Abstract
The TATA binding protein (TBP) is an essential transcription initiation factor in Archaea and Eucarya. Bacteria lack TBP, and instead use sigma factors for transcription initiation. TBP has a symmetric structure comprising two repeated TBP domains. Using sequence, structural and phylogenetic analyses, we examine the distribution and evolutionary history of the TBP domain, a member of the helix-grip fold family. Our analyses reveal a broader distribution than for TBP, with TBP-domains being present across all three domains of life. In contrast to TBP, all other characterized examples of the TBP domain are present as single copies, primarily within multidomain proteins. The presence of the TBP domain in the ubiquitous DNA glycosylases suggests that this fold traces back to the ancestor of all three domains of life. The TBP domain is also found in RNase HIII, and phylogenetic analyses show that RNase HIII has evolved from bacterial RNase HII via TBP-domain fusion. Finally, our comparative genomic screens confirm and extend earlier reports of proteins consisting of a single TBP domain among some Archaea. These monopartite TBP-domain proteins suggest that this domain is functional in its own right, and that the TBP domain could have first evolved as an independent protein, which was later recruited in different contexts.
INTRODUCTION
The architecture of multisubunit RNA polymerases (RNAPs) in the three domains of life is surprisingly well conserved (1,2), and evolutionary analyses indicate that the common RNAP core evolved before the divergence of Bacteria, Archaea and Eucarya (3–5). In contrast, a number of RNAP subunits and associated transcription factors are conserved between Eucarya and Archaea to the exclusion of Bacteria (5–7). One such case is evident in transcription initiation, where promoter-directed transcription of all classes of RNAPs in Eucarya and Archaea is dependent on the TATA-binding protein (TBP) (7–9). In addition to the canonical TBP factor, the two closely related eukaryotic TBP paralogues TBP-related factor 2 and 3 are involved in gene expression during development and differentiation in some metazoans [reviewed in (10)].
TBP is absent from bacteria, and bacterial RNAPs instead initiate transcription using the evolutionarily unrelated sigma factors (11). This has led to the suggestion that TBPs and sigma factors emerged independently in the archaeal/eukaryotic and bacterial lineages, respectively. Consequently, the RNAP of the Last Universal Common Ancestor (LUCA) may have initiated transcription in a factor-independent manner (7).
A related question, and the focus of this work, is the origin of the TBP domain itself. TBP consists of a highly conserved core comprising two symmetric repeats (TBP domains) and an N-terminal extension that is less well conserved and whose function remains poorly understood (12,13). Even though the bipartite symmetry of TBP suggests that it was generated by duplication, the origin of the TBP domain and of TBP protein remains uncertain.
The TBP domain consists of a five-stranded anti-parallel β-sheet, which binds the target DNA, and two α-helices that cover the opposite surface of the sheet (14,15). As summarized in Table 1, the TBP domain has been found in a number of protein families, notably RNase HIII and DNA glycosylases, though sequence similarity between these is low. Although these proteins bind nucleic acids, functional and structural studies do not implicate the DNA glycosylase TBP domain in DNA binding, in contrast to RNase HIII and TBP. Given the small size of the TBP domains, differing DNA-binding capacity and low sequence similarity between them, one could question whether these domains are homologous. Indeed, the TBP domain has been identified as a member of a broad fold group, dubbed the helix-grip fold, members of which vary in the number of strands and helices (16,17). Within the broader context of homology versus convergence for protein folds (18,19), we were therefore interested in examining whether the aforementioned examples of TBP domains could be established as definitively having evolved from a common ancestor and exploring the evolutionary scenarios giving rise to the different protein contexts in which it is found.
Table 1.
Distribution and features of key TBP-domain proteinsa
| Protein | Context | DNA binding | Distribution | Pfam family | Pfam clan |
|---|---|---|---|---|---|
| TBP | Bipartite | Yes | AE | PF00352 | CL0407 |
| RNase HIII | N-term | (Yes) | BAb | PF11858 | - |
| DNA glycosylases | N-term | No | BAE | PF07934 | CL0407 |
| PF06029 |
aData derived from Pfam. Other proteins carrying TBP-domain proteins have been reported, as discussed in the text.
bArchaeal distribution investigated in this study.
Among the cases noted in Table 1, DNA glycosylases are notable for their broad distribution, spanning all three domains of life. This is a group of homologous enzymes that recognize and remove damaged bases in DNA by flipping out the base and hydrolyzing the bond between deoxyribose and base. The resulting abasic lesion is subsequently repaired by the base excision repair pathway (20).
Members of the RNase H family are ribonucleases that have a substrate specificity for RNA–DNA hybrids and play an important role during DNA replication (21). Unlike RNAPs that can initiate RNA synthesis using nucleotide triphosphates, DNA polymerases (DNAPs) require a primer to initiate DNA synthesis (22). During replication, primases provide these short RNA primers that subsequently are removed from the de novo synthesized DNA strand by ribonucleolysis, which is facilitated by members of the RNase H family (21). There are three families of RNase H termed type I, II and III, but although their substrate specificities have been characterized, and vary considerably, the biological relevance of the subtypes remains uncertain (21).
In contrast to TBPs and DNA glycosylases, and the ubiquitously distributed RNase HII family, RNase HIII has a comparatively narrow phylogenetic distribution, being sparsely scattered across bacteria, with only a few potential archaeal homologues annotated in sequenced genomes. What then is the origin of the TBP domain in RNase HIII?
To improve our understanding of the evolutionary origin of the TBP domain, we examined a combination of sequence, phylogenetic and structural data. Our analyses indicate that the TBP domains across a range of proteins are homologous, and that the TBP domain was a likely constituent of the LUCA. Two possible evolutionary scenarios are presented for the subsequent distribution of the TBP domain. One scenario proposes the initial presence of a monopartite TBP domain in LUCA that was later co-opted into new roles, with domain duplication leading to the emergence of TBP, and fusion of a monopartite TBP-domain protein with other domains giving rise to RNase HIII and DNA glycosylase. In the alternative scenario, the TBP domain originated from within DNA glycosylase, evolving into a modular domain through truncation, this modular domain later giving rise to TBP via duplication and fusion, and to RNase HIII through fusion with RNase HII.
Although the primary focus of this work was to establish whether an evolutionary relationship existed between the TBP domains in TBP protein, DNA glycosylase and RNase HIII, our searches also revealed other proteins containing the TBP domain, some of which are thought to bind DNA, lending some weight to the hypothesis that this domain may have served a DNA binding role in LUCA.
MATERIALS AND METHODS
Assessing remote homologies using sequence analysis
To evaluate remote homologies between the TBP domains in TBP, DNA glycosylase and RNase HIII, we searched existing domain family classifications—SCOP (23), CATH-Gene3D (24) and Pfam (25). We also used sensitive sequence-based search methods, i.e. the FFAS server (26) and the HHpred server (27), to detect very remote homologies not captured in SCOP, CATH or Pfam. Structural comparisons were also performed and are described later in the text.
SCOP, CATH and Gene3D
The CATH (24) and SCOP (23) databases classify homologous protein domain structures in superfamilies on the basis of structural, sequence and functional similarities.
Gene3D is a sister resource of CATH containing sequence relatives for each domain structure superfamily. Hidden Markov Models (HMMs) are built using HMMer for representative sequences from each CATH domain structure family and used to scan UniProt and ENSEMBL to identify sequence relatives. We searched SCOP, CATH and Gene3D for all protein sequences containing TBP domains.
We also performed HMM–HMM comparisons using PRC (28) for all HMMs from domain superfamilies within the CATH helix-grip fold, i.e. the fold group containing the TBP domain superfamily. This identified other putative homologous relationships between all superfamilies in the helix group fold. We considered that all matches with e-values <0.01 indicated putative homologies. As the fold in total is represented by 20 of the 11 330 models in the CATH HMM library, we regarded the relatively high e-value of 0.01 as significantly indicating homology.
Pfam analysis
Pfam groups clearly homologous protein domains in families and uses pairwise comparisons of profile HMMs derived from alignments of family members to group individual families into broader clans (25). In Pfam clans, e-values for pairwise relationships are calculated based on scoring a given pHMM against the complete library of pHMMs, using Profile Comparer (PRC) (29). Pfam analyses were performed on the Pfam server (http://pfam.sanger.ac.uk/). Information on the TBP-like clan, including precomputed e-values for clan relationships, was retrieved from Pfam. Pairwise HMM logos showing relationships between Pfam families (Supplementary Figures S1–S3) were visualized using LogoMat-P (http://www.sanger.ac.uk/cgi-bin/software/analysis/logomat-p.cgi) (30).
FFAS scan
The Fold and Functional Assignment System (FFAS) takes a query sequence as input, builds a profile of that sequence and a set of close homologues found using PSI-BLAST (31) and then uses that profile to compare against a pre-built library of profiles (26). Profile–profile alignment scores are compared against a distribution of scores obtained between pairs of unrelated sequences to produce the final FFAS score. A FFAS score below −9.5 is usually indicative of a significant similarity between the two profiles. The input sequences we used for FFAS searches were the same as those used for structural comparisons (see later in the text), i.e. the N-terminal TBP domain of Sulfolobus acidocaldarius TBP, the TBP domain of Bacillus stearothermophilus RNase HIII and the TBP domain of Escherichia coli 3-methyladenine DNA glycosylase. All FFAS scans were performed against a library of profiles derived from Pfam families.
HHpred scan
HHpred compares a profile HMM derived from an input sequence with profile HMMs derived from a number of alignment databases (27). In this work, we searched all source alignment databases provided by HHpred. HHpred reports the probability ranging from 0 to 100 for any putative homologous relationship it detects. Here, only hits with a probability ≥95.0 are considered. The input data used for HHpred searches were the multiple sequence alignments of Pfam family seed members for each TBP domain.
Structural comparisons
Comparisons between structural representatives of the different TBP domains were performed using the Sequential Structure Alignment Program, (SSAP) (32). For any pair of structures being superimposed, SSAP produces a structural similarity score that can range from 0 to a maximum of 100 for identical structures. SSAP scores above 80 are usually associated with highly similar structures (32).
For each SSAP score, an associated Z-score was computed based on the distribution of all SSAP scores between one of the domains being compared and all non-redundant domains in CATH at the 95% sequence identity level. The Z-score helps in evaluating whether an SSAP score obtained between two domains is likely to be due to chance structural similarity.
The boundaries of TBP domains used in this study were defined manually from PDB entries. The following boundaries were used: for TBP, we used residues 15–97 of chain A of PDB structure 1mp9; for DNA glycosylase, we used residues 1–84 of chain A of PDB structure 1 mpg; finally, for the N-terminal domain of RNase HIII, we considered residues 2–67 of chain A in PDB structure 2d0a.
Structural prediction
Structural predictions of the putatively single TBP-domain proteins were performed using the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/) (33).
Phylogenetic analyses
All multiple sequence alignments were performed with the Kalign software version 2.04 (34) and default settings. Alignments were subsequently manually inspected for alignment errors.
Bayesian trees were inferred using PhyloBayes 3.2e (35) using two parallel chains with the CAT + Gamma + I model (36) until sufficient convergence was achieved, corresponding to ∼10 million generations with sampling every 100 generations and 2.5 million generations discarded as burn-in.
Cluster analyses
Cluster analysis of TBP domain-containing proteins was performed using CLANS v. 2 (37). A data set comprising the full complement of sequences as of 2011-11-07 for the Alka-N, OGG_N and TBP families was downloaded from PFAM (http://pfam.sanger.ac.uk/) (25). A representative selection of RNase HIII TBP-domain sequences and putative single TBP-domain proteins were then added to that data set. Accession information for the sequences present in the analysis is given in Supplementary Table S1. PSI-BLAST as implemented in the CLANS software was allowed to run for 10 000 cycles followed by identification of clusters by convex clustering with default values and 100 jack-knife replicates.
Analyses to reveal the evolutionary ancestry of specific proteins
In addition to the generic sequence and structure-based protocols outlined earlier in the text to determine the relationships between the TBP domains in TBP protein, DNA glycosylase and RNase HIII, further, more specific analyses (described later in the text) were performed (1) to determine whether the TBP domain existed as a single-domain protein and (2) to reveal the evolutionary ancestry of the RNase HIII protein.
Search for monopartite TBP-domain proteins
Several complementary approaches were used to find additional proteins consisting of a single TBP domain, over and above those identified previously (16,17,38).
Search of Gene3D
We searched for occurrence of possible monopartite TBP domains in CATH-Gene3D. As mentioned earlier in the text, CATH-Gene3D provides CATH domain predictions for all proteins in the major sequence databases (39,40). In CATH, TBP domains from TBP are classified in superfamily 3.30.310.10, whereas domain structures from DNA glycosylases are classified in superfamilies 3.30.310.20 and 3.30.310.40. TBP domains from RNase HIII are classified in 3.30.310.10, together with those from TBP. We therefore scanned Gene3D v11.0.0 to retrieve sequences with one domain from any of these three superfamilies and no other domain.
This scan was performed programmatically using the Gene3D web services (41). Fragments were then removed from the list of hits. We also excluded any protein with a sequence length of >150 residues, as we assumed such a length would suggest the presence of another, yet undetected domain.
BLAST search
We retrieved several candidates consisting of a monopartite TBP domain from the full alignment of the Pfam TBP-like clan. Genomic context confirmed these were not truncated or misannotated proteins. Specifically, we performed six-frame translations, ran blastp and searched for evidence of larger orfs using blastx against Genbank’s non-redundant database).
HMMer search
An additional search was performed using a more sensitive method [HMMer 3.0 (42)]. MAFFT (43) was used to align the candidate monopartite TBP-domain sequences identified with the alternative searches mentioned earlier in the text. A profile HMM was derived from this multiple sequence alignment using HMMer, and was used to scan a non-redundant sequence database consisting of Ensembl (44), RefSeq (45) and Uniprot (46). The scan was iterated until no new homologues were found (convergence). Subsequently, a highly dissimilar domain identified in the previous iterative search was used to seed a new iterative search. This process was done in total five times. From the resulting set of matches, we then filtered out all with a sequence length of >150 residues and those containing more than one predicted domain in Gene3D (40).
Search for archaeal RNase HIII proteins
A possible scenario for the emergence of RNase HIII in Bacteria is horizontal transfer from Archaea, and therefore we searched for occurrence of this protein in Archaea.
A first set of sequence searches were performed using BLASTP against the NCBI non-redundant database, and complete archaeal genomes in Genbank (www.ncbi.nlm.nih.gov), as of June 2011.
Additionally, a semi-automated iterative procedure using in-house developed Perl-scripts was developed using more sensitive HMM-based protocols to explore the presence/absence of putative RNase HIII candidates in sequenced archaeal genomes. First, HMMer 3.0 was used to construct HMM profiles using proteins identified as the target protein (in this case, the RNase HIII protein from B. stearothermophilus). The profile was calibrated and used in an iterative search against all sequenced archaeal genomes (www.ncbi.nlm.nih.gov) as of June 2011.
As putative domain matches were identified they were manually inspected for previous annotation and e-value. If the putative hit was determined to represent a true hit, it was included in a new HMM profile, and the procedure repeated until no further hits were detected.
RESULTS AND DISCUSSION
Sequence and structure indicate that TBP domains are related by common descent
Available data indicate that TBP domains from TBP, DNA glycosylase and RNase HIII display diverse sequence and structure and are associated with a broad range of functions, spanning DNA repair, transcription and replication (Table 1 and Figure 1). We therefore looked at both sequence and structural data to examine evidence for common ancestry. Results are summarized in Table 2. Six lines of evidence were considered, i.e. data from three existing databases of protein classification, results from profile-based and HMM-based sequence searches and structural comparisons.
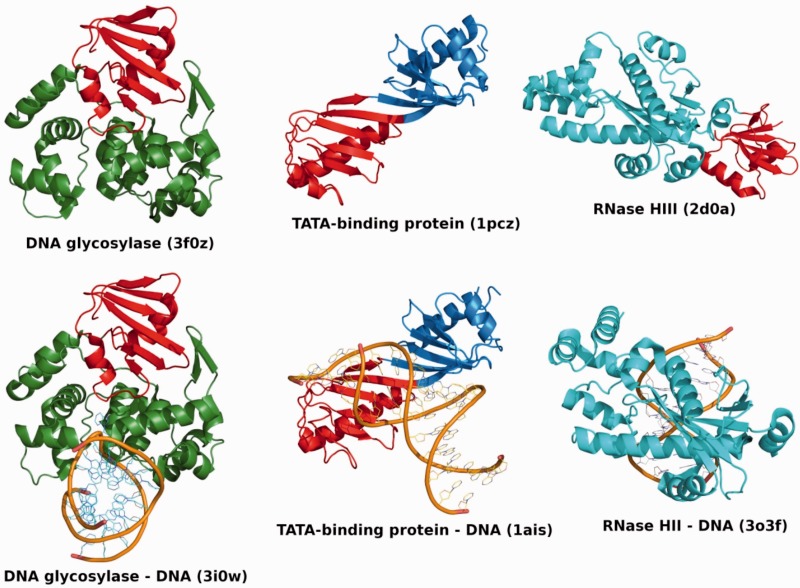
Figure 1.
Structure and DNA binding of TBP domain-containing proteins. Overview of DNA glycosylase (left), TBP (centre) and RNase HII/HIII (right) structures in the free forms (top) and DNA-bound forms (bottom). The free and DNA-bound forms are shown in the same orientation for each protein. In all cases, one TBP domain is coloured in red, whereas the rest of the protein chain is coloured green for DNA glycosylase, blue for TBP and cyan for RNase HII/HIII. For RNase HIII, no DNA-bound structure is available in the PDB; thus, a structure of DNA-bound RNase HII is shown instead to illustrate the conserved RNase domains in the two classes of enzymes. The RNase domain of RNase HII is in the same orientation as the equivalent RNase domain of RNase HIII. DNA backbones are represented as orange traces. The identifiers of the PDB entries used in this figure are indicated. Figures 1 and 2 were generated using PyMol (The PyMol Molecular Graphics System, version 1.3, Schrödinger LLC).
Table 2.
Evidence of homology between TBP domainsa
| Protein | TBP | DNA glycosylase | RNase HIII |
|---|---|---|---|
| TBP | Scop: superfamily | CATH: fold | |
| Pfam: clan | FFAS: −13.0 | ||
| CATH: fold | HHpred: 95.0 (PIRSF 037748) | ||
| SSAP: 82.49 (3.14) | SSAP: 79.70 (2.82) | ||
| DNA glycosylase | Scop: superfamily | CATH: fold | |
| Pfam: clan | SSAP: 75.34 (2.42) | ||
| CATH: fold | |||
| SSAP: 82.49 (3.27) | |||
| RNase HIII | CATH: fold | CATH: - fold | |
| FFAS: −13.0 | SSAP: 75.34 (2.34) | ||
| HHpred: 97.8 (PF00352) | |||
| SSAP: 79.70 (2.80) |
aThis table describes the evidence gathered in this work to confirm remote homology of TBP domains in TBPs, DNA glycosylases and RNase HIII. Each cell in the table shows the evidence for each pair of TBP domains. Six lines of evidence were considered, i.e. presence of TBP domains in the same category of the SCOP, CATH and Pfam databases, FFAS search, HHpred search and structural comparison using the program SSAP. For SCOP, Pfam and CATH, we simply indicate the classification level at which the two domains being compared are found together. For FFAS, a score lower than −9.5 is usually indicative of remote homology. For HHpred, the score returned is a probability of homology, and we consider any probability >95 as a positive hit. A SSAP score >80.0 is considered as a hit. SSAP associated Z-scores are provided next to each SSAP score (see ‘Materials and Methods’ section). Rows provide evidence for domains used as search queries. Therefore, using RNase HIII TBP domain as query in HHpred returns a hit to Pfam family PF00352 (TBP) with a probability of 97.8, whereas the reciprocal search with TBP as query returns a hit to PIR family PIRSF037748 (RNase HIII) with a probability of 95.0. See ‘Materials and Methods’ section for details on how searches were performed.
First, we examined evidence of homology of TBP domains in existing databases of protein classification. TBP domains from TBP and DNA glycosylase belong to the same superfamily in SCOP and to the same Clan (CL0407) in Pfam.
In contrast, FFAS and HHpred searches do not return significant matches between TBP domains of TBPs and DNA glycosylases but seem to suggest a significant similarity between the N-terminal domain of RNase HIII and the TBP domains of TBP, as previously reported (47,48).
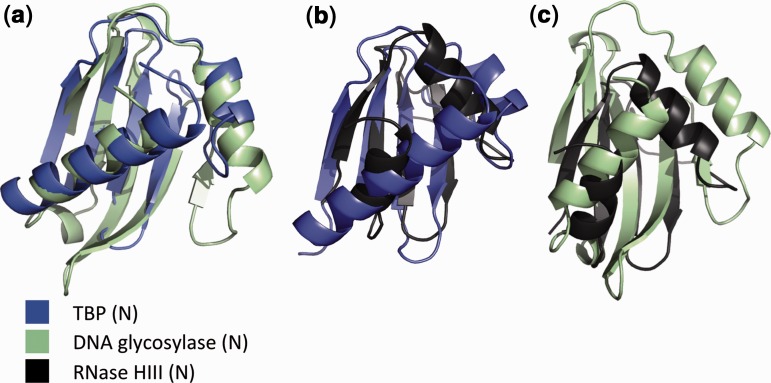
We next performed structural superpositions of TBP-domain structures using the structural comparison program SSAP. SSAP comparison results are listed in Table 2, and structural superpositions are shown in Figure 2. The observed structural similarity between TBP domains of TBP and DNA glycosylase supports the notion of a remote yet significant homology between these domains (see Figure 2 and Table 2).
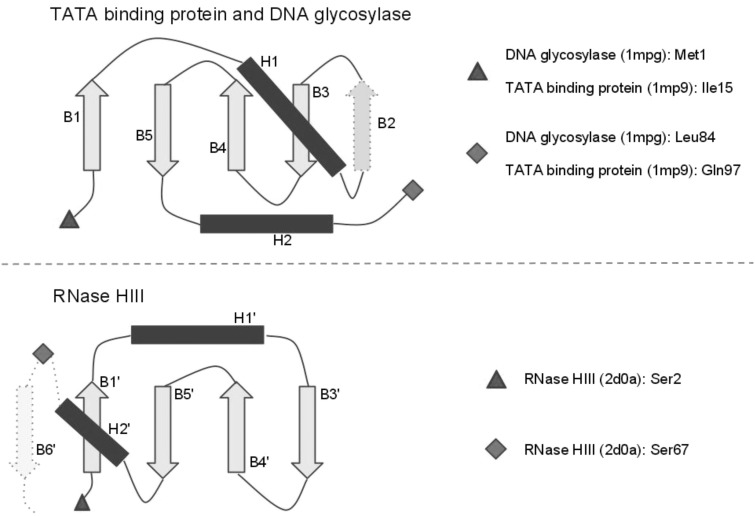
Figure 2.
Structural topology comparison of TBP-domains. The Top panel shows the common topology of TBP-domains from DNA glycosylases and TATA-binding proteins, which belong to the same Pfam clan. The Bottom panel shows the topology of the TBP-domain of RNase HIII. The β-strands in all structures are represented as light arrows, whereas the α-helices are represented as dark rectangles. Strands B2 and B6', which are present in only a subset of the structures, have a dotted outline. The N-terminal ends of the domains are represented as triangles, and C-terminal ends are represented as diamonds. The N- and C-terminal residues for each domains are indicated on the right of the Figure.
The SSAP score obtained between the TBP domain of RNase HIII and the N-terminal TBP domain of TBP is 79.70, which is marginally <80.00 threshold (see ‘Materials and Methods’ section). It is noteworthy that the N-terminal domain of RNase HIII is classified in Pfam as a domain of unknown function (DUF3378/PF11858) and falls outside the CL0407 clan; despite a comparable level of profile–profile similarity apparent across all families (Supplementary Figures S1–S3), the resulting e-values are above the cut-off for inclusion in the CL0407 TBP-like clan. As small domains like these may get such scores just by random similarities, we also ran SSAP comparisons between the RNase HIII N-terminal domain and all 35 583 non-redundant domains in CATH v3.3. Of all these domains, 99.95% have a SSAP score <79.70 (Z-score of 2.82), suggesting that the similarity obtained between RNase HIII N-terminal domain and the N-terminal domain of TBP is not just obtained randomly because of the small size of the domain. Similarly, the structural similarity between the TBP domains of TBP and DNA glycosylase is also supported by a Z-score of 3.27. In addition, the structure of the TBP domain comprises a rather unusual topology. Although it contains a common β-meander motif, this comprises less than one-third of the structure and is therefore unlikely to be the explanation for the considerable structural similarity between the domains.
Comparison of the structural topology of TBP domains from TBP and DNA glycosylases with the TBP domain of RNase HIII illuminates the clear structural similarity (Figure 3), despite high levels of sequence divergence. Secondary structure elements B1′, H1′, B3′, B4′, B5′, H2′ and the intervening loops from RNase HIII appear structurally equivalent to secondary structure elements B1, H1, B3, B4, B5, H2 from the other TBP domains. As indicated in Figure 3, the primary difference between the TBP domains of RNase HIII and the other TBP domains is the insertion/deletion of strand B2. Of note, strand B2 is absent in the structure of TBP from Archaea (PDB 1mp9), where it is replaced by a very long loop.
Figure 3.
Structural topology comparison of TBP domains. The Top panel shows the common topology of TBP domains from DNA glycosylases and TBP, which belong to the same Pfam clan. The Bottom panel shows the topology of the TBP domain of RNase HIII. The β-strands in all structures are represented as light arrows, whereas the α-helices are represented as dark rectangles. Strands B2 and B6′, which are present in only a subset of the structures, have a dotted outline. The N-terminal ends of the domains are represented as triangles, and C-terminal ends are represented as diamonds. The N- and C-terminal residues for each domains are indicated on the right of the figure.
Although the structure data or sequence data may not be strong enough on its own to indicate homology, the combination of these two different types of evidence is extremely unlikely to have occurred by chance and is therefore compelling evidence of homology. This clearly suggests that the TBP domains in the different proteins analysed here have a single evolutionary origin. It is worth pointing out that the current evidence points at homology between the TBP domains of TBPs and DNA glycosylases, and between the TBP domains of TBPs and RNase HIII, but not directly between the TBP domains of DNA glycosylase and RNase HIII.
DNA binding is observed in some proteins possessing the TBP domain
Although the extant TBP domains appear to have diverged from a common ancestor, it is harder to attribute DNA binding to that ancestral TBP domain [see also (17)]. As evident from Figure 1, which shows TBP domain containing proteins in complex with their nucleic acid target or substrate, TBP is the only one for which available structural data show that the TBP domain is involved in DNA binding. Both RNase HIII and DNA glycosylases also bind DNA, but it is yet uncertain whether the TBP domain participates in the interaction of those proteins with DNA. However, recent evidence seems to suggest that in RNase HIII at least, the TBP domain might also be involved in DNA binding (49).
The function of the TBP domain in RNase HIII is unclear, but it has been suggested to serve as a nucleic acid-binding domain that expands versatility of the HIII subtype by providing additional interactions with the DNA–RNA template (48). Indeed, a recent mutagenesis study indicates that key conserved residues in the RNase HIII TBP domain do contribute to nucleic acid binding, and the structure is consistent with a model wherein a flexible linker domain enables the TBP domain to swing round to bind the DNA–RNA substrate (49).
According to that mutagenesis study, the TBP domain of RNase HIII binds DNA via a surface equivalent to that of the TBP domains of TBP. This similar way of binding DNA further reinforces the notion that the TBP domains of TBP and RNase HIII are related by common descent.
Although DNA glycosylases must of course recognize dsDNA, the X-ray structure of a complex formed by E. coli DNA glycosylase AlkA and its template DNA (PDB ID 1diz) shows that the TBP domain is not in proximity of, and therefore not directly involved in, interactions with the DNA template. To our knowledge, there are no currently available data to suggest that the TBP domain of DNA glycosylases interacts with the substrate DNA.
Origin of RNase HIII and its TBP domain
RNase HIII consists of an N-terminal TBP domain and of a C-terminal domain related to RNase HII (48). RNase HIII has only been characterized in a handful of bacterial genomes, whereas RNase HII is found across all three domains of life (21). The origin of the TBP domain of RNase HIII is enigmatic. TBPs could be the source of this domain, although that is difficult to reconcile with the fact that TBPs are not found in bacteria, whereas RNase HIII seems mostly limited to them.
In an attempt to decipher the origins of the TBP domain in RNase HIII, we first checked whether homologues of RNase HIII could be found in Archaea and Eukaryotes. To that end, we first performed BLASTp-searches against the NCBI non-redundant protein database with the RNase HIII sequence from B. stearothermophilus as seed. Although the distribution of RNase HIII among the bacterial domain is patchy, the protein is present in taxonomically very divergent bacterial clades. Searches also yielded two putative RNase HIII sequences in archaeal genomes.
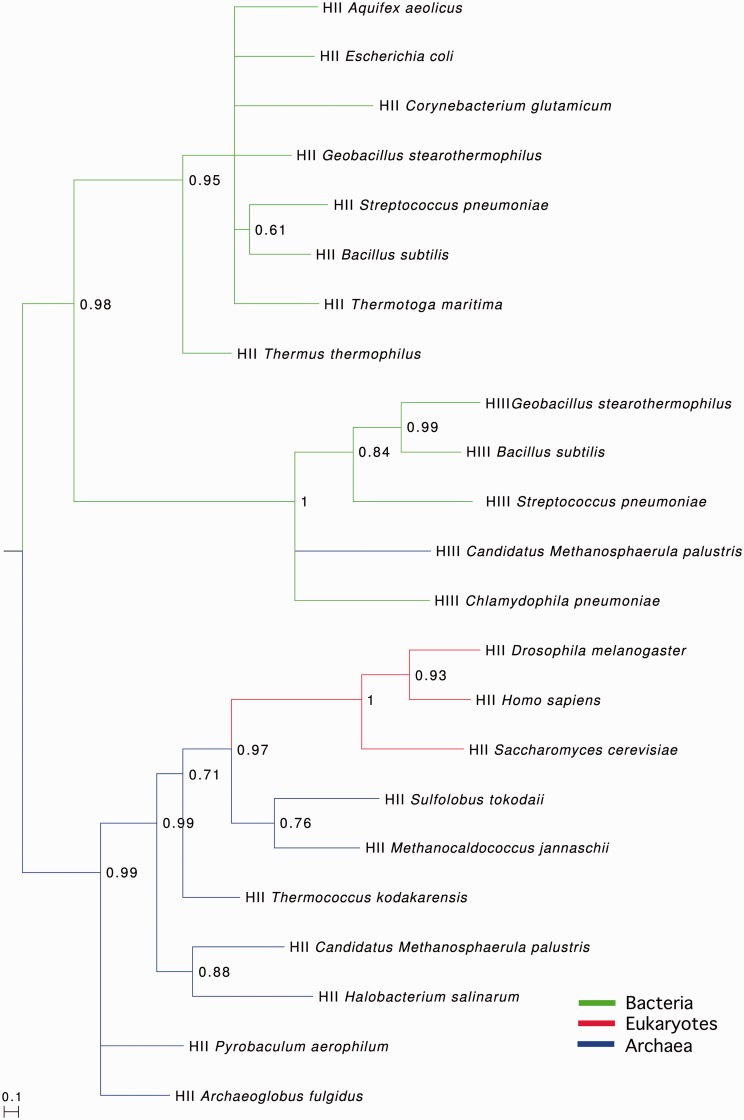
We next examined whether the origin of RNase HIII could be unravelled using phylogeny. We therefore performed a Bayesian phylogenetic analysis with RNase HII and HIII sequences from a variety of bacteria, archaea and eukaryotes. Although there was insufficient signal in the TBP domain on its own for reliable phylogenies, we were able to able to perform phylogenetic analyses using the RNase HII-like domain.
As shown in Figure 4, we uncovered three well-separated and largely well-resolved clades consisting of archaeal/eukaryotic RNases HII, bacterial RNases HII and bacterial/archaeal RNases HIII. The RNase HIII clade is positioned as a sister clade to the bacterial RNase HII clade. This is consistent with the relatively high sequence similarity between the RNase HIII C-terminal domain and bacterial RNase HII and suggests that RNase HIII may have evolved from a bacterial RNase HII through acquisition of the TBP domain.
Figure 4.
Phylogenetic analysis of RNases HII/HIII. The tree indicates that RNase HIII sequences most likely derive from RNase HII, with the archaeal RNase HIII from Candidatus Methanosphaerula palustris possibly being the result of a bacterial to archaeal horizontal gene transfer event. Colour coding of branches refers to taxonomic placement of sequence, with green corresponding to bacterial sequences, blue to archaeal sequences and red to eukaryotic sequences. HII/HIII indicates whether the sequence is an RNase HII or HIII. The tree was generated using PhyloBayes (see ‘Materials and Methods’ section) and is mid-point rooted.
Regarding the emergence of RNase HIII in archaea, a recent transfer of RNase HIII from bacteria into archaea seems difficult to reconcile with the high level of divergence evident between the TBP domains from bacterial and archaeal RNases HIII (Supplementary Figure S4). Such divergence seems more consistent with an ancient evolutionary origin, not a recent transfer from bacteria to archaea. Therefore, we cannot formally rule out the possibility that RNase HIII entered archaea via an ancient horizontal gene transfer. Assuming the position of the root is correctly placed between archaeal and bacterial RNases HII, an ancient transfer event is plausible.
Together, these data suggest that RNase HIII evolved from an ancestral RNase HII. The differences between the TBP domain from RNase HIII and other known TBP domains indicate that RNase HIII is most likely evolutionarily ancient, despite a limited distribution, and that the TBP domain architecture is older still. We note that, under evolution by horizontal transfer and natural selection, there is no requirement that evolutionarily old sequences must have a broader distribution than evolutionarily younger sequences.
Genomic evidence for a novel class of monopartite TBP-domain genes in Euryarchaea
Although our phylogenetic analyses shed some light on the origin of RNase HIII, the origin of the TBP domain within RNase HIII still remained unclear, and the absence of an obvious donor for the TBP domain in RNase HIII led us to examine whether the RNase HIII TBP domain evolved from a single copy TBP-domain protein. Several candidates exist. TbpA, just such a protein with a single TBP domain, has been identified in Halobacteria (38), and a number of computationally identified cases are also known (16). TbpA appears functional but is not essential for cell viability (50–52).
We performed a screen of the Gene3D database (39,40) for monopartite TBP domains, which recovered three of these TbpA homologues, all of which proved very similar to full-length TBP sequences from the same species (50).
We also retrieved all mono-partite candidates from the full Pfam TBP family (PF00352) alignments, as well as computationally identified candidates (16) and blasted these against Genbank to ascertain whether these were in fact TBPs (with only half the protein annotated in the genome) or whether genomic data supported a monopartite structure.
Examination of adjacent sequences, blast hits and genomic context revealed that the majority of cases were likely to be TBPs that had been misannotated. However, some show no evidence of an adjacent domain, suggesting they are not part of a larger protein. Next, we generated a profile HMM of the monopartite candidates and screened the Uniprot database and all publicly available archaeal genomes (as of November 2011). All resulting cases are from Euryarchaea and are significantly diverged from the sequences of TBPs in these genomes, ruling out misannotation (Table 3).
Table 3.
Candidate single copy TBP domains in euryarchaeal genomes are distantly related to TBPs
| Species | Monopartite TBP-domain candidates |
TBP |
Pairwise similaritya | ||
|---|---|---|---|---|---|
| Genbank accession | Length (aa) | Genbank accession | Length (aa) | ||
| Pyrococcus furiosus DSM 3638 | NP_578195 | 96 | NP_579024 | 182 | 14/25 |
| Pyrococcus sp. NA2 | YP_004424005 | 95 | YP_004424451 | 191 | 19/41 |
| Pyrococcus yayanosii CH1 | YP_004624794 | 95 | YP_004623880 | 191 | 38/73 |
| Thermococcus barophilus MPb | YP_004071110c | 92 | YP_004071510 | 190 | 11/19 |
| YP_004071638c | 91 | YP_004072056 | 174 | 20/37 | |
| Thermococcus gammatolerans EJ3 | YP_002959676 | 97 | YP_002958812d | 192 | 31/62 |
| YP_002959195d | 182 | 29/61 | |||
| Thermococcus kodakarensis KOD1 | YP_184404 | 97 | YP_182545 | 190 | 18/33 |
| Thermococcus onnurineus NA1 | YP_002306670 | 97 | YP_002307696 | 192 | 32/64 |
| Thermococcus sibiricus MM 739 | YP_002993507e | 136 | YP_002993555 | 185 | 5/9 |
| YP_002993607e | 82 | 16/42 | |||
| Average Pairwise identity | 49.6% | 70.9% | |||
aAs established with blast2seq. Numerator refers to number of ‘positives’ (identities and similarities) across the length (denominator) of the local alignment (denominator). The presence of two TBP domains in TBP results in two possible local alignments; the fraction of positives for the hit with lowest e-value was used in this table.
bBest local similarity score given for each monopartite candidate versus both annotated TBPs.
cPairwise similarity between these proteins: 61/91.
dPairwise similarity between these proteins: 130/183.
ePairwise similarity between these proteins: 53/81.
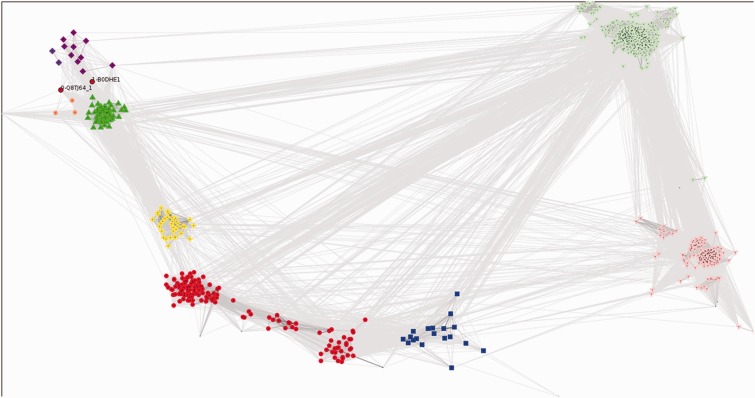
As shown in Figure 5, cluster-based analyses of sequence similarities indicate that the TBP domains of RNase HIII (yellow crosses), TBP domains of TBP (green triangles) and the monopartite TBP domains from Table 3 (purple diamonds) fall into distinct, statistically supported (≥95%) clusters. Notably, the monopartite TBP domains from Halobacterial genomes all cluster with TBP domains of TBPs (green triangles), not the other monopartite TBP domains identified via Pfam (purple diamonds). Figure 6 shows in detail the key multidomain architectures associated with the TBP domain superfamily and relates these to the cluster analysis presented in Figure 5.
Figure 5.
Cluster analysis of TBP domains. Groups identified by convex clustering and their jack-knife support are as follows: DNA glycosylase group 1 (91 members, red circles, left cluster) 97%, DNA glycosylase group 2 (46 members, red circles, right cluster) 40%, TBPs and halobacterial single TBP-domain proteins (61 members including 58 TBPs and 3 halobacterial single TBP-domain proteins, green triangles) 100%, RNase HIII (33 members, yellow crosses) 100%, DNA glycosylase group 3 (18 members, blue squares) 87%, single TBP-domain protein group 1 (10 members, purple diamonds, left cluster) 100%, single TBP-domain protein group 2 (two members, purple diamonds, right cluster) 87%, ThiF-MoeZ-MoeB group (three members, orange circles) 100%, COPG group (302 members, light green inverted triangles) 78%, AP appendage group (152 members, pink Y-shapes) 73% and a number of smaller loosely associated satellite clusters associated with either the COPG or AP appendage group and coloured accordingly. Domain architectures for all clusters are depicted in Figure 6. Coloured symbols are equivalent across both figures. See ‘Materials and Methods’ section for full description of analysis and settings.
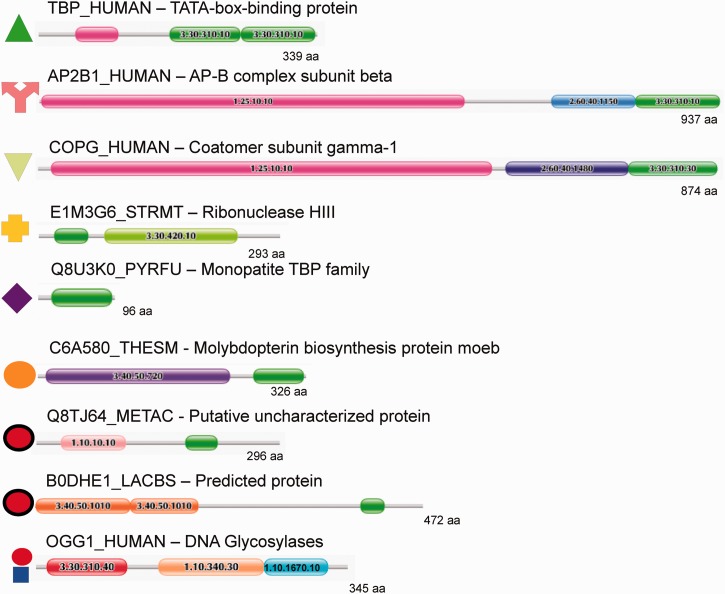
Figure 6.
Example domain architectures for TBP domain containing protein families. For the most common and some example rare domain architectures an example protein is shown using the Pfam ‘beads-on-a-string’ representation. Domains from the same superfamily are coloured identically on each protein, and CATH IDs are given for each domain. Note that the N-terminal extension (pink) associated with the human TBP is a 1.25.10.10 domain. These extensions are interaction sites for eukaryote-specific TBP-associated factors (see text). The symbols that correspond to the clusters in Figure 5 are given adjacent to each protein. TBP-domain superfamilies 3.30.310.10 and 3.30.310.30 are light and dark green, respectively. Note that under the current classification of CATH, this superfamily is split into two architectures; however, PRC results along with previous observations (see text) show they are anciently related, and both are universal to eukaryotes. Two unique architectures with no known full-length homologues (or function) appear to be isolated examples of domain recombination events (red circles with black outline). Both have potential DNA-binding domains at their N-termini and show substantial sequence divergence from the monopartite and TBP families (Figure 5). DNA Glycosylase group III is not shown but has essentially the same domain architecture as groups I and II. The difference is the N-terminal domain is classified as 3.30.310.20 instead of 3.30.310.40 (the latter is coloured red).
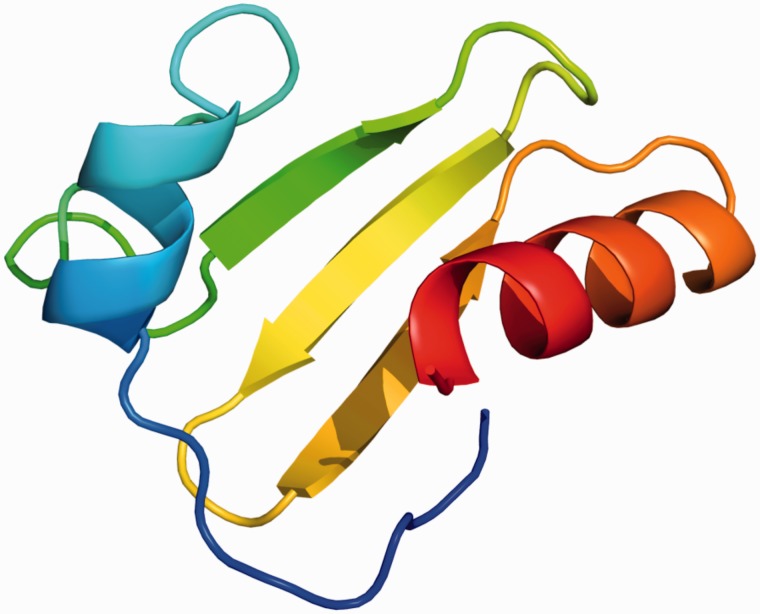
Given low levels of sequence similarity, we sought to examine whether the putative single copy TBP-domain proteins from Table 3 can fold into the characteristic TBP-domain 3D structure. We therefore submitted these sequences to the Phyre2 server to examine putative structure. Figure 7 shows a representative example (NP_578195) from Pyrococcus furiosus. The top 10 predicted structures were all TBP domains, predicted with >90% confidence, with similar results obtained for all sequences in Table 3 (data not shown). Although these data do not establish the function of these single copy TBP-domain proteins and do not shed light on the origin of the RNase HIII TBP domain, they nevertheless indicate that this group of proteins are not misannotated TBPs, are distinct from the Halobacterial monopartite TBP-domain proteins and may therefore have a distinct function in their own right.
Figure 7.
Predicted structure of single TBP-domain protein from P. furiosus (NP_578195), generated using the Phyre2 server (33).
Other multi-domain contexts in which the TBP domain occurs
Although carrying out the iterative HMM searches to search for monopartite TBP domains and determine the full extent of the TBP-domain family, we found the TBP domain in several other domain contexts (Figure 6). Among those shown in Figure 6, the most common TBP domain containing architectures are associated with the adaptin (AP) and COPG (coatomer protein complex, gamma subunit) families from eukaryotes. COPG is a constituent of the coatomer protein complex I (COPI) involved in transport of proteins from the Golgi to the endoplasmic reticulum, and adaptins enable selection and packaging of cargo for vesicles entering the endocytic system. Both COPI and APs have been shown to trace to the Last Eukaryotic Common Ancestor (53–55), and moreover, COPG and APs share clear sequence and structural similarities [(56); Figure 6], indicating that they evolved and diversified before the radiation of eukaryote lineages (57). COPG and alpha and beta APs all carry an appendage domain, of which the platform sub-domain is structurally homologous to the TBP domain (58–62) (Figure 6). Experimental and structural data indicate that these appendage domains, including the platform sub-domain, are involved in mediating a diverse number of protein interactions, acting as interaction hubs for clathrin coat vesicle assembly and cargo selection (63,64).
A previously unrecognized family are homologues of MoeB (molybdopterin synthase sulfurylase, orange circles, Figures 5 and 6), which is essential for Molybdopterin biosynthesis. This architecture is found in both archaea and bacteria, though most MoeB proteins do not contain the TBP-domain region at the C-terminus of the protein. Currently, there is a lack of experimental information associated with these proteins, preventing any determination of how the TBP-domain affects the function of these proteins.
A small number of rare architectures (only one or a few proteins identified) were also found in diverse lineages, including archaea and fungi. Little functional annotation exists for these specific proteins, but it is notable that the associated domains appear to be normally involved in DNA binding. For instance B0DHE1_LACBS contains a repeated endonuclease domain at the N-terminus, whereas Q8TJ64_METAC has a winged helix repressor domain (red circles with black outline, Figures 5 and 6).
A tentative evolutionary history of TBP domains
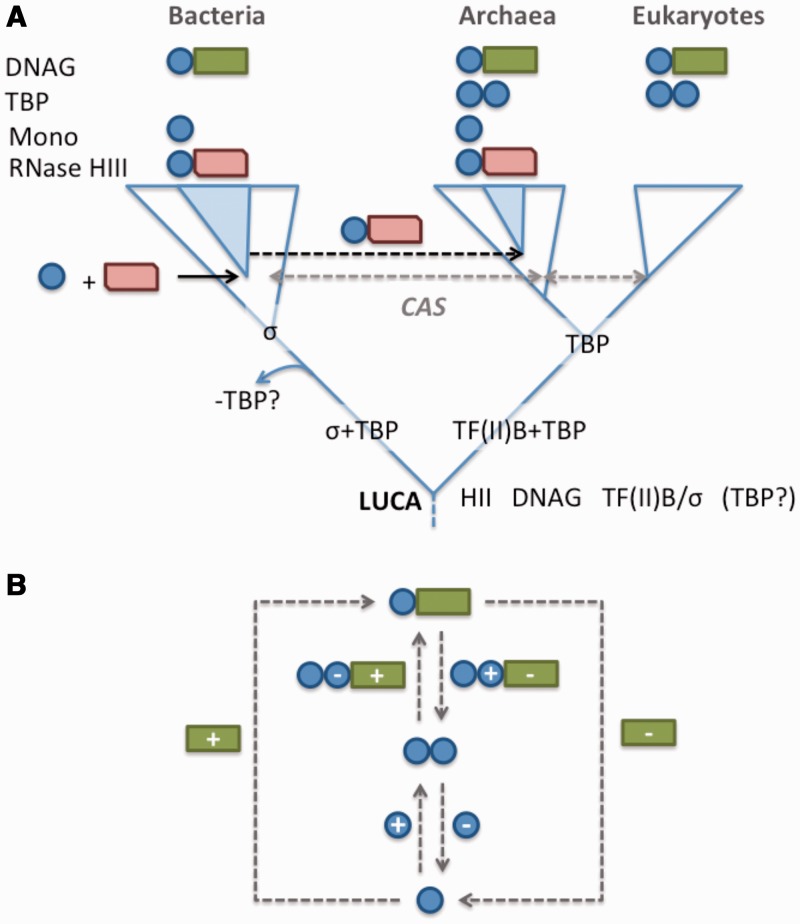
Available data on the distribution and function of the TBP domain do not make it straightforward to unravel its evolutionary history (Figure 8A). The pertinent facts are that (i) the TBP domain is found in all three domains of life; (ii) it functions in DNA binding in some but not all contexts; (iii) it exhibits extensive sequence divergence but (iv) clear structural similarity; and (v) it is found in a variety of protein contexts. These include dual repeats in TBP, as a fusion with other domains (DNA glycosylases and RNase HIII and others) and as a single TBP-domain protein in some euryarchaea.
Figure 8.
Evolutionary history of the TBP-domain. (A) Evolutionary distribution of the TBP-domain containing proteins across the three domains of life. DNAG: DNA glycosylase; Mono: single TBP-domain proteins; CAS: a TBP-domain protein distributed across the three kingdoms, identified by Iyer et al. (17). The authors concluded that the distribution of this protein is likely the outcome of horizontal gene transfer, though this has yet to be confirmed using phylogenetic methods. Speculation that TF(II)B and σ-factors are evolutionarily related (65) could be consistent with the replacement of TBP in the lineage leading to bacteria (placing TBP in LUCA) or with evolution of TBP in the lineage leading to archaea and eukaryotes. The tree depicts a three-kingdom architecture to aid in reading. A proposed archaeal origin for eukaryotes (66) does not alter the interpretation given here. (B) All possible fission and fusion events that could account for the evolution of single TBP-domain proteins (single blue circle), TBP (two blue circles) and DNA glyosylases (blue circle + green rectangle). Plus signs (+) indicate domain fusion events, minuses (−) represent fission (domain loss).
Based on these data, we consider two possible evolutionary scenarios.
One scenario proposes the initial existence of a monopartite TBP domain in LUCA, which subsequently duplicated to give rise to TBP, and fused with RNase HII to give rise to the HIII variant. The second scenario proposes that the TBP domain evolved through truncation from DNA glycosylase. Figure 8B shows a schematic representation of the alternative scenarios.
The DNA glycosylases are an evolutionarily ancient protein family that likely evolved before the divergence of the three domains of life (67,68). That the DNA glycosylases are the most broadly conserved TBP domain containing proteins suggests that the TBP domain may have evolved within this context. Although it seems difficult to envision the precise excision of the TBP domain from a DNA glycosylase, protein evolution can occur via both domain fusion and fission (69–72). Given the antiquity of DNA glycosylases, and no evidence for single TBP-domain proteins of similar antiquity, this model has the advantage of being compatible with extant protein distributions. As single copy TBP-domain proteins appear functional, this suggests a fission model is biologically plausible.
Conversely, proteins consisting exclusively of a single TBP domain, such as TbpA, also lend credence to a possible monomeric origin, as this indicates an ancestral monomer may have been functional in its own right. Single TBP-domain proteins thus provide a conceptually important stepping stone from the integral TBP domain in DNA glycosylases or the duplicate domain in TBP itself, to emergence of multidomain proteins such as RNase HIII. Although sequence divergence makes it difficult to establish the single TBP-domain factors as the ultimate source of this domain in multidomain proteins, they do lend this scenario biological credibility.
The diversification of the TBP domain into other roles may have occurred from a single TBP-domain protein (scenario 1) or following the emergence of a discrete TBP domain from the DNA glycosylase protein (scenario 2). Subsequent duplication of the TBP domain would then have given rise to TBP in the archaeo-eukaryotic lineage. Alternatively, the loss of TBP in the bacterial lineage is also plausible, and of interest in light of recent speculation that TFIIB—which acts in concert with TBP to facilitate transcription initiation—may be evolutionarily related to sigma factors (65) (Figure 8A). Following this line of speculation, the evolution of a hypothetical sigma-TFIIB precursor into sigma in the bacterial lineage may have made TBP functionally redundant for transcription initiation, leading to its loss from the bacterial lineage.
Both single copy TBP domains and the TBP domain of RNase HIII share very low levels of sequence similarity with TBP domains of TBP or DNA glycosylase. This is suggestive of deep evolutionary origins. In our analyses, RNase HIII can be concluded to have evolved from RNase HII. But the sequence divergence between the N-terminal TBP domain of RNase HIII and any other known TBP domains makes it difficult to identify the TBP-domain donor among these. It therefore seems more plausible that RNase HIII evolved via fusion of RNase HII with a single copy TBP domain at its N-terminus, although this is difficult to establish. The sequence divergence between the TBP domain of RNase HIII and other known TBP domains suggests that this fusion would have been a very ancient event.
Provided that the TBP domain originated within the DNA glycosylases (scenario 2), this suggests that the universal DNA glycosylase TBP domain and possibly other ancestral forms of the TBP domain did not necessarily facilitate interactions with nucleic acids. The DNA-binding activity may have emerged later in evolution following the duplication and fusion event that gave rise to the double repeat structure of all contemporary TBP variants. In this regard, it is worth noting that the role for the TBP domains of eukaryotic COPG and adaptins in mediating diverse protein–protein interactions indicates this domain fulfils a broad range of cellular functions.
Finally, the large N-terminal extensions present in many eukaryotic TBPs are likely to have emerged much later in evolution, as did the duplications of TBP in metazoa and vertebrates, which gave rise to the TBP paralogues TBP-related factor 2 and 3 (10). This diversification went hand-in-hand with the multiplication of RNAPs into 3–5 distinct classes each with specific transcription initiation factors, which in extant eukaryotes transcribe distinct and non-overlapping subsets of genes.
CONCLUSIONS
We conclude that the TBP domain traces back to before the divergence of the three domains of life, but that it most probably only took on a role as a transcription initiation factor (TBP) later in evolution. It may have been present as a single domain in LUCA or originated within DNA glycosylases, evolving through truncation. Whatever scenario is correct the modular fold could subsequently be co-opted into new function through duplication and fusions with other domain partners. This has clearly happened a number of times as revealed by the different multi-domain proteins containing the TBP domain. The considerable sequence divergence between similar TBP domains across a narrow range of species hints at a much older evolutionary origin for the TBP domain in RNase HIII, despite a sparse distribution relative to RNase HII, which, like DNA glycosylases, likely traces to the LUCA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–4.
FUNDING
A.M.P. acknowledges past support as a Research Fellow of the Royal Swedish Academy of Sciences via a grant from the Knut and Alice Wallenberg Foundation, and current support from the Royal Society of New Zealand through a Rutherford Discovery Fellowship and the New Zealand Marsden Fund. B.H.D. acknowledges the European Union FP7 Impact grant for financial support and also NIH funding in connection with the PSI structural genomics initiative. Research in the RNAP laboratory is generously funded by a Wellcome Trust Investigator Award WT096553MA, and BBSRC project BB/H019332/1 (to F.W.). Funding for open access charge: Royal Society of New Zealand.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Werner F. Structural evolution of multisubunit RNA polymerases. Trends Microbiol. 2008;16:247–250. doi: 10.1016/j.tim.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol. 2010;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris JK, Kelley ST, Spiegelman GB, Pace NR. The genetic core of the universal ancestor. Genome Res. 2003;13:407–412. doi: 10.1101/gr.652803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tourasse NJ, Gouy M. Accounting for evolutionary rate variation among sequence sites consistently changes universal phylogenies deduced from rRNA and protein-coding genes. Mol. Phylogenet. Evol. 1999;13:159–168. doi: 10.1006/mpev.1999.0675. [DOI] [PubMed] [Google Scholar]
- 5.Poole AM, Logan DT. Modern mRNA proofreading and repair: clues that the last universal common ancestor possessed an RNA genome? Mol. Biol. Evol. 2005;22:1444–1455. doi: 10.1093/molbev/msi132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cramer P. Multisubunit RNA polymerases. Curr. Opin. Struct. Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- 7.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 8.Hausner W, Wettach J, Hethke C, Thomm M. Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J. Biol. Chem. 1996;271:30144–30148. doi: 10.1074/jbc.271.47.30144. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 10.Akhtar W, Veenstra GJ. TBP-related factors: a paradigm of diversity in transcription initiation. Cell Biosci. 2011;1:23. doi: 10.1186/2045-3701-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 12.Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu. Rev. Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 13.Hahn S. The role of TAFs in RNA polymerase II transcription. Cell. 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- 14.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 16.Burroughs AM, Iyer LM, Aravind L. Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2009;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 2001;43:134–144. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 18.Gough J. Convergent evolution of domain architectures (is rare) Bioinformatics. 2005;21:1464–1471. doi: 10.1093/bioinformatics/bti204. [DOI] [PubMed] [Google Scholar]
- 19.Setiyaputra S, Mackay JP, Patrick WM. The structure of a truncated phosphoribosylanthranilate isomerase suggests a unified model for evolution of the (betaalpha)8 barrel fold. J. Mol. Biol. 2011;408:291–303. doi: 10.1016/j.jmb.2011.02.048. [DOI] [PubMed] [Google Scholar]
- 20.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 22.Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim. Biophys. Acta. 2010;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuff AL, Sillitoe I, Lewis T, Clegg AB, Rentzsch R, Furnham N, Pellegrini-Calace M, Jones D, Thornton J, Orengo CA. Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Res. 2011;39:D420–D426. doi: 10.1093/nar/gkq1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaroszewski L, Li Z, Cai XH, Weber C, Godzik A. FFAS server: novel features and applications. Nucleic Acids Res. 2011;39:W38–W44. doi: 10.1093/nar/gkr441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 28.Madera M. Profile comparer: a program for scoring and aligning profile hidden Markov models. Bioinformatics. 2008;24:2630–2631. doi: 10.1093/bioinformatics/btn504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madera M. Profile comparer: a program for scoring and aligning profile hidden Markov models. Bioinformatics. 2008;24:2630–2631. doi: 10.1093/bioinformatics/btn504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuster-Bockler B, Bateman A. Visualizing profile-profile alignment: pairwise HMM logos. Bioinformatics. 2005;21:2912–2913. doi: 10.1093/bioinformatics/bti434. [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orengo CA, Taylor WR. SSAP: sequential structure alignment program for protein structure comparison. Methods Enzymol. 1996;266:617–635. doi: 10.1016/s0076-6879(96)66038-8. [DOI] [PubMed] [Google Scholar]
- 33.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 34.Lassmann T, Sonnhammer EL. Kalign, Kalignvu and Mumsa: web servers for multiple sequence alignment. Nucleic Acids Res. 2006;34:W596–W599. doi: 10.1093/nar/gkl191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 36.Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 2007;7(Suppl. 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frickey T, Weiller G. Analyzing microarray data using CLANS. Bioinformatics. 2007;23:1170–1171. doi: 10.1093/bioinformatics/btm079. [DOI] [PubMed] [Google Scholar]
- 38.Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, et al. Genome sequence of Halobacterium species NRC-1. Proc. Natl Acad. Sci. USA. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lees J, Yeats C, Redfern O, Clegg A, Orengo C. Gene3D: merging structure and function for a thousand genomes. Nucleic Acids Res. 2010;38:D296–D300. doi: 10.1093/nar/gkp987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lees J, Yeats C, Perkins J, Sillitoe I, Rentzsch R, Dessailly BH, Orengo C. Gene3D: a domain-based resource for comparative genomics, functional annotation and protein network analysis. Nucleic Acids Res. 2012;40:D465–D471. doi: 10.1093/nar/gkr1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeats C, Lees J, Carter P, Sillitoe I, Orengo C. The Gene3D Web Services: a platform for identifying, annotating and comparing structural domains in protein sequences. Nucleic Acids Res. 2011;39:W546–W550. doi: 10.1093/nar/gkr438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eddy SR. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.UniProt_Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aravind L, Koonin EV. A natural classification of ribonucleases. Methods Enzymol. 2001;341:3–28. doi: 10.1016/s0076-6879(01)41142-6. [DOI] [PubMed] [Google Scholar]
- 48.Chon H, Matsumura H, Koga Y, Takano K, Kanaya S. Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus. J. Mol. Biol. 2006;356:165–178. doi: 10.1016/j.jmb.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 49.Miyashita S, Tadokoro T, Angkawidjaja C, You DJ, Koga Y, Takano K, Kanaya S. Identification of the substrate binding site in the N-terminal TBP-like domain of RNase H3. FEBS Lett. 2011;585:2313–2317. doi: 10.1016/j.febslet.2011.05.064. [DOI] [PubMed] [Google Scholar]
- 50.Bleiholder A, Frommherz R, Teufel K, Pfeifer F. Expression of multiple tfb genes in different Halobacterium salinarum strains and interaction of TFB with transcriptional activator GvpE. Arch. Microbiol. 2012;194:269–279. doi: 10.1007/s00203-011-0756-z. [DOI] [PubMed] [Google Scholar]
- 51.Coker JA, DasSarma S. Genetic and transcriptomic analysis of transcription factor genes in the model halophilic Archaeon: coordinate action of TbpD and TfbA. BMC Genet. 2007;8:61. doi: 10.1186/1471-2156-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teufel K, Bleiholder A, Griesbach T, Pfeifer F. Variations in the multiple tbp genes in different Halobacterium salinarum strains and their expression during growth. Arch. Microbiol. 2008;190:309–318. doi: 10.1007/s00203-008-0383-5. [DOI] [PubMed] [Google Scholar]
- 53.Dacks JB, Poon PP, Field MC. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc. Natl Acad. Sci. USA. 2008;105:588–593. doi: 10.1073/pnas.0707318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PloS One. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schledzewski K, Brinkmann H, Mendel RR. Phylogenetic analysis of components of the eukaryotic vesicle transport system reveals a common origin of adaptor protein complexes 1, 2, and 3 and the F subcomplex of the coatomer COPI. J. Mol. Evol. 1999;48:770–778. doi: 10.1007/pl00006521. [DOI] [PubMed] [Google Scholar]
- 56.Duden R, Griffiths G, Frank R, Argos P, Kreis TE. Beta-COP, a 110 kd protein associated with non-clathrin-coated vesicles and the Golgi complex, shows homology to beta-adaptin. Cell. 1991;64:649–665. doi: 10.1016/0092-8674(91)90248-w. [DOI] [PubMed] [Google Scholar]
- 57.Field MC, Dacks JB. First and last ancestors: reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr. Opin. Cell Biol. 2009;21:4–13. doi: 10.1016/j.ceb.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Hoffman GR, Rahl PB, Collins RN, Cerione RA. Conserved structural motifs in intracellular trafficking pathways: structure of the gammaCOP appendage domain. Mol. Cell. 2003;12:615–625. doi: 10.1016/j.molcel.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 59.Watson PJ, Frigerio G, Collins BM, Duden R, Owen DJ. Gamma-COP appendage domain - structure and function. Traffic. 2004;5:79–88. doi: 10.1111/j.1600-0854.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- 60.Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR. The structure and function of the beta 2-adaptin appendage domain. EMBO J. 2000;19:4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Traub LM, Downs MA, Westrich JL, Fremont DH. Crystal structure of the alpha appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly. Proc. Natl Acad. Sci. USA. 1999;96:8907–8912. doi: 10.1073/pnas.96.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Owen DJ, Vallis Y, Noble ME, Hunter JB, Dafforn TR, Evans PR, McMahon HT. A structural explanation for the binding of multiple ligands by the alpha-adaptin appendage domain. Cell. 1999;97:805–815. doi: 10.1016/s0092-8674(00)80791-6. [DOI] [PubMed] [Google Scholar]
- 63.Edeling MA, Mishra SK, Keyel PA, Steinhauser AL, Collins BM, Roth R, Heuser JE, Owen DJ, Traub LM. Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev. Cell. 2006;10:329–342. doi: 10.1016/j.devcel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 64.Schmid EM, Ford MG, Burtey A, Praefcke GJ, Peak-Chew SY, Mills IG, Benmerah A, McMahon HT. Role of the AP2 beta-appendage hub in recruiting partners for clathrin-coated vesicle assembly. PLoS Biol. 2006;4:e262. doi: 10.1371/journal.pbio.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iyer LM, Aravind L. Insights from the architecture of the bacterial transcription apparatus. J. Struct. Biol. 2012;179:299–319. doi: 10.1016/j.jsb.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc. Natl Acad. Sci. USA. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol. Biol. Evol. 2003;20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 68.Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pasek S, Risler JL, Brezellec P. Gene fusion/fission is a major contributor to evolution of multi-domain bacterial proteins. Bioinformatics. 2006;22:1418–1423. doi: 10.1093/bioinformatics/btl135. [DOI] [PubMed] [Google Scholar]
- 70.Kummerfeld SK, Teichmann SA. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005;21:25–30. doi: 10.1016/j.tig.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 71.Wang W, Yu H, Long M. Duplication-degeneration as a mechanism of gene fission and the origin of new genes in Drosophila species. Nature Genet. 2004;36:523–527. doi: 10.1038/ng1338. [DOI] [PubMed] [Google Scholar]
- 72.Snel B, Bork P, Huynen M. Genome evolution. Gene fusion versus gene fission. Trends Genet. 2000;16:9–11. doi: 10.1016/s0168-9525(99)01924-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.