Abstract
Chromatin insulators are functionally conserved DNA–protein complexes situated throughout the genome that organize independent transcriptional domains. Previous work implicated RNA as an important cofactor in chromatin insulator activity, although the precise mechanisms are not yet understood. Here we identify the exosome, the highly conserved major cellular 3′ to 5′ RNA degradation machinery, as a physical interactor of CP190-dependent chromatin insulator complexes in Drosophila. Genome-wide profiling of exosome by ChIP-seq in two different embryonic cell lines reveals extensive and specific overlap with the CP190, BEAF-32 and CTCF insulator proteins. Colocalization occurs mainly at promoters but also boundary elements such as Mcp, Fab-8, scs and scs′, which overlaps with a promoter. Surprisingly, exosome associates primarily with promoters but not gene bodies of active genes, arguing against simple cotranscriptional recruitment to RNA substrates. Similar to insulator proteins, exosome is also significantly enriched at divergently transcribed promoters. Directed ChIP of exosome in cell lines depleted of insulator proteins shows that CTCF is required specifically for exosome association at Mcp and Fab-8 but not other sites, suggesting that alternate mechanisms must also contribute to exosome chromatin recruitment. Taken together, our results reveal a novel positive relationship between exosome and chromatin insulators throughout the genome.
INTRODUCTION
The exosome is a multisubunit complex conserved from archaea to humans that is the major cellular 3′ to 5′ RNA degradation machinery. Involved in the turnover of normal as well as aberrant RNAs, the exosome additionally plays a major role in RNA processing and maturation [reviewed in (1)]. The exosome consists of a core complex including a hexameric ring of RNase PH homology domain-containing subunits (Ski6/Rrp41, Rrp42, Rrp43, Rrp45, Rrp46 and Mtr3) capped by a trimer of S1/KH domain-containing subunits (Csl4, Rrp4 and Rrp40) [reviewed in (2)]. It has been shown that yeast exosomes channel RNA through the center of the core complex (3), but it is the association of either of the hydrolytic RNases Dis3/Rrp44 and Rrp6 with the yeast and human core exosome that provides enzymatic activity of the complex. A contrasting view in Drosophila suggests that exosome subunits can function independently or form a continuum of various functional complexes (4).
Although the core exosome and Dis3 localize to both the nucleus and the cytoplasm, the Rrp6 component is predominantly nuclear, suggesting specialized activities for the exosome in the nucleus. Rrp6 alone or the entire exosome have been implicated in several nuclear RNA quality control and surveillance pathways [reviewed in (5)]. Depletion of exosome levels or mutation of exosome components leads to stabilization of cryptic unstable transcripts (CUTs) in yeast, antisense promoter transcripts in mammals (6,7), as well as other aberrant RNAs. In yeast, Nrd1-dependent transcription termination of certain non-coding genes and CUTs from intergenic regions also involves recruitment of the exosome to promote transcript degradation (8–10), raising the possibility of chromatin proximal exosome activity. In these cases, it is not known whether the exosome associates with chromatin in order to carry out its surveillance activities. Toward this end, an overexpressed tagged version of Rrp6 was shown to associate with chromatin of yeast protein coding genes using whole open reading frame cDNA mircroarrays (11). In Drosophila, it was demonstrated that certain exosome subunits associate with at least some actively transcribed genes (12,13) and may be recruited to chromatin through interaction with RNA polymerase II (Pol II) elongation factors Spt5 and Spt6 (14). Nevertheless, a high-resolution genome-wide study of exosome chromatin association has yet to be performed in any organism.
Distributed throughout the genome, chromatin insulators are DNA–protein complexes that organize chromatin into independent transcriptional domains. In Drosophila, there are at least five chromatin insulator families that can be categorized by the association of particular DNA-binding proteins. These classes include the CCCTC-binding factor (CTCF), Suppressor of Hairy-wing (Su(Hw)), GAGA factor (GAF), Zeste-white 5 (Zw5) and boundary element-associated factor (BEAF-32) [reviewed in (15,16)]. For example, the hsp70 genes at 87A7 are located between the scs and scs′ boundary elements, bound by Zw5 and BEAF-32, respectively, (17,18). Zw5 and BEAF-32 interact with each other, and this interaction may promote chromatin looping observed between scs and scs′ (19). In addition, the Fab-8 insulator is a well-characterized cis-regulatory region of the Abdominal-B (Abd-B) locus in the bithorax complex that harbors binding sites for the zinc-finger DNA-binding protein CTCF (20,21). CTCF is required for Fab-8 insulator function and looping interactions among insulators, enhancers and promoters at Abd-B (22–25). Multiple insulator complexes share a common component, Centrosomal protein 190 (CP190), a zinc-finger and BTB/POZ domain-containing protein that may play a global role in chromatin organization (22,26). Since enhancers must often activate their target promoters from long distances, it is likely that insulators act as tethering sites for chromosomal loops that can constrain enhancer–promoter interactions and possibly protect a region from its surrounding chromatin environment.
Depending on their context, chromatin insulators could either repress or promote transcription based on the nature of the higher order chromatin interactions to which they contribute. Recent studies show that several insulator proteins, particularly CP190 and BEAF-32, associate with certain transcriptionally active promoters (27–29). In fact, BEAF-32 appears to be required for transcription at a number of the promoters to which it binds (28). How insulator proteins are targeted to specific promoters is unknown; however, insulator protein recruitment correlates with specific transcription initiation patterns (30). The precise role of insulator proteins at the promoter is still unclear, but these findings suggest that insulator proteins may regulate transcription in a more direct manner than previously suspected.
Earlier work suggests that the catalog of insulator-associated factors and potential regulators is not yet complete. The CP190/CTCF class of insulators interacts with Argonaute2, which promotes Fab-8 activity as well as insulator-dependent looping at this site (25). Moreover, the gypsy class of insulator proteins has been shown to physically interact with the ubiquitin and SUMO ligase Topors and Lamin, a major component of the nuclear matrix (31). Recent work also showed the association of Top2 with gypsy insulator complexes, and this factor appears to be important for the stability of the Mod(mdg4)2.2 protein (32). Of particular interest to this study is the finding that the Rm62 helicase interacts with CP190 in an RNA-dependent manner, suggesting that RNA may be a component of the gypsy insulator complex (33). Finally, the RNA-binding protein Shep was recently identified as the first known tissue-specific regulator of gypsy insulator activity (34). Here, we sought to identify additional RNA-related chromatin insulator factors and examine their potential functional significance.
In this study, we identified the exosome as a novel interactor of CP190 insulator complexes. Genome-wide profiling of exosome components compared with insulator proteins by staining of polytene chromosomes as well as high-resolution ChIP-seq in two different cell lines reveals highly overlapping genome-wide association profiles of the exosome with CP190, BEAF-32 and CTCF insulator proteins. Unexpectedly, we find that the majority of exosome binding sites are situated at the promoters of transcribed genes but not throughout gene bodies or 3′ ends. Exosome chromatin association correlates with active transcription; however, transcript levels at exosome binding sites are not significantly changed in response to exosome depletion. Finally, we found that depletion of BEAF-32 or CP190 has no effect on exosome chromatin association, but reduction of CTCF levels reduces exosome association with the CTCF-dependent Mcp and Fab-8 insulators. These results reveal a previously unknown association between exosome and chromatin insulators throughout the genome.
MATERIALS AND METHODS
Chromatin immunoprecipitation and ChIP-seq
Cells were fixed by addition of 1% formaldehyde to cell media for 10 min at R.T. with gentle agitation. Formaldehyde was quenched by addition of glycine to 0.125 M with gentle agitation for 5 min at R.T. Cells were pelleted at 400 rcf, washed twice in PBS, and resuspended in 1 ml ice–cold cell lysis buffer (5 mM PIPES, pH 8, 85 mM KCl, 0.5% NP-40) supplemented with Complete protease inhibitors (Roche). Nuclei were released by Dounce homogenization with pestle B and pelleted by centrifugation at 9190 rcf for 5 min at 4°C. Nuclei and chromatin were further processed as described previously (35). Immunoprecipitations were performed with 3 ul rabbit α-Rrp6 (14), rabbit α-Rrp40 (36), rabbit α-BEAF-32 (37), rabbit α-CP190 (25), rabbit α-CTCF (22), rabbit α-Su(Hw) (37) or rabbit IgG (Santa Cruz Biotechnology) coupled to rProtein A agarose beads (GE Healthcare). Primers used are listed in Supplementary Table S9.
Samples for ChIP-seq from input DNA, Rrp6 ChIP and Rrp40 ChIP were prepared according to the manufacturer’s protocol with standard or TruSeq adapters (Illumina). DNA was sequenced on an Illumina Genome Analyzer II or HiSeq 2000 at the NIDDK Genomics Core Facility. Exosome ChIP-seq data are available at Gene Expression Omnibus (GSE41950).
dsRNA knockdowns
Amplicons used for dsRNA knockdowns were designed based on recommendations from the Drosophila RNAi Screening Center. Templates were PCR amplified from genomic DNA using primers containing the T7 promoter sequence (Supplementary Table S9). DsRNAs were produced by in vitro transcription of PCR templates using the MEGAscript T7 kit (Ambion) and purified using NucAway Spin Columns (Ambion). S2 and S3 cells were grown at 25°C in Shields and Sang M3 Insect medium (Sigma) supplemented with 0.1% yeast extract, 0.25% bactopeptone and 10% fetal bovine serum (HyClone). Transfections of 1 × 107 cells using 2 µg of dsRNA were performed using Cell Line Nucleofector Kit V (Amaxa Biosystems) transfection reagent using the G-30 protocol. On day 2, cells were diluted with normal media ∼1:5. Five days after transfection, cells were collected, and knockdown efficiency was confirmed by western blotting.
RNA purification and RNA-seq
Total RNA was isolated from S2 and S3 cells using Trizol Reagent (Invitrogen) using the recommended protocol. Polyadenylated RNA and ribosomal (rRNA) depleted RNA was purified from total RNA using the Poly(A)Purist MAG or MicroPoly(A)Purist Kits (Ambion) and the RiboMinus Eukaryote Kit for RNA-Seq (Invitrogen), respectively. Depletion of rRNA was ∼99% by qPCR measurement of 5S rRNA. Sequencing libraries were prepared from Poly(A)+ and rRNA depleted RNA samples according to the manufacturer’s protocol (Illumina), and sequencing was performed on an Illumina Genome Analyzer II or HiSeq 2000 at the NIDDK Genomics Core Facility. Exosome knockdown RNA-seq data are available at Gene Expression Omnibus (GSE41950).
CP190 immunoaffinity purification
CP190 complex immunoaffinity purification was performed from nuclear extracts from 0–24 h OR embryos as described previously (33). Columns were used only once for western blotting analyses but reused multiple times for RNase activity assays to conserve material, which led to slightly altered elution profiles.
RNase activity assay
RNA substrate was synthesized by in vitro transcription using the pAWG-su(Hw) vector containing the su(Hw) cDNA. Primers used are listed in Supplementary Table S9. The resulting 99 nt RNA was end labeled with [5′-32P]pCp by incubation with T4 RNA ligase at 4°C O/N and purified by running on a 12% polyacrylamide gel (8 M urea, 1x TAE) followed by extraction of RNA bands by crushing with a motorized pestle and soaking in 0.3 M NaOAc pH 5.2. RNA was ethanol precipitated with 20 µg glycogen. RNA substrate was incubated with buffer alone, Csl4–Flag exosome complexes, or immunoaffinity column fractions in 10 mM Tris-HCl pH 8.0, 50 mM KCl, 5 mM MgCl2 and 10 mM DTT for 2 h at 37°C. Csl4–Flag complexes were purified from transiently transfected S2 cells as described previously (36). Samples were then heated to 65°C for 15 min, immediately cooled on ice, and separated on a 15% polyacrylamide gel (8 M urea, 1x TAE) at 15 W for 45 min. Gels were then exposed to phosphorimager plates overnight and scanned on a Fuji FLA5000 imager.
Immunofluorescence of polytene chromosomes
Polytene chromosome spreads were prepared essentially as described previously (38). Rabbit α-Rrp6, rabbit α-Rrp40, mouse α-BEAF (19) (Developmental Studies Hybridoma Bank), guinea pig α-CP190 (25), guinea pig α-CTCF [generated similarly as in (22)], guinea pig α-Su(Hw) (35) and guinea pig α-Mod(mdg4)2.2 (35) were used for staining.
Western blotting
Western blotting was performed with guinea pig α-CP190, rabbit α-CTCF, guinea pig α-Mod(mdg4)2.2, mouse α-Lamin (39) (ADL67.10, Developmental Studies Hybridoma Bank), mouse α-BEAF, rabbit α-Rrp6, rabbit α-Rrp40, guinea pig α-Rrp4 (36), guinea pig α-Rrp46 (36), guinea pig α-Csl4 (36) and rat α-Ski6 (14).
Computational analyses
ChIP-seq
Reads were mapped with Bowtie 0.12.7 (40) to the dm3 assembly, excluding chrUextra. Only the best uniquely mapping reads allowing two mismatches were kept (parameters –best –strata -m1 -n2 –tryhard -k1). Libraries from technical replicates were merged. Duplicate reads were collapsed with Picard's MarkDuplicates (http://picard.sourceforge.net/command-line-overview.shtml), and reads falling in repetitive regions were removed. Peaks were called with SPP using z.thr = 3, window.size = 1000, an FDR of 1% and otherwise default parameters. Similar results were obtained using the MACS algorithm (41).
Binary heat maps
Supervised hierarchical clustering of overlap by at least 1 bp was performed as in (42), using the multi-intersect program from BEDTools (43) (with the cluster option) and the binary_heatmap() function from pybedtools (44).
Overlap enrichment heat maps
Data files containing called peaks were downloaded and converted to BED files (25,45–55). Endogenous siRNA cluster coordinates were used directly as reported in supplementary tables, or raw reads from Czech et al. and Fagegaltier et al. were clustered de novo as previously defined (25).
Enrichment scores were calculated as described previously (25). The full enrichment matrix was hierarchically clustered using correlation as a distance metric and complete linkage clustering as implemented in SciPy, with rows clustered identically as columns. Selected rows from the full clustered matrix in Supplementary Figure S1 are shown in Figure 4B. For the active regions heatmap, active regions were first defined as any region bound by both Pol II and H3K4me3 using data for these factors in S2 cells from modENCODE (http://intermine.modencode.org/). Genome-wide features used in the full heat map were then subsetted so that only those features overlapping an active region by at least 1 bp were considered. Enrichment scores were generated similarly to the full heat map, except that when features were shuffled, the new random locations were required to fall within an active region. The same row/column ordering in Figure 4B was applied in order to facilitate comparison.
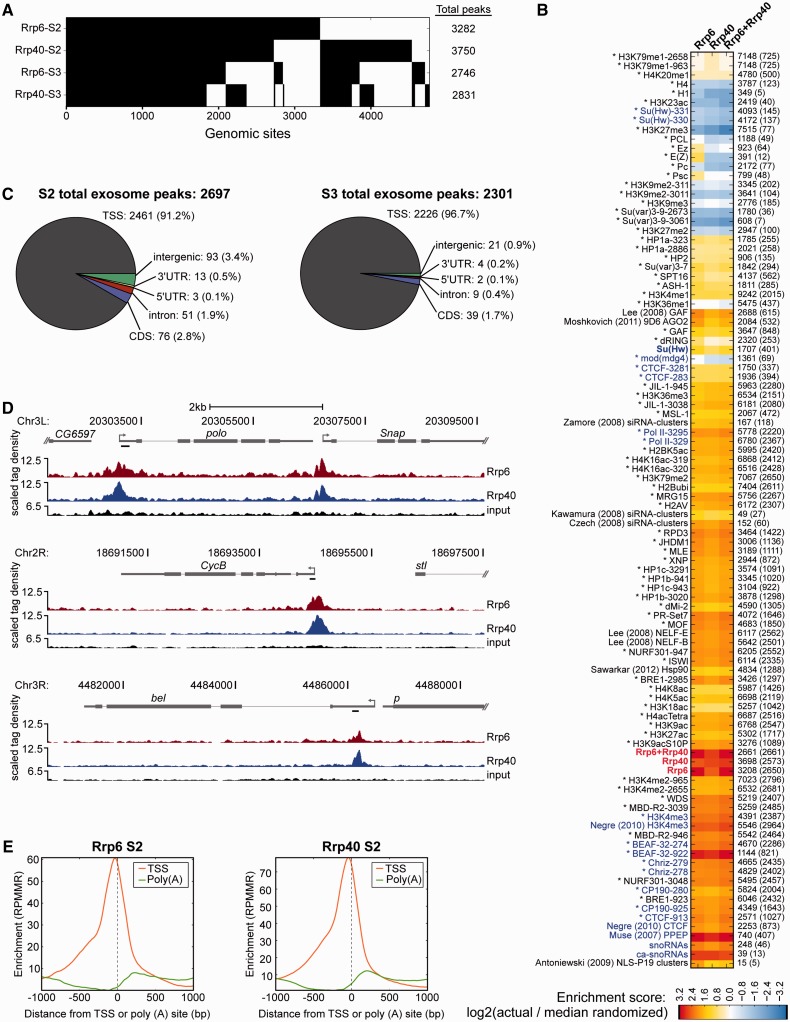
Figure 4.
ChIP-seq analysis of exosome components in two embryonic cell lines. (A) Binary heat map of Rrp6 and Rrp40 binding sites in S2 and S3 cells ordered by supervised hierarchical clustering. Each column represents a single genomic location, and a black mark in a row represents the presence of a particular factor at that site. Total peak counts are indicated for each factor (right). (B) Heat map of log2 enrichment scores for pairwise comparisons of binding sites for Rrp6, Rrp40 and exosome with additional data sets in S2 cells. Color scale corresponding to enrichment value is indicated (bottom). Positive values indicate significant enrichment whereas negative values indicate significant negative enrichment. Self–self comparisons are indicated in gray, and pairwise comparisons that are not statistically significant (P > 0.001) are indicated in white. Numbers to the right of each row indicate the total number of features in each data set, and the number of sites overlapping with exosome are indicated in parentheses. Data sets in bold were generated in this study. Data generated by modENCODE consortium is marked with an asterisk. Other data are from studies as indicated (25,48–55). The ca-snoRNA data set was defined in embryos (62). Full heat map with hierarchical clustering is shown in Supplementary Figure S1. (C) Classification of Rrp6 and Rrp40 ChIP-seq peaks in S2 and S3 cells with respect to annotated genes. Number of sites and percentage of total in parentheses corresponding to TSS, transcription start site; CDS, coding sequence; 5′ UTR, 5′ untranslated region; 3′ UTR, 3′ untranslated region. See methods for classification hierarchy of overlapping categories. (D) Screenshots of input, Rrp6 and Rrp40 ChIP-seq signals at the polo, CycB and bel loci in S3 cells. The bottom of each scale bar indicates 0. Black bars indicate primer sets used for directed ChIP in Figure 6B. (E) Genome-wide profiles of Rrp6 and Rrp40 S2 input normalized ChIP-seq tag density around transcription start sites (orange) or poly (A) sites (green) for all genes.
Pie charts
Feature classes [TSSs (1 bp transcript start position), CDSs, introns, 5′UTRs and 3′UTRs] were extracted from all annotated isoforms of all annotated genes in FlyBase release 5.33. Intergenic regions were defined as the remainder of dm3. Since a ChIP-seq peak can fall in more than one class, we classified a peak by its highest priority annotation class, where the priorities from highest to lowest are TSS, CDS, intron, 5'UTR, 3'UTR and intergenic.
TSS matrices
Each row in the matrix in Figure 5B corresponds to the genomic region +/− 1 kb around each TSS split into 20 bp bins. Reads were extended 3' to a total length of 200 bp to represent the fragment size. For each column i in the matrix representing a 20 bp-wide genomic location, the input-normalized value in reads per million mapped reads (RPMMR) was calculated as (IPi / IPtotal) − (inputi / inputtotal) where IPi and inputi are the numbers of reads overlapping that region in the IP and input, respectively, and IPtotal and inputtotal are the total number of mapped reads, in millions, in the libraries. Active genes were defined as those with both Pol II and H3K4me3 within a window extending 250 bp upstream and 750 bp downstream of the TSS. We defined ambiguous genes as having either Pol II or H3K4me3 but not both in this window and inactive genes as having neither Pol II nor H3K4me3 in this window. Within each of the three categories, rows are sorted by row means. Line plots show the column sums of the TSS matrix (Figure 5B) or the column sums of a similarly constructed poly (A) site matrix in which each row is centered on the 3'-most coordinate of each isoform.
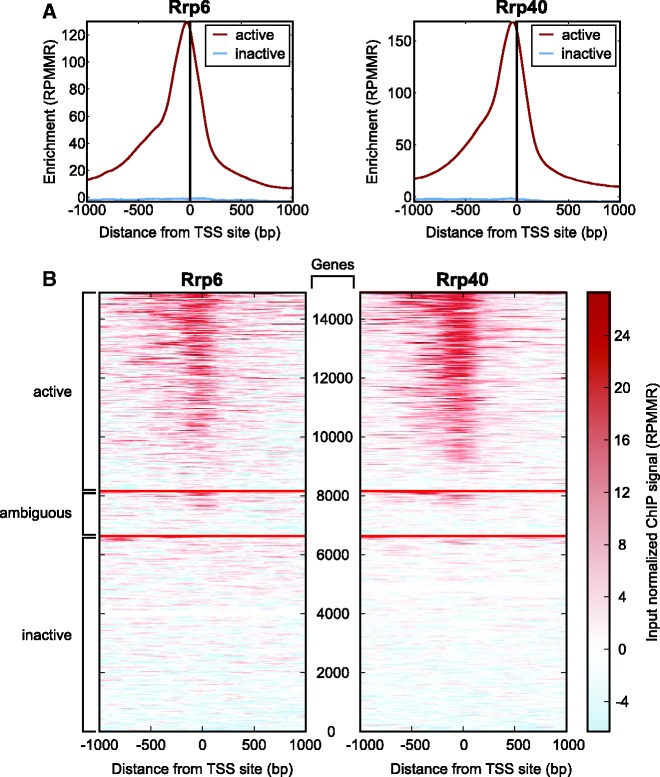
Figure 5.
Genome-wide exosome chromatin association correlates strongly with active transcription. (A) Exosome associates preferentially with active promoters. Profile of S2 Rrp6 and Rrp40 input normalized ChIP-seq tag density around TSSs associated (red) or not associated (blue) with H3K4me3 and Pol II 250 bp upstream or 750 bp downstream of the TSS. (B) Matrices of Rrp6 and Rrp40 S2 ChIP signal across all annotated TSSs +/− 1 kb. Each row corresponds to an annotated gene, and red lines indicate grouping of rows into active, ambiguous or inactive TSSs. Rows are ordered by mean signal from highest to lowest within each subgroup. Active genes were defined as genes with Pol II and H3K4me3 within 250 bp upstream or 750 bp downstream of the TSS, ambiguous genes had one of these marks within this window, and inactive genes had neither mark in the same window. Signal intensity is shown in input-normalized enriched RPMMR. Color scale corresponding to enrichment is indicated (right).
Note that modENCODE supplies several data sets for Pol II (accessions 3295 and 329), CTCF (283, 3281 and 913), CP190 (925, 280) and BEAF-32 (922, 274). In the enrichment analysis heatmap, these data sets were treated separately. In all other analyses, data sets have been combined by first concatenating all data sets for a factor and then merging any features that overlap by at least 1 bp into a single feature.
Hypergeometric overlap tests
To assess the enrichment of insulators and exosome specifically at active TSSs, we used the hypergeometric overlap test (56) where n is the total number of non-redundant annotated isoforms of all active genes; n1 is the number of isoforms with an exosome peak at an active TSS, n2 is the number of isoforms with an insulator peak at an active TSS and m is the number of isoforms with both exosome and insulator peak at an active TSS. Active TSSs were identified as those with both Pol II and H3K4me3 overlapping the TSS. For each factor, the bound active TSSs consisted of the set of active TSSs that had at least 1 bp overlap with at least one binding site for that factor. Similar results were obtained when considering the full set of TSSs, regardless of transcriptional status.
Divergent promoters
Divergent promoters were identified from coding and non-coding genes and were defined as a minus-strand TSS followed by a plus-strand TSS separated by no more than threshold N bp with no intervening genic sequence between them. Several different values of N (100, 250, 500 bp) were used for analyses.
GO analysis
Gene ontology was assessed using DAVID (57,58) with the genome as the background gene set and ‘high’ stringency setting for clustering. We considered all genes with a Rrp6 or Rrp40 peak anywhere in the gene. DAVID scaled enrichment values are reported.
RNA-seq and differential expression analysis
Libraries were sequenced on both GAII and HiSeq 2000 machines in either single-end and paired-end mode. Due to small insert sizes, only the first end of the PE libraries was used. Adapters were clipped using cutadapt 1.0 (59). Clipped reads were mapped with TopHat 1.4.1 (60) to the dm3 assembly, excluding chrUextra. Mapped reads were counted in all annotated coding and non-coding genes in FlyBase r5.33 using HTSeq in ‘union’ mode, which only includes reads that map unambiguously to a single gene. Counts from technical replicates were summed. Counts from replicates were used in DESeq using ‘local’ fit type and ‘fit-only’ sharing mode (61). Comparisons used the ‘per-condition’ method for estimating dispersions. Differentially expressed genes were considered those with adjusted P-values < 0.05.
Analysis of intergenic transcription, snoRNA and snoRNA precursors
To evaluate changes in upstream regions upon knockdown, reads were counted with HTSeq in 0–500 bp upstream of gene TSSs, all intergenic exosome peaks, and selected cis-regulatory regions of Abd-B. Subregions that also overlapped annotated genes were subtracted in order to focus on changes in intergenic RNA. Regions <100 bp were also removed. Bound upstream regions were defined as gene-level TSS intersecting an exosome peak. Upstream and intergenic peak regions were merged such that overlapping features were considered a single feature.
Annotated snoRNAs from FlyBase r5.33 were considered and classified as being chromatin-associated based on (62). Putative precursor regions for snoRNA were defined as the largest intron overlapping each snoRNA or ca-snoRNA. Ten intergenic ‘orphan’ snoRNAs, which do not have an obvious precursor, were not considered. Precursors were then merged such that each would be represented only once in the analysis even though multiple snoRNAs could be in one precursor, or both ca-snoRNAs and snoRNAs could come from the same precursor. For each precursor, the number of processed snoRNA-specific reads in the region was subtracted from the total number of reads in the region in order to isolate precursor-specific reads. These final precursor read counts, as well as counts for all mature snoRNAs including the ‘orphan’ snoRNAs, were examined.
Regions containing zero reads in all RNAseq libraries were removed. For each rRNA depletion knockdown experiment, DESeq was run as described above for RNA-seq, and all regions were reported.
RESULTS
Exosome and RNase activity copurify with chromatin insulator proteins
Given previous evidence that RNA and RNA-binding proteins associate with chromatin insulators, we sought to determine whether the exosome physically associates with CP190 chromatin insulator complexes. Using a well-established protocol to isolate CP190 insulator components and associated factors such as CTCF (22,25,33), embryonic nuclear extracts were immunoaffinity purified over control preimmune or α-CP190 antibody columns by washing and subsequent elution with increasing MgCl2 concentrations (Figure 1A). Eluates were examined for the presence of insulator proteins as well as exosome subunits. Western blotting verifies the presence of CP190 and Mod(mdg4)2.2 insulator proteins in the α-CP190 eluates as well as the exosome components Rrp6, Rrp4, Rrp46, Csl4 and Ski6 but not in preimmune eluates. In contrast, a negative control protein, HP1a, is not identified as a specific interactor of CP190 (data not shown). Therefore, multiple exosome components copurify with CP190 insulator complexes, suggesting that the intact exosome may be stably associated with CP190 chromatin insulator complexes in the nucleus.
Figure 1.
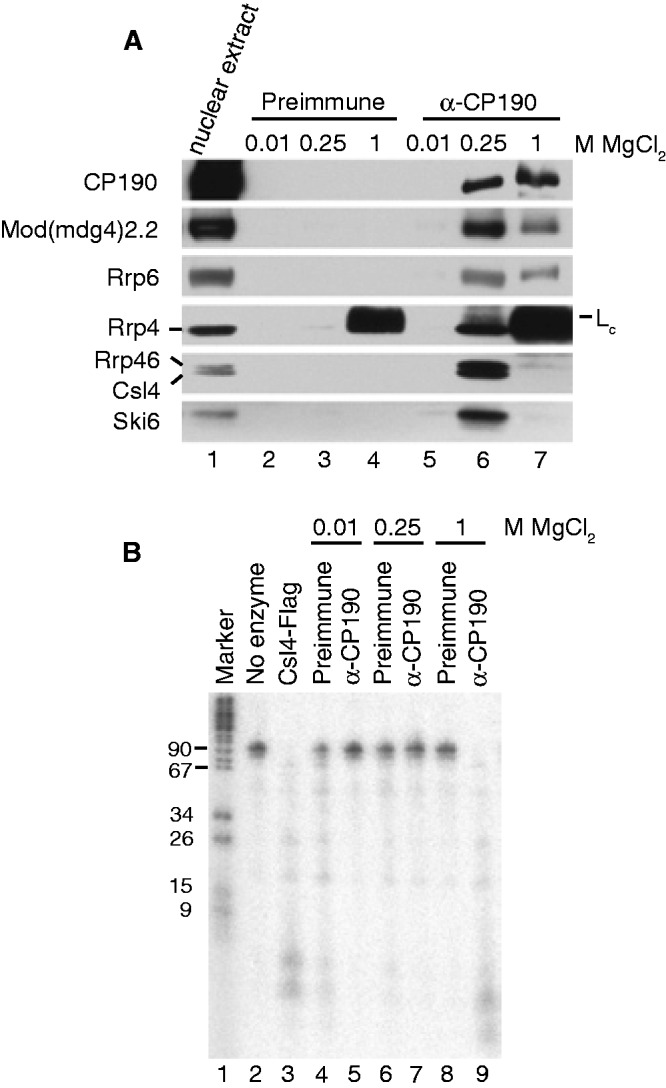
CP190 insulator complexes associate with the nuclear exosome and copurify with RNase activity. (A) Exosome subunits physically associate with CP190 insulator complexes. Western blotting of embryonic nuclear extracts (lane 1, 0.4% of input) bound to a control preimmune column (lanes 2–4) or α-CP190 column (lanes 5–7) and step-eluted with increasing MgCl2 concentrations as indicated. Note the presence of the IgG light chain (Lc) in both 1 M MgCl2 elutions in the α-Rrp4 blot. For CP190 blots, 4% of the eluates were probed. For all other blots, 96% of the eluates were probed. (B) RNase activity copurifies with CP190 insulator complexes. Shown are DNA marker (lane 1), intact in vitro transcribed RNA substrate (lane 2), RNA substrate after incubation with Csl4–Flag purified control complex (lane 3), or RNA substrate after incubation with complexes step-eluted from either preimmune antibody column (lanes 4, 6, 8) or α-CP190 column (lanes 5, 7, 9) at MgCl2 concentrations as indicated. Samples were run on a 15% PAGE urea denaturing gel and exposed to a phosphorimager plate.
In order to assess whether physical association between the exosome and CP190 chromatin insulator complexes is functionally relevant, we examined whether immunopurified CP190 complexes harbor ribonuclease activity. To this end, a radiolabeled 99 nt in vitro transcribed RNA was used as a substrate in in vitro degradation assays. As a control, RNA was incubated with exosome complexes immunopurified from S2 cells stably transfected with Csl4-Flag using α-Flag (Figure 1B). Examination of the RNA on a high percentage urea polyacrylamide gel indicates extensive degradation of the RNA as result of incubation with exosome complexes compared with buffer alone. Incubation of the RNA substrate with preimmune or α-CP190 immunoaffinity column fractions results in extensive degradation specifically in the 1 M α-CP190 elution fraction, while the majority of RNA substrate remains intact in other fractions tested. Lack of activity in the 250 mM CP190 fraction was unexpected but may be due to absence of a critical factor, presence of a repressor or minor differences in purification procedures used for western blotting and RNase activity assays (see Materials and Methods). These results suggest that CP190 chromatin insulators may associate with active exosome complexes in vivo.
Exosome subunits colocalize with certain chromatin insulator proteins on polytene chromosomes
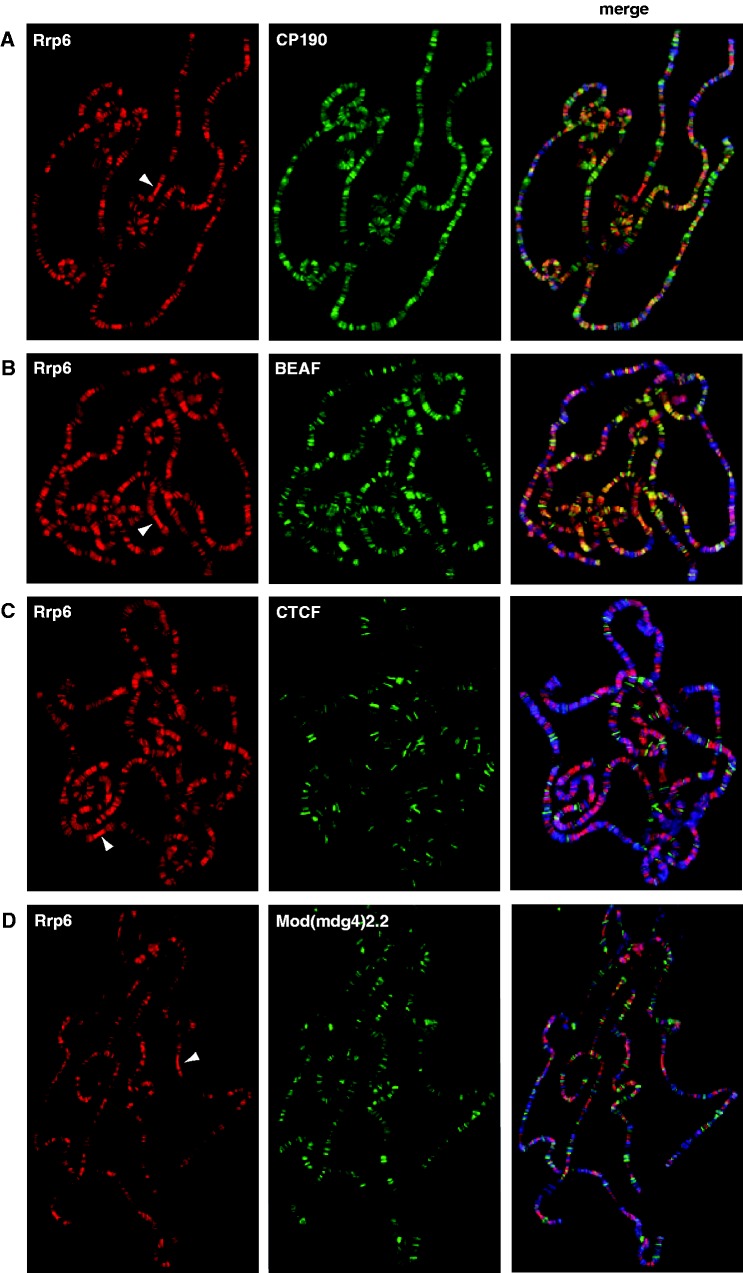
Given physical interaction between CP190 and the exosome, we wished to examine the genome-wide binding profiles of the exosome compared with insulator proteins. To this end, we performed immunostaining of highly replicated interphase polytene chromosomes from third instar larval salivary glands. Wildtype chromosomes were stained with either α-Rrp6 or α-Rrp40 and costained for α-CP190, α-BEAF-32, α-CTCF, α-Mod(mdg4)2.2 or α-Su(Hw) insulator proteins (Figures 2–3). Both Rrp6 and Rrp40 localize to DAPI interbands, indicative of localization to transcribed regions of the genome, similar to a previous study of the exosome component Ski6 (14). Since both exosome antibodies were generated in the same organism, double staining of Rrp6 and Rrp40 was not possible, but the localization patterns of both proteins are highly similar. For example, both antibodies intensely label the actively transcribed ‘gooseneck’ region at cytological position 31. Extensive colocalization is observed for Rrp6 compared with CP190 (Figure 2A) and to a higher extent with BEAF-32 (Figure 2B). In contrast, at this level of resolution, Rrp6 does not appear to colocalize considerably with the insulator proteins CTCF or Mod(mdg4)2.2 (Figure 2C and D). Furthermore, no appreciable overlap was observed between Rrp40 and Su(Hw) (Figure 3). The same results were obtained with both α-Rrp6 and α-Rrp40 antibodies (data not shown). These results suggest that the exosome colocalizes with actively transcribed regions and specific classes of insulator proteins throughout the genome.
Figure 2.
Exosome colocalizes extensively with CP190 and BEAF-32 insulator proteins on polytene chromosomes. Rabbit α-Rrp6 was detected with α-rabbit conjugated Alexa-594 (red). Guinea pig α-CP190 (A), mouse α-BEAF-32 (B), guinea pig α-CTCF (C) or guinea pig α-Mod(mdg4)2.2 (D) were detected with α-guinea pig or α-mouse conjugated Alexa-488 (green) as appropriate. DAPI-stained DNA is shown in blue in the merged image (right). White arrowheads point to the transcriptionally active ‘gooseneck’ region at cytological position 31. Yellow signal in the merged images indicates a high degree of overlap of Rrp6 with CP190 and BEAF-32.
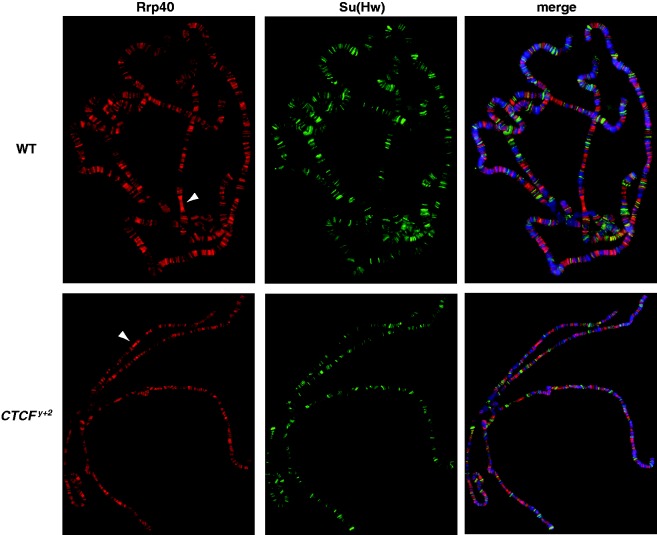
Figure 3.
Exosome chromatin association is not globally affected in CTCFy+2 mutants. Localization of Rrp40 (red) and the Su(Hw) insulator protein (green) on wildtype (top) or CTCFy+2 mutant (bottom) larval salivary gland polytene chromosomes. Rabbit α-Rrp40 was detected with α-rabbit conjugated Alexa-594. Guinea pig α-Su(Hw) was detected with α-guinea pig conjugated Alexa-488. DAPI-stained DNA is shown in blue in the merged image (right). White arrowheads point to the transcriptionally active ‘gooseneck’ region at cytological position 31.
High-resolution genome-wide mapping of exosome subunits by ChIP-seq
In order to obtain a high-resolution map of exosome genome-wide chromatin association, we performed ChIP-seq of exosome components in two embryonic cell lines, S2 and S3 cells. We determined Rrp6 and Rrp40 ChIP-seq profiles using Illumina sequencing and peak calling using the SPP algorithm at 1% FDR (63). Greater than 2700 exosome peaks were observed for Rrp6 or Rrp40 in either S2 or S3 cells (Figure 4A). High correspondence was observed between Rrp6 and Rrp40 in the same cell type; 82% of Rrp6 in S2 overlaps with Rrp40, and 84% of Rrp6 in S3 overlaps with Rrp40. Similar profiles obtained with antibodies to two distinct exosome components suggests that the intact exosome may associate with chromatin at these sites and furthermore verifies the specificity of the antibodies. Antibodies directed against other exosome components did not successfully detect signal on either polytene chromosomes or by ChIP or were not tested due to limited availability (data not shown). We subsequently refer to either specific subunit profiles or a common set of ‘exosome’ binding sites for a particular cell type at which both Rrp6 and Rrp40 are bound.
We next examined the overlap of exosome components with that of insulator proteins and other chromatin-associated factors or other features previously profiled in S2 cells. We calculated enrichment scores for two-way overlaps between all factors based on comparison with random shuffling of sites within the same chromosome. As expected, Rrp6 and Rrp40 profiles display among the highest levels of enrichment between one another compared with all other tested factors (Figure 4B). Consistent with polytene chromosome staining, statistically significant levels of enrichment are observed for both exosome factors with CP190 and BEAF-32 (78% and 84% of exosome sites, respectively, Supplementary Table S1). Substantial enrichment is also observed with CTCF (41%), whereas little to no enrichment is observed between either exosome component compared with Su(Hw) and Mod(mdg4)2.2 (20% and 3%, respectively). Unsupervised hierarchical clustering was performed in order to group factors based on overall similarity of enrichment profiles (Supplementary Figure S1). This analysis further indicates correspondence of exosome components with the CP190 and BEAF-32 insulator proteins. Notably, we identified high levels of overlap enrichment as well as hierarchical clustering with Chromator/Chriz, a factor recently implicated in boundary formation in a genome-wide chromatin conformation study (64). In summary, exosome colocalizes significantly with specific insulator proteins on a genome-wide level.
Exosome recruitment correlates with active transcription
Our comparative analyses also revealed extensive overlap of exosome components with chromatin-associated factors and marks indicative of active transcription. Strong enrichment between Rrp6 and Rrp40 is observed with factors such as RNA Pol II and H3K4me3 (Figure 4B). These results were not unexpected given previous work indicating physical interaction between exosome and transcription elongation factors (14). Based on this work as well as another study suggesting that the Rrp4 exosome component associates with gene bodies (12), we anticipated that exosome peaks would be observed at the middle and/or 3′-end of genes. In striking contrast, we found that the distribution of exosome peaks with respect to genes is heavily biased to the TSS and not gene bodies (Figures 4C and D). Plots of average Rrp6 or Rrp40 signal in S2 cells normalized to input within 1 kb of an annotated TSS reveal a strong enrichment of exosome components slightly upstream of TSSs, whereas little to no enrichment is observed in the vicinity of polyadenylation sites (Figure 4E). Similar TSS enrichment is also observed for exosome binding in S3 cells (data not shown).
Based on preferential binding to the TSS and the fact that the exosome is the major RNA degradation machinery, we hypothesized that exosome recruitment to chromatin may occur in a transcription-dependent manner. When exosome signal in S2 cells is examined at TSSs separated into transcriptionally active or inactive based on the presence of Pol II and H3K4me3, enrichment is observed only for active TSSs (Figure 5A). In order to explore the relationship of binding with gene expression level in more detail, matrices were generated for input normalized ChIP signal in 100 bins +/− 1 kb of annotated TSSs (Figure 5B). As expected, a clear bias toward expressed genes is observed for exosome ChIP signal at the TSS. Strong enrichment is seen for exosome at a set of promoters that display promoter-proximal enriched Pol II (PPEP, Figure 4B), suggesting that exosome preferentially associates with genes with paused Pol II. Taken together, we conclude that exosome chromatin association correlates with active transcription.
Overlap of exosome with insulator proteins at specific transcriptionally active sites
Given that exosome and certain insulator proteins are enriched at transcriptionally active regions and TSSs, we wanted to verify that enrichment of overlap observed throughout the genome is not due simply to a general preference for active regions. We first assessed the overlap of exosome with insulator proteins at genomic sites corresponding to active TSS, inactive/ambiguous TSS and non-TSS sites in S2 cells (Supplementary Figure S2, Supplementary Table S1). As expected, the majority of sites at which exosome and insulator proteins overlap corresponds to active TSSs. However, it is apparent that many active TSSs are not co-occupied by exosome and insulator proteins. Therefore, we calculated the statistical probabilities of two-way overlap between exosome and CP190, BEAF-32 or CTCF sites by hypergeometric tests considering only active TSSs of all annotated gene isoforms and indeed observed highly significant overlap over expectation (P < 3.7 × 10−110 for each comparison). We also determined that enrichment scores for two-way overlaps between exosome and insulator proteins are similar when considering only actively transcribed regions of the genome (Supplementary Figure S3) compared to when transcriptionally inert regions of the genome are also considered (Figure 4B). We conclude that colocalization of exosome and CP190, BEAF-32 and CTCF occurs at specific regions of the genome and is not solely due to active transcription status.
Exosome preferentially associates with divergently transcribed genes as well as specific functional gene classes similar to BEAF-32
Like BEAF-32, as well as CP190 and CTCF, exosome binding is enriched at TSS of divergently transcribed genes. Previous work showed that BEAF-32 preferentially associates with the promoters of ‘head-to-head’ or divergent gene pairs (28). Thus, we performed a series of Fisher’s Exact tests to examine whether exosome-bound and insulator-bound regions associate preferentially with divergent versus non-divergent TSSs. We confirmed the enrichment of BEAF-32 at divergent genes using different threshold distances and also observed enrichment for CP190, CTCF and exosome (Supplementary Figure S4). In contrast, Su(Hw) and Mod(mdg4)2.2 display minimal or negative enrichment for divergent promoters, respectively. Interestingly, we found that divergent genes display notably higher expression than non-divergent genes (Supplementary Figure S5). Indeed, Pol II is also highly enriched at divergent TSSs (Supplementary Figure S4), raising the possibility that enrichment at divergent promoters may be at least partially due to elevated transcriptional activity at these regions.
In an attempt to better understand the biological significance of exosome chromatin association, we performed gene ontology analysis to identify functional classes of genes enriched for exosome binding. DAVID analysis (57) on exosome binding sites reveals extensive overrepresentation of cell cycle and ribosomal protein genes in both S2 and S3 cells (Supplementary Table S2). Consistent with our finding that exosome colocalizes with BEAF-32 genome-wide, BEAF-32 has previously been shown to associate with cell cycle genes (29). Overall, these results demonstrate that the genome-wide exosome chromatin binding profile shares features with that of CP190, BEAF-32 and CTCF insulator proteins.
Lack of stabilized transcripts at exosome-bound regions
We wished to address whether chromatin regions bound by the exosome correspond directly to sites of active RNA degradation. A previous study in human cells revealed the stabilization of an ∼1.5 kb polyadenlyated divergent transcript in the opposite orientation at promoters of many genes (6). In order to determine whether a similar phenomenon occurs in Drosophila, we performed dsRNA knockdown of GFP as a control, as well as Rrp6 and Rrp40 in S3 cells, harvested total RNA, and performed rRNA depletion followed by non-directional RNA-seq. Efficient knockdown of exosome components was confirmed by western blotting (Supplementary Figure S6), although residual protein could be detected by ChIP (data not shown). Visual inspection of our RNA-seq data did not uncover obvious evidence of upstream divergent transcription in either control or exosome knockdown libraries (data not shown). RNA-seq libraries sequenced to a depth of over 20 M reads from two biological replicates each of control versus knockdown cell lines were analysed for differential expression specifically in the 500 bp intergenic region upstream of exosome-bound TSSs using the DESeq algorithm, and no statistically significant upregulation of these exosome-bound upstream regions was observed (Supplementary Tables S3 and S4).
As an alternate approach, we also examined distributions of genome-wide RNA fold changes upon exosome subunit knockdown at sites upstream of TSSs either bound or unbound by exosome. For exosome-bound upstream regions, an approximately equal number of sites show an increase or decrease of RNA-seq reads in Rrp6 or Rrp40 depleted cells compared with control cells (Supplementary Figure S7). In contrast, regions upstream of TSSs unbound by exosome actually display slightly increased median expression in Rrp6 depleted cells. Furthermore, no statistically significant changes in expression were detected at intergenic exosome peaks by DESeq analysis (Supplementary Tables S3 and S4). Similar results were obtained with oligo-dT selected RNA-seq libraries from either S2 or S3 cells (data not shown). Therefore, we conclude that exosome chromatin recruitment does not necessarily correspond to regions of transcript degradation, and we find no clear evidence for extensive divergent transcription even when the major cellular RNA degradation machinery is depleted.
Given that exosome is implicated in snoRNA maturation in yeast, we examined whether depletion of exosome subunits leads to stabilization of snoRNA or putative precursors. In Drosophila, the majority of snoRNAs are derived from introns of protein coding or non-coding genes (65). We used DESeq expression analysis to examine mature snoRNAs and intron regions harboring snoRNAs in rRNA depleted exosome knockdown versus control S3 libraries. We were unable to detect evidence for genome-wide stabilization of mature snoRNA or precursors (Supplementary Tables S3 and S4); however, we did observe accumulation of two likely snoRNA precursors with 3′ extensions when either Rrp6 or Rrp40 is knocked down (Supplementary Figure S8). Both of these snoRNA genes are bound by exosome, in addition to 61 of 249 annotated snoRNAs genome-wide considering either S2 or S3 cells. Two-way overlap between exosome and snoRNA genes is enriched over random expectation (Figure 4B), and interestingly, exosome preferentially associates with a class of chromatin-associated snoRNAs (ca-snoRNAs, 21 of 40) suggested to be involved in maintaining open chromatin structure (62). These results suggest that the Drosophila exosome machinery affects processing of at least a subset of snoRNA transcripts and might perform this function in association with chromatin.
Exosome binding at the TSS does not correlate with exosome-dependent transcript regulation genome-wide
We wondered whether exosome may be recruited to genes to enhance transcript degradation. In order to address this possibility, we performed mRNA expression profiling using control, Rrp6, and/or Rrp40 knockdown RNA-seq libraries from S2 and S3 cells using the DESeq algorithm, this time at the gene-level for annotated genes. Similar to a previous study (66), we identified hundreds of transcripts with altered expression profiles upon knockdown, the majority of which are stabilized upon exosome depletion (Supplementary Tables S5–S7). Strikingly, we found that transcripts that increase in exosome knockdowns are actually less likely to harbor an exosome peak at the TSS compared with transcripts unaffected by knockdown (Supplementary Table S8). Negative enrichment is likely due to the fact that exosome associates with higher expression genes whereas transcript stabilization is biased toward lower expression transcripts (Supplementary Figure S9). Hence, no obvious genome-wide correlation is apparent between exosome-dependent effects on gene expression and exosome chromatin association.
Curiously, depletion of exosome in S3 cells led to stabilization of an aberrant transcript at the Abd-B locus. The Abd-B gene is expressed in posterior cells later in embryonic development and is regulated by a host of downstream cis-regulatory elements including the CTCF-dependent boundary elements Mcp and Fab-8. Upon close visual inspection, we noted stabilization of an ∼20 kb polyadenylated transcript extending between downstream of the RE isoform promoter to beyond the RA isoform promoter particularly in Rrp6 but also in Rrp40 depleted cells (Supplementary Figure S10). We verified by directional RT-PCR that the transcript is transcribed in the same orientation as the Abd-B gene (data not shown), which is normally expressed in S3 but not S2 cells. Transcription in this region was not observed in exosome depleted S2 cells, suggesting that the transcript requires the same factors needed for normal Abd-B expression. We also observed evidence for expression of a similar transcript during the 4–10 h window of embryonic development based on modEncode oligo-dT selected RNA-seq data from whole embryos (Supplementary Figure S10). Aberrant expression of this nature was not observed extensively throughout the genome, and the functional or mechanistic significance of this stabilized transcript is not known.
Exosome subunits are essential in Drosophila
To date, specific mutations in exosome subunit genes have not been reported in Drosophila, precluding in vivo analyses of exosome function in the context of organismal development. We obtained a panel of UAS-inducible transgenic double-stranded RNA (dsRNA) hairpin lines directed against exosome subunit genes from the Vienna Drosophila RNAi Center and tested the effects of knockdowns using various Gal4-drivers on general development and adult viability. As expected, expression of RNAi hairpins against Dis3, Mtr3, Rrp6, Rrp42 or Ski6 using the strong ubiquitous Act5C::Gal4 driver results in adult lethality (Table 1), indicating that individual exosome subunits are essential for viability and are non-redundant. We additionally tested RNAi hairpins in limited subsets of tissue and observed necrosis of presumptive head tissue in pupae when driving with the GMR::Gal4 eye-restricted driver, although this phenotype was not fully penetrant or observed with all knockdown lines. Furthermore, driving of all hairpins using the vestigial wing margin enhancer vgM::Gal4 resulted in necrosis and severe wing blistering as well as loss or disrupted arrangement of scutellar bristles. Based on these results, we were unable to utilize exosome hairpin lines to assay insulator-dependent phenotypes in vivo. Nevertheless, these findings confirm the essential role of the exosome in general fly development, similar to the insulator proteins CP190 (26) and CTCF (22,23).
Table 1.
Summary of phenotypes observed for exosome RNAi hairpin lines using indicated Gal4 drivers
| VDRC stock no. | Gene | Act5C::Gal4 | vgM::Gal4 | GMR::Gal4 |
|---|---|---|---|---|
| 35 090 | Dis3 | Lethal | Blistered wings, scutellar bristles missing | 50% show necrosis of presumptive head tissue at pupal stage |
| 26 038 | Mtr3 | Lethal | Blistered wings, scutellar bristles missing | Unaffected |
| 35 199 | Rrp6 | Lethal | Blistered wings, scutellar bristles missing | 20% show necrosis of presumptive head tissue at pupal stage |
| 24 305 | Rrp42 | Lethal | Blistered wings, scutellar bristles missing | Not tested |
| 32 612 | Ski6 | Lethal | Blistered wings, scutellar bristles missing | Unaffected |
CTCF is required for exosome recruitment to certain sites including Fab-8
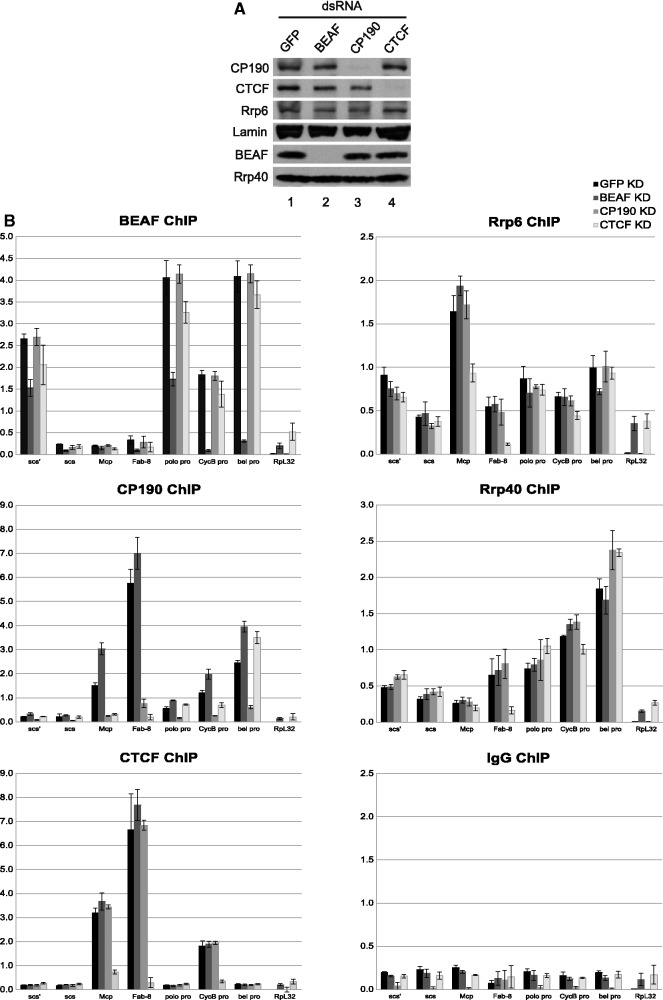
Given physical association between the exosome and CP190 insulator complexes as well as extensive genome-wide overlap, we next examined whether insulator proteins are required for exosome chromatin association. In order to deplete insulator proteins, S3 cells were transfected with dsRNAs to GFP as a control, BEAF-32, CP190 or CTCF. Western blotting verified highly efficient knockdown of insulator proteins but no change in levels of the exosome components Rrp6 or Rrp40 (Figure 6A). Directed ChIP followed by quantitative PCR was performed for a variety of well-characterized chromatin insulator sites, such as scs, scs′, Mcp and Fab-8 as well as several non-divergent cell cycle gene promoters strongly enriched for exosome binding. We also examined the highly transcribed RpL32 coding region, as a control site.
Figure 6.
CTCF is specifically required for exosome chromatin association at the Fab-8 insulator. (A) Western blotting of lysates from S3 cells transfected with GFP (lane 1), BEAF-32 (lane 2), CP190 (lane 3) or CTCF (lane 4) dsRNA. (B) S3 cells transfected with GFP (black), BEAF-32 (dark gray), CP190 (gray) or CTCF (light gray) dsRNA were subjected to ChIP using α-BEAF-32, α-CP190, α-CTCF, α-Rrp6, α-Rrp40 or control rabbit IgG as indicated. Percent input DNA immunoprecipitated is shown for each primer set, and error bars indicate standard deviation of quadruplicate PCR measurements. Similar results were obtained for independent biological replicates.
We first examined the chromatin association profiles of Rrp6 and Rrp40 compared with that of insulator proteins in control cells. In GFP-treated cells, Rrp6 associates with all regions except RpL32 substantially over the IgG control (Figure 6B). For Rrp40, signal is strongest at promoters, lower at Fab-8 and scs′, and essentially at background levels at scs and Mcp. The BEAF-32 insulator protein is also very highly enriched at these promoter regions as well as its characterized binding site scs′. We note that the scs′ insulator overlaps the aurora promoter, which is also part of a divergently transcribed gene pair. However, BEAF does not associate with scs, Mcp or Fab-8 insulators. In contrast, CP190 and CTCF insulator proteins preferentially associate with characterized binding sites Mcp and Fab-8 and to a lesser extent with certain promoters. Therefore, exosome subunit binding partially mirrors that of insulator proteins at these insulator and promoter sites.
As a control for knockdown efficiency, we next verified that BEAF-32, CP190 and CTCF knockdowns reduce chromatin association of the respective target protein at all binding sites. We thus examined interdependence of insulator proteins to their respective binding sites. BEAF-32 is not significantly affected by knockdown of either CP190 or CTCF at scs′ or scs insulator sites or at the polo, CycB or bel promoters (Figure 6B). Furthermore, CP190 chromatin association at these sites is unaffected by BEAF-32 depletion, consistent with a recent study showing independence of BEAF-32 and CP190 binding despite extensive genome-wide overlap (67). Confirming previous work, CP190 ChIP is reduced in CTCF depleted cells at the insulator sites Mcp and Fab-8, (25) but is mainly unaffected at the polo, CycB and bel promoters (Figure 6B). Similar to a previous study (67), CTCF binding is unaffected by either BEAF-32 or CP190 depletion at all sites tested including the Fab-8 insulator, which contrasts with dependence on CP190 previously identified in S2 cells (25). This result may be due to differences in cell type or expression state of the Abd-B locus. Interestingly, Rrp6 chromatin association is substantially reduced at Fab-8 and at Mcp and weakly reduced at the CycB promoter uniquely in CTCF depleted cells, whereas binding to all other loci tested remains constant under all knockdown conditions (Figure 6B). Similarly, Rrp40 ChIP signal is specifically reduced at Fab-8 in CTCF depleted cells.
In order to determine whether CTCF plays a genome-wide role in recruitment of the exosome to chromatin, we examined Rrp40 localization in polytene chromosomes of wildtype compared with CTCFy+2 mutants. This mutant produces little or no CTCF protein (22) and displays strongly reduced Abd-B looping interactions (25). We found no significant differences at this level of resolution (Figure 3), and Rrp6 was similarly unaffected (data not shown). These results indicate dependence on CTCF for exosome recruitment to the Mcp and Fab-8 insulators and possibly the CycB promoter or other untested sites. We were unable to assess whether insulator protein chromatin association is dependent on exosome components using the depletion strategy because sufficient levels of knockdown could not be achieved in order to fully remove Rrp6 or Rrp40 from chromatin (data not shown). Our results indicate that the exosome is at least partially dependent on CTCF for chromatin association.
DISCUSSION
Here, we present the first high-resolution map of exosome chromatin association in any organism. In contrast to expectations based on the prior literature, we find that exosome binds mainly promoters but not other genic regions of actively transcribed genes. Our results support the conclusion that exosome associates with chromatin in a transcription-dependent manner; however, exosome is not recruited to all actively transcribed genes. At the genome-wide level, exosome associates extensively with the CP190, BEAF-32 and CTCF insulator proteins. In support of this finding, our results uncover a previously unknown physical association between exosome and chromatin insulators. At the Mcp and Fab-8 insulators, exosome recruitment is reduced by CTCF depletion. However, at most other sites tested, insulator proteins are dispensable for exosome chromatin association. Our results provide new insights into potential functions of the nuclear exosome on chromatin.
Genome-wide profiling of exosome chromatin association
Our findings demonstrate that exosome associates physically with CP190 insulator complexes, and both exosome and specific insulator factors colocalize extensively throughout the genome. We found that exosome copurifies with CP190 insulator complexes isolated from embryonic nuclear extracts. Since CP190 is a component of multiple chromatin insulator complexes, we relied on parallel genome-wide colocalization approaches in a variety of cell types in order to ascertain the specificity of the interaction. Comparison of genome-wide profiles of exosome and insulator proteins on larval salivary gland polytene chromosomes revealed extensive overlap between exosome and the insulator proteins CP190 and BEAF-32, but low correspondence with CTCF, Su(Hw) and Mod(mdg4)2.2. Similarly, high-resolution ChIP-seq profiling in S2 cells confirmed the specific overlap of exosome with CP190 and BEAF-32 and was also able to detect significant overlap with CTCF that was not apparent in polytene chromosomes. Differential detection of the positive correlation between exosome and CTCF using the two techniques could either be due to differences in cell type or developmental stage examined, or alternatively, the higher resolution of the ChIP-seq method compared with the cytological approach. We analysed our ChIP-seq data in greater detail in order to take advantage of genome-wide profiling data of chromatin-associated factors in the same cell type compiled by other studies and the modENCODE consortium.
We found that exosome colocalizes with CP190, BEAF-32 and CTCF insulator proteins at a specific subset of active promoters. It is currently unclear how this specificity of recruitment is achieved. Previous work in Drosophila showed that exosome interacts physically with transcription elongation factors and colocalizes extensively with these factors on polytene chromosomes (14). Another study demonstrated that transfected epitope tagged Rrp4 and Rrp6 can crosslink to the 3′-end of several genes tested, in support of a cotranscriptional RNA surveillance model (12). However, our high-resolution ChIP-seq data clearly shows that exosome recruitment to genes occurs in sharp peaks preferentially at promoters compared with coding regions. Likewise, localized binding is also observed for CP190, BEAF-32 and CTCF to promoters.
Importantly, overlap between exosome and insulator proteins is also observed at well-characterized insulator sequences. Exosome ChIP signal is apparent at many known CP190, CTCF or BEAF-32-dependent insulator sites such as scs, scs′, Mcp and Fab-8, and these were further confirmed by directed ChIP. Despite extensive overlap with CP190, BEAF-32 and CTCF insulator proteins, we found that only CTCF is required for exosome recruitment to any sites examined in this study. Depletion of CTCF reduces exosome recruitment to both Mcp and Fab-8 whereas other insulator proteins are dispensable. Since reduction of CTCF levels reduces Abd-B expression (23,25), it is possible that reduced exosome chromatin association is a consequence of a change in transcription state in the general vicinity. Finally, considering that insulator proteins do not appear to be required for exosome association with other sites tested including co-occupied promoters, additional alternate mechanisms must exist to recruit exosome to chromatin.
Exosome chromatin association is dependent on active transcription
Our results suggest that exosome is recruited to chromatin in a transcription-dependent manner. Previous work showed that exosome is recruited to heat shock genes upon transcriptional induction (14). Likewise, we found that exosome associates preferentially with active over inactive promoters. These findings would be consistent with an RNA-based mechanism of recruitment to chromatin. However, specific association of exosome with the promoter and not gene bodies implies that exosome is not simply cotranscriptionally recruited to and stably associated with an elongating transcript since its ChIP signal would likely extend further 3′ into the gene as has been observed for a variety of RNA-binding proteins (68,69). If exosome is indeed recruited through direct RNA interaction, nascent RNAs may only be accessible to the exosome during a brief window, such as prior to capping, before assembly into protein-rich ribonucleoparticles. However, even if recruited to a transcript cotranscriptionally, exosome may not be able to load on the majority of transcripts for degradation since the 3′-end would not be accessible. Another possibility is that exosome specifically recognizes short RNAs associated with stalled Pol II. Finally, exosome recruitment could depend on assembly of the transcriptional apparatus but not the RNA itself. In this case, the full or partially formed exosome complex could be poised for degradation at promoters but not actively engaged.
Stabilization of transcripts as a result of exosome depletion
We were unable to obtain evidence for substantial transcript stabilization corresponding to regions to which exosome associates with chromatin. We did observe exosome at a subset of snoRNA genes, but we only observed a few cases of snoRNA precursor accumulation in exosome knockdowns. Given the strong enrichment of exosome at the TSS and previous reports that divergent transcription is stabilized at human and yeast promoters in exosome depleted cells (6,70,71), we looked for evidence of stabilized transcripts using both oligo-dT selected and rRNA depleted RNA-seq libraries of exosome knockdown cells. However, we found no evidence for divergent transcription upstream of annotated promoters regardless of exosome depletion, consistent with a previous study of uni-directional genome-wide transcription initiation in Drosophila (72). One possibility is that divergent upstream transcription does not occur in Drosophila and bidirectional transcription from a single promoter may be specific to certain species (73). Other purely technical possibilities are that our RNA-seq libraries were of insufficient depth or failed to capture aberrant transcription or altered stability since a conventional size selection step was used for sequencing, which in effect constrains the lower limit of captured RNA to 80 nt. Perhaps development and use of specialized cloning protocols could successfully identify aberrant transcripts or mild effects on transcript stabilization.
In order to test the possibility that exosome is recruited to chromatin in order to increase the efficiency of RNA surveillance, we also looked for evidence of stabilization of transcripts in exosome knockdown cells at genes at which exosome is associated at the promoter. We note that our knockdowns could not fully remove exosome from chromatin; therefore, sufficient depletion may not have been achieved in order to observe an effect on target RNAs. We did observe stabilization of many transcripts in exosome depleted cells; however these genes do not generally correspond to exosome binding sites. In fact, we observed a negative relationship between the two data sets likely because exosome tends to be recruited to more highly transcribed genes whereas the transcripts stabilized in exosome knockdown cells tend to be lowly expressed in wildtype cells. Although we cannot rule out that cotranscriptional recruitment is a mechanism to promote efficient RNA surveillance on a subset of genes, this does not appear to be a widespread or efficient mechanism. Another possibility is that recruitment of exosome to chromatin poises the degradation machinery for action under specific yet unknown conditions or to degrade transcripts synthesized in trans to binding sites.
Although not a focus of our study, our expression analysis in exosome subunit depleted cell lines revealed mainly stabilization of low expression transcripts. Our results generally confirm a previous study, which methodically depleted each individual exosome subunit in S2 cells and performed gene expression profiling by microarray (66). In that study, substantially varying expression profiles were obtained with individual subunit knockdowns, supporting a previously suggested model that the Drosophila exosome functions not as a singular degradation machine but as independently functioning, partially redundant subunits (36). A genome-wide expression profiling of conditional exosome mutants in Arabidopsis also detected limited subunit-specific effects on transcript levels (74). We did observe markedly different expression profiles between Rrp6 and Rrp40 knockdowns; however, this was likely due to arrest of cell proliferation in Rrp6 but not Rrp40 depleted cells. Our results are consistent with cell cycle arrest specifically observed in Rrp6 knockdown cells as previously reported (36), and these effects substantially confound our expression analyses. Intriguingly, we did find that both exosome subunits preferentially associate with promoters of cell cycle genes, suggesting that exosome could play a direct role in regulation of cell cycle gene transcript regulation. It would be interesting to examine exosome chromatin association at different stages of the cell cycle given that expression of many cell cycle regulators are tightly controlled and regulated in a cell cycle-dependent manner.
Functional relevance of exosome–insulator complex association
Our findings raise the possibility that exosome contributes to insulator activity or regulation, perhaps through an RNA-based mechanism. RNA may contribute to higher order insulator-dependent interactions and has been postulated to be an important component of the gypsy insulator complex in Drosophila (33,34) and the CTCF/cohesin insulator complex in mammals (75). Thus association of exosome with insulator complexes could regulate the abundance or activity of insulator-associated RNAs, which in turn affect RNA-mediated interactions between insulator complexes and factors capable of regulating insulator activity. A feasible scenario is that exosome regulates expression of the Abd-B locus through CTCF-dependent recruitment to the Mcp and Fab-8 insulators. Thus far, no specific RNA has yet been implicated in insulator activity at Abd-B. However, a complex array of intergenic transcripts arising from the cis-regulatory region of Abd-B has been shown to regulate expression of the downstream homeotic gene abd-A (76–78). Therefore, exosome surveillance and degradation activity may be particularly important at this complicated gene region. Isolation of conditional or loss-of-function exosome mutants should help elucidate the precise role of exosome in chromatin insulator activity.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–9 and Supplementary Figures 1–10.
FUNDING
Funding for open access charge: Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases [DK015602-05 to E.L.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank E. Andrulis for generously providing exosome antibodies and the Csl4-Flag cell line, V. Corces for α-BEAF-32 and α-CTCF antibodies, and E. Lai for fly lines. We also thank J. Zhu and members of the Lei laboratory for critical reading of the manuscript.
REFERENCES
- 1.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 2.Lorentzen E, Basquin J, Conti E. Structural organization of the RNA-degrading exosome. Curr. Opin. Struct. Biol. 2008;18:709–713. doi: 10.1016/j.sbi.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 4.Kiss DL, Andrulis ED. The exozyme model: a continuum of functionally distinct complexes. RNA. 2011;17:1–13. doi: 10.1261/rna.2364811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fasken MB, Corbett AH. Mechanisms of nuclear mRNA quality control. RNA Biol. 2009;6:237–241. doi: 10.4161/rna.6.3.8330. [DOI] [PubMed] [Google Scholar]
- 6.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 7.Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc. Natl Acad. Sci. USA. 2011;108:10460–10465. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3' processing of RNA polymerase II transcripts. Mol. Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 9.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 11.Hieronymus H, Yu MC, Silver PA. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 2004;18:2652–2662. doi: 10.1101/gad.1241204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hessle V, Bjork P, Sokolowski M, Gonzalez de Valdivia E, Silverstein R, Artemenko K, Tyagi A, Maddalo G, Ilag L, Helbig R, et al. The exosome associates cotranscriptionally with the nascent pre-mRNP through interactions with heterogeneous nuclear ribonucleoproteins. Mol. Biol. Cell. 2009;20:3459–3470. doi: 10.1091/mbc.E09-01-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hessle V, von Euler A, Gonzalez de Valdivia E, Visa N. Rrp6 is recruited to transcribed genes and accompanies the spliced mRNA to the nuclear pore. RNA. 2012;18:1466–1474. doi: 10.1261/rna.032045.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 15.Gurudatta BV, Corces VG. Chromatin insulators: lessons from the fly. Brief. Funct. Genomic. Proteomic. 2009;8:276–282. doi: 10.1093/bfgp/elp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda RK, Karch F. Making connections: boundaries and insulators in Drosophila. Curr. Opin. Genet. Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 19.Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- 21.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyrchanova O, Toshchakov S, Podstreshnaya Y, Parshikov A, Georgiev P. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol. Cell. Biol. 2008;28:4188–4195. doi: 10.1128/MCB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–1701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang N, Emberly E, Cuvier O, Hart CM. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell. Biol. 2009;29:3556–3568. doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emberly E, Blattes R, Schuettengruber B, Hennion M, Jiang N, Hart CM, Kas E, Cuvier O. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 2008;6:2896–2910. doi: 10.1371/journal.pbio.0060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rach EA, Winter DR, Benjamin AM, Corcoran DL, Ni T, Zhu J, Ohler U. Transcription initiation patterns indicate divergent strategies for gene regulation at the chromatin level. PLoS Genet. 2011;7:e1001274. doi: 10.1371/journal.pgen.1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 32.Ramos E, Torre EA, Bushey AM, Gurudatta BV, Corces VG. DNA topoisomerase II modulates insulator function in Drosophila. PLoS One. 2011;6:e16562. doi: 10.1371/journal.pone.0016562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat. Genet. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 34.Matzat LH, Dale RK, Moshkovich N, Lei EP. Tissue-specific regulation of chromatin insulator function. PLoS Genet. 2012;8:e1003069. doi: 10.1371/journal.pgen.1003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010;6:e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham AC, Kiss DL, Andrulis ED. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol. Biol. Cell. 2006;17:1399–1409. doi: 10.1091/mbc.E05-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 39.Stuurman N, Sasse B, Fisher PA. Intermediate filament protein polymerization: molecular analysis of Drosophila nuclear lamin head-to-tail binding. J. Struct. Biol. 1996;117:1–15. doi: 10.1006/jsbi.1996.0064. [DOI] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 2010;6:e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter C, Oktaba K, Steinmann J, Muller J, Knoblich JA. The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nat. Cell. Biol. 2011;13:1029–1039. doi: 10.1038/ncb2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 52.Lee C, Li X, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang C, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fagegaltier D, Bouge AL, Berry B, Poisot E, Sismeiro O, Coppee JY, Theodore L, Voinnet O, Antoniewski C. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc. Natl Acad. Sci. USA. 2009;106:21258–21263. doi: 10.1073/pnas.0809208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawarkar R, Sievers C, Paro R. Hsp90 globally targets paused RNA polymerase to regulate gene expression in response to environmental stimuli. Cell. 2012;149:807–818. doi: 10.1016/j.cell.2012.02.061. [DOI] [PubMed] [Google Scholar]
- 56.Fury W, Batliwalla F, Gregersen PK, Li W. Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006;1:5531–5534. doi: 10.1109/IEMBS.2006.260828. [DOI] [PubMed] [Google Scholar]
- 57.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 58.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 60.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schubert T, Pusch MC, Diermeier S, Benes V, Kremmer E, Imhof A, Langst G. Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Mol. Cell. 2012;48:434–444. doi: 10.1016/j.molcel.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 63.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 65.Huang ZP, Zhou H, He HL, Chen CL, Liang D, Qu LH. Genome-wide analyses of two families of snoRNA genes from Drosophila melanogaster, demonstrating the extensive utilization of introns for coding of snoRNAs. RNA. 2005;11:1303–1316. doi: 10.1261/rna.2380905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiss DL, Andrulis ED. Genome-wide analysis reveals distinct substrate specificities of Rrp6, Dis3, and core exosome subunits. RNA. 2010;16:781–791. doi: 10.1261/rna.1906710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwartz YB, Linder-Basso D, Kharchenko PV, Tolstorukov MY, Kim M, Li HB, Gorchakov AA, Minoda A, Shanower G, Alekseyenko AA, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012;22:2188–2198. doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 71.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, Ohler U, Zhu J. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat. Methods. 2010;7:521–527. doi: 10.1038/nmeth.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 75.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gummalla M, Maeda RK, Castro Alvarez JJ, Gyurkovics H, Singari S, Edwards KA, Karch F, Bender W. abd-A regulation by the iab-8 noncoding RNA. PLoS Genet. 2012;8:e1002720. doi: 10.1371/journal.pgen.1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bender W. MicroRNAs in the Drosophila bithorax complex. Genes Dev. 2008;22:14–19. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.