Background: Mutations of proteolipid protein 1 (PLP1) induce endoplasmic reticulum (ER) stress.
Results: PLP1 mutants deplete some chaperones from the ER and induce fragmentation of the Golgi apparatus (GA).
Conclusion: These changes affect clinical pathology in disease-causing mutations of PLP1.
Significance: This work provides a novel insight involving global changes of organelles in pathogenesis of ER stress-related diseases.
Keywords: Endoplasmic Reticulum Stress, Molecular Chaperone, Myelin, Neurodegenerative Diseases, Unfolded Protein Response
Abstract
Missense mutations in the proteolipid protein 1 (PLP1) gene cause a wide spectrum of hypomyelinating disorders, from mild spastic paraplegia type 2 to severe Pelizaeus-Merzbacher disease (PMD). Mutant PLP1 accumulates in the endoplasmic reticulum (ER) and induces ER stress. However, the link between the clinical severity of PMD and the cellular response induced by mutant PLP1 remains largely unknown. Accumulation of misfolded proteins in the ER generally leads to up-regulation of ER chaperones to alleviate ER stress. Here, we found that expression of the PLP1-A243V mutant, which causes severe disease, depletes some ER chaperones with a KDEL (Lys-Asp-Glu-Leu) motif, in HeLa cells, MO3.13 oligodendrocytic cells, and primary oligodendrocytes. The same PLP1 mutant also induces fragmentation of the Golgi apparatus (GA). These organelle changes are less prominent in cells with milder disease-associated PLP1 mutants. Similar changes are also observed in cells expressing another disease-causing gene that triggers ER stress, as well as in cells treated with brefeldin A, which induces ER stress and GA fragmentation by inhibiting GA to ER trafficking. We also found that mutant PLP1 disturbs localization of the KDEL receptor, which transports the chaperones with the KDEL motif from the GA to the ER. These data show that PLP1 mutants inhibit GA to ER trafficking, which reduces the supply of ER chaperones and induces GA fragmentation. We propose that depletion of ER chaperones and GA fragmentation induced by mutant misfolded proteins contribute to the pathogenesis of inherited ER stress-related diseases and affect the disease severity.
Introduction
A number of inherited human diseases are caused by missense mutations. These mutations in the membrane and secretary proteins often lead to improper protein folding and accumulation in the endoplasmic reticulum (ER),2 resulting in an induction of ER stress. In cells under ER stress, accumulation of mutant proteins in the ER activates the unfolded protein response (UPR), which initiates a block in translation, increases retrotranslocation and degradation of ER-localized proteins, and bolsters the protein-folding capacity of the ER (1, 2). Through these processes, the UPR functions as a cellular quality control system that essentially protects cells from the toxicity of accumulated proteins in the ER. The UPR is activated by three distinct pathways, activating transcription factor 6 (ATF6), inositol-requiring kinase 1 (IRE1), and protein kinase-like ER kinase (3), all of which are negatively regulated by interaction with the 78-kDa glucose-regulated protein (GRP78, also referred to as BiP/HSPA5). On accumulation of unfolded protein, GRP78 binds to unfolded proteins and dissociates from the ER stress sensors, which trigger the UPR (3). ATF6 induces transcription of major ER chaperones and X box-binding protein 1 (XBP1) (4). The endonuclease activity of IRE1 splices XBP1 (5), which functions as a transcription factor that drives the expression of UPR-related genes (4). The ATF6 and IRE1-XBP1 axes promote the expression of ER chaperones that facilitate the correct folding or assembly of ER proteins and prevent their aggregation, thereby improving cell survival (3, 4, 6). However, when ER stress overwhelms the capacity of this intrinsic quality control, apoptosis is induced by up-regulation of the C/EBP homologous protein (CHOP).
In inherited diseases associated with ER stress, different mutations in the causative genes result in various phenotypes. One representative example, Pelizaeus-Merzbacher disease (PMD), is an X-linked recessive leukodystrophy characterized by diffuse hypomyelination in the central nervous system (CNS) (7). Missense mutations in the proteolipid protein 1 (PLP1) gene cause a wide spectrum of clinical phenotypes from a mild allelic disease, spastic paraplegia type 2 (SPG2) to severe connatal PMD (7). In these diseases, mutant proteins are misfolded and accumulate in the ER, leading to induction of ER stress and apoptosis of oligodendrocytes in the CNS (8, 9). However, little is known about how different mutations in the same gene induce ER stress differently and affect clinical severity. Factors, such as retention of misfolded proteins or the extent of UPR activation, may influence phenotypic variation (9–11). However, can any other factors contribute to the pathology of such ER stress-related disease? Here we focused on ER chaperones as a potential player. ER chaperones are highly conserved proteins that assist in protein folding. Therefore, it is generally believed that accumulation of misfolded proteins in the ER up-regulates chaperones to alleviate ER stress. In terms of its association with disease pathology, interaction between the mutant PLP1 and a major ER chaperone, calnexin (CANX), was shown to inhibit degradation of the misfolded mutant proteins (12). In the mutant superoxide dismutase model of amyotrophic lateral sclerosis (ALS), an ER stress-associated neurodegenerative disease, down-regulation of another chaperone, calreticulin (CALR), was shown to induce ER stress and trigger the death of mutant superoxide dismutase motoneurons (13). A recent study reported up-regulation of protein-disulfide isomerase (PDI, also referred to as P4HB), which is a chaperone in the ER catalyzing the formation and breakage of protein disulfides bonds, in microglia of transgenic mutant superoxide dismutase 1 mice (14). Therefore, we sought to determine whether changes in the expression of ER chaperones alter the accumulation of misfolded protein and ER stress, potentially modifying the cellular and clinical phenotypes.
For this purpose, we used PMD as a model and investigated missense PLP1 mutations (8, 11). PLP1 with an A243V substitution (PLP1msd) is representative of the severe hypomyelination in myelin synthesis deficit (msd) mice (15) and humans (16), whereas two other mutations, W163L and I187T, are representative of the milder condition found in mild PMD/SPG2 patients (17, 18): the latter is also the mutation found in an SPG2 mouse model, rumpshaker (19). We also employed mutants in two other genes responsible for peripheral myelin disorders, a myelin protein zero (MPZ) mutant associated with a severe neuropathy, Dejerine-Sottas neuropathy (20), and two peripheral myelin protein 22 (PMP22) mutants that are associated with a clinically mild neuropathy, Charcot-Marie-Tooth disease (21–23).
In this study, we examined the expression of ER chaperones in response to mutants of PLP1 and two other genes. Unexpectedly, we found that some ER chaperones were depleted rather than up-regulated. In addition, these mutant proteins induced fragmentation of the Golgi apparatus (GA). We also found an association between these changes and phenotypic severity. Furthermore, we proposed potential mechanisms underlying these cellular phenotypes. The results of this study suggest that changes in these subcellular organelles may contribute to the cellular pathogenesis and phenotypic severity of inherited ER stress-related diseases caused by mutant proteins.
EXPERIMENTAL PROCEDURES
Mice
Msd mice, which carry the spontaneous A243V mutation in the Plp1 gene (15), were maintained in a B6C3F1/J background in accordance with the institutional guidelines of the National Center of Neurology and Psychiatry.
Plasmid Construction
Expression vectors for PLP1wt and PLP1msd were reported previously (24). PLP1-W163L and PLP1-I187T genes were generated by site-directed mutagenesis with modifications (25), and subcloned into pCAGGS (24), as fusions with N-terminal FLAG epitopes. Human wild-type and mutant PMP22 and MPZ genes were amplified from cloned cDNAs (kind gift from Dr. JR Lupski) using appropriate primers and inserted into pCAGGS. For construction of an expression vector for the membrane-linked cell surface green fluorescent protein (GFP) as illustrated in Fig. 9E. The GFP gene was inserted into pDisplay (Invitrogen) in an in-frame manner. The cytoplasmic luciferase (Cluc) and immunoglobulin (Ig) κ light chain (Igκ-Rluc) genes were amplified from cloned cDNA (Promega) with appropriate primers, and cloned into pCDNA3.1 (Invitrogen) and pAP-Tag5 (GenHunter) to construct pCMV-Cluc and pCMV-Igκ-Rluc, respectively. To determine subcellular localization, the Rluc gene was inserted in-frame between the Igκ and myc sequences of pAP-Tag5 to make pCMV-Igκ-Rluc-Myc. The mouse myelin oligodendrocyte glycoprotein (MOG) gene was also amplified with appropriate primers using cDNA from postnatal day (P) 14-mouse spinal cord (SC), and cloned into pEGFP-N1 (Takara).
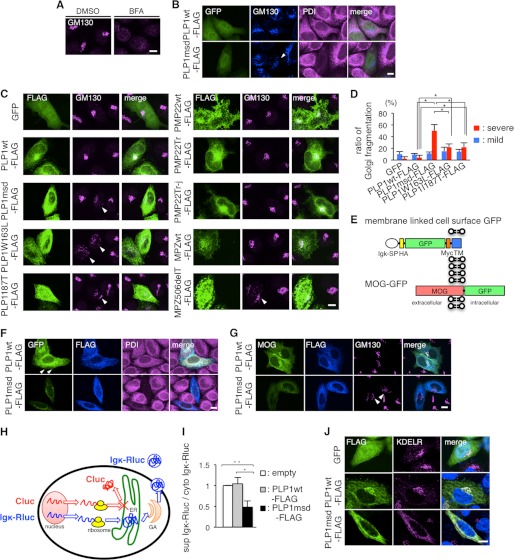
FIGURE 9.
PLP1msd overexpression induces GA fragmentation, retention of MOG in the ER, and reduction of protein secretion. A and B, immunocytochemistry of GM130 in HeLa cells treated with BFA (A) and HeLa cells expressing PLP1wt or PLP1msd (B). HeLa cells were treated with DMSO (as a control) or BFA as described in the legend to Fig. 7A. HeLa cells co-transfected with GFP (to visualize transfected cells) along with PLP1wt-FLAG or PLP1msd-FLAG were immunostained using anti-GM130 (blue) and anti-PDI (magenta) antibodies and were observed with a confocal fluorescence microscope. Cells expressing PLP1msd showed the fragmentation of the GA (arrowhead). Scale bar, 10 μm. C, immunocytochemistry of GM130 in HeLa cells expressing PLP1, PMP22, and MPZ mutants. HeLa cells transfected with the indicated vectors were immunostained with the anti-FLAG (green) and anti-GM130 (magenta) antibodies. Note that cells expressing PLP1 mutants and MPZ506delT showed fragmentation of GA detected by GM130 staining (arrowheads). Scale bar, 10 μm. D, the proportion of cells showing GA fragmentation in HeLa cells transfected with PLP1wt or mutant PLP1 genes, as shown in B and C. FLAG-positive cells were classified into 3 categories based on GA morphology, as normal, mild fragmentation, and severe fragmentation and the number of cells in each class was counted. Data for normal are not shown. The results are represented as the mean ± S.E. from three independent experiments with >100 cells counted in each experiment (*, p ≤ 0.05; **, p ≤ 0.005). E, scheme for the construction of membrane-linked cell surface GFP and MOG-GFP. SP, signal peptide; Igκ, immunoglobulin κ light chain; HA, hemagglutinin; TM, transmembrane of platelet-derived growth factor receptor. F and G, subcellular localization of membrane-linked cell surface GFP (F) or MOG-GFP (G) in HeLa cells expressing PLP1wt-FLAG or PLP1msd-FLAG. HeLa cells co-transfected with membrane-linked cell surface GFP or MOG-GFP along with PLP1wt-FLAG or PLP1msd-FLAG were immunostained using anti-FLAG (blue) and anti-PDI (magenta) antibodies. Cells expressing PLP1wt showed the cell surface GFP expression (arrowhead). Scale bar, 10 μm. H, scheme for the luciferase reporter assay. Secretary Renilla luciferase (Igκ-Rluc, blue), which is fused with Igκ signal peptide, penetrates into the ER and is secreted to the extracellular space through the GA. Firefly luciferase (Cluc, red), which is a cytosolic protein, served as an internal control. This system enables the measurement of secretion of the secretory reporter protein in live cells by comparing the total and supernatant activities of Igκ-Rluc. Co-transfection with the Cluc gene not only detects leakage of these reporter proteins to the supernatant from dead cells, but also compares translation efficacies between cytoplasmic and secretary proteins. I, luciferase reporter assay to evaluate the effect of PLP1msd on the secretory protein transport. HeLa cells were co-transfected with the firefly luciferase (Cluc) and Igκ-Rluc genes along with an empty vector, PLP1wt-FLAG, or PLP1msd-FLAG. Each of Cluc and Igκ-Rluc in the cell lysate and supernatant was simultaneously measured by a luminometer. Efficient of secretion of Igκ-Rluc was calculated as follows: Renilla luciferase activity of supernatant/total (supernatant plus cytosol) Renilla luciferase activity. Results are represented as fold-induction compared with empty vector control experiment. Values are represented as the mean ± S.E. from three independent experiments (*, p ≤ 0.05; **, p ≤ 0.005). J, immunocytochemistry of KDEL receptor in HeLa cells expressing PLP1wt or PLP1msd. HeLa cells transfected with the indicated vectors were immunostained with the anti-FLAG (green) and anti-KDEL receptor (magenta) antibodies and observed with a confocal fluorescence microscope. Scale bar, 5 μm.
Chemicals and Antibodies
The following reagents were purchased from the suppliers indicated: brefeldin A (Wako), tunicamycin (Merck), thapsigargin (Sigma), lactacystin (Wako), and MG132 (Wako). The primary antibodies included mouse anti-PDI (Thermo Scientific, MA3-019), rabbit anti-CALR (Sigma, C4606), rabbit anti-GRP78 (Abcome, ab21685), rabbit anti-CANX (Enzo Life Sciences, ADI-SPA-860), mouse anti-CHOP (Santa Cruz Biotechnology, sc-7351), rabbit anti-GM130 (Abcome, ab52649), mouse anti-FLAG M2 (Sigma, F3165), rabbit anti-FLAG M2 (Cell Signaling, number 2368), mouse anti-c-Myc (Nacalai Tesque, MC045), rabbit anti-PLP (a kind gift from Dr. M. Itoh, NCNP), rabbit anti-Oligo2 (IBL 18953), mouse anti-myelin basic protein (MBP) (Covance, SMI-99P), mouse anti-action (Millipore, MAB1501), rabbit anti-KDEL receptor (Santa Cruz Biotechnology, sc-33806) and mouse anti-ubiquitin (Santa Cruz Biotechnology, sc-8017) antibodies. Alexa Fluor-488, -594, and -647 secondary antibodies were purchased from Invitrogen. Horseradish peroxidase-labeled anti-mouse and rabbit antibodies were purchased from GE Healthcare.
Cell Culture
HeLa cells and human oligodendrocytic cells (MO3.13) were maintained in Dulbecco's modified Eagle's medium (DMEM, Thermo Scientific) supplemented with 20 units ml−1 of penicillin, 20 μg ml−1 of streptomycin, and 10% fetal bovine serum. For transfection, HeLa cells were plated onto 6-well plates or 18-mm round coverslips in 12-well plates, and transfected with the indicated constructs using Lipofectamine 2000 (Invitrogen) or TransIt LT1 (Mirus), respectively, according to the manufacturers' protocols. After 24 h, transfected cells in the 6-well plates or on coverslips were subjected to immunoblotting, quantitative PCR and immunocytochemistry, respectively.
Mixed Glial Culture (MGC) Generated to Oligodendrocyte
MGCs were established from wild-type and msd mice, which were then differentiated into oligodendrocytes, as described by Abematsu et al. (26).
Immunoblot Analysis
HeLa cells and mouse SCs were lysed with TNE(+) lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton-X-100, and 0.1% SDS) supplemented with protease and phosphatase inhibitors on ice for 10 min. For the digitonin fractionation experiment, HeLa cells were permeabilized with phosphate-buffered saline (PBS) containing 0.01% digitonin with protease and phosphatase inhibitors on ice for 10 min. After soluble proteins were collected, insoluble proteins were further treated with the TNE(+) lysis buffer. These extracts were centrifuged at 12,000 × g for 10 min to remove cell debris. Co-immunoprecipitation and cell surface biotinylation were performed as described previously (27). The cell extracts, co-immunoprecipitation and biotinylated samples, were subjected to immunoblotting with primary antibodies and horseradish peroxidase-labeled secondary antibodies. All immunoblot analyses were repeated at least 3 times with similar results. The relative protein expression levels on immunoblotting were quantified by an image analyzer.
Immunocytochemistry
HeLa cells, MO3.13 cells, and primary oligodendrocytes were fixed with 4% paraformaldehyde in PBS for 10 min, permeabilized with 0.1% Triton X-100 for 10 min, and treated with 3% bovine serum albumin to block nonspecific reaction. Detection of cell surface proteins, cells were not permeabilized by 0.1% TritonX-100. Cells were further incubated with the primary antibodies for 60 min at RT followed by visualization using the appropriate secondary antibodies labeled with Alexa-488, -594, or -647 with 4′,6-diamidino-2-phenylindole (DAPI). Apoptotic cells were detected using ApopTag kit (Chemicon), according to the manufacturer's protocol. These stained cells were observed with a confocal fluorescence microscope (FV-1000; Olympus).
Quantitative Reverse Transcriptase-Polymerase Chain Reaction
Total RNA was extracted from HeLa cells and mouse SCs and was converted to cDNA using SuperScript III reverse transcriptase (Invitrogen). Transcript levels were analyzed by a thermal cycler (7900HT; Applied Biosystems) with synthesized cDNA and the following pre-designed TaqMan probes (Applied Biosystems): human GAPDH, Hs99999905; human CHOP, Hs00358796; human P4HB, Hs00168586; human CALR, Hs00189032; human GRP78, Hs99999174; human CANX, Hs00233492; mouse Gapdh, Mm99999915; mouse Chop, Mm00492097; mouse P4hb, Mm01243184; mouse Calr, Mm00482936; mouse Grp78, Mm00517691; and mouse Canx, Mm00500330. Relative transcript levels were calculated by the ΔΔCT method according to the manufacturer's standard protocol.
Luciferase Reporter Assay
HeLa cells were co-transfected with the Cluc and Igκ-Rluc genes along with pCDNA3.1-PLP1wt-FLAG, pCDNA3.1-PLP1msd-FLAG, or the empty vector. Activities for firefly luciferase and Igκ-Rluc in the cell lysate and supernatant were measured using a dual-luciferase assay system (Promega) according to the manufacturer's instructions. Relative Cluc and Igκ-Rluc activities in the supernatant were determined as ratios to cytosolic luciferase activity.
Statistical Analysis
Student's t test and analysis of variance were used for statistical analyses.
RESULTS
PDI, CALR, and GRP78 Are Depleted in the ER of HeLa Cells Expressing PLP1msd
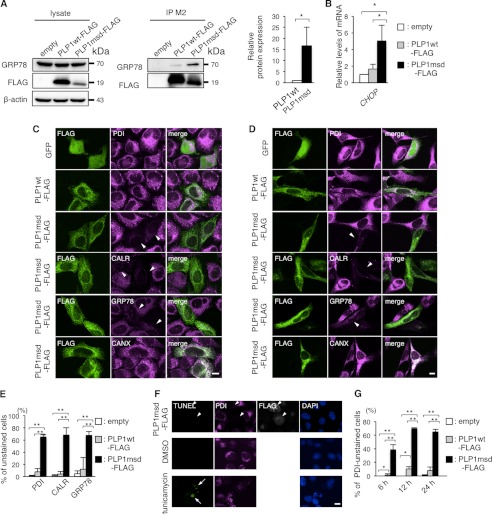
Typically, when cells are under ER stress, ER chaperones are up-regulated as a part of the UPR. ER chaperones improve cell survival by facilitating the correct folding or assembly of misfolded proteins and preventing their aggregation (28). In HeLa cells, FLAG-tagged PLP1msd (PLP1msd-FLAG), a PMD-causing severe mutant known to induce ER stress (8) but not FLAG-tagged wild-type PLP1 (PLP1wt-FLAG), effectively co-immunoprecipitated GRP78 (Fig. 1A) and up-regulated the CHOP gene (Fig. 1B), a well characterized ER stress marker gene, indicating that this transient transfection system is applicable for analyzing cellular pathogenesis of ER stress caused by exogenous mutant proteins. To further analyze the changes in ER chaperone expression induced by this mutant PLP1, transfected HeLa cells were immunostained with antibodies against the FLAG epitope, PDI, CALR, GRP78, and CANX. Unexpectedly, we found that PDI, CALR, and GRP78 were drastically depleted, whereas CANX expression was unchanged (Fig. 1C). These changes were also observed in the human oligodendrocytic cell line MO3.13 (Fig. 1D), the human glioma cell line U-138, and simian kidney cell line COS-7 cells (data not shown). Almost 65% of the cells transfected with PLP1msd-FLAG had faint PDI, CALR, and GRP78 staining. This proportion was significantly higher than in cells transfected with PLP1wt-FLAG (Fig. 1E), suggesting that this phenomenon is due to the mutant PLP1, not the overexpression of PLP1 itself.
FIGURE 1.
PLP1msd overexpression resulted in a negative ER chaperone staining pattern. A, co-immunoprecipitation of PLP1 with GRP78 in HeLa cells. B, quantitative RT-PCR for the CHOP gene in HeLa cells expressing PLP1wt or PLP1msd. The GAPDH gene was used as an internal control. Results are represented as fold-induction compared with empty vector-transfected control experiment. Values are represented as the mean ± S.E. from three independent experiments (*, p ≤ 0.05). C and D, immunocytochemistry of ER chaperones in HeLa cells (C) and human oligodendrocytic cells, and MO3.13 cells (D) expressing PLP1wt or PLP1msd. Cells transfected with the indicated vectors were immunostained with an anti-FLAG antibody (green) together with anti-PDI, anti-CALR, anti-GRP78, or anti-CANX antibodies (magenta) and observed with a confocal fluorescence microscope. Note that cells expressing PLP1msd showed an extremely faint staining pattern (arrowheads) for PDI, CALR, and GRP78, but not for CANX. Scale bar, 10 μm. E, the proportion of unstained cells for PDI, CALR, and GRP78. F, apoptosis of HeLa cells expressing PLP1msd. TUNEL assay combined with immunocytochemical staining using the anti-FLAG (white) and anti-PDI (magenta) antibodies. Tunicamycin treatment served as a positive control for TUNEL (arrow). None of the PLP1msd-positive cells showed positive signals for TUNEL (arrowheads). Scale bar, 10 μm. G, time course of the proportion of PDI negative HeLa cells transfected with the PLP1msd gene. The results are represented as the mean ± S.E. from three independent experiments with >100 cells counted in each experiment (*, p ≤ 0.05; **, p ≤ 0.005).
Next, we determined if reduced chaperone expression is caused by apoptotic cell death due to overwhelming ER stress. Cells expressing PLP1msd that had faint PDI immunostaining had no positive signal in the terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay (Fig. 1F). Furthermore, depletion of PDI occurred as early as 6 h after transfection (Fig. 1G). These results suggest that PLP1msd impairs the ER chaperones independent of apoptosis.
Expression of PLP1msd Translocates the ER Chaperones from the ER
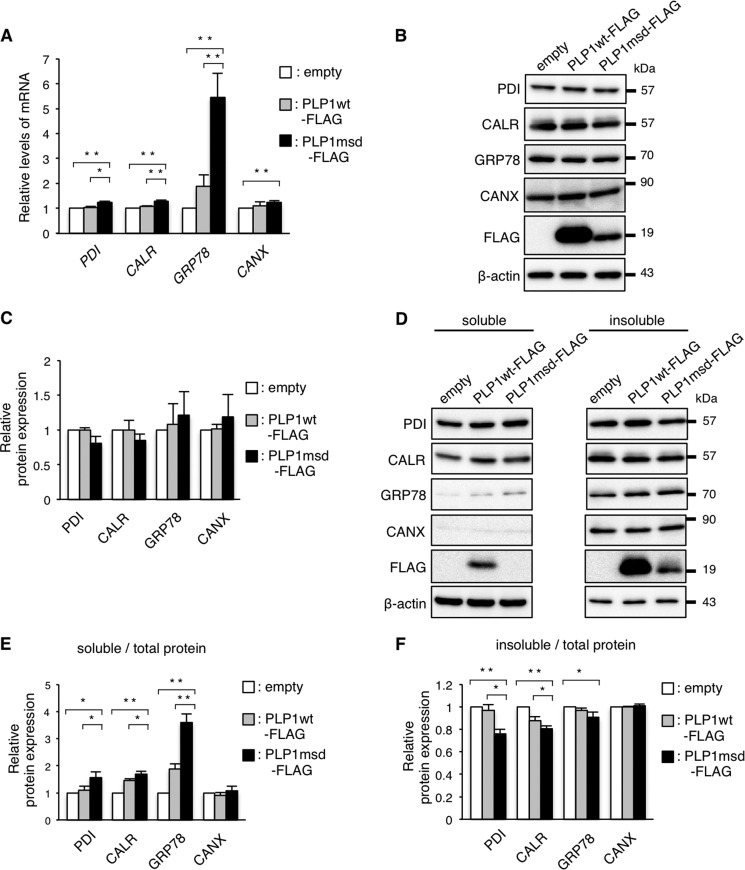
To examine whether PLP1msd depletes the chaperones by inhibiting their transcription in HeLa cells, we performed quantitative real time-polymerase chain reaction (RT-PCR) (Fig. 2A). As we demonstrated previously (24), GRP78 mRNA expression was increased significantly in cells transfected with PLP1msd-FLAG compared with cells transfected with PLP1wt-FLAG. The expression of PDI and CALR was slight, but significantly up-regulated. These results indicate that PDI, CALR, and GRP78 are depleted in the ER without decreasing their transcription. Furthermore, total protein levels of these chaperones were essentially unchanged in cells transfected with PLP1msd-FLAG (Fig. 2, B and C). These results indicate that PDI, CALR, and GRP78 depletion in the ER is not due to a reduction in the total proteins.
FIGURE 2.
PDI, CALR, and GRP78 are depleted from the ER without decreasing their transcripts. A, relative expression of the transcripts of the ER chaperones in HeLa cells transfected with the PLP1 genes. Expression levels of PDI, CALR, GRP78, and CANX mRNA were analyzed by quantitative RT-PCR and normalized to GAPDH. The results are represented as fold-induction against the control experiment (empty vector transfection). B and C, total amount of the ER chaperones in HeLa cells transfected with PLP1wt-FLAG and PLP1msd-FLAG. Protein samples from the cells transfected with the indicated vectors were subjected to immunoblotting with the indicated antibodies (B). The amounts of the proteins were measured by densitometry and normalized to β-actin. The results are represented as fold-induction against the control experiment using the empty vector (C). D–F, subcellular fractionation analysis using 0.01% digitonin. Transfected cells were treated with 0.01% digitonin followed by 0.1% SDS, 1% Triton X-100. The extracts of digitonin soluble (left) and insoluble fraction (right) were subjected to immunoblotting with the indicated antibodies (D). The blots were quantitatively analyzed by densitometry to measure the proportion of soluble fraction (E) and insoluble fraction (F) in each protein. Bar graphs are represented as fold-induction ± S.E. against the mean of control experiment from three independent experiments (*, p ≤ 0.05; **, p ≤ 0.005).
Next, we considered the possibility that PLP1msd affected subcellular localization of the ER chaperones from the ER to another cellular compartment. To test this possibility, transfected cells were treated with 0.01% digitonin, which permeabilizes the plasma membrane but not organelles' membranes, followed by treatment with 0.1% SDS, 1% Triton X-100, which permeabilizes the organelles, including the ER (Fig. 2, D–F). In cells expressing PLP1msd, the proportion of PDI, CALR, and GRP78 in the fraction containing the plasma membrane and cytosol (soluble fraction) was higher than in cells expressing PLP1wt (Fig. 2, D and E). In contrast, the amount of these chaperones was lower in fractions containing the ER (insoluble fraction) in PLP1msd expressing cells (Fig. 2, D and F). Interestingly, the amount of CANX was unchanged in the digitonin-soluble and -insoluble fractions. These results suggest that the decrease in PDI, CALR, and GRP78 in the ER may be due to their translocation from the ER to the plasma membrane or cytosol, but not due to the decrease of total protein.
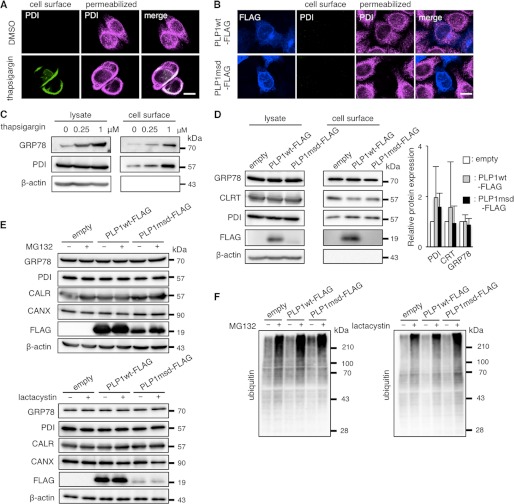
Recently, Zhang et al. (29) reported that ER stress actively promotes GRP78 localization on the cell surface. We confirmed that thapsigargin, a well known ER stressor, increases cell surface expression of PDI in HeLa cells by immunocytochemistry (Fig. 3A) and increases cell surface expression of GRP78 and PDI in HeLa cells by cell surface biotinylation (Fig. 3C). However, we observed no expression of PDI on the cell surface of PLP1msd-transfected cells (Fig. 3B) or no increment of the biotinylated PDI, CALR, and GRP78 in these cells (Fig. 3D). These findings suggest that these mutant proteins induce translocation of the chaperones from the ER to the cytosol, rather to the cell surface.
FIGURE 3.
PLP1msd does not increase cell surface expression of the ER chaperones. A and B, immunocytochemical analysis of cell surface PDI on HeLa cells treated with 1 μm thapsigargin (A) or transfected with the PLP1msd gene (B). Cell surface PDI (green) were stained with the anti-PDI antibody without permeabilization followed by intracellular staining with the same antibody (magenta). Scale bar, 10 μm. C and D, biochemical analysis of cell surface expression of the ER chaperones. HeLa cells were treated with thapsigargin for 16 h (C). Transfection was performed before 24 h of cell surface biotinylation (D). Cell surface proteins were labeled with biotin, precipitated with streptavidin beads followed by immunoblotting with anti-PDI, anti-CALR, and anti-GRP78 antibodies. E and F, protease inhibitors do not increase total amounts of the ER chaperones in HeLa cells expressing PLP1msd. Transfected cells were treated with 5 μm MG132 for 16 h or 1 μm lactacystin for 8 h followed by immunoblotting with the anti-PDI, anti-CALR, anti-GRP78, anti-CANX (E) and anti-ubiquitin antibodies (F). Protein amounts were measured by densitometry. The results are represented as fold-induction against the control experiment using the empty vector. Values are represented as the mean ± S.E. from three independent experiments (D).
We speculated that mislocalized cytoplasmic chaperons are degraded through a ubiquitin-dependent ERAD pathway. However, although the amounts of ubiquitinated proteins increased in the presence of proteasome inhibitors, MG-132 or lactacystin (Fig. 3F), those of the ER chaperons were not affected (Fig. 3E), suggesting that they are not degraded through the ERAD pathway after releasing to the cytosol.
Differences and Similarities among Disease-causing Mutations in Other Myelin Genes
To determine whether the depletion of chaperone proteins from the ER is unique to the mutant PLP1 protein or is a common phenomenon observed with mutant proteins encoded by other disease-causing genes, we also examined PMP22 and MPZ genes. Mutations in these genes cause a spectrum of autosomal dominant peripheral demyelinating neuropathies (30).
First, to determine whether accumulation of misfolded proteins in the ER is sufficient to reduce ER chaperone proteins to undetectable levels (by immunostaining) in HeLa cells, we examined two representative PMP22 mutants, Trembler-J (Tr-J) (an L16P substitution) and Trembler (Tr) (a G150D substitution), both of which accumulate in the ER by associating with CANX, however, do not induce UPR (31). Both of Tr-J and Tr are found in humans (32, 33) and mice (34, 35). First, we found no increase in the immunoreactivity of CHOP, which is one of the universal markers of ER stress (36), in either Tr or Tr-j, confirming that these mutants evoked no ER stress (Fig. 4B). We then performed immunocytochemistry with the anti-PDI (Fig. 4, A and C), anti-CALR, and anti-GRP78 (CALR, GRP78, data not shown) antibodies in HeLa cells transfected with the PMP22wt or mutant PMP22 genes. In contrast to our findings in cells transfected with the mutant PLP1 gene, we observed no depletion of these chaperones from the ER in cells expressing the mutant PMP22. These findings indicate that ER accumulation of these mutant proteins, which trigger no ER stress, is insufficient to induce depletion of ER chaperones from the ER.
FIGURE 4.
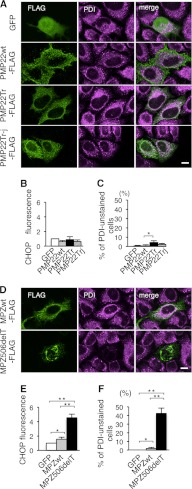
Effect of PMP22 and MPZ mutations on ER chaperones. A and D, immunocytochemistry of PDI in HeLa cells transfected with the PMP22wt and mutant PMP22 genes (A) or the MPZwt and MPZ506delT genes (D). HeLa cells transfected with the indicated vectors were immunostained with the anti-FLAG (green) and anti-PDI (magenta) antibodies followed by observation with a confocal fluorescence microscope. Note that cells expressing MPZ506delT showed a extremely faint staining pattern for PDI (arrowhead). Scale bar, 10 μm. B and E, relative expression of CHOP in HeLa cells transfected with the PMP22wt and mutant PMP22 genes (B) or the MPZwt and MPZ506delT genes (E). HeLa cells transfected with the indicated vectors were stained with the anti-FLAG and anti-CHOP antibodies together with DAPI to visualize the nuclei. The relative fluorescence intensity of CHOP in the nuclei was analyzed by densitometry. C and F, proportion of unstained cells with anti-PDI antibody in HeLa cells transfected with the PMP22wt and mutant PMP22 genes (C) or the MPZwt and MPZ506delT genes (F). Bar graphs are represented as fold-induction ± S.E. against the mean of control experiment from three independent experiments with >100 cells counted in each experiment (*, p ≤ 0.05; **, p ≤ 0.005).
Next, to analyze whether mutations in another gene that triggers ER stress also deplete the chaperones from the ER, we performed the same experiments using an MPZ gene harboring the 506delT mutation (MPZ506delT), which induces ER stress and causes a more severe form of peripheral neuropathy, Dejerine-Sottas neuropathy. This frameshift mutation results in 82 residues of shifted amino acid sequence starting from codon 169 in the MPZ protein (20). CHOP immunofluorescence was increased in the nucleus of HeLa cells transfected with the MPZ506delT gene, but not in cells transfected with the wild-type MPZ gene (MPZwt) (Fig. 4E), confirming that this mutant protein is an ER stressor. Immunocytochemistry (Fig. 4D) showed that the MPZ506delT mutant, but not MPZwt, increased the number of cells unstained with anti-PDI antibody (Fig. 4F). These results strongly suggested that depletion of the chaperones from the ER is not induced solely by protein accumulation in the ER, but instead requires both the accumulation of particular mutant proteins and ER stress. In addition, mutations in disease-causing genes other than PLP1 can elicit the chaperone depletion.
Depletion of PDI, CALR, and GRP78 Is Linked to PMD Severity
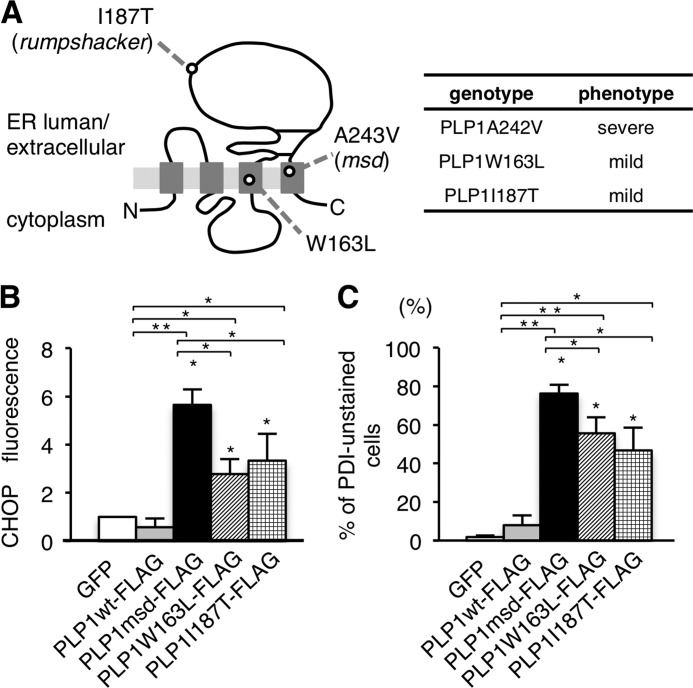
Because ER chaperones are connected with protein folding in the ER, we hypothesized that depletion of the ER chaperones may affect the pathogenesis or severity of PMD. We employed two PLP1 mutants, W163I (11) and I187T (18), both of which result in the mild end of the clinical spectrum of PMD (Fig. 5A). Densitometric analysis of CHOP immunofluorescence in transfected HeLa cells confirmed that these milder PLP1 mutants activated the UPR, but to a lesser extent than cells transfected with the PLP1msd gene (Fig. 5B).
FIGURE 5.
Effect of different PLP1 mutations on ER chaperones. A, schematic diagram of PLP1 with the positions of mutations examined in this study (left) and their associated phenotypes (right). B, expression of CHOP in HeLa cells transfected with the PLP1wt and mutant PLP1 genes. The fluorescence intensity of CHOP in the nuclei was analyzed by densitometry as described in the legend to Fig. 4B. C, proportion of unstained cells with anti-PDI antibody in HeLa cells transfected with PLP1wt and the indicated PLP1 mutant genes as described in the legend to Fig. 4C. Bar graphs are represented as fold-induction ± S.E. against the mean of control experiment from three independent experiments with >100 cells counted in each experiment (*, p ≤ 0.05; **, p ≤ 0.005).
Next, to analyze whether ER chaperone depletion is associated with clinical severity, we compared the expression of PDI in HeLa cells transfected with these PLP1 mutant genes. We found that the proportion of PDI-unstained cells expressing FLAG-tagged PLP1-W163L or PLP1-I187T was significantly lower than that of cells expressing PLP1msd (Fig. 5C). In addition, we observed a similar tendency in cells expressing the MPZ mutants. A mild MPZ allele, MPZS63del, evoked less ER stress and resulted in a smaller proportion of “PDI-unstained cells” than a severe allele, MPZ506delT (data not shown). Together, these results strongly suggest a potential linkage between chaperone depletion and the phenotypic variation in ER stress-related disorders.
Down-regulation of Pdi in the SC and Primary Culture of msd Mice
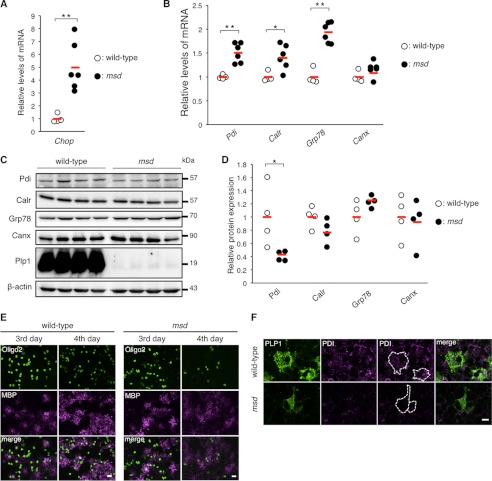
In our in vitro analyses, expression of the PDI, CALR, and GRP78 proteins did not increase in cells expressing PLP1msd, despite the significant increase in their transcripts (Fig. 2A). To determine whether this also occurs in vivo, we further investigated the mRNA and protein expression of these ER chaperones in the SCs isolated from male msd mice, which carry the Plp1A243V allele, on P14, when the Plp1 gene is most strongly expressed in the SCs of mutant mice (8). Expression of the Chop transcript was significantly higher in msd mice than in wild-type mice, suggesting that cells in the SCs of msd mice were under ER stress (Fig. 6A). We then analyzed the expression of these ER chaperone mRNA by quantitative RT-PCR (Fig. 6B). The expression of Grp78 mRNA was significantly increased (2-fold) in msd mice. Pdi and Calr were also up-regulated, but to a lesser extent. However, at protein levels, we observed discordance (Fig. 6, C and D). Pdi protein expression in msd mice was significantly decreased. The expression of Calr showed a similar tendency, although these results did not reach significance (p = 0.06). In contrast, Grp78 and Canx protein expression did not differ significantly when compared with wild-type mice.
FIGURE 6.
PDI is down-regulated in the SCs and primary oligodendrocytes of msd mice. A and B, quantitative analysis of Chop (A), Pdi, Calr, Grp78, and Canx genes (B) in the SCs of msd mice. Quantitative RT-PCR was performed to analyze the expression levels of Pdi, Calr, Grp78, and Canx in the SCs of wild-type (open circle, n = 4) and msd (filled circle, n = 6) mice at P14. GAPDH was used as an internal control. The results are represented as fold-induction against the means of wild-type mice. Red horizontal bars indicate the mean. C and D, relative amounts of ER chaperones in the SCs of msd mice. The SCs of P14 wild-type (open circle, n = 4) and msd (filled circle, n = 4) mice were subjected to immunoblotting with the indicated antibodies (C). The amounts of the proteins were measured by densitometry and normalized to β-actin (D). E, expression of MBP and Olig2 in primary oligodendrocytes from msd mice. Immunocytochemistry of MBP and Oligo2 in primary oligodendrocytes of msd mice. Primary mixed glial cultures were prepared from the forebrains of E14.5 wild-type or msd mice. On the 3rd and 4th days after induction of oligodendrocyte differentiation, the oligodendrocytes were immunostained with anti-MBP (magenta) and anti-Oligo2 (green) antibodies and observed with a confocal fluorescence microscope. Scale bar, 5 μm. F, immunocytochemistry of PDI in primary oligodendrocytes of msd mice. Primary oligodendrocytes prepared from the forebrains of wild-type or msd mice at E14.5 and immunostained with anti-PLP1 (green) and anti-PDI (magenta) antibodies and observed with a confocal microscope. Scale bar, 5 μm. A, B, and D, *, p ≤ 0.05; **, p ≤ 0.005.
We also immunocytochemically examined primary MGCs isolated from the brains of embryonic day (E) 14.5 wild-type or msd mice. On the 4th day after induction of oligodendrocyte differentiation, profound maturation with increased MBP immunoreactivity was evident in the wild-type MGCs (Fig. 6E). In contrast, rapid regression of Oligo2-positive cells and decreased MBP immunoreactivity were observed in the msd MGCs, suggesting that oligodendrocyte maturation induced apoptosis in msd on the 4th day after induction. Pdi was detected in the cell body of Plp1-positive mature oligodendrocytes in wild-type MGCs, whereas, it was only faintly stained in Plp1-positive oligodendrocytes in the msd MGCs. (Fig. 6F). These results suggest that endogenous PLP1msd depletes Pdi from the ER of the oligodendrocytes.
Inhibition of GA to ER Transport Is Associated with the Disappearance of PDI, CALR, and GRP78
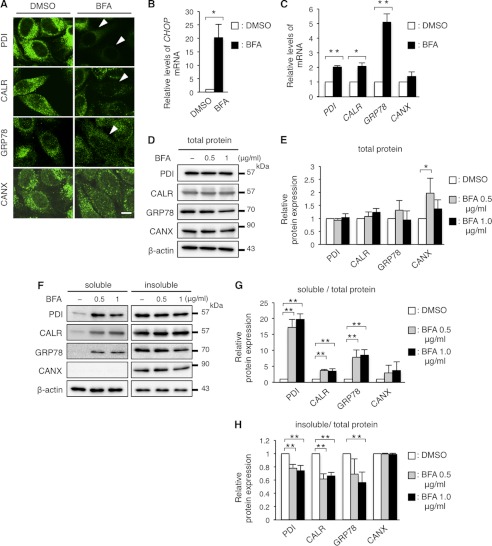
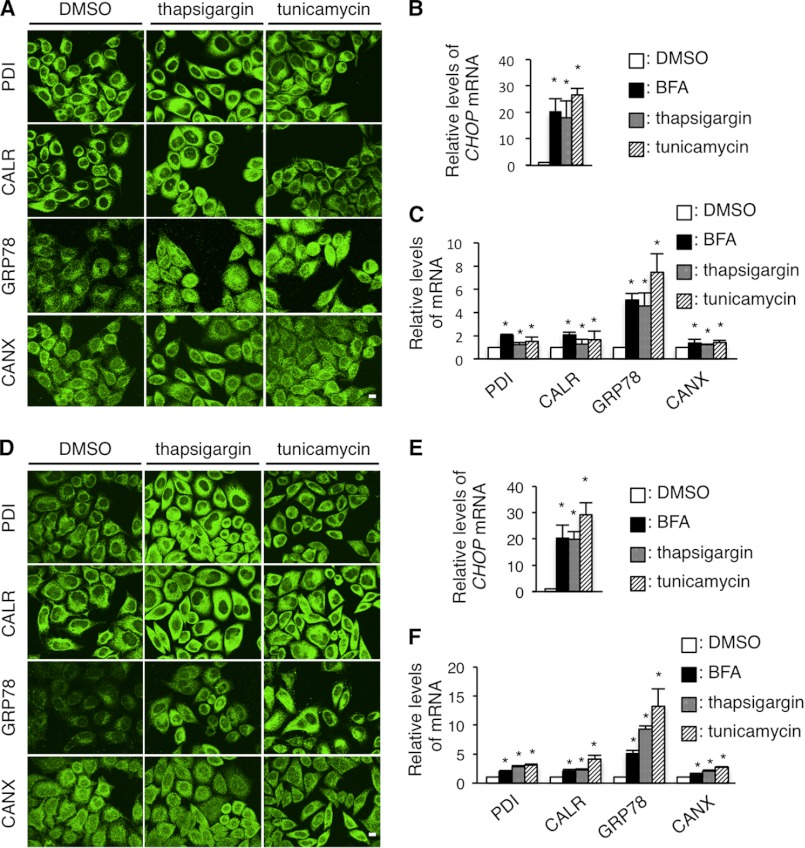
To determine the underlying mechanism for PDI, CALR, and GRP78 depletion from the ER in cells expressing PLP1 mutants, we further analyzed the effects of the following chemical ER stressors on the expression of ER chaperones: thapsigargin, a sarco/endoplasmic reticulum Ca2+-ATPase inhibitor; tunicamycin, an N-glycosylation inhibitor, and brefeldin A (BFA), a GA-ER transport inhibitor. These three compounds greatly up-regulated the transcripts of CHOP and GRP78, confirming that they work as ER stressors (Figs. 7, B and C, and 8, B, C, E, and F). These compounds also slightly but significantly increased the expression of PDI, CALR, and CANX transcripts.
FIGURE 7.
BFA treatment recapitulates the disappearance of PDI, CALR, and GRP78. A, immunocytochemistry of ER chaperones in HeLa cells treated with BFA. HeLa cells were treated with 1 μg/ml of BFA for 8 h were immunostained with the indicated antibodies and observed with a confocal fluorescence microscope. Scale bar, 10 μm. Note that cells treated with BFA showed extremely faint staining (arrowheads) for PDI, CALR, and GRP78. B and C, relative expression of the transcripts of the ER chaperones in HeLa cells treated with BFA. Expression levels of the transcripts of CHOP (B), PDI, CALR, GRP78, and CANX mRNA (C) in HeLa cells treated with BFA were analyzed by qRT-PCR and normalized to GAPDH. D and E, total amounts of PDI, CALR, GRP78, and CANX in HeLa cells treated with BFA. HeLa cells were treated with BFA as in A and subjected to immunoblotting with the indicated antibodies (D). The amounts of the proteins were measured by densitometry and normalized to β-actin (E). F-H, digitonin fractionation of HeLa cells treated with BFA. Digitonin fractionation was performed as described in the legend to Fig. 2D and the extracts were subjected to immunoblotting with the indicated antibodies (F) followed by quantitative analysis (G and H) as in Fig. 2, E and F. Results are represented as fold-induction compared with DMSO control experiment. Values are represented as the mean ± S.E. from three independent experiments (*, p ≤ 0.05, **, p ≤ 0.005).
FIGURE 8.
Thapsigargin and tunicamycin treatments do not cause the depletion of PDI, CALR, and GRP78. A and D, immunocytochemistry of ER chaperones in HeLa cells treated with thapsigargin and tunicamycin. HeLa cells were treated with 1 μm thapsigargin or 2 μm tunicamycin for 8 (A) or 24 h (D), immunostained with the indicated antibodies and observed with a confocal fluorescence microscope. Scale bar, 5 μm. B, C, E, and F, quantitative RT-PCR for CHOP (B and E) PDI, CALR, GRP78, and CANX (C and F) genes in HeLa cells treated with 1 μm thapsigargin or 2 μm tunicamycin for 8 h (B and C) or 24 h (E and F). The GAPDH gene was used as an internal control. Results are represented as fold-induction compared with DMSO control experiment. Values are represented as the mean ± S.E. from three independent experiments (*, p ≤ 0.05; **, p ≤ 0.005).
Next, HeLa cells were treated with these compounds for 8 h, followed by immunocytochemistry with anti-PDI, anti-CALR, anti-GRP78, or anti-CANX antibodies. Thapsigargin and tunicamycin did not alter the expression of the chaperones (Fig. 8A), even after an extended incubation (Fig. 8D). In contrast, BFA treatment clearly diminished PDI, CALR, and GRP78 from the ER; however, CANX expression remained unchanged (Fig. 7A). In BFA-treated HeLa cells, total amounts of the chaperone proteins (except for CANX at 0.5 μg/ml of BFA treatment) were mostly unaffected despite their increased transcripts (Fig. 7, D and E), as was observed in HeLa cells transfected with the PLP1msd gene (Fig. 2, B and C). We performed fractionation experiments with 0.01% digitonin as described in Fig. 2D. In the BFA-treated HeLa cells, the proportion of PDI, CALR, and GRP78 in the digitonin-soluble fraction containing the cytosol and plasma membrane was significantly higher, whereas the proportion of these proteins in the insoluble fraction containing ER proteins was lower than in untreated cells (Fig. 7, F–H); similar to that observed in cells expressing the mutant PLP1 (Fig. 2, D–F). This multitude of common evidence between BFA-treated and PLP1msd-transfected cells suggests that these may have a common mechanism underlying depletion of PDI, CALR, and GRP78.
The Fragmentation of GA in Cells Expressing PLP1msd and Phenotypically Milder PLP1 Mutants
As previously reported (37), BFA induces fragmentation of the GA (Fig. 9A). To further examine whether the mutant proteins affect the structure of the GA, we co-transfected GFP vector and the PLP1msd gene in HeLa cells and immunostained cells with antibodies against GM130, a GA marker, and PDI. We found that the GA was fragmented in GFP+ cells, which are expressing PLP1msd (Fig. 9B). These cells were not stained with anti-PDI antibody. This GA fragmentation was not observed in cells expressing PLP1wt. GM130 co-localized with the dense signal of PLP1wt at the perinuclear structure, presumably staining normal GA. We also found that expression of the MPZ506delT mutation also induced fragmentation of the GA in HeLa cells, whereas MPZwt, PMP22wt, and both Tr and Tr-j PMP22 mutants did not induce any morphological changes of the GA (Fig. 9C). These findings suggest that ER stressor proteins also induce GA fragmentation.
Next, to determine whether fragmentation of the GA is associated with the phenotypic variation in PMD patients, we evaluated the GA structure in HeLa cells transfected with the PLP1msd and two milder PLP1 mutants and classified the GA morphology into three categories, “normal,” “mild fragmentation,” and “severe fragmentation,” as previously reported elsewhere (38). Cells expressing each mild allele showed a higher proportion of “severe” GA fragmentation than those expressing PLP1wt; however, this proportion was lower than in cells expressing PLP1msd (Fig. 9D). These results suggest that GA fragmentation is involved in pathogenesis of disease-causing PLP1 mutations, and that the degree of GA fragmentation is associated with the severity of PMD, in conjunction with depletion of the ER chaperones.
Obstruction of Membrane and Secretory Protein Transport by PLP1msd
Newly synthesized membrane and secretory proteins are transported from the ER to the GA when they undergo post-translational modification (39). ER chaperone proteins are subsequently transported back to the ER from the cis-Golgi. In contrast, other membrane and secretory proteins that reach the trans-Golgi are sorted to carriers for further transport to various cellular destinations (40) (Fig. 10A). We hypothesized that PLP1msd interferes with the maturation of these proteins because it depletes the ER chaperones, which assist with protein folding in the ER, and induces morphological changes in the GA. To test this hypothesis, we created an expression vector encoding membrane-linked cell surface GFP, summarized in Fig. 9E. When HeLa cells were co-transfected with either PLP1wt-FLAG or PLP1msd-FLAG along with the cell surface GFP, we found that expression of the reporter protein was mislocalized in cells co-transfected with the PLP1msd gene (Fig. 9F). On the other hand, co-transfection with the PLP1wt gene showed GFP fluorescence in the cell surface. Such phenomenon was also observed in HeLa cells co-transfected with the MOG gene (41), which is an oligodendrocyte-specific membrane protein, fused with the GFP gene (MOG-GFP, summarized in Fig. 9, E and G). These findings suggested the possibility that PLP1msd impairs the transport of membrane proteins from the ER to the cell surface through the GA.
FIGURE 10.
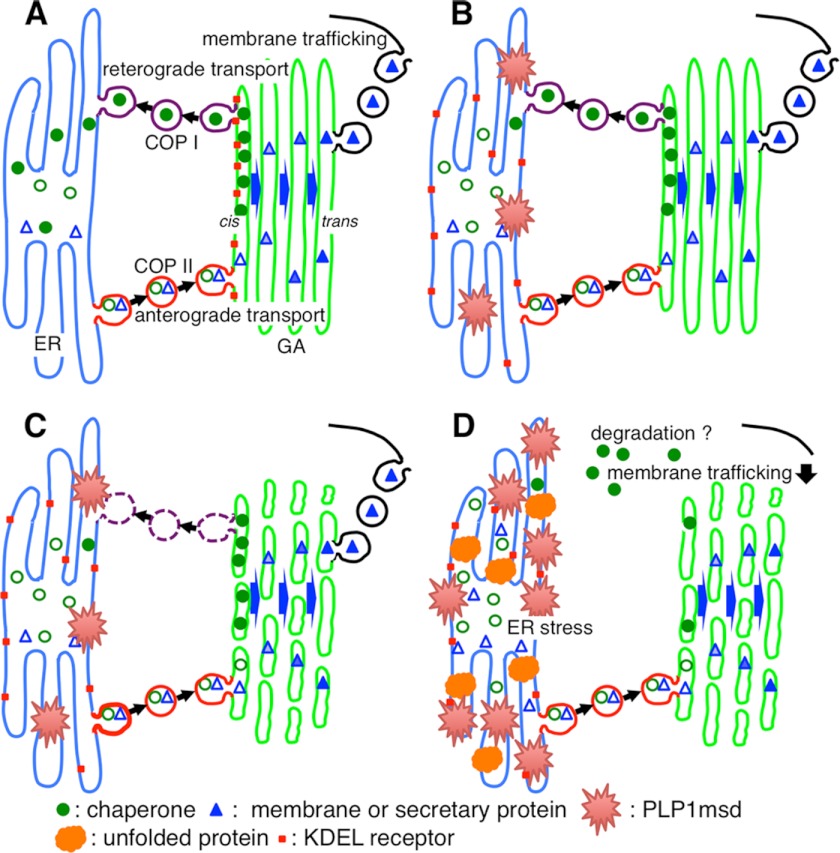
Scheme for the mechanism of ER chaperone depletion and GA fragmentation. A, under physiological conditions, efficient amounts of mature ER chaperones (filled circles) can fold unfolded proteins into their correct conformation. Immature ER chaperones with KDEL motifs (open circles), along with membrane and secretory proteins (triangles), are first transported from ER exit sites to the entry (cis) side of the GA to undergo post-translational modifications (coatomer protein (COPII) II-mediated anterograde transport). The ER chaperones are recycled back to the ER through the interaction with the KDEL receptor (red square) in the GA (coatomer protein I-mediated retrograde transport). In contrast, mature membrane and secretory proteins that reach to the trans-Golgi are further transported to various cellular destinations. B, ER stress-related mutant protein (red spines) induces dysfunction of retrograde transport from the GA to the ER by KDEL receptor mis-localized in the ER, resulting in a reduced supply of ER chaperones. C, this inhibition of retrograde transport, and probably the ER stress itself, may lead to the fragmentation of the GA. D, dysfunction of the ER to GA transport may jam membrane and secretory protein trafficking, leading to the further accumulation of misfolded proteins (orange). These changes in cellular homeostasis triggered by misfolded mutant proteins may further accelerate ER stress.
Furthermore, we analyzed whether intracellular transport of the secretory proteins was also affected by PLP1msd. We created a reporter secretory protein, in which Renilla luciferase (Rluc) was fused at the N terminus with the signal sequence of Igκ light chain (Igκ-Rluc). This fusion protein penetrates into the ER and is secreted to the extracellular space through the GA. HeLa cells were co-transfected with the regular firefly luciferase (cytoplasmic luciferase, Cluc) gene and the Igκ-Rluc gene along with an empty vector, PLP1wt-FLAG or PLP1msd-FLAG gene (Fig. 9H). We then simultaneously measured Cluc and Igκ-Rluc in cell lysate and culture supernatants with a luminometer. Total Rluc activity normalized to Cluc activity did not differ among cells expressing empty vector, PLP1wt, and PLP1msd, confirming a stable translation ratio between ER-mediated and non-ER-mediated processes (data not shown). Of note, we found that expression of the Igκ-Rluc protein in the culture supernatant normalized to intracellular Igκ-Rluc was significantly lower in cells transfected with the PLP1msd gene (Fig. 9I). Immunocytochemistry revealed that the Igκ-Rluc reporter protein is faintly localized in the GA in cells expressing PLP1wt; whereas, the same protein is clearly accumulated in the ER of cells expressing PLP1msd (data not shown). Together, these results suggest that PLP1msd induces obstruction of membrane and secretory protein transport.
PLP1msd Disturbs the Localization of KDEL Receptor in the GA
PDI, CALR, and GRP78 contain a carboxyl-terminal retrieval signal KDEL (Lys-Asp-Glu-Leu) motif (42). The KDEL motif is recognized by the KDEL receptor in the GA after releasing the chaperones from the ER, and then the chaperones are retrogradely transported back to the ER by the receptor in a coatomer protein I-dependent manner (43) (Fig. 10A). In contrast, CANX, which lacks a KDEL motif, was not depleted by PLP1msd transfection. These results promote us to investigate subcellular localization of the KDEL receptor in cells expressing PLP1msd. The KDEL receptor mainly localizes in the GA (44). Surprisingly, in the PLP1msd-transfected cells, the KDEL receptor was displayed as a mesh-like distribution through the cells and co-localized well with PLP1msd (Fig. 9J). In the cells expressing PLP1wt, we observed that the KDEL receptor co-localized with PLP1wt at the perinuclear structure, which was probably localized in the GA as shown in Fig. 9B. These results suggest that PLP1msd induced mislocalization of the KDEL receptor.
DISCUSSION
Involvement of ER stress and the subsequent UPR has been implicated in pathogenesis of multiple human inherited diseases, including cystic fibrosis (45), retinitis pigmentosa (46), CMT (22), and PMD (8, 11). Although there is wide phenotypic variation in each of these diseases, even among the mutations in same genes, little is known about the factors that determine the difference in ER stress and the severity of disease. In this study, we investigated the organelle changes in cells expressing different PLP1 missense mutations associated with a wide-range of clinical severities in PMD. We demonstrated that accumulation of the ER stress-associated mutant PLP1 leads to depletion of some important ER chaperones and GA fragmentation, both of which are more profound in cells expressing mutants associated with more severe phenotypes. We also found that an ER stress-related MPZ mutant also induces these cellular phenotypes; however, two PMP22 mutants, which cannot induce ER stress despite their ER retention, do not induce them. Based on these findings, we suggest that the cellular phenotypes of ER chaperone depletion and GA fragmentation may be involved in the pathogenesis of particular mutations in certain genes in ER stress-related diseases.
We observed that PDI, CALR, and GRP78 were depleted in the ER of HeLa cells transfected with the PLP1msd gene, whereas CANX remained unaffected. Similar phenomenon was also observed in endogenous Pdi and Calr in the SCs of msd mice (Fig. 6, C and D). By contrast, we could not find an obvious decrease of endogenous Grp78 in the mutant SCs, possibly due to the large enhancement of mRNA up-regulation. We considered the features that were either functionally or structurally common among the depleted chaperones. Functionally, each of these chaperones has a distinct role in protein folding and maintenance of ER homeostasis. For example, PDI catalyzes the formation and rearrangement of molecular disulfide bonds for protein folding (47). CALR and CANX are calcium-binding proteins implicated in the trimming of N-glycosylation and storage of calcium in the ER (48). GRP78 controls activation of the UPR, acting as a sensor for misfolded proteins in the ER (49). Based on this evidence, it is unlikely that the depletion is linked to a particular function of these chaperones.
Structurally, PDI, CALR, and GRP78 contain a carboxyl-terminal retrieval signal KDEL motif (42), which is recognized by the KDEL receptor to transport them back to the ER (43). This retrieval mechanism by the KDEL receptor contributes to quality control at the ER (50). We observed that the KDEL receptor was localized in the ER in cells expressing PLP1msd, whereas the same protein was localized in the GA in PLP1wt and control cells (Fig. 9J). Moreover, depletion of PDI, CALR, and GRP78 was also observed in HeLa cells treated with the chemical ER stressor, BFA, which inhibits retrograde transport from the GA to the ER (51, 52). Interestingly, the other chemical ER stressors tested, thapsigargin and tunicamycin, did not recapitulate the findings. These results suggest that misfolded mutant proteins may induce ER chaperone depletion by inhibition of their KDEL receptor-mediated retrograde transport of these chaperones by mis-localizing KDEL receptor.
Our results suggested that PLP1 and MPZ mutants, and possibly other mutant proteins that evoke ER stress, specifically deplete chaperones containing a KDEL motif from the ER. These proteins were unlikely degraded by the ERAD-proteasome system (Figs. 2, B and C, and 3, E and F). We further demonstrated that the proportion of these chaperone proteins in the digitonin-soluble fraction, which contains the plasma membrane and cytosolic proteins, increased in cells expressing PLP1msd (Fig. 2, D and E). However, we found no change in the amounts of these chaperone proteins on the cell surface (Fig. 3, B and D). These results suggest that KDEL-containing ER chaperones mainly translocate from the ER to the cytosol in cells expressing ER stress proteins. However, we could not rule out a possibility that small populations may translocate to the plasma membrane, as described previously (29).
In contrast, the GA fragmentation observed in cells treated with BFA was also observed in cells treated with thapsigargin (data not shown). GA fragmentation has been reported in another ER stress-related disorder, ALS (53). These findings suggest that GA fragmentation may be a common pathology in ER stress-related diseases.
An association between cellular pathology and clinical severity for PLP1 mutations has been reported. Gow and Lazzarini (10) reported a cellular mechanism that the amount of mutant PLP1 gene product accumulated in the ER accounts for disease severity in PMD. Recent studies showed that differences in the UPR (9) and ER quality control (11) have the potential to modulate disease severity. These reports suggest that retention of PLP1 mutants determines the severity of ER stress and clinical outcome. Consistent with these findings, the depletion of ER chaperones and GA fragmentation are closely linked to clinical severity (Figs. 5C and 9D), indicating that these cellular phenotypes are associated with disease pathology. In addition, we also demonstrated that PLP1msd not only induces ER stress, but also inhibits secretion and cell surface expression of proteins, probably due to impairment of ER chaperone transport from the GA to the ER and/or GA fragmentation. These trafficking defects may also contribute to the pathogenesis of disease by preventing cell-to-cell and cell-to-environment communications. Additional studies are required to elucidate how these mutants affect the maturation and trafficking of other membrane and secretory proteins.
Based on our findings, we propose a novel model for mechanisms to explain how mutant misfolded proteins affect intracellular homeostasis, as summarized in Fig. 10. When misfolded proteins accumulate in the ER, they inhibit GA to ER retrograde transport by KDEL receptor mis-localization in the ER (Fig. 10B). Ultimately, inhibition of retrograde transport results in depletion of KDEL-containing ER chaperones from the ER. Blockage of GA to ER retrograde transport also contributes to abnormal accumulation of ER chaperones in the cis-Golgi, which may, in part, contribute to GA fragmentation (Fig. 10C). As a consequence, the ER to GA transport of membrane/secretory proteins is disturbed, and misfolded proteins and other membrane/secretory proteins accumulate in the ER, resulting in a trafficking defect and further acceleration of ER stress (Fig. 10D). This unexpected discovery and new model for the disease mechanism may promote our understanding of how different mutations in the same gene differently evoke ER stress and affect disease phenotype. Our findings may have further implications for ER stress-related diseases in which the UPR modulates pathology. Because practically no effective treatment is available for these diseases, ER chaperone and GA may serve as potential targets for therapeutic intervention.
Acknowledgments
We thank Dr. W. B. Macklin (Cleveland Clinic Foundation) for providing msd mice, Dr. H. Osaka (Kanagawa Children's Medical Center) for the human PLP1 genes, Dr. J. R. Lupski (Baylor College of Medicine) for the human PMP22 genes and MPZ genes, Dr. J. Miyazaki (Osaka University) for pCAGGS, and Dr. M. Itoh (NCNP) for anti-PLP1 antibody, respectively.
This work was supported in part by grants from the Health and Labor Sciences Research Grants, Research on Intractable Diseases H24-Nanchitou-Ippan-072 (to K. I.), a grant from Takeda Science Foundation (to K. I.), and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, KAKENHI: 21390103 and 23659531 (to K. I.) and 23580417 (to T. M.).
- ER
- endoplasmic reticulum
- PLP1
- proteolipid protein 1
- PMD
- Pelizaeus-Merzbacher disease
- msd
- myelin synthesis deficit
- SPG2
- spastic paraplegia type 2
- msd
- myelin synthesis deficit
- GA
- Golgi apparatus
- PDI
- protein-disulfide isomerase
- CALR
- calreticulin
- GRP78
- glucose-regulated protein of 78 kDa
- CANX
- calnexin
- MBP
- myelin basic protein
- UPR
- unfolded protein response
- ATF6
- activating transcription factor 6
- IRE1
- inositol-requiring kinase 1
- XBP1
- X-box protein 1
- CHOP
- C/EBP homologous protein
- ALS
- amyotrophic lateral sclerosis
- MPZ
- myelin protein zero
- PMP22
- peripheral myelin protein 22
- CMT
- Charcot-Marie-Tooth disease
- SC
- spinal cords
- TUNEL
- terminal deoxynucleotidyl transferase dUTP nick end labeling
- MGC
- mixed glial culture
- BFA
- brefeldin A
- MOG
- myelin oligodendrocyte glycoprotein
- GFP
- green fluorescent protein
- luc
- luciferase
- Rluc
- Renilla luciferase
- Igκ
- immunoglobulin κ light chain
- Cluc
- cytoplasmic luciferase
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Kaufman R. J. (2002) Orchestrating the unfolded protein response in health and disease. J. Clin. Invest. 110, 1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lindholm D., Wootz H., Korhonen L. (2006) ER stress and neurodegenerative diseases. Cell Death Differ. 13, 385–392 [DOI] [PubMed] [Google Scholar]
- 3. Szegezdi E., Logue S. E., Gorman A. M., Samali A. (2006) Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7, 880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. (2007) Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev. Cell 13, 365–376 [DOI] [PubMed] [Google Scholar]
- 5. Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 [DOI] [PubMed] [Google Scholar]
- 6. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 7. Inoue K. (2005) PLP1-related inherited dysmyelinating disorders. Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics 6, 1–16 [DOI] [PubMed] [Google Scholar]
- 8. Southwood C. M., Garbern J., Jiang W., Gow A. (2002) The unfolded protein response modulates disease severity in Pelizaeus-Merzbacher disease. Neuron 36, 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gow A., Southwood C. M., Lazzarini R. A. (1998) Disrupted proteolipid protein trafficking results in oligodendrocyte apoptosis in an animal model of Pelizaeus-Merzbacher disease. J. Cell Biol. 140, 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gow A., Lazzarini R. A. (1996) A cellular mechanism governing the severity of Pelizaeus-Merzbacher disease. Nat. Genet. 13, 422–428 [DOI] [PubMed] [Google Scholar]
- 11. Roboti P., Swanton E., High S. (2009) Differences in endoplasmic-reticulum quality control determine the cellular response to disease-associated mutants of proteolipid protein. J. Cell Sci. 122, 3942–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swanton E., High S., Woodman P. (2003) Role of calnexin in the glycan-independent quality control of proteolipid protein. EMBO J. 22, 2948–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bernard-Marissal N., Moumen A., Sunyach C., Pellegrino C., Dudley K., Henderson C. E., Raoul C., Pettmann B. (2012) Reduced calreticulin levels link endoplasmic reticulum stress and Fas-triggered cell death in motoneurons vulnerable to ALS. J. Neurosci. 32, 4901–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jaronen M., Vehvilainen P., Malm T., Keksa-Goldsteine V., Pollari E., Valonen P., Koistinaho J., Goldsteins G. (2012) Protein disulfide isomerase in ALS mouse glia links protein misfolding with NADPH oxidase-catalyzed superoxide production. Hum. Mol. Genet. 22, 646–655 [DOI] [PubMed] [Google Scholar]
- 15. Gencic S., Hudson L. D. (1990) Conservative amino acid substitution in the myelin proteolipid protein of jimpy(msd) mice. J. Neurosci. 10, 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamamoto T., Nanba E., Zhang H., Sasaki M., Komaki H., Takeshita K. (1998) Jimpy(msd) mouse mutation and connatal Pelizaeus-Merzbacher disease. Am. J. Med. Genet. 75, 439–440 [PubMed] [Google Scholar]
- 17. Koizume S., Takizawa S., Fujita K., Aida N., Yamashita S., Miyagi Y., Osaka H. (2006) Aberrant trafficking of a proteolipid protein in a mild Pelizaeus-Merzbacher disease. Neuroscience 141, 1861–1869 [DOI] [PubMed] [Google Scholar]
- 18. Kobayashi H., Hoffman E. P., Marks H. G. (1994) The rumpshaker mutation in spastic paraplegia. Nat. Genet. 7, 351–352 [DOI] [PubMed] [Google Scholar]
- 19. Schneider A., Montague P., Griffiths I., Fanarraga M., Kennedy P., Brophy P., Nave K. A. (1992) Uncoupling of hypomyelination and glial cell death by a mutation in the proteolipid protein gene. Nature 358, 758–761 [DOI] [PubMed] [Google Scholar]
- 20. Khajavi M., Inoue K., Wiszniewski W., Ohyama T., Snipes G. J., Lupski J. R. (2005) Curcumin treatment abrogates endoplasmic reticulum retention and aggregation-induced apoptosis associated with neuropathy-causing myelin protein zero-truncating mutants. Am. J. Hum. Genet. 77, 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. D'Antonio M., Feltri M. L., Wrabetz L. (2009) Myelin under stress. J. Neurosci. Res. 87, 3241–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gow A., Sharma R. (2003) The unfolded protein response in protein aggregating diseases. Neuromolecular Med. 4, 73–94 [DOI] [PubMed] [Google Scholar]
- 23. Warner L. E., Garcia C. A., Lupski J. R. (1999) Hereditary peripheral neuropathies. Clinical forms, genetics, and molecular mechanisms. Annu. Rev. Med. 50, 263–275 [DOI] [PubMed] [Google Scholar]
- 24. Yu L. H., Morimura T., Numata Y., Yamamoto R., Inoue N., Antalfy B., Goto Y., Deguchi K., Osaka H., Inoue K. (2012) Effect of curcumin in a mouse model of Pelizaeus-Merzbacher disease. Mol. Genet. Metab. 106, 108–114 [DOI] [PubMed] [Google Scholar]
- 25. Morimura T., Ogawa M. (2009) Relative importance of the tyrosine phosphorylation sites of Disabled-1 to the transmission of Reelin signaling. Brain Res. 1304, 26–37 [DOI] [PubMed] [Google Scholar]
- 26. Abematsu M., Kagawa T., Fukuda S., Inoue T., Takebayashi H., Komiya S., Taga T. (2006) Basic fibroblast growth factor endows dorsal telencephalic neural progenitors with the ability to differentiate into oligodendrocytes but not γ-aminobutyric acidergic neurons. J. Neurosci. Res. 83, 731–743 [DOI] [PubMed] [Google Scholar]
- 27. Morimura T., Hattori M., Ogawa M., Mikoshiba K. (2005) Disabled1 regulates the intracellular trafficking of reelin receptors. J. Biol. Chem. 280, 16901–16908 [DOI] [PubMed] [Google Scholar]
- 28. Kaufman R. J. (1999) Stress signaling from the lumen of the endoplasmic reticulum. Coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211–1233 [DOI] [PubMed] [Google Scholar]
- 29. Zhang Y., Liu R., Ni M., Gill P., Lee A. S. (2010) Cell surface relocalization of the endoplasmic reticulum chaperone and unfolded protein response regulator GRP78/BiP. J. Biol. Chem. 285, 15065–15075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suter U., Scherer S. S. (2003) Disease mechanisms in inherited neuropathies. Nat. Rev. Neurosci. 4, 714–726 [DOI] [PubMed] [Google Scholar]
- 31. Dickson K. M., Bergeron J. J., Shames I., Colby J., Nguyen D. T., Chevet E., Thomas D. Y., Snipes G. J. (2002) Association of calnexin with mutant peripheral myelin protein-22 ex vivo. A basis for “gain-of-function” ER diseases. Proc. Natl. Acad. Sci. U.S.A. 99, 9852–9857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Valentijn L. J., Baas F., Wolterman R. A., Hoogendijk J. E., van den Bosch N. H., Zorn I., Gabreëls-Festen A. W., de Visser M., Bolhuis P. A. (1992) Identical point mutations of PMP-22 in Trembler-J mouse and Charcot-Marie-Tooth disease type 1A. Nat. Genet. 2, 288–291 [DOI] [PubMed] [Google Scholar]
- 33. Ionasescu V. V., Searby C. C., Ionasescu R., Chatkupt S., Patel N., Koenigsberger R. (1997) Dejerine-Sottas neuropathy in mother and son with same point mutation of PMP22 gene. Muscle Nerve 20, 97–99 [DOI] [PubMed] [Google Scholar]
- 34. Suter U., Moskow J. J., Welcher A. A., Snipes G. J., Kosaras B., Sidman R. L., Buchberg A. M., Shooter E. M. (1992) A leucine-to-proline mutation in the putative first transmembrane domain of the 22-kDa peripheral myelin protein in the trembler-J mouse. Proc. Natl. Acad. Sci. U.S.A. 89, 4382–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suter U., Welcher A. A., Ozcelik T., Snipes G. J., Kosaras B., Francke U., Billings-Gagliardi S., Sidman R. L., Shooter E. M. (1992) Trembler mouse carries a point mutation in a myelin gene. Nature 356, 241–244 [DOI] [PubMed] [Google Scholar]
- 36. Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12, 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sütterlin C., Hsu P., Mallabiabarrena A., Malhotra V. (2002) Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell 109, 359–369 [DOI] [PubMed] [Google Scholar]
- 38. Lane J. D., Lucocq J., Pryde J., Barr F. A., Woodman P. G., Allan V. J., Lowe M. (2002) Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J. Cell Biol. 156, 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee M. C., Miller E. A., Goldberg J., Orci L., Schekman R. (2004) Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20, 87–123 [DOI] [PubMed] [Google Scholar]
- 40. Marie M., Sannerud R., Avsnes Dale H., Saraste J. (2008) Take the “A” train. On fast tracks to the cell surface. Cell. Mol. Life Sci. 65, 2859–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Quarles R. H. (2002) Myelin sheaths. Glycoproteins involved in their formation, maintenance and degeneration. Cell. Mol. Life Sci. 59, 1851–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Munro S., Pelham H. R. (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907 [DOI] [PubMed] [Google Scholar]
- 43. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins. Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 44. Griffiths G., Ericsson M., Krijnse-Locker J., Nilsson T., Goud B., Söling H. D., Tang B. L., Wong S. H., Hong W. (1994) Localization of the Lys, Asp, Glu, Leu tetrapeptide receptor to the Golgi complex and the intermediate compartment in mammalian cells. J. Cell Biol. 127, 1557–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ward C. L., Omura S., Kopito R. R. (1995) Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127 [DOI] [PubMed] [Google Scholar]
- 46. Shinde V. M., Sizova O. S., Lin J. H., LaVail M. M., Gorbatyuk M. S. (2012) ER stress in retinal degeneration in S334ter Rho rats. PloS One 7, e33266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Higa A., Chevet E. (2012) Redox signaling loops in the unfolded protein response. Cell. Signal. 24, 1548–1555 [DOI] [PubMed] [Google Scholar]
- 48. Ellgaard L., Frickel E. M. (2003) Calnexin, calreticulin, and ERp57. Teammates in glycoprotein folding. Cell Biochem. Biophys. 39, 223–247 [DOI] [PubMed] [Google Scholar]
- 49. Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. (2000) Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. cell Biol. 2, 326–332 [DOI] [PubMed] [Google Scholar]
- 50. Yamamoto K., Fujii R., Toyofuku Y., Saito T., Koseki H., Hsu V. W., Aoe T. (2001) The KDEL receptor mediates a retrieval mechanism that contributes to quality control at the endoplasmic reticulum. EMBO J. 20, 3082–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Anders N., Jürgens G. (2008) Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell Mol. Life Sci. 65, 3433–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Citterio C., Vichi A., Pacheco-Rodriguez G., Aponte A. M., Moss J., Vaughan M. (2008) Unfolded protein response and cell death after depletion of brefeldin A-inhibited guanine nucleotide-exchange protein GBF1. Proc. Natl. Acad. Sci. U.S.A. 105, 2877–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakagomi S., Barsoum M. J., Bossy-Wetzel E., Sütterlin C., Malhotra V., Lipton S. A. (2008) A Golgi fragmentation pathway in neurodegeneration. Neurobiol. Dis. 29, 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]