Abstract
Selenoprotein S (SelS), has previously been associated with a range of inflammatory markers, particularly in the context of cardiovascular disease (CVD). The aim of this study was to examine the role of SELS genetic variants in risk for subclinical CVD and mortality in individuals with type 2 diabetes mellitus (T2DM). The association between 10 polymorphisms tagging SELS and coronary (CAC), carotid (CarCP) and abdominal-aortic calcified plaque (AACP), carotid intima media thickness (IMT) and other known CVD risk factors was examined in 1220 European Americans from the family-based Diabetes Heart Study. The strongest evidence of association for SELS SNPs was observed for CarCP; rs28665122 (5′ region; β=0.329, p=0.044), rs4965814 (intron 5; β=0.329, p=0.036), rs28628459 (3′ region; β=0.331, p=0.039) and rs7178239 (downstream; β=0.375, p=0.016) were all associated. In addition, rs12917258 (intron 5) was associated with CAC (β =−0.230, p=0.032) and rs4965814, rs28628459 and rs9806366 were all associated with self reported history of prior CVD (p=0.020–0.043). These results suggest a potential role for the SELS region in the development subclinical CVD in this sample enriched for T2DM. Further understanding the mechanisms underpinning these relationships may prove important in predicting and managing CVD complications in T2DM.
Keywords: genetics, atherosclerosis, calcified plaque, diabetes mellitus
INTRODUCTION
Cardiovascular disease (CVD) is the major cause of mortality in Western industrial countries accounting for ~35% of all-cause mortality [1]. Although there is clear evidence that CVD clusters in families, neither incident CVD, nor any of its associated subclinical signs and symptoms segregate in families as a single Mendelian trait. Clearly, CVD risk arises from complex interactions between multiple genetic and environmental factors. In individuals with type 2 diabetes mellitus (T2DM), the hyperglycemic environment favours atherogenesis, culminating in diabetic macrovascular disease. Individuals with T2DM are at least twice as likely to have coronary artery disease compared to non-diabetic individuals and CVD accounts for >65% of mortality in T2DM patients [1, 2].
With atherosclerosis recognized as involving an inflammatory process within the sub-endothelial tissues, factors involved in the regulation of inflammation are logical candidates when attempting to profile the heritable risk for CVD. Selenoprotein S (SelS) is one recently identified gene that warrants further investigation. Encoded by the SELS gene on chromosome 15q26.3, SelS belongs to a unique family of ~25 proteins, all of which contain selenocysteine residues [3, 4], and is located on both the endoplasmic reticulum (ER) and plasma membranes [5, 6]. SelS participates in the retro-translocation of mis-folded proteins from the ER for degradation [7, 8] thereby protecting the cell from oxidative stress and inflammatory events which accompany the mis-folded protein response [9, 10]. ER stress has recently been considered in the pathogenesis of T2DM and atherosclerosis [11–13]. In this context, a potential regulatory role of SelS cannot be discounted, particularly given the identification of a proposed ER stress response element sequence in the promoter region of the SELS gene [14].
SELS gene expression has been reported to be influenced by a polymorphism in the promoter region [15]. In addition, SELS polymorphisms have also been associated with circulating levels of pro- and anti-inflammatory cytokines [15, 16] providing further evidence supporting a likely role of SelS in the regulation of inflammation. Despite such evidence, there are only a limited number of studies directly examining the role of genetic variation in SELS in CVD risk [5, 6]. To further elucidate a potential role of SelS in the pathogenesis of CVD, we evaluated the association between SELS SNPs and established quantitative measures of subclinical CVD, including vascular calcified plaque [17] and carotid intima-medial thickness [18, 19], in the Diabetes Heart Study (DHS); a study of CVD in a T2DM-enriched families [20, 21].
RESEARCH DESIGN AND METHODS
Study Design and Sample
The study sample consisted of 1220 self-described European American (EA) individuals (1021 T2DM-affected individuals, 199 T2DM-unaffected individuals from 474 families) in the Diabetes Heart Study. Ascertainment and recruitment have been described in detail previously [20, 22–24]. Briefly, siblings concordant for T2DM, but without advanced renal insufficiency were recruited with additional non-diabetic siblings also enrolled when possible. T2DM was defined clinically as diabetes developing after the age of 35 years treated with insulin and/or oral agents, in the absence of historical evidence of ketoacidosis.
Study protocols were approved by the Institutional Review Board at Wake Forest University School of Medicine, and all participants provided written informed consent. Participant examinations were conducted in the General Clinical Research Centre of the Wake Forest University Baptist Medical Centre, and included interviews for medical history and health behaviours, anthropometric measures, resting blood pressure, electrocardiography, fasting blood sampling for laboratory analyses and spot urine collection. Standard laboratory analyses included a blood lipid profile, fasting glucose, glycated haemoglobin (HbA1C) and high-sensitivity C-reactive protein (CRP). Subjects were assessed for the presence of Metabolic Syndrome (MetS) using criteria established in the Third Report of the National Cholesterol Education Program Expert Panel Detection, Evaluation and Treatment in Adults (ATP III) [25].
Carotid artery intima-media thickness (IMT) was measured by high-resolution B-mode ultrasonography with a 7.5-MHz transducer and a Biosound Esaote (AU5) ultrasound machine (Biosound Esaote, Inc., Indianapolis, IN) as previously described [24]. Coronary artery calcified plaque (CAC), carotid artery calcified plaque (CarCP) and infra-renal abdominal aortic calcified plaque (AACP) were measured using fast-gated helical CT scanners, and calcium scores were calculated as previously described [26, 27]. Not all measurements were available for all participants.
For all participants, vital status was determined from the National Social Security Death Index maintained by the United States Social Security Administration. For those participants confirmed as deceased, length of follow-up was determined to date of death. For deceased participants, copies of death certificates were obtained from relevant county Vital Records Offices to confirm cause of death. For all other participants the length of follow-up was determined from the date of the initial study visit to a follow-up telephone interview during the first half of 2011.
Genotyping
Total genomic DNA was purified from whole blood samples obtained from subjects using the PUREGENE DNA isolation kit (Gentra, Inc., Minneapolis, MN). DNA concentration was quantified using standardized fluorometric readings on a Hoefer DyNA Quant 200 fluorometer (Hoefer Pharmacia Biotech Inc., San Francisco, CA). Samples were diluted to a final concentration of 5 ng/μl.
A total of 10 SNPs, spanning the SELS gene (NM_203472.1) were selected for analysis. Of these, six SNPs were found in the HapMap database (http://www.hapmap.org). The additional four SNPs were chosen from the literature [5, 15, 28]. The genotypic variation across the region encompassing the 10 selected SNPs was assessed using the greedy pairwise tagging algorithm implemented in the Tagger program of Haploview v4.2 (Broad Institute, Cambridge, MA).
Genotypes, were determined using a MassARRAY SNP Genotyping System (Sequenom Inc., San Diego, CA) following standard protocols as described previously [29]. This system uses single-base extension reactions to create allele-specific products that are separated and scored in a matrix-assisted laser desorption ionization/time of flight mass spectrometer. Primers for PCR amplification and extension reactions were designed using the MassARRAY Assay Design Software (Sequenom Inc). Genotype calls were reviewed using the Sequenom MassArray Typer v3.4 software (Sequenom Inc).
A total of 41 quality controls samples were included in the genotyping analysis to serve as blind duplicates and allow for evaluation of genotyping accuracy. The concordance rate for these blind duplicates was 100%. For all SNPs the minimum acceptable call frequency was 95%. The average call frequency was 98.3 ± 0.7% (mean ± SD). Samples with genotyping efficiency rates <90% were excluded from further analysis; the average genotyping efficiency rate was 99.5 ± 0.05% (mean ± SD).
Statistical Analysis
Allele and genotype frequencies for each SNP were calculated from unrelated individuals and tested for departures from Hardy-Weinberg equilibrium using a Chi-squared goodness of fit test. Association between each of the 10 SNPs and four primary phenotypes of interest was performed using variance components methods implemented in Sequential Oligogenic Linkage Analysis Routines (SOLAR) v 4.3.1 (Texas Biomedical Research Institute, San Antonio, Tx, USA) to account for the relatedness between subjects [30]. The primary phenotypes of interest were CAC, CarCP, AACP and, IMT. Additional association analysis was undertaken with a number of other phenotypes including, self-reported history of CVD, hypertension, MetS, HbA1c, CRP, total cholesterol, HDL cholesterol, LDL cholesterol and triglyceride concentrations. Continuous variables were transformed prior to analysis to approximate conditional normality and to reduce heterogeneity of residual phenotypic variance across SNP genotypes. Results from the additive model are reported here; results from dominant and recessive models are included in Supplementary Material. All analyses presented herein are adjusted for, age, gender and diabetes affected status. Statistical significance was accepted at p<0.05. Additional models adjusting for (i) age and gender only and (ii) age, gender, diabetes affected status and smoking status (history of current or prior smoking) were also performed, but inferences largely similar (Supplementary Material).
RESULTS
The clinical characteristics of the 1182 EA subjects with available genotyping data are presented in Table 1. As anticipated, a predominance of known CVD risk factors, including high body mass, hypertension and high blood lipids, was evident in this sample. In addition, calcification scores across the different vascular beds reflect a substantial burden of subclinical CVD in these diabetes-affected individuals and their siblings. Determination of vital status revealed 216 (18.2%) individuals were deceased over an average follow-up time of 7.3 years; of these, 95 (8.0%) had a documented CVD-related cause of death.
Table 1.
Clinical characteristics of European American (n=1182) participants in the Diabetes Heart Study.
| Mean ± SD or % | |
|---|---|
| Age (yrs) | 61.5 ± 9.4 |
| Gender (% female) | 53.6% |
|
| |
| Height (cm) | 168.6 ± 9.7 |
| Weight (kg) | 90.6 ± 20.3 |
| BMI (kg/m2) | 31.8 ± 6.5 |
|
| |
| Diabetes affected (%) | 83.5% |
| Diabetes Duration (yrs) | 10.4 ± 7.2 |
|
| |
| Length of Follow-up (yrs) | 7.3 ± 2.3 |
|
| |
| % smoking current or past | 58.8% |
|
| |
| Systolic BP (mmHg) | 139 ± 19 |
| Diastolic BP (mmHg) | 73 ± 10 |
| Hypertension (%) | 85.4% |
|
| |
| Lipid Lowering medication (%) | 43.5% |
| Anti-hypertensive medication (%) | 73.9% |
|
| |
| Laboratory Measures | |
| Cholesterol (mg/dL) | 186.7 ± 42.3 |
| HDL (mg/dL) | 43.0 ± 12.4 |
| LDL (mg/dL) | 105.2 ± 32.7 |
| Triglycerides (mg/dL) | 200.9 ± 131.5 |
| Blood Glucose (mg/dL) | 138.6 ± 54.9 |
| HbA1C (%) | 7.3 ± 1.7 |
| CRP (mg/L) | 6.0 ± 9.8 |
|
| |
| Self reported history of | |
| Angina | 16.2% |
| Stroke | 9.0% |
| Cardiovascular intervention* | 23.6% |
| MI | 19.1% |
|
| |
| Vascular Imaging | |
| CAC | 1650 ± 3167 |
| CarCP | 315 ± 678 |
| AACP | 11112 ± 15889 |
| IMT | 0.694 ± 0.144 |
either coronary bypass grafting, coronary angioplasty or carotid endarterectomy
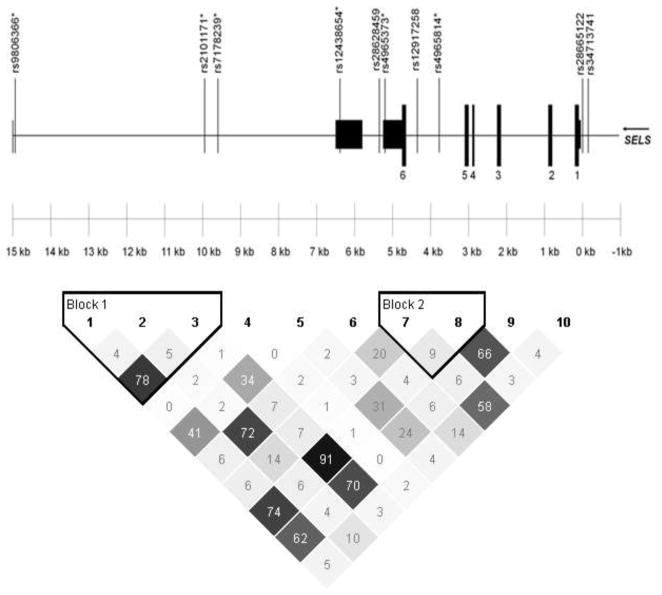
Ten SNPs encompassing a 14.9 kb region containing the SELS gene were chosen for genotyping. These SNPs capture 0.861 of the genetic variation in the selected region as defined by r2. Figure 1 shows the location of the 10 SNPs examined relative to the SELS gene structure and the linkage disequilibrium structure generated from Haploview. Two of the SNPs are 5′ to the coding region including rs28665122, previously reported to influence SELS gene expression. The additional eight SNPs are intronic, 3′ to the coding sequence or downstream of the gene. All SNPs were consistent with Hardy-Weinberg proportions (p=0.42–1.00).
Figure 1.
Location and linkage disequilibrium structure (represented by r2 values) for the 10 SELS SNPs genotyped.
Table 2 summarizes the results of the association analysis with four quantitative traits: CAC, CarCP, AACP and IMT. Overall, five SNPs showed statistically significant evidence of association with at least one of the vascular calcification phenotypes examined. The strongest evidence of association was for CarCP, which was significantly associated with rs28665122, rs4956814, rs28628459 and rs7178239 (Table 2). For rs28665122, rs28628459 and rs7178239 CarCP scores were 10–50% higher in the minor allele homozygotes compared to the major allele homozygotes. For all three of these SNPs similar patterns were noted for CAC and AACP scores (10–60% differences), however these were not statistically significant (Table 2). For rs4986814, CarCP scores were ~8% lower in the minor allele homozygotes. Finally, rs12917258 was significantly associated with CAC; scores were ~30% lower in the minor allele homozygotes. Again, similar patterns were also noted for CarCP and AACP (20–30% differences), although these were not significantly different (Table 2).
Table 2.
Untransformed mean trait values for coronary (CAC), carotid (CorCP) and abdominal aortic calcified plaque (AACP), carotid intima media thickness (IMT) by genotype in European Americans (n=1220) from the Diabetes Heart Study for each of the SELS SNPs and association results assuming an additive model of inheritance with covariates age, gender, diabetes affected status.
| Mean ± Standard Deviation (n) | Additive Model | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| SNP | MAF | Location | Alleles (1/2) | Trait | 1/1 | 1/2 | 2/2 | p-value | B ± SE |
| rs34713741 | 0.283 | 5′ | C/T | CAC | 1844 ± 3713 (575) | 1484 ± 2444 (451) | 1315 ± 2526 (88) | 0.2610 | −0.1255 ± 0.1211 |
| CarCP | 338 ± 698 (570) | 307 ± 691 (447) | 227 ± 460 (88) | 0.6462 | −0.0537 ± 0.1241 | ||||
| AACP | 11964 ± 16827 (434) | 10839 ± 14926 (345) | 7510 ± 14257 (68) | 0.5334 | −2.1309 ± 3.4095 | ||||
| IMT | 0.677 ± 0.135 (567) | 0.676 ± 0.138 (419) | 0.663 ± 0.118 (91) | 0.4407 | 0.0026 ± 0.0034 | ||||
|
| |||||||||
| rs28665122 | 0.127 | 5′ | C/T | CAC | 1650 ± 3271 (857) | 1643 ± 2882 (230) | 2231 ± 2630 (16) | 0.4994 | 0.1039 ± 0.1580 |
| CarCP | 300 ± 630 (849) | 365 ± 823 (230) | 446 ± 809 (15) | 0.0439 | 0.3286 ± 0.1852 | ||||
| AACP | 11012 ± 16226 (648) | 11365 ± 14805 (180) | 15940 ± 19450 (11) | 0.3900 | 4.0626 ± 4.6882 | ||||
| IMT | 0.674 ± 0.138 (823) | 0.682 ± 0.126 (225) | 0.659 ± 0.097 (16) | 0.4149 | 0.0060 ± 0.0074 | ||||
|
| |||||||||
| rs4965814 | 0.164 | Intronic | T/C | CAC | 1702 ± 3373 (778) | 1542 ± 2687 (312) | 1550 ± 2166 (30) | 0.2282 | −0.1255 ± 0.1211 |
| CarCP | 315 ± 659 (772) | 322 ± 733 (310) | 290 ± 625 (27) | 0.0359 | 0.3286 ± 0.1852 | ||||
| AACP | 11467 ± 16763 (590) | 10191 ± 13532 (241) | 12663 ± 16057 (20) | 0.8824 | 0.5930 ± 3.7393 | ||||
| IMT | 0.675 ± 0.140 (756) | 0.675 ± 0.126 (296) | 0.680 ± 0.082 (27) | 0.7844 | 0.0020 ± 0.0075 | ||||
|
| |||||||||
| rs12917258 | 0.315 | Intronic | G/C | CAC | 1816 ± 3704 (540) | 1568 ± 2065 (466) | 1218 ± 2423 (115) | 0.0321 | −0.2297 ± 0.0922 |
| CarCP | 320 ± 619 (533) | 327 ± 774 (465) | 255 ± 522 (114) | 0.3173 | −0.1126 ± 0.1134 | ||||
| AACP | 12099 ± 17037 (408) | 10589 ± 14515 (363) | 8835 ± 15601 (88) | 0.3772 | −2.8824 ± 3.2112 | ||||
| IMT | 0.678 ± 0.134 (514) | 0.679 ± 0.141 (451) | 0.652 ± 0.111 (114) | 0.3104 | −0.0034 ± 0.0033 | ||||
|
| |||||||||
| rs4965373 | 0.304 | 3′UTR | G/A | CAC | 1718 ± 3372 (522) | 1491 ± 2878 (477) | 2055 ± 3391 (112) | 0.6453 | 0.0492 ± 0.1278 |
| CarCP | 342 ± 736 (520) | 290 ± 629 (470) | 307 ± 611 (112) | 0.5764 | −0.0627 ± 0.1052 | ||||
| AACP | 11394 ± 16466 (385) | 10742 ± 15070 (387) | 12065 ± 17267 (74) | 0.8306 | 0.6995 ± 2.8541 | ||||
| IMT | 0.680 ± 0.136 (515) | 0.671 ± 0.130 (447) | 0.672 ± 0.147 (111) | 0.4436 | −0.0026 ± 0.0034 | ||||
|
| |||||||||
| rs28628459 | 0.129 | 3′UTR | T/C | CAC | 1640 ± 3274 (858) | 1692 ± 2864 (228) | 1886 ± 2476 (17) | 0.1930 | 0.1994 ± 0.1178 |
| CarCP | 317 ± 701 (850) | 308 ± 598 (227) | 384 ± 768.1 (17) | 0.0390 | 0.3314 ± 0.9744 | ||||
| AACP | 11022 ± 16241 (646) | 10888 ± 14121 (179) | 17602 ± 19339 (11) | 0.3667 | 4.2446 ± 4.7029 | ||||
| IMT | 0.673 ± 0.139 (829) | 0.683 ± 0.130 (217) | 0.666 ± 0.0.94 (17) | 0.4652 | 0.0058 ± 0.0064 | ||||
|
| |||||||||
| rs12438654 | 0.058 | 3′ UTR | C/G | CAC | 1650 ± 3196 (989) | 1736 ± 3013 (124) | 357 ± 722 (5) | 0.2568 | 0.2387 ± 0.3624 |
| CarCP | 321 ± 683 (981) | 287 ± 654 (123) | 61 ± 99 (5) | 0.4695 | −0.1595 ± 0.2428 | ||||
| AACP | 11304 ± 16201 (759) | 9946 ± 13228 (89) | 3219 ± 1812 (3) | 0.1832 | −8.8350 ± 6.2472 | ||||
| IMT | 0.674 ± 0.135 (950) | 0.688 ± 0.135 (124) | 0.618 ± 0.085 (5) | 0.3595 | 0.0058 ± 0.0064 | ||||
|
| |||||||||
| rs7178239 | 0.154 | downstream | C/G | CAC | 1692 ± 3352 (792) | 1533 ± 2703 (302) | 1872 ± 2332 (22) | 0.2014 | 0.1784 ± 0.1398 |
| CarCP | 310 ± 653 (785) | 328 ± 742 (301) | 338 ± 700 (21) | 0.0155 | 0.3752 ± 0.1325 | ||||
| AACP | 11353 ± 16652 (600) | 10278 ± 13650 (235) | 15480 ± 17704 (14) | 0.7188 | 1.5508 ± 4.2661 | ||||
| IMT | 0.673 ± 0.140 (769) | 0.681 ± 0.125 (286) | 0.671 ± 0.086 (22) | 0.2399 | 0.0099 ± 0.0085 | ||||
|
| |||||||||
| rs2101171 | 0.243 | downstream | T/C | CAC | 1677 ± 3311 (637) | 1556 ± 2887 (406) | 1981 ± 3419 (75) | 0.7374 | −0.0381 ± 0.1040 |
| CarCP | 329 ± 718 (632) | 303 ± 621 (402) | 248 ± 641 (75) | 0.2318 | −0.1429 ± 0.1191 | ||||
| AACP | 11102 ± 16541 (475) | 10813 ± 14388 (322) | 13302 ± 18676 (74) | 0.7269 | 1.2167 ± 3.3197 | ||||
| IMT | 0.677 ± 0.141 (618) | 0.674 ± 0.122 (387) | 0.670 ± 0.147 (73) | 0.5719 | −0.0020 ± 0.0037 | ||||
|
| |||||||||
| rs9806366 | 0.127 | downstream | C/T | CAC | 1639 ± 3266 (858) | 1688 ± 2854 (244) | 1895 ± 2556 (16) | 0.2896 | 0.1613 ± 0.1569 |
| CarCP | 315 ± 690 (850) | 316 ± 632 (243) | 401 ± 790 (16) | 0.0608 | 0.2994 ± 0.2190 | ||||
| AACP | 10973 ± 16157 (646) | 11371 ± 14762 (195) | 16859 ± 20218 (10) | 0.3524 | 4.3521 ± 4.7208 | ||||
| IMT | 0.673 ± 0.138 (829) | 0.683 ± 0.128 (234) | 0.675 ± 0.089 (116) | 1.0000 | 0.0021 ± 0.0047 | ||||
MAF = minor allele frequency. 1=major allele, 2= minor allele.
SELS associations with the secondary phenotypes were less consistent (Table 3). The SNPs rs4965814, rs28628459 (both associated with CarCP) and rs9806366 were all associated with self-reported history of prior CVD. In addition, rs12438654 was associated with blood glucose; rs4965373 and rs2101171 were associated with blood glucose and HbA1c; rs34713741 was associated with HDL and triglyceride concentrations; and rs12917258 (also associated with CAC) was also associated with triglyceride concentrations. SELS was not associated with mortality, CVD mortality, hypertension, MetS, CRP, cholesterol, or LDL.
Table 3.
Association results of SELS SNPs with a range of CVD-related phenotypes assuming an additive model of inheritance with covariates age, sex, diabetes affected status (with additional covariates: *anti-hypertensive medication use; †lipid-lowering medication use).
| rs # | Association p-values
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mortality (n=1181) | CVD-M (n=1181) | CVD1 (n=1181) | Hypertension* (n=1181) | MetS (n=1181) | Glucose (n=1176) | HbA1c (n=1170) | CRP (n=991) | Cholesterol† (n=1162) | HDL† (n=1162) | LDL† (n=1088) | TG† (n=1162) | |
| rs34713741 | 0.4688 | 0.5546 | 0.5253 | 0.3976 | 0.8594 | 0.0631 | 0.1852 | 0.5366 | 0.6886 | 0.0259 | 0.5859 | 0.0307 |
| rs28665122 | 0.6751 | 0.8513 | 0.0818 | 0.0539 | 0.7218 | 0.8890 | 0.4452 | 0.9561 | 0.4316 | 0.6133 | 0.6794 | 0.6004 |
| rs4965814 | 0.5740 | 0.6187 | 0.0204 | 0.0819 | 0.8865 | 0.8290 | 0.5873 | 0.2897 | 0.4674 | 0.2734 | 0.8615 | 0.9302 |
| rs12917258 | 0.6688 | 0.8147 | 0.6793 | 0.5212 | 0.6304 | 0.0748 | 0.1894 | 0.5576 | 0.3845 | 0.9968 | 0.6741 | 0.0383 |
| rs4965373 | 0.2938 | 0.6361 | 0.8042 | 0.8539 | 0.6275 | 0.0023 | 0.0074 | 0.7944 | 0.2636 | 0.7799 | 0.9193 | 0.1725 |
| rs28628459 | 0.4324 | 0.8583 | 0.0427 | 0.1497 | 0.9550 | 0.9007 | 0.8779 | 0.3786 | 0.5318 | 0.9564 | 0.9403 | 0.3241 |
| rs12438654 | 0.4755 | 0.5254 | 0.6397 | 0.8592 | 0.5611 | 0.0420 | 0.5056 | 0.4364 | 0.4400 | 0.5353 | 0.9975 | 0.2273 |
| rs7178239 | 0.5190 | 0.8082 | 0.0574 | 0.0625 | 0.8687 | 0.8276 | 0.6697 | 0.5250 | 0.2090 | 0.2209 | 0.4992 | 0.8915 |
| rs2101171 | 0.3478 | 0.7642 | 0.5632 | 0.4536 | 0.8543 | 0.0047 | 0.0008 | 0.9925 | 0.1424 | 0.8717 | 0.4417 | 0.0982 |
| rs9806366 | 0.3924 | 0.8034 | 0.0372 | 0.2093 | 0.9782 | 0.8586 | 0.9261 | 0.4775 | 0.6040 | 0.9805 | 0.8375 | 0.3947 |
CVD-M = CVD Mortality; CVD1 = self reported history of either: angina, myocardial infarction, stroke, angioplasty, endarterectomy or coronary artery bypass surgery; MetS = metabolic syndrome; TG=Triglycerides.
DISCUSSION
While SELS has been identified as a candidate gene contributing to risk for CVD [5, 28], few studies have examined the association with subclinical CVD-related traits, or in cases with T2DM. The current study evaluated the association of 10 SNPs spanning 14.9 kb of the SELS gene region with vascular calcification, IMT, and other known CVD risk factors in European American families enriched for T2DM. Five SELS SNPs were modestly associated with at least one of the measures of vascular calcification, but not at a level considered statistically significant after correction for the multiple comparisons undertaken. In addition, there was minimal association with IMT, likely the result of the poor correlation between these phenotypes in the DHS [21] reflecting the contribution of different mechanisms to the development of vascular calcification and IMT.
In the current study, the strongest evidence of association for these SELS SNPs was with CarCP. An association between SELS and CVD-related traits extends earlier studies examining relationships of SELS with CVD endpoints. A prospective study examining the incidence of fatal and non-fatal coronary events and ischemic stroke in two independent Finnish cohorts (n~1000 each) reported an association of two SELS SNPs rs8025174 and rs7178239 with coronary heart disease (hazard ratio: 2.95) and ischemic stroke (hazard ratio: 3.35) respectively, in women [5]. In the current study we observed an association between rs7178239 and CarCP (rs8025174 was not genotyped). Consistent with the earlier study in which the minor allele was the risk allele, we also found the minor allele to be associated with increased CarCP scores. However, in contrast to our findings, this earlier investigation did not detect evidence of association for rs28665122 (rs12917258 and rs28628459 were not measured) with the incident CVD phenotypes recorded [5], nor did another study examining the association of rs28665122 with incident ischemic stroke in two smaller independent samples (n~200 each) [28].
These conflicting findings may simply be the result of differences in the sample sizes across studies or may be accounted for by differences in sample ascertainment (i.e. diabetes-affected or unaffected) and the varying phenotypes/disease states examined. While the current study has ~80% power to detect differences as small as 15% for SNPs with a MAF of 0.2, we did observe MAFs as low as 0.06. If the MAFs observed in the current study are consistent across other studies in European samples, the inability of some of the smaller studies to replicate SELS associations with CVD traits is not surprising. As such, and given the limited number of studies addressing these questions, further replication is required to better clarify the association of SELS with incident CVD as well as with subclinical CVD traits and to assess whether these associations are evident in a range of different racial groups or if the diabetic environment enhances any effects of SELS polymorphisms.
In addition to the association between SELS variants and measures of subclinical CVD, we also observed evidence of association between SELS SNPs and self-reported history of prior CVD, in keeping with existing reports of SELS SNP associations with incident CVD. However, these SNPs were not similarly associated with either all-cause or CVD-mortality. This lack of replication across the prior CVD and CVD-mortality phenotypes may simply be the result of the relatively small proportion of DHS participants deceased from documented CVD causes (<10%). That said, the identified shortcomings of using cause of death information obtained from death certificates [31, 32] should be acknowledged as a source of potential heterogeneity in the classification of CVD mortality which may obscure any underlying genetic associations. Alternatively, it is possible that the mechanisms through which SelS may mediate risk for CVD development become irrelevant in the context of mortality where multiple other local and systemic responses may be at play.
The associations between SELS and blood glucose and HbA1C measurements are in keeping with the possibility that SelS may be involved in the mechanisms contributing to diabetic macrovascular disease. Consistent with findings for other known glucose-regulated proteins, in vitro experiments have revealed a decrease in SELS expression with increasing glucose concentrations [33] and an increase in SELS expression under insulin stimulation [6]. The observed association of SELS with HbA1C in the current study suggests SELS as potentially responsive to impaired glycemic control. Association with measures of glucose homeostasis, including the homeostasis model of insulin resistance (HOMA-IR) and β-cell function (HOMA-beta) may also be informative here, but were not available in the current study.
Also of note, alterations in SELS expression have been previously associated with cytokine concentrations, suggesting that SelS may act to link T2DM and inflammation. For example, the minor allele of rs28665122 has been associated with both decreased SELS expression and increased inflammatory cytokine concentrations [15]. Our observation of more severe vascular calcification phenotypes in association with the minor allele of rs28665122 is in keeping with a potential inflammatory contribution to the lesion development in atherosclerosis. However, measured SELS concentrations were not available in the DHS to further examine these relationships. Although we did not see an association between these SELS SNPs and systemic inflammation, reflected by CRP concentrations, other modulators of inflammation were not examined and cannot be discounted as a possible mechanism accounting in part for the SELS association with measures of subclinical CVD. In contrast, we found no association between SELS and a range of other measures including hypertension, blood cholesterol and LDL concentrations, all known to influence CVD risk. This suggests that the contribution of SELS to risk for macrovascular complications in this T2DM enriched sample was unlikely to be mediated through these other known risk factors.
In summary, findings from the current study reveal an association of SELS variants with coronary and carotid calcified plaque in European Americans with T2DM. The observed association of the SELS SNPs with both blood glucose and HbA1C suggests that SELS may provide a link between glycemic control and vascular complications in T2DM. More thorough characterization and improved understanding of these interactions may prove important for the prediction and management of macrovascular complications of T2DM.
Supplementary Material
Acknowledgments
This study was supported in part by R01 HL67348, R01 HL09230, and R01 NS058700 to Dr Donald W Bowden. The authors thank the other investigators, the staff, and the participants of the DHS study for their valuable contributions. We would like to acknowledge the Centre for Public Health Genomics at Wake Forest University for its continued support of our biostatisticians.
Footnotes
CONFLICT OF INTEREST: None
References
- 1.American Heart Association. Heart disease & stroke statistics - 2010 update at-a-glance. American Heart Association; 2010. [Google Scholar]
- 2.Centers for Disease Control and Prevention. Diabetes data & trends. Department of Health and Human Services; 2009. [Google Scholar]
- 3.Lu J, Holmgren A. Selenoproteins. J Biol Chem. 2009;284(2):723–7. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- 4.Hesketh J. Nutrigenomics and selenium: Gene expression patterns, physiological targets, and genetics. Annu Rev Nutr. 2008;28:157–77. doi: 10.1146/annurev.nutr.28.061807.155446. [DOI] [PubMed] [Google Scholar]
- 5.Alanne M, Kristiansson K, Auro K, Silander K, Kuulasmaa K, Peltonen L, Salomaa V, Perola M. Variation in the selenoprotein S gene locus is associated with coronary heart disease and ischemic stroke in two independent finnish cohorts. Hum Genet. 2007;122(3–4):355–65. doi: 10.1007/s00439-007-0402-7. [DOI] [PubMed] [Google Scholar]
- 6.Olsson M, Olsson B, Jacobson P, Thelle DS, Bjorkegren J, Walley A, Froguel P, Carlsson LM, Sjoholm K. Expression of the selenoprotein S (SELS) gene in subcutaneous adipose tissue and SELS genotype are associated with metabolic risk factors. Metabolism. 2011;60(1):114–20. doi: 10.1016/j.metabol.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez A, Santiago JL, Varade J, Marquez A, Lamas JR, Mendoza JL, de la Calle H, Diaz-Rubio M, de la Concha EG, Fernandez-Gutierrez B, Urcelay E. Polymorphisms in the selenoprotein S gene: Lack of association with autoimmune inflammatory diseases. BMC Genomics. 2008;9:329. doi: 10.1186/1471-2164-9-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellinger FP, Raman AV, Reeves MA, Berry MJ. Regulation and function of selenoproteins in human disease. Biochem J. 2009;422(1):11–22. doi: 10.1042/BJ20090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455–62. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci. 2008;65(6):862–94. doi: 10.1007/s00018-007-7383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hotamisligil GS. Endoplasmic reticulum stress and atherosclerosis. Nat Med. 2010;16(4):396–9. doi: 10.1038/nm0410-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hotamisligil GS. Inflammation and endoplasmic reticulum stress in obesity and diabetes. Int J Obes (Lond) 2008;32(Suppl 7):S52–4. doi: 10.1038/ijo.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroder M, Sutcliffe L. Consequences of stress in the secretary pathway: The ER stress response and its role in the metabolic syndrome. Methods Mol Biol. 2010;648:43–62. doi: 10.1007/978-1-60761-756-3_3. [DOI] [PubMed] [Google Scholar]
- 14.Gao Y, Hannan NR, Wanyonyi S, Konstantopolous N, Pagnon J, Feng HC, Jowett JB, Kim KH, Walder K, Collier GR. Activation of the selenoprotein SEPS1 gene expression by pro-inflammatory cytokines in HEPG2 cells. Cytokine. 2006;33(5):246–51. doi: 10.1016/j.cyto.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Curran JE, Jowett JB, Elliott KS, Gao Y, Gluschenko K, Wang J, Abel Azim DM, Cai G, Mahaney MC, Comuzzie AG, Dyer TD, Walder KR, Zimmet P, MacCluer JW, Collier GR, Kissebah AH, Blangero J. Genetic variation in selenoprotein S influences inflammatory response. Nat Genet. 2005;37(11):1234–41. doi: 10.1038/ng1655. [DOI] [PubMed] [Google Scholar]
- 16.Bos SD, Kloppenburg M, Suchiman E, van Beelen E, Slagboom PE, Meulenbelt I. The role of plasma cytokine levels, CRP and selenoprotein S gene variation in OA. Osteoarthritis Cartilage. 2009;17(5):621–6. doi: 10.1016/j.joca.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Budoff MJ. Atherosclerosis imaging and calcified plaque: Coronary artery disease risk assessment. Prog Cardiovasc Dis. 2003;46(2):135–48. doi: 10.1016/s0033-0620(03)00083-5. [DOI] [PubMed] [Google Scholar]
- 18.Sipila K, Kahonen M, Salomaa V, Paivansalo M, Karanko H, Varpula M, Jula A, Kaaja R, Kesaniemi YA, Reunanen A, Moilanen L. Carotid artery intima-media thickness and elasticity in relation to glucose tolerance. Acta Diabetol. 2012;49(3):215–23. doi: 10.1007/s00592-011-0291-z. [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto M, Kotani K, Okada K, Fujii Y, Konno K, Ishibashi S, Taniguchi N. The correlation of common carotid arterial diameter with atherosclerosis and diabetic retinopathy in patients with type 2 diabetes mellitus. Acta Diabetol. 2012;49(1):63–8. doi: 10.1007/s00592-011-0287-8. [DOI] [PubMed] [Google Scholar]
- 20.Bowden DW, Cox AJ, Freedman BI, Hugenschmidt CE, Wagenknecht LE, Herrington D, Agarwal S, Register TD, Maldjian JA, Ng MC-Y, Hsu F-C, Langefeld CD, Williamson JD, Carr JJ. Review of the Diabetes Heart Study (DHS) family of studies: A comprehensively examined sample for genetic and epidemiological studies of type 2 diabetes and its complications. The Review of Diabetic Studies. 2010;7(3):188–201. doi: 10.1900/RDS.2010.7.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowden DW, Lehtinen AB, Ziegler JT, Rudock ME, Xu J, Wagenknecht LE, Herrington DM, Rich SS, Freedman BI, Carr JJ, Langefeld CD. Genetic epidemiology of subclinical cardiovascular disease in the Diabetes Heart Study. Ann Hum Genet. 2008;72(Pt 5):598–610. doi: 10.1111/j.1469-1809.2008.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagenknecht LE, Bowden DW, Carr JJ, Langefeld CD, Freedman BI, Rich SS. Familial aggregation of coronary artery calcium in families with type 2 diabetes. Diabetes. 2001;50(4):861–6. doi: 10.2337/diabetes.50.4.861. [DOI] [PubMed] [Google Scholar]
- 23.Bowden DW, Rudock M, Ziegler J, Lehtinen AB, Xu J, Wagenknecht LE, Herrington D, Rich SS, Freedman BI, Carr JJ, Langefeld CD. Coincident linkage of type 2 diabetes, metabolic syndrome, and measures of cardiovascular disease in a genome scan of the diabetes heart study. Diabetes. 2006;55(7):1985–94. doi: 10.2337/db06-0003. [DOI] [PubMed] [Google Scholar]
- 24.Lange LA, Bowden DW, Langefeld CD, Wagenknecht LE, Carr JJ, Rich SS, Riley WA, Freedman BI. Heritability of carotid artery intima-medial thickness in type 2 diabetes. Stroke. 2002;33(7):1876–81. doi: 10.1161/01.str.0000019909.71547.aa. [DOI] [PubMed] [Google Scholar]
- 25.Expert Panel on Detection Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive summary of the third report of the national cholesterol education program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) JAMA. 2001;285(19):2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 26.Carr JJ, Crouse JR, Goff DC, Jr, D’Agostino RB, Jr, Peterson NP, Burke GL. Evaluation of subsecond gated helical CT for quantification of coronary artery calcium and comparison with electron beam CT. AJR Am J Roentgenol. 2000;174(4):915–21. doi: 10.2214/ajr.174.4.1740915. [DOI] [PubMed] [Google Scholar]
- 27.Carr JJ, Nelson JC, Wong ND, McNitt-Gray M, Arad Y, Jacobs DR, Jr, Sidney S, Bild DE, Williams OD, Detrano RC. Calcified coronary artery plaque measurement with cardiac CT in population-based studies: Standardized protocol of multi-ethnic study of atherosclerosis (MESA) and coronary artery risk development in young adults (CARDIA) study. Radiology. 2005;234(1):35–43. doi: 10.1148/radiol.2341040439. [DOI] [PubMed] [Google Scholar]
- 28.Hyrenbach S, Pezzini A, del Zotto E, Giossi A, Lichy C, Kloss M, Werner I, Padovani A, Brandt T, Grond-Ginsbach C. No association of the -105 promoter polymorphism of the selenoprotein S encoding gene SEPS1 with cerebrovascular disease. Eur J Neurol. 2007;14(10):1173–5. doi: 10.1111/j.1468-1331.2007.01898.x. [DOI] [PubMed] [Google Scholar]
- 29.Buetow KH, Edmonson M, MacDonald R, Clifford R, Yip P, Kelley J, Little DP, Strausberg R, Koester H, Cantor CR, Braun A. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci U S A. 2001;98(2):581–4. doi: 10.1073/pnas.021506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cambridge B, Cina SJ. The accuracy of death certificate completion in a suburban community. Am J Forensic Med Pathol. 2010;31(3):232–5. doi: 10.1097/PAF.0b013e3181e5e0e2. [DOI] [PubMed] [Google Scholar]
- 32.Sington JD, Cottrell BJ. Analysis of the sensitivity of death certificates in 440 hospital deaths: A comparison with necropsy findings. J Clin Pathol. 2002;55(7):499–502. doi: 10.1136/jcp.55.7.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y, Feng HC, Walder K, Bolton K, Sunderland T, Bishara N, Quick M, Kantham L, Collier GR. Regulation of the selenoprotein SELS by glucose deprivation and endoplasmic reticulum stress - SELS is a novel glucose-regulated protein. FEBS Lett. 2004;563(1–3):185–90. doi: 10.1016/S0014-5793(04)00296-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.