Abstract
The 3′-untranslated regions of many plant viral RNAs contain cap-independent translation elements (CITEs) that drive translation initiation at the 5′-end of the mRNA. The barley yellow dwarf virus-like CITE (BTE) stimulates translation by binding the eIF4G subunit of translation initiation factor eIF4F with high affinity. To understand this interaction, we characterized the dynamic structural properties of the BTE, mapped the eIF4G-binding sites on the BTE and identified a region of eIF4G that is crucial for BTE binding. BTE folding involves cooperative uptake of magnesium ions and is driven primarily by charge neutralization. Footprinting experiments revealed that functional eIF4G fragments protect the highly conserved stem–loop I and a downstream bulge. The BTE forms a functional structure in the absence of protein, and the loop that base pairs the 5′-untranslated region (5′-UTR) remains solvent-accessible at high eIF4G concentrations. The region in eIF4G between the eIF4E-binding site and the MIF4G region is required for BTE binding and translation. The data support the model in which the eIF4F complex binds directly to the BTE which base pairs simultaneously to the 5′-UTR, allowing eIF4F to recruit the 40S ribosomal subunit to the 5′-end.
INTRODUCTION
The efficiency of translation initiation is dictated by the sequence and structural elements in the mRNA, primarily in the untranslated regions (UTRs) (1,2). A central role in recognition and interaction with these elements is played by eIF4F, which, in plants, is a heterodimer composed of the small cap-binding subunit, eIF4E and the large scaffold protein, eIF4G. In animals, eIF4F is a heterotrimer consisting of eIF4E, eIF4G and the helicase, eIF4A. In plants, eIF4A may play a similar role as in animals but it does not co-purify with eIF4F (3,4). Cellular mRNAs contain a 5′-m7GpppN cap structure that stabilizes the mRNA and assists in recruitment of the 43S ribosomal pre-initiation complex to the mRNA through binding to eIF4E (5,6). eIF4G further stabilizes this eIF4E-cap interaction by binding to the mRNA directly. The 3′-UTR, like its 5′ counterpart, affects translatability and stability, being adjacent to the poly(A) tail, which is recognized by poly(A)-binding protein (PABP). Both eIF4E and PABP interact with eIF4G, and these interactions circularize the mRNA and enhance recruitment of the ribosomal 40S subunit (7–9).
eIF4G is the central platform for interaction with initiation factors. It is a large (160 kDa), modular protein composed of a long N-terminal domain, containing the PABP-binding site, followed by the eIF4E-binding site. The C-terminal domain of eIF4G contains the key functional region required for scanning, MIF4G, which contains three HEAT repeats (10–12). MIF4G is conserved among all known eIF4Gs and interacts with eIF4A, the mRNA and eIF3 (13,14). The second HEAT domain, MA3, which is the eIF4A-binding site (14,15) is conserved in eIF4Gs in the animal and plant kingdoms, but is absent in yeast. The last HEAT domain, W2, occurs only in mammalian eIF4G and binds Mnk kinase (11,16).
In addition to the eIF4F homolog shared across kingdoms, plants contain an additional isoform, eIFiso4F, a dimer composed of eIFiso4G and eIFiso4E. eIFiso4F is less efficient at facilitating translation initiation, but 10-fold more abundant in the cell (17). eIFiso4G has a similar organization to the conserved domains of eIF4G (14,18). However, at 86 kDa, it is more compact and lacks many of the non-conserved regions of eIF4G to which no functions have been mapped.
The RNA genomes of many plant viruses contain cap-independent translation elements (CITEs) that differ from mammalian internal ribosome entry sites (IRESs) in that they are located in the 3′-UTR, rather than at the 5′-UTR. Moreover, translation of these mRNAs requires scanning from the 5′-end (19–21). These CITEs fall into about seven distinct classes with little conservation of sequence, secondary structure or the component of the eIF4F complex with which they interact (22,23). Among these elements is the well-characterized and highly structured CITE of barley yellow dwarf virus (BYDV). The BYDV 3′ CITE and BYDV-like translation elements (BTEs) from other viruses show remarkable diversity of secondary structure with only a few conserved features. The BTEs have three to six helices protruding from a central hub. Around this hub are a number of non-conserved but essential non-Watson–Crick paired bases that link the radiating helices (19,24–26). All BTEs contain a 17-nt conserved sequence (17 CS) that conforms to the consensus, GGAUCCUGGGAAACAGG. This sequence encompasses stem–loop I (SL-I, underlined bases are paired). BTEs also have a stable stem–loop (SL-III in BYDV) capable of base pairing to complementary sequences in the viral 5′-UTR—with the apparent exception of the BTE of red clover necrotic mosaic virus RNA 1 (RCNMV1) (27).
All tested CITEs have been found to bind a component of the translation initiation machinery. The BYDV BTE binds and requires eIF4F (28). More precisely, the full-length eIF4G subunit or a C-terminal fragment of eIF4G lacking the 4E-binding site stimulated translation of uncapped reporter mRNA containing the 3′-BTE, whereas the eIF4E subunit enhanced the stimulatory activity and BTE-binding affinity of eIF4G (28). However, the precise mechanism of eIF4F binding to the BYDV BTE remains to be determined. Furthermore, the interactions of other BTEs with initiation factors have not been studied.
As the BTE function is tied to its three-dimensional structure, we sought to understand how the BTE folds and utilizes its structural features to recognize and bind to eIF4G and direct it to the 5′-end where initiation ensues. Using structure probing, native (non-denaturing) polyacrylamide gel electrophoresis (PAGE) and footprinting methods, we show that BTE folding involves cooperative uptake of magnesium ions driven primarily by charge neutralization, and this folding is required for function. We also identify an essential region in eIF4G that interacts with all tested BTEs and the eIF4G-binding site as the conserved stem–loop I and an internal loop downstream. The overall fold of the BTE was unaffected by eIF4G fragment binding, indicating that the BTE forms a stable scaffold for protein factor binding prior to protein recognition.
MATERIALS AND METHODS
Plasmids
Preparation of the individual BTE SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) cassettes used for structure probing was described by Wang et al. (26). For trans-inhibition studies, short BTE templates containing an upstream T7 RNA polymerase promoter sequence and BYDV 4814–4918 nt, TNV-D 3573–3664 nt or RCNMV1 3590–3740 genome segments were prepared by PCR amplification. BLucB is a reporter plasmid containing the firefly luciferase gene flanked by the BYDV genomic 5′- and 3′-UTRs (29).
Plasmids pET28a_4G100 and pET28a_4G70 were constructed for expression of the truncated versions of eIF4G (p100 and p70), lacking the N-terminal 586 or 862 aa, respectively. The 2758- and 1878-nt long fragments were generated by PCR using pET3d harboring the eIF4G ORF as a template. The forward primer for both constructs (BamHI_TEV_4G-p100 and BamHI_TEV_4G-p70) includes a BamHI site in the 5′-end, followed by nucleotides coding for the tobacco etch virus (TEV) proteinase cleavage site, and in BamHI_TEV_4G-p100 sequence corresponding to nucleotides 1759–1776 on eIF4G cDNA (in bold and underlined): AAGGATCCGAAAACCTGTATTTTCAGTCTATGCCTA CTGCAGAGCTTGCC; in BamHI_TEV_4G-p70 to nucleotides 2687–2607 on eIF4G cDNA (in bold and underlined): AAGGATCCGAAAACCTGTATTTTCAGTCTATGCACAAAGCCGAGAAAAAG. Reverse primer XhoI-4G-39r contains at its 5′-end an XhoI site followed by sequence complementary to nucleotides 4464–4447 in the eIF4G ORF (in bold and underlined): AACTCGAGTTATTAAGT CAACATGAAGGCATC. The PCR product was cut with BamHI and XhoI and cloned into plasmid pET28a that had been digested with the same enzymes.
RNA synthesis and purification
SmaI-linearized BTE SHAPE cassettes and uncapped PCR-generated BTE templates were transcribed using the Megashortscript kit (Ambion). SmaI-linearized BLucB and BLucBF were transcribed using the MEGAscript kit (Ambion). Capped versions of these transcripts were produced with T7 Message mMachine (Ambion) or T7 mScript (CellScript). Cap-Luc described by Iizuka et al. (30), was linearized with EcoICRI, transcribed with the MEGAscript kit (Ambion) and post-transcriptionally capped using the T7 mScript Standard mRNA Production System (Cell Script). All transcripts were purified by phenol/chloroform extraction and ethanol precipitation. RNA concentrations were determined spectrophotometrically and integrity was verified by 0.8% agarose gel electrophoresis.
Preparation of BY-2 cell extract
Extract from evacuolated BY-2 protoplasts was prepared as described by Komoda et al. (31) with some modifications. To prepare protoplasts, cells from a 3-day-old culture (30–40 ml packed cell volume) were digested with a solution of 0.8% (w/v) Cellulase RS (Yakult, Tokyo, Japan), 0.2% (w/v) Driselase (Sigma, St. Louis, MO, USA) and 0.8% (w/v) hemicellulase in 12.5 mM NaOAc, 5 mM CaCl2, 0.37 M mannitol, pH 5.8, for 3 h. The protoplasts were overlaid on a discontinuous gradient formed from 2 ml layers of 10, 20, 30, 40 and 70% Percoll (GE Healthcare) in 0.6 M mannitol, 20 mM MgCl2 and 5 mM PIPES–KOH (pH 7.0). They were then centrifuged at 10 000g for 1 h at 25°C in an SH-7 rotor (Sorvall). The evacuolated protoplasts were collected from the 40–70% Percoll solution interface. Next, they were suspended in four volumes of 30 mM HEPES–KOH, pH 7.4, 80 mM potassium acetate, 1.8 mM magnesium acetate, 2 mM DTT, complete mini EDTA-free protease inhibitor mixture (Roche Diagnostics, Mannheim, Germany) and disrupted using a Dounce homogenizer (Wheaton Science Products, Millville, NJ, USA). Homogenate was centrifuged for 10 min at 500g at 4°C to remove nuclei. The supernatant was frozen in aliquots at −80°C until use.
Trans-inhibition of translation
Non-saturating amounts (2 nM) of uncapped BLucB transcript, pre-mixed with 400 nM of the designated viral BTEs or control 18S RNA, were translated in wge (Promega) for 1 h at 22°C as described (28). The luciferase activity was measured by addition of 2 µl of the translation reaction to 40 µl of the Luciferase Assay Reagent (Promega) followed by measurement of relative light units in a GloMaxTM20/20 luminometer. Translation in BY-2 cell extracts was performed according to Komoda et al. (31) using 20 nM of BLucB, BLucBBF or capped BLucBBF mRNAs and 2 µM of the BTEs or control human 18S rRNA.
PAGE
About 30 pmol of each of the indicated BTE four-way junction (4wj) mutant RNA were heated at 92°C for 2 min and immediately cooled on ice. To these solutions, triple dye loading buffer (National Diagnostics) was added to final 1× concentration and RNA was analyzed separately by electrophoresis on a 10% non-denaturing polyacrylamide gel (29:1 acrylamide to bisacrylamide ratio) containing 1× TBM buffer (89 mM Tris, 89 mM boric acid and 0–10 mM magnesium chloride) or 10% denaturing polyacrylamide gel (19:1 acrylamide to bisacrylamide ratio) containing 1× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA) at 4°C. RNA was electrophoresed for 4 h at 15 W and then visualized by UV trans-illumination following ethidium bromide staining.
Recombinant protein expression and purification
His-tagged eIF4E in pET23d vector was introduced into Escherichia coli (BL21 cells) and expression was induced at OD600nm = 0.8, with 100 mM IPTG. After 4 h of induction, cells were harvested from 1 l of culture by centrifugation at 10 000 g for 10 min. The cells were frozen at −80°C for at least 1 h and sonicated 12 times for 30 s each with 2 min cooling on ice in binding buffer [25 mM HEPES–KOH at pH 7.6, 100 mM KCl, 2 mM MgCl2, 10% glycerol plus 0.1 mM phenylmethyl–sulphonyl fluoride, 0.1% Soybean trypsin inhibitor and 1 tablet/10 ml of Complete protease inhibitor cocktail, EDTA-free (Roche)]. The homogenate from 1 l of cells was centrifuged at 38 000g for 20 min at 4°C and supernatant was applied to 1 ml of Ni-NTA Superflow Cartridge (Qiagen). The cartridge was washed with 10 volumes of binding buffer plus 10 mM imidazole and then with 10 volumes of binding buffer plus 20 mM imidazole. The his-tagged proteins were eluted with 250 mM imidazol in the same buffer.
Recombinant [wild-type (wt)] wheat eIF4F was expressed from dicistronic constructs in a pET3D vector harboring eIF4G and eIF4E from wheat and purified as described (32). The dicistronic plasmids were introduced into E. coli (BL21 cells) and induced with 0.1 M IPTG. Four hours post-induction, cells were harvested by centrifugation and sonicated prior to purification. The lysates were loaded onto a phosphocellulose column, followed by a m7GTP-Sepharose affinity column and finally, the protein was concentrated on a second phosphocellulose column. The proteins were dialyzed against N′-100 (25 mM HEPES–KOH at pH 7.6, 100 mM KCl, 1 mM MgCl2, 1 mM DTT) to remove excess m7GTP and concentrated on Microcon YM-10 (Amicon) with three changes of N9-100. Recombinant scaffold proteins were expressed from pET3d harboring wheat eIF4G (32) and eIFiso4G (33) and purified on a phosphocellulose column and centrifuged through Microcon YM-100 (eIF4G) or Microcon YM-50 (eIFiso4G). Expression and purification of p86 and p70 were performed as for eIF4G, followed by an additional step on 1 ml of Ni-NTA Superflow Cartridge (Qiagen) as described for his-tagged eIF4E. The purity of all proteins was verified by SDS–PAGE and coomassie brilliant blue staining and concentration determined by Bradford assay (BioRad Protein Assay).
Filter-binding assay
The binding assay was performed essentially as described previously (28,34). Briefly, 32P-labeled BTE (0.4 nM) was incubated with the indicated proteins in a final volume of 50 μl of binding buffer, 25 mM HEPES–KOH (pH 7.6), 100 mM potassium acetate, 30 mM KCl, 2 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, 50 μg/ml tRNA, 50 μg/ml poly(dI-dC) and 2.5% glycerol. Samples were filtered through nitrocellulose and Hybond N++ membranes (Amersham Bioscience) in a 96-well manifold (Schleicher and Schuell) connected to a vacuum aspirator. Both membranes were exposed to a PhosphorImager screen (Amersham Bioscience) and the intensity of obtained spots was quantified using ImageQuant software (Molecular Dynamics). The percent of bound RNA was determined by dividing the value on the nitrocellulose membrane (RNA bound to protein) by the sum of the values on the nitrocellulose (RNA bound to protein) and nylon (unbound RNA). Data were fitted using GraphPad software (GraphPad Software, Inc.). The experiment was repeated at least three times.
RNA structure probing and footprinting
Chemical and enzymatic RNA structure probing was performed as described previously (26,35), except we used benzoyl cyanide (60 mM, final concentration) as the modifying reagent. Briefly, 500 ng of refolded RNA alone or pre-incubated with indicated proteins was treated with 10% (v/v) of benzoyl cyanide (Sigma-Aldrich) and incubated for 30 s at 22°C. As a control, RNA refolded in the presence of 3 mM Mg2+ was treated with 10% (v/v) of DMSO in place of chemical reagent. RNA was then purified by phenol–chloroform extraction and ethanol precipitation. Reactions were resolved on an 8% denaturing polyacrylamide gel and dried following primer extension. Dried gels were exposed to a storage phosphor screen and the band intensity was quantified using Phoretix 1D software. Data were fitted using GraphPad software (GraphPad Software, Inc.) and Hill values were calculated using the best-fitted curve. Each experiment was repeated at least three times.
RESULTS
Cation dependence of BTE folding: perturbation of the BTE helical junction by mutagenesis destabilizes the compact state
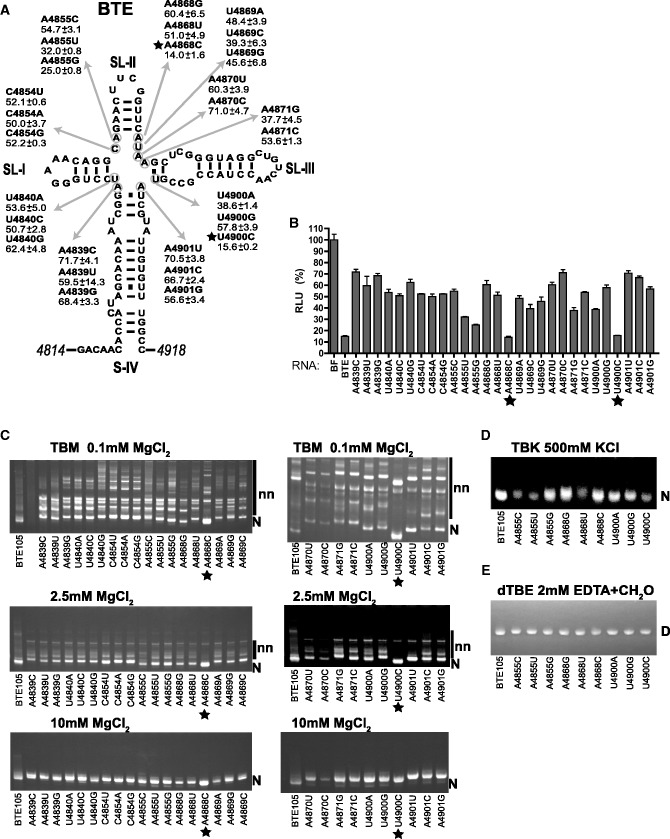
The 105-nt BYDV BTE (BTE105) was shown previously to consist of four helices joined at a central junction (Figure 1A). An RNA junction forms where two or more helices meet in space and is a major structural feature that contributes toward the overall tertiary RNA fold (36–38). Thus, we were concerned with folding at the complex 4wj in the BYDV BTE that has many unpaired bases as indicated by the chemical probing data (26), although in the absence of high-resolution BYDV BTE structure we cannot exclude possible formation of non-Watson–Crick base pairs. If cap-independent translation activity of the BTE were dependent on formation of a compact native structure, then point mutations that destabilize the compact fold would destabilize BTE function. To address this, we altered the BTE helical junction by mutating unpaired bases one at a time into each of the three remaining bases, except where the mutation would introduce a start codon. These 4wj mutants were added in trans to a wheat germ translation extract (wge) containing the very efficient reporter mRNA BlucB, which contains the 5′- and 3′-UTRs of BYDV genomic RNA (including the BTE) and tested for the ability to inhibit translation, as observed previously (19,26,28). This trans-inhibition occurs presumably by the BTE binding to and sequestering eIF4F by the excess BTE RNA. Non-functional BTE mutants would not bind eIF4F and thus not inhibit in trans. At 100-fold excess over BLucB mRNA, all tested BTE mutants reduced translation of BLucB mRNA to various levels, but only 4wj point mutants A4868C and U4900C reduced translation of BLucB mRNA as efficiently as the wt BTE (to <20%, Figure 1B), indicating that these two point mutations did not alter BTE function.
Figure 1.
Effect of magnesium chloride and mutations on BTE function and folding. (A) Relative trans-inhibition activity (±standard error) of 4wj BTE mutants mapped onto the BTE secondary structure. Stars indicate mutants that retained strong trans-inhibition activity. (B) Relative translation levels of 4 nM uncapped BlucB (reporter mRNA with BYDV UTRs flanking firefly luciferase coding sequence) in wheat germ extract (wge) containing 400 nM of the indicated 4wj mutant BTE RNAs. Luciferase expression in the presence of each of the 4wj mutants was normalized to translation of BlucB mRNA in the presence of non-functional BTE-BF (100%). Experiments were performed in triplicate and repeated three times. (C) Native gels (TBM) containing Tris–borate buffer and indicated amounts of MgCl2. (D) Native gel (TBK) containing Tris–borate buffer and 500 mM KCl. (E) Denaturing gel (dTBE) containing Tris–borate buffer and 2 mM EDTA. For details, see ‘Materials and Methods’ section.
To determine whether the function of BTE mutants correlates with the efficiency of adopting the native compact fold (N), we compared the mobilities of wt and the mutant BTEs by native PAGE in the presence of three different Mg2+ concentrations. This is a direct measure of the tendency of the BTE to adopt a single compact species. At the low (0.1 mM) Mg2+ concentration (TBM gels, Figure 1C), all RNAs adopted multiple non-native conformations (nn) manifested as dispersed bands of retarded mobility, but the wt BTE105 and two fully functional 4wj mutants, A4868C and U4900C, existed predominantly as two bands. The rapidly moving band of BTE105 and functional mutants reflects a compact, native fold and a dispersed slower moving band likely reflects fast sampling of near-native states. As the concentration of magnesium ions was increased, the number of bands (three-dimensional conformations) of wt BTE and mutants A4868C and U4900C decreased, with an increase in the dominance of the structure population by the single compact band (TBM gels, 2.5 mM MgCl2; lane 1, Figure 1C). In contrast, at 0.1 mM and 2.5 mM MgCl2, the mutants with reduced or no function showed numerous bands, indicating a preponderance of multiple conformations. At 10 mM MgCl2, all RNAs folded as a single compact band (TBM gels, Figure 1C). It is possible that addition of Mg2+ results in formation of some secondary and tertiary contacts but non-functional mutations probably had disordered local packing of the RNA backbone, inhibiting their ability to adopt the compact fold.
To distinguish between the alternative possibilities that: (i) the cation-induced BTE folding requires magnesium specifically, as expected, if coordination of Mg2+ at a specific site in the BTE is required for folding or (ii) Mg2+ simply neutralizes the negatively charged phosphate backbone to facilitate folding, we monitored BTE105 mobility on native PAGE in the presence of monovalent cations. In the absence of Mg2+, high concentrations of potassium ion (500 mM KCl) also induced the BTE to adopt a single native conformation (TBK gel, 500 mM KCl; lane 1, Figure 1D). Formation of a single conformation via either Mg2+ or K+ indicates folding driven by charge neutralization. The requirement for a significantly higher monovalent than divalent cation concentration for RNA folding is expected, as reported for the Tetrahymena group I ribozyme (39) and the Hepatitis C virus (HCV) IRES (40). This requirement stems from higher charge density of Mg2+, leading to more efficient neutralization of the negative RNA phosphate backbone charge per cation atom (41,42). We also monitored BTE105 and selected mutants folding in the presence of increasing CaCl2 and observed a similar increase in compactation of the BTE structure with increasing salt concentration (Supplementary Figure S1). Folding is not as efficient as with Mg2+ presumably owing to the larger size of Ca2+ ion and lower charge density, and its different coordination geometry, which makes it less effective at shielding phosphate charges (42).
To determine whether the differences in mobility of functional BTEs versus the non- or reduced-functional mutants result from a disrupted native state in the non-functional BTEs, BTE105 and selected mutants were electrophoresed under denaturing conditions and in the presence of the divalent cation chelating agent, EDTA. All analyzed RNAs ran as single sharp bands with the same velocity under denaturing conditions (dTBE gel, Figure 1E), indicating that retarded mobility of mutants is due to secondary and tertiary structure rather than primary structure.
Cooperativity of BTE folding in three virus genera
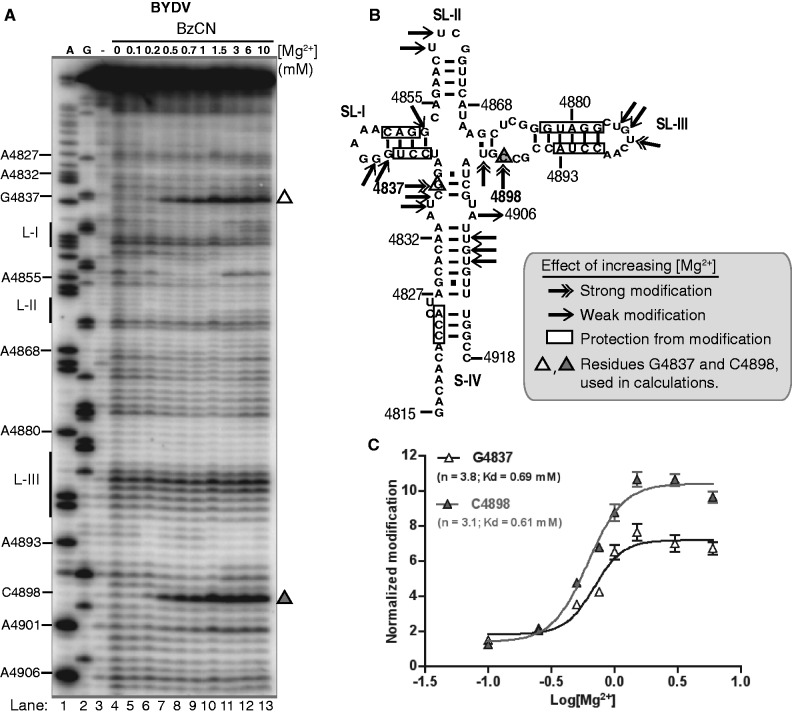
To complement the native gel analysis and further investigate the role of Mg2+ in BTE folding, we probed the BYDV BTE structure in solution using SHAPE technology. SHAPE chemistry interrogates the conformation of each nucleotide by reacting with conformationally unconstrained (unpaired) nucleotides to form a bulky 2′-O-adduct that prevents primer extension by reverse transcriptase (43,44). Specifically, we used a fast-acting reagent, benzoyl cyanide (BzCN) (45) to monitor BTE folding in 10 different Mg2+ concentrations. If BTE folding involves cooperative uptake of Mg2+ then we could expect widely separated regions of sequence to change in modification pattern (structure) as a threshold Mg2+ concentration is reached. Indeed, quantification of the observed benzoyl cyanide-induced modifications at two remote, exposed regions of the BYDV BTE sequence, universally conserved nucleotide G4837 and nucleotide C4898 (open and filled triangles, respectively, Figure 2A), showed an increase in modification at increasing magnesium concentrations. Additional Mg2+-induced reactivity changes mapped on the BTE secondary structure are summarized in Figure 2B. Calculated Hill coefficients for the modification at nucleotides G4837 and C4898 were 3.8 and 3.1, respectively, indicating cooperative binding of Mg2+ to the BTE (open and filled triangles, Figure 2C). Half-folding was observed at 0.6 mM with complete folding at 2 mM magnesium ion. Higher Mg2+ concentrations of up to 20 mM caused no additional structural rearrangements.
Figure 2.
Cation-driven BTE RNA folding. (A) The BYDV BTE modification and cleavage structure probing patterns generated using the SHAPE reagent benzoyl cyanide (BzCN) in the presence of increasing Mg2+ concentration is shown. A and G are dideoxy sequencing lanes, with positions of selected bases indicated at left. Also at left, the positions of loops L-I, L-II and L-III are indicated. Unmodified RNA (-lane) shows background RNA hydrolysis. (B) Positions of Mg2+-induced folding changes on the BTE secondary structure. (C) Hill plot of increase in modification at two widely separated residues G4837 (open triangle) and C4898 (filled triangle) as a function of [Mg2+]. Data were fitted using GraphPad software and Hill values were calculated using the best-fitting curve.
To determine whether cooperative uptake of Mg2+ upon folding is a general behavior of this class of CITEs, we also probed the structures of the tobacco necrosis virus D (TNV-D) and RCNMV1 BTEs with BzCN in the presence or absence of Mg2+. Like the BYDV BTE, all probed BTEs underwent a Mg2+-dependent conformational change at similar low 0.5–1.2 mM Mg2+ concentrations (Figure 3A). The calculated dissociation constant for Mg2+ binding to the TNV-D BTE by monitoring G3591 modification was 0.6 mM and for the G3637 modification was 0.47 mM (Figure 3B). Similarly, the calculated Mg2+ dissociation constant for modification of two widely separated regions of the RCNMV1 BTE are similar, with dissociation constants (KDs) of 1.23 mM and 1.49 mM for modification of G3608 and U3685, respectively (Figure 3B), suggesting that the BTE folds in a single step. Uptake of Mg2+ upon folding of TNV-D and RCNMV1 BTEs is moderately cooperative, with Hill coefficients for the modification at the universally conserved G residue in TNV-D (G3591) of 2.06 and RCNMV1 (G3608) of 2.84, suggesting the presence of multiple Mg2+ binding sites (Figure 3B). Together, the data indicate that all three BTEs adopt a stable, compact native fold at physiological Mg2+ concentrations in the absence of protein factors.
Figure 3.
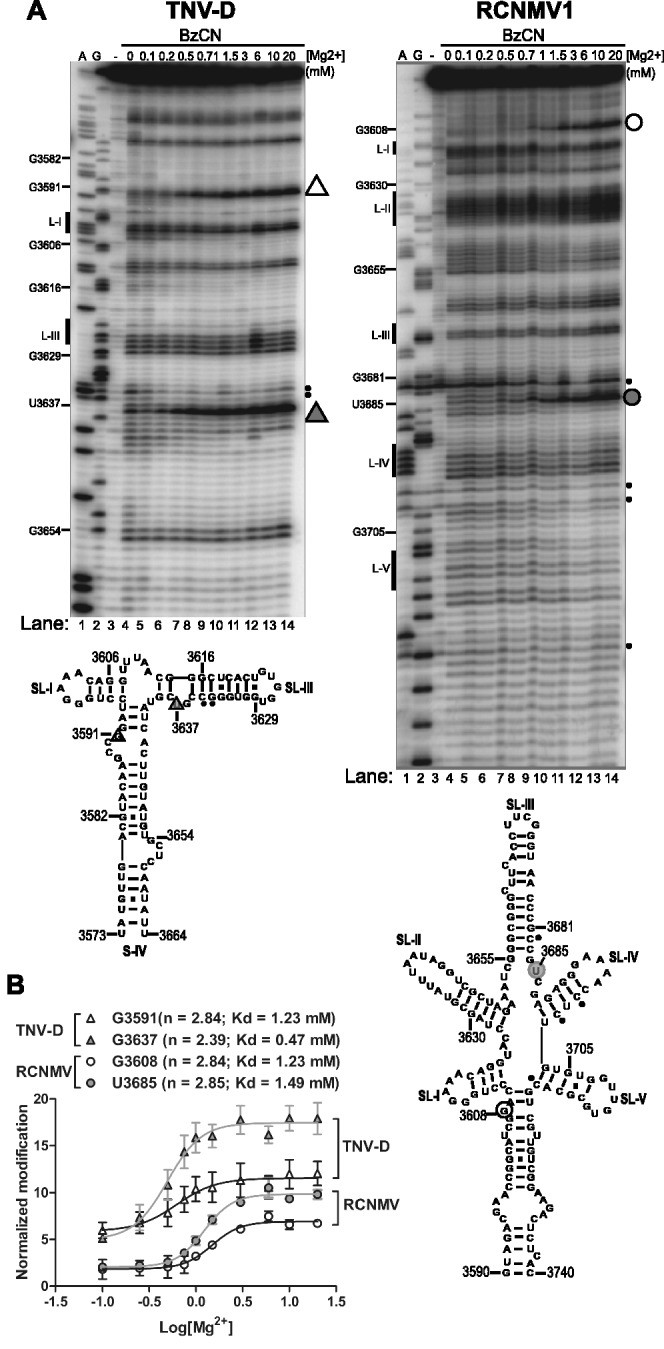
Cooperativity of BTE RNA folding in related viral genera. (A) Treatment of TNV-D and RCNMV1 BTEs with BzCN in the presence of increasing Mg2+ concentrations. A and G are dideoxy sequencing lanes. Unmodified template RNA (-lane) was not treated with BzCN prior to primer extension. Small filled circles at right denote regions of autohydrolysis in the absence of BzCN. Triangles (TNV-D) and large circles (RCNMV1) to the right of each gel indicate positions of modified nucleotides used to calculate Hill plots. (B) BzCN modification was quantified for the conserved highly modified G nucleotide: G3591 in TNV-D (open triangle) and G3608 in RCNMV1 (open circle) and at non-conserved bases G3637 in TNV-D (filled triangle) and U3685 in RCNMV1 (filled circle). Band intensity was plotted as function of log [Mg2+] as described above in Figure 2 legend.
Effect of ion concentration on BTE translation activity
The positive correlation between tendency to form compact structure and translation activity (Figure 1), leads us to predict that BTEs should be more active in translation extracts at higher ionic strength. In fact, cap-independent translation of BYDV RNA is optimal at 130–140 mM K+ (46), whereas optimal K+ concentrations for cap-dependent translation have been reported to be <80 mM (47–49). We compared the level of inhibition of cap-dependent with cap-independent translation by BYDV, TNV-D and RCNMV1 BTEs under low (53 mM) and high (133 mM) K+ concentrations, using a 100-nt fragment of 18S RNA as a negative control. As a model reporter for cap-dependent translation, we used a capped and polyadenylated non-viral mRNA encoding firefly luciferase (cap-Luc) (30) and for cap-independent translation, we used BLucB. Translation efficiencies of both reporter RNAs in the absence of inhibitor were much higher at 133 mM K+ than at 53 mM K+ (Figure 4). Under conditions of low K+, addition of a 200-fold molar excess of BYDV, TNV-D3573–3664 or RCNMV13590–3740 BTEs (numbering is according to position occupied in the viral genome) had only slight negative impact on translation of BLucB mRNA (Figure 4A), while translation of cap-Luc mRNA was inhibited significantly by all three of these elements (Figure 4B). In the presence of BYDV BTE, it dropped to 32%, whereas TNV-D and RCNMV1 elements lowered translation to 67 and 53%, respectively, compared with non-inhibited cap-Luc (Figure 4B). Interestingly, under high K+ conditions (133 mM K+), BYDV, and to a lesser extent, TNV-D BTEs exhibited stronger trans-inhibitory activity (Figure 4B). The BYDV BTE was the most efficient inhibitor and lowered translation of BLucB and cap-Luc, respectively, to 20 and 3%, while TNV-D inhibited BLucB and cap-Luc, respectively to 58 and 46% (Figure 4). The RCNMV1 BTE was the least effective inhibitor and its level of trans-inhibition of BlucB (to ∼80%) or cap-luc mRNA (∼50%) was unaffected by K+ concentration. Perhaps, this is due to its strikingly different six-helix structure. The 18S RNA did not affect BlucB translation and had only a minor impact (<20%) on translation of cap-Luc mRNA at all tested K+ concentrations (Figure 4). In summary, with the exception of the RCNMV1 BTE, higher salt concentrations tended to favor the inhibitory activity of the BTE in trans, consistent with the requirement for the ion-induced fold for BTE function. However, we cannot rule out effects of ionic strength on other components of the translational machinery as contributing to the observed results.
Figure 4.

Effect of potassium concentration on trans-inhibition of translation by the BTEs. Relative translation levels of mRNAs (2 nM) in wge containing 53 mM or 133 mM K+ and supplemented with 400 nM of the indicated BTEs or control 18S RNA. (A) Uncapped BlucB mRNA. (B) Capped luciferase mRNA containing vector-derived UTRs.
Differences in inhibitory activity of BTEs may result from the fact that BYDV infects monocots, whereas TNV-D and RCNMV both infect only dicot plants. Thus, their BTEs may not function as efficiently in wge. Thus, we compared BTE activities in a dicot translation system, using an extract from tobacco (BY2) cells (31). The BY2 extract was programmed with BLucB, BLucBBF and capped BLucBBF mRNAs alone, or in the presence of 100-fold molar excess of BYDV, TNV-D or RCNMV1 BTEs or the 100-nt fragment of 18S rRNA as a negative control (Figure 5). BLucBBF has the same sequence as BLucB, except for a four-base (GAUC) duplication in the 17 CS of the BTE that abolishes translational activity (50). As in wge, uncapped BlucBBF was translated very weakly and capping caused a 10-fold increase in its translation (Figure 5A). BLucB mRNA was translated about half as efficiently as the capped BLucBBF and 5-fold more than uncapped BlucBBF, showing efficient cap-independent translation in the BY2 extract (Figure 5A). The 18S rRNA had little inhibitory activity toward the mRNAs. In trans, all BTEs significantly reduced translation of both BLucB and capBLucBBF but their presence had no influence on the low level of uncapped BLucBBF translation. The inhibitory activities of BYDV and TNV-D BTEs were similar, both reducing the BLucB and the capBLucBBF translation to 20%. The RCNMV1 BTE reduced translation of both mRNAs to 30% (Figure 5A). However, at the lower BLucB mRNA concentration of 2 nM, TNV-D and RCNMV1 BTEs had no significant inhibitory activity and the BYDV BTE inhibited BLucB translation to only 50% (Figure 5B). In general, all BTEs functioned in BY2 extract but this inhibitory activity was observed only if BY2 extract was programmed with higher concentrations (20 nM) of mRNA. These results are consistent with the notion that high RNA concentrations result in more competition for initiation factors.
Figure 5.
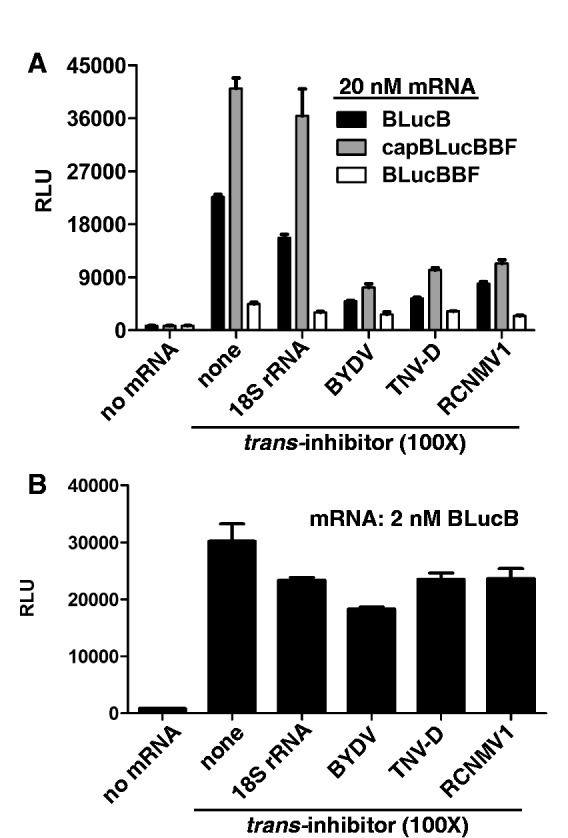
Inhibition of translation by BTEs in BY-2 extracts. (A) Relative translation levels of uncapped 20 nM BlucB, BLucBBF or capped BLucBBF in BY-2 extract supplemented with 2 µM of the indicated BTEs or control 18S RNA. (B) Inhibition of 2 nM BlucB mRNA. The data shown are averages of triplicates from two experiments and error bars represent standard error.
The eIF4G region residing between the eIF4E-binding site and MIF4G is crucial for BTE-dependent translation and interacts with the BTE
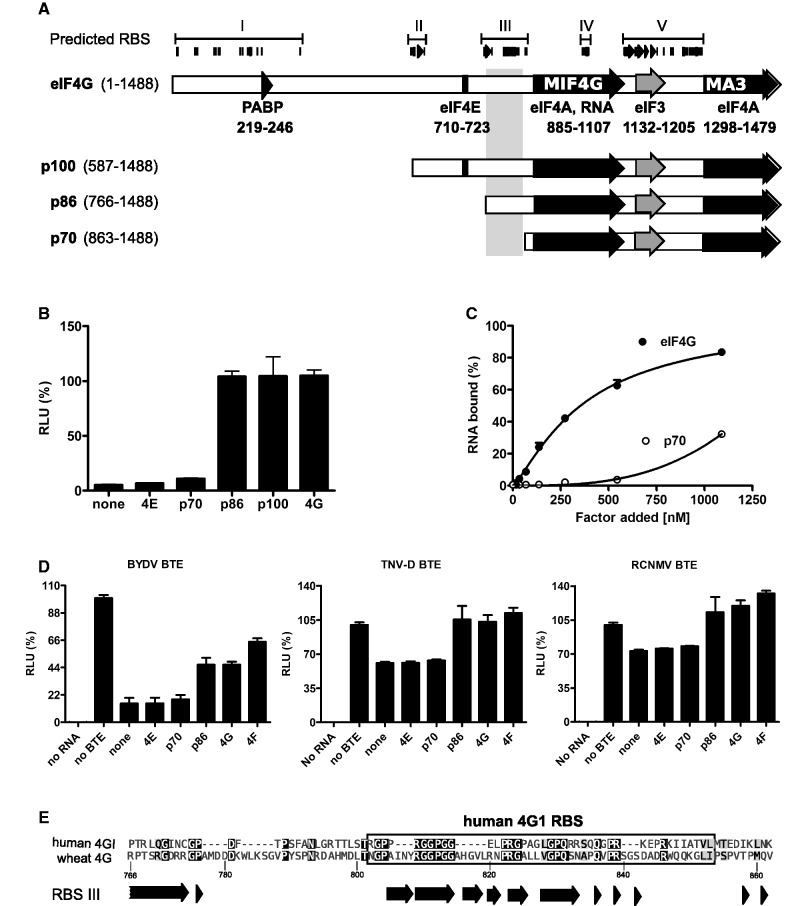
To further understand the BTE mechanism, we focused next on the interaction between the BTE and eIF4G. Previously, we showed that BYDV BTE binds and requires eIF4G (28). To define the eIF4G region that interacts with the BTE, we used the RNABindR program (51) to predict RNA-binding sites (RBSs) in eIF4G. RBSs grouped in five putative RNA-binding domains (RBDs), numbered I to V from the N-terminus (Figure 6A). These predictions agree with previous studies in wheat eIF4G (17,52) and the mammalian eIF4G (53,54) with the exception of RBDIII, which has additional sequence in between three blocks of conserved sequence. Using these predictions as a guide, we prepared N-terminally truncated, 100, 86 and 70 kDa fragments of eIF4G (Figure 6A). Fragment p100 contains the eIF4E-binding site and all predicted RBDs except for RBD I. Mutant p86 starts at the N-terminus of RBD III and lacks the eIF4E-binding site, while p70 starts directly after the C-terminus of RBD III, near the amino terminus of the MIF4G domain.
Figure 6.
Mapping a region of eIF4G that is crucial for eIF4G–BTE interaction. (A). Schematic representation of wheat eIF4G and its deletions mutants. Indicated regions, except for RBSs were found by pairwise alignment with human eIF4G (14). Factor-binding sites and two highly conserved HEAT domains, MIF4G and MA3, are shown, with their respective amino acid positions below the full-length map. Amino acid positions of sequences remaining in deletion constructs are in parentheses. Predicted RBDs are shown as numbered brackets above the predicted RBSs which are shown as filled bars and arrows above the eIF4G map. A region crucial for ability of eIF4G to recover BTE-inhibited translation is highlighted as a gray rectangle underlying the region between the N termini of p86 and p70. (B) Relative light units (RLU) obtained upon translation of uncapped BlucB mRNA in eIF4F-depleted wge in the presence of 200× of the indicated factors. (C) Binding curves of eIF4G and p70 deletion mutant (50 nM) for [α-32P]-labeled BTE (0.4 nM). Both protein-bound and unbound RNAs were measured in a double membrane filter-binding assay as described in ‘Materials and Methods’ section. Each point represents an average from at least three independent experiments. (D) Restoration of BLucB translation by eIF4G and its deletions in wge inhibited by the indicated BTEs. BTEs and 18S rRNA (400 nM) were added to wge, which was next supplemented with 100 nM of indicated factors and programmed with 2 nM of BLucB mRNA. The level of translation is expressed as a percentage of luciferase produced in the uninhibited extract. The data represent averages of triplicates from at least two experiments and error bars show standard error. (E) Amino acid sequence of region highlighted by gray box in panel A, aligned with homologous region of human eIF4G1. Dark arrows represent amino acids strings predicted to bind RNA by RNABindR software (51).
We tested the ability of these eIF4G truncations to restore translation in wge that had been depleted of eIF4F by m7GTP-Sepharose chromatography (28). As we reported previously (28), eIF4E alone did not restore translation, whereas p86 was as active as full-length eIF4G in restoring BLucB translation (Figure 6B). As expected, p100 was also as active as whole eIF4G. However, p70, which is just 97 aa shorter than p86, had no activity (Figure 6B). Thus, a region between aa 766 and 863 in eIF4G, covering RBS III, is required to facilitate BTE-dependent translation.
We reported previously that there is a strong correlation between binding affinity of the factors to the BTE and their ability to stimulate BTE-mediated translation (28). Thus, we measured binding of p70 to the BTE using eIF4G as positive control in a filter-binding assay (Figure 6C). As expected, at nanomolar concentrations, the BTE was bound much more tightly by complete eIF4G than by p70.
To determine the activities of eIF4G truncation mutants, we monitored their ability to restore BLucB translation in wge that had been depleted of available BTE-binding complexes by addition of a large excess of BYDV, TNV-D or RCNMV1 BTEs. In extracts inhibited with a 200-fold molar excess of BYDV BTE, addition of eIF4E or p70 did not restore translation of BLucB, whereas addition of p86, eIF4G or eIF4F boosted translation to 50, 50 and 70%, respectively, of the level in uninhibited extracts (Figure 6D). Lack of full recovery of BLucB translation can be explained by the fact that extracts contained 400 nM of inhibitory BTE and only 100 nM of recovering factors. In extracts inhibited with 200-fold molar excess of TNV-D or RCNMV1 BTE, again eIF4E and p70 did not restore translation, whereas p86, eIF4G and eIF4F boosted BLucB translation slightly above (by 20–30%) the level obtained in the absence of BTEs (Figure 6D). However, inhibition by TNV-D and RCNMV1 BTEs was much less efficient than that of the BYDV BTE. Because eIF4E and p70 did not restore translation, while p86, eIF4G and eIF4F did restore translation in extracts inhibited by all BTEs, we conclude that the region of eIF4G containing RBD III plays a key role in the interaction of eIF4G with the BTE.
Prévôt et al. (53) reported that a 40-aa tract between the eIF4E-binding site and MIF4G of human eIF4GI (aa 681–721) comprises an RBD required for ribosome scanning of mRNAs. Alignment of the wheat RBS III sequence (aa 766–858) with the corresponding human eIF4GI region (aa 647–723) revealed 31% identity and strong similarity of the predicted RBSs in RBD III to the human sequence (Figure 6E). In both proteins, this region is strongly basic (pI > 11) and is rich in glycine (14.7%), proline (14.7%) arginine (11.8%) and leucine (8%) as characteristic of many RNA-binding regions (55,56). Alignment of this region from diverse species revealed that the putative wheat RBD III is well conserved among plant and mammalian eIF4Gs (45% pairwise identity), with two highly conserved strings of amino acids (Supplementary Figure S2). The first spans the N-terminus of RBD III (truncated in p86) and contains plant-specific DR repeats. The second region of homology aligns to human 4G1 RBD and is divided into three blocks, rich in conserved gly, pro and arg, separated by sequences that are uniquely conserved in plants (Supplementary Figure S2). This region is directly upstream of the N-terminus of p70 and upstream of a polyG-specific RBD in wheat eIF4G and eIFiso4G (52).
eIF4G binds stem–loop I and bases around the hub of the BTE
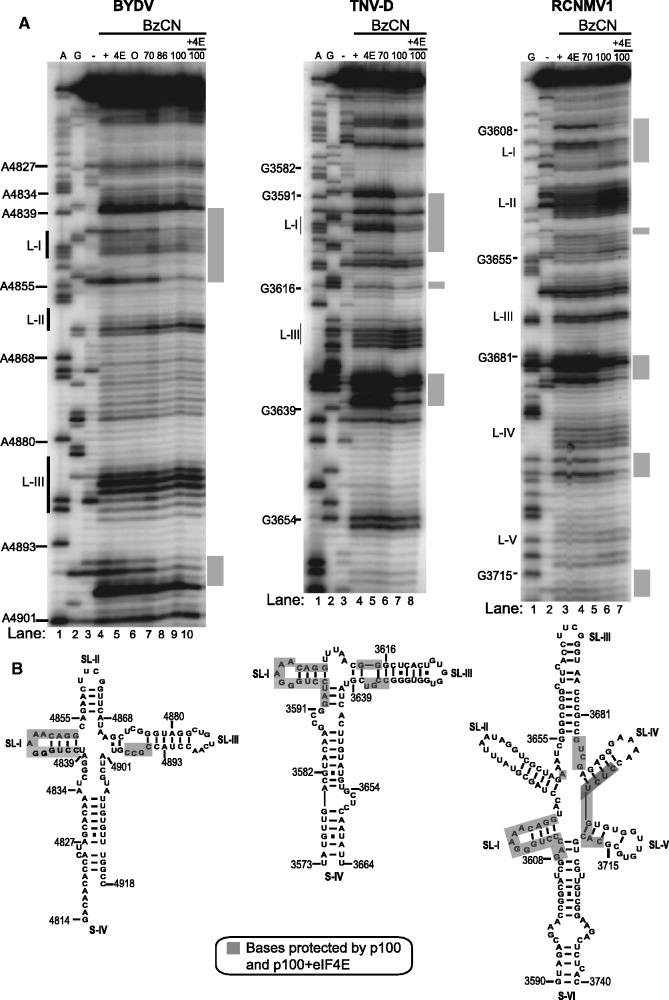
To visualize the precise eIF4G-binding site in the BTE, we probed luteovirus, necrovirus and dianthovirus BTEs with the SHAPE reagent BzCN in the presence and absence of eIF4G fragments. We used fully active p100 and p86 fragments in place of whole eIF4G in our footprinting experiments because whole eIF4G was too unstable during purification. N-terminally truncated forms of eIF4G that facilitate BTE-mediated translation (p86 and p100) protected the region around the conserved SL-I, and at an internal bulge downstream of the long-distance kissing loop in all three BTEs and in RCNMV1 at other positions around the central hub (Figure 7A, BYDV lanes 8–10; TNV-D lanes 7–8; RCNMV1 lanes 6–7). Addition of eIF4E enhanced protection in the presence of p100, the only form that contains the eIF4E-binding site, in both TNV-D and RCNMV1 (Figure 7A, lanes 8 and 7 in TNV-D and RCNMV1 gels, respectively). In particular, added eIF4E in the presence of p100 induced additional protection at the 3′-site of helices 4 and 5 in RCNMV1 RNA (Fig 7A, RCNMV1 lane7). This agrees with the enhancement of eIF4G binding provided by eIF4E in filter-binding studies that showed eIF4E binding to eIF4G increases affinity of eIF4G for the BTE and stimulates eIF4G by 20–30% in BTE-mediated translation (28). Importantly, loop III of the BYDV and TNV-D BTEs and loops 2, 4 and 5 in the RCNMV1 BTE remained exposed and available for chemical modification in all BTEs in the presence of protein (Figure 7). This is significant because base pairing of loop III of BYDV and TNV-D to the 5′-UTR is required for translation initiation at the 5′-end (19,20,25,57).
Figure 7.
Mapping of the eIF4G-binding sites on the BTEs of BYDV, TNV-D and RCNMV1. (A) BzCN footprinting of the indicated BTE in the presence of non-binding proteins eIF4E (4E), ovalbumin (O) and p70 truncation of eIF4G (70) or N-terminally truncated BTE-binding proteins p86 (86) and p100 (100) with or without added eIF4E. All proteins were added to final concentration of 1 μM. A and G are dideoxy sequencing lanes showing positions of A and G residues, respectively, in the RNA template. Unmodified RNA (-lane) shows background RNA hydrolysis. Gray bars to the right show regions protected by protein from BzCN modification. (B) BTE secondary structures below each gel showing sequences (gray bars in panel A) protected by p100 and p100 + eIF4E from chemical modification by BzCN.
As expected, in all BTEs, addition of proteins known not to bind the BTE directly (eIF4E, ovalbumin, or p70) did not change modification patterns of the BTEs (Figure 7A, BYDV lanes 4–7; TNV-D lanes 4–6; RCNMV1 lanes 3–5). Moreover, no clear protection by p100 or p86 was observed on the non-functional BTE-BF RNA, which contains the four-base duplication in the BamHI site of the 17 CS rendering it inactive in trans-inhibition assays (Figure 1 and Supplementary Figure S3) (20,26,29). The presence of p100 or p86 did not change the BTE modification pattern outside of the protected region, indicating that eIF4G binding does not alter the BTE structure, supporting above conclusions that the BTE folds into the functional structure in the absence of protein. These data strongly support our model in which the BTE simultaneously binds eIF4F and base pairs to the 5′-UTR via loop III, in order for eIF4F to recruit the ribosome to the 5′-UTR (20,22).
DISCUSSION
Magnesium is necessary for BTE folding and function
Viral RNAs frequently adopt various folded shapes to recruit specific host factors and facilitate efficient translation of viral mRNA, often at the expense of host protein synthesis. Using a combination of functional and structural assays, we found that CITEs in the 3′-UTRs of three viral genera adopt a stable cation-dependent fold at a Mg2+ concentration in the physiological range and in the absence of initiation factors or any other protein. By comparing functional with poorly functional mutants, we found that the stability of the native state is related directly to functional efficiency and thus reflects the ability to recruit components of the translation initiation machinery.
At very low (0.1 mM) Mg2+ concentrations, only wt BTE105 and two functional 4wj mutants adopted a predominantly compact native fold (Figure 1). In the functional point mutant U4900C, replacement of uridine with cytidine converts a weak guanine-uracil (G-U) pair into a more stable guanine-cytosine (G-C) base pair that should stabilize the base of helix III. The effect of replacing adenine with cytosine in the other functional mutant, A4868C appears not to involve a canonical Watson–Crick-type interaction. One possibility is that replacement of adenine with cytosine results in the formation of non-Watson–Crick-type cytidine-adenine (C-A) base pairing, which has been observed in a high-resolution crystal structure of octameric RNA (58) and is isosteric to a G-U pair (59). Such C-A pairing may stabilize the base of helix II while allowing for flexibility.
At elevated (up to 10 mM) Mg2+ concentrations, migration rates of functional and non-functional 4wj mutants were equal, indicating that even non-functional 4wj mutants collapsed to a compact fold. Such behavior was noted for the bi5core RNA of a group I intron (60) and an Azoarcus ribozyme (61). With regard to bi5core RNA, it was proposed that Mg2+-mediated compact, but non-native, states are biologically relevant intermediates that achieve their complete native fold only in presence of protein (60). This is not the case with the non-functional 4wj mutants because they were unable to bind and sequester initiation factors from reporter mRNA in the non-competitive conditions of wheat germ extract (Figure 1B). We conclude that non-native compact BTE states of the mutants are not biologically relevant and are probably gel-matrix-arrested, destabilized, near-native intermediates, as observed for Azoarcus ribozyme (61).
There are two possibilities for how Mg2+ might stabilize the BTE’s compact native fold. One is coordination of Mg2+ ion at a specific site in the RNA structure, as observed in the crystal structures of the Tetrahymena group I intron (62,63) and the other involves Mg2+ binding in a diffuse manner to neutralize RNA phosphates, as observed for non-coding RNAs (40,64). The latter possibility was indicated by the fact that the wt and mutant BTEs could fold into a single species in 0.5 M K+ (TBK gel, Figure 1D) in the absence of Mg2+. Similarly, the functional BTEs folded as predominantly the native form in 10 mM Ca2+ (Supplementary Figure S1). Thus, BTE RNA folding is driven by neutralization of the negative phosphate charges of RNA backbone. The requirement for a 50-fold greater concentration of K+ ions than Mg2+ ions to complete the BTE folding is expected as a consequence of the lower charge density of K+ compared with Mg2+, leading to less-efficient neutralization of the phosphate charges (42,65).
Chemical modification experiments with a BzCN SHAPE reagent as a function of Mg2+ concentration, revealed that most secondary structural elements form at physiological (0.6–1.0 mM) Mg2+ concentrations. Furthermore, BzCN-mediated base modification showed that base pairing forms less cooperatively in the non-functional 4wj mutants than in functional RNAs, suggesting that destabilization of the BTE helical junction most likely introduces an alternative folding pathway. Similar folding traps were observed for the Azoarcus ribozyme in which disruption of a tetraloop–tetraloop receptor interaction led to formation of slower moving non-native species on the native gel and required higher Mg2+ concentrations to form base pairs compared with the wt ribozyme (66). Most exposed regions of the BTE displayed an increase in modification as a function of Mg2+ concentration and quantification of these folding changes for two distant regions of the sequence are nearly identical (Figure 2), suggesting that the BTE folds in a single step stabilized by cooperative binding of Mg2+ in diffuse manner.
BTE folding correlated strongly with ability to stimulate translation initiation. Mutants with defective helical junction and fold competed poorly with reporter mRNA for the initiation machinery (Figure 1). Reduced inhibition of BLucB relative to the strong inhibition of cap-Luc mRNA may result from the fact that the elements added in trans were competing with the BTE in cis, residing in its native full size 3′-UTR. Cap-dependent translation is usually optimal at K+ concentrations <80 mM whereas cap-independent translation is usually optimal at K+ concentrations >80 mM (47–49). The optimal translation at K+ concentrations >100 mM (up to 200 mM) was also reported for IRES-dependent translation (67–69) suggesting that the activity imparted to the BTE by cation-dependent folding may be a general feature of CITEs.
One might speculate that the lower activities of the TNV-D and RCNMV1 BTEs reflect host specialization of the viruses from which these elements originated. However, in the translation extract prepared from tobacco BY-2 cells, the BYDV BTE trans-inhibited translation at least as efficiently as the TNV-D and RCNMV1 BTEs, even though the latter viruses infect tobacco and BYDV does not (Figure 5).
The RBS III domain between the eIF4E-binding site and MIF4G is essential for interaction of eIF4G with the BTE
We confirm our previous observation (28) that N-terminal truncation mutant p86, missing the PABP and eIF4E-binding sites facilitates BTE-dependent translation as efficiently as full-length eIF4G (Figure 6B). Here, we also show that removing 97 N-terminal amino acids from p86 to form p70 abolishes the ability of the remaining portion of eIF4G to facilitate translation via the BTE (Figure 6C). This truncation disrupts eIF4G binding to BYDV, TNV-D and RCNMV1 BTEs (Figure 7).
In human eIF4G, MIF4G binds globin mRNA and the picornaviral IRES (70). Wheat eIFiso4G and eIF4G contain a highly homologous poly(G)-preferential RNA-binding region starting in close proximity to the N-terminal end of MIF4G (52). Detailed studies of RNA-binding regions in eIFiso4G confirmed that MIF4G alone can bind poly(G) but to bind the poly(A) homopolymer, it requires the entire N-terminal part of the protein (14). As p70 contains MIF4G, including a region of high homology (52), it is clear that the MIF4G RNA-binding region alone can neither support BTE-dependent translation, nor bind to BTEs. These functions require the presence of the additional 97-aa-long N-terminal extension that contains blocks of RBSs forming RBD III as predicted by RNABindR (Figure 6E). A similar RBD was described in human eIF4GI, where a 40-aa tract N-terminally preceeding the MIF4G region was crucial for eIF4G binding to the 5′-end of capped and uncapped mRNAs. This interaction was required for ribosome scanning (53). There is substantial homology between the C-terminal domain of wheat eIF4G RBD III and human eIF4G I RBD (Figure 6E). Moreover, all RBD III domains share previously unreported regions of homology among mammalian and plant eIF4Gs with very conserved glycine, proline and arginine residues (Supplementary Figure S2). Thus, we conclude that the region in question is responsible for the binding of eIF4G to BTE. We predict that it binds the BTE directly but cannot rule out the possibility that it may be required only for correct folding of eIF4G to allow eIF4G to bind the BTE via a different RBD.
eIF4G fragment binding to the BTE does not affect BTE structure and leaves loop III accessible for base pairing to the 5′-UTR
Inspection of individual BTE base interactions with eIF4G by footprinting methods revealed that functional eIF4G fragments bind primarily to the highly conserved SL-I in all BTEs with little re-organization of the BTE structure. The loop of SL-I fits the consensus of a GNRNA pentaloop characterized in the boxB domain of bacteriophage λ RNA (71). This structure resembles the highly stable GNRA fold (72), but the extra base of the pentaloop (N at position 4) protrudes. The boxB RNA is bound by the λ N protein, which in turn fosters binding of host protein NusA via the protruding fourth base of the pentaloop (71). Thus, like boxB, perhaps the BTE forms this structure and recruits additional proteins upon eIF4G binding, such as eIF4E or other unidentified proteins (28). It is possible that SL-I is the core functional sequence and that additional RNA structures (different in each BTE) evolved to position this loop in such an orientation as to enhance interaction with initiation machinery, as observed for certain riboswitches (73). Despite differences in sequence and secondary structure, the diverse BTEs must fold in a similar way to render the SL-I surface exposed and available for protein recognition.
eIF4G can bind the BTE and promote BTE-dependent translation alone (28). However, eIF4F has almost 10-fold higher affinity for the BTE and exhibits 20–30% higher activity than eIF4G in promoting translation in vitro (28). Here, we show that p100, which can bind eIF4E, binds to the same SLI region of BTEs as p86 and in the presence of eIF4E, protection of the BTE from BzCN-directed modification is enhanced in both TNV-D and RCNMV BTEs. Thus, binding of eIF4E to eIF4G locks eIF4F on to the BTE, yet in this process, eIF4E forms few, if any additional contacts with the BTE.
It is interesting to compare the BTE with the completely unrelated HCV IRES. Like the BTE, the HCV IRES pre-folds into a distinct cation-dependent fold prior to recognition by the translation initiation machinery, and mutations that disrupt IRES folding also disrupt its function (40). Like the BTE’s long-distance communication loop, structure probing of HCV IRES highlighted a loop with an embedded start codon that remained solvent accessible in order to interact readily with translation machinery. Thus, it appears that these unrelated CITEs evolved diverse structures capable of accomplishing the same task of recruiting and activating the translation machinery in a similar way. Our data lead us to propose a general model of BTE folding, factor recruitment and delivery (Figure 8) in which the BTE class of CITEs pre-organize in specific cation-dependent 3D scaffolds that allow simultaneous binding of eIF4F to SL-I and base pairing of SL-III to the 5′-UTR, in order for eIF4F to recruit the ribosome to the 5′-UTR. This mechanism is supported by studies of a different 3′-CITE (I-shaped structure) that was shown to physically bind eIF4F and the 5′-UTR simultaneously (21,74).
Figure 8.
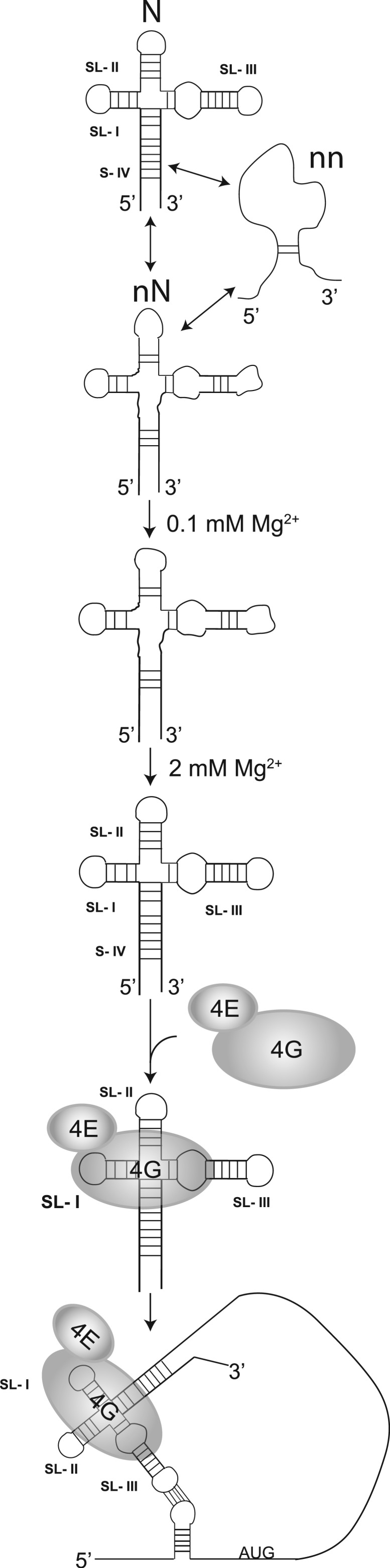
Model for the Mg2+-dependent BYDV BTE folding and eIF4F (eIF4G+eIF4E) binding. (Only the factors studied in this report are shown.) At low magnesium concentrations, the BYDV BTE samples native (N), near-native (nN) and non-native (nn) conformational states. In the presence of increasing magnesium concentrations, BTE folding is shifted towards the native form that is recognized by eIF4F (4G + 4E). eIF4G binds the SL-I of the BTE and a region at the junction-proximal end of SL-III. eIF4E binding to eIF4G further enhances the BTE–eIF4G interaction. This interaction leaves SL-III loop exposed for interaction with the 5′-UTR, thus delivering eIF4F to the 5′-end to facilitate ribosome recruitment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
The National Institutes of Health (NIH) [2R01 GM067104] with an American Recovery and Reinvestment Act supplement; Hatch Act and State of Iowa funds. The U.S. Department of Agriculture [AFRI NIFA Fellowship 2011-67012-30715 to J.J.K.]. Funding for open access charge: NIH [2R01 GM067104].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Zhaohui Wang for construction of the BTE SHAPE cassettes and plasmids for expression of truncated eIF4Gs as well as for advice on RNA structure probing methods. We thank Dr Aurélie Rakatondrafara for graciously providing us with the cap-Luc control. Also, we thank Alice Hui for assistance with BY-2 extract preparation.
REFERENCES
- 1.Dreher TW, Miller WA. Translational control in positive strand RNA plant viruses. Virology. 2006;344:185–197. doi: 10.1016/j.virol.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallie DR. Protein-protein interactions required during translation. Plant Mol. Biol. 2002;50:949–970. doi: 10.1023/a:1021220910664. [DOI] [PubMed] [Google Scholar]
- 4.Gallie DR. Translational Control in Plants and Chloroplasts. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 747–775. [Google Scholar]
- 5.Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 6.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 7.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 8.Tarun SZ, Jr, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl Acad. Sci. USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park EH, Walker SE, Lee JM, Rothenburg S, Lorsch JR, Hinnebusch AG. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1*PABP mRNPs in vivo. EMBO J. 2011;30:302–316. doi: 10.1038/emboj.2010.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CU, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- 11.Marintchev A, Wagner G. eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry. 2005;44:12265–12272. doi: 10.1021/bi051271v. [DOI] [PubMed] [Google Scholar]
- 12.Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK. Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure. 2006;14:913–923. doi: 10.1016/j.str.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 14.Cheng S, Gallie DR. Competitive and noncompetitive binding of eIF4B, eIF4A, and the poly(A) binding protein to wheat translation initiation factor eIFiso4G. Biochemistry. 2010;49:8251–8265. doi: 10.1021/bi1008529. [DOI] [PubMed] [Google Scholar]
- 15.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallie DR, Browning KS. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J. Biol. Chem. 2001;276:36951–36960. doi: 10.1074/jbc.M103869200. [DOI] [PubMed] [Google Scholar]
- 18.Patrick RM, Browning KS. The eIF4F and eIFiso4F complexes of plants: An evolutionary perspective. Comp. Funct. Genomics. 2012;2012:287814. doi: 10.1155/2012/287814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo L, Allen EM, Miller WA. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- 20.Rakotondrafara AM, Polacek C, Harris E, Miller WA. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′-cap-independent translation element. RNA. 2006;12:1893–1906. doi: 10.1261/rna.115606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson BL, Wu B, Chevtchenko I, White KA. Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA. 2010;16:1402–1419. doi: 10.1261/rna.2135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller WA, Wang Z, Treder K. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem. Soc. Trans. 2007;35:1629–1633. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholson BL, White KA. 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr. Opin. Virol. 2011;1:373–380. doi: 10.1016/j.coviro.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Mizumoto H, Tatsuta M, Kaido M, Mise K, Okuno T. Cap-independent translational enhancement by the 3′ untranslated region of red clover necrotic mosaic virus RNA1. J. Virol. 2003;77:12113–12121. doi: 10.1128/JVI.77.22.12113-12121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meulewaeter F, van Lipzig R, Gultyaev AP, Pleij CW, Van Damme D, Cornelissen M, van Eldik G. Conservation of RNA structures enables TNV and BYDV 5′ and 3′ elements to cooperate synergistically in cap-independent translation. Nucleic Acids Res. 2004;32:1721–1730. doi: 10.1093/nar/gkh338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Kraft JJ, Hui AY, Miller WA. Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology. 2010;402:177–186. doi: 10.1016/j.virol.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwakawa HO, Kaido M, Mise K, Okuno T. Cis-acting core RNA elements required for negative-strand RNA synthesis and cap-independent translation are separated in the 3′-untranslated region of Red clover necrotic mosaic virus RNA1. Virology. 2007;369:168–181. doi: 10.1016/j.virol.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 28.Treder K, Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008;14:134–147. doi: 10.1261/rna.777308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo L, Allen E, Miller WA. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA. 2000;6:1808–1820. doi: 10.1017/s1355838200001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komoda K, Naito S, Ishikawa M. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl Acad. Sci. U SA. 2004;101:1863–1867. doi: 10.1073/pnas.0307131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayberry LK, Dennis MD, Leah Allen M, Ruud Nitka K, Murphy PA, Campbell L, Browning KS. Expression and purification of recombinant wheat translation initiation factors eIF1, eIF1A, eIF4A, eIF4B, eIF4F, eIF(iso)4F, and eIF5. Methods Enzymol. 2007;430:397–408. doi: 10.1016/S0076-6879(07)30015-3. [DOI] [PubMed] [Google Scholar]
- 33.van Heerden A, Browning KS. Expression in Escherichia coli of the two subunits of the isozyme form of wheat germ protein synthesis initiation factor 4F. Purification of the subunits and formation of an enzymatically active complex. J. Biol. Chem. 1994;269:17454–17457. [PubMed] [Google Scholar]
- 34.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Parisien M, Scheets K, Miller WA. The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure. 2011;19:868–880. doi: 10.1016/j.str.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klostermeier D, Millar DP. Helical junctions as determinants for RNA folding: origin of tertiary structure stability of the hairpin ribozyme. Biochemistry. 2000;39:12970–12978. doi: 10.1021/bi0014103. [DOI] [PubMed] [Google Scholar]
- 38.Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011;44:1302–1311. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 39.Heilman-Miller SL, Thirumalai D, Woodson SA. Role of counterion condensation in folding of the Tetrahymena ribozyme. I. Equilibrium stabilization by cations. J. Mol. Biol. 2001;306:1157–1166. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
- 40.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 41.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koculi E, Hyeon C, Thirumalai D, Woodson SA. Charge density of divalent metal cations determines RNA stability. J. Am. Chem. Soc. 2007;129:2676–2682. doi: 10.1021/ja068027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 44.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 45.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry. J. Am. Chem. Soc. 2008;130:16178–16180. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- 46.Wang S, Miller WA. A sequence located 4.5 to 5 kilobases from the 5′ end of the barley yellow dwarf virus (PAV) genome strongly stimulates translation of uncapped mRNA. J. Biol. Chem. 1995;270:13446–13452. doi: 10.1074/jbc.270.22.13446. [DOI] [PubMed] [Google Scholar]
- 47.Carrasco L, Smith AE. Sodium ions and the shut-off of host cell protein synthesis by picornaviruses. Nature. 1976;264:807–809. doi: 10.1038/264807a0. [DOI] [PubMed] [Google Scholar]
- 48.Niepmann M. Effects of potassium and chloride on ribosome association with the RNA of foot-and-mouth disease virus. Virus Res. 2003;93:71–78. doi: 10.1016/s0168-1702(03)00067-4. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt-Brauns J. Chloride concentration discriminates between Foot-and-mouth disease virus ires-dependent translation and classical scanning translation: new aspects of the picornavirus shutoff mechanism. Acta Virol. 2003;47:65–72. [PubMed] [Google Scholar]
- 50.Wang S, Browning KS, Miller WA. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terribilini M, Sander JD, Lee JH, Zaback P, Jernigan RL, Honavar V, Dobbs D. RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res. 2007;35:W578–W584. doi: 10.1093/nar/gkm294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim CY, Takahashi K, Nguyen TB, Roberts JK, Webster C. Identification of a nucleic acid binding domain in eukaryotic initiation factor eIFiso4G from wheat. J. Biol. Chem. 1999;274:10603–10608. doi: 10.1074/jbc.274.15.10603. [DOI] [PubMed] [Google Scholar]
- 53.Prévôt D, Décimo D, Herbreteau CH, Roux F, Garin J, Darlix JL, Ohlmann T. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 2003;22:1909–1921. doi: 10.1093/emboj/cdg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinton TM, Coldwell MJ, Carpenter GA, Morley SJ, Pain VM. Functional analysis of individual binding activities of the scaffold protein eIF4G. J. Biol. Chem. 2007;282:1695–1708. doi: 10.1074/jbc.M602780200. [DOI] [PubMed] [Google Scholar]
- 55.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 56.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen R, Miller WA. The 3′ untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J. Virol. 2004;78:4655–4664. doi: 10.1128/JVI.78.9.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jang SB, Hung LW, Chi YI, Holbrook EL, Carter RJ, Holbrook SR. Structure of an RNA internal loop consisting of tandem C-A+ base pairs. Biochemistry. 1998;37:11726–11731. doi: 10.1021/bi980758j. [DOI] [PubMed] [Google Scholar]
- 59.Masquida B, Westhof E. On the wobble GoU and related pairs. RNA. 2000;6:9–15. doi: 10.1017/s1355838200992082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buchmueller KL, Webb AE, Richardson DA, Weeks KM. A collapsed non-native RNA folding state. Nat. Struct. Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- 61.Rangan P, Masquida B, Westhof E, Woodson SA. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc. Natl Acad. Sci. USA. 2003;100:1574–1579. doi: 10.1073/pnas.0337743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 63.Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat. Struct. Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 64.Misra VK, Draper DE. The linkage between magnesium binding and RNA folding. J. Mol. Biol. 2002;317:507–521. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 65.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 66.Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J. Am. Chem. Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jackson RJ. Potassium salts influence the fidelity of mRNA translation initiation in rabbit reticulocyte lysates: unique features of encephalomyocarditis virus RNA translation. Biochim. Biophys. Acta. 1991;1088:345–358. doi: 10.1016/0167-4781(91)90124-5. [DOI] [PubMed] [Google Scholar]
- 68.Borman AM, Bailly JL, Girard M, Kean KM. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 1995;23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jünemann C, Song Y, Bassili G, Goergen D, Henke J, Niepmann M. Picornavirus internal ribosome entry site elements can stimulate translation of upstream genes. J. Biol. Chem. 2007;282:132–141. doi: 10.1074/jbc.M608750200. [DOI] [PubMed] [Google Scholar]
- 70.Lomakin IB, Hellen CU, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 72.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 73.Schultes EA, Spasic A, Mohanty U, Bartel DP. Compact and ordered collapse of randomly generated RNA sequences. Nat. Struct. Mol. Biol. 2005;12:1130–1136. doi: 10.1038/nsmb1014. [DOI] [PubMed] [Google Scholar]
- 74.Nicholson BL, Zaslaver O, Mayberry LK, Browning KS, White KA. Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J. Virol. 2013;87:1872–1883. doi: 10.1128/JVI.02711-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.