Abstract
Although the human mitochondrial genome has been investigated for several decades, the proteins responsible for its replication and expression, especially nucleolytic enzymes, are poorly described. Here, we characterized a novel putative PD-(D/E)XK nuclease encoded by the human C20orf72 gene named Ddk1 for its predicted catalytic residues. We show that Ddk1 is a mitochondrially localized metal-dependent DNase lacking detectable ribonuclease activity. Ddk1 degrades DNA mainly in a 3′–5′ direction with a strong preference for single-stranded DNA. Interestingly, Ddk1 requires free ends for its activity and does not degrade circular substrates. In addition, when a chimeric RNA–DNA substrate is provided, Ddk1 can slide over the RNA fragment and digest DNA endonucleolytically. Although the levels of the mitochondrial DNA are unchanged on RNAi-mediated depletion of Ddk1, the mitochondrial single-stranded DNA molecule (7S DNA) accumulates. On the other hand, overexperssion of Ddk1 decreases the levels of 7S DNA, suggesting an important role of the protein in 7S DNA regulation. We propose a structural model of Ddk1 and discuss its similarity to other PD-(D/E)XK superfamily members.
INTRODUCTION
Mitochondria play a pivotal role both in the life of the cell and its fate. Their unique feature is that their composition and function depend on genes encoded by two physically separate genomes—nuclear and mitochondrial. Most of mitochondrial proteins are encoded in the nuclear genome, synthesized in the cytoplasm and then imported into mitochondria (1). Human mitochondrial DNA (mtDNA) contains 37 genes, of which 13 encode proteins of the oxidative phosphorylation system, whereas expression of the 22 provides tRNA species, and two encode rRNAs necessary for mitochondrial gene translation (2). Physically, the human mitochondrial genome is a circular double-stranded DNA molecule organized in nucleoprotein structures referred to as nucleoids (3,4). The copy number of mtDNA varies depending on cell type and metabolic conditions. The organization of human mitochondrial genetic information is notable for its compactness. Mitochondrial genes lack introns and in most cases are separated by only a few nucleotides, or even overlap. The longest non-coding mtDNA fragment lies between the genes coding for tRNAPro and tRNAPhe. This fragment, called the non-coding region (NCR), encompasses most of the cis-regulatory elements involved in mtDNA transcription and replication (1,2).
Single-stranded DNA (ssDNA) arising from the NCR, 7S DNA, hybridizes to some mtDNA molecules to form a triple-stranded structure (D-loop). Although the role of 7S DNA and the D-loop remains unclear, the D-loop may be a product of stalled or aborted mtDNA replication or could play a role in protein recruitment to the primary control region (5).
The mitochondrial genetic system requires the activity of various RNA and DNA nucleases. Moreover, on stimulation of cell death pathways, mitochondria are known to release several nucleases, including the promiscuous nuclease EndoG, which has both DNase and RNase activity and is involved in apoptosis (6). Some mitochondrial RNases have also been identified, for example, RNase P (7) and tRNase Z (8), which process primary mitochondrial RNA (mtRNA) and subsequently excise tRNAs; PDE12, which deadenylates mt-mRNA (9,10); PNPase, which degrades mtRNA (11); and RNase L, which is involved in stress-induced degradation of mtRNA (12,13). However, it is clear that the list of mitochondrial RNases is far from complete, but their identification using proteomic, enzymatic or bioinformatic approaches has so far been unsuccessful (14).
Although DNA is regarded as a stable, low turnover molecule, its replication, repair and recombination require the activity of nucleases. In humans, the exo-/endonuclease Dna2 was shown to localize to both the nucleus (15) and mitochondria (15,16). In vitro studies indicated that Dna2 is a structure-specific nuclease that preferentially acts on forked and flap DNA substrates (17). This supports the role of hDna2 in mtDNA stability and maintenance. Functional studies showed that RNAi-mediated depletion of the helicase/nuclease hDna2 decreases replication intermediate levels and impairs repair of mtDNA damage induced by hydrogen peroxide treatment (15,16). In nuclei, Dna2 cooperates with the endo-/exonuclease FEN1 to process long flap structures that can form during Okazaki fragment maturation or DNA repair (18–21). The presence of FEN1 in human mitochondria is a subject of debate, with some reports supporting a mitochondrial localization (16,22) and others finding no evidence for mitochondrial FEN1 (23). Moreover, functional studies on the involvement of FEN1 in mtDNA repair are inconsistent. Zheng et al. (16) showed that immunodepletion of FEN1 from mitochondrial lysates impairs the capability of the extract to process substrates that mimic DNA lesions, indicating that FEN1 functions in mtDNA repair. In contrast, Tann et al. (24) suggested that EXOG, but not FEN1, is responsible for long-patch base excision repair in human mitochondria. The former protein was identified as a paralog of the EndoG nuclease (25). EXOG was shown to localize to the mitochondrial intermembrane space and/or inner membrane (25) and has both exo- and endonucleolytic activity that acts in a 5′–3′ direction with a preference for single-stranded DNA substrates (25). Taken together, knowledge of mitochondrial DNases is fragmentary, and there are likely nucleases that are yet to be revealed. For example, 7S DNA has a high turnover rate (26–28), but its degrading enzyme remains unknown.
Many nucleases, including Dna2, belong to the PD-(D/E)XK phosphodiesterase superfamily, which is a large and diverse protein group that encompasses many nucleic acid cleavage enzymes involved in important biological processes such as DNA restriction (29), tRNA splicing (30), transposon excision (31), DNA recombination (32), Holliday junction resolution (33), DNA repair (34) and Pol II termination (35). The common conserved structural core of PD-(D/E)XK proteins consists of a central, four-stranded, mixed β-sheet flanked on either side by an α-helix (with a αβββαβ topology) to form a scaffold that exposes the catalytic residues from the relatively conserved PD-(D/E)XK motif (36). In addition to this motif, other conserved residues often contribute to active site formation and play various catalytic roles that include coordination of up to three divalent metal ion cofactors.
Here, we characterized a novel putative PD-(D/E)XK nuclease named Ddk1 based on its predicted catalytic residues. We show that Ddk1 is a metal-dependent nuclease that localizes to mitochondria and is necessary for proper level of 7S DNA. Ddk1 lacks detectable ribonuclease activity and degrades DNA mainly in a 3′–5′ direction with a strong preference for single-stranded DNA. Interestingly, Ddk1 requires free ends for its activity. Finally, we propose a structural model of Ddk1 and discuss its similarity to other PD-(D/E)XK superfamily members.
MATERIALS AND METHODS
Bioinformatic analysis
A sequence search for Ddk1 homologs was performed against the NCBI non-redundant protein database using PSI-BLAST (37) (two iterations, E-value threshold 0.005). The resulting set of 244 sequences was clustered with CLANS (38) to visualize relationships between Ddk1-like sequences and other closely related PDDEXK_1 (PF12705) protein family members. Multiple sequence alignment of the identified protein sequences was derived using PCMA (39) and manually adjusted. The sequence-to-structure alignment between Ddk1-like proteins and selected distantly related structures was built using a consensus alignment approach and 3D assessment (40) based on the results of Meta-BASIC and 3D-Jury (41), as well as conservation of critical active site residues and hydrophobic patterns. A 3D model of human Ddk1 was generated with Modeller (42) using the Escherichia coli RecE crystal structure (pdb|3h4r) (43) as a template. Cellular localization of Ddk1-like proteins was predicted with MultiLoc (44), TargetP (45) and Mitoprot (46) web services.
Plasmid construction
DNA cloning was performed using procedures described in Supplementary Data. All constructs are listed in Supplementary Table S1.
Ddk1 purification and multiangle light scattering analysis
Ddk1 was overexpressed in E. coli as an N-terminal fusion with a 6xHis and SUMO-containing tag and purified by affinity chromatography. The fusion protein was cleaved using SUMO protease and subjected to gel filtration. A detailed protocol for protein purification and determination of absolute molar mass by multiangle light scattering (MALS) can be found in Supplementary Data.
Substrates
5′- end labelling of substrates was performed using [γ-32P] adenosine triphosphate (Hartmann Analytic) with T4 polynucleotide kinase (NEB) and 3′-labelling was performed using terminal transferase (NEB) with [α-32P] dATP, respectively. After labelling, oligonucleotides were subjected to phenol–chloroform extraction, precipitated and purified by denaturing or native polyacrylamide gel electrophoresis for single- or double-stranded substrates, respectively, as previously described (47). To obtain double-stranded substrates, before electrophoresis, labelled oligonucleotides were mixed with complementary oligonucleotides, heated for 10 min at 95°C and slowly cooled to room temperature. For circularization, 5′ labelled 44DNA oligonucleotides were treated with CircLigase II ssDNA ligase (Epicentre, CL9021K) according to the manufacturer’s instructions and purified by urea–polyacrylamide gel electrophoresis (PAGE). The RNA ladder was obtained by alkaline hydrolysis of 34RNA oligonucleotide using the Alkaline Hydrolysis Buffer (Ambion 9750G) according to manufacturer’s recommendations. The 1–24-nt DNA ladder was prepared by mixing 24 synthetic oligodeoxyribonucleotides: 24DNA and its truncated derivatives missing consecutive 3′ residues. The mixtures were subjected to 5′ end labelling with T4 polynucleotide kinase (NEB). Oligonucleotides were purchased from Metabion (Germany) or FutureSynthesis (Poland), and the sequences are listed in Table 1.
Table 1.
Oligonucleotides used in this study
| Name | Length | Sequence | Producer/reference |
|---|---|---|---|
| v81 | 34 | TTGCCGATGAACTTTTTTTTTTGATCGAGACCTT | FutureSynthesis/(48) |
| 24DNA | 24 | CGACTGGAGCACGAGGACACTGAC | FutureSynthesis/this study |
| 44DNA | 44 | CGACTGGAGCACGAGGACACTGACATGGACTGAAGGAGTAGAAA | FutureSynthesis/this study |
| 44DNAcomp | 44 | TTTCTACTCCTTCAGTCCATGTCAGTGTCCTCGTGCTCCAGTCG | FutureSynthesis/this study |
| 44DNA-RNA | 44 | CGACTGGAGCACGAGGACACTGACAUGGACUGAAGGAGUAGAAA | Metabion/this study |
| 44RNA-DNA | 44 | CGACUGGAGCACGAGGACACUGACATGGACTGAAGGAGTAGAAA | Metabion/this study |
| 44RNA-DNA-RNA | 44 | CGACUGGAGCACGAGGACACTGACATGGACTGAAGGAGUAGAAA | FutureSynthesis/this study |
| 44RNA | 44 | CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA | Metabion/(49) |
| 44RNAcomp | 44 | UUUCUACUCCUUCAGUCCAUGUCAGUGUCCUCGUGCUCCAGUCG | Metabion/this study |
| RNA17-5A | 23 | CCCCACCACCAUCACUUAAAAA | Metabion/(50) |
| RNA17-2A | 19 | CCCCACCACCAUCACUUAA | Metabion/(50) |
| 34RNA | 34 | CGACUGGAGCACGAGGACACUGACAUGGACUGAA | FutureSynthesis/this study |
| 22RNA | 22 | CGACUGGAGCACGAGGACACUG | FutureSynthesis/this study |
| 20RNA | 20 | CCCCACCACCAUCACAAAAA | FutureSynthesis/this study |
Ribonucleotides are underlined.
Nuclease assays
The standard enzymatic assay was performed in a 20 μl of mixture containing 10 pmoles of substrate, 0.5 pmole Ddk1, bovine serum albumin (BSA) (0.1 μg/μl), NaCl (125 mM), MgCl2 (5.12 mM), Tris–HCl pH 8.0 (10 mM) and DTT (1 mM). Mixtures differing from this standard are described in the figure legends. Reactions were incubated at 37°C for the indicated time and terminated by adding an equal volume of formamide loading dye [90% formamide, 20 mM ethylenediaminetetraacetic acid (EDTA), 0.03% bromophenol blue, 0.03% xylene cyanol in 1× TBE] and immediately frozen by immersion in liquid nitrogen. Reaction products were resolved on denaturing 12, 15 or 20% polyacrylamide, 8 M urea, 1× TBE gels and visualized using autoradiography.
Cell culture
HeLa or Flp-In 293 T-REx cells (Invitrogen) were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) at 37°C and 5% CO2. Exogenous gene expression was induced by addition of tetracycline (25 ng/ml) to the culture medium.
siRNA transfection
HeLa cells were plated to reach 40–50% confluence in 24 h and subjected to Stealth RNA (Invitrogen) transfection on the next day. Transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s recommendations. The Stealth RNA were used at a final concentration of 10 nM and are listed in Table 2. When indicated, cells were passaged 2 days after transfection and subjected to a second round of transfection on the next day according to the same protocol and collected on the next 2 and 4 days.
Table 2.
StealthRNA used in this study
| Name | Gene | Oligo ID |
|---|---|---|
| Ddk1A | DDK1 | HSS132389 |
| Ddk1B | DDK1 | HSS132390 |
| EXOGA | EXOG | HSS115057 |
| EXOGB | EXOG | HSS115058 |
| FEN1A | FEN1 | HSS103627 |
| FEN1B | FEN1 | HSS103629 |
| DNA2A | DNA2 | HSS141856 |
| DNA2B | DNA2 | HSS141857 |
| TwinkleA | Twinkle | HSS125596 |
| TwinkleB | Twinkle | HSS125597 |
| Neg | Negative control | Stealth™ RNAi Negative Control Med GC |
Establishing stable cell lines
Stable cell lines were established using Flp-In 293 T-REx cells (Invitrogen) according to the protocol described previously (51).
Localization studies
For localization of transiently expressed FLAG-tagged Ddk1, HeLa cells were cultured on glass cover slips for 24 h, then transfected with Ddk1-FLAG encoding plasmid (pRS570) using the TransIT® LT2020 reagent (Mirus) and subjected to immunofluorescence staining on the next day as described previously (51). Cells were incubated with MitoTracker Orange CMTMRos (100 nM) (Molecular Probes) for 20 min at 37°C, washed 2 times with phosphate buffered saline (PBS) and fixed with 3.7% formaldehyde for 25 min at room temperature. All subsequent steps were carried out at room temperature. The cells were washed three times with PBS and permeabilized with 0.5% Triton X-100 in 10% FBS/PBS for 15 min. After washing with PBS, cells were blocked with PBS solution containing 10% FBS for 30 min. Primary and secondary antibodies were diluted with blocking solution. Cells were incubated with primary anti-FLAG M2 antibody (Sigma, 1:250) for 1 h. Following three washes with PBS, secondary antibodies conjugated with Alexa Fluor 488 were applied at 1:900 (Molecular Probes) for 1 h. Cells were washed three times with PBS, and the nuclei stained by a 5 min incubation in Hoechst 33342/PBS solution (1 μg/ml) followed by a subsequent PBS wash. Cells were mounted in ProLong Gold antifade reagent (Invitrogen) and subjected to microscopy analysis. Microscopy was performed on a FluoView FV10i (Olympus) confocal microscope with a 60× water immersion objective (NA 1.2). The same procedure was used for Ddk1-FLAG localization in stable 293 cell lines except that cells were cultured on poly-D-lysine coated glass cover slips, and slides were analysed using a FluoView1000 confocal microscope (Olympus) with a PLANAPO 60.0 × 1.40 oil objective. To analyse Ddk1-EGFP localization, HeLa cells were transfected with the appropriate DNA construct (pRS572) using the TransIT® LT2020 reagent (Mirus). On the next day, cells were incubated with MitoTracker Orange CMTMRos (100 nM) (Molecular Probes) for 20 min at 37°C, washed once with culturing medium and twice with PBS and fixed with 3.7% formaldehyde for 25 min at room temperature. After washing twice with PBS and staining the nuclei as described earlier in the text, cells were mounted in ProLong Gold antifade reagent (Invitrogen) and subjected to microscopy analysis using a FluoView FV10i (Olympus) confocal microscope with a 60× water immersion objective (NA 1.2).
Isolation of total DNA and Southern blot analysis
Total DNA was isolated by phenol–chloroform extraction. Cells (2–3 × 106) were harvested in RSB buffer (10 mM Tris–HCl pH 7.4, 10 mM NaCl, 10 mM EDTA) containing proteinase K (400 µg/ml, Fermentas) and RNase A (150 µg/ml, Sigma) and lysed by addition of sodium dodecyl sulphate (0.9%). The cells were then incubated for 3.5 h at 37°C with gentle mixing every 15–20 min. After incubation, DNA was extracted by sequential extraction with equal volumes of phenol, phenol–chloroform and chloroform solutions (all from Sigma). DNA was precipitated by mixing with 0.1 volume of 10 M ammonium acetate and 1 volume of isopropanol, incubating for 30 min at room temperature, and centrifugation (16 400g, 30 min, 18°C). The precipitated DNA was then washed with 75% ethanol and dissolved in water. To analyse the level of mtDNA, total DNA was digested overnight with NcoI and DraI (both from Fermentas), and 2 µg were resolved by standard agarose (1%) gel electrophoresis. Subsequently, the gel was immersed and mixed for 0.5 h in each of the following solutions: depurination (0.2 M HCl), denaturation (1.5 M NaCl, 0.5 M NaOH) and neutralization (0.5 M Tris–HCl pH 7.0, 1.5 M NaCl) with brief rinsing with water between solutions. DNA was transferred to a Nytran-N membrane (Whatman Schleicher & Schuell BioScience) by overnight upward capillary transfer using 10× SSC (1.5 M sodium chloride, 150 mM sodium citrate) and immobilized by ultraviolet cross-linking. Hybridizations were performed overnight in PerfectHyb Plus buffer (Sigma) at 64°C. Mitochondrial and nuclear DNA was detected using [α-32P] dATP-labelled probes (Hartmann Analytic) complementary to the ND2 and 28rDNA genes, respectively, and prepared by random priming with the HexaLabel DNA Labeling Kit (Fermentas). Polymerase chain reaction products amplified with the following primers were used as templates for probe preparation: ND2—RSZ266 taatacgactcactatagggtctgagtcccagaggttac and RSZ267 atttaggtgacactatagaattcaggtgcgagatagtag; 28rDNA—RSZ547 gcctagcagccgacttagaactgg and RSZ548 ggcctttcattattctacacctc (28rDNA). To analyse 7S DNA, the same procedure was applied, except that the total DNA was digested overnight with EcoRI (Fermentas) and before electrophoresis, samples were heated for 6 min at 95°C and cooled on ice. To detect 7S DNA, a polymerase chain reaction product spanning bp 16109–16437 of mtDNA was used as a template to prepare radiolabelled probes. Following hybridization, filters were exposed to PhosphorImager screens (FujiFilm) that were scanned using a Typhoon FLA 9000 scanner (GE Healthcare).
Western blot
Protein samples were prepared and processed as described previously (51) using anti-FLAG M2 (1:2000, Sigma, F3165) or anti-Ddk1 (c20orf72) antibodies (1:300, Sigma, HPA040913). Primary antibodies were detected with goat anti-mouse or anti-rabbit peroxidase conjugated secondary antibodies (Calbiochem) and visualized using an Immun-Star WesternC Chemiluminescence Kit (Bio-Rad) according to the manufacturer’s instructions.
RESULTS
Protein identification, structural and phylogenetic analysis
We recently performed exhaustive distant similarity searches to identify new PD-(D/E)XK superfamily members in the human proteome (36). These searches identified a protein of unknown function having two aspartates and lysine (DDK) in the predicted catalytic site. Consequently, the protein encoded by the C20orf72 gene was named Ddk1. In silico prediction of the cellular localization of Ddk1-like proteins suggested that they can be targeted to mitochondria (see later in the text). Thus, we subjected Ddk1 to more detailed studies.
Ddk1-like proteins group together hypothetical and uncharacterized proteins that are present exclusively in Opisthokonta (Monosiga, Capsaspora and Metazoa) but are not observed in all Drosophila species. One-to-one orthologs can be found in most of the sequenced chordate genomes, which indicates that this protein has an evolutionarily conserved function. Ddk1-like proteins are a subgroup of the large PDDEXK_1 protein family (PF12705) that clusters >5000 sequences, including various helicases and exonucleases that are largely involved in double-strand break repair.
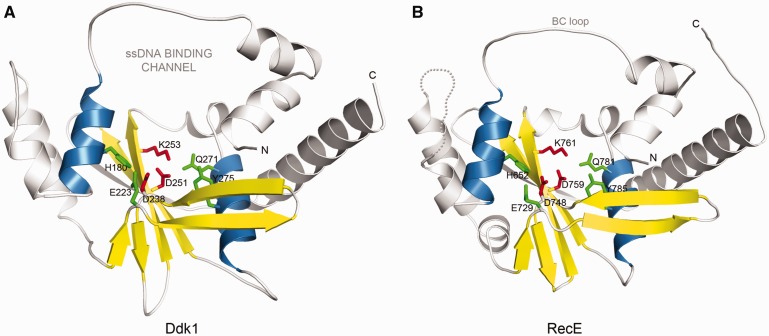
Ddk1 shares significant similarity with several exonucleases of known structure, including E. coli RecE exonuclease (pdb|3h4r) (Figure 1) (43), a putative exonuclease from E. rectale (pdb|3l0a) and the E. coli RecB nuclease (pdb|1w36) (52) as indicated by the distant homology detection method Meta-BASIC that assigned highly reliable scores of 142, 99 and 88, respectively (predictions with a score >40 are expected to have <5% chance of being incorrect). Moreover, human Ddk1 and its homologs conserve sequence motifs described in RecE by Zhang et al. (43). In addition to Motifs II and III that provide the invariant catalytic residues of the PD-(D/E)XK signature (D238, D251, K253 in Ddk1) (Figure 1A and Supplementary Figure S1A), Ddk1-like proteins also retain additional residues that are identical to those in RecE and support the active site: E223 (Motif I), Q271 (Motif IV), Y275 (Motif IV) and H180 (Motif V) (Figure 1A and Supplementary Figure S1A).
Figure 1.
Ddk1 belongs to the PD-(D/E)XK phosphodiesterase superfamily. (A) 3D model for the PD-(D/E)XK nuclease domain of human Ddk1 (gi|14042227). Catalytic PD-(D/E)XK signature residues are shown in red, whereas other potentially important active site amino acids are denoted in green. Secondary structure elements forming the structural core of the fold are coloured yellow (β-strands) and blue (α-helices). (B) E. coli RecE (pdb|3h4r) nuclease domain (the closest homologue of known structure).
Ddk1-like proteins, with the exception of sequences from Tunicata and Nematoda, possess a ∼100 amino acids long variable and mostly unstructured region at the N-terminus. Consequently, most Ddk1-like proteins harbour no additional domains. However, a hypothetical protein from Platynereis dumerilii (gi|83318955) carries a Kazal domain found in Kazal-type serine protease inhibitors (MEROPS inhibitor family I1, clan IA) that are predominantly encoded by metazoan genomes and inhibit S1 family serine proteases (SMART|SM00280).
Interestingly, Ddk1-like nucleases group with Archaeal and phage (prophage) exonucleases (Figure S1B). Most of these phages belong to Caudovirales, which infect cyanobacteria and proteobacteria from which mitochondria are thought to originate. As the mitochondrial proteome is known to have undergone multiple gain, loss and replacement events in early eukaryotic evolution (53), high similarity to phage proteins is a characteristic feature of some nucleic acid processing mitochondrial proteins. For example, RNA polymerase (POLRMT), DNA polymerase (POLG) and helicase Twinkle (3,53–55) have replaced the original α-proteobacterial equivalents. The observed similarity between Ddk1-like proteins and phage proteins may suggest a similar evolutionary scenario.
Ddk1 localizes to mitochondria
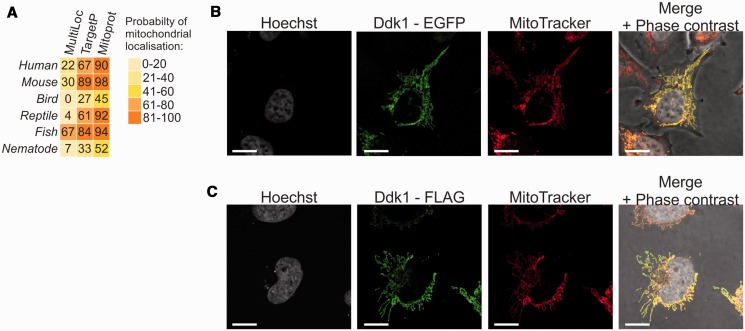
Protein sequence analysis suggested that Ddk1 may be targeted to mitochondria (Figure 2A). To test this possibility, the coding sequence of Ddk1 was cloned in fusion with an EGFP-encoding fragment at the Ddk1 C-terminus, and the fusion protein was transiently expressed in human HeLa cells (Figure 2B). Mitochondria were stained using the mitochondria specific-dye MitoTracker. Microscopy analysis revealed that the subcellular distribution of EGFP-tagged Ddk1 overlaps entirely with the MitoTracker-labelled mitochondria, indicating a mitochondrial localization of Ddk1 (Figure 2B). Because the length of EGFP itself is similar to Ddk1 and thus may affect Ddk1 localization, we confirmed these results using a much shorter FLAG tag (nine amino acids). The C-terminal FLAG-tagged Ddk1 was expressed in HeLa cells and detected using immunofluorescence (Figure 2C) that also showed colocalization solely with MitoTracker-labelled mitochondria. These results show that Ddk1 is a mitochondrial protein.
Figure 2.
Ddk1 localizes to mitochondria. (A) Bioinformatic prediction of Ddk1-like proteins localization. Probability for mitochondrial localization is shown. Web services used for analysis are indicated. Ddk1-like proteins from the following organisms were analysed: Homo sapiens, Mus musculus, Gallus gallus, Anolis carolinensis, Danio rerio, Caenorhabditis elegans. (B and C) Subcellular localization of tagged Ddk1 protein in HeLa cells. Cells were transiently transfected with plasmids encoding C-terminal fusion of Ddk1 with EGFP (B) or FLAG (C). Mitochondria and nuclear DNA was labelled with MitoTracker and Hoechst, respectively. The bar represents 10 μm.
Recombinant Ddk1 has metal-dependent deoxyribonuclease activity in vitro
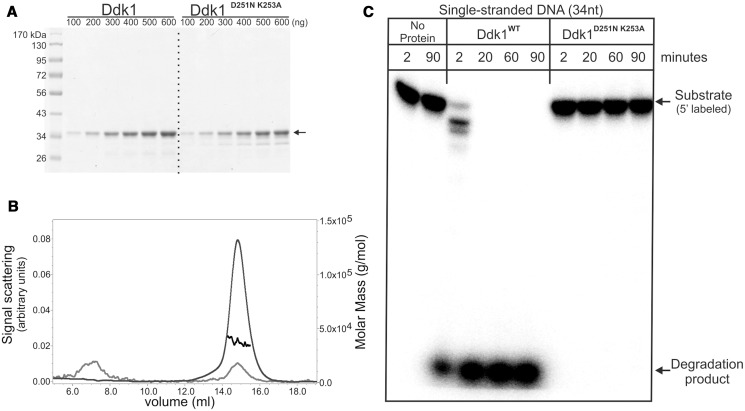
Ddk1 was classified as a member of the PD-(D/E)XK phosphodiesterase superfamily. To determine whether Ddk1 has nuclease activity, it was heterologously overexpressed in E. coli as an N-terminal 6× histidine-SUMO tag fusion protein and purified using a two-step approach with affinity chromatography followed by size exclusion separation. Because the full-length protein was insoluble (data not shown), an N-terminal hydrophobic fragment corresponding to the predicted mitochondrial localization signal was deleted. This truncated Ddk1 lacked the 21 N-terminal amino acids and was purified to near homogeneity (Figure 3A). The protein exists as a monomer as revealed by its migration on size exclusion chromatography (data not shown), and this finding was confirmed by determining its absolute molecular mass using MALS (Figure 3B). The measured mass of Ddk1 (37.2 ± 1.4 kDa) corresponds to that calculated for the monomeric form of the protein (37 kDa). In addition to the wild-type version of Ddk1, a mutated form having substitutions at the predicted catalytic residues (D251N K253A) was also cloned and purified (Figure 3A). The mutated protein behaved as wild-type during purification.
Figure 3.
Purified Ddk1 is active in vitro. (A) Electrophoretic analysis of purified wild-type or mutated Ddk1. Increasing amounts of proteins were loaded. Arrow indicates purified protein. (B) Multiple angle light scattering analysis of purified wild-type Ddk1. Curves represent: measured mass (black), detected signal at 280 nm (grey), voltage (light grey). (C) DNase activity of purified proteins was tested using single-stranded 5′ labelled v81 substrate. Reactions were performed in standard conditions for the indicated time with the exception that a large excess of the enzyme (0.5 pmole) over substrate (0.06 pmole) was used. Samples were analysed using 15% urea–PAGE.
Although both DNases and RNases can be found in the PD-(D/E)XK phosphodiesterase family, most members have deoxyribonuclease activity. Therefore, in the initial experiments with Ddk1, the activity of purified proteins was tested using DNA substrates. Wild-type or mutated Ddk1 was incubated with a 5′-labelled 34 nucleotide ssDNA substrate, and the products of the reactions were resolved using denaturing urea-polyacrylamide gel electrophoresis (Figure 3C). Incubation of substrates with wild-type, but not the mutated form of Ddk1, led to rapid substrate degradation (Figure 3C), indicating that Ddk1 has deoxyribonuclease activity and confirming that the predicted catalytic residues (D251 and K253) are required for this activity.
Subsequently, the optimal conditions for Ddk1 in vitro activity were determined, including the effect of ionic strength, pH and the presence of reducing agent. Ddk1 activity was pH-sensitive with the nuclease activity being weakest at the lowest tested pH (6.8) and increasing with pH (Supplementary Figure S2). Further reactions were performed using pH 8.0 at which Ddk1 has high activity and which corresponds to the estimated pH of the mitochondrial matrix (56,57). Similarly, the presence of reducing agent (DTT or beta-mercaptoethanol) also affected Ddk1 activity, although to a lesser extent than pH (Supplementary Figure S2). Ddk1 is active across a broad range of monovalent cation (Na+ or K+) concentrations (>15 mM, <200 mM) and showed the highest activity ∼125 mM salt concentration (Supplementary Figure S2). To establish what concentration of divalent metal ions is optimal for reaction catalysed by Ddk1, we assessed Ddk1 activity in the presence of various amounts of magnesium or manganese chloride (Supplementary Figure S3), which both supported Ddk1 activity. The activity was higher in the presence of Mg2+ ions, with the optimal Mn2+ concentration (0.16 mM) being one order of magnitude lower than the optimal Mg2+ concentration (≥2.56 mM). Within a range of tested concentrations (0.005–10.24 mM), Ddk1 activity increased concomitantly with the Mg2+ concentration, whereas for Mn2+, deviations from the optimal concentration inhibited the activity (Supplementary Figure S3). The presence of metal ions is a pre-requisite for Ddk1 activity, as their depletion with chelating agent (EDTA) completely abolishes its activity (Supplementary Figure S3). Interestingly, the activity of Ddk1 is temperature sensitive in the absence of BSA. Incubation of the enzyme without BSA for 20 min at 37°C leads to its complete inactivation (Supplementary Figure S4); however, addition of BSA to a reaction mixture stabilizes Ddk1 activity (Supplementary Figure S4).
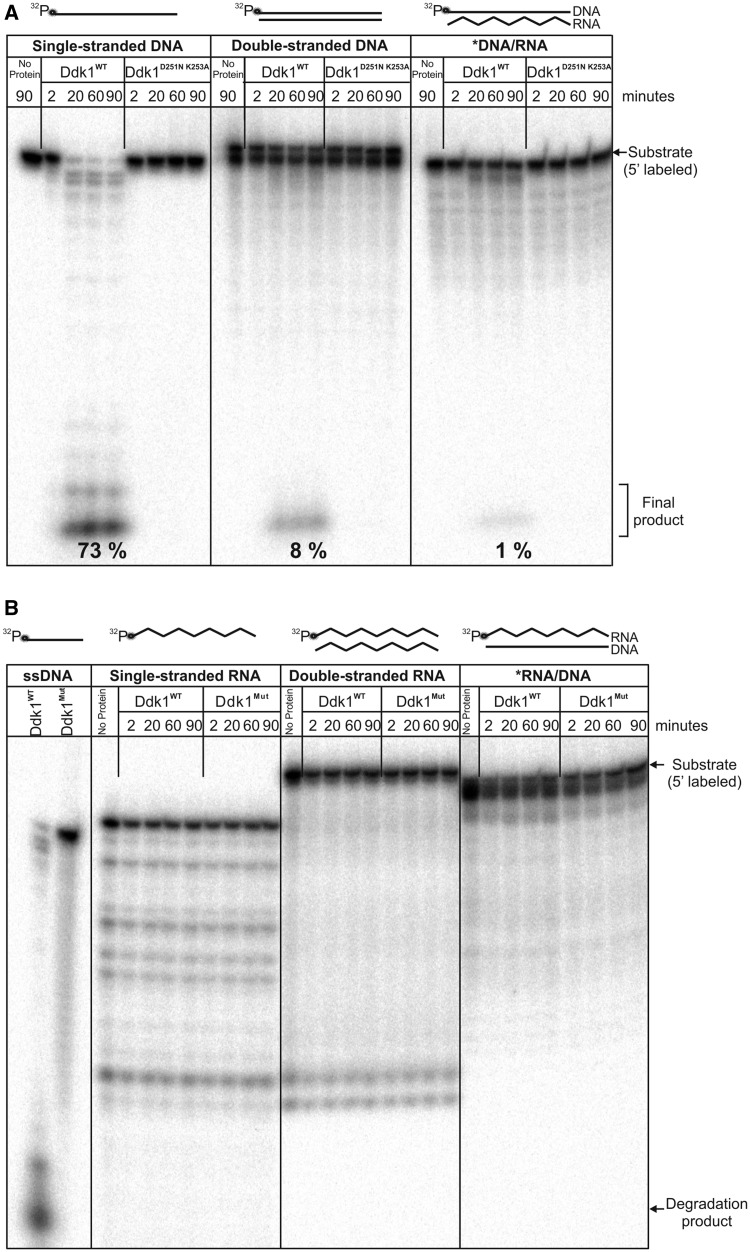
Human mitochondria contain both single-stranded (7S DNA) and double-stranded DNA molecules (full-length mitochondrial genome). Moreover, mammalian mtDNA, including human, can also have stable tracts of DNA–RNA hybrids (R-loops) (58–61). Therefore, we tested whether Ddk1 could be involved in the metabolism of each type of mentioned molecules. Ddk1 was incubated for varying times with the following linear 44-nt/bp length substrates: ssDNA, blunt-ended dsDNA or DNA/RNA hybrid and reactions products were analysed on denaturing PAGE (Figure 4A). Although Ddk1 degraded all tested substrates, it displayed a ∼10-fold higher efficiency towards ssDNA in comparison with dsDNA (Figure 4A). DNA/RNA substrate was even less efficiently degraded than dsDNA (Figure 4A). Reaction products were observed only in samples containing wild-type Ddk1 and not their mutated form, confirming that they arose owing to Ddk1 nucleolytic activity (Figure 4A). In contrast, RNA substrates were not cleaved by Ddk1 (Figure 4B), which was unable to degrade 44-nt/bp single-stranded, double-stranded RNA or RNA hybridized to DNA (Figure 4B), even though an ssDNA substrate was degraded in a reaction performed simultaneously. To exclude the possibility that Ddk1 is specific for short RNAs and/or adenylated molecules, we tested its activity using a 17-nt RNA substrate containing short adenine tails. We detected no Ddk1 activity towards these oligoribonucleotides, although the activity of the protein towards DNA substrates was confirmed at the same time (Supplementary Figure S5).
Figure 4.
Ddk1 has deoxyribonuclease, but not ribonuclease activity. Denoted substrates were incubated with wild-type or mutated (D251N K253A; Mut) Ddk1 in standard conditions for the indicated time and analysed on 15% urea–PAGE. 5′-labelled 44DNA (A) or 44RNA (B) substrate in a single-stranded form or annealed to complementary DNA (44DNAcomp) or RNA (44RNAcomp) was analysed. (A) Numbers represent the amount of the final product generated in samples incubated for 20, 60 and 90 min. Amounts were quantified by dividing the intensity of the signal corresponding to the final product only by the total intensity measured in three entire analysed lanes (i.e. sum of product, degradation intermediates and final product).
We thus concluded that Ddk1 is a deoxyribonuclease in vitro and has no detectable ribonuclease activity.
Ddk1 is a linear substrate-specific endo-/exonuclease that acts from both substrate ends
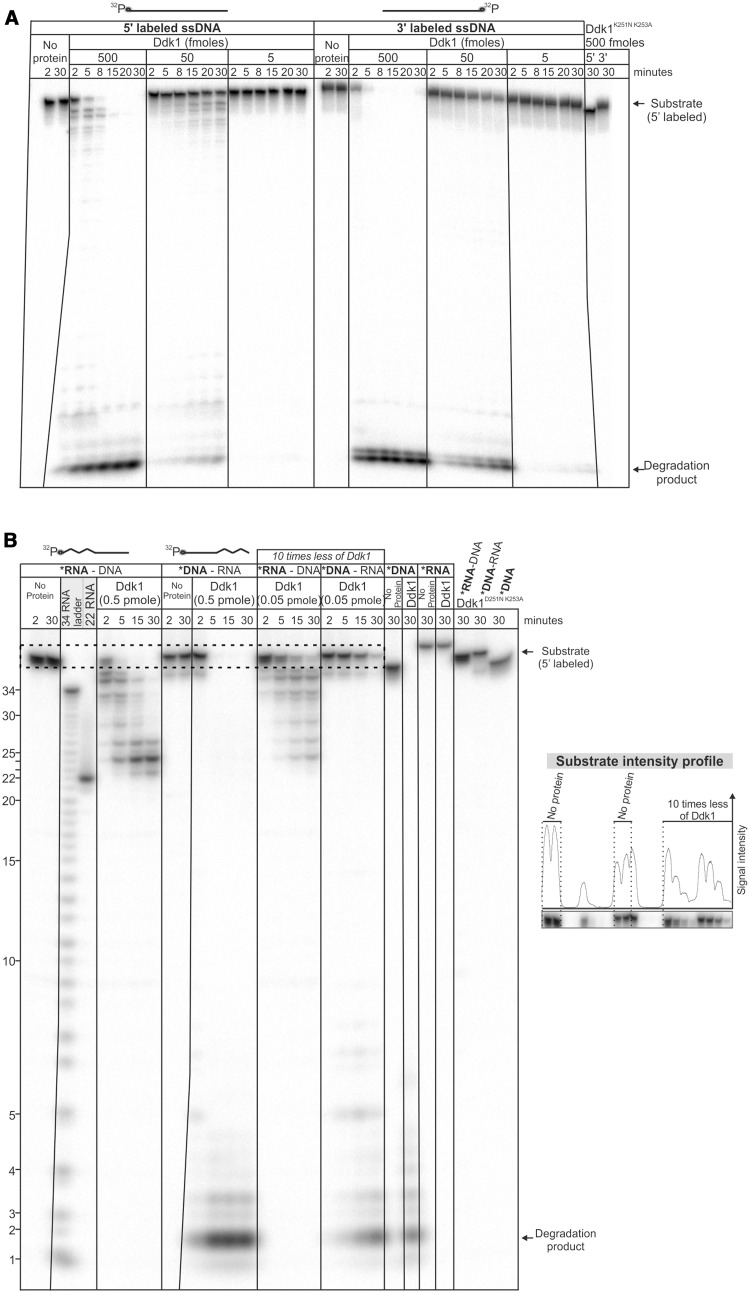
Many known nucleases act only in one direction. To investigate whether Ddk1 manifests such polarity, we tested its activity using 44-nt ssDNA substrates labelled at either the 5′- or 3′-end (similar data were obtained using 80-nt ssDNA, data not shown). We assumed that detection of partially degraded substrates only in the case of 5′- or 3′-labelled substrates would indicate that the enzyme works only in one direction. Detailed analysis revealed the presence of partially degraded substrates in both cases (Figure 5A and Supplementary Figure S6 for longer exposure), suggesting that Ddk1 functions in both directions. However, the amount of degradation intermediates was much higher for 5′- than 3′-labelled substrates (Figure 5A and Supplementary Figure S6), indicating that Ddk1 preferentially acts in the 3′–5′ direction. Moreover, our data suggest that Ddk1 is a processive enzyme, as the final digestion product is clearly visible while a large fraction of the substrate remains intact (Figure 5A).
Figure 5.
Ddk1 can act in both directions. (A) Ddk1 activity was assayed using 5′- or 3′-labelled single-stranded 44DNA substrate and (B) 5′ 44RNA–DNA, 44DNA–RNA chimeric substrates and 44DNA, 44RNA substrates. Dashed rectangle marks the region of the gel subjected to quantitative analysis (graph, right). An RNA ladder and a reference RNA fragment (22RNA) were also resolved. (A and B) Reactions were performed in standard conditions using indicated amounts of the enzyme for indicated times. Products were resolved using 20 (A) or 15% (B) urea–PAGE.
To confirm this directionality, we performed an experiment that takes advantage of Ddk1’s inability to degrade RNA by using chimeric substrates composed of DNA and RNA fragments with labelling at the 5′ end (Figure 5B). Two substrates with RNA at the 5′ or 3′ end were used with the expectation that, if the enzyme has exonucleolytic activity and operates preferentially in one direction, one of the substrates would be degraded with very low efficiency. Unexpectedly, the amount of full-length chimeric substrate was decreased at a similar rate for both cases (Figure 5B). Importantly, no degradation was observed in reactions carried out with inactive Ddk1D251N, K253A (Figure 5B). In agreement with previous data showing that Ddk1 has no ribonuclease activity, the pattern of reaction products indicated incomplete degradation of chimeric substrates with labelled RNA at the 5′ end. Comparison of reaction products with a reference RNA fragment of the same length as the one present in the chimeric RNA–DNA substrate, as well as to an RNA ladder, showed that the major final product contained two deoxyribonucleotides downstream of the RNA fragment (Figure 5B). A similar feature was observed for another PD-(D/E)XK nuclease EXO5 (48). The second substrate (DNA–RNA) seemed fully degraded because the DNA portion of the molecule that was labelled can be hydrolyzed; however, as Ddk1 has no RNase activity, we assume that the RNA portion remains, although not visible in the autoradiogram (Figure 5B). Together, the results suggest that Ddk1 can function in both directions. However, they did not exclude the possibility that this enzyme functions preferentially from the 3′ to 5′ end and has endonucleolytic activity. In this case, a substrate having RNA at the 3′ end would be efficiently degraded even if Ddk1 had no activity in the 5′–3′ direction. Internal cleavage of DNA would thus provide the substrate for exodeoxyribonucleolytic activity.
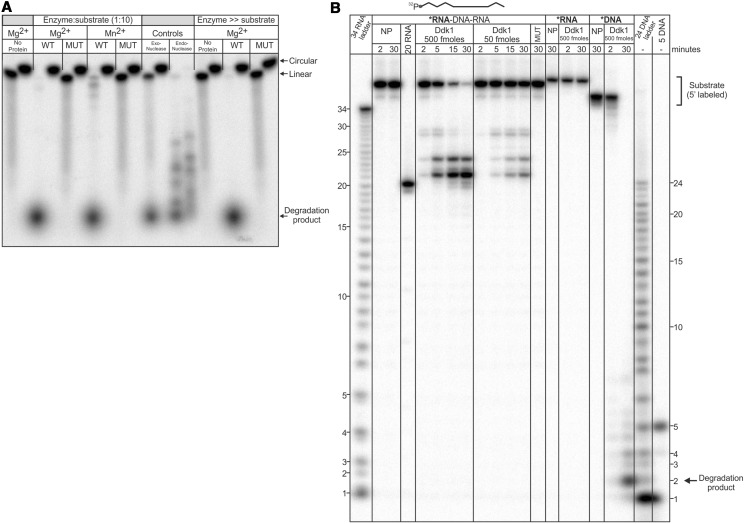
To examine whether Ddk1 has endonuclease activity, the enzyme was incubated with radioactively labelled single-stranded circular DNA or its linear counterpart, and reaction products were analysed by electrophoresis (Figure 6A). The nature of the substrates was confirmed using exonuclease I and endonuclease DNase I (Figure 6A). As the type of reaction catalysed by nucleases may depend on the type of ions present [e.g. the ribonuclease Dis3 catalyses exo- or endonucleolytic reactions in the presence of Mg2+ or Mn2+, respectively (62,63)], reactions were performed in the presence of magnesium or manganese. The analysis of reaction products showed that even excess amounts of Ddk1 are unable to degrade circular substrates (Figure 6A). Under the same conditions, the corresponding linear substrate was efficiently degraded (Figure 6A), indicating that Ddk1 has no endonuclease activity on a circular single-stranded substrate. However, it was possible that Ddk1 could have such activity towards linear substrates.
Figure 6.
Ddk1 requires free ends for activity in vitro. (A) Wild-type or catalytically inactive (D251N K253A) Ddk1 were incubated with linear 5′-labelled single-stranded 44DNA substrate or its circularized form. Indicated ratios of enzyme to substrate were analysed. Reactions were performed in standard conditions using an optimal concentration of magnesium (5.12 mM) or manganese (0.16 mM) and resolved by 12% denaturing PAGE. The nature of the substrates was controlled using exonuclease Exo I (NEB) and endonuclease DNAse I (Roche). (B) Ddk1 activity tested using 5′-labelled 44RNA–DNA–RNA substrate. To control the activity of Ddk1, reactions with single-stranded 44DNA or 44RNA were performed. Standard conditions were used. Products were resolved using 15% urea–PAGE. An RNA ladder and DNA ladder, as well as a reference RNA fragment (20RNA), were also resolved. NP-no protein.
To test this possibility, we analysed Ddk1 activity using a 5′-labelled single-stranded substrate composed of DNA flanked by RNA on both sides (RNA–DNA–RNA) (Figure 6B). As Ddk1 is incapable of RNA hydrolysis, the appearance of degradation products would demonstrate Ddk1 endodeoxyribonuclease activity. Analysis of reaction products showed the presence of a partially degraded RNA–DNA–RNA substrate in which the DNA was degraded while the RNA remained intact, supporting the ability of Ddk1 to catalyse endonucleolytic cleavage (Figure 6B). Importantly, in a parallel control reaction in which a RNA substrate was used, no Ddk1 activity was observed (Figure 6B), which excludes the possibility that the detected degradation of the RNA–DNA–RNA substrate resulted from RNA cleavage (Figure 6B). Similar to the previous analysis (Figure 5B), the presence of ribonucleotides at the end of the RNA–DNA–RNA substrate precludes its complete degradation by Ddk1 (Figure 6B). However, in this case, the enzyme leaves not two (Figure 5B), but one deoxyribonucleotide in the final degradation product.
Finally, we determined the size of the end product of DNAse activity of Ddk1. An ssDNA substrate was treated with Ddk1, and the migration of reaction products was compared with a DNA ladder. This revealed that the major final product is 2 nucleotides in length, which is the same as observed for yeast EXO5 (48) (Figure 6B).
Overall, we conclude that in vitro Ddk1 is a processive endo-/exodeoxyribonuclease that preferentially acts from the 3′ to the 5′ end. Moreover, it requires a free 3′ and/or 5′ end for activity.
Silencing of DDK1 increases the level of 7S DNA
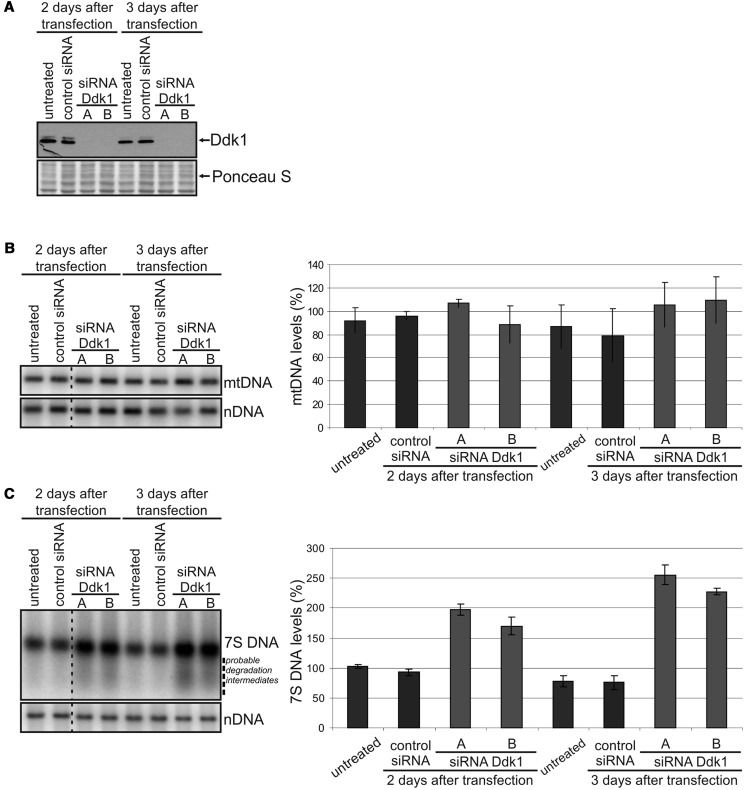
We next used RNA interference methodology to study the function of Ddk1 in vivo. HeLa cells were transfected with two different siRNAs specific for the DDK1 gene or control siRNA designed not to affect expression of any human gene. Silencing of DDK1 gene expression was confirmed using western blot (Figure 7A). As biochemical studies of Ddk1 indicated that the protein might be involved in mtDNA metabolism, we used Southern blotting to measure mtDNA levels after DDK1 silencing (Figure 7B). We observed no significant changes in mtDNA levels 3 days after siRNA transfection (Figure 7B). Because the effect on mtDNA level might require longer periods of Ddk1 depletion, we performed an experiment in which cells were re-transfected with siRNA 3 days after the first transfection and collected after an additional 2 and 4 days. Again, Southern blotting revealed no changes in mtDNA levels even 7 days after siRNA transfection (Supplementary Figure S7A). Using the same experimental procedure, we were able to demonstrate that silencing of the helicase Twinkle, a well-known factor involved in mtDNA replication (64), results in mtDNA depletion, as expected (Supplementary Figure S8). Therefore, we conclude that within the limited timeframe of our experimental strategy, Ddk1 seems not to be directly involved in mtDNA maintenance.
Figure 7.
Silencing of Ddk1 increases the level of 7S DNA and has no substantial effect on mtDNA levels. DNA or RNA was isolated from HeLa cells transfected with control siRNA or two different Ddk1-specific siRNAs and collected after 2 or 3 days. Graphs present the mean values obtained in two independent experiments. Each experiment was performed in three technical repeats [three sets of cultures transfected (or not) at the same time], and an equal number of cells from different repeats was mixed after collection. Error bars represent standard deviation. We did not observe any effect on cell growth within the timeframe of the presented RNAi experiment. (A) Western blot analysis of Ddk1 level. Ponceau S staining of the membrane was performed for standardization. (B and C) The level of mtDNA (B) and 7S DNA (C) was examined using Southern blots. The signal arising from hybridization to the nuclear 28rDNA gene (nDNA) was used as a loading control. Differences are statistically insignificant (B) or significant (C) [analysis of variance (ANOVA) test]. (C) P-values for samples collected 2 and 3 days after transfection are 0.0009 and 0.0001, respectively (ANOVA test). Dashed line indicates probable degradation intermediates.
We then examined whether Ddk1 can participate in the metabolism of mitochondrial ssDNA, which would be in agreement with the in vitro results showing that Ddk1 has strongest activity towards ssDNA. Human mitochondria contain 7S DNA, a fragment ∼570–655 nucleotides in length (65–68), of which pool potentially may exist as ssDNA. Using Southern blotting to examine the levels of 7S DNA (Figure 7C), we found that DDK1 silencing leads to 7S DNA accumulation. Similar changes in 7S DNA were observed in cells transfected with both Ddk1-specific siRNAs (Figure 7C). In comparison with untreated cells, the level of 7S DNA increased twice in cells with depleted DDK1 expression and remained unaffected in cells transfected with control siRNA (Figure 7C). The effect was observed 2 days after transfection (Figure 7C) and was augmented by extended silencing of DDK1 (Figure 7C and Supplementary Figure S7B). Moreover, Ddk1 depletion results in the appearance of a smear below 7S DNA (Figure 7C) that may represent partially degraded 7S DNA molecules accumulated in the absence of Ddk1 nucleolytic activity.
To verify that the accumulation of 7S DNA on Ddk1 silencing is specifically caused by perturbation of Ddk1 function, we tested whether depletion of other mitochondrial DNAses might have a similar effect. We performed single knockdown experiments of EXOG, FEN1 or DNA2 (two different siRNAs were used per gene) and double knockdowns of each of those genes with DDK1 (Supplementary Figure S9). DDK1 alone was silenced for reference. Then, we measured the levels of 7S DNA and mtDNA. None of the four genes had any effect on mtDNA levels when silenced (Supplementary Figure S9B). In agreement with the data presented above, knockdown of Ddk1 resulted in strong accumulation of 7S DNA (Supplementary Figure S9C). Among the other investigated DNAse encoding genes, only silencing of EXOG slightly increased the level of 7S DNA; however, the effect was far less pronounced then for DDK1 silencing (Supplementary Figure S9C). Silencing of FEN1 or DNA2 did not change the level of 7S DNA at all (Supplementary Figure S9C). Moreover, we also found that double depletions of the three DNAses with Ddk1 did not have any additive or synergistic effects on the level of 7S DNA or mtDNA (Supplementary Figure S9). This emphasizes the significance and specificity of Ddk1 in 7S DNA metabolism.
To see whether a cellular role played by Ddk1 is important for maintaining mitochondrial inner membrane potential (ΔΨm) or mitochondrial biogenesis, we studied the influence of DDK1 silencing on ΔΨm mitochondrial potential and overall mitochondrial mass using flow cytometry and found that none of the investigated features were changed after Ddk1 depletion (Supplementary Figure S10). Recently, synthesis of 7S DNA was shown to vary during the cell cycle and occur mainly in the S phase (69). Thus, it was possible that accumulation of 7S DNA after DDK1 silencing may not be related to Ddk1 function but instead results from cell cycle perturbations. Therefore, we examined the cell cycle profile of control and Ddk1-depleted cells. We found that changes in the level of 7S DNA resulted from Ddk1 depletion and were not caused by alterations in the cell cycle (Supplementary Figure S11).
Overexpression of Ddk1 decreases 7S DNA levels
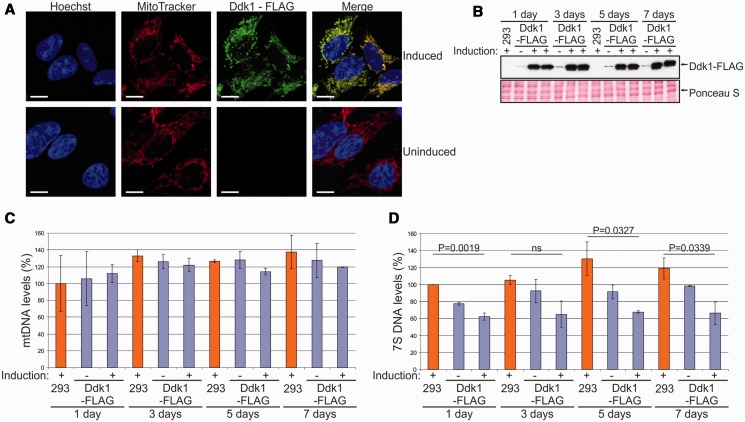
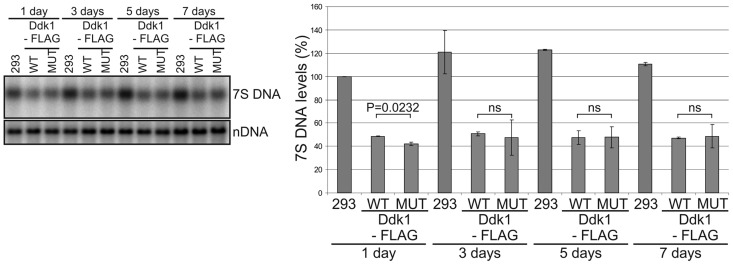
DDK1 silencing resulted in accumulation of 7S DNA and appearance of shorter DNA species that could represent 7S DNA degradation intermediates. We considered that if these changes are related to Ddk1 function, we should see a decrease of 7S DNA levels on Ddk1 overexpression. To verify this possibility, we established a stable human 293 cell line that inducibly expresses C-terminal FLAG-tagged Ddk1. In agreement with data obtained with HeLa cells, we confirmed by immunofluorescence that induced Ddk1-FLAG localizes to mitochondria (Figure 8A). We next overexpressed Ddk1-FLAG and collected cells every 2 days to analyse the level of mtDNA and 7S DNA (Figure 8C and D) while monitoring Ddk1-FLAG levels by western blot, which showed that induction of exogenous gene expression led to Ddk1-FLAG overexpression (Figure 8B). We also noticed weak DDK1-FLAG expression even in the absence of the inducer (Figure 8B). In addition to mtDNA and 7S DNA studies, we examined the effect of Ddk1 overexpression on mitochondrial inner membrane potential and mitochondrial mass (Supplementary Figure S10). As with Ddk1 silencing, we observed no considerable changes in the mtDNA level (Figure 8C), ΔΨm, or the amount of mitochondria (Supplementary Figure S10). Compared with the parental cell line, lower 7S DNA levels were observed in the transgenic uninduced cells (Figure 8D), which is likely owing to leaky expression of Ddk1-FLAG. Similar leakage of exogenous genes in transgenic 293 T-REx cells has also been observed by others (70). However, the key result remains that Ddk1 overexpression caused further decreases in 7S DNA levels (Figure 8D). Overall, these results support the conclusion that Ddk1 is necessary for maintaining the proper level of 7S DNA, which is in agreement with observed Ddk1 in vitro activity. Ddk1 degrades DNA but has no activity towards RNA, which is further manifested by the fact that neither Ddk1 silencing nor overexpression affected the level of tested mtRNAs or induced the appearance of abnormal mtRNAs (Supplementary Figure S12).
Figure 8.
Overexpression of Ddk1 decreases 7S DNA levels and has no substantial effect on mtDNA levels. (A) Subcellular localization of Ddk1-FLAG in a stable inducible 293 cell line. Exogenous gene expression was induced for 24 h, and cells were subjected to immunofluorescence staining and confocal microscopy. The same microscope settings were used for uninduced and induced cells. Mitochondria were labelled with MitoTracker, and Ddk1-FLAG with primary mouse anti-FLAG antibodies followed by detection with secondary AlexaFluor 488 conjugated antibodies. Nuclei were stained with Hoechst dye. The bar represents 10 μm. (B) Western blot analysis of Ddk1-FLAG level in parental 293 or transgenic cell lines. Expression of the Ddk1-FLAG was induced for the indicated times. Samples from two independent induced cultures were analysed. Ponceau S staining of the membrane was performed for standardization. (C and D) Analysis of mtDNA (C) and 7S DNA (D) levels in parental 293 cells and their derivative that overexpresses (or not) Ddk1 for the indicated time. Levels were examined using Southern blots. Graphs present the mean values obtained in two independent experiments. Error bars represent standard deviation. The signal arising from hybridization to the nuclear 28rDNA gene (nDNA) was used as a loading control. (C) Differences are not statistically significant (ANOVA test). (D) The P-values obtained in an ANOVA test are indicated. ns, statistically insignificant; P > 0.05.
This raises the question of whether Ddk1 nuclease activity is directly involved in regulating 7S DNA. To examine this issue, we established a stable 293 cell line that inducibly expresses catalytically inactive Ddk1D251N, K253A -FLAG. Such a fusion protein also localizes to mitochondria (Supplementary Figure S13); however, its overexpression leads to depletion of 7S DNA, similarly to overexpression of its active counterpart (Figure 9), indicating that the Ddk1 nuclease activity may not be directly required for controlling 7S DNA levels.
Figure 9.
Overexpression of inactive Ddk1 affects 7S DNA levels. Analysis of 7S DNA levels in parental 293 cells and their derivatives overexpressing wild-type or mutated (D251N K253A) Ddk1 for the indicated times. Each cell line was cultured in the presence of tetracycline. 7S DNA levels were examined using Southern blots. Graph presents the mean values obtained in two independent experiments. Error bars represent standard deviation. The signal arising from hybridization to the nuclear 28rDNA gene (nDNA) was used as a loading control. The P-values obtained in the Student’s t-test are indicated. ns, statistically insignificant; P > 0.05.
DISCUSSION
Although the human mitochondrial genome has been investigated for several decades, the proteins responsible for its replication and expression, especially nucleolytic enzymes, are not fully described. For example, in vitro experiments using mitochondrial lysates and radioactively labelled substrates suggested the presence of unidentified deoxyribonuclease/s in human mitochondria (23,71). We describe here the identification of a new mitochondrial processive deoxyribonuclease that we named Ddk1.
Some Ddk1 biochemical properties set it apart from previously described human mitochondrial DNases. In contrast to EXOG or EndoG, Ddk1 is a strict sugar-specific nuclease, as it had no activity towards RNA substrates, regardless of their length and structure. Moreover, neither Ddk1 depletion nor overexpression affected mtRNA metabolism. This indicates that Ddk1 is not directly involved in mtRNA metabolism.
Ddk1 is unique among previously characterized human mitochondrial DNases in that it is fully active at physiological salt concentrations. Whereas EXOG, EndoG, FEN1 and hDna2 are sensitive to monovalent cations and have strongly reduced activity at NaCl concentrations >50 mM (72–77), Ddk1 exhibits a broad Na+/K+ concentration optimum with the highest activity at 125 mM. The reason for this discrepancy between these proteins is not clear. But, for example, in the case of EndoG, it was suggested that its low activity at physiological ionic strength implies existence of additional co-activators (73). Thus, it may mean that Ddk1 does not require such factors for its maximal activity in vivo.
Noticeably, Ddk1 preferentially degrades substrates from the 3′ to 5′ end. Such polarity was also detected for human Dna2 (76), but it also has strong 5′–3′ activity (76). In vivo, degradation of ssDNA in the 3′–5′ direction may be required to process equilibrating flaps (76) that can arise during the removal of RNA primers from newly synthesized DNA strands, regardless of whether they formed via strand-asynchronous or strand-coupled mechanisms (5,58,78,79).
In addition to the sugar bond, the physical form of substrates is also important for Ddk1 activity. Whereas EndoG, EXOG and Dna2 (25,80) cleave circular DNA molecules, Ddk1 has no such activity. However, Ddk1 can cleave substrates in an endonucleolytic manner provided they have free ends. The linear chimeric substrate composed of RNA–DNA–RNA was internally cleaved within the DNA fragment, suggesting that, as for Dna2 and FEN1 (81,82), Ddk1 may use a tracking mechanism where the enzyme binds the free end of the substrate and moves along it until a hydrolysable fragment is reached. Taken together, our results demonstrate that Ddk1 differs in biochemical properties from other human mitochondrial DNases, which may suggest its involvement in different mtDNA-related transactions.
The monomeric form and some predicted structural features of Ddk1 distinguish it from evolutionarily related exonucleases such as RecE, alkaline exonuclease and lambda exonuclease (83), which form funnel-shaped multimers to direct dsDNA recognition and consequently perform ssDNA cleavage. Importantly, RecE, the closest homologue having a known structure, carries several features necessary for tetramerization and dsDNA binding, which, according to our prediction, are not present in Ddk1. Specifically, Ddk1 lacks the extended loop corresponding to RecE residues 665–698, a long C-terminal tail and the equivalents of critical residues important for dsDNA binding (e.g. K704, R858 and W859 in RecE). This observation is consistent with our experimental results indicating that Ddk1 does not form multimers and has much higher activity towards ssDNA.
An important structural feature of Ddk1-like proteins that is also present in RecE, as well as alkaline and lambda exonucleases, is a conserved loop corresponding to the ‘BC loop’ in RecE or ‘DE loop’ in lambda exonuclease, which covers the active site and forms a narrow channel for ssDNA (Figure 1). This channel is wide enough to allow passage of one DNA strand, but not the entire duplex or single-stranded circular DNA.
The yeast genome encodes a mitochondrialy localized nuclease DEM1/EXO5 that, as Ddk1, belongs to the PD-(D/E)XK superfamily, and although a highly distant relative in terms of sequence, exhibits similar catalytic properties to Ddk1 (36,48). EXO5, which lacks RNase activity, can degrade a RNA–DNA chimeric substrate by sliding across RNA and cleaving within the DNA. Like Ddk1, EXO5 does not digest circular substrates, but in contrast to Ddk1, it acts from the 5′ to 3′ end (48). The role of EXO5 has not been fully elucidated but has been shown to be essential for mitochondrial genome maintenance, as its inactivation generates petites with unstable mitochondrial genomes that continuously undergo rearrangements (48). Thus, it was proposed that EXO5 may participate in replication and/or recombination of yeast mtDNA (48). In our experiments, we did not see significant changes in mtDNA levels in response to Ddk1 depletion or overexpression. Therefore, it seems that the enzyme is not essential for maintaining mtDNA, and/or its depletion is compensated by the activity of other proteins. The considerable similarity of Ddk1 to phage proteins suggests that the enzyme was adapted for mitochondrial function at similar stages as other phage-like mitochondrial proteins, that is, mtRNA polymerase, mtDNA polymerase and replicative helicase Twinkle. Those proteins are vital for maintaining mtDNA.
Our results indicate that the function played by Ddk1 is necessary to maintain 7S DNA levels, which would be consistent with its biochemical properties. The enzyme preferentially degrades ssDNA and thus could target a possible pool of single-stranded 7S DNA that is unhybridized to mtDNA. Moreover, kinetic studies of mtDNA replication intermediates showed a high turnover rate of 7S DNA (26–28), implying the existence of undescribed 7S DNA degradation factor(s). Therefore, we assumed that 7S DNA could act as an in vivo substrate of Ddk1. However, we found that 7S DNA depletion is caused by overexpression of wild-type Ddk1 and a catalytically inactive mutant alike, which argues this hypothesis and suggests that the amount of Ddk1 rather than its activity is important in maintaining the levels of 7S DNA. This effect seems to be caused specifically by overexpression of Ddk1 and not just any protein, as 7S DNA levels are unaffected by overexpression of mitochondrially targeted EGFP, whereas EGFP-tagged Ddk1 (wild-type or mutant) do cause 7S DNA depletion (data not shown). Similar relationship between the level of the 7S DNA and a protein was observed for TFAM (84), a main component of mitochondrial nucleoid. Taken together, these results suggest that Ddk1 nuclease activity may be involved in other unrevealed aspects of mtDNA metabolism; however, the level of Ddk1 is necessary for maintaining proper 7S DNA levels.
The existence of the D-loop and 7S DNA was described a few decades ago, but the role of this structure still remains unknown. Early studies led to precise mapping of 7S DNA (78); however, its further functional studies have been less successful. According to the strand-displacement model of mtDNA replication, the 7S DNA can be regarded as a product of preterminated synthesis of the leading strand (78). It was postulated that regulation of pretermination can play a primary role in controlling the rate of mtDNA replication (85). However, owing to the rapid turnover of 7S DNA, its involvement in mtDNA replication is considered as unlikely (78). Moreover, it was found that levels of 7S DNA and mtDNA are not dependent on each other (69). Instead, it was proposed that D-loop formation enables mtDNA to associate with other entities or marks the non-coding regulatory region, which helps to recruit proteins (78). Indeed, it was shown that ATAD3p and POLG2, the accessory subunit of mtDNA polymerase gamma, preferentially bind to D-loop containing substrates in vitro (86,87). Both proteins are necessary for formation and/or maintenance of mtDNA multimers in vivo, the assembly of which involves D-loop containing mtDNA fragments (86,87). Silencing of POLG2 or its overexpression decreases the level of 7S DNA (87), but the mechanism of this effect and its functional consequences require further studies. It was shown that POLG2 takes part in mitochondrial nucleoid maintenance; however, the role of 7S DNA in this process remains to be investigated. Although RNAi-mediated depletion of POLG2 increases the number of nucleoids, overexpression of the protein has the opposite effect (87), even though they have the same effect on the level of 7S DNA. Moreover, it was reported that mitochondrial single-stranded binding protein plays an important role in 7S DNA maintenance. Knockdown of mitochondrial single-stranded binding protein severely reduces the synthesis of 7S DNA and has no effect on the organization of mitochondrial nucleoids (88). Taken together, studies of 7S DNA led to identification of some protein factors, which are involved in its metabolism, but the role of 7S DNA itself is still far from clear. An interesting hypothesis, which needs to be experimentally challenged, came from Antes et al. (69), who suggested that degradation of 7S DNA may contribute to regulation of the mitochondrial pool of dNTPs.
Even though elucidation of the exact in vivo function of Ddk1 requires further study, our research adds another interesting and important player in mtDNA metabolism and a potential candidate for disease-related genes, of which many have been found in the PD-(D/E)XK superfamily (36).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–13 and Supplementary Methods.
FUNDING
Ministry of Science and Higher Education of Poland [0542/IP1/2011/71 to R.J.S., Iuventus programme] and co-supported by [0376/IP1/2011/71]; grants from the Foundation for Polish Science [TEAM/2010-6] and National Science Centre [2011/02/A/NZ2/00014]. R.J.S. and K.S. were the recipients of the Stipend for Young Researchers from the Foundation for Polish Science. R.J.S. is the recipient of a TEAM scholarship from the Foundation for Polish Science. Experiments were carried out with the use of CePT infrastructure financed by the European Union—the European Regional Development Fund (Innovative economy 2007–13, Agreement POIG.02.02.00-14-024/08-00). Funding for open access charge: Ministry of Science and Higher Education of Poland [0542/IP1/2011/71].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Katarzyna Kowalska and Krystian Stodus for help in cloning, protein purification and MALS analysis. They are also grateful to Aleksander Chlebowski for his help with the manuscript. R.J.S. performed all biochemical studies, localization studies of EGFP-tagged Ddk1, established stable cell lines and obtained most of DNA constructs. R.J.S. and M.S.H. analysed mtDNA and 7S DNA levels as well as investigated mitochondrial mass, mitochondrial membrane potential, cell cycle profile and mitochondrial transcripts. L.S.B. analysed the localization of tagged Ddk1 in stable cell lines and measured genes expression. K.S. performed initial bioinformatic identification of Ddk1 and together with AM carried out in silico phylogenetic analysis. K.G. designed and supervised bioinformatic research. R.J.S. and A.D. designed the experimental research. R.J.S., K.G. and A.D. obtained funding. R.J.S., K.G. and A.D. wrote the article with contributions of others. The article was written mostly by R.J.S. A.D. supervised and coordinated the project. All authors approved the final version of the manuscript.
REFERENCES
- 1.Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp. Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 4.Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta. 2012;1819:914–920. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Holt IJ. Mitochondrial DNA replication and repair: all a flap. Trends Biochem. Sci. 2009;34:358–365. doi: 10.1016/j.tibs.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 7.Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 8.Brzezniak LK, Bijata M, Szczesny RJ, Stepien PP. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol. 2011;8:616–626. doi: 10.4161/rna.8.4.15393. [DOI] [PubMed] [Google Scholar]
- 9.Rorbach J, Nicholls TJ, Minczuk M. PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Res. 2011;39:7750–7763. doi: 10.1093/nar/gkr470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poulsen JB, Andersen KR, Kjaer KH, Durand F, Faou P, Vestergaard AL, Talbo GH, Hoogenraad N, Brodersen DE, Justesen J, et al. Human 2′-phosphodiesterase localizes to the mitochondrial matrix with a putative function in mitochondrial RNA turnover. Nucleic Acids Res. 2011;39:3754–3770. doi: 10.1093/nar/gkq1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2012;41:1223–1240. doi: 10.1093/nar/gks1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Roy F, Bisbal C, Silhol M, Martinand C, Lebleu B, Salehzada T. The 2-5A/RNase L/RNase L inhibitor (RLI) [correction of (RNI)] pathway regulates mitochondrial mRNAs stability in interferon alpha-treated H9 cells. J. Biol. Chem. 2001;276:48473–48482. doi: 10.1074/jbc.M107482200. [DOI] [PubMed] [Google Scholar]
- 13.Le Roy F, Silhol M, Salehzada T, Bisbal C. Regulation of mitochondrial mRNA stability by RNase L is translation-dependent and controls IFNalpha-induced apoptosis. Cell Death Differ. 2007;14:1406–1413. doi: 10.1038/sj.cdd.4402130. [DOI] [PubMed] [Google Scholar]
- 14.Bruni F, Gramegna P, Lightowlers RN, Chrzanowska-Lightowlers ZM. The mystery of mitochondrial RNases. Biochem. Soc. Trans. 2012;40:865–869. doi: 10.1042/BST20120022. [DOI] [PubMed] [Google Scholar]
- 15.Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart JA, Campbell JL, Bambara RA. Dna2 is a structure-specific nuclease, with affinity for 5′-flap intermediates. Nucleic Acids Res. 2010;38:920–930. doi: 10.1093/nar/gkp1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budd ME, Campbell JL. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao HI, Bambara RA. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit. Rev. Biochem. Mol. Biol. 2003;38:433–452. doi: 10.1080/10409230390259382. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L, Shen B. Okazaki fragment maturation: nucleases take centre stage. J. Mol. Cell Biol. 2011;3:23–30. doi: 10.1093/jmcb/mjq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gloor JW, Balakrishnan L, Campbell JL, Bambara RA. Biochemical analyses indicate that binding and cleavage specificities define the ordered processing of human Okazaki fragments by Dna2 and FEN1. Nucleic Acids Res. 2012;40: 6774–6786. doi: 10.1093/nar/gks388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair. 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczesny B. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5′-EXO/endonuclease) in their repair. J. Biol. Chem. 2011;286:31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cymerman IA, Chung I, Beckmann BM, Bujnicki JM, Meiss G. EXOG, a novel paralog of Endonuclease G in higher eukaryotes. Nucleic Acids Res. 2008;36:1369–1379. doi: 10.1093/nar/gkm1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robberson DL, Clayton DA. Pulse-labeled components in the replication of mitochondrial deoxyribonucleic acid. J. Biol. Chem. 1973;248:4512–4514. [PubMed] [Google Scholar]
- 27.Bogenhagen D, Clayton DA. Mechanism of mitochondrial DNA replication in mouse L-cells: kinetics of synthesis and turnover of the initiation sequence. J. Mol. Biol. 1978;119:49–68. doi: 10.1016/0022-2836(78)90269-3. [DOI] [PubMed] [Google Scholar]
- 28.Gensler S, Weber K, Schmitt WE, Perez-Martos A, Enriquez JA, Montoya J, Wiesner RJ. Mechanism of mammalian mitochondrial DNA replication: import of mitochondrial transcription factor A into isolated mitochondria stimulates 7S DNA synthesis. Nucleic Acids Res. 2001;29:3657–3663. doi: 10.1093/nar/29.17.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orlowski J, Bujnicki JM. Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res. 2008;36:3552–3569. doi: 10.1093/nar/gkn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belfort M, Weiner A. Another bridge between kingdoms: tRNA splicing in archaea and eukaryotes. Cell. 1997;89:1003–1006. doi: 10.1016/s0092-8674(00)80287-1. [DOI] [PubMed] [Google Scholar]
- 31.Hickman AB, Li Y, Mathew SV, May EW, Craig NL, Dyda F. Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell. 2000;5:1025–1034. doi: 10.1016/s1097-2765(00)80267-1. [DOI] [PubMed] [Google Scholar]
- 32.Dahlroth SL, Gurmu D, Schmitzberger F, Engman H, Haas J, Erlandsen H, Nordlund P. Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus. FEBS J. 2009;276:6636–6645. doi: 10.1111/j.1742-4658.2009.07374.x. [DOI] [PubMed] [Google Scholar]
- 33.Aravind L, Makarova KS, Koonin EV. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ban C, Yang W. Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J. 1998;17:1526–1534. doi: 10.1093/emboj/17.5.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L. Structure and function of the 5′–>3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steczkiewicz K, Muszewska A, Knizewski L, Rychlewski L, Ginalski K. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012;40:7016–7045. doi: 10.1093/nar/gks382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 39.Pei J, Sadreyev R, Grishin NV. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 40.Ginalski K, Rychlewski L. Protein structure prediction of CASP5 comparative modeling and fold recognition targets using consensus alignment approach and 3D assessment. Proteins. 2003;53(Suppl. 6):410–417. doi: 10.1002/prot.10548. [DOI] [PubMed] [Google Scholar]
- 41.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 42.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Xing X, Herr AB, Bell CE. Crystal structure of E. coli RecE protein reveals a toroidal tetramer for processing double-stranded DNA breaks. Structure. 2009;17:690–702. doi: 10.1016/j.str.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoglund A, Donnes P, Blum T, Adolph HW, Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics. 2006;22:1158–1165. doi: 10.1093/bioinformatics/btl002. [DOI] [PubMed] [Google Scholar]
- 45.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 46.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 47.Malecki M, Stepien PP, Golik P. Assays of the helicase, ATPase and exoribonuclease activities of the yeast mitochondrial degradosome. Methods Mol. Biol. 2010;587:339–358. doi: 10.1007/978-1-60327-355-8_24. [DOI] [PubMed] [Google Scholar]
- 48.Burgers PM, Stith CM, Yoder BL, Sparks JL. Yeast exonuclease 5 is essential for mitochondrial genome maintenance. Mol. Cell Biol. 2010;30:1457–1466. doi: 10.1128/MCB.01321-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 50.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38:279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 53.Huynen MA, Duarte I, Szklarczyk R. Loss, replacement and gain of proteins at the origin of the mitochondria. Biochim. Biophys. Acta. 2013;1827:224–231. doi: 10.1016/j.bbabio.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Filee J, Forterre P. Viral proteins functioning in organelles: a cryptic origin? Trends Microbiol. 2005;13:510–513. doi: 10.1016/j.tim.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 55.Shutt TE, Gray MW. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006;22:90–95. doi: 10.1016/j.tig.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 56.Abad MF, Di Benedetto G, Magalhaes PJ, Filippin L, Pozzan T. Mitochondrial pH monitored by a new engineered green fluorescent protein mutant. J. Biol. Chem. 2004;279:11521–11529. doi: 10.1074/jbc.M306766200. [DOI] [PubMed] [Google Scholar]
- 57.Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 58.Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell. 2002;111:495–505. doi: 10.1016/s0092-8674(02)01075-9. [DOI] [PubMed] [Google Scholar]
- 59.Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pohjoismaki JL, Holmes JB, Wood SR, Yang MY, Yasukawa T, Reyes A, Bailey LJ, Cluett TJ, Goffart S, Willcox S, et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown TA, Tkachuk AN, Clayton DA. Native R-loops persist throughout the mouse mitochondrial DNA genome. J. Biol. Chem. 2008;283:36743–36751. doi: 10.1074/jbc.M806174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 63.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 65.Brown WM, Shine J, Goodman HM. Human mitochondrial DNA: analysis of 7S DNA from the origin of replication. Proc. Natl Acad. Sci. USA. 1978;75:735–739. doi: 10.1073/pnas.75.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gillum AM, Clayton DA. Displacement-loop replication initiation sequence in animal mitochondrial DNA exists as a family of discrete lengths. Proc. Natl Acad. Sci. USA. 1978;75:677–681. doi: 10.1073/pnas.75.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tapper DP, Clayton DA. Mechanism of replication of human mitochondrial DNA. Localization of the 5′ ends of nascent daughter strands. J. Biol. Chem. 1981;256:5109–5115. [PubMed] [Google Scholar]
- 68.Walberg MW, Clayton DA. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981;9:5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Antes A, Tappin I, Chung S, Lim R, Lu B, Parrott AM, Hill HZ, Suzuki CK, Lee CG. Differential regulation of full-length genome and a single-stranded 7S DNA along the cell cycle in human mitochondria. Nucleic Acids Res. 2010;38:6466–6476. doi: 10.1093/nar/gkq493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell. Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kieper J, Lauber C, Gimadutdinow O, Urbanska A, Cymerman I, Ghosh M, Szczesny B, Meiss G. Production and characterization of recombinant protein preparations of Endonuclease G-homologs from yeast, C. elegans and humans. Protein Expr. Purif. 2010;73:99–106. doi: 10.1016/j.pep.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 73.Kalinowska M, Garncarz W, Pietrowska M, Garrard WT, Widlak P. Regulation of the human apoptotic DNase/RNase endonuclease G: involvement of Hsp70 and ATP. Apoptosis. 2005;10:821–830. doi: 10.1007/s10495-005-0410-9. [DOI] [PubMed] [Google Scholar]
- 74.Widlak P, Li LY, Wang X, Garrard WT. Action of recombinant human apoptotic endonuclease G on naked DNA and chromatin substrates: cooperation with exonuclease and DNase I. J. Biol. Chem. 2001;276:48404–48409. doi: 10.1074/jbc.M108461200. [DOI] [PubMed] [Google Scholar]
- 75.Harrington JJ, Lieber MR. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Masuda-Sasa T, Imamura O, Campbell JL. Biochemical analysis of human Dna2. Nucleic Acids Res. 2006;34:1865–1875. doi: 10.1093/nar/gkl070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim JH, Kim HD, Ryu GH, Kim DH, Hurwitz J, Seo YS. Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res. 2006;34:1854–1864. doi: 10.1093/nar/gkl102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 79.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 80.Fortini BK, Pokharel S, Polaczek P, Balakrishnan L, Bambara RA, Campbell JL. Characterization of the endonuclease and ATP-dependent flap endo/exonuclease of Dna2. J. Biol. Chem. 2011;286:23763–23770. doi: 10.1074/jbc.M111.243071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murante RS, Rust L, Bambara RA. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 82.Kao HI, Campbell JL, Bambara RA. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 2004;279:50840–50849. doi: 10.1074/jbc.M409231200. [DOI] [PubMed] [Google Scholar]
- 83.Zhang J, McCabe KA, Bell CE. Crystal structures of lambda exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc. Natl Acad. Sci. USA. 2011;108:11872–11877. doi: 10.1073/pnas.1103467108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pohjoismaki JL, Wanrooij S, Hyvarinen AK, Goffart S, Holt IJ, Spelbrink JN, Jacobs HT. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–5828. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kai Y, Miyako K, Muta T, Umeda S, Irie T, Hamasaki N, Takeshige K, Kang D. Mitochondrial DNA replication in human T lymphocytes is regulated primarily at the H-strand termination site. Biochim. Biophys. Acta. 1999;1446:126–134. doi: 10.1016/s0167-4781(99)00015-9. [DOI] [PubMed] [Google Scholar]
- 86.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Di Re M, Sembongi H, He J, Reyes A, Yasukawa T, Martinsson P, Bailey LJ, Goffart S, Boyd-Kirkup JD, Wong TS, et al. The accessory subunit of mitochondrial DNA polymerase gamma determines the DNA content of mitochondrial nucleoids in human cultured cells. Nucleic Acids Res. 2009;37:5701–5713. doi: 10.1093/nar/gkp614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ruhanen H, Borrie S, Szabadkai G, Tyynismaa H, Jones AW, Kang D, Taanman JW, Yasukawa T. Mitochondrial single-stranded DNA binding protein is required for maintenance of mitochondrial DNA and 7S DNA but is not required for mitochondrial nucleoid organisation. Biochim. Biophys. Acta. 2010;1803:931–939. doi: 10.1016/j.bbamcr.2010.04.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.