Abstract
Intensive research interest has focused on small RNA-processing machinery and the RNA-induced silencing complex (RISC), key cellular machines in RNAi pathways. However, the structural mechanism regarding RISC assembly, the primary step linking small RNA processing and RNA-mediated gene silencing, is largely unknown. Human RNA helicase A (DHX9) was reported to function as an RISC-loading factor, and such function is mediated mainly by its dsRNA-binding domains (dsRBDs). Here, we report the crystal structures of human RNA helicase A (RHA) dsRBD1 and dsRBD2 domains in complex with dsRNAs, respectively. Structural analysis not only reveals higher siRNA duplex-binding affinity displayed by dsRBD1, but also identifies a crystallographic dsRBD1 pair of physiological significance in cooperatively recognizing dsRNAs. Structural observations are further validated by isothermal titration calorimetric (ITC) assay. Moreover, co-immunoprecipitation (co-IP) assay coupled with mutagenesis demonstrated that both dsRBDs are required for RISC association, and such association is mediated by dsRNA. Hence, our structural and functional efforts have revealed a potential working model for siRNA recognition by RHA tandem dsRBDs, and together they provide direct structural insights into RISC assembly facilitated by RHA.
INTRODUCTION
RNA silencing refers to a conserved sequence-specific gene regulation mechanism mediated by small RNA molecules (1). RNA silencing plays a fundamental role in many important biological processes in eukaryotes, including host gene regulation and defense against invading foreign nucleic acids (2,3). The small RNA-mediated gene silencing process is mainly divided into two steps, carried out by two super RNA–protein complexes: small RNA-processing machinery and the RNA-induced silencing complex (RISC) (4). Small RNA-processing machinery dices the endogenous primary microRNA precursors or exogenous long double-stranded RNAs (dsRNA) into microRNAs or siRNAs of ∼21–27 nucleotides (5), whereas RISC is responsible for slicing or repressing the translation of the mRNA targets in a sequence-specific manner (6,7). Structural and functional data have been analysed to dissect the detailed small RNA-processing mechanisms by dicer-like proteins and their partner proteins (8–12). Similarly, the detailed molecular mechanisms of guide strand selection, passenger strand cleavage (13) and small RNA-guided mRNA target cleavage have been well visualized using high resolution structures of Argonaute proteins from different species (14–17).
However, limited structural data are available regarding the RISC assembly process, which is the essential step connecting small RNA processing and small RNA-mediated gene regulation. Although virtually all RISCs are assembled following cleavage of the passenger strand of the loaded small RNA duplex (18), the structural mechanisms of small RNA duplex loading are still largely unknown.
Many proteins in humans have been shown to associate with RISC (19,20). Among them, Argonaute2 (Ago2), Dicer and Trans-activation-responsive RNA-binding protein 2 (TRBP2) have been reported to participate in siRNA/miRNA loading (21,22). In addition, a number of other proteins, such as Dicer, protein activator of the interferon-induced protein (PACT), Moloney leukemia virus 10 (MOV10) and Importin-8 (IMP8), are involved in RISC assembly and RISC-mediated gene regulation enhancement (23–26).
Recently, RNA helicase A (RHA) was shown to interact with Ago2, TRBP and Dicer in human cells (27). RHA was identified as a novel component of Ago-containing RISC using co-IP approaches (28). Notably, RHA functions as a small RNA-loading factor involved in RISC assembly, indicated by the fact that RHA depletion in human cells reduced RISC formation. This evidence suggests that RHA functions in the RNA silencing pathway by promoting the formation of active RISC (27).
RHA is a DEIH-motif-containing helicase (also named as DHX9), which shares high sequence similarity to Drosophila maleless protein (MLE), a protein that regulates dosage compensation in Drosophila (29). RHA contains a catalytic helicase core (DEIH and Helicase domains) flanked by two tandem dsRNA-binding domains (dsRBDs) at its N-terminus and one RGG motif at its C-terminus (Figure 1A) (30). Interestingly, the two dsRBD domains are indispensable for interaction with RISC while the helicase core is not absolutely needed to facilitate the formation of active RISC in humans (27).
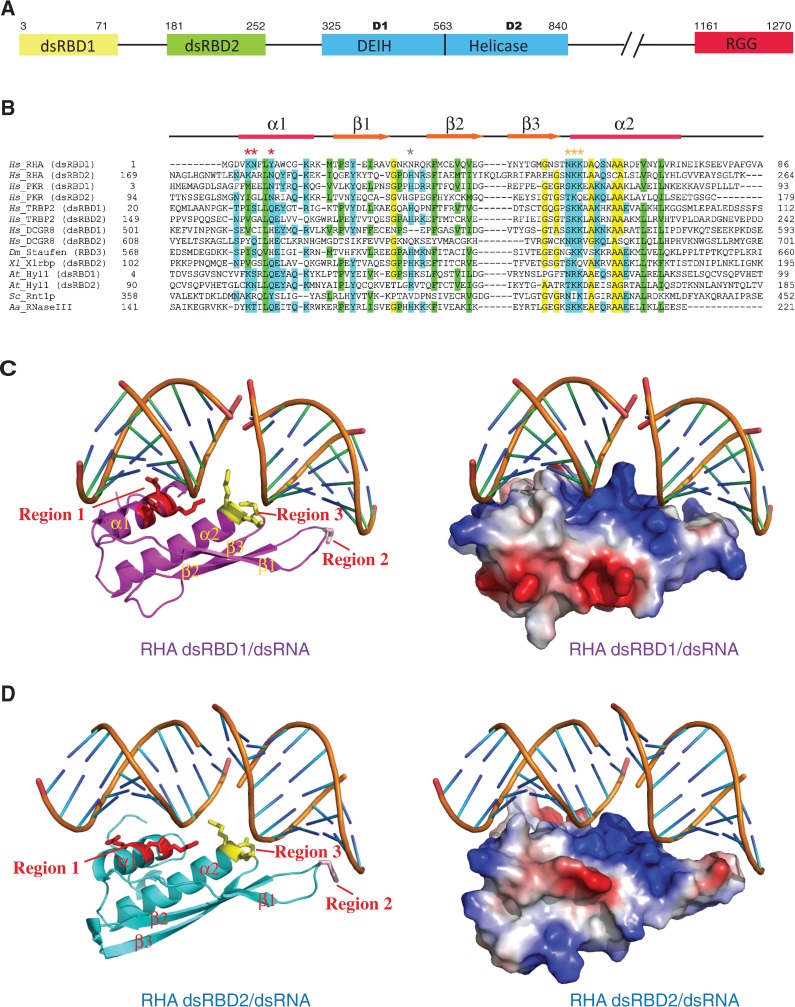
Figure 1.
Sequence alignment and overall structures of RHA dsRBD domains in complex with GC10 duplex. (A) Schematic diagram of human RNA Helicase A (DHX9). RHA comprises N-terminal tandem dsRBD domains, an ATPase-dependent helicase core (DEIH and Helicase domains or D1 and D2) and a C-terminal RGG motif. (B) Sequence alignment of RHA dsRBDs with various dsRBDs proteins from different organisms which are as follows: Hs stands for Homo sapiens, Dm stands for Drosophila melanogaster, Xl stands for Xenopus laevis, At stands for Arabidopsis thaliana, Sc stands for Saccharomyces cereviase and Aa stands for Aquifex aeolicus. Identical or similar residues are shaded in green, yellow or cyan. Asterisks indicate the critical residues involved in protein–RNA interactions in our structural complexes. The secondary structure elements are shown above the sequence. (C) Overall structure of RHA dsRBD1 in complex with GC10 duplex. (Left panel) Cartoon view of the complex structure with RHA dsRBD1 coloured in magenta and GC10 duplex coloured in orange. The critical residues belonging to region 1, 2 and 3 required for dsRNA recognition are shown in stick mode and coloured in red, wheat and yellow, respectively. (Right panel) Electrostatic surface view of the complex structure with the same view as that in the left panel. Blue, red and white colours represent positive, negative and non-polar patches on the protein, respectively. (D) Structure of RHA dsRBD2 in complex with GC10 duplex. (Left panel) Cartoon view of the complex structure with RHA dsRBD2 coloured in cyan and GC10 duplex in orange. The critical residues belonging to region 1, 2 and 3 required for dsRNA recognition are shown in stick mode and coloured in red, wheat and yellow, respectively. (Right panel) Electrostatic surface view of the complex structure with the same colour code as that in (C).
Although RHA is proposed to facilitate RISC assembly by virtue of RHA’s ability to bind siRNA duplex and its interactions with Ago2, TRBP and Dicer (27), the structural and functional features of RHA in dsRNA binding and RISC assembly are largely unknown. To gain the structural insights into siRNA duplex recognition and RISC assembly facilitated by RHA dsRBD domains, we determine the crystal structures of RHA dsRBD domains in complex with dsRNAs.
Structural comparison of RHA dsRBD1 with RHA dsRBD2 not only confirms the structural principles of dsRNA recognition by dsRBDs but also reveals structural insight into the higher siRNA duplex-binding affinity displayed by dsRBD1 compared with dsRBD2. This observation is further validated by an isothermal titration calorimetric assay. Moreover, by comparing our structure of a crystallographic RHA dsRBD1 pair in complex with dsRNA to the structure of Aquifex aeolicus RNaseIII-dsRBD in complex with dsRNA, we propose a potential working model for siRNA duplex recognition by tandem dsRBD domains. In addition, co-IP assays coupled with mutagenesis assays demonstrate that integrated tandem dsRBD domains with high dsRNA-binding affinity are required for RHA to interact with RISC and that such interaction is mediated by dsRNA. Hence, our structural and functional analysis of dsRNA recognition and RISC interaction via RHA dsRBD domains provides direct structural insight into RISC assembly facilitated by RHA.
MATERIALS AND METHODS
Protein expression and purification
The RHA dsRBD fragments were PCR amplified from a full-length RHA gene (Accession No.: NM_001357.4) purchased from Open Biosystems (Thermo Scientific; www.thermoscientific.com). The expression constructs were generated by insertion of the PCR products into pET SUMO expression vector (Invitrogen; www.invitrogen.com) with an N-terminal His-SUMO tag. Mutant alleles were prepared using QuickChange Site-Directed Mutagenesis Kit (Stratagene; www.genomics.agilent.com) and verified by sequencing.
Recombinant RHA dsRBDs and their mutants were expressed in Escherichia coli (strain BL21/DE3/RILP) overnight at 20°C, and protein expression was induced using 0.4 mM isopropyl β-d-thiogalactoside (IPTG). Cells were harvested by centrifugation, and the pellets were re-suspended and passed through a cell disruptor (www.avestin.com) five times. After ultracentrifugation at 40 000 rpm for 1 h, the supernatants were loaded onto Ni2+ affinity columns for purification. Pooled fractions were dialysed overnight against dialysis buffer [25 mM Tris (pH 7.4), 100 mM NaCl)] supplemented with Ulp1 proteinase for His-SUMO tag cleavage. The fractions were reloaded onto the Ni2+ affinity column to remove the His-SUMO tag. The flowthrough were then subjected to Heparin column (GE-healthcare) for further purification. The purified proteins were dialysed against the dialysis buffer again and concentrated to ∼15 mg/ml in a Centriprep-30 (Amicon) for subsequent crystallization. RNA oligoribonucleotides were purchased from Dharmacon (Thermo Scientific; www.thermoscientific.com).
Protein crystallization and construct optimization
The RNA oligos were directly added to the concentrated RHA proteins with a molar ratio of 1.1:1 and then incubated overnight before crystallization. The final protein concentrations in the mixed solution were approximately 10 mg/ml. Crystallizations were performed using the hanging drop vapor diffusion method at 20°C by mixing 1.0 μl of protein/RNA mixture with 1.0 μl of reservoir solution and equilibrating the mixture against 600 μl of reservoir solution.
Initial attempts to screen the crystallization conditions of RHA dsRBD fragments in complex with GC10 duplex (a self-complementary 10 bp RNA with sequence: 5′-GCGCGCGCGC-3′) by combining multiple-construct approach and incomplete sparse matrix approach failed. Then, limited proteolysis analyses of purified RHA dsRBD1 (residues: 1–130 aa) and dsRBD2 (residues: 131–318 aa) were conducted by varying concentrations of five different proteases (Trypsin, Chymotrypsin, Elastase, Subtilisin and Papain) at 37°C for 30 mins. The proteinase-resistant bands were identified by SDS-PAGE and submitted for Mass Spectrometric analysis.
The gene fragments corresponding to the proteinase-resistant peptides characterized by Mass Spectrometric analysis (RHA dsRBD1: residues 1–91 aa, RHA dsRBD2: residues 165–264 aa) were cloned, and the recombinant proteins were expressed and purified according to the protocol described above.
Crystals of RHA dsRBD1 (residues 1–91 aa) in complex with GC10 duplex were grown against crystallization buffer containing 13–17% PEG 8000, 0.2–0.3 M CaAc2 and 0.1 M Sodium Cacodylate (pH 6.5), whereas the crystals of RHA dsRBD2 (residues 165–264 aa) in complex with GC10 duplex were grown against crystallization buffer containing 15–17% PEG 4000, 0.6–0.8 M KNO3 and 0.1 M Tris (pH 7.0). These crystals grew to a maximum size of 0.3 mm × 0.1 mm × 0.1 mm over the course of 7 days.
Data collection and structure determination
Crystals were flash frozen (100 K) in the above reservoir solution supplemented with 30% glycerol. A total of 360 frames of a native data set with 1° oscillation at 1.075 Å wavelength were collected on each RHA dsRBD1/GC10 duplex crystal, whereas a total of 180 frames of a native data set with 1° oscillation at 0.9792 Å wavelength were collected on each RHA dsRBD2/GC10 duplex crystal. The data were processed by HKL2000 (www.hkl-xray.com). The structure of RHA dsRBD1 in complex with GC10 duplex (space group C2) was determined by molecular replacement using the NMR structure of the N-terminal dsRBD domain of hypothetical protein BAB28848 (PDBID: 1WHQ), and the structure of RHA dsRBD2 in complex with GC10 duplex (space group P43212) was determined by molecular replacement using the NMR structure of the second dsRBD domain of hypothetical protein BAB28848 (PDBID: 1UIL). The model was built by using the program O (http://xray.bmc.uu.se/alwyn) and refined using REFMAC/CCP4 (www.ccp4.ac.uk). The crystallographic statistic details of these structures are listed in Table 1.
Table 1.
Data collection, phasing and refinement statistics
| RHA dsRBD1 | RHA dsRBD2 | |
|---|---|---|
| Data collection | ||
| Synchrotron beamline | X29A, NSLS | U17, SSRF |
| Space group | C2 | P43212 |
| Cell dimensions | ||
| a, b, c (Å) | 52.252, 78.529, 112.242 | 63.198, 63.198, 107.617 |
| β (°) | 98.805 | 90 |
| Wavelength (Å) | 1.007 | 0.9792 |
| Resolution (Å)a | 50∼2.9 (2.95∼2.9) | 50∼2.3 (2.34∼2.3) |
| Rsym (%) | 8.6 (25.4) | 9.5 (29.9) |
| I/σ(I) | 27.7 (5.8) | 32.2 (9.2) |
| Completeness (%)a | 99.5 (94.7) | 100.0 (100.0) |
| Redundancy | 3.2 (3.7) | 13.8 (12.5) |
| Refinement | ||
| Resolution range (Å) | 50∼2.9 (3.0∼2.9) | 50∼2.3 (2.35∼2.3) |
| No. reflections | 9511 | 9800 |
| Rwork (Rfree) (%) | 21.9/26.2 (27.2/36.8) | 19.3 /26.3 (21.8/36.2) |
| No. atoms | ||
| Protein | 1356 | 765 |
| RNA | 1060 | 432 |
| Water | 82 | |
| B-factors (Å2) | ||
| Protein | 48.5 | 30.6 |
| RNA | 44.1 | 29.0 |
| Water | 36.1 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.011 | 0.015 |
| Bond angles (°) | 1.65 | 1.72 |
| % favoured (disallowed)in Ramachandran plot | 87.9 (0) | 94.0 (0) |
aValues for the highest-resolution shell are in parentheses.
Isothermal titration calorimetric (ITC) assay
All experiments were performed on the MicroCal VP-ITC calorimeter (Northampton, MA, USA) at 25°C. Protein concentrations were determined by spectrophotometric assay and dialysed against a dialysis buffer containing 12.5 mM Tris (pH 7.4) and 100 mM NaCl. Synthesized siRNA duplex (a self-complimentary 21 bp RNA with sequence: 5′-P-AGACAGCAUUAUGCUGUCUUU-3′) was dissolved in the dialysis buffer. Both protein (20 μM) and siRNA duplex (200 μM) were filtered and degassed before titration. Protein samples were loaded into the cell, and RNA samples were loaded into the syringe. Data were collected in the high feedback mode with a filter period of 2 s and a stirring speed of 310 r.p.m. The calorimetric data were processed and fitted into one-site binding model for both RHA dsRBD1 and dsRBD2 as well as their mutants, and two-site-binding model for RHA dsRBD1 + 2 using Microcal Origin (version 5.0).
Co-immunoprecipitation (Co-IP) assay
293FT cells were maintained under standard culture conditions in DMEM medium containing 10% FCS and 100 μg/ml Penicillin/Streptomycin. The pcDNA5-FRT-6 × Flag vector was used to make N-terminal 6 × Flag-tagged protein constructs, whereas pXJ40-myc vector was used to make N-terminal c-Myc-tagged protein constructs. Plasmids were co-transfected into 293FT cells using a linear polyethylenimine (PEI)-mediated approach (31). Cells were harvested after 48 hrs and lysed on ice using lysis buffer [50 mM Tris (pH 8.0), 100 mM NaCl, 10% glycerol, 0.2% NP-40 and Protease inhibitor cocktail (Cat# P8304)]. After centrifugation, the supernatants were incubated with pre-equilibrated 15 μl Anti-Flag Affinity Gel (Sigma, Cat# A2220) for 3 h. The beads were washed 5 times with washing buffer [50 mM Tris (pH 8.0), 100 mM NaCl, 10% glycerol, 0.1% NP-40], and the proteins were eluted using 60 μl 250 μg/ml 3 × FLAG peptides (Sigma, Cat# F4799). Both the input and anti-flag immunoprecipitants were loaded onto 15% SDS-PAGE gels and visualized by western blot using standard protocols. For RNase treatment assay, 4 μl of RNase was added into 100 μl of beads slurry during washing steps. The mixture was incubated in 37°C water bath for 30 min.
RESULTS
Overall structures of human RHA dsRBD domains in complex with dsRNA
As revealed by our in vitro pull-down assay, both individual RHA dsRBDs and tandem dsRBDs were capable of siRNA duplex binding (Supplementary Figure S3A). To investigate the structural principles of RNA duplex recognition by human RHA dsRBD domains, we systematically screened hundreds of crystallization conditions for various RHA fragments together with RNA duplexes of different lengths and sequences by combining a multi-construct approach (32) and limited proteolysis approach (33,34).
In the end, we were able to crystallize and determine the crystal structures of both RHA dsRBD1 (residues: 1–91) and RHA dsRBD2 (residues: 165–264) in complex with a self-complementary 10 bp RNA (GC10: 5′-GCGCGCGCGC-3′). The structure of RHA dsRBD1 was determined by molecular replacement using the NMR structure of the N-terminal dsRBD domain of hypothetical protein BAB28848 (PDBID: 1WHQ) as the search model. The structure of RHA dsRBD2 was determined using the NMR structure of the second dsRBD domain of hypothetical protein BAB28848 (PDBID: 1UIL) as the search model.
The RHA dsRBD1 in complex with GC10 duplex was crystallized in space group C2 and refined to 2.9 Å, whereas the RHA dsRBD2 in complex with GC10 duplex was crystallized in space group P43212 and refined to 2.3 Å. There are two RHA dsRBD1 domains, two GC10 duplexes and one single-stranded GC10 per asymmetric unit in the RHA dsRBD1/dsRNA structure. There are one RHA dsRBD2 and one GC10 duplex per asymmetric unit in the RHA dsRBD2/dsRNA structure. Notably, in the RHA dsRBD1/dsRNA structure, the two protein-bound GC10 duplexes stack end-to-end, and the single-stranded GC10 pairs with its symmetric-related single-stranded GC10 to form a GC10 duplex that stacks end-to-end to the two GC10 duplexes to form a continuous A-form-like helix. Similarly, in the RHA dsRBD2/dsRNA structure, the bound GC10 duplex stacks end-to-end to its symmetry-related GC10 duplex to form a continuous elongated A-form-like helix. The detailed crystallographic statistics are summarized in Table 1.
Similar to other dsRBDs, RHA dsRBD domains contain the core structure with ∼70 amino acids in length sharing a common α-β-β-β-α fold, where the two α helices lie on one surface of the three-stranded anti-parallel β sheets (35). However, RHA dsRBD1 has an additional 13 a.a. random coil structure at its C-terminus, whereas RHA dsRBD2 has an additional 12 a.a. random coil at its N-terminus and a 10 a.a. random coil at its C-terminus (Figure 1B).
Structural analysis of RHA dsRBD domains in complex with dsRNAs showed that the three conserved regions (36) in the α-β-β-β-α core structure are crucial for dsRNA binding (Figure 1C and D). In the RHA dsRBD1 structure, residues K5, N6 and Y9 at α1 interact with the minor groove, and residue N30 at the loop connecting β1 -β2 interacts with the successive minor groove, whereas residues N53 at the loop connecting β3-α2, and K54, K55 at α2 interact with the intervening major groove of the bound dsRNA duplex directly (Figure 2A and B, left panel). Similarly, in the RHA RBD2 structure, residues N186 and Q187 interact with the minor groove, and residue H207 interacts with the successive minor groove, whereas residues N234, K235 and K236 interact with the intervening major groove of the bound dsRNA duplex directly (Figure 2A and B, right panel). Notably, these residues are surface exposed and highly conserved in dsRBDs (Figure 1B). Consistent with our structural observation, the conserved lysine residues in RHA (K54, K55 in dsRBD1 and K236 in dsRBD2) were reported to be involved in selective recognition and translation of mRNA substrates (37).
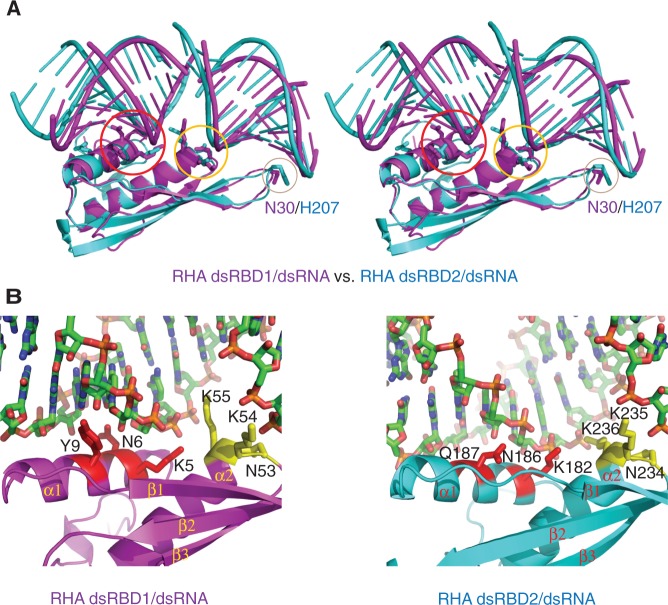
Figure 2.
Structural comparison of RHA dsRBDs in complex with dsRNAs. (A) Stereoview of structural superimposition of RHA dsRBD1/GC10 duplex (magenta) with RHA dsRBD2/GC10 duplex (cyan). The three protein–RNA interaction regions are highlighted in the circles coloured in red, yellow and wheat, respectively. The key residues (N30 in dsRBD1 and H207 in dsRBD2) involved in dsRNA binding in region 2 are shown in stick mode. (B) Close-view of the molecular details of dsRNA recognition by RHA dsRBDs. (Left panel) Structural details of dsRNA recognition by RHA dsRBD1. Residues involved in dsRNA recognition in region 1 and 3 are coloured in red and yellow, respectively. (Right panel) Structural details of dsRNA recognition by RHA dsRBD2. Residues involved in dsRNA recognition in region 1 and 3 are coloured in red and yellow, respectively.
To further validate whether these conserved residues in both RHA dsRBD domains are critical to siRNA duplex binding in vitro, we introduced alanine mutations into these residues and performed ITC assays to measure the binding affinities of individual RHA dsRBD domains and their mutants to a self-complimentary 21 bp siRNA duplex (siRNA: 5′-P-AGACAGCAUUAUGCUGUCUUU-3′). As expected, individual RHA dsRBD domains showed significant binding to siRNA duplex (Figure 4A and B, and Table 2–1, 6). Not surprisingly, introduction of a Y9A single mutation or N53A/K54A/K55A triple mutations into RHA dsRBD1 or introduction of N234A/K235A/K236A triple mutations into RHA dsRBD2 completely abolished siRNA duplex binding (Supplementary Figure S1B, D, H and Table 2–3, 5, 10). Similarly, introduction of K5A/N6A double mutations into RHA dsRBD1 or a K182A single mutation, N186A/Q187A double mutation or H207A single mutation into RHA dsRBD2 significantly reduced siRNA duplex binding (Supplementary Figure S1A, E–G and Table 2–2, 7, 8, 9). By contrast, introduction of an N30A single mutation into RHA dsRBD1 showed comparable siRNA duplex-binding affinity to that of wild-type RHA dsRBD1 (Supplementary Figure S1C and Table 2–4). Circular dichroism analysis of conformational differences between wild-type and these mutant proteins showed that the mutations did not perturb the overall structure of RHA dsRBD domains (Supplementary Figure S2).
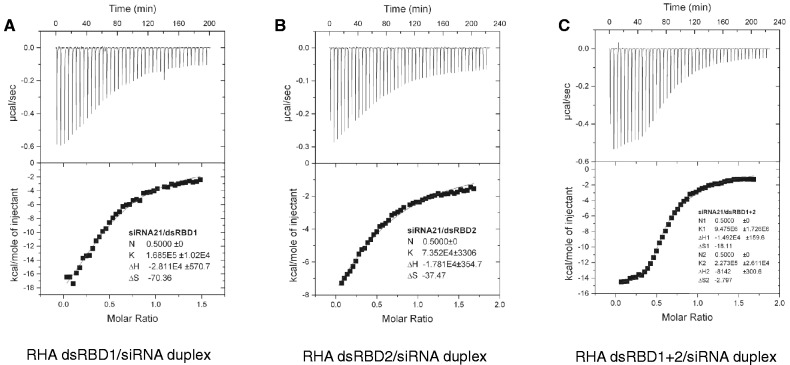
Figure 4.
ITC data of titrating siRNA duplex into dsRBD1 (A), dsRBD2 (B) and dsRBD1 + 2 (C). Raw data are shown in the upper panel, whereas fitted curves and binding parameters after integration are shown in the lower panel. Equilibrium constant K and stoichiometry factor N are shown within each profile. RHA dsRBD1 and RHA dsRBD2 were fitted into one-site binding model, whereas RHA dsRBD1 + 2 was fitted into two-site binding model.
Structural comparison of RHA dsRBD1 with RBD2
Comparison of crystal structures of RHA dsRBD1/dsRNA with dsRBD2/dsRNA showed that these two dsRBD domain structures are very similar with an r.m.s.d. value of 2.1 Å (Z score 10.9, 81 Cα atoms), although these two dsRBD domains only share 21% sequence similarity. In addition to the different orientations of N- and C-terminal random coils, the only other significant difference between these two dsRBD structures is at the loop region connecting β2-β3 (Figure 1B). Compared with the RHA dsRBD1 structure, the RHA dsRBD2 structure has a longer loop with a 4 a.a. insertion; however, this loop is located at the other side of the surface and is not involved in dsRNA binding (Figure 2A).
In contrast, comparison of the RHA dsRBDs/dsRNA complex structures demonstrated ∼5 Å movement and ∼15° rotation between the two bound pseudo-continuous dsRNA helices (Figure 2A). The pseudo-continuous dsRNA helices bound to the RHA dsRBD1 domain adopt a standard A-form dsRNA helix, whereas the pseudo-continuous dsRNA helices bound to the RHA dsRBD2 domain adopt a relatively elongated structure with broader minor and major grooves (Figure 2A). Notably, the dsRNAs used for crystallization of both RHA dsRBD1 and dsRBD2 are from the same batch with identical sequence. The significant structural deviation of the bound dsRNA suggests that the ‘induced-fit’ mechanism (38) may be applicable for dsRNA binding by dsRBD domains in general. Consistent with this notion, within the RHA dsRBD1 structure, the N-terminal half of helix α1 moves ∼5 Å towards the bound dsRNA to secure tight binding between RHA dsRBD1 and dsRNA (Figure 2A).
Strikingly, the 21% sequence similarity between the RHA dsRBD1 and dsRBD2 mainly reside on the α-helices (Figure 1B). In contrast, there is poor sequence homology among the β-sheets (Figure 1B). Hence, even though the overall structures of RHA dsRBD1 and dsRBD2 are almost identical at the α-β-β-β-α core motif, significant differences are observed in the electrostatic surface charges on both sides of the protein surfaces (Figure 1C right panel and D right panel). Notably, the side chain of the corresponding residue of K55 on RHA dsRBD1, which is protruding from the surface and is involved in dsRNA recognition by RHA dsRBD1, is rotated ∼70° away from the bound dsRNA in the RHA dsRBD2 structure (Figure 2B). Consistently, ITC data showed that the dissociation constant between RHA dsRBD1 and siRNA duplex was 5.93 × 10−6 M, whereas that of dsRBD2 with siRNA was 1.36 × 10−5 M (Table 2–1, 6 and Figure 4A and B), demonstrating that the binding affinity of RHA dsRBD1 to siRNA duplex is 2.3-fold higher than that of RHA dsRBD2 to siRNA. Interestingly, the higher binding affinity displayed by RHA dsRBD1 is mainly facilitated by basic residues from α1 and α2, while the residue N30 residing at the loop connecting β1-β2 has an insignificant contribution to binding affinity as revealed by ITC data (Supplementary Figure S1C and Table 2–4). In contrast, all the basic residues residing at regions 1, 2 and 3 in RHA dsRBD2 play a role in siRNA-duplex binding. Consistent with the ITC data, the structure of RHA dsRBD1 in complex with dsRNA showed that basic residues K5, N6, Y9 form a cluster to enable tight dsRNA binding (Figure 2B, left panel), whereas the structure of RHA dsRBD2 in complex with dsRNA showed that basic residues K182, N186, Q187 are spread out in helix α1 (Figure 2B right panel). This structural difference could be the reason for weaker binding affinity to siRNA duplex displayed by RHA dsRBD2 compared with RHA dsRBD1. The lack of significant overall sequence conservation in RHA dsRBDs suggests that the unique conformation and electrostatic distribution of each RHA dsRBD may determine selection of unique RNA or protein for interactions.
Structural model of siRNA duplex recognition by RHA RBD1+2 domains
RHA contains two tandem dsRBD domains at its N-terminus, which is a common feature of many RNA-binding proteins (36). A structural and functional analysis of human TRBP protein showed that both TRBP dsRBD1 and dsRBD2 domains exhibited dsRNA binding affinity; however, the full-length TRBP and its fragment containing dsRBD1 and dsRBD2 exhibited much stronger dsRNA binding affinity than individual domain alone (39). Similarly, both dsRBDs of human PKR protein are required for high binding affinity to TAR RNA (40). Strikingly, both dsRBDs of Arabidopsis HYL1 protein are required for high binding affinity to microRNA precursors, although HYL1 dsRBD2 only showed very weak dsRNA binding affinity (41). Nevertheless, structural and biochemical evidence demonstrated that the tandem dsRBD domains of these proteins simultaneously bind to siRNA duplex to achieve high affinity, whereas the flexible linker between these two dsRBD domains might wrap around the siRNA duplex to enhance the binding affinity (42).
Interestingly, structural analysis of RHA dsRBD1 in complex with dsRNA showed that both faces of the bound dsRNA helix are recognized by the RHA dsRBD1 domain, which suggests that the two RHA dsRBD domains may adopt a similar binding mode in order to recognize the bound siRNA duplex and achieve higher binding affinity, as was observed in the crystal structure of A. aeolicus RNaseIII-dsRBD complexed with dsRNA (38) (Figure 3A and B). This proposal of dsRBD1 and dsRBD2 may bind to the opposing faces of dsRNA was further validated by the co-IP experiment (Supplementary Figure S3B). In this case, the long flexible linker (∼110 a.a.) of RHA might wrap around the bound siRNA duplex. Remarkably, structural superimposition of these two structures showed striking structural similarities and positioning for both the bound dsRNA duplex and the dsRBD domains (Figure 3C). Our structural observation was consistent with the report that RHA dsRBD1+2 exhibit high binding affinity to poly(rI.rC)-agarose column by adopting a cooperative binding mode (30).
Figure 3.
Structural models of siRNA duplex recognition by RHA dsRBD1 + 2. (A) (Left panel) Cartoon view of crystal structure of RHA dsRBD1 in complex with dsRNA. In this structure, the dsRNA is recognized by a pair of RHA dsRBD1 from both sides. The dsRNA is shown in stick mode and coloured in orange, whereas two RHA dsRBD1 domains are shown in cartoon view. One copy of RHA dsRBD1 is coloured in magenta and the second copy is coloured in cyan. Our structure might represent a structural model of siRNA duplex recognition by RHA dsRBD1 + 2. In this model, siRNA duplex is recognized by RHA dsRBD1 and RHA dsRBD2 simultaneously from both sides, whereas the flexible linker may wrap around siRNA duplex. (Right panel) Cartoon view of the crystal structure of A. aeolicus RNaseIII-dsRBD in complex with dsRNA. In this structure, the dsRNA is recognized by a pair of dsRBD domains from both sides. The dsRNA is shown in stick mode, whereas two dsRBD domains are shown in cartoon view. (B) Electrostatic surface view of (A). (C) Structural alignment of RHA dsRBD1s/dsRNA complex with A. aeolicus dsRBDs/dsRNA complex. (Left panel) Superimposition of the structures by alignment of one dsRBD domain. Two RHA dsRBD1 domains and the bound dsRNA are coloured in magenta, whereas two A. aeolicus dsRBD domains and the bound dsRNA are coloured in cyan. (Right panel) Rotation of (left panel) along X-axis by 180°.
In an effort to validate the structural observation that RHA dsRBD1 + 2 recognizes the bound siRNA duplex in a cooperative manner, we performed the ITC assay to measure the binding affinity between siRNA duplex and dsRBD1 + 2, and compared it with that of individual dsRBDs with siRNA duplex. As expected, RHA dsRBD1 + 2 exhibited much stronger binding affinity to siRNA duplex compared with RHA dsRBD1 alone or RHA dsRBD2 alone (Figure 4 and Table 2–1, 6, 11). Our ITC experiments indicated that RHA dsRBD1 + 2 binds to siRNA duplex in a two-site binding mode. The fitted dissociation constants between RHA dsRBD1 + 2 and siRNA duplex demonstrated a tight binding site for dsRBD1 (Kd = 1.06 × 10−7 M) and a relatively weaker binding site for dsRBD2 (Kd = 4.40 × 10−6 M) (Table 2–1, 6, 11). Remarkably, the binding affinities between the siRNA duplex and each RHA dsRBDs are dramatically elevated when the two dsRBDs are fused together. Compared with the corresponding individual dsRBDs, the binding affinity of dsRBD1 as part of RHA dsRBD1 + 2 to siRNA duplex increased 56-fold from 5.93 × 10−6 to 1.06 × 10−7 M (Table 2–1, 11). Similarly, the binding affinity of dsRBD2 as part of RHA dsRBD1 + 2 to siRNA duplex increased 3-fold from 1.36 × 10−5 to 4.40 × 10−6 M (Table 2–6, 11). Therefore, we concluded that RHA dsRBD1 functions as the dominant binder to siRNA duplex, and RHA dsRBD1 and dsRBD2 work cooperatively to simultaneously bind both faces of the siRNA duplex to enhance binding affinity.
Molecular insights into RISC assembly mediated by RHA dsRBD domains
RHA was reported as an RISC-loading factor interacting with RISC in human cells. Importantly, N-terminal tandem dsRBD domains (dsRBD1 + 2) are critical for this function, as the RHA dsRBD deletion mutant, lacking the N-terminal 272 a.a., is not capable of co-IPing human RISC components (27).
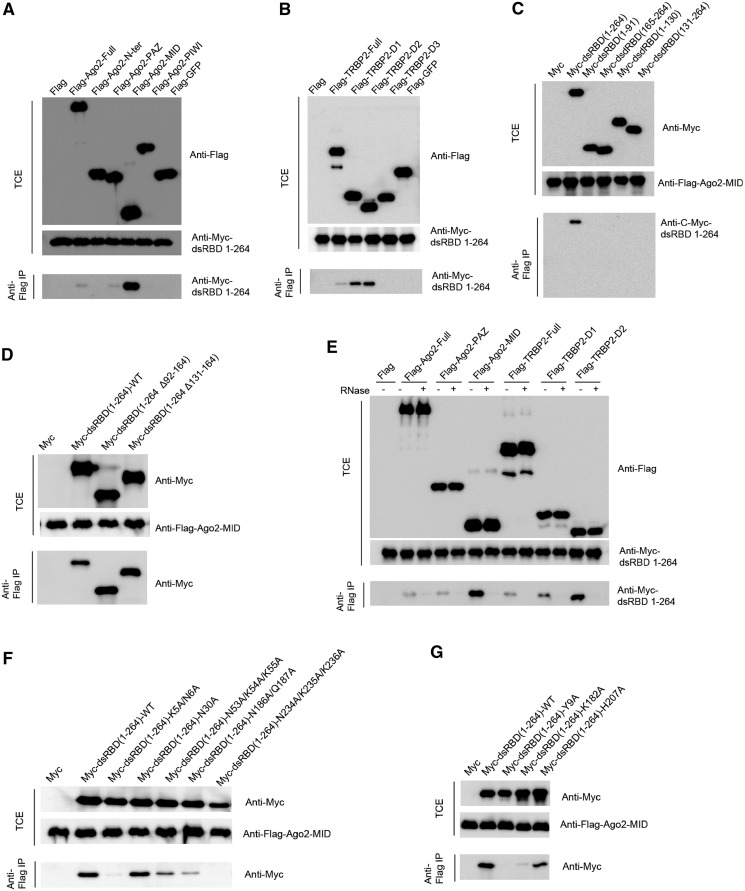
To investigate whether RHA dsRBD1 + 2 directly interacts with RISC components and what the exact functional roles of the tandem dsRBDs are in RISC assembly, we performed co-IP experiments to assay for interactions between RHA dsRBD1 + 2 and human RISC components. As expected, Myc-tagged RHA dsRBD1 + 2 (residues: 1–264) successfully co-IPed the Flag-tagged full-length Ago2 and TRBP2 when they were co-transfected into human 293FT cells (Figure 5A and B). A follow-up co-IP assay showed that Myc-tagged RHA dsRBD1 + 2 displays a strong association with the Ago2 MID domain and the first and second dsRBD domains of TRBP2. A weaker association was observed with the Ago2 PAZ domain, and no interaction was observed with the N-terminal and PIWI domains of Ago2 or the third dsRBD domain of TRBP2 (Figure 5A and B).
Figure 5.
Molecular insights into RISC assembly facilitated by RHA dsRBD domains. (A) Co-IP of RHA dsRBD1 + 2 (residues: 1–264) by Ago2 and its domains. RHA dsRBD1 + 2 displays weak interactions with full-length Ago2 and the Ago2 PAZ domain and a strong association with the Ago2 MID domain. The upper panel shows the expression of Flag-tagged Ago2 and its domains and the middle panel shows the loading control of Myc-tagged RHA dsRBD1 + 2. The lower panel shows RHA dsRBD1 + 2 co-IPed by Ago2 or its domains and detected by anti-myc antibody. Flag- only and Flag-GFP protein were used as negative controls. (B) Co-IP of RHA dsRBD1 + 2 (residues: 1–264) by TRBP2 and its domains. RHA dsRBD1 + 2 displays weak interaction with full-length TRBP2 and strong associations with TRBP2 dsRBD1 and dsRBD2. The upper panel shows the expression of Flag-tagged TRBP2 and its domains and the middle panel shows the loading control of Myc-tagged RHA dsRBD1 + 2. The lower panel shows RHA dsRBD1 + 2 co-IPed by TRBP2 or its domains and detected by anti-myc antibody. Flag only and Flag-GFP protein were used as negative controls. (C) Co-IP of RHA dsRBD1 + 2 (residues: 1–264) and individual dsRBDs by the Ago2 MID domain. Unlike RHA dsRBD1 + 2, RHA dsRBD1 (residues: 1–91 or residues: 1–131) or RHA dsRBD2 (residues: 165–264 or residues: 131–264) lack detectable interaction with the Ago2 MID domain. The upper panel shows the expression of Myc-tagged RHA dsRBD1 + 2 and individual dsRBDs, and the middle panel shows the loading control of Flag-tagged Ago2 MID domain. The lower panel shows RHA dsRBD1 + 2 and individual dsRBDs co-IPed by the Ago2 MID domain and detected by anti-myc antibody. Myc only was used as a negative control. (D) Co-IP of RHA dsRBD1 + 2 (residues: 1–264) and its mutants with loop deletion by the Ago2 MID domain. Both RHA dsRBD (1–264 Δ92–164) and RHA dsRBD (1–264 Δ131–164) display stronger interactions with the Ago2 MID domain comparable with wild type RHA dsRBD1 + 2. The upper panel shows the expression of Myc-tagged RHA dsRBD1 + 2 and its loop deletion mutants, and the middle panel shows the loading control of the Flag-tagged Ago2 MID domain. The lower panel shows RHA dsRBD1 + 2 and its loop deletion mutants co-IPed by the Ago2 MID domain and detected by anti-myc antibody. Myc only was used as a negative control. (E) Co-IP of RHA dsRBD1 + 2 (residues: 1–264) by Ago2, Ago2-PAZ, Ago2-MID, TRBP2, TRBP2-dsRBD1 and TRBP2-dsRBD2 after RNase treatment. RNase treatment completely abolished interactions between dsRBD1–264 and Ago2, Ago2-PAZ, Ago2-MID, TRBP2, TRBP2-dsRBD1 and TRBP2-dsRBD2. ‘+’ stands for RNase treatment that was introduced into the washing steps during co-IP, whereas ‘−’ stands for no RNase treatment. The upper panel shows the expression of Flag-tagged Ago2, Ago2-PAZ, Ago2-MID, TRBP2, TRBP2-dsRBD1 and TRBP2-dsRBD2 in both experimental and control samples, and the middle panel shows the loading control of Myc-tagged RHA dsRBD1 + 2. The lower panel shows RHA dsRBD1 + 2 co-IPed by Flag-tagged proteins and detected by anti-myc antibody. (F) Co-IP of RHA dsRBD1 + 2 (residues: 1–264) and its mutants by the Ago2 MID domain. Introduction of triple N234A/K235A/K236A mutations into RHA dsRBD1 + 2 (residues: 1–264) completely abolished its interaction with the Ago2 MID domain. Introduction of double K5A/N6A mutations, N186A/Q187A mutations or triple N53A/K54A/K55A mutations into dsRBD1 + 2 (residues: 1–264) substantially reduced their interactions with the Ago2 MID domain, whereas introduction of single N30A mutation into dsRBD1 + 2 (residues: 1–264) retained its binding strength. The upper panel shows the expression of Myc-tagged RHA dsRBD1 + 2 and its mutants as indicated, and the middle panel shows the loading control of the Flag-tagged Ago2 MID domain. The lower panel shows RHA dsRBD1 + 2 and its mutants co-IPed by the Ago2 MID domain and detected by anti-myc antibody. Myc only was used as a negative control. (G) Co-IP of RHA dsRBD1 + 2 (residues: 1–264) and additional mutants by the Ago2 MID domain as an extension of (F). Introduction of single Y9A mutation into RHA dsRBD1 + 2 (residues: 1–264) completely abolished its interaction with the Ago2 MID domain and introduction of single K182A or H207A mutation into dsRBD1 + 2 (residues: 1–264) severely reduced its interaction with the Ago2 MID domain. The upper panel shows the expression of Myc-tagged RHA dsRBD1 + 2 and its mutants, and the middle panel shows the loading control of the Flag-tagged Ago2 MID domain. The lower panel shows RHA dsRBD1 + 2 and its mutants co-IPed by the Ago2 MID domain and detected by anti-myc antibody. Myc only was used as a negative control.
Next, we tested whether both RHA dsRBD1 and dsRBD2 domains are needed for strong interactions with RISC components. We dissected RHA dsRBD1 + 2 into dsRBD1 and dsRBD2, and tested the interactions of these dsRBDs with the Ago2 MID domain, respectively. To our surprise, individual dsRBD domains did not show detectable interactions with the Ago2 MID domain (Figure 5C), suggesting that either the specific loop region connecting RHA dsRBDs or the higher dsRNA-binding affinity displayed by RHA dsRBD1 + 2 is important for Ago2 interaction.
To determine whether the flexible region is important for tight interaction between RHA dsRBD1 + 2 and Ago2, we made two loop deletion mutants, RHA dsRBD1 + 2 (Δ92–164) and dsRBD1 + 2 (Δ131–164), and repeated the co-IP experiments. Co-IP results showed that the RHA dsRBD1 + 2 loop deletion mutants display strong association with the Ago2 MID domain, which was comparable with that between wild-type RHA dsRBD1 + 2 and the Ago2 MID domain (Figure 5D). This observation suggests that these specific parts of the loop region do not favour Ago2 binding and underscores the fact that the higher dsRNA binding affinity displayed by RHA dsRBD1 + 2 is important for Ago2 interaction.
Finally, we asked whether the interactions between dsRBDs of RHA and RISC components depend on its dsRNA-binding activity. To this end, we performed RNase treatment assay during the washing steps in co-IP experiments. As expected, RNase treatment completely abolished the interactions between RHA dsRBD1 + 2 and Ago2 or TRBP2 (Figure 5E). This result demonstrates that the interactions between RHA dsRBD1 + 2 and RISC components depend on the dsRNA-binding activity of RHA dsRBD1 + 2.
To further validate that the interactions between RHA dsRBD1 + 2 and RISC components are dsRNA dependent, we mutated those critical residues for dsRNA binding (Table 2) and repeated co-IP assay. Remarkably, there are high correlations between the ITC data (Table 2) and the co-IP results (Figure 5F and G). Most of the alanine mutants showing no or extremely weak binding to siRNA duplex also demonstrated no or severely reduced association with the Ago2 MID domain, such as K5A/N6A, Y9A, K182A, N186A/Q187A, H207A and N234A/K235A/K236A mutants (Figure 5F and G). In contrast, the alanine mutant N30A that showed strong binding affinity to siRNA duplex demonstrated tight association with the Ago2 MID domain (Figure 5F). Interestingly, N53A/K54A/K55A mutants showing no binding affinity with siRNA duplex demonstrated only decreased association with the Ago2 MID domain (Figure 5F).
Table 2.
Binding of RHA dsRBDs and their mutants with a 21 bp small interfering RNA duplex by ITC
| siRNA21 titrated into RHA dsRBD1 WT and 4 mutants |
||||||
|---|---|---|---|---|---|---|
| N = 0.5 | 1 | 2 | 3 | 4 | 5 | |
| D1(WT) | D1(K5A/N6A) | D1(Y9A) | D1(N30A) | D1(N53A/K54A/K55A) | ||
| Kd (M) | (5.93 ± 0.36) × 10−6 | EWB | NB | (1.45 ± 0.08) × 10−5 | NB | |
| ΔH (cal/mol) | (−2.81 ± 0.057) × 104 | EWB | NB | (−4.23 ± 0.11) × 104 | NB | |
| ΔS (cal/mol/deg) | −70.36 | EWB | NB | −119.60 | NB | |
| siRNA21 titrated into RHA dsRBD2 WT and 4 mutants |
||||||
|---|---|---|---|---|---|---|
| N = 0.5 | 6 | 7 | 8 | 9 | 10 | |
| D2(WT) | D2(K182A) | D2(N186A/Q187A) | D2(H207A) | D2(N234A/K235A/K236A) | ||
| Kd (M) | (1.36 ± 0.06) × 10−5 | (3.34 ± 0.018) × 10−4 | EWB | (2.91 ± 0.24) × 10−4 | NB | |
| ΔH (cal/mol) | (−1.78 ± 0.035) × 104 | (−2.33 ± 0.11) × 105 | EWB | (−1.57 ± 0.12) × 105 | NB | |
| ΔS (cal/mol/deg) | −37.47 | −765.20 | EWB | −511.50 | NB | |
| siRNA21 titrated into RHA dsRBD1 + 2 WT |
||||||
|---|---|---|---|---|---|---|
| N1 = N2 = 0.5 | 11 | |||||
| 1st binding site | 2nd binding site | |||||
| Kd (M) | (1.06 ± 0.19) × 10−7 | Kd (M) | (4.40 ± 0.51) × 10−6 | |||
| ΔH (cal/mol) | (−1.49 ± 0.016) × 104 | ΔH (cal/mol) | (−8.14 ± 0.30) × 103 | |||
| ΔS (cal/mol/deg) | −18.11 | ΔS (cal/mol/deg) | −2.80 | |||
1–11 indicates the number of independent ITC experiments performed for different protein fragments.
D1 = dsRBD1 of RNA Helicase A; D2 = dsRBD2 of RNA Helicase A; WT = wild type; NB = no detectable binding; EWB = extremely weak binding; Kd = dissociation constants, which were manually calculated from K (the binding constants shown in the fitting curve panels) based on equation Kd = 1/K.
The estimated errors for K, ΔH were the sum of the squares of the deviations of the fitting curve from the experimental data.
Nevertheless, these results demonstrate that RHA N-terminal tandem dsRBDs are the primary functional domains for RISC interaction, which is mediated by siRNA duplex. The cooperative siRNA duplex binding mode by RHA may facilitate siRNA duplex transfer from another RISC component, such as TRBP, to Ago2 for incorporation.
DISCUSSION
In humans, the core components of the RISC are Dicer, Ago2 and TRBP proteins, which can be assembled in vitro to fulfil the dicing, Ago2-loading, guide-strand selection and slicing activities observed in endogenous RISC (43,44). Within the RISC, Dicer is the catalytic engine for microRNA processing, whereas Ago2 bound with a microRNA forms the minimal functional RISC, which tends to release from the holo-RISC once it is formed (45,46).
Although Dicer itself is sufficient to cleave, reposition, pre-select and handover small RNAs to Ago2 in vitro, other RISC components, such as TRBP, PACT and RHA, are required for stabilization of Dicer, recruitment of Ago2, repositioning of RNA duplexes, facilitation of small RNA production and loading small RNAs onto Ago2 (5,9,27,47–50).
RHA comprises several recognizable domains/motifs: N-terminal tandem dsRBD domains, an ATPase-dependent helicase core (DEIH and helicase domains) and a C-terminal RGG motif (30). Notably, all these three domains/motifs are observed widely among RNA-interactive proteins. Many dsRBD-containing proteins, including Dicer and its partners TRBP2 and PACT, Drosha and its partner DCGR8, participate in RNA silencing pathway. Among them, both TRBP2 and PACT contain three dsRBDs in tandem and show cooperation between their N-terminal tandem dsRBDs to achieve higher binding affinity to RNA duplex.
The N-terminal dsRBDs of RHA play important role in enabling RHA to interact with the essential components of different cellular processes, including transcriptional regulation (51), RNA processing (52) and RNA exporting (53). Moreover, RHA enhances HIV transcription in vivo by Trans-activation response element (TAR) binding via its N-terminal dsRBDs, and K236 was shown to be essential for TAR binding (54); RHA promotes DNA processing activity of Werner Syndrome Helicase (WRN) by directly interacting with WRN via its N-terminal dsRBD2 and C-terminal RGG box (55). In addition, RHA dsRBDs are necessary for selective recognition and translation of complex mRNAs and that conserved lysine residues (K54, K55 and K236) are necessary to engage structure features of its substrates (37). Taken together, N-terminal dsRBDs of RHA are multiple functional domains, which enable RHA to target both protein substrates and RNA substrates.
Our structural and functional efforts have demonstrated that the N-terminal tandem dsRBDs play the important role in siRNA duplex recognition and association with Ago2 for siRNA duplex loading. However, the biological functions of the ATP-dependent helicase core domains and RGG motif of RHA in RISC assembly are still unknown. Interestingly, the RHA helicase core was also suggested to be involved in human RISC assembly, as a K417R mutation on the RHA helicase core, which disrupts ATP binding and abolishes RNA helicase activity, led to negative impacts on RHA interactions with RISC (27).
Although the crystal structure of D1 (DEIH domain) of RHA helicase core provides the structural basis for unselective nucleotide base binding (56), the structural principles of siRNA duplex binding by RHA helicase core domains are still unknown. Notably, a recent crystal structure of yeast Mss116p helicase provides remarkable structural details on short dsRNA recognition and unwinding by RNA helicase in general. This structure reveals that D2 (helicase domain) of Mss116p helicase core recognizes short RNA duplex, whereas D1 (DEAD domain) of Mss116p helicase core promotes the unwinding of bound substrate where only a few RNA base pairs are separated (57).
Comparison of the Mss116p D2/dsRNA structure with our RHA dsRBD1/dsRNA structure through structural superimposition suggests that both the RHA helicase core domains and the RHA dsRBD1 + 2 domains are able to bind to the same short RNA duplex by bracketing from two opposite faces (Figure 6A). Interestingly, such a binding mode was observed in the crystal structure of A. aeolicus RNaseIII-dsRBD in complex with short RNA duplex. In this A. aeolicus structure, the bound short RNA duplex is sandwiched between two dsRBD domains on one side and two RNaseIII domains on the other side (Figure 6B) (38). We speculate that such a binding mode is applicable to siRNA duplex recognition by full-length RHA (Figure 6A). Therefore, as an essential human RISC component, RHA might play significant roles in transferring the siRNA duplexes processed from the Dicer–TRBP complex and facilitating siRNA loading onto Ago2 via interactions with siRNA duplex, TRBP and Ago2. During this process, RHA N-terminal tandem dsRBD1 + 2 might first bind to siRNA duplex simultaneously, trigger the conformational change of RHA, assist siRNA duplex binding via the helicase domain and finally facilitate local unwinding of the bound duplex for Ago2 loading. Consistent with this hypothesis, RHA N-terminal tandem dsRBD1 + 2 is shown to engage in binding with RNA first and then stimulate the ATP-dependent helicase activity necessary for mRNA translation (30). Moreover, the binding of PKR N-terminal tandem dsRBD1 + 2 with dsRNA has been shown to act as a modulator of its C-terminal kinase domain activation for association with other factors involved in RNA transporting (58).
Figure 6.
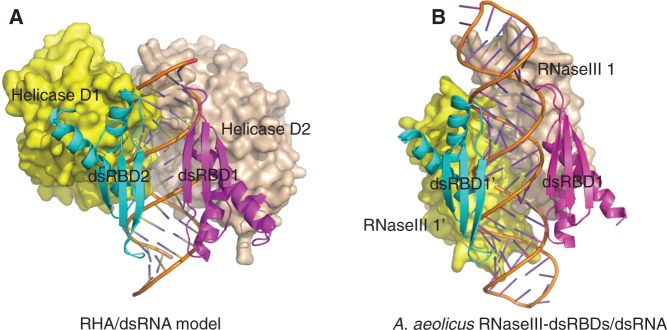
Structural models of siRNA duplex recognition by RHA. (A) Structural model of RHA full protein without C-terminal RGG motif in complex with siRNA duplex. In this model, the siRNA duplex is sandwiched between two dsRBDs in the front and D1 and D2 of RHA helicase core in the back without stereo clashes. The dsRBD domains are coloured in magenta and cyan respectively, whereas D1 and D2 are coloured in yellow and wheat, respectively. The siRNA duplex is coloured in orange. This model might represent a working model for siRNA duplex recognition and partial unwinding by the full-length RHA protein. (B) Crystal structure of A. aeolicus RNaseIII-dsRBD in complex with dsRNA (PDBID: 2EZ6).
Taken together, our structural and functional studies of dsRNA recognition and RISC interaction by RHA dsRBD domains have provided important information for better understanding how RHA facilitates RISC assembly.
ACCESSION NUMBERS
The coordinates have been deposited in the PDB under the accession codes 3VYY (RHA dsRBD1/GC10 duplex) and 3VYX (RHA dsRBD2/GC10 duplex).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3 and Supplementary Methods.
FUNDING
Funding for open access charge: Tier 2 research fund from Singapore Ministry of Education [MOE2010-T2-2-091 to Y.A.Y.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank J. He at Shanghai Synchrotron Radiation Facility (BL17U, SSRF) and H. Robinson at Brookhaven National Lab (X29A, NSLS) for their assistance in data collection.
REFERENCES
- 1.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 2.Plasterk RHA. RNA silencing: the genome's immune system. Sci. Signal. 2002;296:1263. doi: 10.1126/science.1072148. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 7.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 8.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Macrae IJ, Li F, Zhou K, Cande WZ, Doudna JA. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb. Symp. Quant. Biol. 2006;71:73–80. doi: 10.1101/sqb.2006.71.042. [DOI] [PubMed] [Google Scholar]
- 10.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 11.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Leuschner JF, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Sci. Signal. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 18.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 19.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA–protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakurai K, Amarzguioui M, Kim DH, Alluin J, Heale B, Song M, Gatignol A, Behlke MA, Rossi JJ. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Res. 2011;39:1510–1525. doi: 10.1093/nar/gkq846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Min JY, Krug RM, Sen GC. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 26.Weinmann L, Höck J, Ivacevic T, Ohrt T, Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 27.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuroda MI, Kernan MJ, Kreber R, Ganetzky B, Baker BS. The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell. 1991;66:935–947. doi: 10.1016/0092-8674(91)90439-6. [DOI] [PubMed] [Google Scholar]
- 30.Zhang S, Grosse F. Domain structure of human nuclear DNA helicase II (RNA helicase A) J. Biol. Chem. 1997;272:11487–11494. doi: 10.1074/jbc.272.17.11487. [DOI] [PubMed] [Google Scholar]
- 31.Tom R, Bisson L, Durocher Y. Transfection of HEK293-EBNA1 cells in suspension with linear PEI for production of recombinant proteins. Cold Spring Harb. Protoc. 2008;2008 doi: 10.1101/pdb.prot4977. pdb.prot4977. [DOI] [PubMed] [Google Scholar]
- 32.Graslund S, Sagemark J, Berglund H, Dahlgren LG, Flores A, Hammarstrom M, Johansson I, Kotenyova T, Nilsson M, Nordlund P, et al. The use of systematic N- and C-terminal deletions to promote production and structural studies of recombinant proteins. Protein Expr. Purif. 2008;58:210–221. doi: 10.1016/j.pep.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Dong A, Xu X, Edwards AM, Chang C, Chruszcz M, Cuff M, Cymborowski M, Di Leo R, Egorova O, Evdokimova E, et al. In situ proteolysis for protein crystallization and structure determination. Nat. Methods. 2007;4:1019–1021. doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wernimont A, Edwards A. In situ proteolysis to generate crystals for structure determination: an update. PLoS One. 2009;4:e5094. doi: 10.1371/journal.pone.0005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle M, Jantsch MF. New and old roles of the double-stranded RNA-binding domain. J. Struct. Biol. 2002;140:147–153. doi: 10.1016/s1047-8477(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 36.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell. Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 37.Ranji A, Shkriabai N, Kvaratskhelia M, Musier-Forsyth K, Boris-Lawrie K. Features of double-stranded RNA-binding domains of RNA helicase A are necessary for selective recognition and translation of complex mRNAs. J. Biol. Chem. 2011;286:5328–5337. doi: 10.1074/jbc.M110.176339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gan J, Tropea JE, Austin BP, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 39.Yamashita S, Nagata T, Kawazoe M, Takemoto C, Kigawa T, Guntert P, Kobayashi N, Terada T, Shirouzu M, Wakiyama M, et al. Structures of the first and second double-stranded RNA-binding domains of human TAR RNA-binding protein. Protein Sci. 2011;20:118–130. doi: 10.1002/pro.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim I, Liu CW, Puglisi JD. Specific recognition of HIV TAR RNA by the dsRNA binding domains (dsRBD1–dsRBD2) of PKR. J. Mol. Biol. 2006;358:430–442. doi: 10.1016/j.jmb.2006.01.099. [DOI] [PubMed] [Google Scholar]
- 41.Yang SW, Chen HY, Yang J, Machida S, Chua NH, Yuan YA. Structure of Arabidopsis HYPONASTIC LEAVES1 and Its Molecular Implications for miRNA processing. Structure. 2010;18:594–605. doi: 10.1016/j.str.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc. Natl Acad. Sci. USA. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 46.Lau PW, Potter CS, Carragher B, MacRae IJ. Structure of the human Dicer-TRBP complex by electron microscopy. Structure. 2009;17:1326–1332. doi: 10.1016/j.str.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kok KH, Ng MHJ, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J.Biol. Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 50.Noland CL, Ma E, Doudna JA. siRNA repositioning for guide strand selection by human Dicer complexes. Mol. Cell. 2011;43:110–121. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 52.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 53.Tang H, McDonald D, Middlesworth T, Hope TJ, Wong-Staal F. The carboxyl terminus of RNA helicase A contains a bidirectional nuclear transport domain. Mol. Cell. Biol. 1999;19:3540–3550. doi: 10.1128/mcb.19.5.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujii R, Okamoto M, Aratani S, Oishi T, Ohshima T, Taira K, Baba M, Fukamizu A, Nakajima T. A role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J. Biol. Chem. 2001;276:5445–5451. doi: 10.1074/jbc.M006892200. [DOI] [PubMed] [Google Scholar]
- 55.Friedemann J, Grosse F, Zhang S. Nuclear DNA helicase II (RNA helicase A) interacts with Werner syndrome helicase and stimulates its exonuclease activity. J. Biol. Chem. 2005;280:31303–31313. doi: 10.1074/jbc.M503882200. [DOI] [PubMed] [Google Scholar]
- 56.Schutz P, Wahlberg E, Karlberg T, Hammarstrom M, Collins R, Flores A, Schuler H. Crystal structure of human RNA helicase A (DHX9): structural basis for unselective nucleotide base binding in a DEAD-box variant protein. J.Mol. Biol. 2010;400:768–782. doi: 10.1016/j.jmb.2010.05.046. [DOI] [PubMed] [Google Scholar]
- 57.Mallam AL, Del Campo M, Gilman B, Sidote DJ, Lambowitz AM. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature. 2012;490:121–125. doi: 10.1038/nature11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waechter C, Pelech S, Flynn A, Proud C, FEBS L, Frederickson R, Montine K, Sonenberg N, Rinker-Schaeffer C, Rhoads R. Role of the double-stranded RNA-activated protein kinase (PKR) in cell regulation. Biochem. Soc. Trans. 1997;25:509–513. doi: 10.1042/bst0250509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.