Abstract
Single-strand break repair (SSBR) and base excision repair (BER) of modified bases and abasic sites share several players. Among them is XRCC1, an essential scaffold protein with no enzymatic activity, required for the coordination of both pathways. XRCC1 is recruited to SSBR by PARP-1, responsible for the initial recognition of the break. The recruitment of XRCC1 to BER is still poorly understood. Here we show by using both local and global induction of oxidative DNA base damage that XRCC1 participation in BER complexes can be distinguished from that in SSBR by several criteria. We show first that XRCC1 recruitment to BER is independent of PARP. Second, unlike SSBR complexes that are assembled within minutes after global damage induction, XRCC1 is detected later in BER patches, with kinetics consistent with the repair of oxidized bases. Third, while XRCC1-containing foci associated with SSBR are formed both in eu- and heterochromatin domains, BER complexes are assembled in patches that are essentially excluded from heterochromatin and where the oxidized bases are detected.
INTRODUCTION
Cellular DNA is continuously exposed to oxidative stress arising from both endogenous and exogenous sources. As a consequence, lesions such as modified bases, abasic (AP) sites and single-strand breaks (SSBs) are generated. Oxidized bases are recognized and excised by specific DNA glycosylases that initiate the base excision repair (BER) pathway. The AP site produced by the DNA glycosylase activity is then incised by the AP endonuclease APE1 resulting in a SSB. In most cases, the subsequent synthesis and ligation steps are carried out by POL β and LIG3, respectively, to restore an intact DNA molecule. AP sites and SSBs can also be directly induced in genomic DNA and some of the enzymatic steps required for their repair are shared by Single Strand Break Repair (SSBR) and BER pathways. Besides the enzymes mentioned earlier, other proteins are also required for efficient repair of modified bases and SSBs. SSBs are swiftly bound by PARP1, which subsequently parylates itself and other targets. PARP1 activity is required for the recruitment of XRCC1 to SSBs (1,2). No enzymatic functions for XRCC1 have been identified and the protein is thought to function as a scaffolding platform for the SSBR and BER activities (3,4). XRCC1 is essential for embryonic development in mice (5) and cells deficient in XRCC1 exhibit increased frequencies of sister chromatid exchanges and chromosomal rearrangements. Once XRCC1 is bound to the SSB, it serves as a scaffolding platform to recruit, activate or regulate the downstream SSBR enzymes. This coordination function of XRCC1 is thought to be important to avoid accumulation of toxic DNA repair intermediates (6). In the case of BER, the mechanism by which XRCC1 is recruited is less clear. While in some models it is assumed that XRCC1 recruitment to BER is mediated by PARP1 after DNA incision and formation of a SSB intermediate (7), the direct interaction of XRCC1 with different glycosylases and APE1 (4,8,9) suggests that XRCC1 could in fact be recruited during the very first steps of BER and would therefore also be required for the coordination of the initial part of the pathway.
In the majority of cases, DNA repair proficiency requires the formation of multi-molecular complexes that are assembled at the site of the damage. This has been clearly demonstrated for some of the more thoroughly studied DNA repair pathways such as homologous recombination, non-homologous end joining and nucleotide excision repair (10–12). In spite of being responsible for the removal of the most abundant class of DNA lesions and the extensive molecular epidemiology literature associated to this pathway, the role and mechanism of protein complex formation in BER, although known to be critical, are much less understood. One of the underlying reasons is that, so far, it has been difficult to dissociate BER from SSBR, which has been quite extensively studied with respect to the formation of complexes.
Another unanswered question in BER is how modified bases or abasic sites are recognized within chromatin in the first place. More generally, whether DNA repair proteins have access to damaged DNA inside heterochromatin has been a matter of debate for a long time. The answer to that question seems to depend on the repair pathway under consideration. Although it is now accepted that DNA double-strand breaks can also arise in heterochromatic DNA regions, it has been considered for a long time that heterochromatin is refractory to formation of DSB repair foci, limiting the DNA repair complex assembly to the periphery of heterochromatin domains (13–16). Recently published results indicate that gammaH2AX foci can indeed be formed in heterochromatin domain and that there is a time-dependent relocation of the breaks to the periphery of heterochromatin (17,18). For the case of BER of 8-oxoG, a major mutagenic base lesion induced by oxidative stress, it has been shown that in vitro activity of OGG1, its cognate DNA glycosylase, is inhibited when the modified base is in nucleosomally organized DNA compared with naked DNA, probably due to impaired accessibility of the enzyme to the lesion. These results led to the suggestion that chromatin remodeling may be required for efficient 8-oxoG repair in chromatinized substrates (19,20). Therefore, it is not clear how the repair of modified bases inducing little or no distortion of the DNA structure can be initiated in the context of chromatin.
Here, by using different methods to induce SSB and 8-oxoG in genomic DNA, we show that PARP is not required for the recruitment of XRCC1 to the BER pathway and that the distributions within the nucleus of the repair complexes involved in SSBR or BER can be distinguished. After the induction of 8-oxoG by the oxidative agent KBrO3, both the lesion and the enzymes dedicated to BER are mainly detected in euchromatin regions, while SSBR complexes are detected both in euchromatin and heterochromatin.
MATERIALS AND METHODS
Plasmid construction
To generate the hOGG1, APE1 and XRCC1 fusions with fluorescent proteins, the open reading frames were amplified and subcloned into pEGFP-NI and pDsRed-Monomer-N1 from Clontech. The mutant proteins OGG1(K249Q) and XRCC1(L360D) were obtained by site-directed mutagenesis using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). The plasmid coding for the LIG3-RFP was a kind gift from H. Leonhardt. For the plasmid expressing OGG1-Flag, the OGG1 coding sequence was amplified by PCR using a primer containing one copy of the Flag sequence and subsequently cloned into pcDNA3.1.
Cell lines, culture and treatments
HeLa and mouse embryo fibroblast (MEF) cells were cultured in DMEM (Lonza) containing 10% of fetal bovine serum at 37°C with 5% CO2. Both parp1+/+ and parp1−/− MEF cells were kindly provided by V. Schreiber (IREBS, Strasbourg).
Cells were grown on coverslips for in situ visualization experiments and on Petri dishes for biochemical extraction. Transient transfections were done with LipoFectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Experiments were performed 24 h after transfection. Cells at ∼80% confluence were treated with 40 mM of potassium bromate (KBrO3) (Sigma) diluted in DPBS (Cambrex), for 30 min at 37°C. After treatment, cells were allowed to recover in DMEM for the indicated periods of time before fixation or extraction. When indicated, PARP inhibitors were added to the medium 1 h before the treatment and kept during all the process including the washing steps. PARP inhibitors AZD-2281 (Olaparib), 1,5-dihydroxyisoquinoline (DIQ) and N-(6-oxo-5,6-dihydro-henanthridin-2-yl)-N,N-dimethylacetamide HCl hydrochloride hydrate (PJ-34) were purchased from Sigma–Aldrich and used at a concentration of 5, 200 and 10 µM, respectively.
For the removal of soluble proteins, cells were washed twice on ice-cold phosphate-buffered saline (PBS) and extracted for 5 min on ice in cytoskeleton (CSK) buffer (100 mM NaCl, 300 mM glucose, 10 mM PIPES pH 6.8, 3 mM MgCl2, 0.5% Triton X-100 and protease inhibitors). Cells were washed twice on ice-cold PBS before fixation in 4% paraformaldehyde (PFA) for 30 min at room temperature. Nuclear DNA was counterstained with 1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI). Coverslips were mounted in Dako Fluorescent Mounting Medium.
Immunofluorescence
For visualization of 8-oxoG in situ, cells grown on coverslips were fixed in acetone:methanol (1:1) and air dried. Cells were hydrated for 15 min in PBS, and DNA was denatured by incubating cells in 2 N HCl for 45 min at room temperature. This step was critical in order to allow access of the antibody to the chromatin. Cells were washed three times in PBS and neutralized with 50 mM Tris–HCl pH 8.8 for 5 min before proceeding to the immunofluorescence protocol, as previously described (21). For the staining of 8-oxoG in microirradiated cells, cells were fixed in 4% PFA and the denaturation step was not required (probably due to high accumulation of 8-oxoG in a small nuclear region), allowing the simultaneous visualization of the fluorescent protein. The anti 8-oxoG mouse monoclonal antibody (N45.1) (Gentaur) was used at a final concentration of 1 µg/ml. Anti-mouse Alexa-488 or anti-mouse Alexa-594 (Life technologies) were used as a secondary antibody. Nuclear DNA was counterstained with 1 μg/ml propidium iodide with 50 μg/ml RNase when indicated. Coverslips were mounted in Dako Fluorescent Mounting Medium. For the detection of PAR polymer formation, cells were fixed in 4% PFA and immunofluorescence was performed using the anti-PAR mouse antibody, clone 10H at a dilution of 1/200.
Microscopy and image treatment analysis
Image acquisition was performed with a Leica confocal microscope SPE (Wetzlar, Germany), using ACS APO 40.0 × 1.15 OIL or ACS APO 63.0 × 1.30 OIL lenses. Plot profiles and image treatment and analysis were done with the ImageJ software (Rasband, 1997, ImageJ, U.S. National Institutes of Health, Bethesda, MD, USA; http://rsb.info.nih.gov/ij/). Correlation coefficients were estimated using the ImageJ JACoP plug-in (22). Three-dimensional image reconstructions and measurements were accomplished using Volocity® software (Perkin Elmer, Waltham, MA, USA). Nuclear protein localization was represented in isosurface rendering and quantification of proteins co-localization was assessed using Pearson’s correlation coefficient (R) calculated with Costes et al. (23) algorithms.
Live-cell microscopy, microirradiation and photobleaching experiments
Cells were grown on 35-mm glass bottom Petri dishes. Live-cell imaging was performed with a Nikon A1 inverted confocal microscope equipped with an environmental chamber allowing the control of temperature, humidity and gas mixture. For excitation, 405 and 561 nm laser diodes and a 488-nm Argon-laser line were used. Confocal image series were typically recorded with a frame size of 512 × 512 pixels. Acquisitions were performed using a PLAN APO 60×/1.4 oil objective.
For fluorescence recovery after photobleaching (FRAP) experiments, a square of 1 µm2 was selected and photobleached for 2.5 s with the 488-nm Argon-laser set to maximum power. Before and after bleaching, confocal image series of one mid z-section were acquired every second with the 488-nm laser set at 3.5% of the power (6 pre-bleaching and 31 post-bleaching frames). Fluorescence intensity on the bleached region was measured and normalized to the intensity of the last pre-bleach value.
Microirradiation was carried out with a 405-nm diode laser set to 10% power. Preselected regions of 1 µm2 were microirradiated for ∼6 s. Six seconds before irradiation and for 1 or 2 min (as indicated) after irradiation, confocal image series of one mid z-section were obtained every 1 s with the 488 and 564 lasers set at 5% of power. For evaluation of the recruitment kinetics, fluorescence intensities of the irradiated region were measured and normalized to the immediately post-irradiation value. Experiments were repeated at least three times and an average of 10 cells from a representative experiment were displayed in the corresponding graphs.
When indicated, the Ro 19-8022 photosensitizer was added to the medium for 5 min before the microirradiation at a final concentration of 5 µM. Ro 19-8022 was a kind gift of Prof. Bernd Epe (University of Mainz).
Immunoprecipitation and western blot analysis
Protein extracts were done in NP40 buffer (0.1% NP40, 1 mM EDTA, 20 mM Tris–HCl pH 8, 150 mM NaCl), sonicated with the Bioruptor® bath (pulses 30″ on/30″ off for 10 min at maximum intensity) and centrifuged for 20 min at 13 000 rpm. For immunoprecipitation, 1 mg of protein extract was incubated with anti-FLAG antibody (Sigma), and protein complexes were recovered using the Dynabeads Goat anti-Mouse IgG (Invitrogen Dynal AS) following the manufacturer’s instructions. Protein extracts and immunoprecipitated proteins were separated by 10% SDS–PAGE gel and blotted with anti-FLAG and anti-GFP (Roche) antibodies.
RESULTS
PARP activity is not required for recruitment of XRCC1 and OGG1 to locally induced base damage
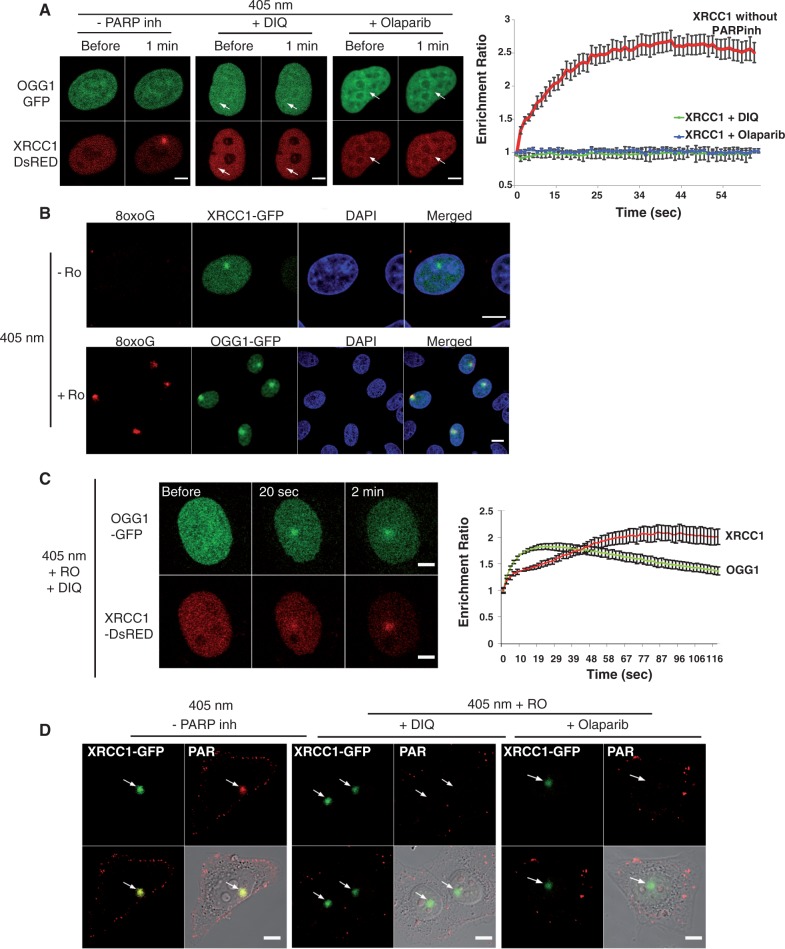
Several studies have indicated that recruitment of XRCC1 to laser-induced DNA damage depends largely on PARP activity (24–26). However, because under certain conditions of irradiation the presence of PARP inhibitors only slightly reduced the recruitment of XRCC1 at the sites of damage, it was suggested that different kinds of damage could be induced under those conditions (27). In order to explore the involvement of PARP in the recruitment of XRCC1 to the first steps of BER, we optimized a microirradiation protocol based on the use of the 405-nm laser in combination with the photosensitizer Ro 19-8022 to promote local formation of oxidative base damage (28,29). In the absence of photosensitizer, while XRCC1 was clearly recruited to the site of damage after irradiation with the laser, we were not able to detect the recruitment of OGG1, the DNA glycosylase responsible for the recognition of 8-oxoG and initiation of its repair. PARP inhibitors DIQ or Olaparib completely abolished recruitment of XRCC1, suggesting that in the absence of photosensitizer SSBs are the major lesion induced at the laser intensity used (Figure 1A). Irradiation with the same laser doses, but this time in the presence of the photosensitizer, resulted in the formation, in addition to SSBs, of 8-oxoG (Figure 1B). Interestingly, in that case, both OGG1 and XRCC1 proteins were recruited to the irradiated site, even when either DIQ or Olaparib were present (Figure 1C and Supplementary Figure S1A). These results were confirmed by using a different microirradiation setup for the induction of local oxidative DNA damage in the presence of the PARP inhibitor PJ34 (30) (Supplementary Figure S1B).
Figure 1.
Recruitment of XRCC1 and OGG1 to SSB or 8-oxoG induced by laser microirradiation. (A) Live-cell imaging of HeLa cells co-expressing OGG1-GFP and XRCC1-DsRED microirradiated with the 405-nm laser in the absence or presence of PARP inhibitors DIQ and Olaparib. Arrows signal the site of microirradiation; scale bar, 5 µm. (B) After microirradiation in the presence of the photosensitizer Ro 19-8022 (Ro) cells were fixed and immunofluorescent detection of 8-oxoG (red) was performed. The irradiated region is detected by the recruitment of XRCC1-GFP and OGG1-GFP. Scale bar, 10 µm. (C) Live-cell imaging of microirradiated HeLa cells co-expressing OGG1-GFP and XRCC1-DsRED, in the presence of the photosensitizer Ro and the PARP inhibitor DIQ. Scale bar, 5 µm. (D) Cells expressing XRCC1-GFP were microirradiated and immediately fixed. Anti-PAR antibodies were used to check for the efficient inhibition of polymer formation when PARP inhibitors were present. Graphs indicating the time course of recruitment represent mean values from 10 cells. Error bars represent the SEM.
In order to verify that the PARP activity was efficiently inhibited in the conditions where base damage was generated and XRCC1 was recruited, we analysed the formation of PAR polymer at the site of damage. A clear accumulation of PAR was observed by immunofluorescence at the microirradiated region in the absence of PARP inhibitors (Figure 1D). The presence of either DIQ or Olaparib in the medium completely abolished PAR formation even in the presence of Ro where the recruitment of XRCC1 could be clearly observed (Figure 1D). Taken together, these results show that BER proteins, including XRCC1, do not require PARP activity to be recruited to the site of base damage.
The sequential arrival of OGG1 and XRCC1 to the microirradiated region in the presence of Ro and PARP inhibitors suggests that the initial recognition step by the glycosylase is required for the recruitment of XRCC1. Ideally, this could be verified by OGG1 silencing or by performing these experiments in Ogg1−/− cells. However, considering that other glycosylases, such as NTH1 (Supplementary Figure S2A), NEIL1 and NEIL2 (25), are also recruited to the microirradiated region and that most of them interact with XRCC1 (8), the interpretation of the results after the silencing/removal of just one of the glycosylases would be very difficult. Thus, in order to circumvent this restriction, and considering that the concentration of OGG1 seems to be a limiting factor in the BER pathway, we opted for analysing the effect of the overexpression of OGG1 on the recruitment of XRCC1. We could indeed observe a higher recruitment of XRCC1 to the site of the damage when OGG1-DsRED was overexpressed (compared with cells expressing endogenous levels of OGG1), suggesting that recruitment of XRCC1 to BER is dependent on the glycosylase (Supplementary Figure S2B). Whether XRCC1 arrives before or after formation of the nick is yet to be determined, however, it is clear that the recognition of the nick as a SSB by PARP1 and the subsequent activation of the protein are not required for the recruitment of XRCC1 to the BER pathway.
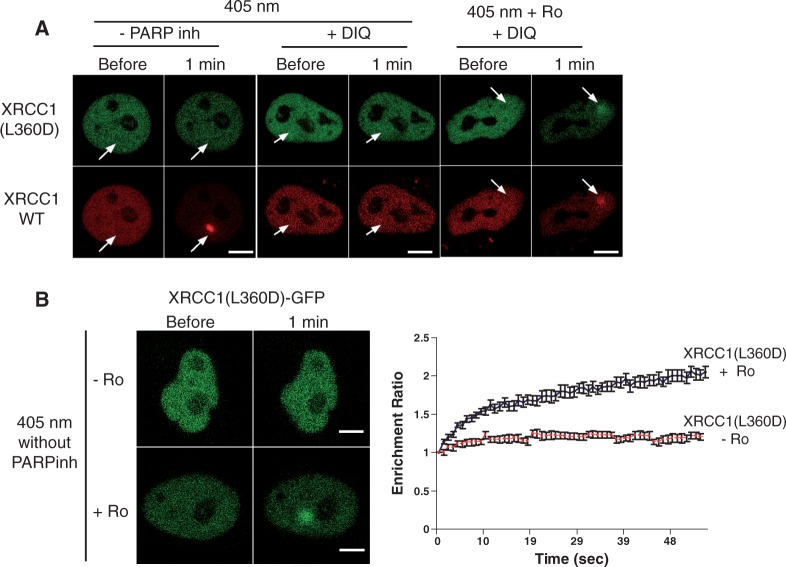
Among the members of the PARP family, at least two of them, PARP-1 and PARP-2, have been implicated in the DNA damage response (31). We chose to use PARP enzymatic inhibition to analyse XRCC1 recruitment to DNA damage after irradiation since, due to the high level of similarity between PARP-1 and PARP-2, most PARP inhibitors affect both enzymes (32). However, the use of PARP inhibitors will affect the enzymatic function of PARP-1, but may also result in the blockage of PARP-1 at the site of damage (33), thus allowing the recruitment of downstream factors via PARP-1-dependent protein–protein interactions. To circumvent this possible limitation of inhibitors, we explored the recruitment of the XRCC1 mutant L360D in which the BRCT1 domain, which is required for the interaction with PARP, is disrupted (34). This mutation results in impaired recruitment of XRCC1 to sites of SSB damage (2). By co-transfecting plasmids expressing either XRCC1-DsRed or XRCC1(L360D)-GFP, we analysed in the same cells the recruitment of both allelic forms to irradiated areas. As expected, induction of SSBs with the 405-nm laser in the absence of photosensitizer failed to trigger the recruitment of the mutant form of XRCC1, while the wild type (WT) version was clearly recruited to the damaged region (Figure 2A and Supplementary Figure S1). In the presence of Ro 19-8022, when oxidized bases are induced (Figure 1B), both the WT and the mutant XRCC1(L360D) were recruited to the site of damage, even in the presence of PARP inhibitors (Figure 2A and Supplementary Figure S1), confirming that the XRCC1 interaction with PARP is not required for its recruitment to base oxidative damage induced by microirradiation. Recruitment kinetics were measured in cells expressing the XRCC1(L360D)-GFP alone. Consistently, Figure 2B shows that in the absence of photosensitizer the mutant protein was not recruited to the irradiation site. In contrast, accumulation of XRCC1(L360D) at the irradiated spot was observed when base damage was generated by addition of Ro-19-8022 at the time of irradiation. Interestingly, the presence of PARP inhibitors in the medium had no impact on the recruitment kinetics of the XRCC1(L360D) (Supplementary Figure S1C) indicating that inhibitors did not represent a physical interference for the arrival of XRCC1. Since neither XRCC1 nor OGG1 recruitments were impeded by PARP inhibition (Supplementary Figure S2C), their use in the microirradiations in the presence of the photosensitizer allows to specifically study the recruitment of proteins to BER without interference of their participation to SSBR. We therefore took advantage of this protocol for the local induction of 8-oxoG to compare the recruitment kinetics for OGG1 and XRCC1 to the site of oxidative DNA damage. Recruitment of the fully functional OGG1-GFP (21) reached a maximum ∼20 s after microirradiation and thereafter the signal rapidly decreased (Figure 1C), as previously described using a different microirradiation system (25). When we used an active site mutant, OGG1(K249Q), which recognizes 8-oxoG but is unable to excise it, the signal at the site of damage persisted for longer periods of time suggesting that the enzymatic activity, and thus the excision of 8-oxoG, is required for the rapid release of OGG1 from the irradiated area (Supplementary Figure S2D). In contrast to the very fast recruitment of XRCC1 to the sites of SSB, which reached maximal intensity after 25 s (Figure 1A), the PARP-independent XRCC1 recruitment to oxidative base damage was slower, reaching its maximum ∼100 s after irradiation (Figure 1C). The earlier results are consistent with the proposed role of XRCC1 in BER (4,8). Indeed, after initiating the repair of the induced 8-oxoG and facilitating the recruitment of XRCC1, OGG1 would be released from the site of damage while XRCC1 would remain to participate in the subsequent BER steps.
Figure 2.
Disruption of the XRCC1 BRCT1 domain results in the abolishment of XRCC1 recruitment to SSB but does not affect the recruitment of the protein to oxidative damage. (A) HeLa cells co-transfected with the WT XRCC1-DsRED monomer (red) and the mutant XRCC1(L360D)-GFP (green) were microirradiated in the presence or absence of Ro. Arrows indicate the site of irradiation. (B) Live-cell imaging of microirradiated HeLa cells expressing the XRCC1 mutant XRCC1(L360D)-GFP, in the absence of PARP inhibitors and in the presence or absence of Ro. Scale bar, 5 µm. Graph corresponds to mean values from 10 cells. Error bars represent the SEM.
OGG1 protein mobility is reduced after global 8-oxoG KBrO3 induction
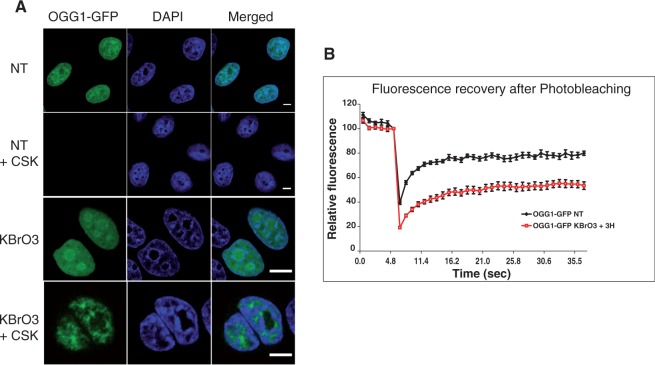
Although microirradiation is a powerful tool to assess the sequence of DNA repair proteins’ recruitment to the site of damage, this technique induces a high local concentration of damage in the nucleus and does not allow to address the question of how a cell responds to an oxidative stress resulting in the induction of 8-oxoG all over the genome and under more physiological conditions. For that purpose, we decided to treat cells with the carcinogenic agent KBrO3 that is known to induce oxidative stress in eukaryotic cells, resulting in the abundant formation of 8-oxoG in genomic DNA. Other DNA lesions such as SSBs, abasic sites and endonuclease III sensitive sites are formed, albeit in a smaller proportion (35). We have previously shown that after exposing cells to KBrO3, OGG1 and other proteins of the BER pathway, such as APE1 and XRCC1, are recruited to a detergent-resistant protein fraction, suggesting their association with chromatin in order to perform repair [(21); Figure 3A]. To confirm that resistance of OGG1-GFP to CSK extraction after KBrO3 treatment was indeed reflecting transient immobilization of this protein, we used single-cell in vivo FRAP to measure protein mobility (36). We compared the dynamics of OGG1 in KBrO3-treated cells 3 h after removal of the genotoxic agent, a time where we observed the recruitment of the glycosylase to chromatin, with that in untreated ones. Fluorescence recovery was clearly slower in bromate-treated cells indicating an immobilization of the protein likely to reflect the transient binding of OGG1 to chromatin during repair (Figure 3B).
Figure 3.
Immobilization of OGG1 after KBrO3 treatment. (A) Distribution patterns of OGG1-GFP in untreated cells (NT) and cells treated with 40 mM of KBrO3 for 30 min. After the treatment, cells were allowed to recover in DMEM at 37°C for 3 h. When indicated, soluble proteins were removed by washing with CSK buffer prior to fixation as described in ‘Materials and Methods’ section. Scale bar, 5 µm. (B) Mobility of OGG1-GFP in NT cells and in cells incubated for 3 h in DMEM after treatment with KBrO3. The mobility of the protein was determined by bleaching a region of the nucleus and measuring the subsequent recovery of fluorescence. Experiments were repeated at least three times and the average of 10 cells from a representative experiment were used to generate graphs. Error bars represent the SEM.
OGG1 and XRCC1 subcellular re-localization after global oxidative DNA damage is PARP-independent
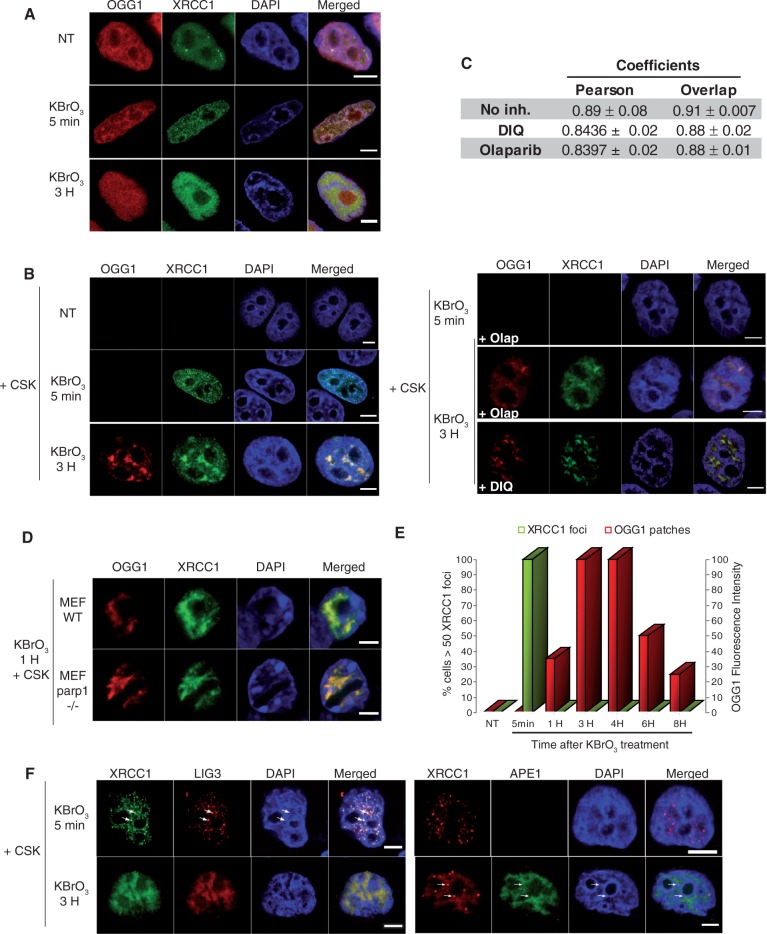
The recruitment of XRCC1 to SSBR has been largely documented. During the few minutes following damage induction by MMS or H2O2, XRCC1 is assembled in small and bright foci (1,27). The rapid and transient formation of PARP1-dependent XRCC1 foci is consistent with SSBR kinetics (1). In order to evaluate the requirement of PARP1 for the recruitment of OGG1 and XRCC1 to BER in KBrO3-treated cells, OGG1-DsRED and XRCC1-GFP were co-expressed and their localization followed in the presence or absence of PARP inhibitors. Similar experiments were carried out in parp1−/− MEFs cell lines. In untreated cells, both proteins were homogeneously distributed in the nucleoplasm and remained soluble as indicated by the fact that they were washed out when cells were treated with detergent prior to fixation (Figure 4A). After KBrO3 treatment, cells were incubated in DMEM in order to allow the assembly of repair complexes. Five minutes after the end of the treatment, hundreds of nuclear XRCC1 foci resistant to detergent extraction were detected in all cells, while no recruitment of OGG1-DsRed was observed at that early time point (Figure 4A and B). These foci rapidly disappeared and recovered basal levels after 1 h, with ∼20% of cells presenting just a few XRCC1 foci. This rapid XRCC1 foci formation is likely to reflect the recruitment of the protein to SSB. Indeed, the XRCC1 foci partially co-localized with the PAR polymer, a characteristic of SSBR, while they did not co-localize with gammaH2AX (Supplementary Figure S3), widely used as a marker for DNA double-strand breaks. Furthermore and most importantly, as expected for SSBR, inhibition of PARP activity by either Olaparib or DIQ resulted in the abolishment of XRCC1 foci formation [Figure 4B; (2)]. Taken together, these results confirm that the XRCC1 foci detected at early times after KBrO3 treatment correspond in their majority to the recruitment of the protein to SSBR.
Figure 4.
Distinct subcellular re-localizations of SSBR and BER proteins after global oxidative DNA damage. (A) HeLa cells expressing XRCC1-GFP and OGG1-DsRED were treated with KBrO3 for 30 min, washed and allowed to recover in DMEM for 5 min or 3 h. (B) 5 min or 3H after KBrO3 treatment, in the presence or absence of PARP inhibitors Olaparib or DIQ, cells were washed with CSK buffer and fixed. (C) As indicators of co-localization, Pearson and Overlap correlation coefficients between XRCC1 (green) and OGG1 (red) signals were determined. Values represent the average of coefficients calculated for 10 cells ± SEM. The Kruskal and Wallis statistical test indicates that there is no significant difference in the absence or presence of PARP inhibitors (DIQ or Olaparib). (D) Co-localization of OGG1 and XRCC1 in the CSK resistant fraction in parp1−/− MEF cells after KBrO3 treatment. (E) Kinetics of XRCC1 foci and OGG1 patches formation after KBrO3 treatment. The percentage of cells presenting >50 foci detected without CSK pre-extraction is represented after substracting those present in non-treated cells (green). Fluorescence intensity was used to quantify OGG1-GFP patches after CSK treatment. (F) HeLa cells expressing XRCC1-GFP and LIG3-RFP or APE1-GFP and XRCC1-DsRED monomer were treated with KBrO3 as above and allowed to recover in DMEM for 5 min or 3 h and washed with CSK before fixation. Arrows indicate XRCC1 foci in which LIG3 but not APE1 proteins could be detected. A good co-localization of both proteins with XRCC1 could be observed 3 h after KBrO3 treatment. Scale bar, 5 µm.
The repair of induced 8-oxoG is a much slower process, taking a few hours to the cells to complete it (21). Finding the lesion in the nuclear genome by the glycosylase is probably the limiting step as suggested by the fact that overexpression of OGG1 accelerates the in vivo repair kinetics (21,37). The correlation between the removal of 8-oxoG and the recruitment of OGG1, together with APE1 and XRCC1, to euchromatin regions (starting to be detected 30 min after the treatment and reaching its maximum at 3 h) suggests that indeed BER takes place in those nuclear regions (21). Consistently, when cells were allowed to recover for 3 h after KBrO3 treatment, the removal of soluble proteins with CSK buffer allowed us to detect the co-localization of XRCC1 and OGG1 in large nuclear regions (patches) (Figures 4B, 6B and C). The recruitment of OGG1 and XRCC1 to these nuclear domains was not affected by the presence of PARP inhibitors (Figure 4B), corroborating the microirradiation results. Indeed, the analysis of correlation coefficients indicates that there is no significant impact on the co-localization between XRCC1 and OGG1 due to the presence of PARP inhibitors (DIQ or Olaparib) indicating that PARP activity is not essential for the recruitment of XRCC1 to BER (Figure 4C). In order to verify that PARP1 protein is dispensable for the recruitment of XRCC1 to BER, parp1−/− MEF cells expressing both XRCC1-GFP and OGG1-DsRED were treated with KBrO3. While no XRCC1 foci were observed in those cells, consistent with previous reports [(1); data not shown], a good co-localization of XRCC1 and OGG1 in nuclear patches was observed 1 h after the KBrO3 treatment (Figure 4D). Taken together, the earlier results demonstrate that neither the presence of PARP1 nor its activity is required for the recruitment of XRCC1 to BER. The implication of the patches in BER was further confirmed by the detection of other repair enzymes. Indeed, Figure 4F shows that the endonuclease APE1, involved essentially in BER, was only detected in patches, whereas LIG3 could be detected in both foci and patches, consistent with its participation in both DNA repair pathways, SSBR and BER.
The distinct PARP dependences of nuclear foci and patches together with their time course of formation and disappearance (Figure 4E) strongly suggest that those structures correspond to the sites of SSBR and BER, respectively.
The BRCT1 domain of XRCC1 is required for recruitment to SSBR but not to BER
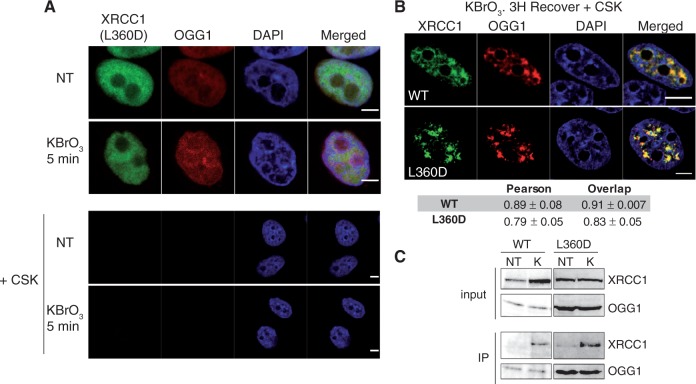
Cells were co-transfected with OGG1-DsRED and XRCC1(L360D)-GFP-expressing plasmids and treated with KBrO3 before following the protein localization by confocal microscopy. As expected for a defect in SSBR complexes assembly, no formation of XRCC1(L360D) nuclear foci was observed 5 min after the treatment (Figure 5A). However, both XRCC1 and XRCC1(L360D) co-localized with OGG1 3 h after the treatment (Figure 5B). These results are consistent with a PARP-independent recruitment of XRCC1 to BER, since, as mentioned earlier, the L360D mutation disrupts the BRCT1 domain of XRCC1 required for the interaction with PARP.
Figure 5.
BRCT1 domain is required for XRCC1 SSBR foci formation but not for its recruitment to BER and interaction with OGG1. (A) HeLa cells co-transfected with plasmids XRCC1(L360D)-GFP and OGG1-DsRED were treated with KBrO3 or not (NT) and further incubated in DMEM for 5 min. Cells were either immediately fixed (upper panel) or pre-extracted with CSK buffer prior to fixation (lower panel). (B) HeLa cells co-transfected with XRCC1-GFP or the mutant form XRCC1(L360D)-GFP and OGG1-DsRED were treated with KBrO3 and 3 h after the treatment pre-extracted with CSK prior to fixation. Pearson and Overlap correlation coefficients for XRCC1 (green) and OGG1 (red) signals were calculated. The indicated values represent the average of 10 cells ± SEM. The Kruskal and Wallis statistical test shows no significant difference. (C) HeLa cells co-transfected with OGG1-Flag and either XRCC1-GFP or the mutant form XRCC1(L360D)-GFP were treated with KBrO3 (K) or not (NT) and protein extracts were used for immunoprecipitation (IP) with anti-FLAG antibody. Immunoprecipitated proteins were analysed by western blot using either anti-FLAG (OGG1) or anti-GFP antibodies (XRCC1). The upper panels (input) show the levels of the proteins in the extracts (10% of the input). Scale bar, 5 µm.
Previous in vitro and in vivo experiments (4,8) indicated that XRCC1 recruitment to the initial steps of BER could be mediated by its interaction with the DNA glycosylases. Consistently, in co-immunoprecipitation (IP) assays with an anti-FLAG antibody performed on cells expressing OGG1-Flag and XRCC1-GFP, while almost no interaction between OGG1 and XRCC1 could be detected in untreated cells, 3 h after KBrO3 treatment both proteins co-purified (Figure 5C). This is in agreement with the idea that BER complexes are assembled in the cell in a regulated manner when repair of a particular kind of lesion is required, thus resulting in the stabilization of DNA repair proteins (38). Noteworthy, the IP experiments clearly showed that the L360D mutation in XRCC1 did not affect the interaction with OGG1, confirming that the BRCT1 is not essential for the interaction with the glycosylase (4). These results indicate that different domains of XRCC1 are required for its recruitment to SSBR and BER DNA repair pathways. While recruitment of XRCC1 to SSBR requires the BRCT1 domain, probably by its role in the interaction with PARP1, this is not the case for BER.
SSBR and BER take place in distinct nuclear sub-domains
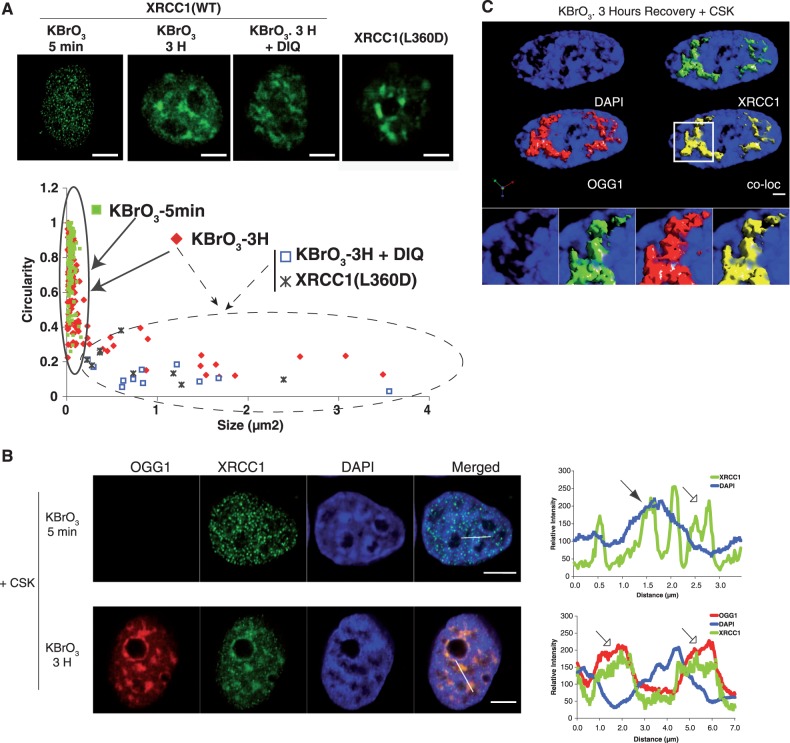
As mentioned earlier, a feature that distinguished the XRCC1 patterns observed at earlier (SSBR) or later (BER) time points after KBrO3 treatment was their difference in size and shape. Thus, while early assembled XRCC1 foci were small with an average size of 0.05 µm2 and with a circularity close to 1, patches appearing 3 h after the treatment, in which XRCC1 co-localized with OGG1, covered larger regions of the cell nucleus with areas of up to 3 µm2 and were much more heterogeneous in shape (circularity ∼0.2). When a correlation between size and shape was displayed in a scatter plot (Figure 6A), the difference between both patterns is clearly distinguishable with larger domains detected only at later time points. Although a few XRCC1 small foci can still be detected 3 h after the KBrO3 treatment, only the larger patches with low circularity were detected in the presence of the PARP-inhibitor DIQ or with the XRCC1 mutant L360D (area within the dashed line in the graph). This observation suggests that patches did not result from the association of smaller ones, since the latter are not formed in the presence of PARP inhibitors. Therefore, different XRCC1 localization patterns (foci and patches) are likely to correspond to the recruitment of the protein to different DNA repair pathways (namely, SSBR and BER). The fact that 1 h after the treatment the percentage of cells presenting XRCC1 foci reached the basal levels observed in NT cells suggests that foci detected at later time points may correspond to spontaneously induced SSB or to the presence of the protein in replication foci (39).
Figure 6.
Visualization of nuclear sub-domains involved in SSBR and BER. (A) HeLa cells expressing XRCC1-GFP or the mutant XRCC1(L360D) were treated with KBrO3 for 30 min and allowed to recover in DMEM for 5 min or 3 h in the presence or absence of the PARP1 inhibitor DIQ. Cells were extracted with CSK buffer prior to fixation. Scatter plots based on size and circularity of the XRCC1-GFP signal are shown. Two patterns can be distinguished: small foci with high circularity (continuous line), and bigger patches with very low circularity (dashed line). (B) Plot profile of fluorescence intensities of DAPI, XRCC1-GFP and OGG1-DsRED, 5 min (upper panel) or 3 h (lower panel) after KBrO3 treatment and CSK pre-extraction were generated through the indicated lines in the image. Weak (hollow arrow) and intense (filled arrow) DAPI stained regions are indicated. (C) Three-dimensional image reconstruction with isosurface rendering for OGG1 and XRCC1, showing enrichment of the proteins in less condensed DAPI regions of the nucleus. The co-localization panel (co-loc) shows in yellow only pixels in which both red and green signals are detected. Pearson’s correlation coefficient between OGG1 and XRCC1 was of 0.95. Scale bar, 5 µm.
Another major difference between SSBR- and BER-associated recruitment of XRCC1 is to be found with respect to the chromatin domains where it takes place. As shown in Figure 6B, while 3 h after exposure to KBrO3 XRCC1 patches co-localizing with OGG1 were excluded from highly DAPI-stained heterochromatin regions (lower panel), at earlier time points, when only PARP1-dependent XRCC1 foci were observed, foci were distributed in both weak (empty arrow) and intense (full arrow) DAPI-stained regions (Figure 6B, upper panel). Previous work showed by using euchromatin- and heterochromatin-specific markers as well as by chromatin fractionation that 3 h after treating the cells with KBrO3 OGG1 becomes insoluble and is completely excluded from heterochromatin-rich regions (21). The 3D reconstruction presented in Figure 6C shows a perfect co-localization of OGG1 and XRCC1 3 h after the treatment in regions of the nucleus presenting a low-density DAPI signal. Neither OGG1 nor XRCC1 were detected in the nucleoli after CSK pre-extraction. It is important to note that the described exclusion of OGG1 from heterochromatin in KBrO3-treated cells corresponds to the CSK-resistant fraction of the protein. This is not to say that the protein cannot be localized to heterochromatin, in fact, and as expected for a protein with high affinity for DNA, OGG1 is enriched in heterochromatin regions as well as in mitotic chromosomes (Supplementary Figure S4), but no signal is detected in those regions after removal of soluble proteins.
These observations indicate that the assembly of repair complexes involved in SSBR and BER takes place in distinct regions of the cell nucleus and suggest that while SSBR can process strand breaks present in either eu- or heterochromatin regions, the repair of damaged bases is preferentially achieved in euchromatin.
Nuclear distribution of 8-oxoG after global induction of the lesion
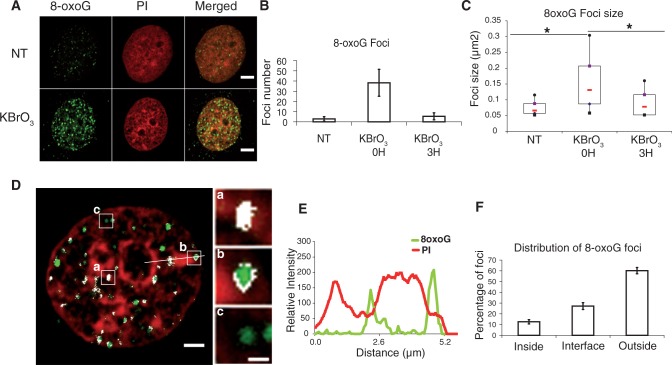
Earlier results raised the question of the distribution of induced 8-oxoG within the genome. The use of an antibody against 8-oxoG on mitotic chromosomes had revealed that the lesion is not distributed homogeneously through the genome (40). Using the same antibody in interphase cells, we found that, indeed, after KBrO3 treatment, the 8-oxoG signal was not homogeneous throughout the nucleus (Figure 7A). Distinct and bright foci were observed soon after treating cells, while very few and smaller foci were detected in non-treated cells, probably reflecting endogenous damage. Noteworthy, 3 h after the treatment, a time where the repair rate for 8-oxoG is at its maximum in OGG1-overexpressing cells (21), the number of foci and their size were greatly reduced (Figure 7B and C), reflecting repair of the lesion. Analysis of 3D images revealed that at early times after KBrO3 treatment, even though a few foci were detected inside heterochromatin domains indicating that the antibody can have access to the lesion even when chromatin is highly compacted, most of them were excluded from heterochromatin or found at the interface in between euchromatin and heterochromatin domains (Figure 7D and F). The Plot profile presented in Figure 7E shows an example of 8-oxoG foci situated at the interface of heterochromatin domains.
Figure 7.
Distribution of 8-oxoG induced by KBrO3 treatment. (A) HeLa cells overexpressing the glycosylase OGG1-GFP, treated with KBrO3 or not (NT) were immunostained with anti-8oxoG antibody. Scale bar, 5 µm. (B) Foci number and (C) size in at least 50 cells were quantified using ImageJ. *P < 0.05 using Kruskal and Wallis test. (D) Distribution of 8-oxoG foci with respect to heterochromatin domains labelled with propidium iodide (scale bar, 2 µm), representative foci located inside (a), at the interface (b) or outside (c) of the heterochromatin domains are shown (scale bar, 0.5 µm). Pixels in which green and red signals co-localize are depicted in white. (E) Plot profile of 8-oxoG (green) and PI (red) signals along the line shown in (D), showing the localization of 8-oxoG foci at the periphery of heterochromatin domains. (F) 8-oxoG foci located inside, at the interface and outside heterochromatin domains, were counted in 3D images using the ImageJ software (10 cells were used for the analysis).
The denaturing conditions required for detection of 8-oxoG preclude the simultaneous localization of proteins by either GFP fusion fluorescence or immunofluorescence. However, comparison of Figures 6B and 7D shows that 8-oxoG and BER proteins OGG1 and XRCC1 coincide in the nuclear space, being mostly excluded from heterochromatin regions and enriched in euchromatin.
DISCUSSION
Our results revealed specific characteristics of the assembly of BER and SSBR complexes within the cell nucleus. Using either laser microirradiation, in the presence or absence of a photosensitizer, or KBrO3 to induce localized and global damage, respectively, we unveiled differences between the recruitment of XRCC1 to either BER or SSBR based on the PARP1 dependence, the kinetics of recruitment to and the sub-nuclear distribution of repair sites (Table 1).
Table 1.
Different XRCC1 patterns are observed in KBrO3-treated cells
| Kinetics | Size | Circularity | PARP dependent | Co-localization with OGG1 | Excluded from HC | |
|---|---|---|---|---|---|---|
| SSBR | Fast (0–5′) | 0.05 μm2 | 1 | Yes | No | No |
| BER | Slow (max 3H) | 3 µm2 | 0.2 | No | Yes | Yes |
Kinetics of formation, morphology, PARP dependency, co-localization with the BER protein OGG1 or exclusion from heterochromatin (HC) domains allows to distinguish XRCC1 participation in SSBR or BER.
PARP-independent recruitment of XRCC1 to BER
Because it is clear that PARP1 is required for the recruitment of XRCC1 to SSBR, it has been postulated that XRCC1 participation in BER also requires PARP1, which would itself be recruited to the SSB intermediate during the repair of damaged bases (41). Others have proposed that PARP1 is recruited to BER through its direct interaction with a glycosylase and in that way it allows recruitment of XRCC1 (42). However, there is also evidence that efficient BER can take place in the absence of PARP-1 (43) and that in most cases the repair of damaged bases or abasic sites can be accomplished in a single, PARP-independent, coordinated step (33,44). Here, by coupling of 405-nm laser microirradiation with a photosensitizer allowing the local induction of 8-oxoG, we show that PARP1 is dispensable for the recruitment to BER of both, the initiating DNA glycosylase, a result in agreement with that found for NTH1 (25) and, more importantly, XRCC1. The same PARP1-independent recruitment of BER proteins is observed after non-targeted induction of 8-oxoG by KBrO3. The PARP1 independence of XRCC1 recruitment was confirmed by the use of the L360D XRCC1 mutant. Indeed, this mutant leads to an unstructured BRCT1 domain that impairs the interaction of XRCC1 with PARP1 (34) and consequently impedes its recruitment to SSBR but not to BER since it preserves its capacity to interact with the DNA glycosylase (Figure 5). This, together with the higher recruitment of XRCC1 in situations in which OGG1 is overexpressed (Supplementary Figure S2B), is in agreement with the proposal that XRCC1 recruitment to BER is normally achieved through its interaction with the BER-initiating enzymes, DNA glycosylases or APE1 (4,8,9,45).
Our results therefore support the view that during BER, PARP1 is not normally essential (33,44). We cannot rule out, however, that in situations where unusual intermediates such as modified DNA ends are generated during BER, a persistent SSB could be formed, requiring in that case PARP1 intervention to recruit other factors necessary to complete the repair process (46).
The bimodal kinetics of XRCC1 recruitment reflects rapid SSBR and slower BER
Induction of localized DNA damage through the use of laser microirradiation is an invaluable tool for following the recruitment of DNA repair proteins in vivo. However, it remains an invasive technique, and several studies have reported a local expansion in chromatin structure after microirradiation (18,47). Depending on the kind of treatment used to induce DNA damage (drugs and laser irradiation), the type of lesion induced, and therefore the DNA repair pathway analysed, the kinetics of recruitment of DNA repair proteins to the sites of repair can vary from a few seconds to a few hours (25,48–50). When lesions are induced genome-wide, XRCC1, which participates both in SSBR and in BER, is recruited within minutes to SSB (1,2,26), in good correlation with the repair kinetics for this kind of lesion. Moreover, the absence of XRCC1 foci is associated with a SSBR defect probably due to the lack of efficient recruitment of the downstream activities (25), so it is generally assumed that the XRCC1 foci observed after genotoxic treatment of cells correspond to sites of SSBR. After treatment with KBrO3, the PARP-independent patches where OGG1 and XRCC1 co-localize appear much later than the SSBR foci (Figure 4), in agreement with the repair kinetics of the oxidized guanine (21). The slow recruitment kinetics observed after global induction of 8-oxoG with KBrO3 contrasts with the very fast recruitment of OGG1 (detectable after 20 s) to the 8-oxoG induced by microirradiation. The huge amount of 8-oxoG induced in a reduced nuclear space, as well as the local chromatin relaxation triggered by the laser microirradiation technique, probably facilitate the recognition of the damage by the glycosylase. However, the distribution of the lesion all over the genome, as it is the case after KBrO3 treatment, and the fact that 8-oxoG does not generate a distortion of the DNA double helix suggest that the finding of the lesion within the chromatin structure is a hard task for the DNA glycosylase. It is accepted that the initial recognition of the damage by the DNA glycosylase is the limiting step in BER (21,33,37,51). The later steps of the pathway would then be, as in the case of SSBR, very rapid to avoid the accumulation of toxic DNA intermediates (3).
BER takes place in low-density chromatin regions
We described here two very distinct patterns of XRCC1 localization in response to KBrO3. The first one, consisting of bright circular foci, is observed a few minutes after DNA damage induction and is PARP-dependent, characteristics of the sites of SSBR (1,2). These foci are found in both eu- and heterochromatin domains (Figure 5B, upper panel), indicating that at least the first steps of SSBR can take place in heterochromatin. This is consistent with the role of PARP1 in providing a flexible and reversible mechanism for chromatin relaxation allowing the repair of SSBs in the heterochromatic regions of the genome (7). Similarly, DNA damage induced by ion-irradiation in living cells, likely to comprise both SSB and DSB, results in a fast recruitment of XRCC1 to both euchromatin and heterochromatin, thus independently of chromatin density (18).
Our results show that the situation for BER is different. Indeed, the large patches where XRCC1 co-localizes with OGG1, observed at later times after the KBrO3 treatment, are excluded from heterochromatin (Figure 5). This, together with the fact that the 8-oxoG lesions are detected mainly in euchromatin regions or at the interface with heterochromatin domains, suggests that BER takes place preferentially in less compacted chromatin regions. In contrast with the very well defined and bright 8-oxoG foci observed after KBrO3 treatments, BER proteins were instead accumulated in larger regions of the nucleus. Moreover, a mutant form of OGG1 that is not able to recognize the 8-oxoG is also recruited to the same nuclear regions (21), suggesting that the 8-oxoG itself is not the signal for the recruitment of BER proteins to euchromatin regions. Although in vitro studies show that DNA glycosylases are able to diffuse along DNA (52) and suggest that those enzymes regularly interrogate DNA by flipping the bases out of the double helix in order to find the damage (53), the efficiency with which lesions are repaired in the nuclear context together with the pattern of BER complexes unveiled here suggest that the repair takes place in open chromatin domains. The mechanisms driving the relocalization of the proteins to these regions remain to be elucidated.
The enrichment of the damaged base in euchromatin was unexpected. It could be presumed that some genome regions are more sensitive to oxidation. It has been previously suggested that DNA compacted in heterochromatin would be protected against oxidation due to the presence of histones that may trap reactive oxygen species (54). However, it was shown that the same levels of 8-oxoG were induced after KBrO3 treatment in conditions in which chromatin is condensed by hypotonic stress (21), suggesting that oxidized base lesions can be generated even in highly condensed chromatin. Therefore, in analogy with the finding that DSB initially generated and processed in heterochromatin are re-localized to the periphery of heterochromatin (17,18), it is tempting to speculate that the 8-oxoG is dynamically relocalized to euchromatin regions or to the interspace between euchromatin and heterochromatin, precisely where BER proteins accumulate, thus facilitating repair of the damage. It has recently been shown that DSB clustering occurs rapidly after irradiation, suggesting the existence of DNA repair centers in which the repair machinery would be assembled (55). The fact that the 8-oxoG foci detected are bigger after KBrO3 treatment supports the idea of lesion clustering. The mechanism driving the movement of damaged DNA to the periphery of heterochromatin is not yet clear and both enzymatic and physical mechanisms, the last one involving local relaxation of chromatin around the damage, have been evoked for other kinds of lesions (17,18). On the other hand, we cannot rule out the possibility that base lesions in heterochromatin are simply less efficiently repaired. This possibility would imply preferential repair of active genome regions. In support of such a scenario, a recent study showed, through analysis of the correlation between single-nucleotide variations and genetic or epigenetic features across cancer genomes, that somatic mutation rates consistently accumulate in heterochromatin-like domains and are reduced in open chromatin regions (56). The reduced efficiency of DNA repair in heterochromatin due to limited access of BER complexes provides a possible explanation for this observation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
Association pour la Recherche contre le Cancer [SFI20111203981 to J.P.R.]; CEA Radiobiology Programme [to J.P.R. and A.C.] and Marie Curie [PIEF-GA-254858 to H.M.]. Funding for open access charge: Commissariat à l’Energie Atomique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Bernd Epe and Eva Moritz (University of Mainz), Vincent Pennaneach (Institut Curie) and the members of our laboratory for fruitful discussions. We also thank the IRCM microscopy facility and Heinrich Leonhardt (University of Munich) for kindly sending us the LIG3-RFP plasmid.
REFERENCES
- 1.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair. 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 4.Marsin S, Vidal AE, Sossou M, Menissier-de Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J. Biol. Chem. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 5.Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 6.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat. Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 7.Caldecott KW. Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair. 2007;6:443–453. doi: 10.1016/j.dnarep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Campalans A, Marsin S, Nakabeppu Y, O’Connor TR, Boiteux S, Radicella JP. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair. 2005;4:826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair. 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vermeulen W. Dynamics of mammalian NER proteins. DNA Repair. 2011;10:760–771. doi: 10.1016/j.dnarep.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 13.Costes SV, Ponomarev A, Chen JL, Nguyen D, Cucinotta FA, Barcellos-Hoff MH. Image-based modeling reveals dynamic redistribution of DNA damage into nuclear sub-domains. PLoS Comput. Biol. 2007;3:e155. doi: 10.1371/journal.pcbi.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 16.Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA Repair. 2010;9:1273–1282. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakob B, Splinter J, Conrad S, Voss KO, Zink D, Durante M, Lobrich M, Taucher-Scholz G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011;39:6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Mol. Cell. Biol. 2007;27:5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menoni H, Shukla MS, Gerson V, Dimitrov S, Angelov D. Base excision repair of 8-oxoG in dinucleosomes. Nucleic Acids Res. 2012;40:692–700. doi: 10.1093/nar/gkr761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amouroux R, Campalans A, Epe B, Radicella JP. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res. 2010;38:2878–2890. doi: 10.1093/nar/gkp1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 23.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godon C, Cordelieres FP, Biard D, Giocanti N, Megnin-Chanet F, Hall J, Favaudon V. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanssen-Bauer A, Solvang-Garten K, Sundheim O, Pena-Diaz J, Andersen S, Slupphaug G, Krokan HE, Wilson DM, 3rd, Akbari M, Otterlei M. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ. Mol. Mutagen. 2011;52:623–635. doi: 10.1002/em.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lan L, Nakajima S, Komatsu K, Nussenzweig A, Shimamoto A, Oshima J, Yasui A. Accumulation of Werner protein at DNA double-strand breaks in human cells. J. Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- 29.Will O, Gocke E, Eckert I, Schulz I, Pflaum M, Mahler HC, Epe B. Oxidative DNA damage and mutations induced by a polar photosensitizer, Ro19-8022. Mutat. Res. 1999;435:89–101. doi: 10.1016/s0921-8777(99)00039-7. [DOI] [PubMed] [Google Scholar]
- 30.Soriano FG, Virag L, Szabo C. Diabetic endothelial dysfunction: role of reactive oxygen and nitrogen species production and poly(ADP-ribose) polymerase activation. J. Mol. Med. 2001;79:437–448. doi: 10.1007/s001090100236. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell. Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 32.Yelamos J, Farres J, Llacuna L, Ampurdanes C, Martin-Caballero J. PARP-1 and PARP-2: new players in tumour development. Am. J. Cancer Res. 2012;1:328–346. [PMC free article] [PubMed] [Google Scholar]
- 33.Strom CE, Johansson F, Uhlen M, Szigyarto CA, Erixon K, Helleday T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011;39:3166–3175. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballmaier D, Epe B. DNA damage by bromate: mechanism and consequences. Toxicology. 2006;221:166–171. doi: 10.1016/j.tox.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 36.Houtsmuller AB, Vermeulen W. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem. Cell Biol. 2001;115:13–21. doi: 10.1007/s004180000234. [DOI] [PubMed] [Google Scholar]
- 37.Hollenbach S, Dhenaut A, Eckert I, Radicella JP, Epe B. Overexpression of Ogg1 in mammalian cells: effects on induced and spontaneous oxidative DNA damage and mutagenesis. Carcinogenesis. 1999;20:1863–1868. doi: 10.1093/carcin/20.9.1863. [DOI] [PubMed] [Google Scholar]
- 38.Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, Dianov GL. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell. 2008;29:477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 39.Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM., 3rd XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohno M, Miura T, Furuichi M, Tominaga Y, Tsuchimoto D, Sakumi K, Nakabeppu Y. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 2006;16:567–575. doi: 10.1101/gr.4769606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caldecott KW. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 42.Noren Hooten N, Kompaniez K, Barnes J, Lohani A, Evans MK. Poly(ADP-ribose) polymerase 1 (PARP-1) binds to 8-oxoguanine-DNA glycosylase (OGG1) J. Biol. Chem. 2011;286:44679–44690. doi: 10.1074/jbc.M111.255869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vodenicharov MD, Sallmann FR, Satoh MS, Poirier GG. Base excision repair is efficient in cells lacking poly(ADP-ribose) polymerase 1. Nucleic Acids Res. 2000;28:3887–3896. doi: 10.1093/nar/28.20.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol. Oncol. 2011;5:387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chou WC, Wang HC, Wong FH, Ding SL, Wu PE, Shieh SY, Shen CY. Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. EMBO J. 2008;27:3140–3150. doi: 10.1038/emboj.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 47.Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagy Z, Soutoglou E. DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol. 2009;19:617–629. doi: 10.1016/j.tcb.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 50.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8: 213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 51.Cappelli E, Degan P, Frosina G. Comparative repair of the endogenous lesions 8-oxo-7, 8-dihydroguanine (8-oxoG), uracil and abasic site by mammalian cell extracts: 8-oxoG is poorly repaired by human cell extracts. Carcinogenesis. 2000;21:1135–1141. [PubMed] [Google Scholar]
- 52.Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39:7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen L, Haushalter KA, Lieber CM, Verdine GL. Direct visualization of a DNA glycosylase searching for damage. Chem. Biol. 2002;9:345–350. doi: 10.1016/s1074-5521(02)00120-5. [DOI] [PubMed] [Google Scholar]
- 54.Ljungman M, Hanawalt PC. Efficient protection against oxidative DNA damage in chromatin. Mol. Carcinog. 1992;5:264–269. doi: 10.1002/mc.2940050406. [DOI] [PubMed] [Google Scholar]
- 55.Neumaier T, Swenson J, Pham C, Polyzos A, Lo AT, Yang P, Dyball J, Asaithamby A, Chen DJ, Bissell MJ, et al. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proc. Natl Acad. Sci. USA. 2012;109:443–448. doi: 10.1073/pnas.1117849108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.