Abstract
Helicobacter pylori is a Gram-negative bacterium that colonizes human stomach and causes gastric inflammation. The species is naturally competent and displays remarkable diversity. The presence of a large number of restriction–modification (R–M) systems in this bacterium creates a barrier against natural transformation by foreign DNA. Yet, mechanisms that protect incoming double-stranded DNA (dsDNA) from restriction enzymes are not well understood. A DNA-binding protein, DNA Processing Protein A (DprA) has been shown to facilitate natural transformation of several Gram-positive and Gram-negative bacteria by protecting incoming single-stranded DNA (ssDNA) and promoting RecA loading on it. However, in this study, we report that H. pylori DprA (HpDprA) binds not only ssDNA but also dsDNA thereby conferring protection to both from various exonucleases and Type II restriction enzymes. Here, we observed a stimulatory role of HpDprA in DNA methylation through physical interaction with methyltransferases. Thus, HpDprA displayed dual functional interaction with H. pylori R–M systems by not only inhibiting the restriction enzymes but also stimulating methyltransferases. These results indicate that HpDprA could be one of the factors that modulate the R–M barrier during inter-strain natural transformation in H. pylori.
INTRODUCTION
Helicobacter pylori is a Gram-negative bacterium that colonizes the human gut and infects more than half of the world’s human population (1). It is a bacterial pathogen responsible for gastrointestinal diseases such as atrophic gastritis, gastric adenocarcinoma, peptic ulcers and mucosa-associated lymphomas (2). Helicobacter pylori is the most abundant phylotype present in the bacterial microbiota of the human stomach (3). Helicobacter pylori has a remarkably high level of genetic variation that reflects its ability to adapt gastric habitats (4,5). This high genetic diversity is believed to contribute towards the success of H. pylori in colonizing the human gastric mucosa where many different microenvironment changing conditions are likely to be encountered (6,7).
The transformation system of H. pylori is fundamentally different from other competent Gram-negative bacteria. The structural core of H. pylori translocation system is related to the bacterial Type IV secretion systems, rather than like a pili (8,9). Natural transformation in H. pylori involves a two-step DNA uptake mechanism (10). The first step involves uptake of double-stranded DNA (dsDNA) from the outer environment to the periplasm. The second step involves conversion of dsDNA to single-stranded DNA (ssDNA) and then transport from periplasm to cytoplasm through the inner membrane. These two steps are temporally and spatially segregated in H. pylori (10).
Lateral transfer of genetic information between bacteria of different species, and even between different strains of the same species, is often limited by one or more restriction modification (R–M) systems (11,12). Although inter-strain transformation is limited by R–M systems, similar methylation patterns enable intra-strain transformation. Incorporation of DNA fragments of small size (on average 1300 bp) through recombination in H. pylori again indicates the role of R–M barrier during horizontal gene transfer (13). A Type III-like restriction endonuclease has been shown to be a major barrier to horizontal genetic transfer in clinical Staphylococcus aureus strains (11). Similarly, a Type I R–M system in S. aureus has been described as a barrier for all the three major mechanisms of lateral genetic transfer, i.e. conjugation, transformation (via electroporation) and transduction (14). In H. pylori, Type II R–M systems act as the main barrier against natural transformation (15,16). A 30-fold higher transformation frequency was observed for DNA from other strains when four Type II restriction enzymes were deleted in H. pylori strain 26695 (17). The inter-strain transformation frequency is reduced but not completely blocked by R–M systems indicating its regulation during horizontal gene transfer (16).
A number of studies have shown the role of a DNA-binding protein ‘DNA processing protein A (DprA)’ in high-frequency uptake and translocation of exogenous DNA (18,19). DprA is a conserved bacterial protein which was first identified in Haemophilus influenzae (20). Knockout of dprA in H. pylori results in reduced transformation efficiency for both chromosomal and plasmid DNA (19). However, dprA knockout in H. influenzae resulted in a reduction of transformation efficiency with chromosomal DNA, but not with plasmid DNA (20). This indicates a different mechanistic role for DprA in the natural transformation pathways of different organisms. DprA expression was shown to be dependent on ComK protein as it could not be detected in comK knockouts in Bacillus subtilis (21). DprA is localized at cell poles as a part of the eclipse complex, suggesting that it gains access to the incoming DNA before other cellular factors (21). DprA from Gram-positive bacteria has been reported to bind and protect ssDNA but not dsDNA (22). These observations collectively suggest that DprA is crucial in the protection of incoming foreign DNA.
In this study, we have analysed the biochemical and molecular properties of HpDprA to understand its functional role in bacterial natural transformation. We demonstrate that HpDprA binds and protects both ssDNA and dsDNA. This observation led us to investigate further role of HpDprA in protecting dsDNA from restriction enzymes. We noticed that dsDNA was not only protected from restriction enzymes but could also be methylated with greater efficiency in the presence of HpDprA. Our findings shed light on a novel role of H. pylori DprA in alleviating the restriction barrier in the host bacterium.
MATERIALS AND METHODS
Bacterial strains and plasmids
Helicobacter pylori J99 strain (cagA+ iceA1 vacAs1am1) genomic DNA was obtained as a gift from New England Biolabs (Beverley, MA, USA). Escherichia coli strain DH5α [F′-end A1 hsd R17 (rk− mk−) glnV44 thi1 recA1 gyrA (NalR) relA1 Δ (lacIZYA – argF) U169 deoR {Φ80dlac Δ (lacZ)M15}] was used as a host for preparation of plasmid DNA. Escherichia coli strain ER2566 [F-fhuA2 ompT lacZ::T7gene1 gal1 sulA11 (mcrC-mrr)114::IS10 (R9mcr-73::mini-Tn10-Tets)2 R(zgb-210::Tn10) (Tets) endA−] (obtained as a kind gift from New England Biolabs) was used for expression and purification of HpDprA.
Reagents
Restriction endonucleases and T4 polynucleotide kinase were obtained from New England Biolabs. T4 DNA ligase and 1 kb DNA ladder were obtained from Fermentas Life Sciences. Phusion DNA polymerase was obtained from Finnzymes. Coomassie Brilliant blue R-250, proteinase K, Tris(hydroxymethyl) aminomethane (Tris), heparin Sepharose, protease inhibitor cocktail and isopropyl β-D-1-thiogalactopyranoside (IPTG) were procured from Sigma Aldrich Ltd (USA). Ni+-NTA agarose and glutathione Sepharose were obtained from GE Healthcare (Sweden). [γ-32P]ATP (3500 Ci/mmol) was obtained from BRIT (India). All other reagents used were of analytical or ultrapure grade.
Oligonucleotides and radiolabeling
All oligonucleotides used in this study were synthesized by Sigma Genosys. The concentrations of oligonucleotides were determined by UV absorbance at 260 nm. Extinction coefficient of oligonucleotides was calculated using the sum of the extinction coefficients of the individual bases. The oligonucleotides (Table 1) were labelled at the 5′-end with [γ-32P] ATP using T4 polynucleotide kinase and purified by Qiagene nucleotide removal kit. For experiments with ssDNA, oligo 1 (50 mer) and oligo 3 (110 mer) were labelled (Table 1) as described above. Duplex dsDNA was formed by first labelling oligo 1, oligo 3 and oligo 5 individually and subsequently annealing the labelled oligos with excess of oligo 2, oligo 4 and oligo 6, respectively (Table 1). Annealing reactions were carried out in 1× saline sodium citrate buffer (23). Sequences of oligonucleotides of increasing length (32–110 mer) are shown in Table 1.
Table 1.
Sequences of the oligonucleotides used
| Oligo 1 50 mer | CGAACCGTAATCGTACCTAGCAGTGACCGGTACCGTTCGGTAATATTCCG |
| Oligo 2 50 mer | CGGAATATTACCGAACGGTACCGGTCACTGCTAGGTACGATTTGGGTTCG |
| Oligo 3 110 mer | TCGCGCGTTTCGGTGATGACGGTGAAAACCTCTGACACATGCAGCTCCCGGAGA |
| CGGTCACAGCTTGTCTGTAAGCGGATGCCGGGAGCAGACAAGCCCGTCAGGGCGCG | |
| Oligo 4 110 mer | CGCGCCCTGACGGGCTTGTCTGCTCCCGGCATCCGCTTACAGACAAGCTGTGACCGT |
| CTCCGGGAGCTGCATGTGTCAGAGGTTTTCACCGTCATCACCGAAACGCGCGA | |
| Oligo 5 60 mer | GCAAAACATTGGATCCGCGAATATTCAAATTTTCAAAGCAAAACATTCTTCAAAA |
| CAAGG | |
| Oligo 6 60 mer | CGTTTTGTAACCTAGGCGCTTATAAGTTTAAAAGTTTCGTTTTGTAAGAAGTTTT |
| GTTCC | |
| Oligo 7 32 mer | TCGATAGTCGGATCCTCTAGACAGCTCCTTTT |
| Oligo 8 40 mer | TCGATAGTCGGATCCTCTAGACAGCTCCTTTTTTTTTTTT |
| Oligo 9 48 mer | TCGATAGTCGGATCCTCTAGACAGCTCCTTTTTTTTTTTTTTTTTTTT |
| Oligo 10 63 mer | ATCGATAGTCGGATCCTCTAGACAGCTCCATGTAGCAAGGCACTGGT |
| AGAATTCGGCAGCGTC | |
| Oligo 11 73 mer | ACGCGATCGCCCAGGTGGCAGGCCCTAGGGTGGAGGGGAGGCCGCC |
| GGCATGGGGACGCGATGGGCGGAGGCG | |
| Oligo 12 90 mer | Poly-dT (dT90) |
Polymerase chain reaction amplification and cloning of H. pylori dprA
The 801-bp hpdprA gene was amplified by polymerase chain reaction (PCR) from H. pylori J99 genomic DNA template using primers (forward primer, 5′-GTCGGATCCATGAAAAGCAATTTCCAATAC-3′ and reverse primer, 5′-CTTCTCGAGTCATGCTAACACCACGAGATG-3′) carrying the sites for BamHI and XhoI. The primers were designed with the help of gene sequence obtained from the annotated complete genome sequence of H. pylori J99 deposited at The Institute for Genomic Research. The amplified PCR fragment was gel purified and digested with restriction enzymes. The DNA was extracted with phenol–chloroform, precipitated by ethanol and ligated into BamHI–XhoI sites of pET28a vector with a hexa-histidine (His)6 tag at the N-terminus of the expressed protein. The DNA construct containing the dprA gene was confirmed by restriction digestion and sequencing.
Overexpression and purification of HpDprA
HpDprA protein was overexpressed in E. coli strain ER2566 harbouring the DNA construct pET28a-hpdprA. The recombinant bacteria were grown in LB with kanamycin selection (50 µg/ml) at 37°C to A600 of 0.6. HpDprA was induced by the addition of 0.5 mM IPTG, and the cultures were incubated for 4 h at 37°C. Cells were collected by centrifugation, resuspended in Buffer A [50 mM Tris–HCl (pH 7.4), 300 mM NaCl, 2 mM β-mercaptoethanol, 10% (v/v) glycerol and 10 mM imidazole] and lysed by sonication. 1× Protease inhibitor cocktail and 0.05% TritonX-100 were added to the cell suspension before sonication. The cell lysate was centrifuged at 16 000 rpm for 1 h at 4°C. The supernatant containing HpDprA protein was loaded onto a Ni-NTA column that had been previously equilibrated with Buffer A. The column was washed with 30 column volumes of Buffer A containing 30 mM imidazole. The protein was eluted with 10 ml of Buffer A containing 300 mM imidazole. The eluate of Ni-NTA column was dialyzed against 50 mM Tris buffer pH 7.4 containing 2 mM β mercaptoethanol, 10% glycerol and 75 mM NaCl. The dialyzed eluate was loaded on a heparin Sepharose column. The column was washed with the same buffer and protein was eluted using a salt gradient of 0.1–1 M. The purified protein was dialyzed at 4°C against Buffer A containing 200 mM NaCl. The purity of the protein preparation was judged on sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) with Coomassie Brilliant blue staining (24) and silver staining (25). Protein concentration was estimated by Bradford assay using bovine serum albumin (BSA) as a standard (26). Polyclonal antiserum against HpDprA was generated and used for western blot analysis. HpDprA antibody was purified from rabbit serum using a Protein A Sepharose affinity column. M.HpyAVIA was as described (27).
Mass spectroscopy and peptide mass fingerprinting
Peptide mass fingerprint analysis of trypsin-treated HpDprA was performed as described (28). Briefly, MALDI-MS data were acquired on an Ultraflex TOF/TOF spectrometer (Bruker Daltonics, Billerica, MA, USA and Bremen, Germany), equipped with a 50-Hz pulsed nitrogen laser (l¼337 nm), operated in positive ion reflectron mode using a 90-ns time delay and a 25-kV accelerating voltage. The samples were prepared by mixing an equal amount of peptide (0.5 ml) with matrices dihydroxybenzoic acid/α-cyano-4-hydroxycinnamic acid saturated in 0.1% trifluoroacetic acid and acetonitrile (1:1, v/v). Masses <500 m/z were not considered as a result of interference from the matrix.
Electrophoretic mobility shift assays
The DNA-binding activity of HpDprA was measured in a 20-µl reaction mixture containing 0.5 nM DNA substrate (32P-ssDNA or 32P-dsDNA) in 1× TAM reaction buffer [50 mM Tris pH 7.4, 50 mM NaOAc, 10 mM MgOAc and 1 mM dithiothreitol (DTT)] with indicated concentrations of HpDprA. For the competition experiments, excess of unlabelled competitor was also included in the reaction mixture. After 30 min incubation at 4°C, free DNA was resolved from the DNA–protein complex by electrophoresis through 8% non-denaturing PAGE in 1× TAME buffer [6 mM Tris–HCl (pH 7.8), 10 mM NaOAc, 4 mM MgOAc and 1 mM ethylenediaminetetraacetic acid (EDTA)]. For ionic strength analysis of the HpDprA interaction with DNA, DNA substrates were incubated with 1 µM protein in reaction buffer 50 mM Tris pH 7.4, 10 mM MgCl2, 1 mM DTT and increasing concentrations of NaCl (10–1000 mM). The samples were electrophoresed on an 8% polyacrylamide gel in 45 mM Tris/borate (pH 8.3) containing 0.4 mM EDTA (0.5 × TBE). A constant voltage of 7 V/cm was applied for 6 h at 4°C. The gel was transferred on to Whatmann 3 MM paper and dried under vacuum at 75°C for 30 min. The gels were visualized by phosphorimaging and quantified using Image Gauge (Version 3.0). For DNA-binding assays with closed circular pUC19 DNA, 20 µl reaction mixtures containing the indicated concentrations of purified HpDprA and 1 × TAM reaction buffer were incubated with DNA for 30 min at 4°C. Free DNA was resolved from the DNA–protein complex by electrophoresis through 1.5% agarose gel in 0.5 × TBE.
Nuclease cleavage assays
Nuclease cleavage assays were performed in a 20 µl reaction mixture containing 0.5 nM DNA substrates (32P-ssDNA or 32P-dsDNA) in reaction buffer (recommended NEB buffer for each respective nuclease) and the indicated concentrations of HpDprA. The cleavage reaction was started with the addition of respective exonucleases (1 U/reaction). Digestion was performed for 30 min at 37°C. The reaction was stopped with the addition of 10 mM EDTA and the samples deproteinized by the action of Proteinase K (10 μg/reaction) in the presence of 0.05% SDS for 15 min at 65°C. Degraded DNA was separated from protected DNA on a denaturing (7 M urea) 8% polyacrylamide gel (0.5 × TBE). A constant voltage of 14 V/cm was applied for 3 h at room temperature. The gel was visualized by phosphorimaging analysis of the dried gel (Fujifilm FLA-9000).
Restriction endonuclease cleavage assays
The 110-bp duplex DNA (0.5 nM) containing one site each for HpyCH4V and Hpy188I and 50-bp dsDNA (0.5 nM) containing two sites for HpyCH4III were incubated with increasing concentrations of HpDprA in NEB reaction buffer 4 for 10 min at 37°C. Further incubation with respective restriction enzymes (1 U/reaction) was carried for 1 h. The reaction was stopped and samples deproteinized as explained for the nuclease cleavage assay. Cleaved DNA product was separated from protected DNA on non-denaturing 8% polyacrylamide gel in 0.5 × TBE.
In vitro methylation assay
Methylation assays were carried out to monitor the incorporation of tritiated methyl groups into DNA by using a modified ion exchange filter-binding assay (29). Methylation assays were performed in a reaction mixture (20 µl) containing supercoiled pUC19 plasmid DNA, [3H]AdoMet (specific activity 66 Ci/mmol) and purified protein (M.HpyAVIA and/or HpDprA) in the 1 × TAM reaction buffer. DNA was first pre-incubated with HpDprA for 10 min at 37°C in an appropriate reaction buffer. The methylation reaction was started with the addition of MTase. After incubation at 37°C for 1 h, reactions were stopped by snap freezing in liquid nitrogen. Background counts were measured at zero-time incubation. The reaction mixture incubated in the absence of enzyme was taken as control and the data were analysed. All methylation experiments were carried out in triplicate and the average values reported. Standard deviations of the average methylation rates were <10%.
Sensitivity to restriction endonuclease MnlI
Methylation of pUC19 DNA (1200 nM site concentration) was carried out with purified proteins (M.HpyAVIA and/or HpDprA) as described above in the presence of 8 µM AdoMet in 1 × TAM reaction buffer for 1 h at 37°C. This was followed by inactivation of both the proteins by heating at 75°C for 30 min. DNA was further incubated with MnlI (1 U/reaction) at 37°C for 1 h. Reactions were stopped and deproteinized as described earlier. Products were analysed by electrophoresis on a 1.2% agarose gel in 0.5 × TBE.
Far Western
Far Western studies were performed by a similar method as described earlier (30). Briefly, the indicated concentrations of M.HpyAVIA in 5 µl volume were spotted on a nitrocellulose membrane followed by blocking with 5% (w/v) skimmed milk in phosphate-buffered saline (PBS) buffer containing 0.05% (v/v) Triton X-100 (1 × PBST) for 2 h at 4°C. The reaction membrane was incubated with HpDprA (1 µM) in 1 × TAM buffer (4°C, O/N), while the control membrane was incubated with 1 × TAM buffer alone. Bound HpDprA was detected with the anti-HpDprA antibody and goat anti-rabbit immunoglobulin G (IgG) horseradish-peroxidase conjugate. The blot was further processed using ECL plus western blot analysis kit from GE Healthcare (UK).
Glutathione Sepharose pull down assay for analysis of M.HpyAVIA–HpDprA interaction
Glutathione Sepharose beads (100 µl) were incubated with glutathione transferase (GST)-tagged M.HpyAVIA (25 µg) at 4°C for 3 h. Beads were pulled down by centrifuging at 3000 rpm. M.HpyAVIA–GST-bound beads were washed with PBST. Each time the wash was collected by centrifuging at 3000 rpm. MTase-bound beads were further incubated with HpDprA (25 µg) in PBS. This was followed with three washes with PBST. Bound MTase was eluted using 25 and 50 mM glutathione. The eluate was probed with anti-His antibody (1:10 000) for the presence of HpDprA.
Enzyme-linked immunosorbent assay for analysis of protein–protein interactions
The interaction between HpDprA and M.HpyAVIA was tested by modified enzyme-linked immunosorbent assay (ELISA) as described (31). Briefly, purified M.HpyAVIA was adsorbed to the wells of an ELISA plate (2.0 µg/well) by overnight incubation at 4°C, and the wells were blocked with 5% skimmed milk in PBS. The indicated concentrations of HpDprA were incubated in 1 × TAM buffer with the previously coated wells for 2 h at 37°C. Detection of bound HpDprA was scored using anti-HpDprA antibody as primary antibody and goat anti-rabbit IgG horseradish–peroxidase conjugate as secondary antibody. Wells were washed between incubations with three washes of 1 × PBST. BSA was used as a control. All experiments were performed in triplicate and standard deviations were calculated.
RESULTS
HpDprA binds ssDNA and dsDNA
HpDprA was expressed as a soluble recombinant protein with a N-terminal (His)6 tag in E. coli and purified to near-homogeneity (Supplementary Figure S1A). A minor protein band of ∼30 kDa was detected just below the purified 33 kDa protein in silver stained SDS–PAGE (Supplementary Figure S1A, lane 2). This protein band was seen in all the purified fractions and in all subsequent protein preparations. The mass spectra of purified HpDprA revealed a sharp peak corresponding to the calculated molecular weight of 33 kDa for the recombinant protein (Supplementary Figure S1B). A short peak corresponding to 30 kDa was also observed (Supplementary Figure S1B). Peptide mass fingerprinting was performed to confirm the identity of the purified protein and the co-purified minor protein. An analysis of the obtained fingerprints revealed several matches for the expected fingerprint of HpDprA confirming the authenticity of the purified protein (Supplementary Figure S1C). Two bands corresponding to peptide ions from the C-terminal region of HpDprA (encircled peaks in Supplementary Figure S1C) were found to be missing in the peptide fingerprint map of the 30 kDa band (Supplementary Figure S1D). These bands correspond to a loss of nearly 30 amino acids from the C-terminus which correlates with a mass difference of 3 kDa. It was confirmed that the cleavage was not at the N-terminus as both bands were picked up in the western blot using an anti-His antibody (data not shown).
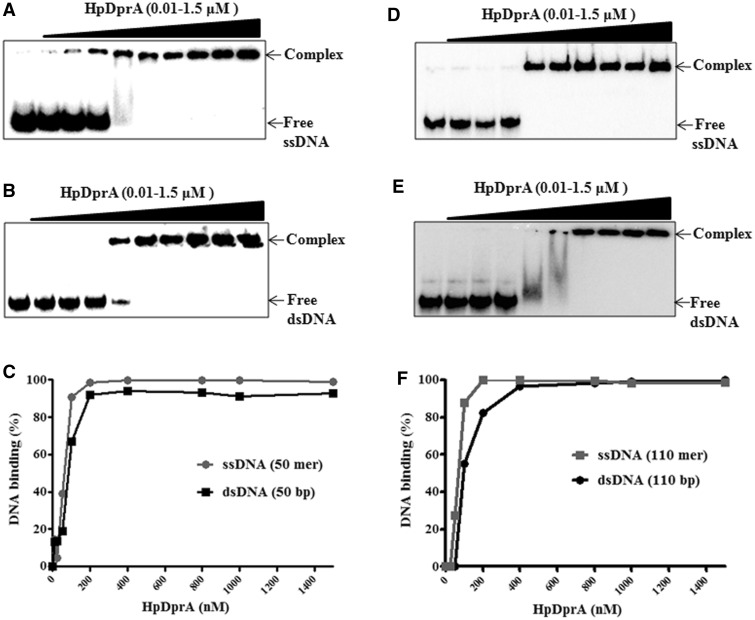
DprA from Streptococcus pneumoniae (SpDprA) and Bacillus subtillis (BsDprA) has been reported to bind and protect ssDNA but not dsDNA (22). HpDprA binds not only ssDNA (Figure 1A) but interestingly to dsDNA too (Figure 1B), resulting in a retardation of both complexes on native PAGE. However, HpDprA showed a higher affinity towards ssDNA as evidenced by the fact that for ssDNA a near complete shift of free DNA was observed at 100 nM concentration of protein. This protein concentration was 2-fold lower than the concentration required for a similar shift with free dsDNA (Figure 1C).
Figure 1.
Binding of HpDprA to ssDNA and dsDNA. Electrophoretic mobility shift assays were performed by incubating different concentrations of HpDprA (0, 10, 25, 50, 100, 200, 400, 800, 1000 and 1500 nM) with 0.5 nM 32P-labelled DNA substrates. Samples were electrophoresed on native PAGE and visualized by autoradiography, as described in ‘Materials and Methods’ section. [(A) 50 mer ssDNA (B) 50 mer dsDNA (D) poly-dT (dT110) (E) poly (dT:dA)110 bp (0.5 nM)]. Quantitation of HpDprA binding to ssDNA and dsDNA of 50 bp length (C) or 110 bp length (F). The data points in Figure 1C and F were obtained from densitometric analysis of EMSA results.
Binding of SpDprA to ssDNA was shown to be sequence non-specific (22). To determine whether binding of HpDprA protein is sequence independent or not, gel shift assays were carried out with homopolymeric ssDNA poly-dT (dT110) and dsDNA poly (dT:dA)110 bp. HpDprA showed a similar binding pattern with both these homopolymers (Figure 1D and E) as it showed for random sequences (Figure 1A and B). A quantitative analysis of binding of HpDprA with ssDNA [poly dT (dT110)] and with dsDNA [poly (dT:dA)110 bp] shown in Figure 1F clearly suggests that HpDprA binds both ssDNA and dsDNA in a sequence-independent manner.
The sensitivity of protein–DNA complex to salt has been shown as a relative measure of its binding affinity (32). To further characterize the interaction of HpDprA with ssDNA and dsDNA, binding assays were performed in the presence of increasing concentration of NaCl. The binding of HpDprA with both ssDNA and dsDNA was stable upto 200 mM salt concentration (Supplementary Figure S2A). However, at salt concentrations >200 mM, the dissociation of dsDNA from its bound complex was more than that of the ssDNA–protein complex. At 300 mM NaCl, ∼60% HpDprA–dsDNA complex and ∼75% HpDprA–ssDNA complex were retained. This indicates that the HpDprA–ssDNA complex is more stable than the HpDprA–dsDNA complex.
SpDprA has been shown to bind to supercoiled ΦX174 DNA, indicating that it does not need a free end to bind ssDNA (22). To determine whether HpDprA binds dsDNA lacking a free end, electrophoretic mobility shift assay was performed with pUC19 plasmid DNA as a substrate. When increasing concentrations of the protein were added to covalently closed circular form of pUC19, its mobility in native agarose gel decreased progressively (Supplementary Figure S2B). It has been demonstrated that increasing the size of the transforming DNA substrate from 50 bp to longer chromosomal DNA resulted in an increase in transformation frequency of H. pylori (15). This has also been observed in the case of SpDprA where the binding affinity of the protein increased with the increase in size of ssDNA and becomes optimal with 50 mer (33). In order to determine whether the DNA length affects its interaction with HpDprA, an analysis of HpDprA interaction with varying lengths of ssDNA (32–110 mer) was carried out. The affinity of HpDprA for ssDNA did not vary significantly with increasing length of ssDNA from 40 to 110 mer. However, a reduced affinity for 32 mer ssDNA was observed (Supplementary Figure S2C). This shows that binding of HpDprA becomes optimal with 40 mer size of ssDNA.
These results together demonstrate that the binding of HpDprA to DNA is sequence independent. The ability of the protein to bind dsDNA indicates the possibility of a wider role for DprA in H. pylori.
HpDprA has a higher affinity for ssDNA than dsDNA
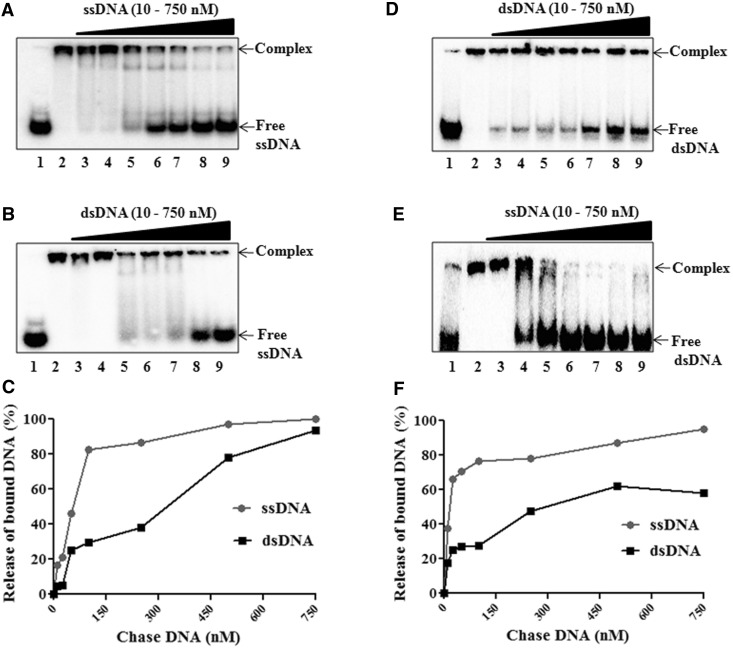
DNA-binding studies with EMSA and ionic strength analysis of the HpDprA–DNA complex indicated a higher affinity of the protein for ssDNA over dsDNA. The higher affinity of HpDprA for ssDNA was ascertained in competition assays. The HpDprA–ssDNA complex was chased with excess of cold ssDNA. A release of labelled DNA from the complex was observed at a ∼200-fold higher concentration of cold competitor DNA (Figure 2A), indicating that HpDprA forms a strong but reversible complex with ssDNA. Next, the HpDprA–ssDNA complex was chased with excess of cold dsDNA. The release of free ssDNA was less with dsDNA than with ssDNA (Figure 2A and B). Quantification of the HpDprA–ssDNA complex chased with ssDNA and dsDNA revealed that 50% complex was competed out by a ∼6-fold higher concentration of cold competitor dsDNA than with cold competitor ssDNA (Figure 2C). Similarly, the HpDprA–dsDNA complex was chased with an excess of cold dsDNA and with cold ssDNA. The complex was more efficiently dissociated by cold competitor ssDNA than by cold dsDNA (Figure 2D and E). A quantitative comparative analysis of competitor assay of the HpDprA–dsDNA complex shows that release of 50% bound dsDNA was observed with a ∼10-fold lower concentration of competitor ssDNA than with dsDNA (Figure 2F). These results indicate a higher preference for ssDNA by HpDprA.
Figure 2.
HpDprA has higher affinity for ssDNA than dsDNA. Chase of preformed 32P-labelled 50 mer ssDNA–HpDprA complex (0.5 nM DNA and 400 nM of HpDprA) with unlabelled (A) ssDNA or (B) dsDNA. Similarly, the chase of 50-bp dsDNA–HpDprA complex with addition of cold (D) dsDNA or (E) ssDNA. Lane 1: DNA alone, lanes 2–9: 0, 10, 25, 50, 100, 250, 500 and 750 (nM) unlabelled competitor DNA. A comparative quantitative analysis of chase with cold competitor DNA (ssDNA and dsDNA) of ssDNA–HpDprA complex (C) and dsDNA–HpDprA complex (F) are shown.
Next, the HpDprA–ssDNA (50 mer) complex was chased with cold ssDNA (50 mer of same sequence) and poly-dT (dT110) separately. ssDNA was competed out from complex with similar efficiency by both types of DNA substrates (Supplementary Figure S3). A similar result was observed for the HpDprA–dsDNA complex (data not shown). These results additionally confirm that HpDprA–DNA binding is not substantially affected by changes in length or sequence of substrate DNA above 40 mer length.
HpDprA–DNA complex is protected from exonucleases and sequence-independent endonucleases
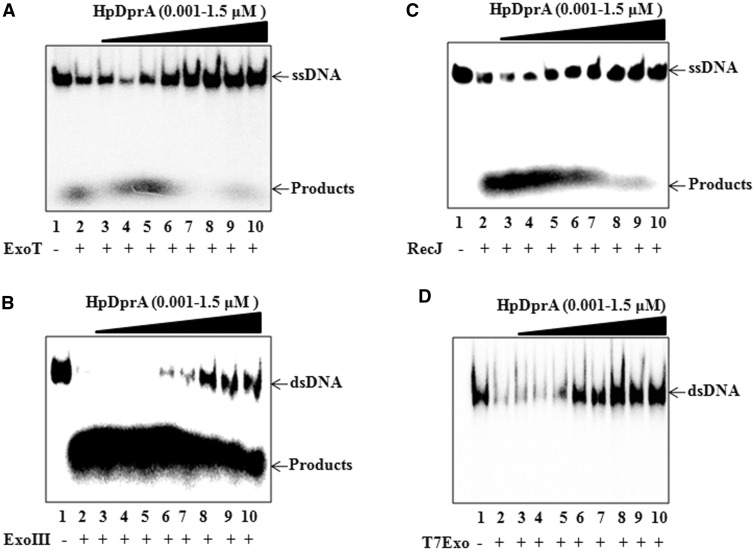
To analyse the nature of interaction of HpDprA with DNA, nuclease protection assays were carried out. The HpDprA–ssDNA complex was subjected to cleavage with ssDNA-specific 3′-exonuclease, ExoT. HpDprA conferred protection to ssDNA from ExoT (Figure 3A) and the protected DNA was of the same size as that of full-length DNA (110 mer). Similarly, protection of dsDNA from the 3′-exonuclease, ExoIII was observed in the presence of HpDprA (Figure 3B), indicating that HpDprA binds and protects both ssDNA and dsDNA. The HpDprA–ssDNA complex was further probed with RecJ, a 5′-exonuclease. As can be seen from Figure 3C, protection of ssDNA from RecJ was observed similar to that with ExoT. The HpDprA–dsDNA complex was found to be resistant to T7Exo (dsDNA-specific 5′-exonuclease) (Figure 3D). Furthermore, the HpDprA–DNA complex was found to be protected from non-specific endonucleases (mung bean endonuclease for ssDNA and DNase1 for dsDNA) as well (data not shown). Protection of the HpDprA–DNA complex from exonuleases as well as sequence non-specific endonucleases indicate that HpDprA coats DNA molecules (both ssDNA and dsDNA) completely and thus prevents access of various nucleases to DNA. Earlier, electron micrographs for interaction of S. pneumoniae DprA with ΦX174 ssDNA showed tightly packed discrete complexes that include numerous protein molecules (22). Such a complex would prevent the access of nucleases to the DNA molecule. Thus, these results for HpDprA are in accordance with the earlier observation.
Figure 3.
Nuclease protection assay. 32P-labelled ssDNA or dsDNA (0.5 nM) either alone (lane 2) or pre-bound with increasing concentrations of HpDprA [1, 5, 10, 50, 100, 500, 1000 and 1500 (nM), lanes 3–10] was incubated for 30 min with 1 unit of (A) ExoT (B) ExoIII (C) RecJ (D) T7Exo. Lane 1: DNA alone.
HpDprA protects dsDNA from Type II restriction endonucleases
Restriction enzymes cleave incoming DNA during inter-strain natural transformation due to their different pattern of methylation and thus act as a transformation barrier in H. pylori (16,34). As inter-strain transformation frequency in H. pylori is reduced but not completely inhibited by R–M systems, the cleavage of incoming DNA should be only partial and limited to only a fraction of restriction sites (16). DNA-binding proteins have been hypothesized to have a role in limiting the accessibility of restriction endonucleases to the DNA molecule and thus preventing cleavage (35). As shown earlier, DprA from H. pyori can bind and protect dsDNA (Figures 1 and 3). Both R–M systems and DprA have been shown to play an early role in natural transformation (16,36). Taken together a functional interaction between DprA and R–M system can be deduced as they participate in the same spatial and temporal events during the process of natural transformation.
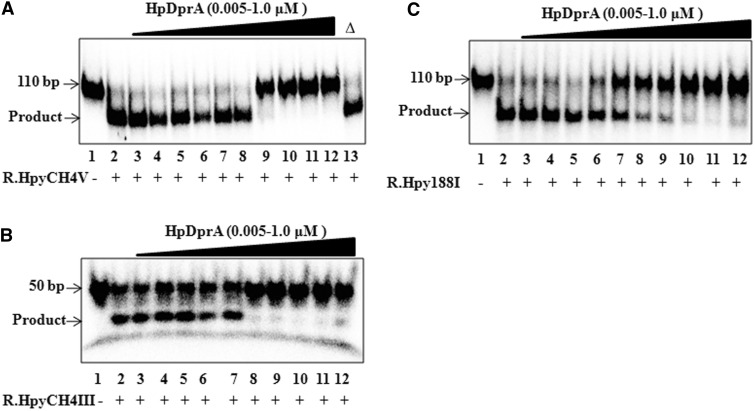
To investigate the ability of HpDprA to confer protection from Type II restriction endonucleases, in vitro protection assays were performed. Three different Type II restriction enzymes R.HpyCH4V, R.HpyCH4III and R.Hpy188I from H. pylori were used in this analysis. The dsDNA substrate (110 bp) with one site for R.HpyCH4V was incubated with increasing concentrations of HpDprA and the reaction was initiated by addition of the restriction enzyme. As the restriction site is 69 bp away from the labelled end, a successful cleavage of 110 bp dsDNA will result in 69 bp labelled dsDNA and 41 bp unlabelled dsDNA. Cleaved DNA was separated from protected DNA (110 bp) on 8% native PAGE. HpDprA was found to protect dsDNA from R.HpyCH4V in the concentration range at which HpDprA showed complete binding with DNA (Figure 4A). Heat inactivated HpDprA failed to protect DNA from restriction cleavage confirming the specificity of the interaction (Figure 4A). A similar protection was observed when the HpDprA–dsDNA complex was probed with R.HpyCH4III (Figure 4B) and R.Hpy188I (Figure 4C).
Figure 4.
HpDprA protects dsDNA from Type II restriction enzymes. 5′-end-labelled dsDNA (0.5 nM) either alone (lane 2) or pre-bound with increasing concentrations of HpDprA [5, 10, 15, 25, 50, 75, 150, 300, 500 and 1000 (nM), lanes 3–12] was incubated with 1 U of (A) R.HpyCH4V (B) R.HpyCH4III (C) R.Hpy188I for 60 min at 37°C. Lane 1: DNA alone. Figure 4A, lane 13: Δ indicates heat inactivated HpDprA (1000 nM).
The effect on restriction enzymes is general. To test this, the HpDprA–dsDNA complex was subjected to restriction activity by MboII. Protection of dsDNA from R.MboII shows that the HpDprA–dsDNA complex is resistant to restriction enzymes from other bacterial species (Supplementary Figure S4). These results indicate a protection of dsDNA from REases in the presence of HpDprA.
HpDprA stimulates the activity of H. pylori MTase
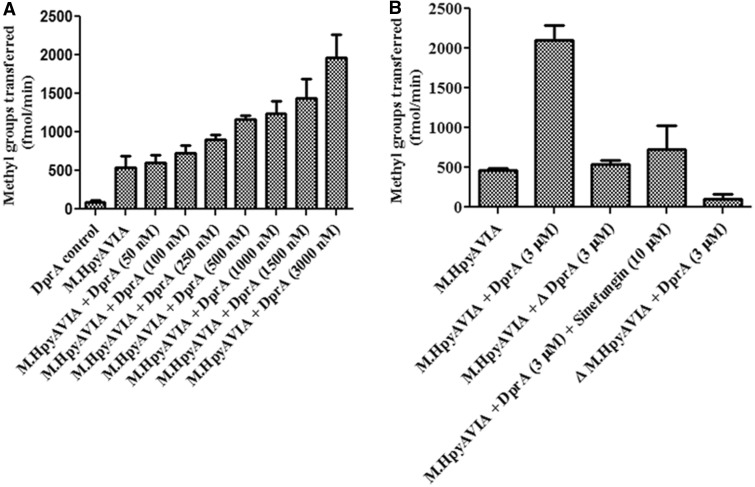
Inhibition of DNA cleavage by Type II restriction enzymes in the presence of HpDprA could be attributed to the ability of DprA to coat dsDNA thus occluding the restriction sites from restriction enzymes. This hypothesis is in agreement with the earlier observation of nuclease protection of DNA by HpDprA (Figures 3 and 4) as well as by SpDprA (22). Having demonstrated HpDprA involvement in protection of DNA from the REases-mediated cleavage, it was reasonable to assess interaction of it with the MTases in H. pylori. An assay was carried out to probe the ability of M.HpyAVIA (a solitary N6 adenine MTase) to methylate pUC19 in the presence and absence of HpDprA. Surprisingly, with increasing concentrations of HpDprA, an increase in activity of the MTase was observed (Figure 5A). Nearly a 4-fold stimulation of MTase activity was observed at 3 µM concentration of HpDprA (Figure 5A). No stimulation was observed when heat inactivated HpDprA was added to the reaction (Figure 5B). When sinefungin (a universal competitive inhibitor of MTases) was added to the reaction in the presence of HpDprA, the activity of MTase was reduced significantly (Figure 5B). Proteins (E. coli RecA, H. pylori SSB and H. influenzae DprA) that bind to ssDNA such as HpDprA had no effect on MTase activity (data not shown), suggesting that stimulation of H. pylori MTases is a unique and specific property of HpDprA.
Figure 5.
Effect of HpDprA on MTase activity. (A) Increasing concentrations of HpDprA were incubated with 1 µM AdoMet and pUC19 DNA (1000 nM site concentration) and 100 nM of M.HpyAVIA. Incorporation of tritiated methyl groups on DNA was monitored. (B) Stimulation of MTase activity of M.HpyAVIA in presence of HpDprA. Δ indicates heat inactivated protein.
HpDprA confers increased protection from restriction enzymes due to stimulation of MTase activity
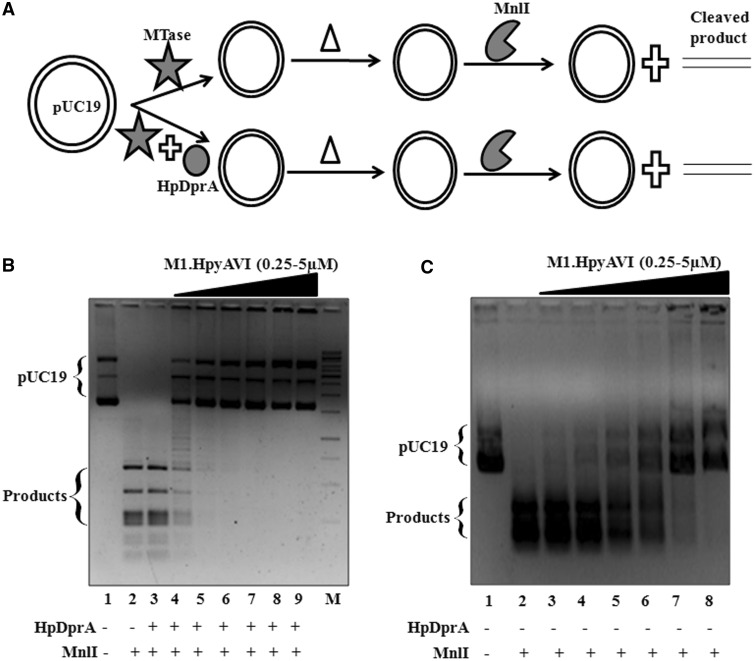
In the case of Type II R–M systems, DNA methylation results in a proportionate protection from the cognate restriction enzyme. Therefore, stimulation of a MTase in the presence of HpDprA should be accompanied with an increased protection from the cognate restriction enzyme. While M.HpyAVIA methylates both GAGG and GGAG sites, MnlI restriction enzyme recognizes and cleaves at GGAG(N)6 site (27). As shown in Figure 6A, pUC19 was methylated with M.HpyAVIA in the presence and absence of HpDprA following which the MTase and/or HpDprA were heat inactivated at 70°C for 30 min. Next, DNA cleavage was initiated by addition of MnlI. Complete protection of pUC19 DNA was observed at a 10-fold lower concentration of the MTase in the presence of HpDprA when compared with that in the absence of DprA (Figure 6B and C). It must be noted that in this assay, methylation of DNA was followed by heat denaturation ensuring inactivation of all DprA and MTase molecules. Figure 6B, lane 3, represents a reaction in which only HpDprA (3 µM) was added and no MTase was added. The reaction mixture was heat inactivated (in a similar manner as for other reactions containing MTase and DprA) and cleavage reaction started with addition of MnlI. A similar cleavage pattern as that of MnlI alone (Figure 6B, lanes 2 and 3) shows that the heat inactivation step inactivated all the HpDprA molecules and the increased protection observed was solely due to increased methylation in the presence of HpDprA. These observations clearly demonstrate that HpDprA stimulates methylation activity of MTase on dsDNA.
Figure 6.
Comparison of restriction digestion patterns of DNA in the presence and absence of HpDprA. (A) Schematic illustration of the experimental design. Δ indicates heat inactivation step. (B) pUC19 either unmethylated (lane 2) or methylated with increasing concentrations of M.HpyAVIA [250, 500, 1000, 2000, 4000 and 5000 (nM), lanes 4–9] in presence of 1 µM of HpDprA was treated with Mnl1 (1 U) for 60 min at 37°C. Lane 3: pUC19 incubated with 1 µM of HpDprA followed by treatment with MnlI. M: 1 kb DNA ladder (C) pUC19 either unmethylated (lane 2) or methylated with increasing concentrations of M.HpyAVIA [250, 500, 1000, 2000, 4000 and 5000 (nM), lanes 3–8] was treated with MnlI. Lane 1: pUC19 DNA alone.
HpDprA shows physical interaction with MTase
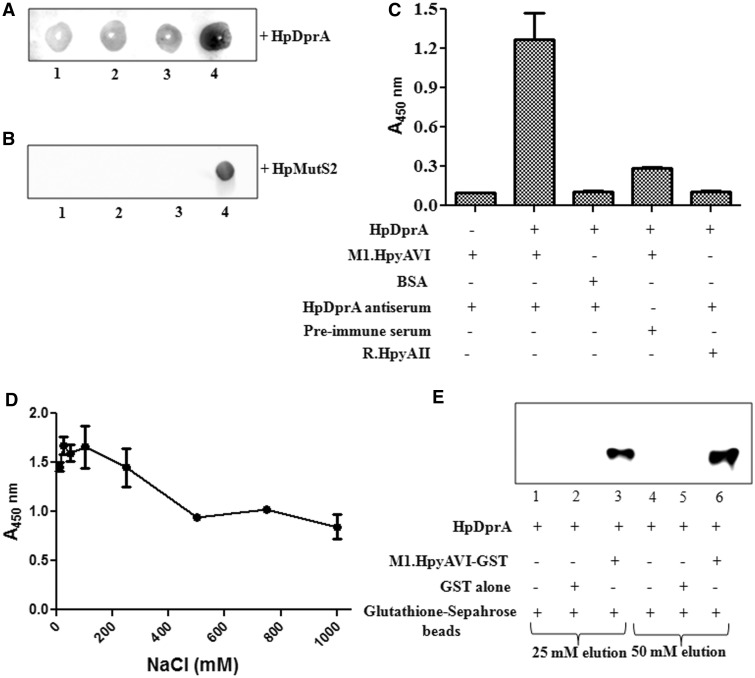
To ascertain whether stimulation of MTases by HpDprA is due to physical or functional interaction, Far Western analysis was carried out. Increasing concentrations of M.HpyAVIA (1–4 µg) were immobilized on a nitrocellulose membrane. After blocking with 5% skimmed milk, the membrane was further incubated with 1 µM HpDprA. Interaction between MTase and HpDprA was probed with HpDprA antiserum as described in ‘Materials and Methods’ section. A greater interaction was observed with the increasing concentration of the immobilized MTase (Figure 7A). However, no signal was obtained when immobilized M.HpyAVIA was directly incubated with HpDprA antiserum (data not shown), indicating that the signal obtained was due to the interaction between HpDprA and MTase. Similarly, when the MTase was tested for interaction with MutS2 of H. pylori (HpMutS2—an antirecombinase protein), no signal was observed (Figure 7B) confirming the specificity of HpDprA–MTase interaction.
Figure 7.
Analysis of in vitro interaction of M.HpyAVIA with HpDprA. (A) Far western analysis of interaction between HpDprA and M.HpyAVIA. Increasing amount of M.HpyAVIA [0.5, 1.0, 2.0 and 4.0 (µg), lanes 1–4] was immobilized on a nitrocellulose membrane. Interaction was studied by incubation of membrane with 1 µM HpDprA and immunoblotting with HpDprA antiserum to detect any bound HpDprA on the membrane. (B) Far western analysis of interaction between HpMutS2 and M.HpyAVIA. Increasing amount of M.HpyAVIA [0.5, 1.0, 2.0 (µg), lanes 1–3] was immobilized on a nitrocellulose membrane. Interaction was studied by incubation of membrane with 1 µM HpMutS2 followed by immunoblotting with HpMutS2 antiserum. Lane 4: 2.0 µg of HpMutS2 immobilized on membrane as positive control. (C) ELISA-based protein–protein interaction assay. ELISA plates coated with M.HpyAVIA and blocked with 10% skimmed milk in PBS were incubated with HpDprA. The remaining steps of reaction were performed as described in ‘Materials and Methods’ section. (D) Effect of salt on HpDprA–M.HpyAVIA interaction. M.HpyAVIA (2 µg) was immobilized on ELISA plates and incubated with 1 µM HpDprA in presence of increasing concentrations of NaCl [10, 25, 50, 100, 250, 500, 750 and 1000 (mM)]. (E) Interaction between HpDprA and M.HpyAVIA by co-elution. HpDprA (25 µg) was incubated with GST (25 µg)-bound glutathione beads (lanes 1 and 4), with glutathione beads alone (lanes 2 and 5) or with GST-tagged M.HpyAVIA (25 µg)-bound glutathione beads (lanes 3 and 6). Lanes 1–3 represent elution with 25 mM glutathione. Similarly, lanes 4–6 represent elution with 50 mM glutathione.
Interaction between M.HpyAVIA and HpDprA was also confirmed by ELISA, where a fixed concentration of M.HpyAVIA (2 µg) was immobilized on ELISA plates and probed for interaction with increasing concentrations of HpDprA. A non-linear saturation curve was obtained for interaction of HpDprA with M.HpyAVIA (Supplementary Figure S5). No signal was obtained when BSA was immobilized instead of the MTase or M.HpyAVIA–HpDprA interaction was probed with pre-immune serum in place of HpDprA antiserum confirming the specificity of the interaction (Figure 7C). Similarly, no interaction was observed for HpDprA with R.HpyAII (Type IIS restriction enzyme containing N-terminal (His)6 tag) (Figure 7C). To understand the nature of interaction between HpDprA and M.HpyAVIA, ELISA was performed in presence of varying concentrations of NaCl. Interaction of HpDprA with M.HpyAVIA was stable upto 250 mM NaCl concentration (Figure 7D). This suggests that interaction of HpDprA with M.HpyAVIA is stable even under high salt concentrations.
M.HpyAVIA interaction with HpDprA was further analysed using glutathione Sepharose pull down experiments. GST-tagged M.HpyAVIA was allowed to bind to glutathione Sepharose beads. HpDprA was added to glutathione Sepharose beads bound to M.HpyAVIA–GST and glutathione Sepharose beads alone in 1 × PBS buffer. The beads were washed with 1 × PBS thrice. M.HpyAVIA was eluted from glutathione Sepharose beads using 25 and 50 mM soluble glutathione. The eluate was probed with anti-His6 antibody for the presence of HpDprA. HpDprA was found to co-elute with M.HpyAVIA from glutathione Sepharose beads (Figure 7E, lanes 3 and 6). HpDprA was present in the eluate of M.HpyAVIA-bound glutathione Sepharose beads but not in the eluate of glutathione Sepharose beads alone (Figure 7E, lanes 2 and 5). This shows that HpDprA was retained on the matrix due to interaction with M.HpyAVIA–GST. To confirm that the observed interaction was with M.HpyAVIA and not with GST, GST alone was incubated with glutathione Sepharose beads which were further allowed to interact with HpDprA. The wash and elution steps were performed as described earlier. HpDprA was absent from glutathione–GST eluate (Figure 7E, lanes 1 and 4) confirming that the interaction observed was with M.HpyAVIA and not GST tag. Collectively, Far Western, ELISA and the co-elution experiments confirmed an in vitro physical interaction between M.HpyAVIA and HpDprA.
DISCUSSION
Helicobacter pylori has a panmictic population structure due to high genetic diversity promoted by both inter- as well as intra-strain transformation (37,38). Intergenomic recombination is subject to strain-specific restriction in H. pylori (39). Annotation of the genomes of H. pylori strains 26695 and J99 shows the presence of nearly two dozen R–M systems out of which 16 were postulated to be Type II for J99 (40–42). These R–M systems act as a barrier to transformation (43). On the other hand, restriction barriers do not restrict all transformation, which could be due to some additional regulation of restriction systems. This balance between restriction and transformation in turn regulates the gene flow to equilibrate competition and cooperation between various H. pylori strains.
RecA, DprA and DprB have been shown to be involved in the presynaptic pathway of recombination (44). Our biochemical characterization of HpDprA revealed ability to bind ssDNA and dsDNA (Figure 1). Binding of HpDprA to both ssDNA and dsDNA results in large nucleoprotein complexes that do not enter the native PAGE. However, DNA trapped in the wells could be released by the addition of excess of competitor DNA, illustrating that the complexes are reversible and do not represent dead-end reaction products (Figure 2). A large DNA–protein complex that sits in the well has also been observed with other DNA-binding proteins such as RecA (45). HpDprA interaction with ssDNA and dsDNA was stable under high salt condtion (200 mM NaCl) indicating that these interactions are specific (Supplementary Figure S2A). The interaction of HpDprA with dsDNA is biologically important since dsDNA plays an important role in natural transformation of H. pylori. The pathway of transformation by dsDNA is highly facilitated (nearly 1000-fold) when compared with ssDNA (15). However, dsDNA is a preferred substrate for REases which are a barrier to horizontal gene transfer. This implies that the decision of ‘restriction’ or ‘facilitation for recombination’ of incoming DNA might be made before the conversion of dsDNA into ssDNA. The incoming DNA has been reported to be in the double-stranded form in periplasm and in single-stranded form in the cytoplasm (10). Hence, the temporal and spatial events surrounding endonuclease cleavage remain to be understood. These studies suggest a very important role of dsDNA in natural transformation process in H. pylori. Hence, the binding and protection of dsDNA by HpDprA is possibly of crucial importance in the success of the natural transformation process for this organism.
Since HpDprA binds dsDNA, one would expect that most of the protein will be bound to the chromosomal DNA of H. pylori. DprA shows polar localization along with CoiA, RecA and SsbB (21,46). These four processing proteins show co-localization with ComGA and/or ComFA. Thus, DprA is less abundant in cytoplasm and localized at cell poles thus interacting with incoming foreign DNA. It may be noted that HpDprA has a higher affinity for ssDNA than dsDNA (Figure 2) and therefore, HpDprA will bind preferentially to incoming ssDNA than to chromosomal DNA.
Both R–M systems and DprA have been shown to have a presynaptic role in the natural transformation process (16,22). While DprA has a protective role, the R–M systems have an inhibitory role for incoming DNA. This indicates a functional interaction between both of them. Our results suggest that when HpDprA interacts with dsDNA, it prevents Type II restriction enzymes from acting on it and at the same time stimulates the activity of MTases thereby resulting in increased methylation of bound DNA (Figures 4 and 5). This observation is of significance as the only way a bacterial cell discriminates between self- and non-self-DNA is through the pattern of methylation. Binding of HpDprA to incoming DNA inhibits access to exonucleases, endonucleases and REases but not to MTases. Moreover, HpDprA may promote the methylation activity of the MTases on incoming dsDNA. As a result, the exogenous DNA will be methylated with the same pattern as that of the host cell and will no longer remain a substrate for restriction enzymes. Thus, HpDprA effectively alleviates the restriction barrier. However, it remains to be understood how DNA in complex with HpDprA, while not accessible to REases or other cellular nucleases, is accessible to a MTase? It has been shown that there is an overlap between DprA dimerization and RecA interaction interfaces and in presence of RecA, DprA–DprA homodimer is replaced with DprA–RecA heterodimer allowing RecA nucleation and polymerization on DNA followed by homology search and synapsis with the chromosome (33). A similar scenario may be possible for the interaction of HpDprA with the MTase.
R–M systems play an important role in protection of genomic DNA from bacteriophage DNA. Hence, dampening the restriction enzymes activity by HpDprA may not be desirable by the host during entire life cycle. Therefore, the positive regulation of DprA expression by ComK, which happens only when competence is achieved, is noteworthy (21). In H. pylori, DNA damage induces a genetic exchange via natural competence (47). Direct DNA damage leads to a significant increase in intergenomic recombination (48). Taken together it can be proposed that when genetic competence is induced, R–M systems are down regulated to allow increased genetic exchange and thus, increasing adaptive capacity in a highly selective environment such as that of the gastric mucosa.
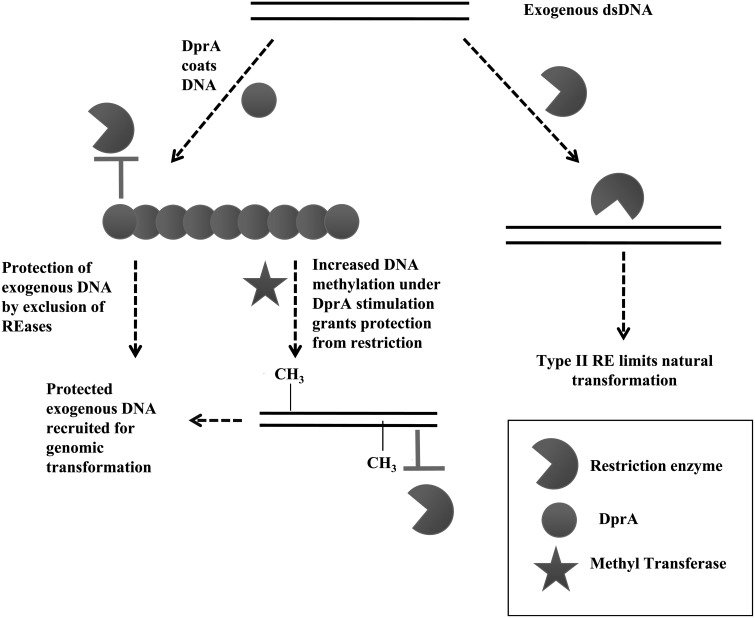
On the basis of the results from this investigation, we propose a model for the modulation of restriction enzymes activity by HpDprA. As Figure 8 describes, during inter-strain transformation, the incoming DNA is cleaved by restriction enzymes, due to recognition of a different pattern of methylation other than host DNA. However, in the presence of HpDprA, incoming DNA is coated by DprA and thus made inaccessible to restriction enzymes and other nucleases. Additionally, the MTase activity on DprA-coated DNA is stimulated and thus the incoming DNA is modified with the same pattern of methylation as that of the host DNA, thereby rendering it resistant to restriction activity.
Figure 8.
Role of HpDprA in alleviating restriction barrier during natural transformation. Diagrammatic illustration of the proposed DprA involvement in alleviating restriction barrier. Coating of DNA by HpDprA occludes restriction enzymes but HpDprA stimulates activity of methyltransferases resulting in protection of incoming DNA.
There is an evolutionary arms race between bacterial genomes and invading DNA molecules. R–M system and anti-restriction systems have co-evolved to maintain an evolutionary balance between the prey and the predator. For example, phage and plasmid employ anti-restriction strategies to avoid restriction barrier by (a) DNA sequence alteration, (b) transient occlusion of restriction sites and (c) subversion of R–M activities (49). The observations of MTase stimulation and site occlusion of restriction sites by HpDprA appear to be analogous to such anti-restriction strategies, otherwise employed by bacteriophages. Thus, HpDprA could be a unique bacterial anti-restriction protein used by the organism for downregulating its own R–M systems to maintain the balance between fidelity and diversity.
In conclusion, we have demonstrated a novel role for H. pylori DprA in the modulation of REase and MTase activity during transformation. HpDprA not only protects incoming DNA from REases but also interacts with MTases and promotes methylation of exogenous DNA to allow it to escape host self-/non-self-recognition. Thus, HpDprA alleviates the R–M barrier and promotes natural transformation in competence-induced conditions. It would be interesting to further study the effects of competence and stress-dependent regulation of DprA and R–M systems in vivo, to understand these mechanisms better.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Funding for open access charge: Department of Atomic Energy, India [DAEO/BBC/DNR/0153].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
G.R.D. thanks CSIR for a senior research fellowship. Genomic DNA of H. pylori strain J99 was kindly provided by New England Biolabs, USA. All members of D.N.R. laboratory are acknowledged for their useful discussions and for critical reading of the article. We thank the DBT mass spectrometry facility at the Indian Institute of Science for peptide mass fingerprinting. D.N.R. acknowledges DST for J.C. Bose Fellowship.
REFERENCES
- 1.Blaser MJ. An endangered species in the stomach. Sci. Am. 2005;292:38–45. doi: 10.1038/scientificamerican0205-38. [DOI] [PubMed] [Google Scholar]
- 2.Basso D, Plebani M, Kusters JG. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2010;15:14–20. doi: 10.1111/j.1523-5378.2010.00781.x. [DOI] [PubMed] [Google Scholar]
- 3.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl Acad. Sci. USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc. Natl Acad. Sci. USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Israel DA, Lou AS, Blaser MJ. Characteristics of Helicobacter pylori natural transformation. FEMS Microbiol. Lett. 2000;186:275–280. doi: 10.1111/j.1574-6968.2000.tb09117.x. [DOI] [PubMed] [Google Scholar]
- 6.Israel DA, Salama N, Arnold CN, Moss SF, Ando T, Wirth H, Tham KT, Camorlinga M, Blaser MJ, Falkow S, et al. Helicobacter pylori strain specific differences in genetic content, identified by microarray, influence host inflammatory responses. J. Clin. Invest. 2001;107:611–620. doi: 10.1172/JCI11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuipers EJ, Israel DA, Kusters JG, Gerrits MM, Weel J, van der Ende A, van der Hulst RWM, Wirth HP, Hook-Nikanne J, Thompson SA, et al. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host. J. Infect. Dis. 2000;181:273–282. doi: 10.1086/315173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smeets LC, Kusters JG. Natural transformation in Helicobacter pylori: DNA transport in an unexpected way. Trends Microbiol. 2002;10:159–162. doi: 10.1016/s0966-842x(02)02314-4. [DOI] [PubMed] [Google Scholar]
- 9.Hofreuter D, Odenbreit S, Puls J, Schwan D, Haas R. Genetic competence in Helicobacter pylori: mechanisms and biological implications. Res. Microbiol. 2000;151:487–491. doi: 10.1016/s0923-2508(00)00164-9. [DOI] [PubMed] [Google Scholar]
- 10.Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. Composite system mediates two-step DNA uptake into. Helicobacter pylori. Proc. Natl Acad. Sci. USA. 2010;107:1184–1189. doi: 10.1073/pnas.0909955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corvaglia AR, Francois P, Hernandez D, Perron K, Linder P, Schrenzel J. A Type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc. Natl Acad. Sci. USA. 2010;107:11954–11958. doi: 10.1073/pnas.1000489107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Humbert O, Salama NR. The Helicobacter pylori HpyAXII restriction–modification system limits exogenous DNA uptake by targeting GTAC sites but shows asymmetric conservation of the DNA methyltransferase and restriction endonuclease components. Nucleic Acids Res. 2008;36:6893–6906. doi: 10.1093/nar/gkn718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin EA, Zhang XS, Levine SM, Gill SR, Falush D, Blaser MJ. Natural transformation of Helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 2009;5:e1000337. doi: 10.1371/journal.ppat.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waldron DE, Lindsay JA. Sau1: a novel lineage-specific Type I restriction–modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 2006;188:5578–5585. doi: 10.1128/JB.00418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine SM, Lin EA, Emara W, Kang J, Dibenedetto M, Ando T, Falush D, Blaser MJ. Plastic cells and populations: DNA substrate characteristics in Helicobacter pylori transformation define a flexible but conservative system for genomic variation. FASEB J. 2007;21:3458–3467. doi: 10.1096/fj.07-8501com. [DOI] [PubMed] [Google Scholar]
- 16.Humbert O, Dorer MS, Salama NR. Characterization of Helicobacter pylori factors that control transformation frequency and integration length during inter-strain DNA recombination. Mol. Microbiol. 2011;79:387–401. doi: 10.1111/j.1365-2958.2010.07456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XS, Blaser MJ. Natural transformation of an engineered Helicobacter pylori strain deficient in Type II restriction endonucleases. J. Bacteriol. 2012;194:3407–3416. doi: 10.1128/JB.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ando T, Israel DA, Kusugami K, Blaser MJ. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 1999;181:5572–5580. doi: 10.1128/jb.181.18.5572-5580.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smeets LC, Bijlsma JJ, Kuipers EJ, Vandenbroucke-Grauls CM, Kusters JG. The dprA gene is required for natural transformation of Helicobacter pylori. FEMS Immunol. Med. Microbiol. 2000;27:99–102. doi: 10.1111/j.1574-695X.2000.tb01418.x. [DOI] [PubMed] [Google Scholar]
- 20.Karudapuram S, Zhao X, Barcak GJ. DNA sequence and characterization of Haemophilus influenzae dprA, a gene required for chromosomal but not plasmid DNA transformation. J. Bacteriol. 1995;177:3235–3240. doi: 10.1128/jb.177.11.3235-3240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer N, Hahn J, Dubnau D. Multiple interactions among the competence proteins of Bacillus subtilis. Mol. Microbiol. 2007;65:454–464. doi: 10.1111/j.1365-2958.2007.05799.x. [DOI] [PubMed] [Google Scholar]
- 22.Mortier-Barriere I, Velten M, Dupaigne P, Mirouze N, Pietrement O, McGovern S, Fichant G, Martin B, Noirot P, Le Cam E, et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130:824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Chevallet M, Luche S, Rabilloud T. Silver staining of proteins in polyacrylamide gels. Nat. Protoc. 2006;1:1852–1858. doi: 10.1038/nprot.2006.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 27.Kumar R, Mukhopadhyay AK, Rao DN. Characterization of an N6 adenine methyltransferase from Helicobacter pylori strain 26695 which methylates adjacent adenines on the same strand. FEBS J. 2010;277:1666–1683. doi: 10.1111/j.1742-4658.2010.07593.x. [DOI] [PubMed] [Google Scholar]
- 28.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 29.Rubin RA, Modrich P. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J. Biol. Chem. 1977;252:7265–7272. [PubMed] [Google Scholar]
- 30.Sharma R, Rao DN. Orchestration of Heamophilus influenza RecJ Exonuclease by interaction with single stranded DNA-binding protein. J. Mol. Biol. 2009;385:1375–1396. doi: 10.1016/j.jmb.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 31.Leonard S, Plante D, Wittmann S, Daigneault N, Fortin MG, Laliberte J. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 2000;74:7730–7737. doi: 10.1128/jvi.74.17.7730-7737.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menetski JP, Kowalczykowski SC. Interaction of RecA protein with single-stranded DNA quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 33.Quevillon-Cheruel S, Campo N, Mirouze N, Mortier-Barriere I, Brooks MA, Boudes M, Durand D, Soulet A, Lisboa J, Noirot P, et al. Structure–function analysis of pneumococcal DprA protein reveals that dimerization is crucial for loading RecA recombinase onto DNA during transformation. Proc. Natl Acad. Sci. USA. 2012;109:E2466–E2475. doi: 10.1073/pnas.1205638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Q, Morgan RD, Roberts RJ, Blaser MJ. Identification of Type II restriction and modification systems in Helicobacter pylori reveals their substantial diversity among strains. Proc. Natl Acad. Sci. USA. 2000;97:9671–9676. doi: 10.1073/pnas.97.17.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polach KJ, Widom J. Restriction enzymes as probes of nucleosome stability and dynamics. Methods Enzymol. 1999;304:278–298. doi: 10.1016/s0076-6879(99)04017-3. [DOI] [PubMed] [Google Scholar]
- 36.Berge M, Mortier-Barriere I, Martin B, Claverys JP. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol. Microbiol. 2003;50:527–536. doi: 10.1046/j.1365-2958.2003.03702.x. [DOI] [PubMed] [Google Scholar]
- 37.Falush D, Kraft C, Taylor NS, Correa P, Fox JG, Achtman M, Suerbaum S. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl Acad. Sci. USA. 2001;98:15056–15061. doi: 10.1073/pnas.251396098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salama NR, Gonzalez-Valencia G, Deatherage B, Aviles-Jimenez F, Atherton JC, Graham DY, Torres J. Genetic analysis of Helicobacter pylori strain populations colonizing the stomach at different times postinfection. J. Bacteriol. 2007;189:3834–3845. doi: 10.1128/JB.01696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ando T, Xu Q, Torres M, Kusugami K, Israel DA, Blaser MJ. Restriction modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 2000;37:1052–1065. doi: 10.1046/j.1365-2958.2000.02049.x. [DOI] [PubMed] [Google Scholar]
- 40.Lin LF, Posfai J, Roberts RJ, Kong H. Comparative genomics of the restriction–modification systems in Helicobacter pylori. Proc. Natl Acad. Sci. USA 98. 2001:2740–2745. doi: 10.1073/pnas.051612298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 42.Kong H, Lin LF, Porter N, Stickel S, Byrd D, Posfai J, Roberts RJ. Functional analysis of putative restriction–modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res. 2000;28:3216–3223. doi: 10.1093/nar/28.17.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang J, Blaser MJ. Bacterial populations as perfect gases: genomic integrity and diversification tensions in Helicobacter pylori. Nat. Rev. Microbiol. 2006;4:826–836. doi: 10.1038/nrmicro1528. [DOI] [PubMed] [Google Scholar]
- 44.Dorer MS, Sessler TH, Salama NR. Recombination and DNA repair in Helicobacter pylori. Annu. Rev. Microbiol. 2011;65:329–348. doi: 10.1146/annurev-micro-090110-102931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganesh N, Muniyappa K. Mycobacterium smegmatis RecA protein is structurally similar to but functionally distinct from Mycobacterium tuberculosis RecA. Proteins. 2003;53:6–17. doi: 10.1002/prot.10433. [DOI] [PubMed] [Google Scholar]
- 46.Tadesse S, Graumann PL. DprA/Smf protein localizes at the DNA uptake machinery in competent Bacillus subtilis cells. BMC Microbiol. 2007;7:105. doi: 10.1186/1471-2180-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dorer MS, Fero J, Salama NR. DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog. 2010;6:e1001026. doi: 10.1371/journal.ppat.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang JM, Iovine NM, Blaser MJ. A paradigm for direct stress-induced mutation in prokaryotes. FASEB J. 2006;20:2476–2485. doi: 10.1096/fj.06-6209com. [DOI] [PubMed] [Google Scholar]
- 49.Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.