Abstract
A bottleneck in our capacity to rationally and predictably engineer biological systems is the limited number of well-characterized genetic elements from which to build. Current characterization methods are tied to measurements in living systems, the transformation and culturing of which are inherently time-consuming. To address this, we have validated a completely in vitro approach for the characterization of DNA regulatory elements using Escherichia coli extract cell-free systems. Importantly, we demonstrate that characterization in cell-free systems correlates and is reflective of performance in vivo for the most frequently used DNA regulatory elements. Moreover, we devise a rapid and completely in vitro method to generate DNA templates for cell-free systems, bypassing the need for DNA template generation and amplification from living cells. This in vitro approach is significantly quicker than current characterization methods and is amenable to high-throughput techniques, providing a valuable tool for rapidly prototyping libraries of DNA regulatory elements for synthetic biology.
INTRODUCTION
Synthetic biology is an application-driven field attempting to apply a rational engineering approach to the re-design of existing or new biological systems (1). The “bottom-up -approach” represents a shift towards the rational engineering and design of biological systems, where the goal is to design complex biological systems in a hierarchical manner from well-characterized and reusable DNA parts and circuits (1,2). One foundational principle of this approach is that the function of a DNA part in isolation aids the prediction of how combinations of DNA parts will function when assembled into larger genetic circuits (1). Coupled with mathematical modelling and in silico simulation, this allows potential genetic designs to be rapidly assessed and also allows for identification of designs with the desired function, bypassing the need for time-consuming alteration and optimization of genetic circuit design by molecular biology techniques (3).
A current bottleneck in this approach is the limited number of well-characterized DNA parts from which to build new biological circuits (4–8). Collaborative efforts have been made to generate and annotate DNA part libraries [for example, the iGEM Registry of Standard Biological Parts (partsregistry.org)], although the majority of DNA parts, particularly DNA regulatory elements, remain inadequately characterized to guide in silico simulation (4,5). A contributing factor to this limitation has been that current techniques to generate and characterize libraries of DNA regulatory elements require living systems, the transformation and culturing of which are inherently time-consuming and low-throughput (5,9,10).
Cell-free synthetic biology offers an alternative approach, whereby complex biological systems are studied in a reduced in vitro experimental system, omitting the limitations and complexity of living systems, resulting in an increase in engineering flexibility (11–14). Escherichia coli extract cell-free expression systems that are based on the transcription and translation of DNA templates in a buffered lysate solution have recently been used to study DNA regulatory elements and genetic circuits in vitro (15–19). Indeed, it has been suggested that cell-free systems could serve as a platform to rapidly characterize DNA parts and genetic circuits to fast-track implementation in living systems (15). Importantly, Karig et al. (15) showed that E. coli extract cell-free systems could be used as a platform for genetic device optimization. The behaviour of a negative-feedback circuit was optimized for expression in E. coli extract cell-free systems by modifications of the promoters, ribosome-binding site (RBS), transcriptional terminators and genetic composition of the design. Interestingly, this final optimized genetic negative-feedback circuit was shown to be functional in live E. coli, although the overall circuit behaviour in live E. coli did not appear to be directly reflective of the behaviour in E. coli extract cell-free systems. As such, this raises the fundamental question: Is the characterization of DNA parts derived in cell-free systems directly comparable with that collected in vivo?
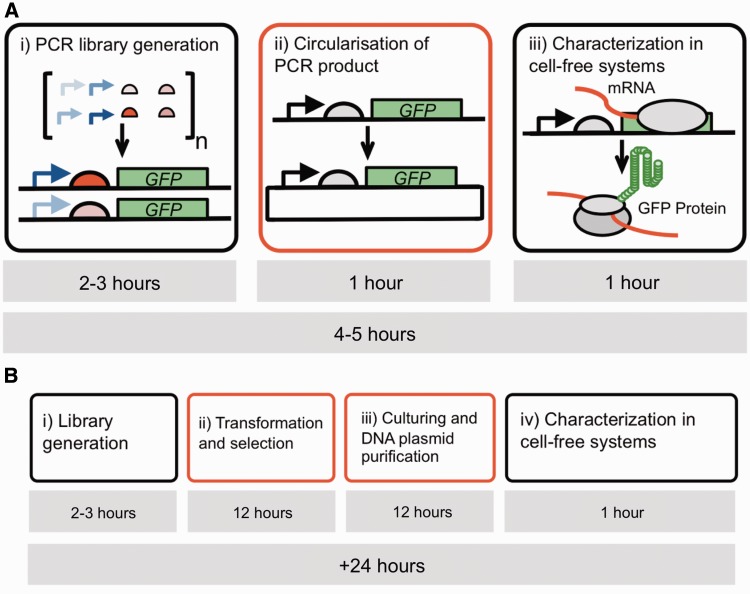
Furthermore, to our knowledge, all previous studies using E. coli extract cell-free systems have used plasmids derived from living systems as DNA templates, negating the time benefit of using cell-free systems (15–19). As such, current cell-free methodology has not yet realized its full potential to rapidly explore the vast landscape of natural and synthetic gene variants. In this study, we have validated a completely in vitro approach to generate and characterize libraries of DNA regulatory elements in cell-free systems to aid more efficient bottom-up engineering of biological systems. First, we demonstrate a clear correlation of the characterization data obtained from expression of plasmid DNA templates in E. coli extract cell-free systems and exponential growing E. coli cells for frequently used DNA regulatory elements. Furthermore, we have investigated in vitro-generated DNA templates as an alternative to purified DNA plasmids for cell-free systems. We used a USER enzyme and T4 DNA ligase (USER–ligase) reaction to efficiently generate closed-circular DNA from polymerase chain reaction (PCR)-generated DNA that is suitable for characterization in cell-free systems. By omitting the need for transformation, culturing and purification of DNA templates from living cells, our approach is significantly quicker than current approaches, with both the DNA template generation and characterization being performed by a three-step procedure within 5 h as shown in schematic Figure 1. Taken together, this work shows the potential for rapid prototyping of DNA regulatory elements in cell-free systems.
Figure 1.
(A) Schematic of suggested approach for DNA part library assembly and characterization in vitro. (i) Generation of a DNA part library is achieved by PCR amplification of DNA regulatory elements upstream of a GFP reporter gene. (ii) These PCR-amplified linear DNA products are circularized by intramolecular ligation to form closed-circular DNA. (iii) Closed-circular DNA products are used as DNA templates for characterization in cell-free systems by fluorescence measurements. Characterization data can be used to annotate DNA part libraries, and closed-circular DNA is stored. (B) Example of current methodology for DNA part assembly and characterization in cell-free systems.
MATERIALS AND METHODS
Plasmids assembly and strains
All plasmids used in this study were constructed by restriction enzyme cloning using DNA obtained from iGEM Registry of Standard Biological Parts (partsregistry.org), oligonucleotides or PCR of Pseudomonas aeruginosa (PAO1) genomic DNA using standard protocols (Supplementary Methods S1). Assembled plasmids were verified by restriction digestion and DNA sequencing. DNA plasmids used for DNA templates in cell-free systems were obtained from cultures of E. coli XL1-Blue strain (Agilent Genomics) transformed with plasmids and prepared using Qiagen Plasmid Midi or Maxi Prep kits. Cells were cultured for plasmid purification in Luria-Bertani (LB) media with 100 µg/ml of ampicillin at 37°C, shaking at 225 r.p.m. overnight. For experiments in live E. coli cells, plasmids were transformed into the E. coli BL21-Gold (DE3) strain (Agilent Genomics). All plasmids used are described in Supplementary Information (Supplementary Tables S1–S3).
Characterization in live E. coli cells
Plasmids were transformed into E. coli BL21-Gold (DE3) chemical competent cells (Agilent Genomics) according to the manufacturer’s protocol and selected on LB agar containing 100 µg/ml of ampicillin. Single colonies were used to inoculate 5 ml of M9 minimal media (1 × M9 minimal salts, 1 mM thiamine hydrochloride, 0.4% glycerol, 0.2% casamino acids, 2 mM magnesium sulfate, 0.1 mM calcium chloride and 100 µg/ml of ampicillin) and grown overnight at 37°C, shaking at 225 r.p.m. Overnight cultures were diluted 1:100 in 5 ml of pre-warmed M9 minimal media and grown for 3 h. After 3 h of growth, cultures were diluted to an optical density at 600 nm (O.D. 600) of 0.07 in 1 ml of pre-warmed M9 minimal media and grown for 1 h at 37°C, shaking at 225 r.p.m.
For the constitutive promoter and RBS libraries, 200 µl of E. coli culture was transferred in triplicate into a 96-well plate (Griener Bio-One). The plate was transferred to a POLARstar Omega plate reader (BMG Labtech) and fluorescence (excitation at 485 nm, emission at 520 nm) and O.D. 600 measured for 30 min at 5-min intervals at 37°C, shaking at 225 r.p.m. For every characterization assay, E. coli transformed with the backbone vector containing the green fluorescent protein (GFP) gene without a promoter (pSB1A2:GFPmut3b) was measured in parallel to determine background fluorescence. The data shown represent the averages and standard deviations of three independent measurements.
For the inducible promoters, 196 µl of E. coli culture was transferred in triplicate into a 96-well plate (Greiner Bio-One) and 4 µl of acylhomoserine lactone (AHL) inducer added to give the final concentrations shown in Figure 3B–D. The plate was transferred to a POLARstar Omega plate reader (BMG Labtech) and fluorescence (excitation at 485 nm, emission at 520 nm) and O.D. 600 measured for 6 h at 5-min intervals at 30°C, shaking at 225 r.p.m. AHL [N-(3-oxododecanoyl)-l-homoserine lactone] (Sigma-Aldrich) solutions were prepared from powder in dimethyl sulfoxide (DMSO). For every characterization assay, E. coli transformed with pSB1A2:GFPmut3b was measured in parallel. The data shown represent the averages and standard deviations of three independent measurements.
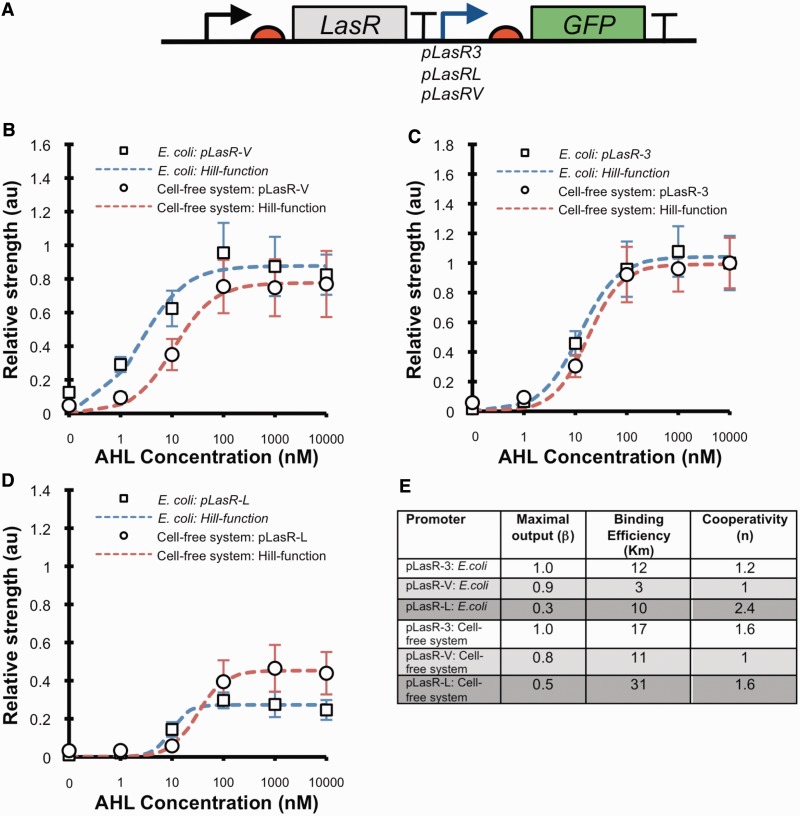
Figure 3.
Characterization of inducible promoters in cell-free systems and E. coli. (A) Diagram of the LasR responsive promoter test constructs. The relative promoter strength of the inducible promoters in response to a range of AHL concentrations were calculated from measurements in E. coli and cell-free systems for the LasR responsive promoters (B) pLasRV, (C) pLasR3 and (D) pLasRL. For experiments in E. coli, fluorescence and absorbance at 600 nm were measured in the mid-log phase of growth to calculate the relative promoter strength. Each promoter was normalized to the strength of pLasR3 at 10 µM AHL. The data represent the mean ± the SD of three independent cultivations. For experiments in cell-free systems, 1 µg of plasmid DNA was added to each reaction and fluorescence measured. The relative promoter strength was calculated from the rate of fluorescence increase in the maximal phase of GFP expression, 30–45 min after initiation, and normalized to the strength of pLasR3 at 10 µM AHL. The data represent the mean ± the SD of three independent reactions. (E) Summary of the fitted Hill function equation parameters for the three LasR responsive promoters calculated from in vivo and cell-free systems characterization data.
For detailed explanation of the data analysis for the characterization in live E. coli, please see Supplementary Methods S3. Briefly, the background fluorescence as determined from E. coli containing the pSB1A2:GFPmut3b was removed from all fluorescence measurements. In addition, the O.D. 600 background from media only was removed from all absorbance measurements. For constitutive promoter and RBS libraries, the fluorescence normalized to O.D. 600 measurements was calculated and averaged across the 30 min of measurements. For inducible promoters, the rate of fluorescence increase was calculated when promoter activity was maximal, and these values normalized to O.D. 600 measurements. Each of these values was normalized to a member of the DNA part library to give relative strength: J23101 for the σ70 constitutive promoters, B0034 for the RBS and pLasR3 at 10 µM AHL for the inducible promoters. Finally the average and standard deviation of the three independent repeats were calculated to give final relative strengths.
Characterization in cell-free systems
The E. coli S30 circular extract cell-free system (Promega) was used for experiments in cell-free systems using DNA plasmids templates and the USER–ligase DNA templates. Reactions were performed according to the manufacturer’s protocol. For characterization of constitutive promoter and RBS libraries, a total volume of 25 µl reactions volume was used containing 1 µg of DNA plasmid. For inducible promoters, a total reaction volume of 27.5 µl was used containing 1 µg of DNA plasmid and 0.5 µl of AHL prepared in DMSO. For characterization of USER–ligase DNA products, a total reaction volume of 30 µl was used containing 1 µg of DNA template. For characterization of linear DNA templates, the E. coli S30 linear extract cell-free system (Promega) for linear DNA templates was used in a total reaction volume of 30 µl containing 1 µg of linear DNA template. For every characterization assay, a reaction with the backbone vector containing the GFP gene without a promoter (pSB1A2:GFPmut3b) was measured in parallel to determine background fluorescence. For experiments with linear DNA templates and USER–ligase DNA templates, templates from pSB1A2:GFPmut3b were processed in parallel to the DNA part libraries to generate both linear DNA templates and PCR-generated products for the USER–ligase reactions. All reactions were prepared in a 384-well plate (Griener Bio-One) and transferred to a POLARstar Omega plate reader (BMG Labtech) and fluorescence (excitation at 485 nm, emission at 520 nm) measured for up to 4 h at 15-min intervals at 30°C, shaking at 225 r.p.m. for 10 s before each measurement. The data shown represent the averages of three independent measurements.
For detailed explanation of the data analysis for the characterization in cell-free systems, please see Supplementary Methods S4. Briefly, the background fluorescence from a cell-free reaction with the pSB1A2:GFPmut3b was removed from all fluorescence measurements and the rate of fluorescence increase calculated between 30 and 45 min (Supplementary Figures S1 and S2). These values were normalized to a member of the DNA part library: J23101 for the σ70 constitutive promoters, B0034 for the RBS and pLasR3 at 10 µM AHL for the inducible promoters. Finally, the average and standard deviation of the three independent repeats were calculated to give final relative strengths.
Assembly of DNA templates by USER–ligase reaction
Templates for the coupled USER–ligase reaction were prepared with PfuTurbo Cx (Agilent Genomics). PCR reactions containing 1× PfuTurbo Cx reaction buffer (Agilent Technologies), 5 U of PfuTurbo Cx (Agilent Technologies), 250 µM dNTPs (Roche Applied Science), 100 ng of forward and reverse primers (Supplementary Table S4) and 10 ng of DNA template in a volume of 50 µl. PCR reactions were performed using a protocol of 5 min at 95°C, 10 cycles of 30 s at 95°C, 45 s at 55°C and 1 min at 72°C, followed by 20 cycles of 30 s at 95°C and 2 min at 72°C. A final extension of 20 min at 72°C was performed to ensure fully blunt PCR products. Desired products were purified by either Gel or PCR MinElute extraction kits (Qiagen). All primers used in this study are described in Supplementary Information (Supplementary Table S4).
A typical USER–ligase reaction was performed containing 1 µg of PCR generated product, 1× T4 DNA ligase buffer (New England Biolabs, NEB), 1 U of USER enzyme (NEB) and 1000 U of T4 DNA ligase (NEB) in a volume of 100 µl. The reaction was incubated for 1 h at 37°C. Assembled products were concentrated using MinElute PCR purification columns (Qiagen) and analysed on a 1% agarose gel stained with GelRed (Cambridge Biosciences). Assembled products were identified by digestion with exonuclease III (NEB) performed according to manufacturer’s protocol.
RESULTS
A correlation between the characterization of constitutive DNA regulatory elements in vivo and in cell-free systems
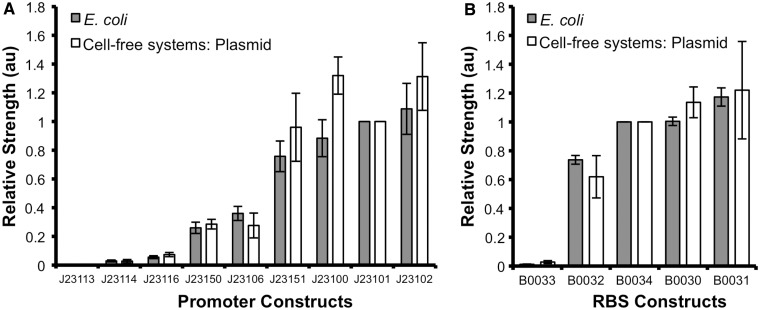
To assess whether the in vivo functionality of gene regulatory elements can be recapitulated in a cell-free system, we tested a routinely used library of minimal σ70 constitutive promoters (5). These promoters have previously been shown to vary in relative promoter strength, with each promoter containing variations within the −10 and −35 hexamer (5,20). Each member of the library was assembled upstream of a GFP reporter gene to enable the indirect quantification of transcription rates and promoter strengths. E. coli cells were transformed with each member of the library and DNA plasmids purified for testing in vivo and in E. coli extract cell-free systems, respectively. GFP expression was determined by fluorescent measurements and promoter strength for each promoter was calculated in exponentially growing E. coli and cell-free systems (please see Material and Methods). Briefly, the GFP expression was determined for each member of the promoter library and normalized to the strength of the promoter J23101.
The minimal σ70 constitutive promoter library was shown to be functional in both live E. coli cells and cell-free systems, able to initiate the transcription of the GFP coding region (Figure 2A). In agreement with previous studies, alterations in the −10 and −35 hexamer sequences of the promoters resulted in divergent levels of transcriptional activity both in vivo and in cell-free systems (Figure 2A and Supplementary Figure S3) (5,20). Interestingly, a comparison of the measurements in cell-free system and exponentially growing E. coli shows a clear correlation (Figure 2A). Although some variation is observed around the stronger promoter strengths such as J23100 and J23102, linear regression of the measurements in cell-free systems and in vivo shows an R2 of 0.946 (Supplementary Figure S4). Comparison of the standard deviation of the relative strength measurements shows a similar level of reproducibility for characterization in cell-free systems and in vivo.
Figure 2.
Characterization of constitutive transcriptional and translational regulatory elements in cell-free systems and E. coli. The relative promoter strength of the (A) minimal σ70 constitutive promoter and (B) RBS libraries were calculated from measurements in E. coli and E. coli extract cell-free systems. For experiments in E. coli, fluorescence and absorbance at 600 nm were measured in the mid-log phase of growth to calculate the relative promoter and RBS strength. Each promoter and RBS was normalized to the strength of the J23101 promoter and B0034 RBS, respectively. The data represent the mean +/− the SD of three independent cultivations. For experiments in cell-free systems, 1 µg of plasmid DNA was added to each reaction and fluorescence measured. The relative promoter and RBS strength were calculated from the rate of fluorescence increase in the maximal phase of GFP expression, 30–45 min after initiation, and normalized to the strength of the J23101 promoter and B0034 RBS, respectively. The data represent the mean +/− the SD of three independent reactions.
To test whether this correlation is observed for translational regulatory elements, a library of RBS expressing GFP coding region was placed downstream of the σ70 constitutive promoter J23101. Each member of the RBS library contained variations in the Shine-Dalgarno (SD) sequence and length that directs recruitment of ribosomes to messenger RNA (mRNA) and has been shown to be a key determinant of the rate of translation (21). Each member of the library was transformed into E. coli cells and DNA plasmids purified for testing in vivo and in cell-free systems, respectively, and the relative RBS strength determined (see Materials and Methods). Here we show that the RBS library is functional in both E. coli and in cell-free systems, initiating the translation of constitutively transcribed GFP mRNA (Figure 2B). We also show that variations in the SD sequence result in divergent strengths of translation both in cell-free systems and in vivo (Figure 2B). Comparison of the relative RBS strengths measured in exponentially growing E. coli and in a cell-free system also shows a clear correlation, with linear regression of the measurements showing an R2 of 0.968 (Supplementary Figure S4). As before, the standard deviation of the relative strength measurements in cell-free systems and exponentially growing E. coli are comparable.
These results clearly demonstrate that despite the significant contextual differences between E. coli and E. coli extract cell-free system, simple constitutive DNA regulatory elements can have the same relative effect on the strength of transcription and translation.
A correlation between characterization of inducible promoters in vivo and in cell-free systems
In genetic circuits, inducible promoters are essential to enable the control of both the level and the dynamics of gene expression by the addition of external inducer signals and are routinely used as nodes to interface genetic devices for engineering complex biological systems, such as multi-cellular communication systems and edge detection (22,23). Dose-response curves have been an effective method to describe and enable the prediction of the input–output characteristics of inducible promoters (8,10). To determine whether the dose-response curves of inducible promoters can be measured in cell-free systems, and whether this would correlate to in vivo measurements, we studied the family of LasR-regulated promoters.
The LasR promoter family is involved in the quorum sensing control of gene regulation in Pseudomonas aeruginosa, where binding of a small ligand AHL to the LasR transcription factor positively up-regulates transcription of LasR responsive promoters (24). The three LasR responsive promoters of PA3904 (pLasR3), VqsR (pLasRV) and LasB (pLasRL) genes were constructed upstream of a GFP coding region and downstream of constitutively expressed LasR transcription factor (Figure 3A) (25–27). E. coli cells were transformed with each member of the library and DNA plasmids purified for testing in vivo and in E. coli extract cell-free systems, respectively. The relative promoter strength was determined across a range of AHL concentrations in exponentially growing E. coli and E. coli extract cell-free system (Figure 3B–D). All three LasR responsive promoters activated transcription of GFP in response to AHL, demonstrating that both the LasR transcription factor and LasR responsive promoters are compatible with the endogenous transcriptional machinery of E. coli. The in vivo and in vitro data were fitted with a Hill function equation (Supplementary Methods S5) to gain an approximation of the dose-response behaviour of the promoters (Figure 3B–D). A comparison of the fitted parameters shows a strong correlation for the calculated maximal transcriptional output of the three LasR responsive promoters when measured both in exponentially growing E. coli and in a cell-free system (Figure 3E). The cooperativity (n) of the three promoters also generally correlates with the pLasRL showing a higher level of cooperativity than pLasRV and pLasR3 both in vitro and in vivo (Figure 3E). The determined binding efficiency shows less correlation, with binding efficiency values measured in a cell-free system higher than in vivo, with the pLasRL showing the largest difference of 21 nM. Binding efficiencies from different inducible promoters in E. coli have been shown to range from nM to mM (three orders of magnitude) (28). Because the difference we observed in binding efficiencies between in vivo and in vitro measurements are within <1 order of magnitude, we believe that cell-free systems provide a good approximation of the observed behaviour in vivo (28). These results show that the in vivo dose-responses of inducible promoters can be effectively determined in a cell-free system.
Characterization using linear DNA templates in cell-free systems
The study of genetic circuits and DNA regulatory elements in E. coli extract cell-free systems have to date been limited to the use of DNA plasmid amplified and purified from E. coli. Therefore, the use of in vitro-generated templates could increase throughput of studies in cell-free systems by omitting time-consuming transformation and plasmid purification procedures. Although linear DNA templates obtained from PCR have been successfully used in cell-free systems to express proteins for structural and functional studies, it remains unclear whether these DNA templates would be suitable for the study of DNA regulatory elements (29,30).
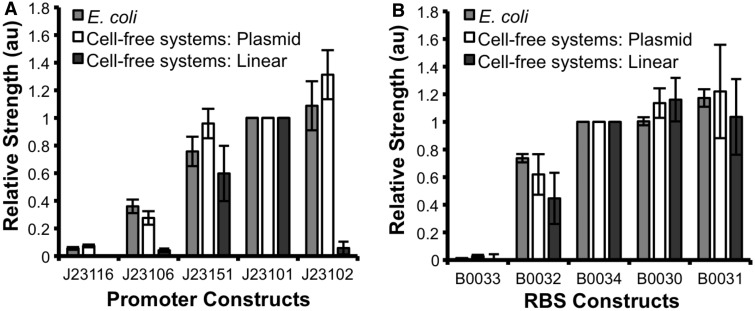
Five members of the σ70 constitutive promoter and RBS library as described above were amplified by PCR and these linear DNA templates tested in a cell-free system. Initial experiments with linear DNA templates showed no detectable expression of GFP across the promoter library. It was thought that the linear DNA templates were unstable in the cell-free system, being rapidly degraded before significant GFP expression occurred. Three modifications were made to increase stability of the DNA templates: the use of modified cell-free systems that lack major proteases and exonucleases (linear E. coli extract); biotinylatation of the 5′-end of DNA to prevent degradation by exonucleases and introduction of 500-bp buffer region to limit degradation of DNA regulatory elements and GFP coding region. In addition, the 500-bp buffer region was from the vector backbone used for in vivo experiments and so ensures that any upstream sequences that might contribute to the strength of downstream regulatory elements are included within the DNA templates. These modifications successfully increased GFP expression to detectable levels, allowing relative promoter and RBS strengths to be calculated as before (Supplementary Figure S5).
The relative strengths calculated from linear DNA templates were compared with the relative strengths determined before in vivo and in cell-free systems using purified DNA plasmids (Figure 4A and B). It can be seen that the relative promoter strength calculated from cell-free systems using linear DNA templates shows no correlation to either the relative strength calculated in vivo or in cell-free systems using DNA plasmids (Figure 4A). While J23151 and J23101 relative promoter strengths match, the other three members of the promoter library repeatedly showed no significant expression of GFP. However, comparison of the relative RBS strengths from the linear DNA templates in cell-free systems does show a correlation to both the characterization in vivo and in cell-free systems using DNA plasmids (Figure 4B). This suggests that linear DNA templates generated as described here are suitable to determine the relative strength of translational regulatory elements but not transcriptional regulatory elements. Although the reason for this is unknown, previous literature has documented a sequence-dependent synergism between DNA template conformation (for example, supercoiling) and optimal transcription activity of promoters in vitro (31–33). As all transcriptional initiation requires unwinding of the DNA double helix to form an open complex between the RNA polymerase and a locally denatured single-stranded DNA segment within the promoter region, it is possible that sequence determinants of promoters have a different relative effect on transcription when present on linear DNA templates (31). Such an effect would not have been seen for the RBS library because all contain the same promoter and should be uniformly transcribed. Furthermore, the translation of mRNA is determined by the RBS sequence and mRNA structure, thus is independent of DNA template structure.
Figure 4.
Characterization of minimal σ70 constitutive promoters by linear PCR products in cell-free systems. The relative promoter strength of five (A) σ70 constitutive promoters and (B) RBS were calculated using linear DNA templates in a modified E. coli extract cell-free systems optimized for linear DNA templates. For experiments, 1 µg of biotinylated linear templates containing a buffer region (linear) was added to each reaction and fluorescence measured. The relative promoter strength was calculated from the rate of fluorescence increase in the maximal phase of GFP expression, 30–45 min after initiation, and normalized to the strength of the J23101 promoter and B0034 RBS. The data represent the mean +/− the SD of three independent reactions. The relative strengths were compared with the relative strengths calculated from experiments in E. coli and E. coli extract cell-free systems using DNA plasmids.
We, therefore, aimed to explore alternative in vitro templates that would be functional for both transcriptional and translation regulatory elements. Comparison of the physical difference between non-functional linear DNA templates and functional DNA plasmids suggests that DNA template should be circular to be effectively coupled for characterization in cell-free systems.
The in vitro generation of closed-circular DNA templates for characterization in cell-free systems
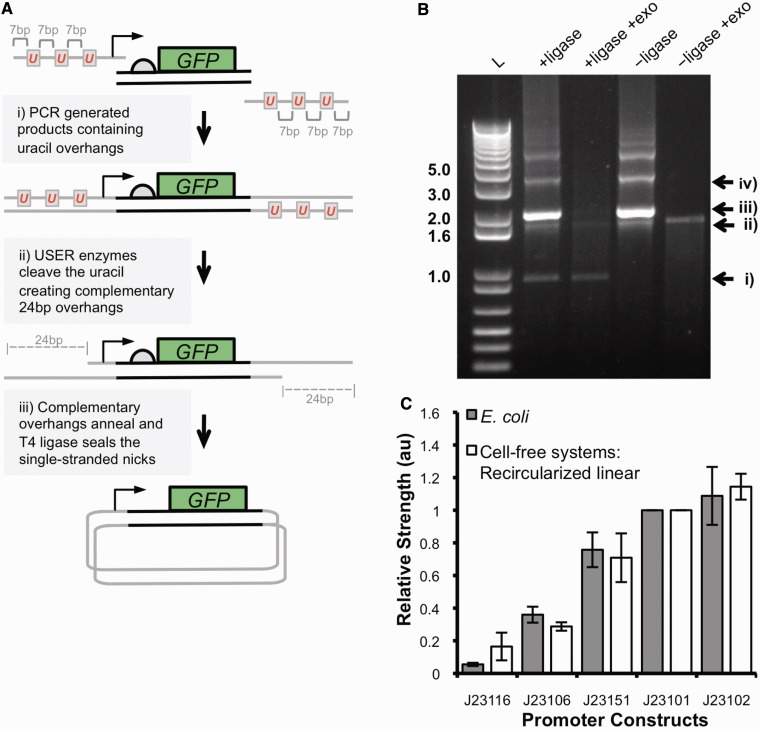
To develop an efficient assembly strategy for generating testable DNA constructs for cell-free systems, a number of different protocols were explored. The aim was to use PCR to generate and amplify DNA regulatory elements upstream of a GFP reporter gene, and either during PCR amplification or by post-processing, ligate and circularize the linear PCR product. An important consideration is the yield of closed-circular product, as a typical cell-free reaction requires 0.2–1 µg of DNA template. We tested multiple assembly methods using PCR products including blunt-end ligations, restriction enzyme digestion and ligation, ligation-dependent amplification PCR and circular polymerase extension cloning (34,35). Although these methods were successful in generating closed-circular DNA products, yields were too low to couple to expression in cell-free systems. To overcome this, we developed a modified USER and T4 DNA ligase (USER–ligase) protocol (see Materials and Methods) where multiple PCR products can be covalently joined by exploiting the incorporation and excision of uracil to create complementary single-stranded overhangs (36). We used longer complementary overhangs of 24 bp containing three uracil nucleotides 7-bp apart (Figure 5A). These overhangs are designed so that once uracil nucleotides have been excised, the remaining 7-bp oligomers (Tm of 22°C) should rapidly dissociate, leaving the 24-bp single-stranded overhangs (Tm of 65°C) to efficiently anneal and ligate.
Figure 5.
(A) A schematic of the USER–ligase assembly strategy. (i) PCR is performed with PfuTurbo Cx DNA polymerase to amplify DNA regulatory elements upstream of a GFP coding region with overhangs containing three uracil nucleotides 7-bp apart. The PCR product is purified, and a USER–ligase reaction is performed. (ii) The USER enzymes excise uracil nucleotides creating 24-bp complementary single-stranded overhangs that anneal. (iii) T4 DNA ligase forms a phosphodiester at the single-stranded nicks of the annealed overhangs, forming closed-circular DNA. (B) Products and digestion of a USER–ligase and USER-only reaction analysed on GelRed strained agarose gel. L: 1 kb plus DNA ladder; + ligase and − ligase indicate the presence or absence of T4 DNA ligase in the USER reaction; + exo indicates where digestion by exonuclease III has been performed. Enzymatic digestions identify the products of the USER–ligase reaction as follows: (i) closed-circular DNA, (ii) linear DNA products, (iii) annealed DNA containing single-stranded nicks and (iv) concatemers from ligation of multiple PCR products. + ligase and − ligase lanes contain 300 ng of DNA products. Enzymatic digestion with exonuclease III contained 300 ng of DNA products. (C) The relative promoter strength of five minimal σ70 constitutive promoters was calculated from cell-free systems using 1 µg of USER–ligase products for each reaction and fluorescence measured. The relative promoter strength was calculated from the rate of fluorescence increase in the maximal phase of GFP expression, 30–45 min after initiation, and normalized to the strength of the J23101 promoter. The data represent the mean +/− the SD of three independent reactions. The relative promoter strength was compared with the relative strengths calculated from experiments in E. coli.
To test our intramolecular ligation strategy, a constitutive promoter GFP coding region was amplified with PfuTurbo Cx DNA polymerase. A USER–ligase and a USER-only reaction were performed on the PCR-generated substrate (Supplementary Figure S6). Analysis showed the formation of a ligase-dependent product thought to be closed-circular DNA (Figure 5B). Digestion by exonuclease III, which is unable to initiate digestion of closed-circular DNA, confirmed that this ligase-dependent product was a closed-circular DNA product (Figure 5B). No further conversion of annealed DNA product to closed-circular DNA was achieved by either blunting of PCR products by T4 DNA polymerase or by increasing T4 ligase and USER concentration. The entire products from the USER–ligase and USER-only reactions were tested in the E. coli extract cell-free system (Supplementary Figure S7). It was shown that only the USER–ligase reaction products containing the closed-circular DNA product expressed detectable levels of GFP. As before, linear DNA templates in this unmodified E. coli extract showed no significant GFP expression (Supplementary Figures S5 and S7), suggesting that the linear DNA templates do not contribute to the observed GFP expression.
We next tested whether this assembly method would be suitable for generating libraries of DNA regulatory elements for characterization in a cell-free system. Five members of the σ70 constitutive promoter library expressing the GFP coding region were amplified by PCR, circularized by the USER–ligase reaction and tested in cell-free systems. Again, the relative promoter strengths measured across the library were compared with the data collected in vivo (Figure 5C). It can be seen that the relative strengths measured in vitro using the USER–ligase assembled templates show a correlation to the relative strengths measured in vivo. In addition, the efficiency of conversion from linear to closed-circular was shown to be reproducible and independent of the DNA part assembled. These results show that the USER–ligase method can be used to generate functional DNA templates for the characterization of DNA regulatory elements in cell-free systems, the results of which are comparable with characterization in vivo.
DISCUSSION
To develop a completely in vitro approach for the characterization of DNA regulatory elements in a cell-free system, we firstly validated the correlation between characterization measurements in cell-free systems using DNA plasmids with that obtained in exponentially growing E. coli for the most commonly used types of DNA regulatory elements. Secondly, we validated a completely in vitro method to generate DNA templates bypassing the use of living cells for the amplification and purification of DNA plasmids.
In our study, we found a clear relevance for the in vitro and in vivo measurements for constitutive DNA transcription and translation control elements. Comparison of relative strengths of σ70 promoters and RBS showed a clear correlation between the relative strengths in E. coli and E. coli extract cell-free system using DNA plasmids purified from E. coli. Furthermore, the dose-response curves of inducible promoters were also shown to be comparable between E. coli and cell-free systems using DNA plasmids. To the authors knowledge, this is the first demonstration of a correlation between E. coli extract cell-free systems and live E. coli for the characterization of the DNA regulatory elements: σ70 constitutive promoters, RBS and inducible promoters. This correlation might be surprising when considering the different environmental contexts in which the transcriptional and translational events occur in E. coli and in cell-free systems. The concentration of cytosolic proteins, including the transcriptional and translational machinery, are typically 10-fold lower in cell-free systems than E. coli (37). Indeed, it has been shown that the absolute rates of translation initiation and elongation reflect this and are an order of magnitude lower in cell-free systems than in vivo (38,39). In addition, the growth and division of cells introduce dynamics into gene expression that are not present in cell-free systems. For example, extensive transcription of DNA plasmids in E. coli has been shown to reduce the plasmid copy number per cell and eventually leads to the loss of plasmid in a population (40,41). Finally, cell-free systems are void of genomic DNA and expression, and contain only a basic metabolism compared with live cells. Nevertheless, our data shows that for simple DNA regulatory elements, a clear correlation is observed between in vitro and in vivo measurements.
To establish a semi high-throughput platform for DNA part characterization, we investigated alternatives to plasmid DNA templates derived from E. coli. We demonstrate that the characterization of σ70 promoters in cell-free systems with linear DNA templates would not be suitable if we are correlating in vitro with in vivo measurements. Further modifications of DNA linear templates not considered here could enable characterization in cell-free systems although we were unable to identify such modifications. Therefore we aimed to devise a method to generate closed-circular DNA templates for characterization of DNA regulatory elements in cell-free systems. As PCR has been used successfully to generate diverse libraries of DNA parts, we aimed to validate a two-step assembly strategy, where PCR is used to generate diverse libraries of DNA parts and then circularized to functionalize them for characterization in a cell-free system. The circularization was achieved by adapting a USER–ligase assembly method that was shown to efficiently drive the intramolecular ligation of linear PCR product to form closed-circular DNA. Our comparison of five promoters amplified by PCR and circularized using USER–ligase method showed reproducible measurements of relative promoter strengths in a cell-free system that correlated with the relative strengths in vivo. We also found that no further purification of the closed-circular DNA product from the other non-desired products of the USER–ligase reaction is required, offering the potential for coupling to high-throughout platforms.
Although the PCR-generated products used for the USER–ligase reaction were amplified from DNA plasmids, the USER–ligase methodology described here could be adapted for in vitro high-throughput DNA library generation. The only prerequisite is that the DNA templates contain complementary overhangs and uracil nucleotides, which can be easily added by PCR amplification. As such, multiple methods to generate libraries of DNA parts can be used as an entry point into the USER–ligase reaction. For example, constitutive DNA regulatory elements are typical short DNA sequences and can be added to reporter genes by PCR. Use of primers containing randomized DNA sequences can be used to create DNA part libraries of diverse sequence and behaviour. For inducible regulatory elements or genetic devices, combinations of DNA parts are required to be assembled for overall behaviour. De novo synthesis or genomic templates can be used in a metagenomic approach to explore diverse sources of genes and DNA regulatory elements. Moreover, utilization of assembly methods such as Gibson assembly would enable rapid multiple DNA part assembly in a single reaction, the products of which can be used as templates for PCR to amplify and add the USER overhangs (42).
The study described here does not represent an exhaustive study of all sequence determinants of DNA regulatory elements; for example, the strength of constitutive promoters is known to be influenced by other sequence determinants such as promoter clearance, proximal supercoiling, UP elements and the sequence and length of the spacer region between the −10 and −35 hexamer (33,43–47). Some determinants such as the abortive transcription of promoter have previously been shown to be comparable between in vivo and in vitro transcription-only reactions (48). Further work is required to determine whether the correlation between characterization measurements in vivo and in cell-free systems is maintained when other more complex sequence determinants of DNA regulatory elements are tested. However, our study does provide compelling evidence that relative activity measurements can be correlated between in vivo and in vitro systems for commonly used DNA regulatory elements in synthetic biology design.
Furthermore, it would be of interest to determine at what complexity of genetic devices this correlation between in vivo and in vitro breaks down. As complexity increases, the number of parameters that effect overall behaviour increases, for example, the rate of transcription and translation, the rate of mRNA and protein degradation, the rate of protein folding and the kinetics of DNA-binding events. Conceivably, to replicate genetic device behaviour in cell-free systems would require these parameters to be carefully balanced. Although a non-trivial task, important progression to controlling parameters such as protein and mRNA degradation has been achieved in cell-free systems (19).
An important advantage of in vitro characterization approach described here is that it is directly adaptable to high-throughput platforms such as microfluidics (49,50). Microfluidic devices have successfully compartmentalized both PCR amplification of DNA and kinetic quantification of in vitro gene expression, offering the potential to develop lab-on-chip devices integrating both generation and characterization of DNA part libraries (49,50). Such techniques would allow thousands of discrete reactions to be homogenously formed, stored and measured from only a few microlitres of the cell-free systems, reducing the cost of such measurements (51). Furthermore, in theory, cell-free systems should provide a more reproducible platform for gene expression measurements because many sources of extrinsic noise that contribute to the overall stochasticity of gene expression are removed. Although we observed comparable reproducibility between in vivo and in vitro measurements of gene expression, the use of defined cell-free systems such as PURE systems composed of purified transcription and translation proteins could provide an alternative cell-free system with increased reproducibility (52). Although PURE systems are currently costly and time-consuming to prepare, the development of genome engineering tools such as Multiplex Automated Genome Engineering (MAGE) have enabled efficient and rapid reconstruction of PURE systems and will soon offer a practical alternative to extract-based cell-free systems (53).
In summary, the work described here provides the first evidence that DNA regulatory element behaviour in cell-free systems is reflective of behaviour in vivo. Moreover, we have validated a completely in vitro approach to generating and characterizing DNA regulatory elements in cell-free systems. By avoiding the use of living systems for either characterization measurements or DNA template generation, this approach is significantly quicker than current approaches and provides a valuable tool to rapidly annotate libraries of characterized DNA parts for synthetic biology design.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–7 and Supplementary Methods.
FUNDING
The Centre for Synthetic Biology and Innovation (CSynBI) is sponsored by Engineering and Physical Sciences Research Council (EPSRC). Funding for open access charge: Engineering and Physical Sciences Research Council [EP/G036004/1]; Center for Synthetic Biology and Innovation at Imperial College London. J.C. was supported by a Medical Research Council (MRC) studentship.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
J.C. and P.S.F. would like to thank the Medical Research Council (MRC) for PhD bursarship. P.S.F. acknowledges support from the Engineering and Physical Sciences Research Council (EPSRC) for The Centre for Synthetic Biology and Innovation (CSynBI). They would like to thank all our colleagues in CSynBI and in particular Geoff Baldwin, Arturo Casini, Tom Ellis, Vincent Rouilly and Richard Kitney for their support and input.
REFERENCES
- 1.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 2.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100073. 2006.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald JT, Barnes C, Kitney RI, Freemont PS, Stan GB. Computational design approaches and tools for synthetic biology. Integr. Biol. (Camb) 2011;3:97–108. doi: 10.1039/c0ib00077a. [DOI] [PubMed] [Google Scholar]
- 4.Kwok R. Five hard truths for synthetic biology. Nature. 2010;463:288–290. doi: 10.1038/463288a. [DOI] [PubMed] [Google Scholar]
- 5.Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, de Mora K, Glieberman AL, Monie DD, Endy D. Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Eng. 2009;3:4. doi: 10.1186/1754-1611-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blount BA, Weenink T, Vasylechko S, Ellis T. Rational diversification of a promoter providing fine-tuned expression and orthogonal regulation for synthetic biology. PLoS One. 2012;7:e33279. doi: 10.1371/journal.pone.0033279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu TK. Engineering scalable biological systems. Bioeng. Bugs. 2010;1:378–384. doi: 10.4161/bbug.1.6.13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin L, Che A, Endy D. Gemini, a bifunctional enzymatic and fluorescent reporter of gene expression. PLoS One. 2009;4:e7569. doi: 10.1371/journal.pone.0007569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 11.Harris DC, Jewett MC. Cell-free biology: exploiting the interface between synthetic biology and synthetic chemistry. Curr Opin Biotechnol. 2012;23:672–678. doi: 10.1016/j.copbio.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgman CE, Jewett MC. Cell-free synthetic biology: thinking outside the cell. Metab. Eng. 2012;14:261–269. doi: 10.1016/j.ymben.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simpson ML. Cell-free synthetic biology: a bottom-up approach to discovery by design. Mol. Syst. Biol. 2006;2:69. doi: 10.1038/msb4100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster AC, Church GM. Synthetic biology projects in vitro. Genome Res. 2007;17:1–6. doi: 10.1101/gr.5776007. [DOI] [PubMed] [Google Scholar]
- 15.Karig DK, Iyer S, Simpson ML, Doktycz MJ. Expression optimization and synthetic gene networks in cell-free systems. Nucleic Acids Res. 2012;40:3763–3774. doi: 10.1093/nar/gkr1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noireaux V, Bar-Ziv R, Libchaber A. Principles of cell-free genetic circuit assembly. Proc. Natl Acad. Sci. USA. 2003;100:12672–12677. doi: 10.1073/pnas.2135496100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin J, Noireaux V. An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 2012;1:29–41. doi: 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
- 18.Shin J, Noireaux V. Efficient cell-free expression with the endogenous E. coli RNA polymerase and sigma factor 70. J. Biol. Eng. 2010;4:8. doi: 10.1186/1754-1611-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin J, Noireaux V. Study of messenger RNA inactivation and protein degradation in an Escherichia coli cell-free expression system. J. Biol. Eng. 2010;4:9. doi: 10.1186/1754-1611-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis JH, Rubin AJ, Sauer RT. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 2011;39:1131–1141. doi: 10.1093/nar/gkq810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shultzaberger RK, Bucheimer RE, Rudd KE, Schneider TD. Anatomy of Escherichia coli ribosome binding sites. J. Mol. Biol. 2001;313:215–228. doi: 10.1006/jmbi.2001.5040. [DOI] [PubMed] [Google Scholar]
- 22.Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 24.Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 2006;30:274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 25.Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl Acad. Sci. USA. 2004;101:15833–15839. doi: 10.1073/pnas.0407229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juhas M, Wiehlmann L, Huber B, Jordan D, Lauber J, Salunkhe P, Limpert AS, von Götz F, Steinmetz I, Eberl L, et al. Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa. Microbiology. 2004;150 (Pt 4):831–841. doi: 10.1099/mic.0.26906-0. [DOI] [PubMed] [Google Scholar]
- 27.Rust L, Pesci EC, Iglewski BH. Analysis of the Pseudomonas aeruginosa elastase (lasB) regulatory region. J. Bacteriol. 1996;178:1134–1140. doi: 10.1128/jb.178.4.1134-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J. Biol. Eng. 2011;5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 30.Rungpragayphan S, Nakano H, Yamane T. PCR-linked in vitro expression: a novel system for high-throughput construction and screening of protein libraries. FEBS Lett. 2003;540:147–150. doi: 10.1016/s0014-5793(03)00251-5. [DOI] [PubMed] [Google Scholar]
- 31.Travers A, Muskhelishvili G. DNA supercoiling - a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 32.Meiklejohn AL, Gralla JD. Activation of the lac promoter and its variants. Synergistic effects of catabolite activator protein and supercoiling in vitro. J. Mol. Biol. 1989;207:661–673. doi: 10.1016/0022-2836(89)90236-2. [DOI] [PubMed] [Google Scholar]
- 33.Borowiec JA, Gralla JD. All three elements of the lac ps promoter mediate its transcriptional response to DNA supercoiling. J. Mol. Biol. 1987;195:89–97. doi: 10.1016/0022-2836(87)90329-9. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Ruffner DE. Amplification of closed circular DNA in vitro. Nucleic Acids Res. 1998;26:1126–1127. [PMC free article] [PubMed] [Google Scholar]
- 35.Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein V, Hollfelder F. An efficient method to assemble linear DNA templates for in vitro screening and selection systems. Nucleic Acids Res. 2009;37:e122. doi: 10.1093/nar/gkp589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noireaux V, Maeda YT, Libchaber A. Development of an artificial cell, from self-organization to computation and self-reproduction. Proc. Natl Acad. Sci. USA. 2011;108:3473–3480. doi: 10.1073/pnas.1017075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iskakova MB, Szaflarski W, Dreyfus M, Remme J, Nierhaus KH. Troubleshooting coupled in vitro transcription-translation system derived from Escherichia coli cells: synthesis of high-yield fully active proteins. Nucleic Acids Res. 2006;34:e135. doi: 10.1093/nar/gkl462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Underwood KA, Swartz JR, Puglisi JD. Quantitative polysome analysis identifies limitations in bacterial cell-free protein synthesis. Biotechnol. Bioeng. 2005;91:425–435. doi: 10.1002/bit.20529. [DOI] [PubMed] [Google Scholar]
- 40.Popov M, Petrov S, Nacheva G, Ivanov I, Reichl U. Effects of a recombinant gene expression on ColE1-like plasmid segregation in Escherichia coli. BMC Biotechnol. 2011;11:18. doi: 10.1186/1472-6750-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stueber D, Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1:1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 43.Estrem ST, Gaal T, Ross W, Gourse RL. Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl Acad. Sci. USA. 1998;95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoyama T, Takanami M. Supercoiling response of E. coli promoters with different spacer lengths. Biochim. Biophys. Acta. 1988;949:311–317. doi: 10.1016/0167-4781(88)90157-1. [DOI] [PubMed] [Google Scholar]
- 45.Mulligan ME, Brosius J, McClure WR. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J. Biol. Chem. 1985;260:3529–3538. [PubMed] [Google Scholar]
- 46.Hook-Barnard IG, Hinton DM. The promoter spacer influences transcription initiation via sigma70 region 1.1 of Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA. 2009;106:737–742. doi: 10.1073/pnas.0808133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu M, Tolstorukov M, Zhurkin V, Garges S, Adhya S. A mutant spacer sequence between -35 and -10 elements makes the Plac promoter hyperactive and cAMP receptor protein-independent. Proc. Natl Acad. Sci. USA. 2004;101:6911–6916. doi: 10.1073/pnas.0401929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldman SR, Ebright RH, Nickels BE. Direct detection of abortive RNA transcripts in vivo. Science. 2009;324:927–928. doi: 10.1126/science.1169237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Courtois F, Olguin LF, Whyte G, Bratton D, Huck WT, Abell C, Hollfelder F. An integrated device for monitoring time-dependent in vitro expression from single genes in picolitre droplets. Chembiochem. 2008;9:439–446. doi: 10.1002/cbic.200700536. [DOI] [PubMed] [Google Scholar]
- 50.Wu N, Zhu Y, Brown S, Oakeshott J, Peat TS, Surjadi R, Easton C, Leech PW, Sexton BA. A PMMA microfluidic droplet platform for in vitro protein expression using crude E. coli S30 extract. Lab Chip. 2009;9:3391–3398. doi: 10.1039/b911581a. [DOI] [PubMed] [Google Scholar]
- 51.Gulati S, Rouilly V, Niu X, Chappell J, Kitney RI, Edel JB, Freemont PS, deMello AJ. Opportunities for microfluidic technologies in synthetic biology. J. R. Soc. Interface. 2009;6(Suppl. 4):S493–S506. doi: 10.1098/rsif.2009.0083.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 53.Wang HH, Huang PY, Xu G, Haas W, Marblestone A, Li J, Gygi S, Forster AC, Jewett MC, Church GM. Multiplexed in vivo his-tagging of enzyme pathways for in vitro single-pot multienzyme catalysis. ACS Synth. Biol. 2012;1:43–52. doi: 10.1021/sb3000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.