Abstract
Several paradigm shifting advances have recently been made on the composition and function of the chromosomal DNA replication machinery. Replisomes appear to be more fluid and dynamic than ever imagined, enabling rapid and efficient bypass of roadblocks and template lesions while faithfully replicating chromosomal DNA. This fluidity is determined by many layers of regulation, which reach beyond the role of replisome components themselves. In fact, recent studies show that additional polymerases, post-transcriptional modifications and chromatin structure are required for complete chromosome duplication. Many of these factors are involved with the more complex events that take place during lagging strand synthesis. These, and other recent discoveries, are the focus of this review.
Keywords: DNA replication, Okazaki fragment synthesis, single molecule analysis, post-translational modifications, transcription, collisions
Conservation of the replisome stops at the fork
The numerous proteins required to advance a replication fork act together as a machine referred to as the replisome [1]. The basic enzymatic activities of cellular replisomes are common to all domains of life and include the DNA polymerases, proofreading 3’–5’ exonucleases, a hexameric helicase, primase, and a heteropentameric clamp loader that assembles ring shaped sliding clamps onto primed sites to tether polymerases to DNA for high processivity [1–3]. The leading strand is synthesized in the direction of DNA unwinding, but the antiparallel structure of duplex DNA requires the lagging strand to be extended in the opposite direction of fork movement, as a series of Okazaki fragments. As leading strand synthesis progresses, the lagging strand template accumulates single-strand (ss) DNA, which is tightly bound by ssDNA binding proteins. Connection between the leading and lagging strand polymerases results in a growing replication loop during Okazaki fragment extension [4]. After the Okazaki fragment is finished, the loop dismantles, the RNA primer is processed and individual fragments are sealed together by ligase.
Although the conservation of these replisome components is substantial, this is where the similarity between eukaryotic and prokaryotic replisomes stop. This review outlines recent advances that change our view of the prokaryotic replisome structure, and reveal a more dynamic machine than previously thought possible. Several new findings in the eukaryotic replication field reveal a replisome that is very different from the prokaryotic counterpart, and is highly regulated by posttranslational modifications. Eukaryotes package the DNA into chromatin, with nucleosomes as the most basic unit. Recent findings reveal a role of the nucleosome in eukaryotic lagging strand synthesis in a fascinating mechanism that controls Okazaki fragment length and regulates removal of the RNA/DNA hybrid primer made by the low fidelity Pol α/primase.
PROKARYOTIC REPLISOMES
Bacterial replisomes contain three DNA polymerases
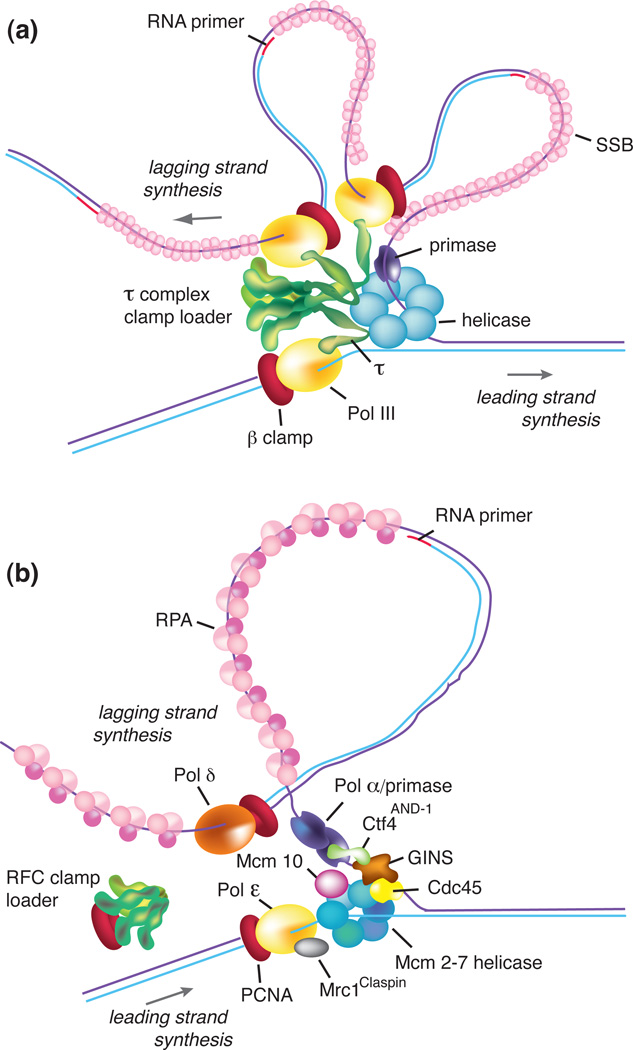
An illustration of the Escherichia coli replisome is shown in Figure 1a. The E. coli DnaB hexameric helicase encircles and tracks along the lagging strand to unwind the parental duplex. DnaG primase transiently interacts with DnaB helicase to initiate primer synthesis, thereby localizing the synthesis of lagging strand RNA primers to the forked junction [1]. Circular β processivity clamps are then loaded onto newly synthesized RNA primers by a multiprotein complex clamp loader (τ-complex (τ3δδ’ψχ)) [2]. In prokaryotes, the τ subunit of the clamp loader interacts with the catalytic α subunit of the DNA polymerase III (Pol) core (α, ε, θ), coupling the polymerases that operate simultaneously on the separated strands of the parental duplex. The τ complex also connects to the helicase, which greatly stimulates the rate of duplex unwinding when the leading strand Pol III is active [5].
Figure 1. Organization of proteins at a moving replication fork.
(a) Bacterial replication fork. The parental duplex is unwound by the DnaB helicase (blue) that encircles the lagging strand and unwinds DNA ahead of Pol III (yellow). Primase (purple) synthesizes short RNA primers to initiate Okazaki fragment synthesis on the lagging strand. The lagging strand ssDNA is coated by single strand DNA binding protein SSB (pink). Three Pol III enzymes are coupled through the three τ subunits of the clamp loader (green), which uses ATP to assemble β processivity clamps (red) onto primed sites. The simultaneous action of two polymerases is a speculative scenario and the frequency with which all three polymerases are used simultaneously is not certain. (b) Eukaryotic replication fork. The Mcm2-7 helicase (blue) is assembled on the leading strand ssDNA. The active form of the helicase is the CMG complex that includes Mcm2-7, the GINS tetramer (brown), and Cdc45 (yellow). Ctf4 (green) interacts with Polα-primase (purple) and GINS. Mcm10 (purple) binds to Mcm2-7. Mrc1 binds the leading strand Pol ε (yellow). Pol ε and the lagging strand Pol δ (chestnut) both make tight contacts to the PCNA (proliferating cell nuclear antigen) processivity clamps (red). The unwound lagging strand ssDNA is coated by RPA (pink). The RFC (replication factor C) clamp loader (green) uses ATP to assemble PCNA clamps onto DNA, but a connection of RFC to the replisome is not known.
It has been thought that replication forks may function in the context of a replication factory, in which two interconnected replisomes remain in a fixed position of the cell while the DNA moves though them[6–8]. Recent data using fluorescently labeled proteins and in vivo imaging have challenged this view. E. coli has a single origin from which two replication forks proceed in a bidirectional fashion around the entire 4.4 Mb genome. Use of high resolution fluorescence microscopy to visualize replisomes in the cell show two separate replication forks at ¼ and ¾ positions in a dividing cell, indicating that the two replication forks are distinct entities, and do not remain coupled in a replication factory [9, 10]. Recent single molecule microscopy studies on replicating λ DNA in Xenopus leavis nuclear extracts suggest that replication forks are uncoupled in eukaryotes as well [11].
Besides a different view on the organization of individual replication forks within the cell, a surprising observation of the organization within the replisome itself has recently come to light. DNA polymerases have long been thought to act in pairs, where one polymerase synthesizes the leading strand and the second polymerase replicates the lagging strand. This view has recently changed. Reconstitution of the E. coli DNA polymerase III holoenzyme from purified proteins results in a particle that contains three active Pol III cores, each connected to one of the three identical τ subunits within the clamp loader [12]. Furthermore, a study in the T4 replication system (Glossary Box) using electron microscopy and nanoscale DNA biopointers (Glossary Box) revealed the presence of three polymerases in about 6% of phage T4 replisomes [13]. Until recently however, it was not known if this tri-polymerase architecture indeed represents the functional unit at the replication fork in vivo. This has now been addressed by the recent development of powerful single molecule high resolution microscopy methods that have enabled the determination of protein stoichiometries in the cell and observations of dynamic processes in vivo (Box 1). Using fluorescently labeled proteins and photobleaching techniques, a new study has revealed the presence of three Pol III cores and three τ subunits within the E. coli replisome in vivo [14] (Box 1), confirming the tri-polymerase replisome structure in the cell. Subsequent studies, both in vitro and in vivo, conclude that all three Pol III cores are active during replisome function [10, 15]. But why should the replisome contain three polymerases when there are only two DNA strands to replicate? Single-molecule Total Internal Reflection Fluorescence (TIRF) microscopy (Glossary Box) studies of the E. coli replisome in vitro show that the additional polymerase functions on the lagging strand, enhances replisome processivity, and supports more complete Okazaki fragment synthesis [9]. In other words, with numerous RNA primers to extend, two polymerases devoted to the lagging strand are better than one.
Box 1. High resolution single-molecule microscopy reveals the stoichiometry of the bacterial replisome in vivo.
Traditional biochemical and biophysical methods provide invaluable information on the structural and enzymatic activities of proteins. However, these methods utilize purified components, which are removed from the context of the functional nanomachine inside the cell. Moreover, the results reflect the average behavior of a population, which might mask rare or short-lived intermediates at the single-molecule level. Powerful advances in single-molecule microscopy have recently facilitated the measurements of dynamic biological processes in real time, both in vitro and in vivo. Direct information on binding and dissociation constants, dynamic stoichiometry and heterogeneous behavior can be deducted from the measurements. These recent advances have shed light on the dynamics of bacterial DNA replication in its natural environment. Other, excellent reviews have covered the biophysical details of single molecule fluorescent microscopy and its applications in other cellular processes [71–74].
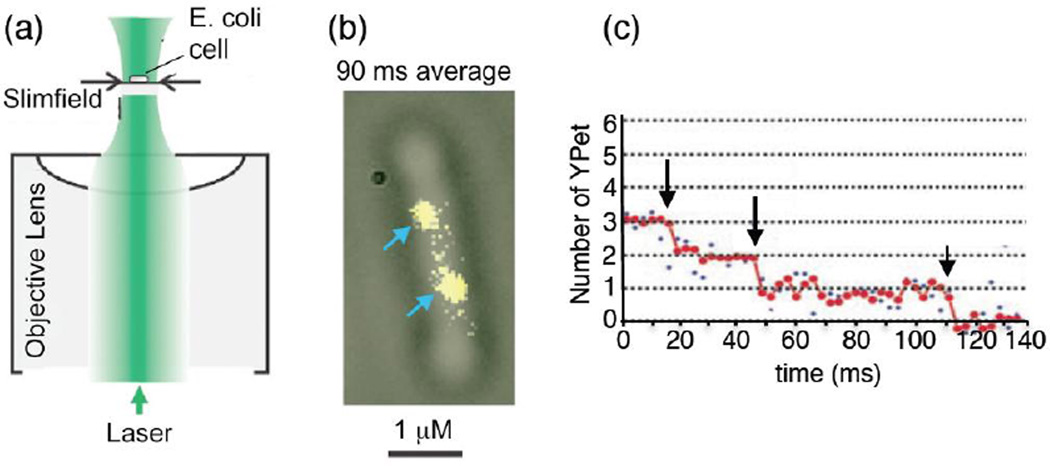
One study by Reyes-Lamothe and colleagues [14] aimed to determine the stoichiometry of replisome components in an E. coli cell in vivo. A difficult challenge in performing single molecule microscopy of cytoplasmic proteins is the high rate of protein diffusion, which requires very short imagining times (in the ms range) to avoid blurred images. To reduce imaging time, the authors applied slimfield microscopy, which concentrates the excitation light into a small area (~ 30 µm2) and illuminates the site of interest with excitation intensities that are ~ 100 times larger than conventional wide-field fluorescence microscopy (Figure Ia). This approach enabled the authors to measure signal intensities at a 3 ms temporal resolution. The investigators fluorescently labeled each component of the replisome with yellow fluorescent protein (YPet) and applied step-wise photobleaching in combination with Fourier spectral analysis to estimate the average fluorescence intensity of a single spot (Figure Ib,c). Photobleaching of a fluorophore occurs in a stochastic fashion, due to formation of radicals in the water surrounding the fluorophore that destroy it. Using this intricate approach, the authors revealed the stoichiometry of the bacterial replisome and found that the replisome contains three polymerases, three proofreading exonucleases, three τ subunits (within the clamp loader), six DnaB subunits (comprising the hexameric helicase) and one δ subunit in the clamp loader. This tri-polymerase stoichiometry confirmed recent biochemical studies [12], and is a sharp departure from the classic view of a replisome with only two polymerases. The presence of a tri-polymerase replisome in vivo was recently confirmed by another intriguing study, where stroboscopic illumination was used to image live cells, and revealed a dynamic turnover of Pol III within the replisome (see text for details) [10].
In overview, the recent developments of highly sensitive in vivo live cell imaging techniques and in vitro single molecule microscopy have revealed fascinating new insights into the dynamic organization of replisomes. Replication forks now are found to progress independently, outside of spatially restricted replication factories [10–12]. Within replisomes, three polymerases are required for efficient replication with two enzymes working on the lagging strand [8–10], indicating that lagging strand synthesis appears to be more complex as previously thought.
Okazaki fragments are not always fully extended
The high processivity of chromosomal replicases could be problematic for the lagging strand polymerase, which must dissociate and recycle to a new primer after completing each 1–2 kb Okazaki fragment. Study of this process using model templates in the E. coli and phage T4 systems has shown that the polymerase disengages from its clamp upon “colliding” with a duplex region (i.e. the 5’ terminus of the previous Okazaki fragment), leaving only a nick [16–18]. This “collision release” mechanism leaves the clamp on DNA and frees the polymerase to bind a new clamp on the next RNA primer.
Suprisingly, study of the lagging strand polymerase within the context of a T4 phage replisome found that not all Okazaki fragments are extended to completion [19]. Subsequent work has shown that this generalizes to phage T7 and E. coli replisomes [15, 19–21]. This premature release of the lagging strand polymerase occurs about half the time and leaves ssDNA gaps on the lagging strand. This action seems incompatible with the extraordinarily high processivity of the replicases, which is much greater than the 1–2 kb Okazaki fragments [22–24]. The observations that polymerase undergoes collision release on model templates, but produces incomplete Okazaki fragments in the context of a full replisome, suggests that the replisome signals the lagging strand polymerase to release from DNA prematurely, a process referred to as “signal release”. Recent studies suggest the signal may be either primase, or clamp loading on a new RNA primer, signaling that a new primed site is ready for action [19–21]. Leaving ssDNA gaps along the lagging strand seems counterintuitive because accumulation of ssDNA can induce an SOS DNA damage response (Glossary Box) [25]. However, ssDNA gaps can probably be rapidly filled-in by soluble polymerases. The purpose of signal release is unknown, but might be beneficial to prevent replication fork arrest in situations where the polymerase encounters a roadblock, including RNA polymerases or template lesions. Overall, accumulating evidence suggests that polymerase release and exchange at the replication fork appears to be a frequent process, in particular on the lagging strand.
Dynamic polymerase exchange during lagging strand synthesis
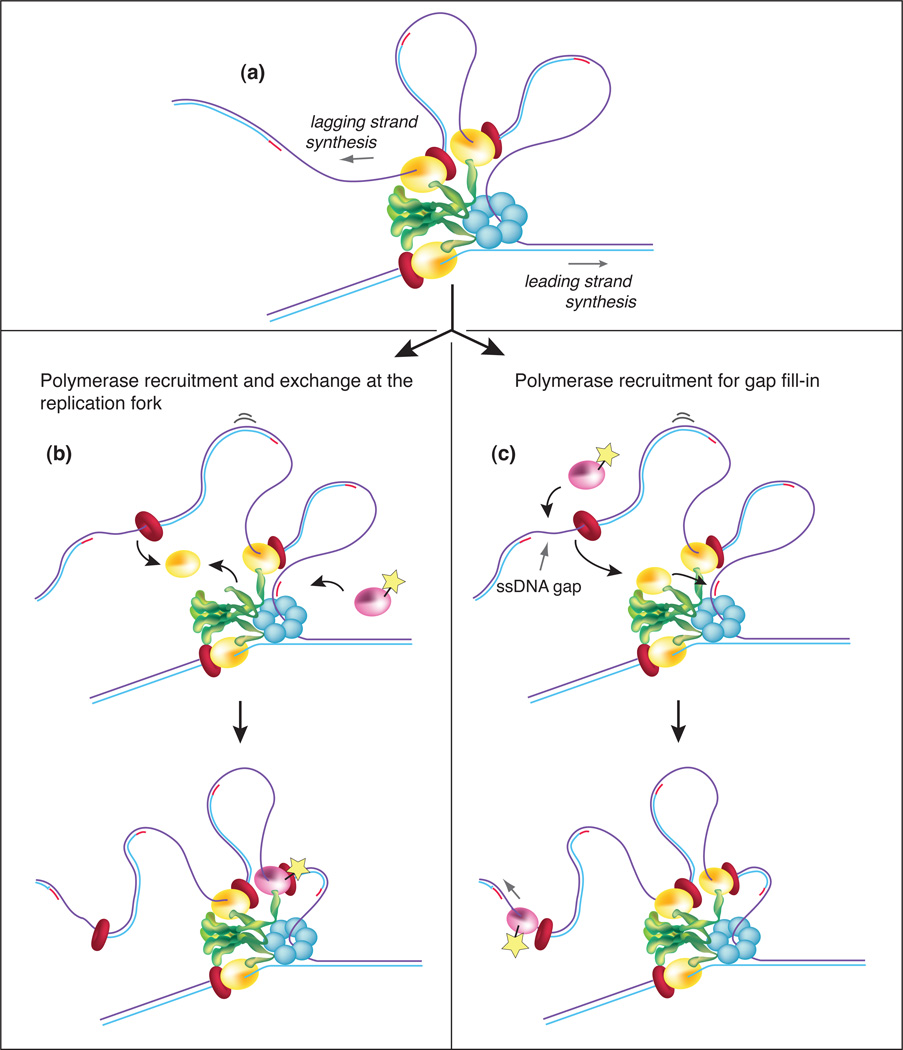
A recent high resolution microscopy study of the E. coli replisome in live cells made an unexpected observation that sheds new light on the dynamic processes involved in Okazaki fragment synthesis (Figure 2) [10]. Lia et al. utilized applied localization techniques with illumination to monitor the dynamics of individual replisome components during replication fork movement. Stroboscopic illumination (Glossary box) allows for measuring the dynamics of Pol III over longer periods. The study observed that new Pol III core molecules associate with, or near replication forks during movement, as determined by the presence of new fluorescent flashes long after the initial tri-polymerase replisome has photobleached [10]. Interestingly, photobleached τ subunits are not replenished, leading to the conclusion that even though the τ-to-Pol III core connection is exceedingly tight (<1nM) [26, 27], this connection is often broken, perhaps with each Okazaki fragment, enabling the photobleached Pol III core to be replaced by a new fluorescent Pol III core fusion protein (Figure 2b,c). It is proposed that Pol III core exchanges only on the lagging strand because the leading strand polymerase is more firmly held into the replisome structure. However, the authors point out that Pol III core exchange conflicts with several biochemical studies showing that the E. coli replisome can replicate both strands of DNA in the absence of extra Pol III core in solution [22]. The polymerase exchange model also conflicts with the very tight Pol III core-τ contact [26, 27]. Now that we know Okazaki fragments are often not completed and leave ssDNA gaps, there is an alternative explanation that nicely rationalizes the biochemical studies and the cellular observations (Figure 2c): the soluble Pol III cores that are recruited to fill-in the ssDNA gaps left by the replisome would be in close proximity to the fork and therefore indistinguishable, by microscopy, from the moving replisome. In this explanation the tri-polymerase replisome remains intact at the fork but is photobleached and thus invisible; only new Pol III cores that fill-in ssDNA gaps are observed.
Figure 2. Polymerase dynamics during Okazaki fragment synthesis in E. coli.
(a) The lagging strand polymerase(s) often dissociate before completing an Okazaki fragment, leaving a ssDNA gap with the β clamp attached. This “signal release” process is thought to be sensed by a replisome component that recognizes a new RNA primer and triggers release of the lagging strand polymerase. Recent findings using fluorescent fusion proteins demonstrate that new Pol III core molecules enter the replication fork on the lagging strand in vivo [10]. Two possible explanations for how new Pol III cores function are proposed. In pathway (b), the lagging strand Pol III core dissociates from the τ subunit at the end of an Okazaki fragment, and exchanges with a new Pol III core that is recruited from the cytoplasm. The new Pol III core is colored pink, and the “star” indicates that it is fluorescent. In pathway (c), the lagging strand Pol III core remains attached to the replisome through its tight interaction with the τ subunit of the clamp loader, and the new (fluorescent) Pol III core is recruited from solution by the β clamp to fill-in the ssDNA gap. Proteins are shown as in Figure 1a.
Regardless of whether Pol III cores exchange on τ, or extra Pol III cores are recruited to fill-in ssDNA gaps, the cellular observations are fascinating. In one case, the proposal that Pol III cores exchange on τ reveal an even greater degree of fluidity to the replisome. The alternative explanation implies that excess Pol III core is recruited to fill-in ssDNA gaps. Perhaps the β clamp that is left on an Okazaki fragment after Pol III dissociates attracts a new Pol III core in preference over other soluble polymerases. The observation that polymerases have the ability to dissociate or exchange rapidly during replication might also be beneficial during encounters of the replisome with lesions or protein bound roadblocks.
The replisome circumvents roadblocks through dynamic fluidity
Unlike RNA polymerase or ribosomes, the replisome cannot fail and “try over”. It has one job, in one lifetime, and it must do it from beginning to end; failure results in genomic instability, mutagenesis or cell death [28, 29]. Considering the length of the chromosome, the replisome is likely to encounter many obstacles, including DNA-bound repressors, transcribing RNA polymerases, and sites of DNA damage. Efficient resolution of collisions is therefore essential for faithful and complete chromosomal replication.
Although understanding the outcome of these “collisions” is still in its infancy, recent progress has made startling discoveries. In vitro studies demonstrate that when the replisome encounters an RNA polymerase transcribing the leading strand in the same direction as fork movement, it overcomes the barrier by rapidly displacing the RNA polymerase and using the RNA transcript as a primer to continue leading-strand synthesis [30]. The replisome does not dissociate from DNA, implying the DNA polymerase hops over the mRNA without being lost from the replication fork. In a cellular context, replisome-RNA polymerase encounters can result in replication fork collapse, and the replisome must reassemble to continue DNA synthesis [31]. Therefore, the cell likely takes a variety of paths depending on the exact circumstances.
Lesions on the lagging strand are circumvented by premature dissociation (e.g. signal release) of the lagging strand polymerase, leaving the lesion in a ssDNA gap to await repair [32]. A recent finding shows a surprising mechanism by which the E. coli replisome can circumvent a lesion on the leading strand, underscoring the remarkable fluidity inherent in replisome action [33]. In this in vitro study using reconstituted E. coli replisomes, the replisome does not collapse when the leading strand polymerase encounters a lesion. Instead, primase makes a new RNA primer ahead of the lesion for continued replisome progression, leaving the lesion behind in a ssDNA gap [33]. That the replisome hops over lesions on both leading and lagging strands is supported by in vivo studies showing that DNA damage results in ssDNA gaps on both leading and lagging strands [34, 35].
DNA lesions can also be bypassed by specialized DNA polymerases. E. coli has five different DNA polymerases (Pols I, II, III, IV and V). Pols II, IV and V are translesion (TLS) polymerases, that are specialized for extending DNA past template lesions [36–38]. Pols II and IV are quickly induced upon DNA damage, which enables them to rapidly take over the replisome from Pol III yet retain the β clamp and DnaB helicase [39, 40]. Pols II and IV are much slower than Pol III, and replisomes containing these TLS polymerases move even slower than the reported intrinsic speed of DnaB helicase [5]. These findings suggest that different polymerases can regulate the speed of the helicase and slow the replisome. A slower replisome may give repair enzymes more time to fix damaged DNA and thereby lower the frequency of replisome encounters with DNA lesions.
Collectively, the bacterial replisome has emerged to be a highly dynamic complex that has evolved to cope with the selective pressure on rapid and efficient DNA replication in a remarkable way. The complex events during Okazaki fragment synthesis for instance, are facilitated by engaging two polymerases in a cooperative manner. Surprisingly, however, Okazaki fragments are often left incomplete, indicating that imperfections are an integral part of DNA replication and are dealt with in the wake of the fork. Replisomes also display a dynamic process that enables them to release from DNA, and to exchange with other polymerases during Okazaki fragment synthesis. This dynamic and fluid nature also provides routes by which the replisome can solve encounters with roadblocks or lesions and continue forward progression while maintaining replication fork integrity. In eukaryotes, replication forks move approximately 15–30 times slower than prokaryotic replication forks. Perhaps the evolutionary pressure on eukaryotes does not require them to perform rapid replication. However, eukaryotic replisomes face additional challenges not present in bacteria, including the need for cell cycle dependent regulation of replication initiation and termination, as well as the need to deal with nucleosomes during passage of the replication fork.
EUKARYOTIC REPLISOMES
The eukaryotic replisome is highly regulated by post-translational modification
Many significant advances have been made in the past few years regarding the composition and function of the eukaryotic replisome (Figure 1b). Of particular interest is the organization of the helicase. The Mcm2-7 helicase is a heterohexamer that surrounds the leading strand and is inactive without certain accessory factors and post-translational modifications. The active helicase is a tightly associated eleven membered complex containing Cdc45, the catalytic Mcm2-7 ring, and the GINS heterotetramer (consisting of four proteins (Sld5-Psf1-Psf2-Psf3) named for the Japanese ‘go-ichi-ni-san’, which means 5-1-2-3); the 11-membered complex is referred to as the CMG (Cdc45, Mcm2-7, GINS) complex [41, 42] (Figure 1b). EM reconstruction of the CMG complex suggests that while the MCM2-7 ring encircles the leading strand, CMG has a second cavity that might encircle the lagging strand [43]. Another recent advance revealed that the chromosome is replicated by distinct leading and lagging strand polymerases, Pol ε and Pol δ, respectively [44, 45]. Both Pol ε(4 subunits), and Pol δ(3–4 subunits) function with the PCNA (proliferating cell nuclear antigen) clamp, which is loaded onto DNA by the RFC (replication factor C) clamp loader. The eukaryotic primase is a four subunit complex (Pol α/primase), unlike single subunit prokaryotic primases. The two small subunits of Pol α/primase synthesize 7–10 nt RNA primers, after which the primer terminus is passed to the polymerase subunit for further extension by approximately 20 nucleotides, resulting in a hybrid RNA/DNA primer [46]. Many of these proteins lack homologues in bacterial systems (e.g. Cdc45, GINS, and all accessory subunits of Pols α, δ and ε). Also in contrast to prokaryotes, the eukaryotic replisome requires specific cell cycle-dependent modifications by multiple kinases for assembly, and is also regulated by modification during ongoing DNA synthesis [47].
Some components of the eukaryotic replisome might not yet be identified. Purification of a large complex from cells, using specific tags fused to genes encoding subunits of the CMG complex, identified a large assembly referred to as the replication progression complex (RPC) [48]. Recent studies using these methods indicated the presence of additional replisome components, including Mcm10, Ctf4, Mrc1, and possibly other proteins [49]. None of these proteins have homologues in bacteria and their function remains to be determined.
Several exciting studies have recently identified specific posttranslational modifications that regulate the assembly and activity of the eukaryotic replisome, including phosphorylation, ubiquitylation, proteolytic degradation and nuclear exclusion. A detailed discussion of this topic is outside the scope of this review, and the reader is directed to excellent reviews [47, 50, 51]. Here, we briefly discuss recent findings that pertain to replisome assembly and progression. First, as the cells enter G1, origin activation starts with assembly of two Mcm2-7 complexes that encircle duplex DNA at an origin [52, 53]. Assembly of the CMG complex requires the S phase kinases Dbf4-dependent Cdc7 kinase (DDK) and cyclin dependent kinase (CDK), as well as the Sld2/3, Dpb11, Sld7 and Mcm10 proteins that facilitate assembly of the CMG complex around the unwound ssDNA leading strand [54–58]. It is thought that DDK is first required for the Sld3 dependent recruitment of Cdc45 to DNA bound MCM; subsequent CDK activity promotes recruitment of GINS [59]. Mcm10 appears to be stimulate activation of the MCM helicase after recruitment and assembly of the CMG complex [60–62]. The activated CMG complex provides unwound DNA for priming and assembly of the polymerases for replisome progression. The DDK targets Mcm4, and the CDK targets Sld2/3. How these proteins function is currently unknown, but the Sld2/3/7 and Dpb11 proteins do not appear to remain as an integral part of the moving replisome.
The replisome is also regulated by post-translational modifications in response to DNA damage, the extent of which is only now coming to light. One of the key targets for regulation is PCNA, which can serve as a binding platform for various enzymes involved in DNA repair, chromatin assembly, and cell cycle control [63]. In response to DNA damage, yeast PCNA is ubiquitylated at a highly conserved lysine residue, K164, which might facilitate recruitment of TLS polymerases and their exchange with the replicative polymerase for lesion bypass [64]. Other replisome components are also phosphorylated in response to DNA damage. A recent study from budding yeast demonstrates that replication forks arrested by hydroxyurea treatment contain phosphorylated forms of the Psf1 subunit of the GINS complex [65]. Another example is Mrc1, which is phosphorylated by the checkpoint kinase Mec1 at stalled replication forks [65]. The consequences of checkpoint induced phosphorylation of replisome components are largely unknown. One possible effect is to recruit factors that stabilize the replication fork upon arrest.
The complexity of eukaryotic replication is reflected by a large number of additional factors not present in bacterial replisomes. To date, the eukaryotic replication machinery involves 48 polypeptides, and most likely additional factors that are yet to be identified. In addition, individual subunits are highly regulated by post-translational modifications in a cell cycle dependent manner. This complex organization is needed to ensure the timely and spatial control of the individual events required for DNA replication and its orchestration with other cellular pathways including transcription and DNA repair. DNA damage for instance introduces additional components at the replication fork and triggers modifications of replisome components that are needed to arrest and stabilize the replication fork and recruit DNA repair machineries. In fact, recent new data indicate that the regulation of DNA replication appears not only to occur on the level of replication proteins and their modifications, but also on how the DNA is organized by nucleosomes.
Nucleosomes solve a lagging strand problem in eukaryotes
Recent findings indicate that nucleosomes are harnessed to solve a long-standing problem in the eukaryotic replication field. Namely, Pol α/primase lacks proofreading activity and thus the DNA portion of the hybrid RNA/DNA primer might contain mistakes. The DNA section of the primer, along with mistakes, could be ligated to another Okazaki fragment. This problem is compounded by the very short length of eukaryotic Okazaki fragments (100–200 bp) [66]. Thus Pol α/primase synthesizes a significant portion of genomic DNA. The RNA/DNA primer is removed by strand displacement synthesis by the high fidelity Pol δ, working in concert with Fen1 nuclease (and/or Dna2), which produces a nick for ligation by DNA ligase I. Both Fen1 and Dna2 nucleases are regulated by acetylation [67, 68] but how Pol δ strand displacement activity is regulated has been an open question. A recent discovery reveals that nucleosomes direct and limit strand displacement activity by Pol δ [69].
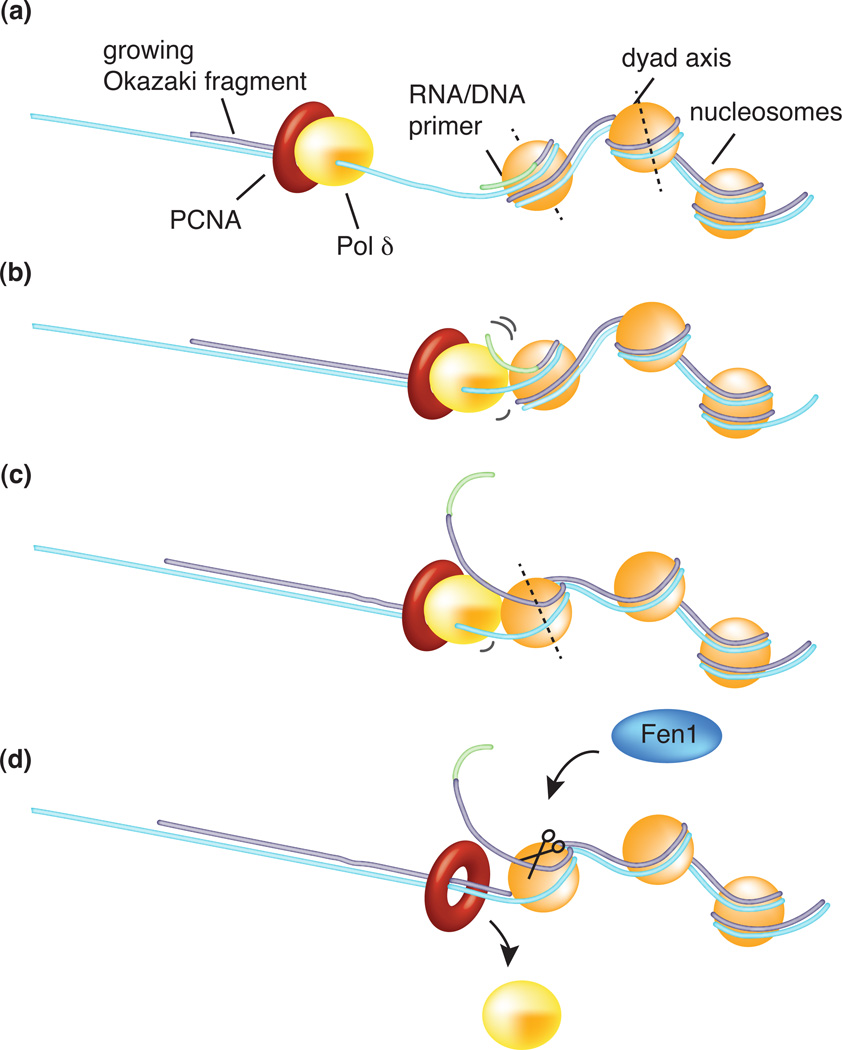
Nucleosomes wrap 150–200 bp of duplex DNA nearly two times around the octamer (Figure 3). Upon advance of a replication fork, nucleosomes are displaced from DNA but rapidly reassemble on newly synthesized DNA in a process that is aided by CAF-1, a nucleosome assembly factor [70]. new report elegantly demonstrates that the small 100–200 bp size of eukaryotic Okazaki fragments closely correlate with the amount of DNA occupied by a nucleosome and suggest a role of nucleosome positioning in the regulation of Okazaki fragment length [69]. Mutation of CAF-1 abrogates the nucleosome-like size of Okazaki fragments, providing compelling evidence that nucleosomes are assembled onto Okazaki fragments as soon as they are formed. The study also provides evidence that nucleosomes regulate strand displacement activity by Pol δ. Using a clever combination of a ligase mutant and deep sequencing, the authors identified the end points of the Pol δ strand displacement reaction (and excision of the displaced DNA by Fen1 and/or Dna2), prior to ligation, over the entire genome. The results were then compared to known nucleosome positions in the yeast genome. The observed Okazaki fragment junctions are enriched at the nucleosome midpoint, called the region of dyad symmetry (Figure 3). The entry and exit sections of DNA bound to nucleosomes is less tightly bound and easily unwound compared to DNA within the dyad symmetry region. The authors conclude that Pol δ displacement synthesis is stopped by the nucleosome on the preceding Okazaki fragment at the tightly bound dyad symmetry position, providing a plausible regulatory mechanism that limits Pol δ/Fen1 action to an amount sufficient to excise the error-prone RNA/DNA primer.
Figure 3. Nucleosome assembly regulates Okazaki fragment length.
(a) A new nucleosome rapidly assembles onto a new Okazaki fragment after passage of the replication fork. (b) During Okazaki fragment extension, the lagging strand Pol δ performs strand displacement synthesis of the previous Okazaki fragment until it reaches the tightly bound region of dyad symmetry (c), the midpoint of the DNA segment wrapped around the nucleosome. At this point nucleosomes become a roadbloack for Pol δ and trigger its dissociation (d). The displaced ssDNA region, encompassing the RNA/DNA primer made by the low fidelity Pol α/primase, is excised by the Fen1 flap-endonuclease. This process provides an elegant mechanism for Okazaki fragment length regulation and the precise excision of the error prone hybrid primer [69].
Concluding remarks
New studies have shown that the bacterial replisome has three DNA polymerases; one for the leading strand and two that synthesize Okazaki fragments. How the DNA polymerases coordinate actions for lagging strand synthesis is not yet understood. Suprisingly, bacterial lagging strand polymerases are often signaled to release DNA before completing an Okazaki fragment, leaving ssDNA gaps. The nature of the “signal” for polymerase release is not clearly understood, nor is the mechanism by which the ssDNA gaps are filled. The replisome is much more fluid and dynamic than ever expected. Low fidelity TLS polymerases trade places with the high fidelity replicase, and control the speed of helicase unwinding. How polymerases exchange and how the helicase is controlled are largely unknown. The eukaryotic replisome contains two different polymerases for the leading and lagging strands, along with a third that performs limited extension of RNA primers. How these polymerases are relegated to their specific strands is unknown. Eukaryotes also contain a very different type of helicase, along with many proteins that have no apparent structural homologue in bacteria. The function of these proteins, and the overall architecture of the eukaryotic replisome, remains to be determined. The evidence that the replisome is targeted for regulation is accumulating at a rapid pace. The field of replication has entered a time of rapid expansion, and holds the promise of understanding basic mechanisms that, gone awry, lead to genetic instability and the accumulation of mutations, which underlie various human diseases including cancer. Clearly, new questions outpace those that have been answered, and the future is certain to bring new and important discoveries.
Figure I. High resolution slimfield microscopy of replisomes in a cell.
(a) In slimfield microscopy the laser beam is narrowed to nearly the size of a cell, intensifying the beam. (b) Replisomes in a cell are visualized using yellow fluorescent protein (YPet) fused to ε, the proofreading subunit of Pol III core. The image is a 90 ms frame averaged fluorescent image (yellow) overlayed with a brightfield image of the whole cell (gray). Arrows point to the two replisomes in the cell. (c) Photobleaching analysis shows that, for a putative single replisome spot, there is a stoichiometry of three bleaching events (arrows). Raw intensity data (blue) and filtered data (red) for a putative single replisome spot. Figure is adapted, with permission, from Figure 1 of reference [8].
Acknowledgment
The authors are grateful for a grant from the NIH (GM38839) that supported the writing of this review. We thank Dr. Lance Langston for critical reading of the manuscript and helpful comments. We also apologize to the researchers whose work could not be referenced due to space limitations.
Glossary
- T4 system
The replication system of the T4 phage serves as a relatively simple system for studying DNA replication in vitro. Reconstituted from purified components it recapitulates formation and propagation of DNA replication vitro. The eight essential components include a DNA polymerase harboring both nucleotide incorporation and 3’–5’ proofreading exonuclease activities (gp43), a ring-shaped homotrimeric processivity factor (gp45), loaded on a primed template by the clamp loader (gp44/62). The polymerase-clamp complex is processive and displaces ssDNA binding protein (gp32) from DNA. A helicase (gp41) unwinds the duplex DNA and primase (gp61) binds the helicase to initiate primer synthesis. A helicase accessory factor (gp59) assists helicase loading.
- TIRF microscopy
Total Internal Reflection Fluorescence (TIRF) microscopy is based on the principle that a thin, exponentially decaying, evanescent field of excitation can be generated at the interface of two mediums of different refractive index (RI) (e.g., the glass coverslip and the biological specimen). This enables the selective excitation of fluorophores in a thin optical plane above the light beam (<200nm), thereby preventing fluorescence emission from molecules that are not in the primary focal plane (e.g. molecules that are not bound to DNA that is restricted to the focal plane).
- Nanoscale DNA biopointers
Nanoscale DNA biopointers consist of short (<200 bp) DNA duplexes labeled with biotin. They bind the multivalent streptavidin molecule, which can also bind a biotinylated protein, thereby coupling the DNA molecule to the protein, through the strepavidin bridge. Binding of biopointers to proteins is used to map the localization of proteins in electron microscopy (EM). Streptavidin labeled biopointers have a high specificity for its target and are small enough not to obscure the target, yet are large enough to visualize in the EM.
- Stroboscopic illumination
A frequent challenge for fluorescence microscopy that continuous high intensity illumination is production of reactive oxygen species that are toxic to cells (phototoxicity) and also bleaching of fluorophores during the course of extended or repeated measurements (photobleaching). Use of a high power light-emitting diode (LED), which can emit short pulses of light (0.5–2 ms) (stroboscopic illumination) to excite fluorophores, maximizes signal intensities and minimizes illumination time, thus reducing phototoxicity and photobleaching.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg A, Baker TA. DNA replication. W.H. Freeman; 1992. [Google Scholar]
- 2.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 3.McHenry CS. DNA replicases from a bacterial perspective. Annu Rev Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- 4.Sinha NK, et al. Efficient in vitro replication of double-stranded DNA templates by a purified T4 bacteriophage replication system. J Biol Chem. 1980;255:4290–4293. [PubMed] [Google Scholar]
- 5.Kim S, et al. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 6.Migocki MD, et al. The midcell replication factory in Bacillus subtilis is highly mobile: implications for coordinating chromosome replication with other cell cycle events. Mol Microbiol. 2004;54:452–463. doi: 10.1111/j.1365-2958.2004.04267.x. [DOI] [PubMed] [Google Scholar]
- 7.Lemon KP, Grossman AD. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 8.Kitamura E, et al. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–1308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes-Lamothe R, et al. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lia G, et al. Polymerase exchange during Okazaki fragment synthesis observed in living cells. Science. 2012;335:328–331. doi: 10.1126/science.1210400. [DOI] [PubMed] [Google Scholar]
- 11.Yardimci H, et al. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McInerney P, et al. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 13.Nossal NG, et al. Architecture of the bacteriophage T4 replication complex revealed with nanoscale biopointers. J Biol Chem. 2007;282:1098–1108. doi: 10.1074/jbc.M606772200. [DOI] [PubMed] [Google Scholar]
- 14.Reyes-Lamothe R, et al. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgescu RE, et al. Single-molecule studies reveal the function of a third polymerase in the replisome. Nat Struct Mol Biol. 2012;19:113–116. doi: 10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stukenberg PT, et al. An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell. 1994;78:877–887. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- 17.Georgescu RE, et al. Mechanism of polymerase collision release from sliding clamps on the lagging strand. The EMBO journal. 2009;28:2981–2991. doi: 10.1038/emboj.2009.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benkovic SJ, et al. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, et al. The control mechanism for lagging strand polymerase recycling during bacteriophage T4 DNA replication. Mol Cell. 2006;21:153–164. doi: 10.1016/j.molcel.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 20.Hamdan SM, et al. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature. 2009;457:336–339. doi: 10.1038/nature07512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Marians KJ. Two distinct triggers for cycling of the lagging strand polymerase at the replication fork. J Biol Chem. 2000;275:34757–34765. doi: 10.1074/jbc.M006556200. [DOI] [PubMed] [Google Scholar]
- 22.Yao NY, et al. Single-molecule analysis reveals that the lagging strand increases replisome processivity but slows replication fork progression. Proc Natl Acad Sci U S A. 2009;106:13236–13241. doi: 10.1073/pnas.0906157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamdan SM, Richardson CC. Motors, switches, and contacts in the replisome. Annu Rev Biochem. 2009;78:205–243. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- 24.Perumal SK, et al. Single-molecule studies of DNA replisome function. Biochim Biophys Acta. 2010;1804:1094–1112. doi: 10.1016/j.bbapap.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlacher K, Goodman MF. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat Rev Mol Cell Biol. 2007;8:587–594. doi: 10.1038/nrm2198. [DOI] [PubMed] [Google Scholar]
- 26.Downey CD, McHenry CS. Chaperoning of a replicative polymerase onto a newly assembled DNA-bound sliding clamp by the clamp loader. Mol Cell. 2010;37:481–491. doi: 10.1016/j.molcel.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leu FP, et al. Mechanism of the E. coli tau processivity switch during lagging-strand synthesis. Mol Cell. 2003;11:315–327. doi: 10.1016/s1097-2765(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 28.Vilette D, et al. Transcription-induced deletions in plasmid vectors: M13 DNA replication as a source of instability. Mol Gen Genet. 1996;252:398–403. doi: 10.1007/BF02173004. [DOI] [PubMed] [Google Scholar]
- 29.Srivatsan A, et al. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pomerantz RT, O'Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456:762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrikh H, et al. Co-directional replication-transcription conflicts lead to replication restart. Nature. 2011;470:554–557. doi: 10.1038/nature09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McInerney P, O'Donnell M. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J Biol Chem. 2004;279:21543–21551. doi: 10.1074/jbc.M401649200. [DOI] [PubMed] [Google Scholar]
- 33.Yeeles JT, Marians KJ. The Escherichia coli replisome is inherently DNA damage tolerant. Science. 2011;334:235–238. doi: 10.1126/science.1209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopes M, et al. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 35.Callegari AJ, et al. Postreplication gaps at UV lesions are signals for checkpoint activation. Proc Natl Acad Sci U S A. 2010;107:8219–8224. doi: 10.1073/pnas.1003449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel M, et al. A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V. Crit Rev Biochem Mol Biol. 2010;45:171–184. doi: 10.3109/10409238.2010.480968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarosz DF, et al. Proficient and accurate bypass of persistent DNA lesions by DinB DNA polymerases. Cell Cycle. 2007;6:817–822. doi: 10.4161/cc.6.7.4065. [DOI] [PubMed] [Google Scholar]
- 38.Friedberg EC, et al. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 39.Indiani C, et al. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Uchida K, et al. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol Microbiol. 2008;70:608–622. doi: 10.1111/j.1365-2958.2008.06423.x. [DOI] [PubMed] [Google Scholar]
- 41.Moyer SE, et al. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ilves I, et al. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nick McElhinny SA, et al. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansson E, Macneill SA. The eukaryotic replicative DNA polymerases take shape. Trends Biochem Sci. 2010;35:339–347. doi: 10.1016/j.tibs.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Mossi R, et al. DNA polymerase switching: II. Replication factor C abrogates primer synthesis by DNA polymerase alpha at a critical length. J Mol Biol. 2000;295:803–814. doi: 10.1006/jmbi.1999.3395. [DOI] [PubMed] [Google Scholar]
- 47.Diffley JF. Quality control in the initiation of eukaryotic DNA replication. Philos Trans Soc Lond B Biol Sci. 2011;366:3545–3553. doi: 10.1098/rstb.2011.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 49.Gambus A, et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. The EMBO journal. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 55.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 56.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka T, et al. Sld7, an Sld3-associated protein required for efficient chromosomal DNA replication in budding yeast. The EMBO journal. 2011;30:2019–2030. doi: 10.1038/emboj.2011.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heller RC, et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanke M, et al. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. Embo J. 2012;31:2182–2194. doi: 10.1038/emboj.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Deursen F, et al. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. Embo J. 2012;31:2195–2206. doi: 10.1038/emboj.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watase G, et al. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol. 2012;22:343–349. doi: 10.1016/j.cub.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 63.Chun AC, Jin DY. Ubiquitin-dependent regulation of translesion polymerases. Biochem Soc Trans. 2010;38:110–115. doi: 10.1042/BST0380110. [DOI] [PubMed] [Google Scholar]
- 64.Fox JT, et al. Dynamic regulation of PCNA ubiquitylation/deubiquitylation. FEBS Lett. 2011;585:2780–2785. doi: 10.1016/j.febslet.2011.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Piccoli G, et al. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell. 2012;45:696–704. doi: 10.1016/j.molcel.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 66.DePamphilis ML, Wassarman PM. Replication of eukaryotic chromosomes: a close-up of the replication fork. Annu Rev Biochem. 1980;49:627–666. doi: 10.1146/annurev.bi.49.070180.003211. [DOI] [PubMed] [Google Scholar]
- 67.Balakrishnan L, et al. Acetylation of Dna2 endonuclease/helicase and flap endonuclease1 by p300 promotes DNA stability by creating long flap intermediates. J Biol Chem. 2010;285:4398–4404. doi: 10.1074/jbc.M109.086397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hasan S, et al. Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol Cell. 2001;7:1221–1231. doi: 10.1016/s1097-2765(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 69.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci U S A. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie XS, et al. Single-molecule approach to molecular biology in living bacterial cells. Annu Rev Biophys. 2008;37:417–444. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- 72.van Oijen AM. Single-molecule approaches to characterizing kinetics of biomolecular interactions. Curr Opin Biotechnol. 2011;22:75–80. doi: 10.1016/j.copbio.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 73.Sako Y. Imaging single molecules in living cells for systems biology. Mol Syst Biol. 2006;2:56. doi: 10.1038/msb4100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lenn T, Leake MC. Experimental approaches for addressing fundamental biological questions in living, functioning cells with single molecule precision. Open Biol. 2012;2:120090. doi: 10.1098/rsob.120090. [DOI] [PMC free article] [PubMed] [Google Scholar]