Abstract
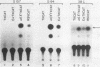
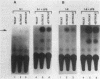
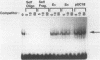
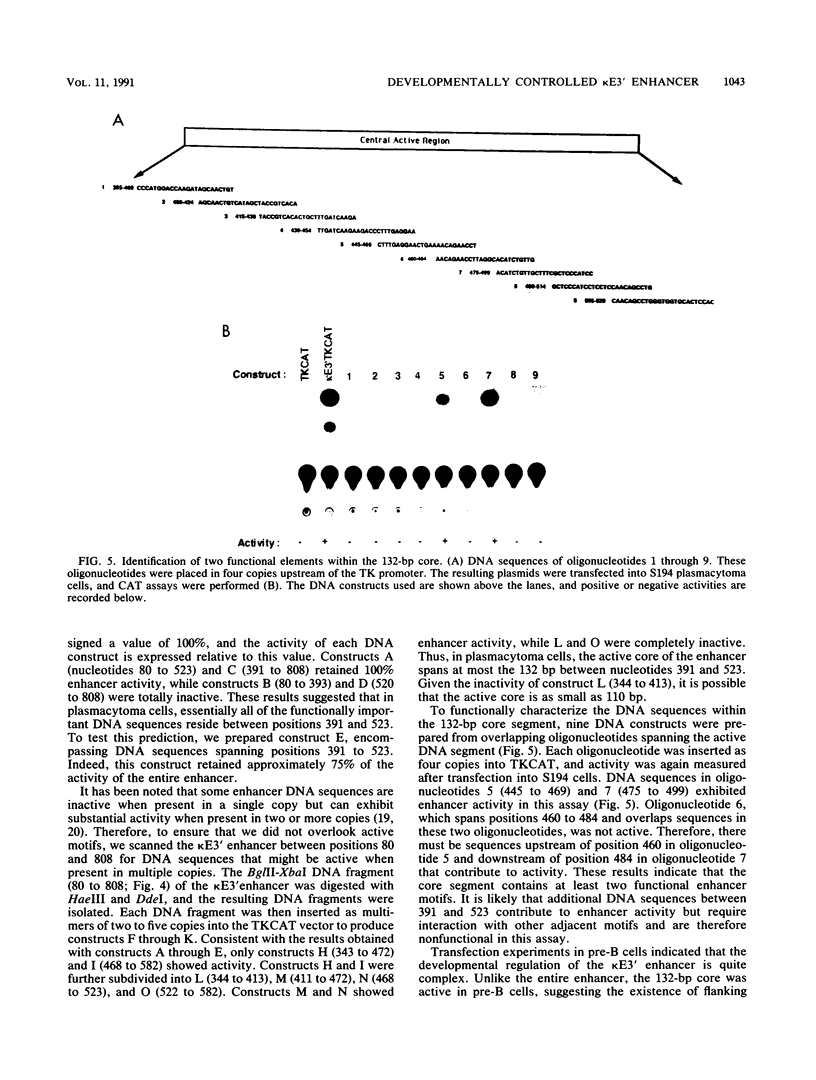
We have functionally characterized an enhancer element (kappa E3') which lies 8.5 kb downstream of the immunoglobulin kappa gene. The activity of this enhancer is developmentally controlled. It is inactive at the pre-B-cell stage but active at the B-cell and plasma cell stages. This enhancer is also functional in S107 plasmacytoma cells, which lack NF-kappa B and therefore intron enhancer activity. The activity of the kappa E3' enhancer therefore provides an explanation for the transcriptional activity of endogenous kappa genes in S107 cells in the absence of intron enhancer function. We have identified a 132-bp segment of the kappa E3' enhancer that retains 75% of the activity of the entire enhancer observed in plasmacytoma cells. Within this 132-bp core, there are at least two functional elements, one of which binds to a B-cell-specific nuclear factor. This element contains a potential binding site for the B-cell- and macrophage-specific transcription factor PU.1. The kappa intron and kappa E3' enhancers were also found to be regulatable by Id, an inhibitor of helix-loop-helix transcription factors. The site of action of Id on the kappa E3' enhancer was mapped to a 25-bp region which contains a potential binding site for a helix-loop-helix transcription factor. A possible model for the developmental control of kappa gene transcription is discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atchison M. L., Perry R. P. Complementation between two cell lines lacking kappa enhancer activity: implications for the developmental control of immunoglobulin transcription. EMBO J. 1988 Dec 20;7(13):4213–4220. doi: 10.1002/j.1460-2075.1988.tb03318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison M. L., Perry R. P. The role of the kappa enhancer and its binding factor NF-kappa B in the developmental regulation of kappa gene transcription. Cell. 1987 Jan 16;48(1):121–128. doi: 10.1016/0092-8674(87)90362-x. [DOI] [PubMed] [Google Scholar]
- Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990 Apr 6;61(1):49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Blasquez V. C., Xu M., Moses S. C., Garrard W. T. Immunoglobulin kappa gene expression after stable integration. I. Role of the intronic MAR and enhancer in plasmacytoma cells. J Biol Chem. 1989 Dec 15;264(35):21183–21189. [PubMed] [Google Scholar]
- Briskin M., Kuwabara M. D., Sigman D. S., Wall R. Induction of kappa transcription by interferon-gamma without activation of NF-kappa B. Science. 1988 Nov 18;242(4881):1036–1037. doi: 10.1126/science.3143155. [DOI] [PubMed] [Google Scholar]
- Cai M., Davis R. W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990 May 4;61(3):437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis H. M., Spann D. R., Posakony J. W. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990 Apr 6;61(1):27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Foster J., Stafford J., Queen C. An immunoglobulin promoter displays cell-type specificity independently of the enhancer. 1985 May 30-Jun 5Nature. 315(6018):423–425. doi: 10.1038/315423a0. [DOI] [PubMed] [Google Scholar]
- Garrell J., Modolell J. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell. 1990 Apr 6;61(1):39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- Goebl M. K. The PU.1 transcription factor is the product of the putative oncogene Spi-1. Cell. 1990 Jun 29;61(7):1165–1166. doi: 10.1016/0092-8674(90)90676-6. [DOI] [PubMed] [Google Scholar]
- Goebl M. K. The PU.1 transcription factor is the product of the putative oncogene Spi-1. Cell. 1990 Jun 29;61(7):1165–1166. doi: 10.1016/0092-8674(90)90676-6. [DOI] [PubMed] [Google Scholar]
- Gopal T. V., Shimada T., Baur A. W., Nienhuis A. W. Contribution of promoter to tissue-specific expression of the mouse immunoglobulin kappa gene. Science. 1985 Sep 13;229(4718):1102–1104. doi: 10.1126/science.2994213. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Merlino G. T., Willingham M. C., Pastan I., Howard B. H. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Baltimore D. Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell. 1985 Jul;41(3):885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- Hall C. V., Jacob P. E., Ringold G. M., Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2(1):101–109. [PubMed] [Google Scholar]
- Henthorn P., Kiledjian M., Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990 Jan 26;247(4941):467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Herr W., Gluzman Y. Duplications of a mutated simian virus 40 enhancer restore its activity. Nature. 1985 Feb 21;313(6004):711–714. doi: 10.1038/313711a0. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kelley D. E., Pollok B. A., Atchison M. L., Perry R. P. The coupling between enhancer activity and hypomethylation of kappa immunoglobulin genes is developmentally regulated. Mol Cell Biol. 1988 Feb;8(2):930–937. doi: 10.1128/mcb.8.2.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990 Apr 6;61(1):113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Meyer K. B., Neuberger M. S. The immunoglobulin kappa locus contains a second, stronger B-cell-specific enhancer which is located downstream of the constant region. EMBO J. 1989 Jul;8(7):1959–1964. doi: 10.1002/j.1460-2075.1989.tb03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. B., Sharpe M. J., Surani M. A., Neuberger M. S. The importance of the 3'-enhancer region in immunoglobulin kappa gene expression. Nucleic Acids Res. 1990 Oct 11;18(19):5609–5615. doi: 10.1093/nar/18.19.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nelson K. J., Kelley D. E., Perry R. P. Inducible transcription of the unrearranged kappa constant region locus is a common feature of pre-B cells and does not require DNA or protein synthesis. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5305–5309. doi: 10.1073/pnas.82.16.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K. J., Mather E. L., Perry R. P. Lipopolysaccharide-induced transcription of the kappa immunoglobulin locus occurs on both alleles and is independent of methylation status. Nucleic Acids Res. 1984 Feb 24;12(4):1911–1923. doi: 10.1093/nar/12.4.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D., Schaffner W. A lymphocyte-specific enhancer in the mouse immunoglobulin kappa gene. Nature. 1984 Jan 5;307(5946):80–82. doi: 10.1038/307080a0. [DOI] [PubMed] [Google Scholar]
- Picard D., Schaffner W. Cell-type preference of immunoglobulin kappa and lambda gene promoters. EMBO J. 1985 Nov;4(11):2831–2838. doi: 10.1002/j.1460-2075.1985.tb04011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983 Jul;33(3):741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- Queen C., Stafford J. Fine mapping of an immunoglobulin gene activator. Mol Cell Biol. 1984 Jun;4(6):1042–1049. doi: 10.1128/mcb.4.6.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986 Dec 26;47(6):921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Singh H., Sen R., Baltimore D., Sharp P. A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986 Jan 9;319(6049):154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- Stafford J., Queen C. Cell-type specific expression of a transfected immunoglobulin gene. Nature. 1983 Nov 3;306(5938):77–79. doi: 10.1038/306077a0. [DOI] [PubMed] [Google Scholar]
- Van Ness B. G., Weigert M., Coleclough C., Mather E. L., Kelley D. E., Perry R. P. Transcription of the unrearranged mouse C kappa locus: sequence of the initiation region and comparison of activity with a rearranged V kappa-C kappa gene. Cell. 1981 Dec;27(3 Pt 2):593–602. doi: 10.1016/0092-8674(81)90401-3. [DOI] [PubMed] [Google Scholar]
- Xu M., Hammer R. E., Blasquez V. C., Jones S. L., Garrard W. T. Immunoglobulin kappa gene expression after stable integration. II. Role of the intronic MAR and enhancer in transgenic mice. J Biol Chem. 1989 Dec 15;264(35):21190–21195. [PubMed] [Google Scholar]