Abstract
Polycyclic aromatic hydrocarbons (PAHs) likely play a role in many cancers even in never-smokers. We tried to find a model to explain the relationship between variation in PAH-related DNA adduct levels among people with similar exposures, multiple genetic polymorphisms in genes related to metabolic and repair pathways, and nucleotide excision repair (NER) capacity. In 111 randomly-selected female never-smokers from the Golestan Cohort Study in Iran, we evaluated 21 SNPs in 14 genes related to xenobiotic metabolism and 12 SNPs in 8 DNA repair genes. NER capacity was evaluated by a modified comet assay, and aromatic DNA adduct levels were measured in blood by 32P-postlabelling. Multivariable regression models were compared by Akaike’s information criterion (AIC). Aromatic DNA adduct levels ranged between 1.7 and 18.6 per 108 nucleotides (mean: 5.8±3.1). DNA adduct level was significantly lower in homozygotes for NAT2 slow alleles and ERCC5 non risk-allele genotype, and was higher in the MPO homozygote risk-allele genotype. The sum of risk alleles in these genes significantly correlated with the log-adduct level (r=0.4, p<0.001). Compared with the environmental model, adding phase I SNPs and NER capacity provided the best fit, and could explain 17% more of the variation in adduct levels. NER capacity was affected by polymorphisms in the MTHFR and ERCC1 genes. Female non-smokers in this population had PAH-related DNA adduct levels 3-4 times higher than smokers and occupationally-exposed groups in previous studies, with large inter-individual variation which could best be explained by a combination of phase I genes and NER capacity.
Keywords: Polycyclic aromatic hydrocarbons, DNA adducts, nucleotide excision repair, polymorphism
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are a class of carcinogens which are produced mainly by incomplete fuel combustion 1. There are several studies documenting a possible role for PAH exposure in the etiology of esophageal squamous cell carcinoma (ESCC) 2-7. In a recent report from Golestan Province, Iran, a high risk area for ESCC, Abedi-Ardekani and colleagues showed a striking dose-response relationship between the PAH content of non-tumoral esophageal biopsies, measured by immunohistochemical staining with antibodies raised against benzo[a]pyrene diol epoxide-I-modified guanosine, and ESCC case status 2. Studies in this area and other high risk areas for ESCC in Brazil and along the central Asian esophageal cancer belt have also shown high exposures to PAHs 3-4, 6. This high exposure has been observed among non-smokers as well as smokers, and food and coal-burning stoves have been suggested as potential sources 3, 5, 7-8. Similarly, exposure to PAH has also been implicated in lung cancer pathogenesis in non-smokers 9.
Many PAHs do not cause biological changes by themselves, but do so only after being converted to active metabolites in the body 10. PAHs are first metabolized to more polar compounds by phase I enzymes. As a result of these reactions, reactive electrophilic intermediates may be formed that can bind to DNA to form DNA adducts, which are responsible for the mutagenic effects of PAHs; or they may be further metabolized by phase II enzymes that conjugate these electrophilic metabolites to form water-soluble compounds that can be excreted. DNA adduct formation is, in fact, an early biological event in the carcinogenic process 11. DNA repair mechanisms, including nucleotide excision repair (NER), can restore the integrity of the damaged parts of DNA 12.
While differences in environmental exposures are partly responsible for variations in DNA adduct formation, inter-individual variations among people with similar exposures may be explained partially by polymorphisms in genes that code for metabolizing or repair enzymes 10, and accumulating evidence suggests a role for genetic polymorphisms in PAH-related DNA adduct formation 13. So far, however, the results have been inconsistent regarding the roles of single polymorphisms, and many researchers now suggest that, due to the complex exposure and nature of PAH metabolism, a combination of genes in this metabolic pathway should instead be considered 10. For example, it has been shown that the sum of risk alleles in multiple PAH-metabolizing genes can affect adduct levels in smokers 14.
In this study, we evaluated a large number of polymorphisms in genes affecting DNA adduct formation and repair, along with NER phenotyping, in Golestan Province, a high-risk area for ESCC in northeastern Iran. Our main purpose was to identify the best models to explain inter-individual variations in adduct levels. Women in this area are not likely to be exposed to PAHs in an occupational setting, so non-smoking women constitute a relatively homogenous group in terms of PAH exposure. We restricted this study to female non-smokers in order to minimize the variation in adduct levels due to differences in environmental exposures.
Materials and Methods
Sampling
One hundred and eleven female non-smokers were randomly selected from subjects recruited in the Golestan Cohort Study (GCS), details of which have been published before 15. Briefly, 50,045 healthy adults from eastern Golestan Province were recruited into this cohort, including 48% men and 52% women, 80% from rural villages, and 74% of Turkmen ethnicity. Questionnaire data were collected about exposure to second-hand smoke at work or home; cooking methods in the household, including frying, boiling, baking, and barbequing; as well as the frequency of coal use for cooking. A standard food frequency questionnaire was administered, and the amounts of total and processed red meat consumption were calculated as potential sources of PAH.
Venous blood was collected from study participants, centrifuged at 800 g for 20 min, and the buffy coat was separated and stored at −70°C. After destroying RBCs using a lysis buffer (155 mmol/L NH4Cl, 10 mmol/L KHCO3, and 10 mmol/L EDTA), genomic DNA was extracted from the buffy coat by standard phenol extraction and stored in −70°C until genotyping and 32P-postlabelling. For the NER assay, lymphocytes were isolated from venous blood using a standard density gradient centrifugation method16 and stored at −70°C. A single spot urine sample was collected from each participant and stored at −20°C.
The study was approved by the Institutional Review Board of the Digestive Disease Research Center of Tehran University of Medical Sciences.
Genotyping
Twenty-one SNPs in 14 genes related to xenobiotic metabolism and 12 SNPs in 8 DNA repair genes were studied (Table 1). SNP’s were selected on the basis of (a) their association with PAH metabolism and/or cancer development or (b) their expected influence on DNA repair activity17. These included genes for phase I metabolizing enzymes: cytochrome P450s (CYP1A1, CYP1A2, CYP1B1, CYP2E1, and CYP3A4), microsomal epoxide hydrolase 1 (mEH), and myeloperoxidase (MPO); genes for phase II enzymes: glutathione peroxidase 1 (GPX1), glutathione S-transferases (GSTM1, GSTP1, GSTT1), N-acetyltransferase 2 (NAT2), NAD(P)H quinone oxidoreductase 1 (NQO1), and manganese superoxide dismutase 2 (MnSOD2); and genes for DNA repair: apurinic-apyrimidinic endonuclease 1 (APEX1), DNA excision repair cross-complementing proteins (ERCC1, ERCC5, ERCC6), methylenetetrahydrofolate reductase (MTHFR), RAD23B, and the xeroderma pigmentosum group (XPA, XPC).
Table 1.
Details of the genes and polymorphisms evaluated in this study.
| Set | gene | SNP ID | mutation | Putative risk allele* (frequency) |
HWE p value |
PCR multiplex |
|---|---|---|---|---|---|---|
| Phase I | CYP1A1 | rs1048943 | Ex7+131A>G | G (0.08) | 0.4 | 2 |
| rs1799814 | Ex7+129C>A | A (0.05) | 0.2 | 2 | ||
| CYP1A2 | rs762551 | IVS1-154C>A | A (0.59) | 0.03 | 1 | |
| CYP1B1 | rs1056836 | Ex3+251G>C | C (0.27) | 0.6 | 2 | |
| rs1800440 | Ex3+315A>G | G (0.20) | 0.5 | 2 | ||
| CYP2E1 | rs6413420 | -70T>G | G (0.10) | 0.3 | 2 | |
| CYP3A4 | rs2740574 | -391A>G | G (0.02) | 0.8 | 2 | |
| mEH | rs2234922 | Ex4+52A>G | A (0.55) | 0.1 | 3 | |
| rs1051740 | Ex3-28T>C | C (0.49) | 0.7 | 3 | ||
| MPO | rs2333227 | -642G>A | A (0.36) | 0.07 | 2 | |
| Phase II | GPX1 | rs1050450 | Ex1-226C>T | T (0.24) | 0.2 | 3 |
| GSTM1 | Ex4+10+>- | -/- (0.52) | - | 1 | ||
| GSTP1 | rs1695 | Ex5-24A>G | G (0.41) | 0.5 | 1 | |
| rs1138272 | Ex6+5C>T | C (0.25) | 0.9 | 1 | ||
| GSTT1 | Ex5-49+>- | -/- (0.89) | - | 1 | ||
| NAT2 | rs1801280 | Ex2+347T>C | C (0.57) | 0.9 | 1 | |
| rs1799931 | Ex2-313G>A | A (0.31) | 0.9 | 1 | ||
| rs1799930 | Ex2-580G>A | A (0.31) | 0.5 | 1 | ||
| NQO1 | rs1800566 | Ex6+40C>T | T (0.09) | 0.4 | 3 | |
| rs1131341 | Ex4-3C>T | T (0.27) | 0.6 | 3 | ||
| MnSOD2 | rs4880 | Ex2+24T>C | C (0.18) | 0.8 | 3 | |
| Repair | APEX1 | rs1130409 | Ex5+5T>G | G (0.34) | 0.07 | 3 |
| ERCC1 | rs11615 | Ex4+33T>C | C (0.29) | 0.6 | 4 | |
| rs3212948 | IVS3+74C>G | G (0.46) | 0.4 | 4 | ||
| rs3212986 | 196+STP T>G | G (0.27) | 0.8 | 4 | ||
| ERCC5 | rs1047768 | Ex2+50T>C | C (0.33) | 0.5 | 4 | |
| ERCC6 | rs2228526 | Ex18+219A>G | G (0.24) | 0.7 | 4 | |
| MTHFR | rs1801131 | Ex8-62A>C | A (0.57) | 0.4 | 1 | |
| rs1801133 | Ex5+79C>T | T (0.18) | 0.2 | 1 | ||
| RAD23B | rs1805329 | Ex7+65C>T | T (0.04) | 0.8 | 4 | |
| XPA | rs1800975 | Ex1+62A>G | G (0.34) | 0.6 | 4 | |
| XPC | rs2228001 | Ex16+211C>A | A (0.50) | 0.8 | 4 | |
| rs2228000 | Ex9-377C>T | T (0.23) | 0.7 | 4 |
SNP: single nucleotide polymorphism; PCR: polymerase chain reaction; MAF: minor allele frequency; HWE: Hardy-Weinberg equilibrium;
alleles which can increase adduct level as evidenced by previous studies or based on their effect on the enzyme activity.
Genotyping was performed by a SNaPshot procedure as described earlier 18 at the Department of Toxicology, Maastricht University (Maastricht, The Netherlands). Polymerase chain reaction (PCR) primers were designed using Primer 3 and Netprimer software, as detailed before 14, 19. PCR was done in four separate multiplex reactions: two 10-plex and two 7-plex (shown as 1-4 in table 1) on a Tgradient 96-well thermal cycler (Biometra, Goettingen, Germany), each in a 10-μL volume using 96-well plates. The annealing temperatures were optimized for each multiplex PCR: 56°C for multiplex 1, 58°C for multiplex 2, 60°C for multiplex 3, and 60°C for multiplex 4. The PCR products were then incubated (37°C for 45 minutes) with 4 μ@L Exo-SAP-IT (Amersham, Roosendaal, The Netherlands) to digest contaminating deoxynucleotide triphosphates and PCR primers. Enzymes were deactivated at 75°C for 15 minutes. Subsequently, the single base extension (SBE) method by SnaPShot (Applied Biosystems, Nieuwekerk, a.d. IJssel, The Netherlands) was used for genotyping. A different multiplex SBE reaction was conducted on each multiplex PCR product 18. After genotyping, SBE products were analyzed on an ABI Prism 3100 genetic analyzer using Genscan Analysis software (version 3.7). Ten percent of the samples were randomly genotyped in a separate experiment and results were in 100% agreement with the first analysis.
Nucleotide Excision Repair
The nucleotide excision repair (NER) assay was performed at the Department of Toxicology, Maastricht University, by a modified single-cell alkaline gel electrophoresis (Comet) assay, which is based on the fact that the rate limiting steps in NER are the recognition and incision of the damaged DNA 20. The capacity of the cells in performing these rate limiting steps can be quantified through formation of single-strand breaks measured by the comet assay. The NER capacity of the cell extracts is reflected by increased percentages of fluorescence in the tail (“comets”) 21. A549 cells (human epithelial lung carcinoma cells) were trypsinized at 80% confluency and diluted to a concentration of 2×106 cells/ml one day before the assay. Aliquots (25 μl) of untreated A549 cells were mixed with 75 μl low melting point agarose and transferred to microscope slides, which were pre-coated with 1.5% normal electrophoresis grade agarose (Sigma–Aldrich, Germany). Slides were lysed overnight in cold (4°C) lysis buffer. On the day of the assay, the slides were washed and the resulting nucleoids were exposed to 1μM benzo[a]pyrene diol expoxide (BPDE) (NCI Chemical Carcinogen Reference Standard Repository, Midwest Research Institute, Kansas City, MO, USA) or vehicle control (DMSO, 0.5%) for 30 minutes.
The lymphocyte extracts were prepared using the method developed by Redaelli et al. 22 Fifty μl of this extract was added to each slide containing BPDE-exposed gel-embedded nucleoids, and incubated for 10 minutes at 37°C. Immediately after the incubation, slides were put on ice to stop the enzymatic reaction. The slides were further processed according to the conventional comet assay 23. In brief, for DNA denaturation the slides were immersed in electrophoresis buffer (0.3 M NaOH, 1 mM EDTA, approximately pH 13) for 20 minutes, followed by 20 minutes of electrophoresis (25 V and 300 mA). The slides were neutralized to a pH of 7.4 and dried. The dried slides were stained with ethidium bromide (10 mg/ml), and comets were visualized using a Zeiss Axioskop fluorescence microscope. On each slide 50 random cells were analysed using Comet assay III software (Perceptive Instruments, Haverhill, UK). After subtracting background levels from all data, the final NER capacity was calculated by subtracting tail moments of the BPDE-/extract+ (non-adduct-containing nucleoids incubated with protein extract) and BPDE+/extract-(adduct-containing nucleoids exposed to BPDE only) slides from the BPDE+/extract+ (adduct-containing nucleoids incubated with protein extract) slides.
The NER capacity could be measured in only 81 subjects, because in 30 samples, either the amount of material provided was not sufficient to reliably measure DNA repair, or there was visible contamination with red blood cells, which made it impossible to make a representative cell extract.
Urinary 1-hydroxypyrene glucuronide concentration
1-hydroxypyrene glucuronide (1-OHPG) was measured in spot urine specimens at the Department of Environmental Health Sciences, Johns Hopkins Bloomberg School of Public Health (Baltimore, MD, USA) 24. Synchronous fluorescence spectroscopy (Perkin Elmer LS50B Luminescence spectrometer, Norwalk, CT, USA) was used with a wavelength difference of 34 nm between excitation and emission. The limit of detection was 0.01 ng 1-OHPG/ml urine and the assay recovery was 95-100%.
Adduct 32P-postlabelling
32P-postlabelling analysis was carried out using the nuclease P1 enrichment with some modifications at the Institute of Cancer Research (Sutton, UK), as described previously 25. Briefly, DNA (4 μg) was digested using micrococcal endonuclease (0.25 U/ml) and spleen phosphodiesterase (2 mg/ml) for 3.5 hours at 37°C. DNA digests were treated with nuclease P1 (0.96 μl, 1.25 mg/ml) for one hour at 37°C. To stop the nuclease P1-reaction, 1.92 ml Tris base (pH 9.6) was added. Labelling of PAH-modified nucleotides was done with [γ-32P]ATP plus polynucleotide kinase (PNK) (6 U) for 30 min at 37°C. The radiolabelled adducted nucleotide bisphosphates were separated on PEI-cellulose sheets (Machery Nagel, Duren, Germany) by multidirectional thin layer chromatography (TLC). A standard with known levels of benzo[a]pyrene diol-epoxide(BPDE)-DNA adducts was analyzed in parallel for quantification purposes. Quantification was performed using an InstantImager (Canberra Packard, Dowers Grove, IL, USA). Assays were done in duplicate (or in triplicate when necessary) and the averages are reported. The assay coefficient of variation (CV) was 11.7% for the raw data and 10.2% for the log-transformed values.
Statistical analysis
Descriptive data are presented as mean ± standard deviation, and median (interquartile range: IQR). Allele and genotype frequencies were calculated for all SNPs and tested for Hardy-Weinberg equilibrium (Table 1). Since adduct levels, NER capacity, and the urinary 1-OHPG did not have normal distributions and were positively skewed, they were logarithmically transformed before being used in the analyses.
SNPs in the same gene were in linkage disequilibrium, as most D’ statistics were >0.9 (Supplementary Table). Polymorphism data were coded using a gene-based approach. First, for each SNP, using the functional information in the literature 17, 26, the alleles potentially enhancing adduct formation (risk alleles) were determined (table 1). The “risk-allele score” was calculated for each gene in the following way: if at least one of the SNPs in a gene was homozygous for the risk allele, the gene was coded 2; if at least one of the SNPs was heterozygous and none were homozygous for the risk allele, it was coded 1; and if all the SNPs in the same gene had low-risk alleles, it was coded as having 0 risk alleles. For the NAT2 gene, the same scoring was used based on having a genotype with zero, one or two slow acetylation alleles in any of the three SNPs studied 27. For GSTM1 and GSTT1, deletion, which results in the absence of the enzyme, was coded 2 (as opposed to 0 for those with the enzyme present).
In order to analyze the effect of individual genes, both univariate and multivariate methods were used. For univariate analyses, we used one-way analysis of variance to compare log-transformed adduct levels and NER capacity between individuals with different numbers of risk alleles. For multivariate analyses, a backward stepwise regression analysis was used. Log-transformed adduct levels were modeled against xenobiotic and repair genes (coded as described above) and the environmental factors (age, ethnicity, place of residence, passive smoking, red and processed meat intake, cooking method and coal use). Each group of genes (phase I, phase II, and repair) was also adjusted for the sum of risk allele scores in the other groups.
Full models were compared to determine the best model to explain inter-individual variation in DNA adduct levels. The base model included only the selected environmental factors. Other models were built by adding one component at a time to the base model (1-OHPG, phase I genes, phase II genes, repair genes, and NER capacity). Models were compared using Akaike’s information criteria (AIC):
AIC=−2 log L(θ)+2k
where L(θ) is the maximized likelihood function and k is the number of parameters in the model. In other words, AIC adds twice the number of parameters to the likelihood of the model as a penalty for overfitting. Lower AIC indicates a better fit for the model 28.
Spearman correlation was used to assess the linear correlation between the number of risk alleles and the log-adduct levels. HaploStats R package was used to build regression models with haplotypes as predictors and log-adducts or log-NER as outcomes. Haplotypes which met these two criteria were used in the models: high linkage disequilibrium (D’>0.9 or r2>0.8) among the alleles, and a haplotype prevalence of at least 2%. We used Stata 11.2 (StataCorp LP, Tx) and snpStats R package version 1.2.1 for analyses. A two-sided p-value of less than 0.05 was considered to be significant.
Results
Sixty-seven (60%) of the 111 non-smoking women selected for this study were from rural villages, and 44 (40%) lived in the city. The mean age of the study population was 48.5±6.7 years, and 86 (78%) were of Turkmen ethnicity.
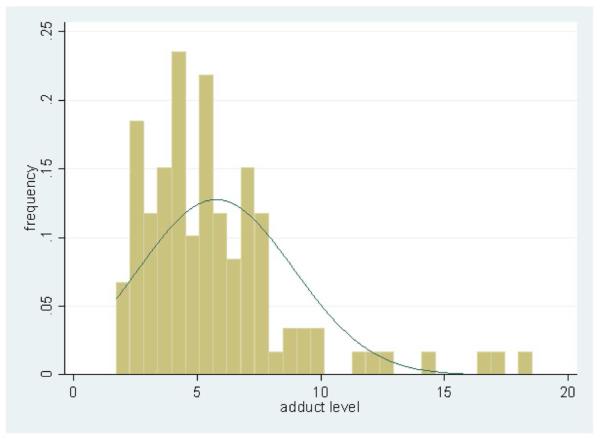
Adduct levels ranged from 1.7 to 18.6 adducts per 108 nucleotides (Figure 1) with a median of 5.1 adducts per 108 nucleotides (IQR: 3.7, 7.1). The mean level of aromatic DNA adducts was 5.8±3.1 adducts per 108 nucleotides.
Figure 1.
Distribution of PAH-related DNA adduct levels among 111 female non-smokers in the Golestan Cohort Study
As Table 2 shows, in the univariate analyses, the levels of aromatic DNA adducts were significantly lower in homozygotes for at least one N-acetyltransferase 2 (NAT2) slow allele and in individuals with the excision repair cross-complementing 5 (ERCC5) non-risk-allele genotype, and were higher in individuals with the myeloperoxidase (MPO) homozygous risk-allele genotype (homozygous carriers of the −463G allele). The multivariate model to predict adduct level as the outcome included all of the genes in one group (phase I, phase II, or repair), and was adjusted for the total of risk-allele scores for the other groups, plus the environmental variables. None of the environmental factors showed a significant association with adduct levels in the multivariate model. Creatinin-adjusted 1-OHPG, a short-term marker of environmental exposure, also did not show a significant association with adduct levels (data not shown). In the adjusted model, the same genes had a significant effect on the adduct level (Table 2): MPO (β=0.21; p=0.01), NAT2 (β=-0.24; p=0.01), and ERCC5 (β=0.16; p=0.04). The sum of the risk allele scores in these three genes also had a significant linear correlation with the log-adduct level (r=0.4, p<0.001).
Table 2.
Univariate and multivariate analyses of the association between the combined risk allele score in each gene and PAH-DNA adduct levels.
| Set | gene | Median adduct (IQR) by risk allele scorest† |
adjusted model †† |
|||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | β | P value | ||
| Phase I | CYP1A1 | 5.0 (3.7,7.1) | 5.3 (4.0,6.5) | 9.2 (9.2,9.2) | −0.04 | 0.73 |
| CYP1A2 | 4.4 (3.6,6.0) | 5.4 (4.4,7.1) | 5.2 (3.3,7.3) | −0.05 | 0.54 | |
| CYP1B1 | 5.4 (4.1,7.0) | 5.2 (3.7,7.2) | 4.4 (3.6,5.3) | −0.13 | 0.15 | |
| CYP2E1 | 5.1 (3.8,7.1) | 5.7 (3.7,7.1) | 4.3 (3.3,5.4) | 0.10 | 0.48 | |
| CYP3A4 | 5.2 (3.8,7.1) | 4.4 (4.0,5.5) | − | −0.20 | 0.43 | |
| mEH | 5.5 (3.0,7.1) | 4.5 (3.9,6.1) | 5.2 (3.3,7.6) | 0.01 | 0.91 | |
| MPO | 4.3 (2.7,5.9) | 4.4 (3.2,6.0) | 5.3 (4.2,7.3)* | 0.21 | 0.01 | |
| Phase II | GPX1 | 5.1 (3.8,7.4) | 5.2 (3.7,6.3) | 4.3 (2.3,6.7) | −0.08 | 0.36 |
| GSTM1 | 5.1 (3.3,7.1) | − | 5.1 (4.2,7.1) | 0.01 | 0.84 | |
| GSTT1 | 5.2 (3.7,7.0) | − | 5.1 (3.8,7.1) | 0.01 | 0.88 | |
| GSTP1 | 5.3 (3.7,7.1) | 5.0 (3.8,7.1) | 5.5 (3.9,6.0) | −0.09 | 0.29 | |
| NAT2 | 6.9 (2.9,7.6) | 5.3 (4.1,7.2) | 4.2 (2.7,5.3)* | −0.24 | 0.01 | |
| NQO1 | 5.3 (3.8,7.2) | 5.1 (3.7,6.7) | 4.6 (2.3,7.0) | −0.12 | 0.20 | |
| MnSOD2 | 4.2 (3.7,5.3) | 6.0 (4.4,7.7) | 4.4 (3.3,5.6) | −0.04 | 0.65 | |
| Repair | APEX1 | 5.1 (3.7,5.7) | 5.2 (4.0,7.1) | 5.1 (3.2,7.1) | −0.01 | 0.97 |
| ERCC1 | 5.0 (4.5,5.5) | 5.1 (3.6,7.1) | 5.3 (4.1,7.1) | 0.02 | 0.86 | |
| ERCC5 | 4.4 (2.8,6.1)* | 5.2 (4.0,7.1) | 6.0 (4.1,7.3) | 0.16 | 0.04 | |
| ERCC6 | 5.0 (4.0,7.0) | 5.3 (3.6,7.1) | 5.0 (2.7,7.7) | −0.09 | 0.31 | |
| MTHFR | 5.3 (3.3,7.1) | 4.8 (3.7,6.7) | 5.4 (3.9, 7.1) | 0.10 | 0.18 | |
| RAD23B | 5.2 (3.8,7.1) | 4.5 (3.6,6.7) | 5.1 (3.7,8.9) | −0.12 | 0.26 | |
| XPA | 4.5 (3.5,6.4) | 5.5 (4.0,7.6) | 5.4 (3.7,6.7) | 0.06 | 0.50 | |
| XPC | 4.7 (4.1,5.7) | 5.1 (3.9,7.1) | 5.3 (3.6,7.0) | −0.01 | 0.90 | |
IQR: inter-quartile range.
2: at least one SNP in the gene was homozygous for the risk allele; 1: at least one SNP in the gene was heterozygous and none were homozygous for the risk allele;0: all of the SNPs in the gene had low-risk alleles.
The model for polymorphisms in each group was adjusted for environmental exposures and the sum of risk allele scores for other groups;
p<0.05 comparing log-transformed adduct levels in one risk score category versus the other two.
NER capacity could be measured in 81 individuals. In these individuals, mean NER capacity was 0.83±0.68 (arbitrary units) and ranged between 0 and 2.49. As Table 3 shows, the homozygote non-risk-allele MTHFR genotype was significantly associated with lower NER capacity. MTHFR CA haplotype also showed a significant association with NER capacity (Supplementary table 2). Also, homozygotes for risk-allele genotype in ERCC1 had a higher NER capacity. NER capacity itself did not have a significant association with the DNA adduct level (data not shown).
Table 3.
Association between the combined risk allele score in repair genes and nucleotide excision repair (NER) capacity.
| gene | Median NER capacity (IQR) by risk allele scorestδ |
||
|---|---|---|---|
| 0 | 1 | 2 | |
| APEX1 | 1.13 (0.73,1.26) | 0.50 (0.29,1.04) | 0.87 (0.42,1.42) |
| ERCC1 | 0.80 (0.56,2.03) | 0.49 (0.17,1.10) | 0.99 (0.60,1.20)* |
| ERCC5 | 0.56 (0.07,1.10) | 0.75 (0.33,1.27) | 0.57 (0.49,1.04) |
| ERCC6 | 0.61 (0.27,1.04) | 0.61 (0.29,1.26) | 1.19 (0.52,2.03) |
| MTHFR | 0.36 (0.17,0.86)* | 0.80 (0.44,1.20) | 0.81 (0.33,1.42) |
| RAD23B | 0.80 (0.27,1.42) | 0.61 (0.33,1.10) | 0.78 (0.37,1.19) |
| XPA | 0.50 (0.14,0.99) | 0.88 (0.42,1.42) | 0.85 (0.40,1.04) |
| XPC | 1.33 (0.44,2.21) | 0.59 (0.32,1.20) | 0.81 (0.30,1.12) |
IQR: inter-quartile range.
2: at least one SNP in the gene was homozygous for the risk allele; 1: at least one SNP in the gene was heterozygous and none were homozygous for the risk allele; 0: all of the SNPs in the gene had low-risk alleles;
p<0.05 comparing log-transformed NER capacity in one risk score category versus the other two.
In general, the models including phenotypically assessed NER capacity had a considerably better fit than the models without it. As Table 4 shows, the model that best fit the data (as evidenced by the lowest AIC) was the one including the environmental factors, phase I enzymes and NER capacity. This model had an r2 of 0.24, and compared with the 0.07 r2 of the model including environmental model, it could explain 17% more of the variation in DNA adduct levels. The model deteriorated slightly when the phase II genes were added. Also, the models had a better fit when phenotypically assessed NER was used instead of the polymorphisms in repair genes.
Table 4.
Comparison of different models in predicting DNA adduct levels
| Variables in the Model* | AIC |
|---|---|
| a. Environmental | 156.33 |
| b. Environmental + 1-ohpg | 157.84 |
| c. Environmental+ phase I polymorphisms | 151.19 |
| d. Environmental+ phase II polymorphisms | 157.10 |
| e. Environmental + phase I and phase II polymorphisms | 154.57 |
| f. Environmental + repair genes | 160.68 |
| g. Environmental + phase I polymorphisms + repair genes | 157.49 |
| h. Environmental + phase I and phase II polymorphisms + repair genes | 158.79 |
| i. Environmental + NER capacity | 102.66 |
| j. Environmental + phase I polymorphisms + NER capacity | 100.07 |
| k. Environmental + phase II polymorphisms + NER capacity | 105.02 |
| l. Environmental + phase I and phase II polymorphisms + NER capacity | 106.50 |
AIC: Akaike's information criterion (lower AIC indicates a better fit of the model); NER: Nucleotide excision repair;
The base model included only environmental factors; other models were built by adding one group of variables at a time to the base model.
Discussion
In this study we observed high levels of PAH-related DNA adducts, with substantial inter-individual variation. The model including phase I gene variant scores and measured NER capacity seemed to explain this variability better than other models, although individual SNPs from phase I, phase II and repair genes also showed significant associations with adduct levels.
In our study, mean aromatic DNA adduct level was 5.8±3.1 adducts per 108 nucleotides. These findings are in line with previous findings of a very high level of exposure to PAH in this region 3. The source of this high PAH exposure is not very well understood, but studies suggest that food, and its method of preparation might play a role8. In any case, we ruled out external contamination by careful quality control of sample collection and processing. In Dutch smokers, the mean DNA adduct level has been reported to be 1.40±0.79 adducts per 108 nucleotides, with a range between 0.25 and 3.90 14. Binkova et al. reported PAH adducts in several occupationally-exposed groups, such as Czech and Slovak coke-oven workers, and even among the smokers in these groups, they observed levels much lower than those in the present study 29-30. In another study of occupationally-exposed workers from China, another country with a high rate of esophageal squamous cell carcinoma, the highest mean PAH-related DNA adduct level, among coke oven workers, was 1.66±0.21 adducts per 108 nucleotides 31. All of these adduct levels were measured using 32P-postlabelling, and variations in postlabelling results from different laboratories may contribute to some of these differences. Nevertheless, both our measurements and those in the Dutch study were done in laboratories which participated in a study to make postlabelling protocols more uniform across Europe, which significantly improved assay reliability in the participating laboratories 32. In general, 32P-postlabelling has proven to be a relatively reliable method for measuring aromatic DNA adducts, and has improved over the years 33, and in our study, we observed a good assay CV (10.2%) in the duplicate samples.
Measurement of DNA adducts can be considered as an intermediate marker of cancer risk, which reflects the biologically effective dose of PAH exposure 30, 34. As in our study, however, SNPs in some individual genes may not show significant associations with DNA adduct levels, in spite of the crucial role of these genes in coding enzymes which metabolize PAHs. This low level of association may be caused by several factors including a large degree of variation in environmental exposures and unmeasured factors which affect adduct levels, and insufficient power of the study due to the low frequency of the studied polymorphisms. In this setting, model comparison using information criteria such as AIC offers two advantages over traditional hypothesis-testing: firstly it does not rely on a predefined significance level, and its power in determining the best model is not dependent on study power or sample size 35. In general, if the difference between the AIC of the “best” model and other models is more than 2, the model has considerable support, and if this difference is above 10, this support is very strong 35. In our study models with NER capacity had the highest AIC difference, followed by those including phase 1 enzymes. To the best of our knowledge, this is the first study showing the importance of NER phenotyping in predicting aromatic DNA adduct levels. Compared to genotyping, repair phenotyping provides a better indication of the cells’ enzymatic repair capacity 17.
The second advantage of AIC, is that it assesses the whole model rather than its individual components. In the present study we found significant results for only one of the phase I genes, and the effect of NER capacity on DNA adduct levels was not significant by itself, but these two together comprised the best model. This underlines the importance of a comprehensive approach to evaluating pathways affecting cancer risk or carcinogenic metabolism. One of the approaches proposed for such analysis is the consideration of simultaneous genetic variations in multiple genes 36. In a study on 170 healthy volunteers, Butkiewicz et al. 37 showed that although a GSTP1 polymorphism alone was not associated with a change in DNA adduct level, when combined with GSTM1 deletion, it was associated with increased adduct formation. Georgiadis and colleagues 38 also used a similar combination method, and showed a synergistic effect between phase I and phase II genes. The limitation of such an approach is that with increasing numbers of genes, and polymorphisms within each gene, interpretation becomes very complicated as the number of different possible combinations increases dramatically.
In a different approach, the total number of risk alleles can be used to summarize the genetic risk profile of an individual. Ketelslegers et al. used this method and observed an association between the number of risk alleles and DNA adduct levels in smokers 14. The sum of three risk alleles in their study (GSTM1, mEH and GPX) was associated with a two-fold increase in adduct level. We used the same approach for three different genes (MPO, NAT2 and ERCC5) and observed a linear association. We also adopted a gene-based approach by combining the polymorphic alleles in each gene, which were known to act in the same direction and were in linkage disequilibrium, and used modeling to identify the combination of different alleles that could best explain variability in adduct levels. In this way, SNP’s in phase I genes appeared to influence DNA adduct formation more than SNP’s in phase II genes. Agudo and colleagues 39 also used a multivariate regression model in the EPIC-Spain cohort and defined “imputed phenotypes” based on the combinations of genotypes in defined pathways. They observed significant changes in DNA adduct levels caused by combined oxidation-hydrolysis (phase I enzymes), acetylation (NAT2), and oxidation-acetylation pathways, which is in line with our findings. Abnet and colleagues 40 studied a group of individuals from another ESCC high risk area in Brazil, using a multivariate approach. They showed that adding phase I polymorphisms to environmental exposures increased the ability of the model to predict 1-OHPG concentration by 12%. 1-OHPG is a biomarker that has been extensively used to evaluate PAH exposure in general population and occupational groups. This marker, however, has a short half-life and may not be suitable for studying the long-term carcinogenic effects of PAHs and their metabolites 40. In our study, 1-OHPG was not associated with the adduct level and did not improve adduct formation model fit.
We found several interesting associations with individual genes. The finding that MPO genotype influences PAH-related DNA adducts is consistent in the literature and is related both to its important role in the benzo-[a]pyrene (BaP) activation as a result of oxidation of B[a]P to BaP-7,8-diol and ultimately BPDE41-42, and to its ability to diminish DNA repair via the local production of hypochlorous acid (HOCl) at inflammation sites43-44. Rojas et al.45 showed that BPDE-DNA adduct formation was decreased in the skin of coal-tar treated patients carrying the MPO mutant allele. In another study, this polymorphism was shown to reduce DNA adduct levels in bronchoalveolar lavage specimens from smokers 46.
Studies on the effects of NAT2 polymorphisms on adduct formation have had different results. Godschalk et al. 10 showed a lower level of DNA adducts in the fast-acetylation group. In contrast, in the EPIC-Spain cohort, slow acetylation by NAT2 was associated with a significantly decreased adduct level in healthy adults 39, similar to our results. Lee et al. 47 showed that slow acetylation was associated with increased DNA adducts in the lung, but a non-significant reduction of adducts in the blood monocytes of lung cancer patients. Substrate-specific action of NAT has been proposed as a possible reason for such differences 39.
Some of the repair genes also showed associations with either DNA adduct level or NER capacity. ERCC-5 was the only repair gene in our data which had a significant effect on DNA adduct level. The ERCC-5 C/C genotype which was associated with increased adduct level in our study, has been shown to increase the risk of lung cancer 48. We also showed that NER capacity itself is increased in individuals with MTHFR and ERCC1 polymorphisms. Two of the SNPs we studied in the ERCC1 gene (rs11615 T allele and rs3212986 C allele) have been shown to increase DNA adduct levels 49, but this is the first study to show the association between this gene and NER capacity.
Although the role of MTHFR in DNA repair may not be straightforward, it has been shown that the rs1801133 MTHFR T allele could increase 5,10-methylenetetrahydrofolate levels, which are necessary for DNA repair 50. Choi and colleagues51 have shown reduced excision repair in folate-depleted rats. However, unlike our assay, which quantifies the comet formation due to single-strand breaks, this reduced repair was reflected by the slowed recovery of the damage-induced comets as a result of insufficient thymidylate and purine production. Besides it is not clear how much dietary intake, in the ranges higher than the depletion investigated in Choi’s study, can affect DNA repair. We did not measure folate intake in our study to further investigate the possibility of such an association, and it is unlikely that our study population had a very heterogeneous pattern of folate intake.
This study has one of the largest sets of PAH metabolizing and repair genes that have been studied together, and it combines repair phenotyping and genotyping in one study. Also, almost all previous studies, unlike the present one, have included only smokers or groups with occupational exposures. Given the importance of PAH exposure in non-smokers and its relevance to ESCC and lung cancer in this group of individuals, this study can contribute to our understanding of PAH-related carcinogenesis in this group. One of the limitations of this study is its relatively small sample size. So, as in many similar studies, the sample size did not allow more sophisticated pathway analyses, and may have been the reason for the lack of association between NER capacity and adduct levels. We also did not study more genes in the pathway, which might explain some of the remaining 76% of the variation in the adduct level, and had to assume linearity of effects when using regression models, which is not necessarily the case.
In conclusion, we have observed high levels of PAH-related DNA adducts in female non-smokers in this population, and found interesting associations between MPO, NAT2 and ERCC5 polymorphisms and DNA adduct levels. We have also shown the importance of studying combined genetic polymorphisms and NER phenotyping when studying cancer risk in relation to PAH exposure. Finally, we have shown that inter-individual variation in adduct levels could best be explained by a combination of polymorphisms in phase I genes and NER capacity.
Supplementary Material
Novelty and Impact.
In a population at high risk of esophageal squamous cell carcinoma (ESCC), female non-smokers had exceptionally high aromatic DNA adducts, with large inter-individual variation best explained by a combination of phase I genes and nucleotide excision repair capacity. This study has one of the largest sets of PAH metabolizing and repair genes studied together along with repair phenotyping, and uses novel methods in modeling an important biomarker in the carcinogenesis of ESCC and lung cancer.
Acknowledgment
This work was supported by the Digestive Disease Research Center of Tehran University of Medical Sciences and the International Agency for Research on Cancer for data and sample collection; ECNIS (Environmental Cancer Risk, Nutrition and Individual Susceptibility), a network of excellence operating within the European Union 6th Framework Program [Priority 5: “Food Quality and Safety” (Contract No. 513943)] for laboratory assessments; the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health; and Tehran University of Medical Sciences [grant No 88-04-37-10130].
References
- 1.Baan R, Grosse Y, Straif K, Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V. A review of human carcinogens--Part F: chemical agents and related occupations. Lancet Oncol. 2009;10:1143–4. doi: 10.1016/s1470-2045(09)70358-4. [DOI] [PubMed] [Google Scholar]
- 2.Abedi-Ardekani B, Kamangar F, Hewitt SM, Hainaut P, Sotoudeh M, Abnet CC, Taylor PR, Boffetta P, Malekzadeh R, Dawsey SM. Polycyclic aromatic hydrocarbon exposure in oesophageal tissue and risk of oesophageal squamous cell carcinoma in north-eastern Iran. Gut. 2010;59:1178–83. doi: 10.1136/gut.2010.210609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamangar F, Strickland PT, Pourshams A, Malekzadeh R, Boffetta P, Roth MJ, Abnet CC, Saadatian-Elahi M, Rakhshani N, Brennan P, Etemadi A, Dawsey SM. High exposure to polycyclic aromatic hydrocarbons may contribute to high risk of esophageal cancer in northeastern Iran. Anticancer Res. 2005;25:425–8. [PubMed] [Google Scholar]
- 4.Fagundes RB, Abnet CC, Strickland PT, Kamangar F, Roth MJ, Taylor PR, Dawsey SM. Higher urine 1-hydroxy pyrene glucuronide (1-OHPG) is associated with tobacco smoke exposure and drinking mate in healthy subjects from Rio Grande do Sul, Brazil. BMC Cancer. 2006;6:139. doi: 10.1186/1471-2407-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wornat MJ, Ledesma EB, Sandrowitz AK, Roth MJ, Dawsey SM, Qiao YL, Chen W. Polycyclic aromatic hydrocarbons identified in soot extracts from domestic coal-burning stoves of Henan Province, China. Environ Sci Technol. 2001;35:1943–52. doi: 10.1021/es001664b. [DOI] [PubMed] [Google Scholar]
- 6.Roth MJ, Qiao YL, Rothman N, Tangrea JA, Dawsey SM, Wang GQ, Cho SH, Kang D, Taylor PR, Strickland PT. High urine 1-hydroxypyrene glucuronide concentrations in Linxian, China, an area of high risk for squamous oesophageal cancer. Biomarkers. 2001;6:381–6. doi: 10.1080/13547500110044780. [DOI] [PubMed] [Google Scholar]
- 7.Roth MJ, Strickland KL, Wang GQ, Rothman N, Greenberg A, Dawsey SM. High levels of carcinogenic polycyclic aromatic hydrocarbons present within food from Linxian, China may contribute to that region’s high incidence of oesophageal cancer. Eur J Cancer. 1998;34:757–8. doi: 10.1016/s0959-8049(97)10071-5. [DOI] [PubMed] [Google Scholar]
- 8.Hakami R, Mohtadinia J, Etemadi A, Kamangar F, Nemati M, Pourshams A, Islami F, Nasrollahzadeh D, Saberi-Firoozi M, Birkett N, Boffetta P, Malekzadeh R. Dietary intake of benzo(a)pyrene and risk of esophageal cancer in north of Iran. Nutr Cancer. 2008;60:216–21. doi: 10.1080/01635580701684831. [DOI] [PubMed] [Google Scholar]
- 9.Samet JM, Avila-Tang E, Boffetta P, Hannan LM, Olivo-Marston S, Thun MJ, Rudin CM. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15:5626–45. doi: 10.1158/1078-0432.CCR-09-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godschalk RW, Dallinga JW, Wikman H, Risch A, Kleinjans JC, Bartsch H, Van Schooten FJ. Modulation of DNA and protein adducts in smokers by genetic polymorphisms in GSTM1,GSTT1, NAT1 and NAT2. Pharmacogenetics. 2001;11:389–98. doi: 10.1097/00008571-200107000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Phillips DH. DNA adducts as markers of exposure and risk. Mutat Res. 2005;577:284–92. doi: 10.1016/j.mrfmmm.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa T, Zhang SS, Qin X, Takahashi Y, Oda H, Nakatsuru Y, Ide F. DNA repair and cancer: lessons from mutant mouse models. Cancer Sci. 2004;95:112–7. doi: 10.1111/j.1349-7006.2004.tb03190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godschalk RW, Van Schooten FJ, Bartsch H. A critical evaluation of DNA adducts as biological markers for human exposure to polycyclic aromatic compounds. J Biochem Mol Biol. 2003;36:1–11. doi: 10.5483/bmbrep.2003.36.1.001. [DOI] [PubMed] [Google Scholar]
- 14.Ketelslegers HB, Gottschalk RW, Godschalk RW, Knaapen AM, van Schooten FJ, Vlietinck RF, Kleinjans JC, van Delft JH. Interindividual variations in DNA adduct levels assessed by analysis of multiple genetic polymorphisms in smokers. Cancer Epidemiol Biomarkers Prev. 2006;15:624–9. doi: 10.1158/1055-9965.EPI-05-0431. [DOI] [PubMed] [Google Scholar]
- 15.Pourshams A, Khademi H, Malekshah AF, Islami F, Nouraei M, Sadjadi AR, Jafari E, Rakhshani N, Salahi R, Semnani S, Kamangar F, Abnet CC, et al. Cohort Profile: The Golestan Cohort Study--a prospective study of oesophageal cancer in northern Iran. Int J Epidemiol. 2010;39:52–9. doi: 10.1093/ije/dyp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyum A. Isolation of lymphocytes, granulocytes and macrophages. Scand J Immunol. 1976;(Suppl 5):9–15. [PubMed] [Google Scholar]
- 17.Hirvonen A. State of the art of genotype vs. phenotype studies Environmental Cancer Risk, Nutrition and Individual Susceptibility (ECNIS) Lodz: Nofer Institute of Occupational Medicine. 2008:98. [Google Scholar]
- 18.Knaapen AM, Ketelslegers HB, Gottschalk RW, Janssen RG, Paulussen AD, Smeets HJ, Godschalk RW, Van Schooten FJ, Kleinjans JC, Van Delft JH. Simultaneous genotyping of nine polymorphisms in xenobiotic-metabolizing enzymes by multiplex PCR amplification and single base extension. Clin Chem. 2004;50:1664–8. doi: 10.1373/clinchem.2004.034058. [DOI] [PubMed] [Google Scholar]
- 19.Langie SA, Wilms LC, Hamalainen S, Kleinjans JC, Godschalk RW, van Schooten FJ. Modulation of nucleotide excision repair in human lymphocytes by genetic and dietary factors. Br J Nutr. 2010;103:490–501. doi: 10.1017/S0007114509992066. [DOI] [PubMed] [Google Scholar]
- 20.Hansen WK, Kelley MR. Review of mammalian DNA repair and translational implications. J Pharmacol Exp Ther. 2000;295:1–9. [PubMed] [Google Scholar]
- 21.Langie SA, Knaapen AM, Brauers KJ, van Berlo D, van Schooten FJ, Godschalk RW. Development and validation of a modified comet assay to phenotypically assess nucleotide excision repair. Mutagenesis. 2006;21:153–8. doi: 10.1093/mutage/gel013. [DOI] [PubMed] [Google Scholar]
- 22.Redaelli A, Magrassi R, Bonassi S, Abbondandolo A, Frosina G. AP endonuclease activity in humans: development of a simple assay and analysis of ten normal individuals. Teratog Carcinog Mutagen. 1998;18:17–26. [PubMed] [Google Scholar]
- 23.Knaapen AM, Schins RP, Borm PJ, van Schooten FJ. Nitrite enhances neutrophil-induced DNA strand breakage in pulmonary epithelial cells by inhibition of myeloperoxidase. Carcinogenesis. 2005;26:1642–8. doi: 10.1093/carcin/bgi116. [DOI] [PubMed] [Google Scholar]
- 24.Strickland PT, Kang D, Bowman ED, Fitzwilliam A, Downing TE, Rothman N, Groopman JD, Weston A. Identification of 1-hydroxypyrene glucuronide as a major pyrene metabolite in human urine by synchronous fluorescence spectroscopy and gas chromatography-mass spectrometry. Carcinogenesis. 1994;15:483–7. doi: 10.1093/carcin/15.3.483. [DOI] [PubMed] [Google Scholar]
- 25.Phillips DH, Arlt VM. The 32P-postlabeling assay for DNA adducts. Nat Protoc. 2007;2:2772–81. doi: 10.1038/nprot.2007.394. [DOI] [PubMed] [Google Scholar]
- 26.Ketelslegers HB, Gottschalk RW, Koppen G, Schoeters G, Baeyens WF, van Larebeke NA, van Delft JH, Kleinjans JC. Multiplex genotyping as a biomarker for susceptibility to carcinogenic exposure in the FLEHS biomonitoring study. Cancer Epidemiol Biomarkers Prev. 2008;17:1902–12. doi: 10.1158/1055-9965.EPI-08-0045. [DOI] [PubMed] [Google Scholar]
- 27.Walker K, Ginsberg G, Hattis D, Johns DO, Guyton KZ, Sonawane B. Genetic polymorphism in N-Acetyltransferase (NAT): Population distribution of NAT1 and NAT2 activity. J Toxicol Environ Health B Crit Rev. 2009;12:440–72. doi: 10.1080/10937400903158383. [DOI] [PubMed] [Google Scholar]
- 28.Bozdogan H. Akaike’s information criterion and recent developments in information complexity. J Math Psychol. 2000;44:62–91. doi: 10.1006/jmps.1999.1277. [DOI] [PubMed] [Google Scholar]
- 29.Binkova B, Topinka J, Mrackova G, Gajdosova D, Vidova P, Stavkova Z, Peterka V, Pilcik T, Rimar V, Dobias L, Farmer PB, Sram RJ. Coke oven workers study: the effect of exposure and GSTM1 and NAT2 genotypes on DNA adduct levels in white blood cells and lymphocytes as determined by 32P-postlabelling. Mutat Res. 1998;416:67–84. doi: 10.1016/s1383-5718(98)00061-8. [DOI] [PubMed] [Google Scholar]
- 30.Binkova B, Chvatalova I, Lnenickova Z, Milcova A, Tulupova E, Farmer PB, Sram RJ. PAH-DNA adducts in environmentally exposed population in relation to metabolic and DNA repair gene polymorphisms. Mutat Res. 2007;620:49–61. doi: 10.1016/j.mrfmmm.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, Li G, Xue X, Zhou Z, Li X, Fu J, Cohen B, Roy N, Li D, Sun J, Nan P, Tang MS, et al. PAH-DNA adducts in a Chinese population: relationship to PAH exposure, smoking and polymorphisms of metabolic and DNA repair genes. Biomarkers. 2008;13:27–40. doi: 10.1080/13547500701671895. [DOI] [PubMed] [Google Scholar]
- 32.Phillips DH, Castegnaro M. Standardization and validation of DNA adduct postlabelling methods: report of interlaboratory trials and production of recommended protocols. Mutagenesis. 1999;14:301–15. doi: 10.1093/mutage/14.3.301. [DOI] [PubMed] [Google Scholar]
- 33.Gallo V, Khan A, Gonzales C, Phillips DH, Schoket B, Gyorffy E, Anna L, Kovacs K, Moller P, Loft S, Kyrtopoulos S, Matullo G, et al. Validation of biomarkers for the study of environmental carcinogens: a review. Biomarkers. 2008;13:505–34. doi: 10.1080/13547500802054611. [DOI] [PubMed] [Google Scholar]
- 34.Brandt HC, Watson WP. Monitoring human occupational and environmental exposures to polycyclic aromatic compounds. Ann Occup Hyg. 2003;47:349–78. doi: 10.1093/annhyg/meg052. [DOI] [PubMed] [Google Scholar]
- 35.Burnham KP, Anderson DR. Model selection and multimodel inference: a practical information-theoretic approach. 2nd ed Springer; New York: 2002. [Google Scholar]
- 36.Alexandrov K, Cascorbi I, Rojas M, Bouvier G, Kriek E, Bartsch H. CYP1A1 and GSTM1 genotypes affect benzo[a]pyrene DNA adducts in smokers’ lung: comparison with aromatic/hydrophobic adduct formation. Carcinogenesis. 2002;23:1969–77. doi: 10.1093/carcin/23.12.1969. [DOI] [PubMed] [Google Scholar]
- 37.Butkiewicz D, Grzybowska E, Phillips DH, Hemminki K, Chorazy M. Polymorphisms of the GSTP1 and GSTM1 genes and PAH-DNA adducts in human mononuclear white blood cells. Environ Mol Mutagen. 2000;35:99–105. [PubMed] [Google Scholar]
- 38.Georgiadis P, Demopoulos NA, Topinka J, Stephanou G, Stoikidou M, Bekyrou M, Katsouyianni K, Sram R, Autrup H, Kyrtopoulos SA. Impact of phase I or phase II enzyme polymorphisms on lymphocyte DNA adducts in subjects exposed to urban air pollution and environmental tobacco smoke. Toxicol Lett. 2004;149:269–80. doi: 10.1016/j.toxlet.2003.12.038. [DOI] [PubMed] [Google Scholar]
- 39.Agudo A, Peluso M, Sala N, Capella G, Munnia A, Piro S, Marin F, Ibanez R, Amiano P, Tormo MJ, Ardanaz E, Barricarte A, et al. Aromatic DNA adducts and polymorphisms in metabolic genes in healthy adults: findings from the EPIC-Spain cohort. Carcinogenesis. 2009;30:968–76. doi: 10.1093/carcin/bgp062. [DOI] [PubMed] [Google Scholar]
- 40.Abnet CC, Fagundes RB, Strickland PT, Kamangar F, Roth MJ, Taylor PR, Dawsey SM. The influence of genetic polymorphisms in Ahr, CYP1A1, CYP1A2, CYP1B1, GST M1, GST T1 and UGT1A1 on urine 1-hydroxypyrene glucuronide concentrations in healthy subjects from Rio Grande do Sul, Brazil. Carcinogenesis. 2007;28:112–7. doi: 10.1093/carcin/bgl131. [DOI] [PubMed] [Google Scholar]
- 41.Petruska JM, Mosebrook DR, Jakab GJ, Trush MA. Myeloperoxidase-enhanced formation of (+-)-trans-7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene-DNA adducts in lung tissue in vitro: a role of pulmonary inflammation in the bioactivation of a procarcinogen. Carcinogenesis. 1992;13:1075–81. doi: 10.1093/carcin/13.7.1075. [DOI] [PubMed] [Google Scholar]
- 42.Knaapen AM, Gungor N, Schins RP, Borm PJ, Van Schooten FJ. Neutrophils and respiratory tract DNA damage and mutagenesis: a review. Mutagenesis. 2006;21:225–36. doi: 10.1093/mutage/gel032. [DOI] [PubMed] [Google Scholar]
- 43.Gungor N, Godschalk RW, Pachen DM, Van Schooten FJ, Knaapen AM. Activated neutrophils inhibit nucleotide excision repair in human pulmonary epithelial cells: role of myeloperoxidase. FASEB J. 2007;21:2359–67. doi: 10.1096/fj.07-8163com. [DOI] [PubMed] [Google Scholar]
- 44.Gungor N, Haegens A, Knaapen AM, Godschalk RW, Chiu RK, Wouters EF, van Schooten FJ. Lung inflammation is associated with reduced pulmonary nucleotide excision repair in vivo. Mutagenesis. 2010;25:77–82. doi: 10.1093/mutage/gep049. [DOI] [PubMed] [Google Scholar]
- 45.Rojas M, Godschalk R, Alexandrov K, Cascorbi I, Kriek E, Ostertag J, Van Schooten FJ, Bartsch H. Myeloperoxidase--463A variant reduces benzo[a]pyrene diol epoxide DNA adducts in skin of coal tar treated patients. Carcinogenesis. 2001;22:1015–8. doi: 10.1093/carcin/22.7.1015. [DOI] [PubMed] [Google Scholar]
- 46.Van Schooten FJ, Boots AW, Knaapen AM, Godschalk RW, Maas LM, Borm PJ, Drent M, Jacobs JA. Myeloperoxidase (MPO) −463G->A reduces MPO activity and DNA adduct levels in bronchoalveolar lavages of smokers. Cancer Epidemiol Biomarkers Prev. 2004;13:828–33. [PubMed] [Google Scholar]
- 47.Lee MS, Su L, Christiani DC. Synergistic effects of NAT2 slow and GSTM1 null genotypes on carcinogen DNA damage in the lung. Cancer Epidemiol Biomarkers Prev. 2010;19:1492–7. doi: 10.1158/1055-9965.EPI-09-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiyohara C, Yoshimasu K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: a meta-analysis. Int J Med Sci. 2007;4:59–71. doi: 10.7150/ijms.4.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee MS, Su L, Mark EJ, Wain JC, Christiani DC. Genetic modifiers of carcinogen DNA adducts in target lung and peripheral blood mononuclear cells. Carcinogenesis. 2010;31:2091–6. doi: 10.1093/carcin/bgq208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma J, Stampfer MJ, Giovannucci E, Artigas C, Hunter DJ, Fuchs C, Willett WC, Selhub J, Hennekens CH, Rozen R. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997;57:1098–102. [PubMed] [Google Scholar]
- 51.Choi SW, Kim YI, Weitzel JN, Mason JB. Folate depletion impairs DNA excision repair in the colon of the rat. Gut. 1998;43:93–9. doi: 10.1136/gut.43.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.