included targeted genes that are involved in amino acid metabolism: Sso0684 encodes a glutamate synthase (gltB), Sso0977 a 2-isopropylmalate synthase (leuA-2) and Sso1906 and Sso2043 both encode amino acid transporter-related proteins. We also included the control region of Sso0572 that codes for a conserved hypothetical ATPase of the PiLT family, for which predicted binding motif is located inside the preceding ORF, and of Sso2824, which encodes a formate dehydrogenase alpha subunit (fdhF-2). Two targets for which LysMSs binds into a coding region were also tested: these genes code for a hypothetical protein (Sso2336) and acyl-CoA dehydrogenase (Sso2497).
With exception of Sso2336, predicted binding motifs are located close to peak maxima (Figure 6). In EMSAs using 100- to 200-bp fragments comprising a sequence centred around the predicted motif, LysMSs binds with high affinity to control region fragments of gltB, leuA-2 and Sso2043 (Figure 6B, C and E). Binding of LysMSs to gltB and leuA-2 resulted in concentration-dependent formation of two complexes, whereas three complexes were formed with Sso2043. Furthermore, low-affinity binding yielded two complexes for Sso1906 and Sso0572 and a single complex for the Sso2497 ORF fragment (Figure 6A, D and G). The other two targets (Sso2336 and Sso2824) formed either unstable complexes resulting in a smearing in EMSA or showed no complex formation (Figure 6F and H). In the former case, we also tested binding to a fragment-bearing part of the ORF of Sso2334 that corresponds better to the peak maximum of the cher, but again only weak binding resulting in some smearing at the highest protein concentrations was observed (Figure 6F).
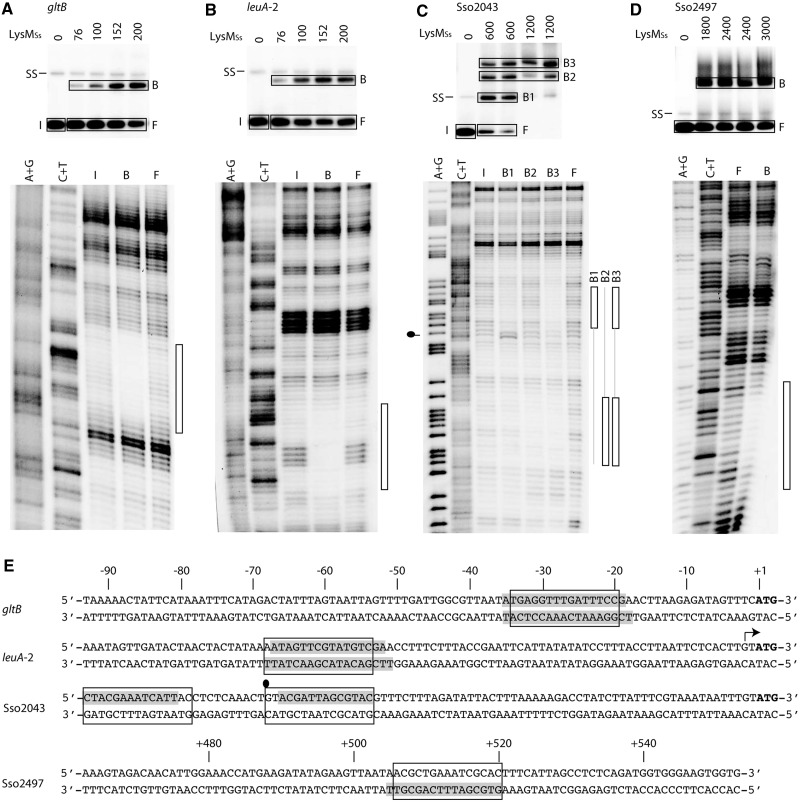
We performed OP–Cu footprinting experiments for four fragments that exhibited high-affinity binding (Figure 7). For binding to gltB and leuA-2 operator fragments and to the Sso2497 ORF fragment, only a single protein–DNA complex was analysed (Figure 7A, B and D). In case of gltB and leuA-2, this complex corresponds to the fastest migrating complex B1 (Figure 6B and C). Without exception, the zone of protection is confirmed to contain the predicted LysM binding motif (Figure 7E).
Figure 7.
In vitro DNA-binding analysis for newly discovered LysMSs targets. (A) In-gel Cu–OP footprinting experiment of LysMSs binding to the control region of gltB. The autoradiograph of the EMSA is shown on top of the figure subpanel, with indication of LysMSs concentrations (in nM), position of single-stranded DNA (SS) and of excised DNA populations (I = input DNA, F = free DNA and B = bound DNA; indicated with rectangles). The autoradiograph of the denaturing gel is shown below the EMSA: A +G and C + T stand for the Maxam–Gilbert sequencing ladders, and I, B and F represent DNA populations. The protected region is indicated with a bar at the right side of the footprint autoradiograph. (B) In-gel Cu–OP footprinting experiment of LysMSs binding to the control region of leuA-2. Notations are the same as in subpanel (A). (C) In-gel Cu–OP footprinting experiment of LysMSs binding to the control region of Sso2043. Notations are the same as in subpanel (A), but in this case, there are three different bound DNA populations: B1, B2 and B3. Hyper-reactivity is indicated by a ball-and-stick symbol on the left side of the autoradiograph. (D) In-gel Cu–OP footprinting experiment of LysMSs binding to the ORF of Sso2497. Notations are the same as in subpanel (A). (E) Sequences of probed regions, with indication of protected regions (grey shade), 15-bp binding motifs (boxed) and position of hyper-reactivity effect (ball-and-stick symbol). For the three control regions, the sequences are aligned according to the translational start (with indication of the positions on top). The transcription start site is indicated with an arrow. For the Sso2497 ORF sequence, positions are indicated on top of the sequence.
Further analysis of three protein–DNA complexes formed with the Sso2043 control region indicated that in complex B1, which exhibits the highest relative mobility and is formed at lower protein concentrations than the two other complexes, the predicted binding motif is bound (Figure 7C and E). This finding confirms the high-affinity nature of this site (predicted KD = 0.3 nM), which is rationalized by full conservation of all specificity-determining residues (Table 1). In complex B2, which migrates with a lower relative mobility, footprinting indicates that LysMSs protects a different, but similarly sized, region. This region is located upstream of the high-affinity site and contains a binding motif with only one good half-site (Figure 7E). As centres of these two sites are 25 bp apart, they are contacted by the protein on opposite faces of the DNA helix. In complex B3, both sites are protected.
A ranking of all in vitro tested binding sites according to their in silico predicted KD values results in a clustering of sites that are efficiently bound both in vitro and in vivo on the one hand (KD < 54 nM) and sites that are only bound in vivo on the other hand (KD > 78 nM) (Table 1). The two categories of binding sites can be found in intergenic regions as well as in ORFs, and ∼24% of all binding regions contain a binding motif with a predicted KD <50 nM, indicating the presence of a true high-affinity LysM binding site (Supplementary Dataset S1). Binding at low-affinity sites might be stabilized in vivo on interaction with other sequence-specific DNA-binding or with nucleoid-associated proteins.
In vivo repression of LysM target genes by l-lysine supplementation
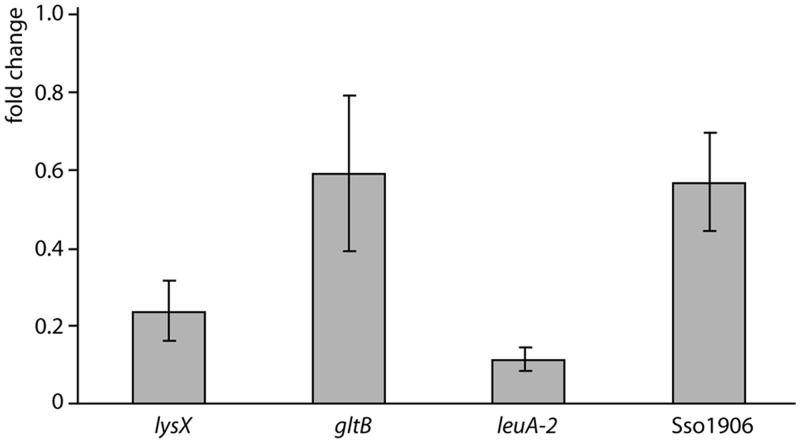
We measured the effect of l-lysine supplementation (5 mM) to growth medium on expression of lysX (second gene of lysWXJK operon), leuA-2, gltB and Sso1906 in wild-type cells with qRT-PCR (Figure 8). Addition of l-lysine results in a significant downregulation of all four LysM target genes tested, with the effect being most pronounced for leuA-2 (10-fold reduction). Therefore, the expression of genes involved in biosynthesis, and possibly also transport, of other amino acids than lysine is significantly modulated in response to changes in intracellular lysine concentration.
Figure 8.
Relative gene expression analysis for LysMSs target genes lysX, gltB, leuA-2 and Sso1906 with qRT-PCR. Relative expression level is given as fold change of the expression level during growth in Brock medium with sucrose and 5 mM lysine versus the expression level during growth in Brock medium with sucrose lacking amino acids. Standard deviations (calculated for two biological replicates) are indicated.
Although differential expression of these genes in presence or absence of lysine is no conclusive proof for LysMSs regulation, it is likely that at least part of this regulatory response is direct and mediated by LysMSs. This hypothesis is supported by the observation that binding characteristics are influenced by lysine concentration. as demonstrated by decreasing ChIP enrichment levels and binding affinities for the lysW operator at increasing lysine concentrations in vivo and in vitro, respectively (Supplementary Figure S5) (22). In conclusion, it appears that LysMSs stimulates expression of all target genes at low intracellular lysine concentration.
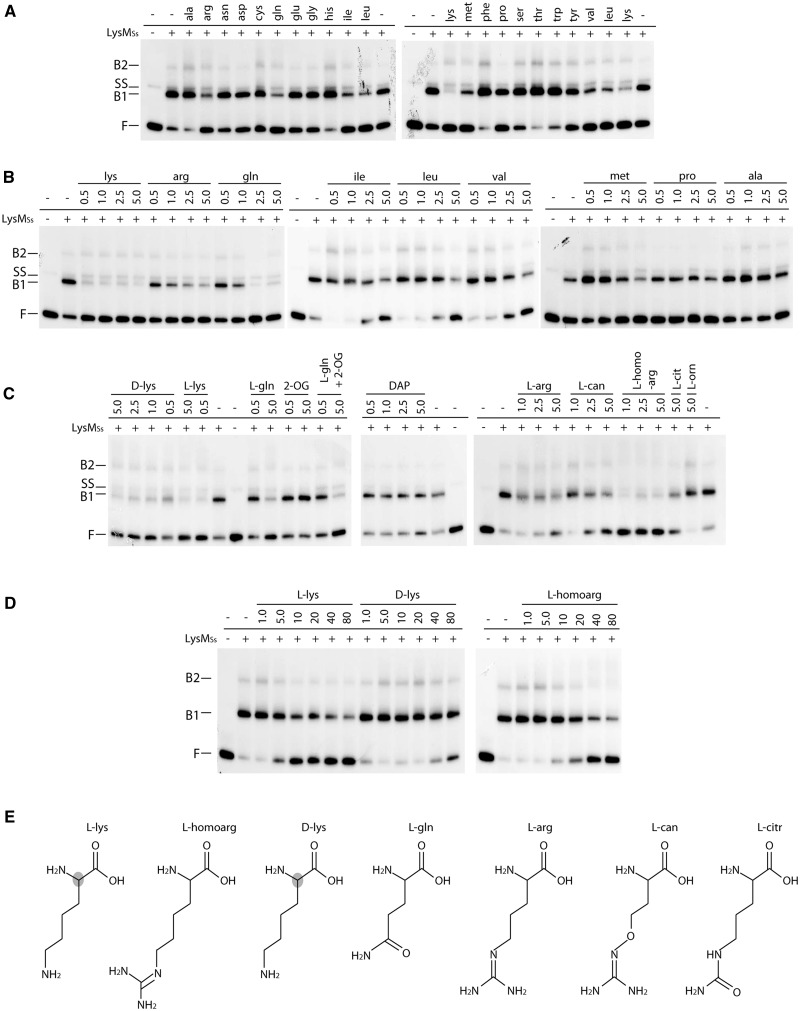
Cofactor specificity of LysM
A systematic EMSA analysis of LysMSs binding to the gltB operator with all 20 naturally occurring l-amino acids as potential effector molecules confirmed that lysine reduces, but does not completely abolish, binding of LysMSs, as observed previously for the lysW operator (22) (Figure 9A and B). Furthermore, arginine, glutamine, isoleucine, leucine, methionine and valine also specifically reduce LysM binding at 5 mM. Similar results were obtained with lysW, leuA-2 and Sso2043 operator regions (data not shown). Similar as for lysine, addition of these six amino acids reduced binding affinity but did not completely abolish complex formation (Figure 9B). l-lysine has the strongest inhibitory effect of all tested potential effector molecules and therefore likely the highest affinity for LysMSs (Figure 9). It was found to exert a rather similar effect from ∼10 µM up to 5 mM, the highest concentration tested (Figure 9B and D).
Figure 9.
Effect of different cofactors on LysMSs DNA binding to the gltB control region on a 167-bp fragment monitored by EMSAs. (A) EMSAs to test binding in presence of each of the 20 l-amino acids. Positions of free DNA (F), single-stranded DNA (SS) and LysMSs–DNA complexes (B1 and B2) are pointed out. In the bottom row above the autoradiograph, presence or absence of LysMSs in the reaction mixture is indicated as + or −, respectively. Used protein concentration is 250 nM. In the top row above the autoradiograph, the three-letter code of the added amino acid is displayed (final concentration 5 mM). (B) EMSAs in which a concentration gradient of a selection of amino acids was tested. Notations are similar as in subpanel A; amino acid concentrations are given in mM. (C) EMSAs to test the effect of the following molecules: d-lys = d-lysine; l-lys = l-lysine; l-gln = l-glutamine; 2-OG = 2-oxoglutarate; DAP = D,l-α,ε-diaminopimelic acid; l-arg = l-arginine; l-can = l-canavanine; l-homoarg = l-homoarginine; l-cit = l-citrulline; l-orn = l-ornithine. Notations are similar as in subpanel (A and B). All cofactor concentrations are displayed in mM. For binding reactions to which l-glutamine and two-oxoglutarate are added simultaneously (indicated by ‘l-gln + 2-OG’), shown concentrations correspond to l-glutamine, whereas 2-oxoglutarate had a final concentration of 5.0 mM in both binding reactions. (D) EMSAs to test low concentration gradients for a selection of cofactors. Notations are similar as in subpanel (A, B and C). However, in this subpanel, cofactor concentrations are displayed in μM. (E) Molecular structures of LysM effector molecules (except for branched-chain amino acids) tentatively ranked according to the strength of their inhibitory effect. The shaded atom in the l-lysine and d-lysine structure indicates a stereochemical difference at this position.
We tested a series of precursors of lysine, arginine and glutamine biosynthesis or analogues of lysine and arginine that show structural similarities with lysine (Figure 9C and E). The EMSA experiments indicate that d-lysine and l-homoarginine also exert a negative effect on LysMSs binding, even in µM range (Figure 9D), and that homoarginine is a more potent cofactor than arginine. The effect of l-canavanine and l-citrulline was less pronounced, whereas l-ornithine and D,l-α,ε-diaminopimelic acid had no significant effect (Figure 9C). Similarly, 2-oxoglutarate had no effect by itself and neither did it interfere with the negative effect of glutamine (Figure 9C).
Combined, these results indicate that both the amino group of l- and d-lysine and the guanidino group of arginine and homoarginine can be accommodated in the cofactor binding-pocket. The optimal length of the cofactor side chain is four CH2-groups.
DISCUSSION
Lrp-like regulators are abundantly present in archaeal genomes (3) and appear to play an important role in adaptation of cellular metabolism to variations in concentration of amino acids as signaling molecules (2,41,42). Some bacterial or archaeal Lrps control only a single or a few target genes, whereas others regulate a vast number of genes involved in various pathways (1,42,43). Here we identify 73 binding sites for LysM and demonstrate that LysM is a much more versatile regulator than originally thought. Binding sites for LysM may occur singly or in combination with an auxiliary more degenerated LysM box, as demonstrated for lysW and Sso2043. LysM is not only involved in transcriptional control of lysine biosynthesis, but it also modulates expression of genes involved in biosynthesis of leucine and glutamate and of genes annotated as amino acid transport-related. Clearly, the control of amino acid metabolism and transport is a primary task of LysM, although the transcription factor also binds in the neighbourhood of promoters expressing genes with a variety of other functions.
Lysine is the main effector molecule of LysM. In vitro DNA binding was invariably reduced in the presence of lysine and in vivo, a significant downregulation was observed on lysine supplementation for all four tested targets (lysX, gltB, leuA-2 and Sso1906). We can assume that this regulatory response originates at least partially from LysM action, and it is strongly suggested that LysM functions as an activator. Although FL11 from Pyrococcus sp. OT3 and LysM have similar cofactor specificity with lysine as the major coregulator, their mode of action is clearly different. Whereas FL11 generally works as a repressor of which the activity is relieved in presence of lysine (9), LysM appears to act as a transcriptional activator in absence of lysine. For lysW, leuA-2, Sso1906 and Sso2043, the main LysM binding site is located just upstream of predicted BRE and TATA box elements (Figure 7E). This is reminiscent of activation by Ptr2, an Lrp-like transcription factor from M. jannaschii that binds at an equivalent relative position and activates transcription by stimulating protein–protein interactions with TBP (20). In the cases of Sso2043 and lysW, binding of LysM at a secondary accessory binding site located either upstream or downstream of the core binding site (Figures 2F and 7E) might contribute to more profound regulatory effects: either further activation or a switch to repression. In contrast, the distance between the main LysM binding site and the translational initiation site in the gltB promoter region suggests that the regulator is positioned in between the promoter region and initiation site.
Often, Lrp-like regulators display a cofactor promiscuity (9,17,44). Here we demonstrate that, besides lysine, several other amino acids (i.e. arginine, glutamine, leucine, isoleucine, methionine and valine) reduce DNA-binding affinity of LysM (Figure 9). The preference for the most potent effector molecules, that is lysine > arginine > glutamine, is identical as for FL11, and the structural basis for this similarity lies in conservation of a glutamine at position 97 and an aspartate at position 121. Side-chain oxygen molecules of these residues form hydrogen bonds with the lysine side chain, thereby determining ligand specificity (45). An arginine-FL11 cocrystal structure demonstrates that arginine is less efficient because of the longer side chain causing conformational changes in the protein (45), and we can assume that a similar situation occurs in LysM. The stronger effect of homoarginine observed for LysM can be explained by its larger structural similarity to lysine (4 CH2-groups preceding an N-atom that may engage in H-bonding) (Figure 9E). Competitive binding of different amino acids, each with a different affinity, will allow an adapted regulatory response to the nutritional state of the cell. This state is reflected by concentration ratios of different amino acids rather than by the absolute concentration of a single amino acid type. Furthermore, a broad cofactor specificity is also rationalized by the observation that LysM regulates not only lysine biosynthesis but also biosynthesis and transport of other amino acids.
All LysMSs targets were bound in vivo in growth conditions with and without lysine supplementation, whereas enrichments quantified by qPCR, in vitro binding affinity of LysM and promoter activities were invariably lower in the presence of lysine. This indicates that ChIP-chip is not a valid quantitative measure of target site occupancy and likely reflects the observation that lysine reduces, but does not completely abolish, binding of LysM, an observation that was made with all six tested intergenic and intragenic binding sites (lysW, gltB, leuA-2, Sso1906, Sso2043 and Sso2497, Figure 9 and data not shown). It is also reminiscent of the effect of l-leucine on DNA-binding capacity of E. coli Lrp (46,47), although in this case, ChIP-chip DNA-binding profiles were clearly sensitive to the presence of exogenous leucine (16). The mechanisms underlying the negative effect of ligand binding on DNA-binding affinity of LysM are not yet understood. In some instances, such as for E. coli Lrp and the archaeal FL11, cofactor binding influences the association state of the Lrp-like protein, thereby shifting the equilibrium between different oligomeric forms that each have different DNA-binding characteristics (45,48). However, this is not a general rule and ligand binding might also induce more subtle conformational changes (6,8). Preliminary data indicate that this is the case for LysM, as lysine does not affect the tetrameric state of the regulator in solution (22). Possibly, effector binding reduces the DNA-binding capacity of LysM by re-orienting two helix-turn-helix motifs with respect to successive major groove segments of the operator. Besides diminishing DNA-binding affinity, this re-orientation could equally modify the position of the C-terminal surface responsible for transcription regulation with respect to elements of the basal transcription apparatus, thereby affecting regulatory outcome.
LysM does not appear to control expression of other transcription factors. Nevertheless, it cannot be excluded that LysM is involved in regulatory networking. This situation might occur at targets that are efficiently bound in vivo but not in vitro, as demonstrated for Sso2824 and Sso2334-2336. Such differential binding is generally ascribed to involvement of co-regulators, other transcription factors or nucleoid-associated proteins, which may enhance binding affinity and/or stability at sites with a low intrinsic affinity for the regulator. In a proteome-wide study of protein–protein interactions in Pyrococcus horikoshii OT3, hetero-interactions have been identified for two Lrp-like proteins (49). Additionally, FL9 and DM1 from Pyrococcus OT3, a full-length and truncated Lrp protein, respectively, have been shown to form hetero-octamers (44). Possibly, LysM is capable of establishing such hetero-oligomeric interactions, thereby largely expanding its regulatory power, both with respect to effector response as to target gene repertoire. Whether LysM exerts autoregulation is not entirely clear. In vivo, LysM associates with the lysYZM promoter region, but we did not detect binding in vitro, and northern blotting indicated that production of lysY and lysM mRNA was not affected on addition of lysine (22).
A large fraction (76%) of LysM-binding sites is located inside translated regions. Eleven of these are predicted to harbour a high-affinity bona fide LysM-binding motif (theoretical KD < 50 nM). Such a high frequency of intragenic binding sites is unusual for bacterial transcriptional regulators (16,50–52), with E. coli RutR as an exception (53). For archaeal transcription factor TrmB of Halobacterium salinarum NRC-1, 40% of binding sites are located inside coding sequences (54). Possibly, intragenic binding is more common for archaeal than for bacterial transcription factors, given their more compact genome organizations and smaller average intergenic region lengths (55). Indeed, some LysMSs targets, for example the lysW and Sso0572 operators, appear to be true promoter-associated regulatory sites while located in the 3′-end of the preceding ORF sequence. Other intragenic sites might also have regulatory functions for as yet undetected transcription units, given the flexible transcriptome architecture of S. solfataricus with a high abundance of conditionally active initiation and termination sites inside operons and of small RNAs (56,57). Alternatively, these binding sites do not function in direct transcription regulation but serve to control the intracellular concentration of free regulatory protein, or they merely occur by chance without any functionality and have not been removed by evolution, as was postulated for RutR (53).
LysMSs is associated with both control regions of genes encoding amino acid transporters (Sso1906/Sso2043) in paralogous highly conserved gene clusters that also contain a glutamate dehydrogenase located downstream of the transporter gene and a divergently transcribed allantoin permease. The Sso1906 gene cluster is flanked by a transposase, suggesting that gene duplication was mediated by a transposition event (58). The intergenic regions are highly conserved without a selective pressure for conservation of the LysM-binding motifs (data not shown). In vitro, LysMSs binds with a significantly higher affinity to the promoter region of the ancestral Sso2043, which contains a binding motif corresponding perfectly to the consensus sequence, than to the Sso1906 promoter, in which two mismatches have originated at specificity-determining positions (Figure 6 and Table 1). Moreover, the accessory binding site identified in the Sso2043 control region is lost in the Sso1906 control region. Therefore, although our ChIP-chip data indicate that this LysMSs regulatory interaction has been inherited after the gene duplication event, differences in binding affinity might lead to differential regulation. As a consequence, the duplicated genes might respond slightly different to metabolic needs, thereby placing a selection pressure on maintenance of the duplicated gene cluster and on further degeneration and possibly even on the future loss of the LysMSs-binding motif in the Sso1906 promoter region. Our analysis has provided a ‘snapshot’ of the evolutionary expansion of the LysMSs regulon.
This work illustrates the power of genome-wide ChIP approaches to obtain a global view of the full-range target site distribution of a transcription factor, but also stresses the necessity to couple such techniques to in-depth studies for a correct interpretation and full understanding of the physiological role of the regulator in the cell.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–5 and Supplementary Datasets 1.
FUNDING
Research Foundation Flanders (FWO-Vlaanderen) [G.0135.08 to D.C., a postdoctoral fellowship to E.P., a pre-doctoral fellowship to L.v.O.]; Research Council of the Vrije Universiteit Brussel (OZR-VUB); Vlaanse Gemeenschapscommissie and the China Scholarship Council-Vrije Universiteit Brussel (CSC-VUB) (scholarship to N.S.). Funding for open access charge: Research Foundation Flanders (FWO-Vlaanderen) [G.0135.08 to D.C.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Phu Nguyen Le Minh for linguistic advice and critical reading of the manuscript and thank Nadine Huysveld for technical assistance. They acknowledge John van der Oost (Wageningen University, The Netherlands) for the gift of plasmid pLUW632.
REFERENCES
- 1.Brinkman AB, Ettema TJG, de Vos WM, van der Oost J. The Lrp family of transcriptional regulators. Mol. Microbiol. 2003;48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- 2.Peeters E, Charlier D. The Lrp family of transcription regulators in archaea. Archaea. 2010;2010:750457. doi: 10.1155/2010/750457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pérez-Rueda E, Janga SC. Identification and genomic analysis of transcription factors in archaeal genomes exemplifies their functional architecture and evolutionary origin. Mol. Biol. Evol. 2010;27:1449–1459. doi: 10.1093/molbev/msq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de los Rios S, Perona JJ. Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J. Mol. Biol. 2007;366:1589–1602. doi: 10.1016/j.jmb.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ettema TJG, Brinkman AB, Tani TH, Rafferty JB, van der Oost J. A novel ligand-binding domain involved in regulation of amino acid metabolism in prokaryotes. J. Biol. Chem. 2002;277:37464–37468. doi: 10.1074/jbc.M206063200. [DOI] [PubMed] [Google Scholar]
- 6.Kumarevel T, Nakano N, Ponnuraj K, Gopinath SCB, Sakamoto K, Shinkai A, Kumar PKR, Yokoyama S. Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding. Nucleic Acids Res. 2008;36:4808–4820. doi: 10.1093/nar/gkn456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard PM, Smits SH, Sedelnikova SE, Brinkman AB, de Vos WM, van der Oost J, Rice DW, Rafferty JB. Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus. EMBO J. 2001;20:990–997. doi: 10.1093/emboj/20.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, Brinkman AB, Akerboom J, van der Oost J, Rafferty JB. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 2006;34:1439–1449. doi: 10.1093/nar/gkl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoyama K, Ishijima SA, Koike H, Kurihara C, Shimowasa A, Kabasawa M, Kawashima T, Suzuki M. Feast/famine regulation by transcription factor FL11 for the survival of the hyperthermophilic archaeon Pyrococcus OT3. Structure. 2007;15:1542–1554. doi: 10.1016/j.str.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Ishihama A. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol. Rev. 2010;34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- 11.Langer D, Hain J, Thuriaux P, Zillig W. Transcription in archaea: similarity to that in eucarya. Proc. Natl Acad. Sci. USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell SD, Jackson SP. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 2001;4:208–213. doi: 10.1016/s1369-5274(00)00190-9. [DOI] [PubMed] [Google Scholar]
- 13.Geiduschek EP, Ouhammouch M. Archaeal transcription and its regulators. Mol. Microbiol. 2005;56:1397–1407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- 14.Hung S-P, Baldi P, Hatfield GW. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 2002;277:40309–40323. doi: 10.1074/jbc.M204044200. [DOI] [PubMed] [Google Scholar]
- 15.Tani TH, Khodursky A, Blumenthal RM, Brown PO, Matthews RG. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl Acad. Sci. USA. 2002;99:13471–13476. doi: 10.1073/pnas.212510999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho B-K, Barrett CL, Knight EM, Park YS, Palsson BØ. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl Acad. Sci. USA. 2008;105:19462–19467. doi: 10.1073/pnas.0807227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hart BR, Blumenthal RM. Unexpected coregulator range for the global regulator Lrp of Escherichia coli and Proteus mirabilis. J. Bacteriol. 2011;193:1054–1064. doi: 10.1128/JB.01183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calvo JM, Matthews RG. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman EB, Lin R. Leucine-responsive regulatory protein: a global regulator of gene expression in E. coli. Annu. Rev. Microbiol. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- 20.Ouhammouch M, Dewhurst RE, Hausner W, Thomm M, Geiduschek EP. Activation of archaeal transcription by recruitment of the TATA-binding protein. Proc. Natl Acad. Sci. USA. 2003;100:5097–5102. doi: 10.1073/pnas.0837150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peeters E, Albers S-V, Vassart A, Driessen AJM, Charlier D. Ss-LrpB, a transcriptional regulator from Sulfolobus solfataricus, regulates a gene cluster with a pyruvate ferredoxin oxidoreductase-encoding operon and permease genes. Mol. Microbiol. 2009;71:972–988. doi: 10.1111/j.1365-2958.2008.06578.x. [DOI] [PubMed] [Google Scholar]
- 22.Brinkman AB, Bell SD, Lebbink RJ, de Vos WM, van der Oost J. The Sulfolobus solfataricus Lrp-like protein LysM regulates lysine biosynthesis in response to lysine availability. J. Biol. Chem. 2002;277:29537–29549. doi: 10.1074/jbc.M203528200. [DOI] [PubMed] [Google Scholar]
- 23.Brock TD, Brock KM, Belly RT, Weiss RL. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Zwieb C, Wu C, Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989;85:15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- 25.Peeters E, Willaert R, Maes D, Charlier D. Ss-LrpB from Sulfolobus solfataricus condenses about 100 base pairs of its own operator DNA into globular nucleoprotein complexes. J. Biol. Chem. 2006;281:11721–11728. doi: 10.1074/jbc.M600383200. [DOI] [PubMed] [Google Scholar]
- 26.Enoru-Eta J, Gigot D, Thia-Toong TL, Glansdorff N, Charlier D. Purification and characterization of Sa-lrp, a DNA-binding protein from the extreme thermoacidophilic archaeon Sulfolobus acidocaldarius homologous to the bacterial global transcriptional regulator Lrp. J. Bacteriol. 2000;182:3661–3672. doi: 10.1128/jb.182.13.3661-3672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peeters E, Wartel C, Maes D, Charlier D. Analysis of the DNA-binding sequence specificity of the archaeal transcriptional regulator Ss-LrpB from Sulfolobus solfataricus by systematic mutagenesis and high resolution contact probing. Nucleic Acids Res. 2007;35:623–633. doi: 10.1093/nar/gkl1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peeters E, Thia-Toong T-L, Gigot D, Maes D, Charlier D. Ss-LrpB, a novel Lrp-like regulator of Sulfolobus solfataricus P2, binds cooperatively to three conserved targets in its own control region. Mol. Microbiol. 2004;54:321–336. doi: 10.1111/j.1365-2958.2004.04274.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Glansdorff N, Charlier D. The arginine repressor of Escherichia coli K-12 makes direct contacts to minor and major groove determinants of the operators. J. Mol. Biol. 1998;277:805–824. doi: 10.1006/jmbi.1998.1632. [DOI] [PubMed] [Google Scholar]
- 30.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 31.Thompson JF, Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Els Conrath K, Lauwereys M, Wyns L, Muyldermans S. Camel single-domain antibodies as modular building units in bispecific and bivalent antibody constructs. J. Biol. Chem. 2001;276:7346–7350. doi: 10.1074/jbc.M007734200. [DOI] [PubMed] [Google Scholar]
- 34.Duc TN, Hassanzadeh-Ghassabeh G, Saerens D, Peeters E, Charlier D, Muyldermans S. Nanobody-based chromatin immunoprecipitation. Methods Mol. Biol. 2012;911:491–505. doi: 10.1007/978-1-61779-968-6_31. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W. Ringo–an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunelle A, Schleif RF. Missing contact probing of DNA-protein interactions. Proc. Natl Acad. Sci. USA. 1987;84:6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawashima T, Aramaki H, Oyamada T, Makino K, Yamada M, Okamura H, Yokoyama K, Ishijima SA, Suzuki M. Transcription regulation by feast/famine regulatory proteins, FFRPs, in archaea and eubacteria. Biol. Pharm. Bull. 2008;31:173–186. doi: 10.1248/bpb.31.173. [DOI] [PubMed] [Google Scholar]
- 42.Schwaiger R, Schwarz C, Furtwangler K, Tarasov V, Wende A, Oesterhelt D. Transcriptional control by two leucine-responsive regulatory proteins in Halobacterium salinarum R1. BMC Mol. Biol. 2010;11:40. doi: 10.1186/1471-2199-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokoyama K, Ishijima SA, Clowney L, Koike H, Aramaki H, Tanaka C, Makino K, Suzuki M. Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS Microbiol. Rev. 2006;30:89–108. doi: 10.1111/j.1574-6976.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- 44.Okamura H, Yokoyama K, Koike H, Yamada M, Shimowasa A, Kabasawa M, Kawashima T, Suzuki M. A structural code for discriminating between transcription signals revealed by the feast/famine regulatory protein DM1 in complex with ligands. Structure. 2007;15: 1325–1338. doi: 10.1016/j.str.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 45.Yamada M, Ishijima SA, Suzuki M. Interactions between the archaeal transcription repressor FL11 and its coregulators lysine and arginine. Proteins. 2009;74:520–525. doi: 10.1002/prot.22269. [DOI] [PubMed] [Google Scholar]
- 46.Chen S, Iannolo M, Calvo JM. Cooperative binding of the leucine-responsive regulatory protein (Lrp) to DNA. J. Mol. Biol. 2005;345:251–264. doi: 10.1016/j.jmb.2004.10.047. [DOI] [PubMed] [Google Scholar]
- 47.Peeters E, Minh PNL, Foulquiã Moreno M, Charlier D. Competitive activation of the Escherichia coli argO gene coding for an arginine exporter by the transcriptional regulators Lrp and ArgP. Mol. Microbiol. 2009;74:1513–1526. doi: 10.1111/j.1365-2958.2009.06950.x. [DOI] [PubMed] [Google Scholar]
- 48.Chen S, Calvo JM. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 2002;318:1031–1042. doi: 10.1016/S0022-2836(02)00187-0. [DOI] [PubMed] [Google Scholar]
- 49.Usui K, Katayama S, Kanamori-Katayama M, Ogawa C, Kai C, Okada M, Kawai J, Arakawa T, Carninci P, Itoh M, et al. Protein-protein interactions of the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Genome Biol. 2005;6:R98. doi: 10.1186/gb-2005-6-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei Q, Le Minh PN, Dötsch A, Hildebrand F, Panmanee W, Elfarash A, Schulz S, Plaisance S, Charlier D, Hassett D, et al. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 2012;40:4320–4333. doi: 10.1093/nar/gks017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho B-K, Federowicz S, Park YS, Zengler K, Palsson BØ. Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat. Chem. Biol. 2011;8:65–71. doi: 10.1038/nchembio.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dillon SC, Espinosa E, Hokamp K, Ussery DW, Casadesús J, Dorman CJ. LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2012;85:1072–1089. doi: 10.1111/j.1365-2958.2012.08162.x. [DOI] [PubMed] [Google Scholar]
- 53.Shimada T, Ishihama A, Busby SJW, Grainger DC. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008;36:3950–3955. doi: 10.1093/nar/gkn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmid AK, Reiss DJ, Pan M, Koide T, Baliga NS. A single transcription factor regulates evolutionarily diverse but functionally linked metabolic pathways in response to nutrient availability. Mol. Syst. Biol. 2009;5:282. doi: 10.1038/msb.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoon SH, Reiss DJ, Bare JC, Tenenbaum D, Pan M, Slagel J, Moritz RL, Lim S, Hackett M, Menon AL, et al. Parallel evolution of transcriptome architecture during genome reorganization. Genome Res. 2011;21:1892–1904. doi: 10.1101/gr.122218.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu N, Li Y, Zhao Y-T, Guo L, Fang Y-Y, Zhao J-H, Wang X-J, Huang L, Guo H-S. Identification and characterization of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. PLoS One. 2012;7:e35306. doi: 10.1371/journal.pone.0035306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Redder P, Garrett RA. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J. Bacteriol. 2006;188:4198–4206. doi: 10.1128/JB.00061-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.