Abstract
Thiamin pyrophosphate (TPP) riboswitches are found in organisms from all three domains of life. Examples in bacteria commonly repress gene expression by terminating transcription or by blocking ribosome binding, whereas most eukaryotic TPP riboswitches are predicted to regulate gene expression by modulating RNA splicing. Given the widespread distribution of eukaryotic TPP riboswitches and the diversity of their locations in precursor messenger RNAs (pre-mRNAs), we sought to examine the mechanism of alternative splicing regulation by a fungal TPP riboswitch from Neurospora crassa, which is mostly located in a large intron separating protein-coding exons. Our data reveal that this riboswitch uses a long-distance (∼530-nt separation) base-pairing interaction to regulate alternative splicing. Specifically, a portion of the TPP-binding aptamer can form a base-paired structure with a conserved sequence element (α) located near a 5′ splice site, which greatly increases use of this 5′ splice site and promotes gene expression. Comparative sequence analyses indicate that many fungal species carry a TPP riboswitch with similar intron architecture, and therefore the homologous genes in these fungi are likely to use the same mechanism. Our findings expand the scope of genetic control mechanisms relying on long-range RNA interactions to include riboswitches.
INTRODUCTION
Riboswitches are metabolite-binding elements that usually are located in noncoding regions of mRNA where they regulate gene expression by conformation changes brought about by binding target ligands (1–6). To date, more than two dozen riboswitch classes, mostly present in diverse lineages of bacteria, have been experimentally validated (7). Each riboswitch representative has sequence and structural features that define its ligand-binding aptamer domain and these features are used to define a riboswitch class. Riboswitch aptamers exhibit far greater conservation than their associated expression platform domains that more directly regulate gene expression (Supplementary Figure S1). In part, this is due to the diverse mechanisms that can be used by riboswitches to control gene expression (6).
Riboswitches that bind the coenzyme thiamin pyrophosphate (TPP) (8–12) are among the most numerous and widespread of all riboswitch classes. TPP riboswitch aptamers are readily identified by using bioinformatics algorithms that seek regions corresponding to the characteristic sequence and structural features of this aptamer class (13,14). These searches have revealed representatives in organisms from all three domains of life, where they commonly control the expression of genes associated with thiamin biosynthesis and metabolite transport. The mechanisms of gene regulation used by TPP riboswitch expression platforms vary greatly depending on the species and the gene contexts in which they are located (15–17). Bacterial TPP riboswitches regulate gene expression almost exclusively via the control of transcription elongation or translation initiation (15) (Supplementary Figure S1), although riboswitch regulatory mechanisms can be far more diverse and intricate (18–21).
Eukaryotic TPP riboswitches typically reside within introns of pre-mRNAs and therefore must make use of regulatory mechanisms that are different than those observed in bacteria. Several recent studies have been conducted to examine the molecular mechanisms of individual TPP riboswitches in fungi, plants and algae (Supplementary Figure S2) (22–26). For example, TPP-mediated regulation of the NMT1 gene in N. crassa involves the ligand-mediated control of alternative aptamer folding to mask or reveal an alternative 5′ splice site in an intron located upstream of the main open reading frame (ORF) (23). One splice product generated when TPP is abundant carries short upstream ORFs (uORFs), which preclude expression of the main ORF. In many plant species, a TPP riboswitch in THIC mRNA controls splicing at a single 5′ splice site to either remove or retain an intron (24,25). Intron removal alters mRNA stability and expression by removing a key processing signal in the 3′ untranslated region (3′ UTR) of the transcript. In the green algae Chlamydomonas reinhardtii, a TPP riboswitch in a THIC intron controls excision of a stop codon that otherwise would yield only truncated THIC protein (26).
The studies noted above begin to reveal the diversity of processes that riboswitches can influence by regulating alternative splicing. Although many molecular mechanisms by which riboswitches could control splicing seem possible, only two mechanisms currently are known to be used. One TPP riboswitch mechanism observed in plants and fungi (23,24) exploits nucleotides of the pyrophosphate-binding domain (pairing elements P4 and P5) (8) to form an alternative base-paired structure that masks a 5′ splice site of an intron (Supplementary Figure S2A and B). Thus, when TPP is absent, the P4-P5 region base pairs to nucleotides flanking the 5′ splice site rather than forming the structure needed to bind the pyrophosphate moiety of the coenzyme. Another mechanism observed in C. reinhardtii (26) involves the alternative folding of the 5′ portion of the P1 stem, which base pairs with a neighboring region to sequester a 3′ splice site, thus resulting in the use of a different 3′ splice site (Supplementary Figure S2C).
In the fungal species N. crassa, the NCU01977 gene encodes a predicted transporter protein whose coding region is interrupted by a long intron carrying a TPP riboswitch. This intron-riboswitch architecture is also found in the homologous genes of many distant fungal relatives, suggesting that gene expression is controlled by TPP levels via a conserved alternative splicing mechanism. Indeed, thiamin-dependent alternative splicing products have been observed when N. crassa was grown in the absence versus the presence of thiamin (23). In the current study, we examined the function of this TPP riboswitch in greater detail to determine if the RNA regulates alternative splicing using a mechanism that is distinct from those published previously. Our findings reveal that this TPP riboswitch regulates alternative splicing by forming long-distance base-paired structures involving nucleotides close to (but not overlapping) the 5′ splice site. This base-pairing interaction, which is conserved among many fungal species, favors complete removal of the large intron possibly by bringing the 3′ splice site into proximity with a distant 5′ donor site, while looping out internal 5′ splice sites that would otherwise be used (27,28). These results and other recent findings (29,30) describing conserved RNA structures within introns suggest that long-distance base pairing to form stable secondary or tertiary structures may be commonly used to regulate alternative splicing.
MATERIALS AND METHODS
DNA oligonucleotides and chemicals
All primers (Supplemental Table S1) were purchased from Sigma-Aldrich. Thiamin and l-histidine were purchased from Sigma-Aldrich. [γ-32P] ATP was purchased from Perkin Elmer.
Plasmids and strains
Plasmid pLL07 (provided by the laboratory of J. C. Dunlap) (31), which carries a luciferase (LUC) reporter gene, was mutated to disrupt the LUC start codon and to insert an XbaI restriction site by using a QuickChange XL Site-Directed Mutagenesis Kit (Stratagene) (23). The promoter for the β-tubulin gene in N. crassa was amplified by polymerase chain reaction (PCR) and inserted in front of the LUC gene as described previously (23) to obtain the plasmid pLL07-2-1. To create in-frame fusions of the LUC reporter to the NCU01977 ORF downstream of intron 2, DNA including 82-nt upstream of the predicted start codon was amplified by PCR from N. crassa genomic DNA and subcloned into a pCR2.1-TOPO vector. After confirmation by sequencing (Keck Foundation Biotechnology Resource Center at Yale University), the inserted fragment was digested with EcoRI and XbaI (NEB) and purified by using a QIAquick Gel Extraction Kit (Qiagen). The purified fragment was inserted into pLL07-2-1 at the EcoRI and XbaI sites. The construct was confirmed by PCR and sequencing. Site-directed mutagenesis and two-step PCR were used to make mutations with the appropriate primers (Supplemental Table S1). Similarly, the mutated fragments were inserted into pLL07-2-1 and confirmed by PCR and sequencing. The constructed plasmids were transformed into N. crassa 87-74 (bd; fqr+ a; his-3) (32) for gene expression assays.
N. crassa transformation and luciferase assay
Electroporation transformation was performed as previously described (33,34) with a few modifications. After electroporation, 1 ml of recovery medium (1× Vogel’s salt, 18.2% sorbitol supplemented with 2% sorbose, 0.05% fructose and 0.05% glucose) was added to the cuvettes and the suspensions were transferred to eppendorf tubes for shaking at 200 rpm for 1 h at 30°C. After incubation, 200 μl of each suspension was mixed with plating medium (recovery medium supplemented with 2.8% agar). The transformants were picked after 3–5 days of incubation at 30°C.
For homokaryon isolation (35), a single colony was picked either from uninucleate microconidia, which were prepared as previous description (36) or from streaked cultures. Insertions of the target genes in the transformants were verified by PCR and sequencing. Transformants were cultured in 2% glucose, 0.5% l-arginine, with or without thiamin (30 μM), and 1× Vogel’s salt.
LUC reporter assays were carried out as described previously (23) using a Luciferase Assay System (Promega) on a multimode detector DTX880 (Beckman Coulter) with a 10-s luminescence assay according to the manufacturer’s instructions. Five micrograms of total protein as determined by Bradford Protein Assay (BioRad) was loaded for each LUC assay. All readings were normalized to the value recorded for the wild-type (WT) transformant. The LUC background activity of the untransformed N. crassa was <0.05% of the transformant without thiamin supplementation.
Reverse transcriptase-polymerase chain reaction
Total RNA was isolated from frozen mycelia using TRIzol reagent (Invitrogen). Five micrograms of RNA were treated with RNase-free DNase (RQI) (Promega) for 1 h at 37°C. Complementary DNA was produced by reverse transcription with a sequence-specific reverse primer for 1 h at 50°C using Superscipt III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. Control reactions using the same RNA preparation without reverse transcription were also performed.
In-line probing of RNA
In-line probing was carried out essentially as described previously (8,23). Briefly, 5′-32P-labeled RNAs were incubated with TPP concentrations from 10−10 M to 10−4 M. The products of spontaneous RNA cleavage were separated by denaturing (8 M urea) 10% polyacrylamide gel electrophoresis.
RESULTS
TPP regulation of alternative splicing products
Alternative splicing products of pre-mRNAs for NCU01977 (Figure 1A) were identified by reverse transcription and PCR (RT-PCR) using total RNA extracted from N. crassa grown in liquid culture either with or without thiamin supplementation. RT-PCR primers (Supplemental Table S1) were used to amplify the region spanning from the beginning of the coding region to the portion of the coding region located immediately following the TPP aptamer. Without splicing, this region encompasses a short intron, exon 2 and a larger intron that contains the TPP riboswitch. Note that the RT-PCR products are different than those reported previously (23). Different primers were used in the current study due to an updated annotation for this region of the N. crassa genome (see below for additional discussion).
Figure 1.

Alternative splicing products of NCU01977. (A) 5′ region of precursor and alternatively spliced mRNAs for NCU01977. S1 through S4 identify alternative 5′ splice sites (GU dinucleotides) within intron 2. Gray, white and black bars represent exons, introns and TPP aptamers, respectively. AG dinucleotides represent 3′ splice sites. Arrows indicate the primer binding sites used for RT-PCR. (B) Agarose gel separation of RT-PCR products from NCU01977 mRNAs of N. crassa grown in the absence (−) or presence (+) of 30 μM thiamin. Bands I through V are DNA products from different splice variants. Asterisks denote RT-PCR products that were not sequenced. RT designates reverse transcription; M designates a lane containing DNA markers.
In the absense of thiamin supplementation, the vast majority of the precursor RNAs are processed to remove both introns in this region, yielding spliced product I (Figure 1B). Numerous longer RNA products were observed when cells were cultured with 30 μM thiamin. Sequencing of the resulting PCR products revealed the existence of 454 nt (I), 675 nt (II), 703 nt (III), 822 nt (IV) and 1138 nt (V) RNAs, which are generated by using several alternative 5′ splice sites located upstream of the TPP aptamer. Alternative splicing products I through IV are generated by the use of 5′ splice sites S1 through S4 residing within the second intron, while product V represents the lack of splicing activity with this second intron. At least two additional minor RNA products are detected by RT-PCR (Figure 1B, asterisks), but these did not yield clones for sequence analysis and were not further investigated.
Product I is the result of the removal of both introns in this region, which yields a contiguous ORF that is expected to produce functional protein. This is consistent with its dominance when thiamin is low in concentration. All other products examined by sequencing most likely do not produce functional protein due to the presence of stop codons in the resulting mRNA templates, which are expected to generate only truncated polypeptides. These results reveal that high thiamin levels reduce splicing at site S1, while splicing is increased at the alternative 5′ splice sites such as S2, S3 and S4.
No single alternative 5′ splice site is critical for gene regulation
Previous studies with the fungal NMT1 (23) and plant THIC (24) TPP riboswitches revealed that regulation by TPP involves unmasking a single proximal 5′ splice site on TPP binding to regulate gene expression (Supplementary Figure S2A and B). In the case of the NCU01977 mRNA, thiamin addition to growth media is expected to reduce gene expression by increasing the number of alternative spliced products that lack a contiguous ORF. Therefore, a regulatory mechanism similar to the known examples in fungi and plants is not likely to be used for alternative splicing control by the NCU01977 intron because the TPP riboswitch presumably would need to simultaneously block at least three other 5′ splice sites to favor use of S1.
To assess the effects of thiamin on gene expression, we created a reporter construct wherein a LUC coding region was grafted onto the exon located immediately downstream of the TPP aptamer (Figure 2A). This WT construct was made without intron 1, which we have observed is constitutively removed regardless of thiamin supplementation in the growth medium (Figure 1). As expected, the absence of supplemented thiamin resulted in a 7.8-fold increase in reporter gene expression for the WT construct compared with that observed when 30 μM thiamin is added (Figure 2B). Mutation of the S1 5′ splice site from GU to GA causes a loss of reporter expression and TPP regulation, which is consistent with the need for splicing at this site to yield gene expression. In contrast, various reporter strains that carry mutations flanking the most proximal splice site (S4) exhibit only modestly altered (∼2-fold) gene expression (Supplementary Figure S3). These data indicate that the S4 5′ splice site may not be used as a major component of the riboswitch expression platform, which is unlike that observed for other fungal and plant TPP riboswitches described previously (23,24).
Figure 2.
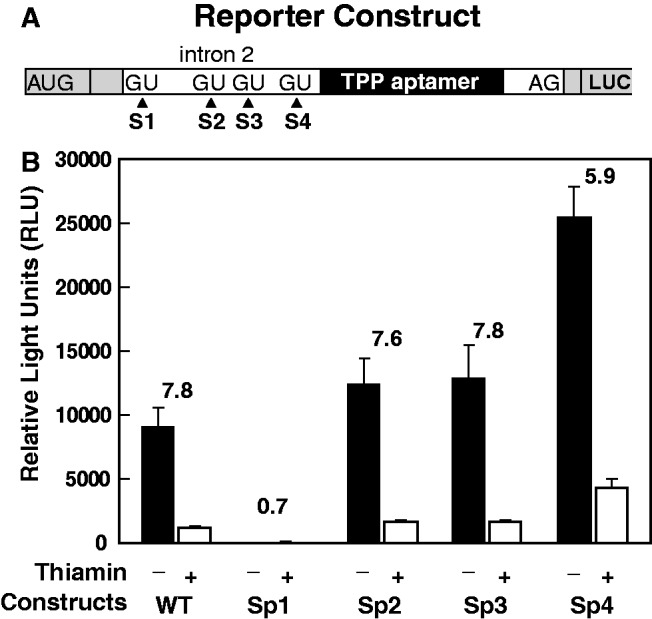
Mutations of splice sites and their effects on TPP riboswitch regulation. (A) Design of reporter constructs with a LUC ORF fused in-frame with the natural ORF located in the exon immediately downstream of the TPP aptamer. Intron 1 has been removed. (B) LUC activities (relative light units, RLU) for WT and various alternative 5′ splice site variant (GU to GA mutations) strains. Strains were cultured in the absence (−) or presence (+) of 30 μM thiamin. Fold modulation values are the ratios of RLU values without or with thiamin supplementation. Ratio averages are the result of three independent replicates, and error bars represent standard deviation.
Likewise, independent disruptions of the 5′ splice sites S2 and S3 via mutation do not diminish reporter gene expression or substantively alter TPP riboswitch control of gene expression. However, a single mutation of a conserved nucleotide in the TPP aptamer causes >5-fold increase in expression over WT and a complete loss of thiamin responsiveness (see below). All of these findings indicate that no single alternative splice site is critical for regulation by thiamin, whereas the aptamer is essential for ligand-mediated gene control.
Long-range base pairing is important for alternative splicing control by TPP
Because interactions between the proximal 5′ splice site (S4) and the TPP aptamer were not evident, we created a series of reporter constructs carrying deletions within intron 2 to search for other regulatory structures. For this series of analyses, we used a construct (Figure 3A) called WT* that is similar to the WT reporter construct (Figure 2A), but carries a truncated aptamer P3 stem (Supplementary Figure S4; see also below). This shortened aptamer was used to facilitate RNA preparation for in vitro TPP binding studies, and we demonstrated that the construct retains TPP binding function when examined by in-line probing (Supplementary Figure S5). Moreover, we observe only a modest (50%) decrease in reporter gene expression and no loss of thiamin-mediated modulation (∼20-fold) during in vivo expression experiments (Supplementary Figure S4). These findings suggest that there are no critical sequences or structural features embedded within the deleted portion of the P3 stem, which is similar to that observed for the fungal NMT1 TPP riboswitch examined previously (23).
Figure 3.
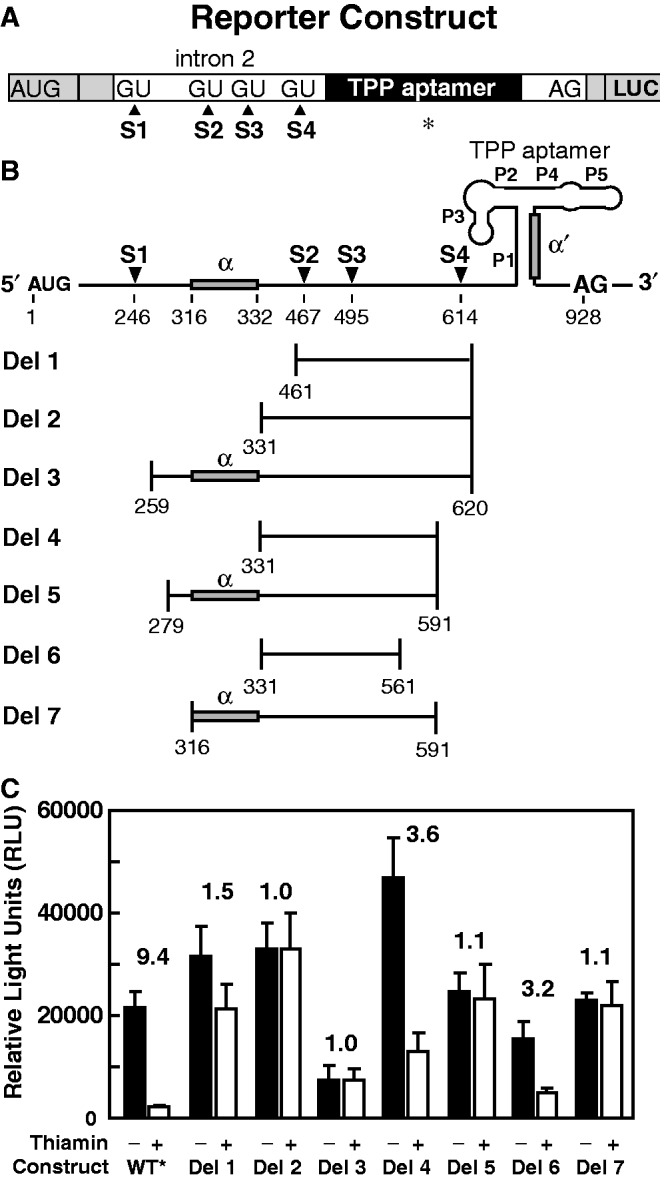
Intron deletions reveal key regulatory elements. (A) Reporter construct design is similar to that depicted in Figure 2A with the exception that the aptamer P3 stem has been shortened (denoted by the asterisk) as depicted in Figure 4A and Supplementary Figure S3A. (B) Depiction of deletion constructs wherein the first and last nucleotides of each region deleted (horizontal lines) are provided. Shaded boxes identify regions (α and α′) that can form alternative base pairing. (C) LUC activities for WT* and various deletion constructs. Other annotations are as described in the legend to Figure 2B.
Within the context of the WT* construct, reporter construct variants (Del 1 through Del 7) were created that lack at least two and in most cases three alternative splice sites and potentially other components that might be critical for riboswitch regulation (Figure 3B). Del 1, Del 2 and Del 3 mutants lack all three alternative splice sites, and the latter two constructs result in the loss of all thiamin-mediated regulation (Figure 3B). However, Del 1 retains a modest amount of regulation on addition of thiamin, which suggests that the alternative splice sites are not essential for gene control and that other features involved in regulation may be present. To determine if the reduced regulation function of Del 1 might be due to the deletion of nucleotides near the TPP aptamer, we created constructs Del 4 and Del 6, which, respectively, carry 29 and 59 additional nucleotides encompassing the proximal alternative splice site S4. Both these constructs retain robust gene expression and robust regulation on thiamin addition. However, it is not clear whether it is S4 or nucleotides flanking this splice site that are critical for robust ligand-responsive gene regulation in these mutant constructs.
Reporter constructs Del 5 and Del 7 fail to generate thiamin-mediated repression activity, despite the fact that they retain S4. Because constructs Del 4 and Del 6 do support thiamin-dependent gene control, the results suggest that an element exists between nucleotides 279 and 331 that is critical for riboswitch regulation. Because bacterial TPP riboswitches commonly use nucleotides that form a part of the aptamer P1 stem to control alternative structure formation (Supplementary Figure S1), we examined the 279 to 331 region for possible base-pairing potential to the nucleotides of P1. Indeed, a GC-rich region from nucleotides 316 to 332 (hereafter called α) can potentially base pair with nucleotides (called α′) in the 3′ shoulder of P1 (Figures 3B and 4).
Figure 4.
Proposed mechanism of alternative splicing regulated by the pairing between α and α′. (A) Portions of α′ can alternatively base pair to form the right shoulder of the aptamer P1 stem or form a base-paired structure with α, which is closest to the 5′ splice site S1. Mutants M1 through M6 involve the shaded nucleotide variations, and their gene expression and alternative splicing activities are presented in Figure 5. (B) Sequence alignments depicting conserved nucleotides and base-pairing potential between the α and α′ elements. Boxed nucleotides can base pair between the two regions. Overlined nucleotides in α′ form the right shoulder of the aptamer P1 stem. Nucleotides depicted in gray are different from the N. crassa sequence but they retain base pairing with the distal complementary site. Ncr, N. crassa; Cgl, Chaetomium globosum; Gze, Gibberella zeae; Fgr, Fusarium graminearum; Fve, Fusarium verticillioides; Val, Verticillium albo-atrum; Vda, Verticillium dahliae; Hca, Histoplasma capsulatum; Pbr, Paracoccidioides brasiliensis; Ure, Uncinocarpus reesii; Cim, Coccidioides immitis; Cpo, Coccidioides posadasii; Mca, Microsporum canis; Mgy, Microsporum gypseum; Ptr, Pyrenophora tritici-repentis; Sno, Stagonospora nodorum; Mgr, Magnaporthe grisea.
Conservation of a long-range base-paired element for riboswitch function
To determine whether the α-α′ base-pairing structure is conserved in the NCU01977 gene of other species, we searched all annotated fungal genome sequences for evidence of this same long-range interaction. Among 34 additional fungal genomes available, 18 species carry an NCU01977 TPP riboswitch representative and 16 of these also appear to exploit the α-α′ base-pairing mechanism for gene control (Figure 4B; Supplementary Figure S6).
Although the central portion of the α element is highly conserved (consensus is RGCGGYRRY, where R is a purine and Y is a pyrimidine), there are several positions that vary among different fungal species. Importantly, when an α mutation occurs relative to the N. crassa sequence, the change is either compatible (retains base pairing) with its corresponding α′ nucleotide, or the α′ nucleotide changes to compensate (Figure 4B). This mutational pattern allows nearly all species to retain at least 11 continuous base pairs in the α-α′ structural element.
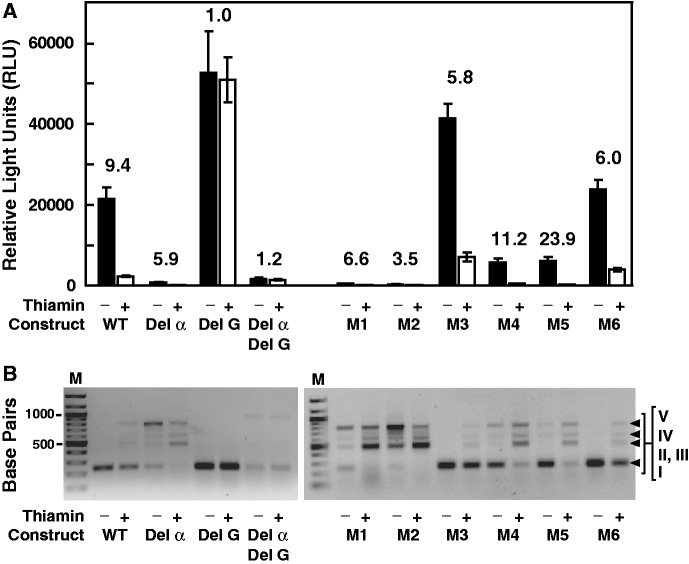
Confirmation of base pairing between α and α′ nucleotides
If α is required for TPP riboswitch function and ultimately for alternative splicing, then its deletion should change the level of gene expression and the pattern of mRNA splicing products. Indeed, deletion of α (construct Del α) causes a dramatic reduction in reporter gene expression (Figure 5A) and causes a large increase in the amount of alternatively spliced products (II through V) relative to WT* (Figure 5B) regardless of thiamin supplementation. However, the addition of thiamin further increases alternative splicing and reduces gene expression, although not to the same level as for WT*. These findings suggest that α-α′ structure formation is an important component of NCU01977 TPP riboswitch control of alternative splicing, but that there is at least one other undiscovered route by which the riboswitch aptamer influences alternative splicing.
Figure 5.
Mutational analysis of the α and α′ interaction in the NCU01977 TPP riboswitch. (A) Plot of LUC reporter gene expression for various constructs in the absence (−) or presence (+) of thiamin supplementation. Reporter constructs for Del α (deletion of the α element) and M1 through M6 (nucleotide changes depicted in Figure 4A) are based on the parent construct depicted in Figure 3A. Del G (has a deletion of a conserved G in the junction of P2 and P3 that disrupts TPP binding) and the double mutant Del α/Del G (deletion of α and G) are based on the original WT construct that retains intron 1 and the natural P3 stem. (B) Agarose gel separation of RT-PCR products from WT and all mutant constructs. Products I through IV are similar to those observed in Figure 1B, except that they are ∼200-nt shorter owing to the use of different primers.
A construct (Del G) that has a deletion of a conserved G (position 53, Supplementary Figure S5) expected to be critical for TPP binding (11,14) completely precludes thiamin-mediated control of gene expression and alternative splicing. The total amount of reporter gene expression increases substantially, which is consistent with formation of the α-α′ structure in the absence of TPP binding. A construct combining both the Del α and Del G mutations yields little gene expression or splice product I and no thiamin-dependent modulation, again highlighting the importance of α-α′ and TPP binding for normal riboswitch function.
To further validate the α-α′ structure and its role in alternative splicing, we created a series of mutant constructs (M1 through M6) based on the WT* sequence (Figure 4A). M1 carries eight mutations within P1 that retain P1 base pairing, and M2 carries four mutations that alter the central portion of α. Both M1 and M2 are expected to preclude formation of a strong α-α′ base-pairing interaction, and both mutant constructs yield gene expression levels and splicing products similar to those observed for Del α. Construct M3 carries all 12 mutations, maintaining P1 formation and restoring full α-α′ base-pairing potential, albeit with different base-pair identities. Importantly, the M3 construct produces splice products and gene expression levels that are similar to those observed with the WT construct, which is consistent with a key role for the α-α′ structure in TPP-mediated regulation.
The results observed for M4, M5 and M6 are similar to those observed for M1, M2 and M3, respectively (Figure 5). The mutations in M4 through M6 alter the nucleotide sequences and the base-pairing interactions near the ends of the P1 and α-α′ stems rather than in the centers (Figure 4A), but with similar effects on splicing and gene expression. Again, these findings are consistent with α-α′ formation as an important component of TPP riboswitch control of alternative splicing and gene expression. Moreover, the mutations within the α elements of constructs M3 and M6 represent significant deviations from the consensus sequence observed in numerous fungal species (Figure 4B), suggesting that the precise sequence in this region is less important than its ability to base pair with α′.
Proximity of key riboswitch structural and functional elements
The distance between the first 5′ splice site (S1) and the 3′ splice site for each species frequently is between 550 and 650 nt, although three species have even more nucleotides separating these elements (Supplementary Figure S7A). Many of these nucleotides reside between α and α′ (Supplementary Figure S7B). In contrast, there commonly are <60 nt separating S1 from the α element (Supplementary Figure S7C) and <60 nt separating α′ and the 3′ splice site (Supplementary Figure S7D). These findings suggest that the TPP riboswitch associated with this gene is under evolutionary selection to maintain relatively short distances between components of its riboswitch expression platform and the 5′ and 3′ splice sites. One outcome of the close arrangement between these key elements is that the formation of α and α′ base pairing brings the 5′ and 3′ splice sites in physical proximity, and this may favor splicing using the first 5′ splicing site.
DISCUSSION
Long-distance RNA secondary structures and alternative splicing
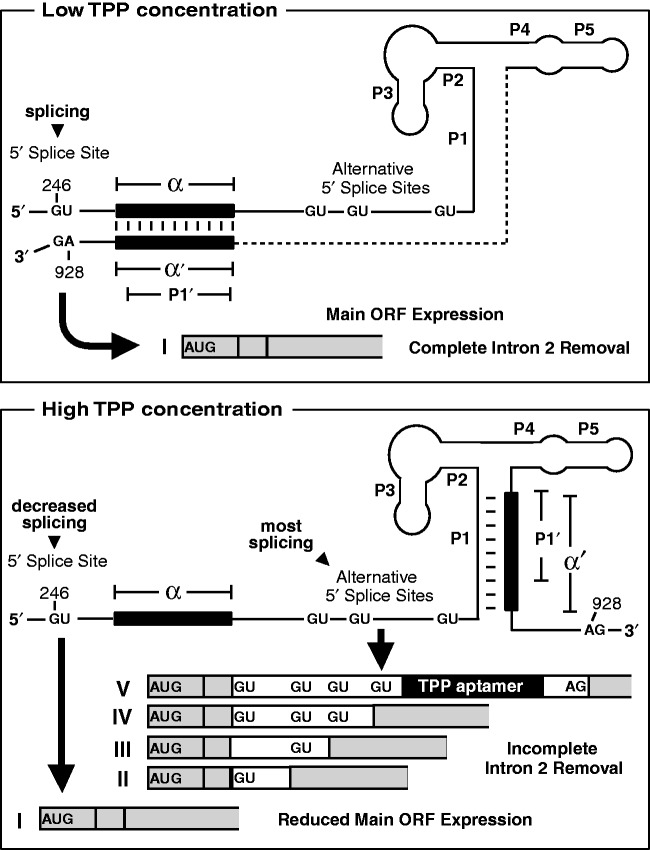
Among eukaryotic TPP riboswitches, those associated with NMT1 and THI4 genes exploit similar mechanisms for alternative splicing control, using aptamer nucleotides from the P4 and P5 stems to block 5′ splice site access when TPP is not bound (Supplementary Figure S2A and B) (23,24). Our data reveal that the TPP riboswitch in the second intron of the NCU01977 gene is using a different mechanism to control splicing that involves the formation of a long-range base-paired structure when TPP is not bound (Figure 6). When TPP is absent, the 3′ side (α′) of the P1 stem of the aptamer alternatively base pairs with a distal RNA sequence (α). In the TPP-bound state, the nucleotides of α′ are sequestered by the formation of a stable P1 stem, which reduces splicing at the distal 5′ splice site (S1) and favors splicing at the more proximal alternative 5′ splice sites (S2, S3 and S4).
Figure 6.
Proposed mechanism for alternative splicing control by the NCU01977 TPP riboswitch. TPP aptamers have at least two structural states: unbound when TPP is low (Top) and ligand-bound when TPP concentrations are high (Bottom). In the unbound state, α′ pairs to α, thereby bringing the otherwise distal 5′ (S1) and 3′ splice sites in proximity to favor their splicing and produce a contiguous ORF. In the TPP-bound state, the nucleotides of α′ are sequestered by the formation of a stable P1 stem, which reduces splicing at the distal 5′ splice site (S1) and favors splicing at the more proximal alternative 5′ splice sites (S2, S3 and S4).
The precise reason why base pairing between α and α′ causes a dramatic increase in splicing at the S1 5′ splice site remains unclear. Formation of the α-α′ structure brings the otherwise distal 5′ (S1) and 3′ splice sites closer in space, and perhaps this proximity favors their splicing to produce a contiguous ORF. Importantly, long-distance RNA secondary structures previously have been found to enhance splicing of large introns in yeast (37–40) and recently have been found to play important roles in the regulation of alternative splicing. For example, the Dscam gene of Drosophila encodes a pre-mRNA that can yield 38 016 splicing isoforms by exploiting long-distance RNA base-pairing interactions between ‘selector’ and ‘docking site’ sequences to direct alternative splicing (41,42).
Also in Drosophila, >200 highly conserved long-distance interactions of at least nine base pairs have been uncovered by using bioinformatics (29). Testing of disruptive and compensatory mutations provided evidence that such interactions are important for alternative splicing. This observation is consistent with an earlier bioinformatics analysis of mRNAs known to undergo alternative splicing, which revealed potential base-pairing interactions within each intron that may be important for exon skipping (43). Findings like these suggest that such long-range base-pairing interactions may be commonly involved in directing splice site choice.
Alternative splicing control by NCU01977 TPP riboswitches
For the TPP riboswitch system under investigation in this study, it seems possible that the long-range α and α′ base-pairing interaction might simply bring two distal splice sites together to favor their splicing. Alternatively, localizing the alternative 5′ splice sites in the loop of a stable stem might reduce their use by the spliceosome, which is a mechanism that previously has been observed with in vitro splicing reactions (27,28). There are other mechanisms known that exploit protein factors such as intronic splicing enhancers and silencers, and exonic splicing enhancers and silencers to regulate splicing (42). Perhaps riboswitch-mediated changes in intron folding either promote or preclude binding of specific protein factors to control splice-site choice.
TPP riboswitch representatives in fungal NMT1 mRNA produce alternative splicing products that contain uORFs, while representatives in plant THIC mRNAs yield alternative splicing products that lack processing sites (Supplementary Figure S2). At high TPP concentrations, these alternative splicing products reduce gene expression either by precluding translation of the main ORF, or by causing degradation of the entire mRNA. Thus, these experimentally validated riboswitches are genetic OFF switches.
Unified interpretation of current and previous data on the NCU01977 TPP riboswitch
Initial analyses of the NCU01977 TPP riboswitch using RT-PCR and reporter-fusion assays (23) suggested that this riboswitch would turn on gene expression under high TPP concentrations. However, these results were generated using primer and reporter-fusion construct designs that were based on a previous misannotation of the N. crassa genome (44). The forward primer for RT-PCR used in the previous work was complementary to a region immediately preceding splice site S4. The two RT-PCR products generated by the RT-PCR reaction using these previously designed primers [see Figure 1 in (23)] only correspond to bands IV and V in the current study (Figure 1B).
In the current study, we used an updated annotation (available in 2009) revealing the existance of at least one additional intron and start codon located upstream of the riboswitch (Supplementary Figures S8 and S9). Thus, the TPP riboswitch is located in a much larger intron than previously recongized. We designed new RT-PCR primers that encompass this longer precursor mRNA to more fully evaluate the distribution of splice variants. Our new RT-PCR data reveals that bands IV and V (observed with the original primers) represent only minor products of RT-PCR (Figure 1B).
Furthermore, an AUG codon located near splice site S4 that is in-frame with a LUC gene in the reporter-fusion construct generated previously [see Figure S9 in (23)] is in the intron and is not the true start codon (Supplementary Figure S8). When a larger reporter-fusion construct that carries the entire intron (from nucleotides 246 to 929) is used to monitor the gene exprerssion, LUC expression increases by ∼100-fold for the WT construct relative to the Del α construct (Figure 5). This finding supports our hypothesis that the α-α′ interaction is a major interaction regulating alternative splicing and this TPP riboswitch turns off gene expression when TPP levels are high.
Control of proximal splice site use by TPP binding
As noted above, the NCU01977 TPP riboswitch is distinct from the fungal and plant TPP riboswitches that make use of base pairing to a proximal 5′ splice site to bring about large changes in alternative splicing and gene expression. Interestingly, the RT-PCR data reported previously the NCU01977 TPP riboswitch (23) revealed that when TPP concentration is high, splicing favored a shorter product versus when TPP concentration is low. This finding can be explained if the P4-P5 region of the TPP aptamer can base pair to the region including splice site S4 to block its use (Supplementary Figure S2). Although this cannot be the source of the majority of the alternative splicing and gene control effects generated by this riboswitch, this mechanism may be the reason why constructs with α-α′ disruptions still produce a small response to TPP binding (Figure 5).
CONCLUSIONS
Nearly all riboswitches characterized to date are found in bacteria where most serve as sensors of metabolites and as simple gene-control elements. This reinforces the generally held view that riboswitches are primitive gene control elements that are not well suited for use by more complex organisms. However, recent findings make it clear that riboswitches responsive to the coenzyme TPP are critical gene control elements in fungi and plants, where they efficiently sense their target metabolite and control gene expression by regulating splicing and processing of specific mRNAs.
Several features of the system we examined in this study are surprising. The riboswitch regulates alternative splicing from at least four different 5′ splice sites. Strikingly, regulation involves the TPP-mediated regulation of an extended base-pairing interaction residing ∼500-nt upstream of the TPP aptamer. The long-range base-pairing interaction curiously does not involve the typical aptamer and splice-site nucleotides, but rather involves base pairing between aptamer P1 nucleotides and a conserved sequence domain located ∼50-nt downstream from the activated 5′ splice site. Because the sequences and architectural features of this type of riboswitch are well conserved among many fungal species, this indicates that this mechanism of metabolite-regulated alternative splicing is likely to be used by many organisms.
It is becoming increasingly apparent that many eukaryotic pre-mRNAs exploit long-distance base-pairing interactions to influence specific splicing events. The recent prediction of large numbers of conserved long-distance interactions in Drosophila (29) suggests that there are numerous opportunities to discover the mechanistic details of alternative splicing regulation that could be mediated by optional pairing of these nucleotides. Our experiments on an intronic TPP riboswitch in fungi reveal that the formation of long-range base-pairing interactions can be controlled by coenzyme binding directly to an intron RNA. This TPP-mediated alternative folding process provides a mechanism by which cells can regulate alternative splicing and gene expression in response to changing concentrations of a small metabolite. It seems reasonable to speculate that numerous protein factors or perhaps some other small molecule ligands remain to be discovered that influence the formation of long-range base-pairing interactions to regulate the splicing and expression of pre-mRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–9.
FUNDING
National Institutes of Health [GM 022778]; Howard Hughes Medical Institute. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. R.R.B. is a cofounder of BioRelix, a biotechnology company that has licensed riboswitch technology from Yale University.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. C. Dunlap for supplying us with N. crassa strains and the luciferase reporter vector. We thank P. Lakin-Thomas and E. Selker for suggestions on electroporation methods and strains for transformation. We also thank Jonathan Perreault and other members of the Breaker lab for helpful comments. The work was supported by the NIH and by the Howard Hughes Medical Institute.
REFERENCES
- 1.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dambach MD, Winkler WC. Expanding roles for metabolite-sensing regulatory RNAs. Curr. Opin. Microbiol. 2009;12:161–169. doi: 10.1016/j.mib.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serganov A. The long and short of riboswitches. Curr. Opin. Struct. Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith AM, Fuchs RT, Grundy FJ, Henkin TM. Riboswitch RNAs: regulation of gene expression by direct monitoring of a physiological signal. RNA Biol. 2010;7:104–110. doi: 10.4161/rna.7.1.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breaker RR. Riboswitches: from ancient gene-control systems to modern drug targets. Future Microbiol. 2009;4:771–773. doi: 10.2217/fmb.09.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012;4:a003566. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breaker RR. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 9.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 10.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 12.Edwards TE, Ferré-D’Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of thiamin biosynthesis in prokaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 14.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wachter A. Riboswitch-mediated control of gene expression. RNA Biol. 2010;7:67–76. doi: 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- 18.Breaker RR. Complex riboswitches. Science. 2008;319:1795–1797. doi: 10.1126/science.1152621. [DOI] [PubMed] [Google Scholar]
- 19.Baird NJ, Kulshina N, Ferré-D’Amaré AR. Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol. 2010;7:328–332. doi: 10.4161/rna.7.3.11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bastet L, Dubé A, Massé E, Lafontaine DA. New insights into riboswitch regulation mechanisms. Mol. Microbiol. 2011;80:1148–1154. doi: 10.1111/j.1365-2958.2011.07654.x. [DOI] [PubMed] [Google Scholar]
- 21.Garst AD, Edwards AL, Batey RT. Riboswitches: structures and mechanisms. Cold Spring Harb. Perspect. Biol. 2011;3:a003533. doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 23.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 24.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc. Natl Acad. Sci. USA. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985;43:667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- 28.Solnick D, Lee SI. Amount of RNA secondary structure required to induce an alternative splice. Mol. Cell Biol. 1987;7:3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raker VA, Mironov AA, Gelfand MS, Pervouchine DD. Modulation of alternative splicing by long-range RNA structures in Drosophila. Nucleic Acids Res. 2009;37:4533–4544. doi: 10.1093/nar/gkp407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pervouchine DD, Khrameeva EE, Pichugina MY, Nikolaienko OV, Gelfand MS, Rubtsov PM, Mironov AA. Evidence for widespread association of mammalian splicing and conserved long-range RNA structures. RNA. 2012;18:10–15. doi: 10.1261/rna.029249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, Dunlap JC. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot. Cell. 2008;7:28–37. doi: 10.1128/EC.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Froehlich AC, Loros JJ, Dunlap JC. Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc. Natl Acad. Sci. USA. 2003;100:5914–5919. doi: 10.1073/pnas.1030057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vann DC. Electroporation-based transformation of freshly harvested conidia of Neurospora crassa. Fungal Genet. Newsl. 1995;42A:53. [Google Scholar]
- 34.Margolin BS, Freitag M, Selker EU. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 1997;44:34–36. [Google Scholar]
- 35.Ebbole D, Sachs MS. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 1990;37:17–18. [Google Scholar]
- 36.Li S, Motavaze K, Kafes E, Suntharalingam S, Lakin-Thomas P. A new mutation affecting FRQ-less rhythms in the circadian system of Neurospora crassa. PLoS Genet. 2011;7:e1002151. doi: 10.1371/journal.pgen.1002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman A. Specific accessory sequences in Saccharomyces cerevisiae introns control assembly of pre-mRNAs into spliceosomes. EMBO J. 1987;6:3833–3839. doi: 10.1002/j.1460-2075.1987.tb02720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker R, Patterson B. Architecture of fungal introns: implications for spliceosome assembly. In: Dudock B, editor. Molecular Biology of RNA: New Perspectives. San Diego, CA: Academic Press; 1987. pp. 133–149. [Google Scholar]
- 39.Libri D, Stutz F, McCarthy T, Rosbash M. RNA structural patterns and splicing: molecular basis for an RNA-based enhancer. RNA. 1995;1:425–436. [PMC free article] [PubMed] [Google Scholar]
- 40.Howe KJ, Ares M., Jr Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc. Natl Acad. Sci. USA. 1997;94:12467–1272. doi: 10.1073/pnas.94.23.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr. Opin. Genet. Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miriami E, Margalit H, Sperling R. Conserved sequence elements associated with exon skipping. Nucleic Acids Res. 2003;31:1974–1983. doi: 10.1093/nar/gkg279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.