Abstract
Ribonuclease H2 (RNase H2) protects genome integrity by its dual roles of resolving transcription-related R-loops and ribonucleotides incorporated in DNA during replication. To unlink these two functions, we generated a Saccharomyces cerevisiae RNase H2 mutant that can resolve R-loops but cannot cleave single ribonucleotides in DNA. This mutant definitively correlates the 2–5 bp deletions observed in rnh201Δ strains with single rNMPs in DNA. It also establishes a connection between R-loops and Sgs1-mediated replication reinitiation at stalled forks and identifies R-loops uniquely processed by RNase H2. In mouse, deletion of any of the genes coding for RNase H2 results in embryonic lethality, and in humans, RNase H2 hypomorphic mutations cause Aicardi–Goutières syndrome (AGS), a neuroinflammatory disorder. To determine the contribution of R-loops and rNMP in DNA to the defects observed in AGS, we characterized in yeast an AGS-related mutation, which is impaired in processing both substrates, but has sufficient R-loop degradation activity to complement the defects of rnh201Δ sgs1Δ strains. However, this AGS-related mutation accumulates 2–5 bp deletions at a very similar rate as the deletion strain.
INTRODUCTION
RNA and DNA worlds are not entirely separated and frequently come together during replication and transcription, forming transient RNA–DNA structures that when become stable can jeopardize genome integrity. These structures can be divided into two groups: (i) RNA, consisting of one or more ribonucleotides, covalently attached to DNA and opposite to DNA (RpD/DNA) and (ii) RNA opposite to DNA (RNA/DNA, later referred as RpR/DNA), in vivo as part of an R-loop or reverse transcripts (1–3). The former are present during replication, as part of the primers for lagging strand synthesis, or when ribonucleotides are erroneously incorporated into DNA by the replicative polymerases, which, if not removed, leads to mutagenesis (4–6). R-loops are formed during transcription when the RNA exiting the RNA polymerase fails to associate with the posttranscriptional machinery and instead remains annealed to the DNA (7–9). R-loops have the template DNA strand annealed to the RNA, whereas the other DNA strand is in a single-stranded form. Stable R-loops constitute a barrier to transcription and replication fork progression; stalling replication and inducing fork collapse (10,11).
Ribonucleases H (RNase H) comprise a group of enzymes devoted to the removal of both types of RNA–DNA structures. There are two main classes of RNases H, grouped by primary sequence and substrate specificity (12). Type 1 RNases H are able to cleave R-loops (RpR/DNA), but because the enzyme recognizes the RNA via contacts with the 2′-OH residues of four ribonucleotides, they are unable to hydrolyze single rNMPs (13). Type 2 RNases H recognize the transition from ribonucleotide to deoxyribonucleotide (RpD), hydrolyzing at the 5′-end of the ribonucleotide, leaving it attached to the 5′-end of the DNA (14). Eukaryotic RNases H2 cleave both RpD/DNA and RpR/DNA structures with similar efficiencies, in contrast to bacterial RNases H2, which have a clear preference for RpD substrates (15). Both types of RNases H are dispensable for viability in bacteria and single-cell eukaryotes (16,17) although genomic instability increases in their absence (18). In higher eukaryotes, both enzymes are essential. RNase H1 is present in nuclei and mitochondria, and it is necessary for mitochondrial DNA replication (19). RNase H2 has been associated with ribonucleotide removal from genomic DNA in yeast and mouse, where it is required for embryonic development (20,21). Mutations in the three genes encoding human RNase H2 (Hu-RNase H2) leads to Aicardi–Goutières syndrome (AGS), a severe neuroinflammatory disorder, with characteristics similar to in utero viral infection (22).
The crystal structure of Thermotoga maritima RNase H2 (Tm-RNase H2) in complex with a DNA containing a single ribonucleotide revealed the crucial role of a conserved tyrosine in the recognition and hydrolysis of the RpD structure (23). This residue displaces the strand to be cleaved and positions the phosphate between the ribose and deoxyribose to coordinate one of the two divalent metal ions required for catalysis. This displacement could not occur efficiently with an RpR structure, and accordingly Tm-RNase H2 cleaves RNA/DNA hybrids very poorly. The conserved Tyr in eukaryotic RNases H2 may have a slightly different position allowing both RpD and RpR cleavages.
Some organisms, such as Bacillus stearothermophilus, have two proteins with primary amino acid sequences (Figure 1A) and three-dimensional structures similar to type 2 RNases H (24). However, one (RNase H2) has enzymatic properties similar to bacterial RNases H2, recognizing an RpD transition and cleaving 5′ of the ribonucleotide attached to DNA, whereas the other (RNase H3) has properties similar to RNases H1, requiring a short string of ribonucleotides for hydrolysis. We reasoned that we could use the cocrystal structure of Tm-RNase H2 as a guide to superimpose the structure of B. stearothermophilus RNase H3 (Bst-RNase H3) and thereby identify the residues involved in single and multiple rNMPs recognition. Using our model, we made changes in the catalytic subunit of Saccharomyces cerevisiae RNase H2 (Sc-Rnh201) to create an enzyme that retains the ability to hydrolyze RpR/DNA hybrids but is unable to cleave RpD substrates. Constructing such a mutant allowed us to unlink the two activities of RNase H2 and assign in vivo phenotypes to each activity.
Figure 1.
Comparison of RNase H2 and H3. (A) Alignment was generated for human RNase H2A (Hu2A), S. cerevisiae RNase H2A (Sc2A), T. maritima RNase H2 (Tm2), B. stearothermophilus RNase H3 (Bst3), Streptococcus pneumonia RNase H3 (Spn3) and Thermovibrio ammonificans HB-1 RNase H3 (Tam3). α-Helices are indicated with pink letters, and β-sheets are indicated with orange letters. Active site residues are highlighted in yellow. Amino acid substitutions in S. cerevisiae RNase H2A are noted above the first row of alignment D39A, G42S, P45D, S or E and Y219A—along with the conserved DSK triplet. The ‘GRG’ motif, DSK and the conserved Tyr of RNase H2 are also highlighted in grey. (B) The 5′-32P-labeled 12 mer substrates indicated above the gels were digested by yeast and Tm-RNase H2 in the presence of 10 mM MgCl2. The lanes marked with 0 contained no enzyme, and lanes marked with triangle contained increasing amount of the proteins (0.16, 1.6, 16 and 160 nM in the case of Tm-RNase H2, 0.011, 0.11, 1.1 and 11 nM in the case of yeast RNase H2 and 0.011, 0.11, 1.1 and 11 nM in the case of Bst-RNase H3). Products of the hydrolysis were analyzed by 20% TBE-urea gels. The sizes of products were measured based on molecular size markers indicated as M (products of digestion of 32P-labeled strands without complementary strand by phosphodiesterase I). Major cleavage sites of the substrates are summarized on the bottom of each gel. (C) Relative activity of Bst-RNase H3 D103N and D103A mutants compared with the wild-type protein was analyzed by liquid RNase H assay using poly-rA/poly-dT substrate as previously done (15).
MATERIALS AND METHODS
Modeling the structure of Bst-RNase H3 in complex with substrates
To model a structure of Bst-RNase H3 in complex with a substrate, we used the crystal structures of Bst-RNase H3 (PDB: 2D0B), human RNase H1 in complex with RNA/DNA hybrid (PDB: 3PUF) and Tm-RNase H2 in complex with duplex DNA containing single ribonucleotide (PDB: 3O3G). The structures of Bst-RNase H3 and human RNase H1-substrate complexes were superimposed onto that of Tm-RNase H2-substrate complex using DaliLite pairwise comparison (http://www.ebi.ac.uk/Tools/dalilite/). In addition, the substrate in human RNase H1 was moved to fit on the substrate of Tm-RNase H2 based on the two nucleotides around the cleavage sites using Pymol (http://www.pymol.org/). This program was also used to display structures in designing mutants.
Purification of yeast RNase H2
Previously, we coexpressed and purified the three subunits of yeast RNase H2 with all the subunits C-terminally fused to an His-tag (25). In this study, we constructed an expression system to facilitate purification of the trimeric complex, in which only Rnh201p subunit was C-terminally tagged with 6XHis. The polycistronic coexpression system (pET-yH2ABC) was constructed as described previously for Hu-RNase H2 (15). For overexpression, Escherichia coli BL21(DE3) was transformed with pET-yH2ABC and cultured in Luria-Bertani (LB) media at 30°C. When the optical density at 600 nm (OD600) reached around 0.5, 1 mM IPTG was added to the culture medium and cultivation was continued for additional 3 h. Cells were harvested by centrifugation at 5000 rpm for 10 min, resuspended in buffer A (20 mM Tris–HCl, pH 7.5, 1 mM EDTA and 1 mM DTT), disrupted by sonication and centrifuged at 15 000 rpm for 30 min. The supernatant was loaded onto a Hitrap Heparin HP column (1 ml) (GE Healthcare) equilibrated with buffer A. The protein was eluted from the column with a linear gradient of 0–1 M NaCl. The fractions containing yeast RNase H2 trimeric complex were collected and applied to the Histrap crude column (1 ml) (GE Healthcare) equilibrated with buffer B (20 mM Tris–HCl pH 7.5, 500 mM NaCl and 15 mM imidazole). The protein was eluted from the column with 60 mM imidazole, dialyzed against 20 mM Tris–HCl pH 7.5 containing 1 mM EDTA, 1 mM DTT and 150 mM NaCl and concentrated. The protein purity was confirmed by SDS–PAGE, followed by staining with Coomassie brilliant blue R250. Expression and purification of yeast RNase H2 mutants were carried out as described for wild-type protein.
RNase H assays
Enzymatic reactions were performed in buffer containing 15 mM Tris–HCl (pH 7.9), 50 mM NaCl, 1 mM DTT, 100 µg/ml BSA, 5% glycerol and 10 mM MgCl2. Activity assays using a uniformly α32P-ATP-labeled poly-rA/poly-dT substrate and the 5′γ-32P-labeled short RNA/DNA (the sequences of the oligonucleotides are given in Figure 1 and Supplementary Figure S1) were performed as described previously (15).
Crystallization and structure solution
Tm-RNase H2 G21S was purified as previously described (23). Nucleic acid for crystallization was prepared by annealing high-performance liquid chromatography–purified oligonucleotides: cleaved strand 5′-GACACcTGATTC (single ribonucleotide in small caps) and complementary DNA strand 5′-GAATCAGGTGTC (both purchased from Metabion, Martinsried, Germany). Before crystallization, Tm-RNase H2 G21S (2 mg/ml final concentration) was mixed with the oligos at 1.2:1 substrate:protein molar ratio. The complexes were mixed with the reservoir solution [50 mM CaCl2, 45% PEG 200 and 0.1 M MES (pH 6.0)] at equal volume and crystallized by the sitting drop vapor diffusion method at 18°C. For data collection, the crystals were flash frozen in liquid nitrogen. Diffraction data were collected at Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung synchrotron at beamline MX-14 (26) on a Mar225 CCD detector at 100 K (Supplementary Table S2) and processed in HKL2000 (27). The structure was solved using molecular replacement in Phaser (28) and refined using Coot (29) and Phenix suite (30). Figures and analyses were prepared in PyMol (www.pymol.org). Atomic coordinates and structure factors have been deposited in Protein Data Base with accession code, 4HHT.
Yeast expression plasmid for Rnh201 expression from its native promoter
C-terminally FLAG tagged RNH201 gene coding for WT, D39A, G42S or P45D-Y219A, flanked with 5′-terminal 608 bp and 3′-terminal 404 bp genomic sequence, was cloned into HindIII/NgoMIV site of ycplac111 containing ARS/CEN and LEU2 marker to make ycNPH2-FL2 plamid expressing WT and mutant RNH201. RNH202 wt and ΔPIP (with a stop codon that makes a protein 11 amino acids shorter eliminating the PIP sequence) was also expressed from plasmid ycplac111 using its own promoter.
Construction of yeast strains
Strain Cy8338 (W303-1A but sgs1::HIS3) (31) was used to first replace the RNH201 and RNH202 genes with the CORE-I-SceI cassette from pGSHU (32). Polymerase chain reaction (PCR) products with the cassette flanked by 60 nucleotides of sequence upstream and downstream of the genes open reading frames were introduced in the strain, generating deletion/replacement of the corresponding gene by homologous recombination (HR). Proper replacement was confirmed by PCR and sequencing. Then, to introduce the desired mutations, WT and mutant genes from plasmid ycNPH2-FL2 were PCR amplified and introduced in the deletion strains (SMC306 and SMC307, Supplementary Table S1), which were growing in the presence of galactose to facilitate HR by I-SceI–mediated double-stranded break. Transformants were selected in 5-FOA plates and confirmed by PCR analysis and sequencing.
Yeast expression plasmid for Rnh1 overexpression under TPI promoter
Yeast RNase H1 gene was cloned into EcoRI and SalI site of pYX242 (16) to make pYX242-YH1. KanMX4 cassette was cloned into SfoI site of pYX242 and pYX242-YH1.
Growth rate determination and microscopic analysis
Overnight cultures grown in liquid Yeast Extract Peptone Dextrose (YPD) were inoculated into fresh YPD medium for OD600 to be about 0.1. The OD600 was measured every hour for 10 h to determine the doubling times. Multiple isolates were analyzed for each strain. Yeast cells fixed with 70% ethanol were stained with 4′,6-diamidino-2-phenylindole (DAPI). The cells were immobilized on polylysine-coated slides and photographed in a fluorescent microscope.
Analysis of spontaneous mutation in pol2-M644G rnh201Δ strain transformed with plasmid expressing rnh201 WT and mutants
The pol2-M644G rnh201Δ yeast strain (5) was transformed with ycNPH2-FL2 WT and derivatives. Cells were grown in SD-Leu medium and plated on SD-Leu lacking arginine and supplemented with 60 µg/ml canavanine. Genomic DNA was isolated from independent canavanine-resistant mutants, PCR amplified and sequenced. Spontaneous mutation rates were measured by fluctuation analysis (33), using FALCOR (Fluctuation Analysis CalculatOR) program (http://www.mitochondria.org/protocols/FALCOR.html). Rates of individual mutation classes were calculated by multiplying the proportion of each mutation type by the total mutation rate.
RESULTS
How RNase H3 recognizes RpR, but not RpD in duplex substrates
We first compared the activities of yeast RNase H2, Tm-RNase H2 and Bst-RNase H3 using RpD and RpR substrates (Figure 1B). Sc-RNase H2 showed the same substrate preference as Hu-RNase H2 (34), cleaving RNA12, RNA6-DNA6 and DNA5-RNA1-DNA6 strands with similar efficiencies when annealed to complementary DNA, although it preferred RpD over RpR sequences when present in the same substrate, cleaving 5′ of the ribonucleotide attached to DNA (Figure 1B, left). In contrast, Tm-RNase H2 only recognized RpD sequences, cutting 5′ of the ribo (Figure 1B, middle), and Bst-RNase H3 hydrolyzes regions containing a stretch of several ribonucleotides and could not process a single ribonucleotide embedded into DNA (Figure 1B, right), even when a large excess of enzyme (110 nM) was used, or when Mn2+ replaced Mg2+ (data not shown). Hu-RNase H2 recognizes RpR or RpD structures only when hybridized to DNA (34), whereas some bacterial enzymes can cleave the strand containing RpD junction with RNA or DNA in the opposite strand (35). Sc-RNase H2 acted like the human enzyme cleaving only in regions of RNA/DNA (Supplementary Figure S1, left); however, Tm-RNase H2 could cleave RpD substrates even when the noncleaved strand was RNA (Supplementary Figure S1, right), similar to other bacterial RNases H2 from E. coli and Thermus thermophilus (2).
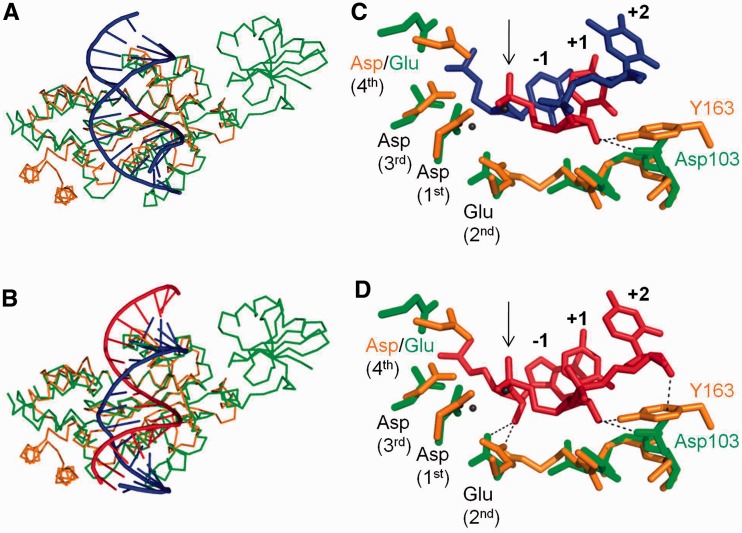
To determine the reason for the different specificity between RNase H2 and RNase H3, we examined which residues of RNase H3 are involved in catalysis by superimposing the structure of Bst-RNase H3 (PDB: 2D0B) and the structure of Tm-RNase H2 in complex with a substrate containing a RpD junction (PDB: 3O3G) (Figure 2A and C). We then replaced the RpD junction substrate with the structure of the RNA/DNA hybrid co-crystallized with Hu-RNase H1 (PDB: 3PUF), and determined the interactions of the Bst-RNase H3/Tm-RNaseH2 superimposition (Figure 2 B and D). Instead of the Tm-RNase H2 Tyr residue essential for catalysis, Bst-RNase H3 has an Asp (D103) that, according to the modeled structure, was within 3.3 and 3.2 Å from the 2′-OH of the ribose residues at positions +1 and +2, respectively. Moreover, the second catalytic residue (Glu-98 of Bst-RNase H3 and Glu-19 of Tm-RNase H2) could interact with the 2′-OH at position -1, suggesting that RNase H3 interacts with at least three consecutive ribonucleotides of the cleaved strand. We tested the activity of Bst-RNase H3 with D103N or D103A substitutions and found that these enzymes have reduced RNase H activity (Figure 1C); especially Bst-RNase H3-D103A that could not form hydrogen bonds with either of the 2′-OH at position +1 or +2, confirming the important role of D103 in substrate recognition and interaction with two consecutive 2′-OH.
Figure 2.
Models of Bst-RNase H3 and Tm-RNase H2 in complex with substrates. The ribbon models of Bst-RNase H3 (in green) and Tm-RNase H2 (in orange) are superimposed, and the complexes with (A) duplex DNA containing single ribonucleotide (DNA in blue and RNA in red) and (B) RNA/DNA hybrid are shown. Active site residues (DEDD motif in Tm-RNase H2 and DEDE motif in Bst-RNase H3) and residues involved in recognition of (C) DpRpD and (D) RpRpR substrates (GRG/Y motif in Tm-RNase H2, GTGD motif in Bst-RNase H3) are shown. Possible hydrogen bonds are indicated by dotted lines. The nucleotides are numbered relative to the scissile phosphate, which is indicated with an arrow. Four catalytic residues (DEDD for Tm-RNase H2 and DEDE for Bst-RNase H3) are also indicated in numerical order.
Creating an Sc-RNase H2 that cleaves RpR not RpD sequences
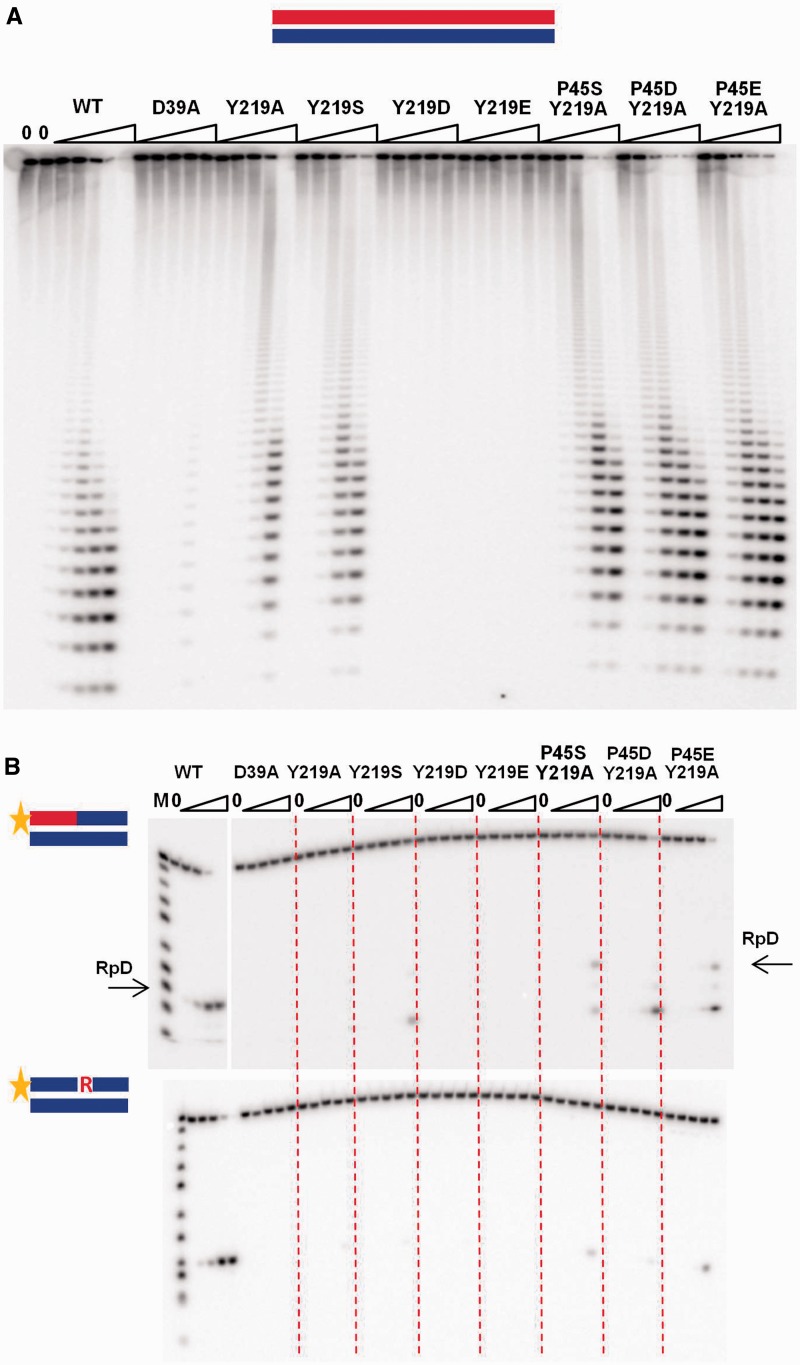
In RNase H3, the residue corresponding to the conserved junction-binding tyrosine from RNase H2 is an alanine (Figure 1A). Tyr to Ala in Sc-Rnh201, the catalytic subunit, resulted in an enzyme with less than 0.1% residual activity with all substrates (Figure 3 and Table 1), as previously shown for Hu-RNase H2, indicating that the Tyr is important for cleavages of both RpD and RpR substrates. To see whether substitution of Y219 by amino acids with OH moieties could provide interaction with the RNA 2′-OH, we replaced Y219 with S, D or E. Rnh201-Y219D and Rnh201-219E were inactive with all substrates, whereas Rnh201-Y219S was inactive for the cleavage of RpD substrates but has higher activity than Rnh201-Y219A with hybrids containing RpR junctions (Figure 3A and B). The structure modeled in Figure 2 suggested that D103 in Bst-RNase H3 plays a role similar to Tm-Y163 in contacting the ribose 2′-OH at position +1. D103 of Bst-RNase H3 is in the same spatial location as P45 of Sc-Rnh201 (Figure 1A, 2C and 2D). Because D103 is a Ser or Glu in other RNases H3 (Figure 1A), we changed P45 residue to D, E or S in the Sc-Rnh201-Y219A. Sc-RNases H2 containing Rnh201-P45D-Y219A or Rnh201-P45E-Y219A had about 40% activity of the wt enzyme for the cleavage of poly-rA/poly-dT substrate, whereas Sc-RNase H2 with Rnh201-P45S-Y219A substitution was less efficient (Figure 3A). In contrast, RpD molecules were essentially not hydrolyzed by yeast RNase H2 enzyme containing Rnh201-Y219A-P45D (Figure 3B). With substrates containing RpR and RpD junctions in the same hybrid (top panel of Figure 3B), the mutant Rnh201- P45D-Y219A showed about 3.1% of the wt activity on RpR structures, while it was less than 0.1% as active as the wt for the RpD cleavage (Table 1). Actually, activity on the 6R substrate may be low due to length of RNA, sequence preferences, or poor stability related to the sequence or the proximity to the end (see Figure 1B). Thus, the accuracy of our model was confirmed. Y219A mutation in Sc-Rnh201 eliminated recognition of RpD and RpR, but the additional replacement of P45 with D provided a key interaction with a 2′-OH of two consecutive ribonucleotides conferring specificity for RpR/DNA cleavage. Therefore, we have converted Sc-RNase H2 into an enzyme that solely cleaves RpR/DNA substrates.
Figure 3.
Cleavage of poly-rA/poly-dT substrate and oligo substrates with Sc-RNase H2 WT and mutants. (A) The uniformly 32P-labeled poly-rA/poly-dT substrate (1 µM) was digested by the Sc-RNase H2 WT and mutants indicated above the gel in the presence of 10 mM MgCl2. The lanes marked with 0 contained no enzyme, and lanes marked with triangle contained increasing amount of the proteins (1 pM, 11 pM, 110 pM, 1.1 nM and 11 nM). Products of the hydrolysis were analyzed by 12% TBE-urea gels and visualized by Phosphorimager. (B) The 5′-32P-labeled 12 mer substrates indicated on the left of the gels were digested by Sc-RNase H2 in the presence of 10 mM MgCl2. The lanes marked with 0 contained no enzyme, and lanes marked with triangle contained increasing amount of the proteins (0.011, 0.11, 1.1 and 11 nM). Products of the hydrolysis were analyzed by 20% TBE-urea gels. The sizes of products were measured based on molecular size markers indicated as M. Cleavage at the 5′ of RpD junction is indicated by an arrow.
Table 1.
Relative activities of yeast RNase H2 derivatives
| 1Ra | 6Ra | 12Ra | poly-rA/ poly-dT | |
|---|---|---|---|---|
| WT | 100 | 100 | 100 | 100 |
| Y219A | <0.1 | <0.1 | <0.1 | <0.1 |
| P45D Y219A | <0.1 | 3.1b (<0.1)c | 25 | 40 |
| G42S | 2.2 | 5.1 | 4 | 8 |
| D39A | <0.1 | <0.1 | <0.1 | <0.1 |
| Rnh202-ΔPIP | 109 | 85 | 112 | 118 |
a12 mer substrates shown in Figure 1B.
bEfficiency for the cleavage at RpR.
cEfficiency for the cleavage at RpD.
Tm-RNase H2 substrate specificity
To examine whether we could change the specificity of Tm-RNase H2 to cleave RpR sequences similarly to eukaryotic RNases H2, we substituted C24 by D and Y163 by A in Tm-RNase H2, which are the equivalents to the Sc-Rnh201 P45D-Y219A substitutions. The Tm-RNase H2 WT enzyme in the presence of Mg2+ cleaves exclusively at the 5′-end of the ribonucleotide of the RpD junction. In the presence of Mn2+, the WT enzyme is able to cleave RpR structures although with lower affinity than RpD sequences. Tm-RNase H2-Y163A had undetectable activity with Mg2+ and very low activity in the presence of Mn2+ with all the substrates (Supplementary Figure S2). Tm-RNase H2-C24D-Y163A was inactive with Mg2+ and only partially active in Mn2+. For the cleavage of RNA6-DNA6/DNA12 substrate, this enzyme cleaves at the 5′-end of both the RpD and RpR sequences, whereas the WT cuts only 5′ of the RpD, suggesting that the mutant lost selectivity for the RpD junction. This indicated that we have identified in the yeast and Tm-RNases H2 the amino acids that confer selectivity for RpD and RpR hydrolysis.
The Gly residue, which in the Tm-RNase H2 co-crystal is part of the GRG motif interacting with the 2′-OH of the RpD substrate, is important for substrate specificity (23). This glycine in the catalytic Hu-RNase H2A is particularly interesting because it is substituted for Ser in patients presenting a severe form of AGS (22). Tm-RNase H2-G21S has low Mn-dependent activity on all substrates and reduced RpD processing (23) in the presence of Mg2+. To understand the effect of the G37S-equivalent mutation in the prokaryotic protein, we solved the crystal structure of Tm-RNase H2-G21S in complex with a duplex DNA containing a single ribonucleotide (Supplementary Figure S3). Surprisingly, in the crystal structure, interactions between the substrate and protein were only modestly perturbed. The main chain of the GRG motif was slightly deformed, still allowing full recognition of the 2′-OH (Supplementary Figure S3C). Moreover, the role of the conserved Tyr in the recognition of the 2′-OH of the scissile phosphate and in the stacking interaction with D of RpD junction was intact, suggesting Tm-RNase H2-G21S retains the ability to recognize RpD junctions. The mutation affected the dihedral angle of the loop containing G21S and the surrounding residues due to the presence of the side chain of Ser21, which needs to be accommodated at the center of protein–nucleic acid interface. These small perturbations around the active site disturbed the binding of metal ion B. The structure suggested that G21S substitution in the GRG loop results in a general defect in catalysis, not a selective loss of RpD or RpR hydrolysis. We made the corresponding change in the catalytic subunit of Sc-RNase H2 and found that the G42S substitution showed reduced activity with the different substrates, having about half the activity with the single-ribonucleotide embedded duplex DNA sequence than with longer stretches of ribonucleotides (Table 1). Similar results have been reported for the mouse (36–38), human (15,34), yeast (25) and Tm-RNase H2 (23).
Recognition of single ribonucleotide by RNase H2 is essential for in vitro ribonucleotide excision repair assay
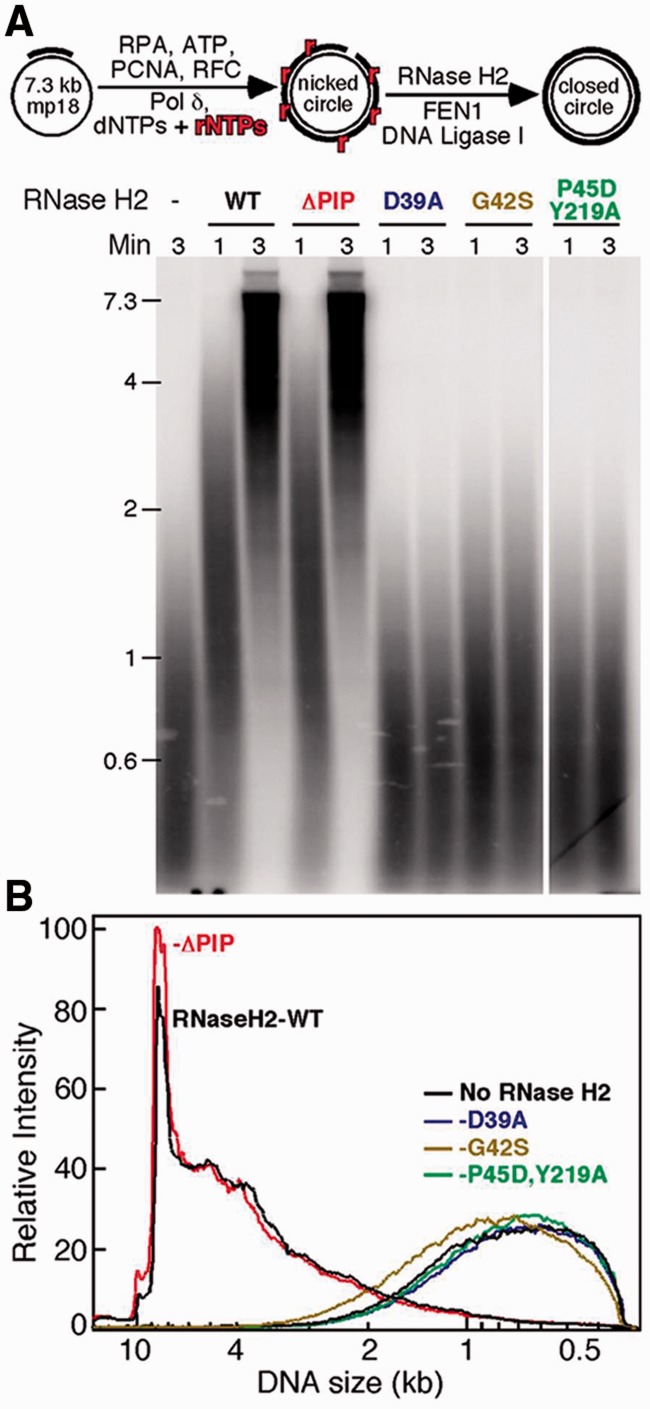
We examined the ability of WT and mutant forms of Sc-RNase H2 to perform in an in vitro ribonucleotide excision repair (RER) assay. Three independent experiments reproducibly gave the results shown in Figure 4. This replication assay relies on several proteins to produce mp18 dsDNAs devoid of misincorporated ribonucleotides as shown in Figure 4A. Alkali treatment results in DNA strand scissions at the ribonucleotide locations as seen in Figure 4A. When Sc-RNase H2 is present, the majority of dsDNA becomes insensitive to alkali treatment (Figure 4A, WT lane after 3 min incubation) indicating the misincorporated ribonucleotides are repaired in an RNase H2-dependent manner. Proliferating Cell Nuclear Antigen (PCNA) is loaded onto the circular DNA by RFC and increases the activities of DNA polymerase δ, FEN1 and ligase1 by interacting with their PCNA-interacting peptide (PIP). However, as we have shown before (39), interaction with PCNA is not necessary for the repair activity of RNase H2, because an RNase H2 with an Rnh202 subunit missing the last 11 amino acid including the PCNA-interacting peptide (PIP) was as active as the wild-type enzyme (Figure 4A, ΔPIP lane after 3 min incubation). Accordingly, the in vitro activity of the ΔPIP protein with different substrates is equivalent to the activity of the WT enzyme (Table 1).
Figure 4.
In vitro RER assay. (A) In vitro RER assay is schematically shown. The 7.3-kb mp18 single-stranded circle DNA was replicated by Polδ and cofactors in the presence of the physiological levels of dNTPs, rNTPs and 32P-labeled dATP at 30°C. After 12 min incubation, FEN1, Ligase1 and wild-type or mutant RNase H2 were added, and the reactions were continued. After 1 or 3 min, aliquots were taken and treated with NaOH for hydrolysis at RpD linkage due to misincorporation of ribonucleotide during replication. DNA was extracted from the aliquots and loaded onto 1% alkali agarose gel. The alkali agarose gel was visualized by Phosphorimager. (B) The radioactivity distribution was scanned and divided by the size distribution to obtain a normalized product distribution as described before (39).
The RNase H2 containing the catalytic subunit Rnh201-P45D-Y219A mutation, which could not cleave RpD substrates in vitro (Figure 3 and Table 1), also did not function to repair misincorporated ribonucleotide in this assay (Figure 4A). Sc-RNase H2-G42S exhibited low activity in single ribonucleotide cleavage (Table 1) and in the RER assay showed a modestly larger distribution of products compared with the catalytically dead Sc-RNase H2-D39A and the Sc-RNase H2-P45D-Y219A mutants (Figure 4B). The in vitro activity of these mutants correlates well with their capacity to process rNMPs in the RER assay.
Mutants defective in RpD cleavage have the same mutator effect as deletion of RNH201
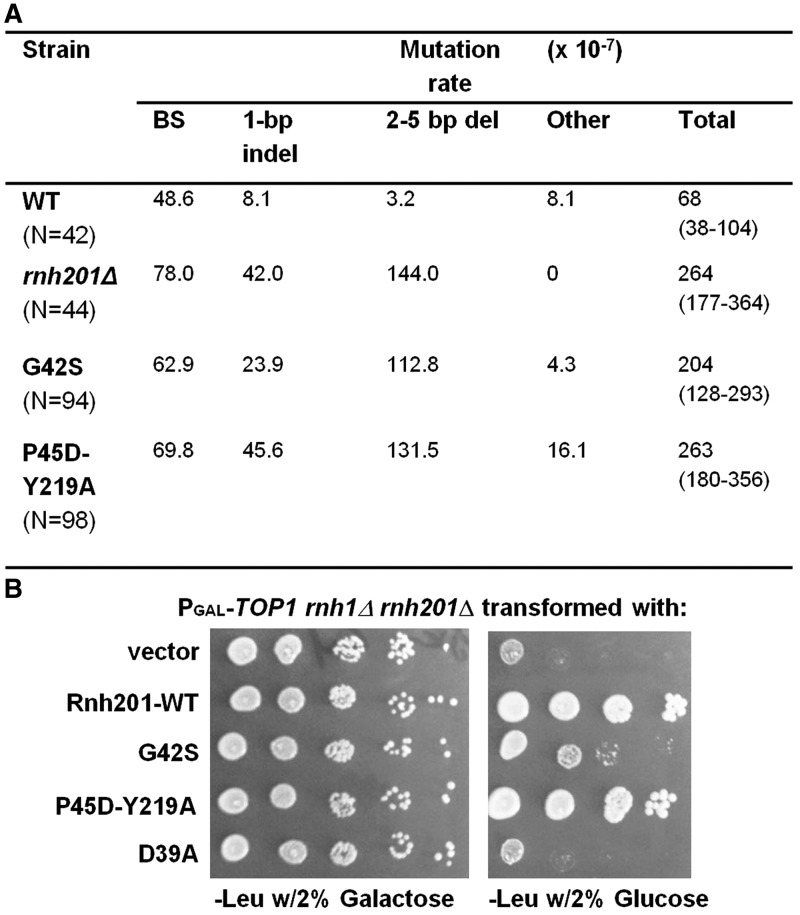
Yeast cells lacking RNH201 accumulate 2–5 bp deletions within short tandem repeats in a topoisomerase 1-dependent manner (6). In combination with error-prone replicative DNA polymerase ε mutant (pol2-M644G), deletion of RNH201 increased the spontaneous mutation rate and the frequency of the 2-5 bp deletions (5). We transformed the double mutant strain, pol2-M644G rnh201Δ, with single copy plasmids expressing RNH201-WT, rnh201-G42S or rnh201-P45D-Y219A driven by their own promoter and a control empty vector. The transformants were analyzed for the spontaneous mutation rate and mutational specificity at the CAN1 locus (Figure 5A). Plasmid-expressed WT RNH201 decreased the mutation rate and showed a mutation spectrum mostly of base substitutions, similar to wild-type cells. However, pol2-M644G rnh201Δ cells expressing rnh201-P45D-Y219A had similar mutation rates and spectra as the deletion strain pol2-M644G rnh201Δ harboring an empty vector (Figure 5A). Cells expressing rnh201-G42S had a slightly reduced rate of 2–5 bp deletions, compared with pol2-M644G rnh201Δ cells expressing rnh201-P45D-Y219A, but still had a much higher accumulation of deletions than wild-type cells. Western analyses demonstrated that WT, G42S and P45D-Y219A Rnh201p were expressed at similar levels in these strains (Supplementary Figure S4).
Figure 5.
In vivo phenotypes of strains rnh201Δ carrying plasmids harboring RNH201 WT and mutants. (A) Saccharomyces cerevisiae strain pol2-M644G rnh201Δ was transformed with plasmid expressing RNH201WT and mutants and empty vector. Strains were plated in canavanine plates and the rates of individual mutations types at CAN1 determined. N, number of mutants sequenced; indel, insertion/deletion/; BS, base substitution. 95% confidence intervals are in parenthesis below total rates. (B) Saccharomyces cerevisiae strain YAEH275 (BY4741 but PGAL1-3HA-TOP1 (KanMx6) rnh201Δ (NatMx6) rnh1Δ (HphMx6)) was transformed with plasmids expressing RNH201-WT, rnh201-G42S, -P45D-Y219A and -D39A and also an empty vector. The transformants were grown at 30°C in liquid SD-Leu medium containing 2% galactose, and series of dilutions were spotted on SD-Leu plates with 2% galactose or 2% glucose and incubated for 3 days at 30°C.
Mutants defective in RpD cleavage can participate in R-loop processing
Next, we determined whether these enzymes can function in vivo in the processing of RNA/DNA hybrids. We chose a system that requires R-loop processing for viability. Loss of Top1 in combination with RNH1 and RNH201 deletions leads to R-loop accumulation and consequent cell death in yeast (40,41). We used yeast strain YAEH275 (PGAL-TOP1 rnh1Δ rnh201Δ), in which the TOP1 gene is expressed from the galactose-inducible GAL1 promoter. Expression is repressed when cells are grown on glucose with associated lethality. YAEH275 was transformed with plasmids expressing wild-type or mutant forms of RNH201 or a control empty vector, and cultures were grown and spotted on selection plates containing either galactose or glucose (Figure 5B). RNH201-WT and rnh201-P45D-Y219A rescued lethality, whereas rnh201-G42S was only partially effective, in accordance with the lower activity on RNA/DNA hybrid substrates of the RNase H2 enzyme containing this mutant protein (Table 1). The catalytically dead rnh201-D39A protein did not rescue lethality in glucose. These results indicate that R-loop removal in vivo parallels that seen in vitro (Table 1).
RNase H2 R-loop processing is sufficient to correct rnh201Δsgs1Δsynthetic sickness
In S. cerevisiae, Sgs1 is crucial for HR repair of double-stranded DNA breaks and for re-establishing of replication after fork collapse (42,43). Deletion of any of the three genes that code for the subunits of the heterotrimeric RNase H2 together with deletion of SGS1 confers a synthetic-sickness phenotype (44), suggesting that the loss of RNase H2 creates a need for recombinational repair.
To examine whether RER or R-loop processing is necessary in the absence of SGS1, we analyzed a set of isogenic strains that were both sgs1Δ and mutated in RNH201 to either eliminate catalytic activity completely (D39A) or eliminate RER but retain R-loop processing (P45D-Y219A or G42S). The growth rate was examined by colony size on YPD plates and by the analysis of doubling time in liquid medium. The doubling time was determined at least four times for each strain. The error bars represent standard deviation (Figure 6A). Chromosomal RNH201-WT, -rnh201-P45D-Y219A or -G42S suppressed the slow growth phenotype, while the strain with active site mutant rnh201-D39A showed the same poor growth as the parental sgs1Δ rnh201Δstrain (Figure 6A). These data were confirmed using plasmid complementation assays (Supplementary Figure S5).
Figure 6.
Effect of RNase H2-mutations in sgs1Δ yeast strain. (A) Doubling time of the yeast strains with RNase H2-mutations in genomic DNA was determined at 30°C in YPD medium. Error bars are standard deviations with P < 0.1 measured by the Ttest in Excel. (B) Representatives of normal cells and abnormal cells from sgs1Δ rnh201Δ background stained with DAPI are shown. (C) Fraction of abnormal cells from log-phase culture was analyzed by gating enlarged cells by FACS, which was correlated with the ratio of abnormal cells observed by microscope (data not shown).
To eliminate the possibility that an RNase H2-independent suppressor mutation had restored the normal growth of the sgs1Δ strain expressing rnh201-G42S and -P45D-Y219A, we replaced the rnh201-G42S and -P45D-Y219A alleles by KanMX-cassette, recreating the rnh201Δ and thereby reproducing the slow growth rate (data not shown). Taken together, these results suggest that the growth defect of the sgs1Δ rnh201Δstrain is caused by R-loops that become stable in the absence of RNase H2 and Sgs1 and that the residual activity of the enzymes containing P45D-Y219A and, surprisingly, G42S mutants is sufficient to complement these defects.
In the sgs1Δ rnh201Δstrain, there was an increase of nonproliferative abnormal cells arrested in G2/M phase (Figure 6B), that seem to be unable to complete the cell cycle, presumably due to replication and repair impairments. By microscopic analysis, we initially noticed increased levels of enlarged dumbbell-shaped cells or cells with abnormal shape as shown in Figure 6B. Next, analysis of the cells for DNA content (propidium iodide (PI) staining) and cell size (side scattering) by Fluorescence-activated cell sorting (FACS) (data not shown) suggested an increased portion of enlarged G2/M-arrested cells in sgs1Δ rnh201Δ mutant, which is in agreement with the microscopic analysis. In Figure 6C, enlarged cells observed by side scattering of FACS data were used to determine the fraction of abnormal cells.
The strain with sgs1Δ rnh201-D39A had elevated level of abnormal cells as sgs1Δ rnh201Δ mutant, whereas strains sgs1Δ rnh201-G42S and sgs1Δ rnh201-P45D-Y219A had the same amount of abnormal cells as single mutant sgs1Δ Figure 6C, indicating that RNase H2 R-loop resolution prevents increased formation of abnormal cells.
To determine whether interaction with PCNA was important in this genetic background, we mutated RNH202 by gene replacement in the strain carrying sgs1Δ to make an RNase H2 enzyme devoid of the PIP sequence. RNase H2-ΔPIP complemented the deletion of RNH202 (Figure 6A), indicating that interaction with PCNA is not necessary for R-loop processing.
RNase H1 overexpression did not complement sgs1Δ rnh201Δsynthetic sickness
R-loop processing can be accomplished by either RNases H1 or H2. This seems to be the case for the R-loops in nucleoli (40). To determine whether RNase H1 can also participate in the processing of the substrates that accumulate in the sgs1Δ rnh201Δ mutant, we transformed the sgs1Δ rnh201Δ strain with a plasmid that carries the RNH1 gene under the strong triosephosphate isomerase (TPI) promoter and looked for complementation of the growth defect of the double mutant. RNase H1 was overexpressed and active (data not shown), but these high levels of RNase H1 did not restore the growth of sgs1Δ rnh201Δ mutant strain to the rate of the single mutant sgs1Δ (Supplementary Figure S6), suggesting that RNase H1 cannot act on or have access to the RNase H2 substrates that accumulate in the absence of Sgs1.
DISCUSSION
Eukaryotic RNases H1 and H2 are important participants in maintaining genome stability by resolving R-loops that form during transcription and by initiating the removal of rNMPs in DNA. Only RNase H2 can process single ribonucleotides in DNA but both enzymes are capable of eliminating RNA/DNA hybrids. Roles of RNase H1 or RNase H2 are usually examined by deleting one or both enzymes, which in S. cerevisiae leads to modest increases in sensitivity to DNA damaging agents (16), but in mouse has devastating consequences, causing embryonic lethality (1,20,21). R-loops naturally form due to transcription errors and are removed by RNase H1 and RNase H2. However, under high transcription (40) and when RNA biogenesis factors are defective (8,9,18), R-loop formation is increased, inducing genome instability. Stable R-loops constitute a block to replication fork progression causing fork collapse, recombination and chromosomal breaks (45,46). The deleterious consequences of the R-loops can be at least partially reversed by overexpression of RNase H1, indicating that normal levels of RNase H1 and RNase H2 are insufficient when there are more abundant R-loops.
In contrast to transcription-induced R-loop formation, single ribonucleotides in DNA occur during genome duplication, and RNase H2 is essential to initiate their removal. In the absence of RNase H2, rNMPs in the genome are aberrantly processed by Top1 (6), originating 2–5 base pairs deletions within short tandem repeats. R-loops and single ribonucleotides in DNA are quite distinct structures that created different challenges to genome integrity, and loss of RNase H2 may result in an accumulation of defects in transcription, replication and repair. Therefore, deletion of the RNH201 gene does not permit the specific assignment of contributions of R-loops and single rNMPs to genome instability.
To assess the separate roles of RNase H1, and particularly RNase H2 in R-loop degradation and RER, we designed a Sc-RNase H2 variant with only two amino acid substitutions in the catalytic subunit (Rnh201-P45D-Y219A), which behaves like a bacterial RNase H3, i.e. the enzyme can participate in R-loop processing but not in the removal of single rNMPs misincorporated in DNA. This mutant constituted an invaluable tool to understand the functions of RNase H2 under normal and pathological condition.
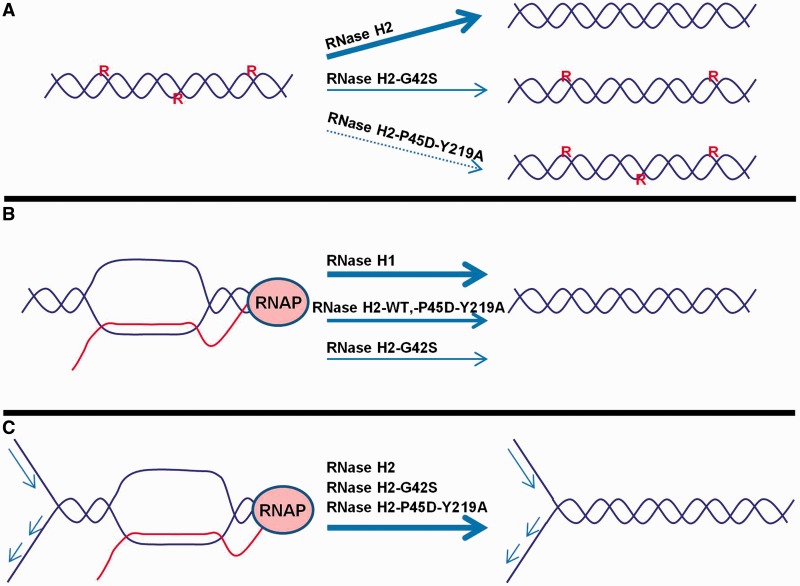
This mutant allowed us to conclusively correlate the 2–5 bp deletions observed in rnh201Δ strains (6) with single ribonucleotide misincorporation in the genome. It also suggested a connection between R-loops and Sgs1-mediated replication reinitiation at stalled forks, as well as identifying R-loops that can be processed by RNase H2, but not by RNase H1 (Figure 7).
Figure 7.
Effects of RNase H2 mutations in single ribonucleotide processing and R-loop resolution. (A) Single ribonucleotides incorporate in the genome can be processed by RNase H2. RNase H2-P45D-Y219A mutant cannot remove single rNMPs and the AGS-related RNase H2-G42S has low but detectable activity. Both mutations lead to the accumulation of 2–5 bp deletions in genomic DNA, although RNase H2-G42S at a lower rate. (B) R-loops that form during transcription are processed primarily by RNase H1. RNase H2 WT and P45D-Y219A mutant can process equally well these structures, whereas RNase H2-G42S is not as effective. (C) R-loops involved in replication fork collapse have unique access to RNase H2. The WT enzyme and the mutants, RNase H2-G42S and RNase H2-P45D-Y219A, are equally active on these substrates.
Genome instability in yeast is greater when both RNH1 and RNH201 genes are deleted than when single deletions are present, although in combination with defects of RNA biogenesis factors, the deletion of RNH1 is more deleterious than that of RNH201 (18). This seems to indicate that RNases H1 and H2 have partially overlapping specificities in the resolution of R-loops, but RNase H1 is the major player in removing transcription-associated R-loops (Figure 7B). Conversely, our results suggest that there are R-loops that are uniquely processed by RNase H2. These loops might be involved in DNA transactions during replication/repair processes (Figure 7C), and RNase H2 could have access to them by interaction with PCNA and the replication machinery via the PIP motif of the Rnh202 subunit. Although no phenotype has been associated thus far with the deletion of the PIP motif, it is likely than under stress circumstances or other conditions not yet tested, the PIP might be required for the function of RNase H2. Moreover, the PIP sequence may not be the only means of RNase H2 interaction with DNA replication/repair machines.
Both RNases H complemented the lethal defects caused by the accumulation of R-loops created under high transcription in a topoisomerase-defective rnh1Δ rnh201Δ mutant, as did the Rnh201-P45D-Y219A mutant that still processes R-loops (Figure 5B). However, only RNase H2 was effective in correcting the slow growth of sgs1Δ rnh201Δ strains (Figure 6 and Supplementary Figure S6). The synthetic defect of sgs1Δ rnh201Δ could be due to misincorporated rNMPS in DNA, especially because sgs1Δ rnh1Δ strains do not exhibit a slow growth phenotype. However, the fact that the mutant completely devoid of single-ribonucleotide processing activity could reverse the slow growth defect of sgs1Δ rnh201Δ (Figure 6A) indicated that persistent R-loops have deleterious effects in this strain.
R-loops present from bacteria to humans constitute a block to replication leading to replication fork stalling (10). Replication after fork arrest can be reinitiated by HR (46), for which Sgs1 is essential (47). Sgs1 recruits the intra-S phase checkpoint kinase Rad53 to stalled forks and contributes to replication fork stability by dissolving fold-back structures and Holliday junctions formed at collapsed forks (48). Our study establishes a link between stable R-loops and Sgs1-mediated replication restart.
The forms of RNase H2 containing the AGS-related mutation Rnh201-G42S, which in vitro has low activity on RNA/DNA hybrids, could reverse the slow growth and accumulation of abnormal cells in the sgs1Δ background, indicating that even a very impaired enzyme has sufficient activity to process the small number of hybrids formed in this strain. However, the mutant RNase H-G42S was only partially effective in YAEH275 (PGAL-TOP1 rnh1Δ rnh201Δ), possibly related to the higher abundance of R-loops in this strain, which might exceed the capacity of the defective RNase H2-G42S. Alternatively, for the resolution of R-loops in the sgs1Δ strain, RNase H2 could be part of a larger complex that may confer stability to the mutant enzyme and consequently increasing enzymatic activity.
In vitro, the mutant RNase H2-G42S had low but detectable activity on single rNMP (Table 1) and was partially effective in the RER assay (Figure 4). In vivo, although the RNase H2-Rnh201-P45D-Y219A mutant strain exhibited a 3.9-fold increase over the wt in mutation rate, the same as the Rnh201 deletion, the Sc-RNase H2 containing Rnh201-G42S has an increase slightly lower (3-fold over wt) mutation rate, and fewer 2–5 bp deletions (Figure 5A), indicating that this mutant and presumably the human enzyme with the corresponding mutation are able to remove single rNMPs in the genome at some low frequency (Figure 7A).
In agreement with the in vitro and in vivo results, the Tm-RNase H2 G21S structure in complex with a single ribonucleotide–substituted duplex DNA showed preserved interactions of the RpD junction with the conserved Tyr and with the GRG (SRG in the mutant) motif, indicating the enzyme has reduced enzymatic activity while maintaining substrate specificity. These data are important to understand the defects associated with the corresponding mutation in the human enzyme, which is responsible for a severe form of AGS, providing a note of caution in interpreting data when both activities are decreased or absent.
Recently, defects in mouse due to deletion or low expression of RNase H2 (20,21) have been attributed to genome instability associated with the presence of rNMPs in DNA, without considering the possible effects of unresolved R-loops, and RNA/DNA hybrids formed during retroelements reverse transcription. However, the deletion of another AGS-inducing gene, Trex1, has been shown to cause the accumulation of single-stranded DNA derived from endogenous retroelements (49). In addition, antiretroviral drugs prevented autoimmune response and restored the viability of Trex1-knockout mice (50). Also SAMHD1, a more recently described AGS-associated protein (51), has been found to restrict the infection of dendritic and other myeloid cells by human immunodeficiency virus type 1 (HIV-1) by hydrolyzing intracellular dNTPs (52). Low concentrations of dNTPs, due to the action of SAMHD1, inhibit the synthesis of the viral RNA by reverse transcriptase (RT). In the absence of SAMHD1, higher levels of dNTPs could facilitate reverse transcription of endogenous retroelements. AGS substrates arising via reverse transcription of endogenous RNAs would lead to standard RNA/DNA duplexes that could be resolved by RNase H2 and RNase H1. The fact that RNase H1 does not cause AGS suggests that RNase H2 has unique access to these substrates. The mutant described in this study that could degrade RNA/DNA hybrids formed by RT but could not remove single rNMPs in DNA will be enormously important to address the defects associated with AGS.
Here, we have shown that the multiple activities of RNase H2 can provide quite different functions in vivo. Our mutant RNase H2 lacking RpD substrate cleavage allows us and others to precisely analyze the contributions of R-loops and single rNMPs in DNA to the in vivo phenotypes until now associated with deletion of RNase H2. It will be particularly interesting to see the pattern of changes in gene expression when the rnh201-P45D-Y219A gene is present compared with the 349 changes when RNH201 is deleted (53), and how the loss of single-ribonucleotide activity of RNase H2 affects the general genomic instability described by Wahba et al. (18).
ACCESSION NUMBERS
RCSB ID code rcsb075483 and PDB ID code 4HHT.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6, Supplementary Tables 1–3 and Supplementary References [5,31,40,53–55].
FUNDING
Intramural Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (in part); National Institutes of Health [GM032431, in part] to P.M.B.; Wellcome Trust International Senior Research Fellowship [081760 to M.N.]. M.N. is an HHMI International Early Career Scholar. Funding for open access charge: Intramural Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Foiani for strain Cy8338, T. Kunkel for strain Δ|(-2)|-7B-YUNI300 and A. El Hage and D. Tollervey for strain YAEH275.
REFERENCES
- 1.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 3.Champoux JJ, Schultz SJ. Ribonuclease H: properties, substrate specificity and roles in retroviral reverse transcription. FEBS J. 2009;276:1506–1516. doi: 10.1111/j.1742-4658.2009.06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundström EB, Burgers PMJ, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl Acad. Sci. USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundström EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim N, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast Topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl Acad. Sci. USA. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutta D, Shatalin K, Epshtein V, Gottesman M, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M, Kanaya S. Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry. 1998;38:605–618. doi: 10.1021/bi982207z. [DOI] [PubMed] [Google Scholar]
- 13.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 14.Eder PS, Walder RY, Walder JA. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 15.Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 2009;37:96–110. doi: 10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arudchandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch RJ. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells. 2000;5:789–802. doi: 10.1046/j.1365-2443.2000.00373.x. [DOI] [PubMed] [Google Scholar]
- 17.Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl Acad. Sci. USA. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahba L, Amon J, Koshland D, Vuica-Ross M. RNase H and Multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 20.Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiller B, Achleitner M, Glage S, Naumann R, Behrendt R, Roers A. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J. Exp. Med. 2012;209:1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat. Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 23.Rychlik MP, Chon H, Cerritelli SM, Klimek P, Crouch RJ, Nowotny M. Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol. Cell. 2010;40:658–670. doi: 10.1016/j.molcel.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chon H, Matsumura H, Koga Y, Takano K, Kanaya S. Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new Type 2 RNase H with TBP-like substrate-binding domain at the N terminus. J. Mol. Biol. 2006;356:165–178. doi: 10.1016/j.jmb.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 25.Rohman MS, Koga Y, Takano K, Chon H, Crouch RJ, Kanaya S. Effect of the disease-causing mutations identified in human ribonuclease (RNase) H2 on the activities and stabilities of yeast RNase H2 and archaeal RNase HII. FEBS J. 2008;275:4836–4849. doi: 10.1111/j.1742-4658.2008.06622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller U, Darowski N, Fuchs MR, Förster R, Hellmig M, Paithankar KS, Pühringer S, Steffien M, Zocher G, Weiss MS. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J. Synch. Rad. 2012;19(Pt 3):442–449. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology. New York: Academic Press; 2007. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallog. D, Biol. Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D, Biol. Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storici F, Resnick MA. The Delitto Perfetto approach to in vivo site−directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. In: Judith LC, Modrich P, editors. Methods in Enzymology.DNA Repair, Part B. New York: Academic Press; 2006. pp. 329–345. [DOI] [PubMed] [Google Scholar]
- 33.Shcherbakova PV, Kunkel TA. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell. Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Figiel M, Chon H, Cerritelli SM, Cybulska M, Crouch RJ, Nowotny M. The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutieres syndrome defects. J. Biol. Chem. 2011;286:10540–10550. doi: 10.1074/jbc.M110.181974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtani N, Tomita M, Itaya M. Junction ribonuclease activity specified in RNases HII/2. FEBS J. 2008;275:5444–5455. doi: 10.1111/j.1742-4658.2008.06673.x. [DOI] [PubMed] [Google Scholar]
- 36.Perrino F, Harvey S, Shaban N, Hollis T. RNaseH2 mutants that cause Aicardi-Goutieres syndrome are active nucleases. J.Mol. Med. 2009;87:25–30. doi: 10.1007/s00109-008-0422-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaban NM, Harvey S, Perrino FW, Hollis T. The structure of the mammalian RNase H2 complex provides insight into RNA·DNA hybrid processing to prevent immune dysfunction. J. Biol. Chem. 2010;285:3617–3624. doi: 10.1074/jbc.M109.059048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coffin SR, Hollis T, Perrino FW. Functional consequences of the RNase H2A subunit mutations that cause Aicardi-Goutières syndrome. J. Biol. Chem. 2011;286:16984–16991. doi: 10.1074/jbc.M111.228833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El Hage A, French SL, Beyer AL, Tollervey D. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.French SL, Sikes ML, Hontz RD, Osheim YN, Lambert TE, El Hage A, Smith MM, Tollervey D, Smith JS, Beyer AL. Distinguishing the roles of topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol. Cell. Biol. 2011;31:482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, Foiani M, Branzei D, Rothstein R. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J. 2009;28:915–925. doi: 10.1038/emboj.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hegnauer AM, Hustedt N, Shimada K, Pike BL, Vogel M, Amsler P, Rubin SM, van Leeuwen F, Guenole A, van Attikum H, et al. An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 2012;31:3768–3783. doi: 10.1038/emboj.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ooi SL, Shoemaker DD, Boeke JD. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 45.Aguilera A, Garcia-Muse T. R Loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat. Rev. Genet. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA Helicases in DNA Repair. Annu. Rev. Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hegnauer AM, Hustedt N, Shimada K, Pike BL, Vogel M, Amsler P, Rubin SM, van Leeuwen F, Guenole A, van Attikum H, et al. An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 2012;31:3768–3783. doi: 10.1038/emboj.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beck-Engeser G, Eilat D, Wabl M. An autoimmune disease prevented by anti-retroviral drugs. Retrovirology. 2011;8:91. doi: 10.1186/1742-4690-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arana ME, Kerns RT, Wharey L, Gerrish KE, Bushel PR, Kunkel TA. Transcriptional responses to loss of RNase H2 in Saccharomyces cerevisiae. DNA Repair. 2012;11:933–941. doi: 10.1016/j.dnarep.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 55.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.