Background: Brassinosteroids (BRs) are plant steroids that signal through the inhibition of GSK3/Shaggy-like kinases such as BIN2.
Results: We show here that BIN2 phosphorylates MKK4, which inhibits its activity against MPK6, in a MAPK module that controls stomata patterning.
Conclusion: BRs control cellular patterning via BIN2-mediated suppression of MKK4 activity.
Significance: Novel cross-talk of GSK3 and MAPK signaling is revealed.
Keywords: Glycogen Synthase Kinase 3, MAP Kinases (MAPKs), Phosphorylation, Signaling, Steroid Hormone, Brassinosteroids, Patterning
Abstract
Brassinosteroids (BRs) are steroid hormones that coordinate fundamental developmental programs in plants. In this study we show that in addition to the well established roles of BRs in regulating cell elongation and cell division events, BRs also govern cell fate decisions during stomata development in Arabidopsis thaliana. In wild-type A. thaliana, stomatal distribution follows the one-cell spacing rule; that is, adjacent stomata are spaced by at least one intervening pavement cell. This rule is interrupted in BR-deficient and BR signaling-deficient A. thaliana mutants, resulting in clustered stomata. We demonstrate that BIN2 and its homologues, GSK3/Shaggy-like kinases involved in BR signaling, can phosphorylate the MAPK kinases MKK4 and MKK5, which are members of the MAPK module YODA-MKK4/5-MPK3/6 that controls stomata development and patterning. BIN2 phosphorylates a GSK3/Shaggy-like kinase recognition motif in MKK4, which reduces MKK4 activity against its substrate MPK6 in vitro. In vivo we show that MKK4 and MKK5 act downstream of BR signaling because their overexpression rescued stomata patterning defects in BR-deficient plants. A model is proposed in which GSK3-mediated phosphorylation of MKK4 and MKK5 enables for a dynamic integration of endogenous or environmental cues signaled by BRs into cell fate decisions governed by the YODA-MKK4/5-MPK3/6 module.
Introduction
One of the major challenges in cell and developmental biology is to understand how cells integrate multiple signaling pathways to regulate specific cell differentiation events. Intercellular communication is essential in the coordination of cell fate specification and patterning, and especially in sessile organisms such as plants, cell fate decisions are made to integrate environmental signals with internal developmental programs (1–3).
Plant developmental plasticity is governed by plant hormones, which also play decisive roles in the regulation of cellular patterning. One class of plant hormones that are considered necessary for the regulation of adaptive growth and development are the brassinosteroids (BRs).2 BRs are polyhydroxylated steroid hormones similar in their structure to mammalian steroid hormones and ecdysteroid from insects (4). BRs are synthesized from sterols, and in Arabidopsis thaliana, signal through a phosphorylation-dependent signal transduction cascade that is initiated by perception of the biologically active BRs brassinolide or castasterone by a cell surface receptor complex containing the receptor-like kinase BRI1 (brassinosteroid-insensitive 1). In response to BR binding, BRI1 autophosphorylates, interacts with BAK1 (BRI1-associated receptor kinase 1), and facilitates an interaction of BSK1 (BR signaling kinase 1), a receptor-like cytoplasmic kinase with BSU1 (BRI1 suppressor 1), a serine/threonine phosphatase. BSU1 mediates dephosphorylation and thereby inactivation of the GSK3 (glycogen synthase kinase 3)/Shaggy-like kinase BIN2 (brassinosteroid-insensitive 2) and its redundantly acting homologues such as ASKθ (A. thaliana Shaggy-like kinase θ) that act to regulate transcription factors of the BES1/BZR1 family and likely also of a set of basic helix loop helix transcription factors including CESTA and BEE1/BEE3 (5) (brassinosteroid enhanced expression 1/3), which in turn control the expression of BR target genes. Thus, at present it is thought that BRs confer their effect through the inhibition of GSK3/Shaggy-like kinases, which phosphorylate transcription factors to alter their activity on BR-responsive promoters (6, 7).
BRs play well defined roles in cell division and cell elongation (8, 9). In addition it is becoming increasingly evident that BRs also control the balance between proliferation and cell fate specification; however, these roles are not well established and the underlying molecular mechanisms have remained ill defined. The observation that inhibition of BR biosynthesis prevented differentiation of mesophyll cells into tracheary elements (10, 11), whereas application of brassinolide had a promotive effect (12), provided first evidence for a role of BRs in xylem differentiation. Recently, BRs were shown to participate in controlling root meristem size (13, 14) and to impact on the specification of cell fate and patterning in the root epidermis (15). In the leaf epidermis stomatal distribution follows the one-cell spacing rule; that is, adjacent stomata are spaced by at least one intervening pavement cell (16, 17). Interestingly, this rule is disrupted in the BR biosynthesis mutant cpd (constitutive photomorphogenesis and dwarfism) where stomatal duplications were reported (18), indicating that BRs may also impact on cellular patterning in stomata development.
Correct stomata patterning in A. thaliana requires the YODA-MKK4/5-MPK3/6 MAP kinase cascade (19–21). In this cascade YODA (YDA), a mitogen-activated protein (MAP) kinase kinase kinase (MAPKKK), phosphorylates the MAPKKs MKK4/MKK5, which in turn control the activity of the MAP kinases MPK3/MPK6 (19–22). Three basic helix loop helix transcription factors, SPCH (SPEECHLESS), MUTE, and FAMA, act directly downstream of MAP kinases to regulate specific events in the entry into and progression through the stomata development pathway (20, 22). MAPK signaling networks are found in all eukaryotic organisms and regulate fundamental aspects of biology including but not limited to cell division, initiation of developmental pathways, and responses to abiotic and biotic stresses (23–25). Via a phosphorelay mechanism these cascades, minimally composed of a MAPKKK, a MAPKK, and a MAPK, link upstream receptors to downstream targets (23). MAPK pathways usually signal multiple stimuli, and also the YDA-MKK4/5-MPK3/6 module not only participates in cellular patterning but also transmits abiotic and biotic stress signals perceived by the receptor kinase flagellin sensing 2 (FLS2) (24).
In the present study, we demonstrate that BR signaling governs cell fate specification in A. thaliana stomata patterning. Our results show that BIN2 phosphorylates MKK4 in its activation loop and that BIN2-mediated phosphorylation severely reduces MKK4 activity against MPK6 in vitro. An overexpression of MKK4 and its homologue MKK5 rescued stomata patterning defects of BR-deficient plants, providing in planta evidence for a function of BIN2 upstream of MKK4/5 in stomata development. A model in which BR signaling impacts on stomata patterning by suppressing BIN2-mediated phosphorylation of MKK4/5 at Thr-234 and thus activating the YDA-MKK4/5-MPK3/6 module is presented and discussed.
EXPERIMENTAL PROCEDURES
Plant Growth Conditions and Treatments
A. thaliana ecotype Columbia-0 (Col-0) was the wild type used for generation of all plant material described in this study. The seeds were sterilized using the chlorine vapor method (26) and plated on A. thaliana salts medium (27). After stratification at 4 °C for 2 days, seeds were placed in a growth chamber at 21 ± 2 °C and incubated in long day growth conditions (16 h, 80 μmol × m−2 s−1 cool white light/8-h dark). For analyzing stomata patterning, seeds of Col-0 and other lines were grown vertically on A. thaliana salts medium without or with 1 μm brassinazole (Brz) (TCI Europe, Eschborn, Germany) for 10 days.
Molecular Cloning and Generation of Transgenic Plant Lines
For generation of plants overexpressing MKK4-YFP or MKK5-YFP, the coding sequences of the genes were amplified with primers and inserted as NcoI+NotI fragments into the corresponding cloning sites of the pGreen derivative 0029 (28) downstream of the constitutive Cauliflower Mosaic Virus 35S promoter. A YFP (yellow fluorescent protein) tag was subsequently inserted in-frame as a NotI+NotI fragment. For the 35S:MKK2-MYC construct, the coding region of MKK2 was PCR-amplified, and was introduced as ApaI+NotI fragment into pGreen0029. The Myc tag was cloned into the NotI restriction site. For the 35S:MKK4T234A-YFP construct, a T234A point mutation was introduced into MKK4 by site-directed mutagenesis and was cloned as NcoI+NotI fragment into the binary plant expression vector pGWR8 (29) downstream of the constitutive cauliflower mosaic virus 35S promoter. The YFP tag was inserted as a NotI+NotI fragment. The integrity of the constructs was verified by sequencing. The resulting plasmids were transformed into Agrobacterium tumefaciens GV3101 carrying the Ti helper plasmid pSOUP. Transgenic plants were generated using the floral dip method (26). Homozygous lines expressing the 35S:MKK4-YFP, the 35S:MKK5-YFP, or the 35S:MKK2-MYC construct to high levels where used in Brz response experiments as described.
To analyze MKK4-YFP localization under its own promoter, the promoter and coding sequence of MKK4 were PCR-amplified and cloned as XhoI+BamHI fragment into the vector described above, to replace the 35S promoter, yielding pMKK4:MKK4-YFP. For cloning of 35S:BIN2-YFP, the coding sequence of BIN2 was amplified by PCR, cloned as a NcoI+NotI fragment into pGWR8, and tagged C-terminally with YFP. For ASKθ and ASKβ, published 35S promoter-driven constructs were used (29, 30), and a YFP or cYFP tag was inserted using the NotI sites.
For protein expression, the coding regions of MKK2 (32), MKK4, MKK5, MKK7, or MKK4T234A were cloned in-frame with the GST coding sequence into pGEX-4T2 (GE Healthcare, Buckinghamshire, UK). To obtain a His6-Myc-tagged version of MKK4, the coding region was cloned as a NcoI+NotI fragment into the pMAL vector (New England Biolabs, Frankfurt am Main, Germany), in which a Myc tag was introduced with specific primers in-frame in front of the His6 tag. The S6A, S230A, S231A, T234D, and T234E point mutations were introduced into the pGEX-MKK4 construct by site-directed mutagenesis. Similarly, the K69R loss of function mutation of BIN2 (BIN2lof) was introduced into pGEX5X3-BIN2 (33) by in vitro mutagenesis. The coding sequence of MPK6 was amplified from cDNA by using gene-specific primers and cloned as a NcoI+NotI fragment into the Gateway-intermediate vector pENTRTM (Invitrogen, Darmstadt, Germany). The entry clone was then introduced into the pDESTTM17 (Invitrogen) vector by site-specific recombination.
Recombinant Protein Purification from Escherichia coli
GST- and His6-tagged proteins were expressed in E. coli BL21 and purified using glutathione-Sepharose beads (GE Healthcare) or nickel-cellulose beads (Carl Roth, Karlsruhe, Germany) as recommended by the suppliers.
Western Blotting
Western blot analysis was performed as described previously (5). Membranes were probed with either a mouse anti-GFP antibody (Roche Diagnostics) or a mouse anti-c-Myc antibody (Santa Cruz Biotechnology). Alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma, Steinheim, Germany) was used as secondary antibody and detected by enhanced chemiluminescence using CDP-Star reagent (Amersham Biosciences).
Microscopy
For investigation of stomata clustering, leaves of 10-day-old seedlings were fixed on a metal support rack with Tissue-Tek (Sakura Finetek, Torrance, CA) and shock-frozen in liquid nitrogen, and leaf epidermal cells were subsequently visualized with a Hitachi T-1000 scanning electron microscope (Hitachi High-Tech, Tokyo, Japan). Alternatively, leaves were incubated in clearing solution (20 g of chloral hydrate, 4.6 ml of water, 2 ml of glycerol, 87%) at room temperature prior investigation by optical bright field microscopy. Leaves of stably transformed A. thaliana seedlings expressing the 35S:MKK4-YFP construct were investigated with a Zeiss LSM Meta confocal microscope for YFP reporter expression. The images were assembled using the Zeiss LSM image browser software version 4.2.0.121.
For the analysis of YFP reporter constructs, plasmid DNA of pMKK4:MKK4-YFP, 35S:MKK4-YFP, 35S:BIN2-YFP, 35S:ASKθ-YFP, or 35S:ASKα-YFP were transiently transformed into A. thaliana protoplasts using a PEG-mediated transformation protocol (34). Fluorescence was investigated using an Olympus Bx61 fluorescence microscope.
For split YFP analysis, cDNAs of MKK4 and ASKθ were cloned in pGWR8 downstream of the 35S promoter and subsequently tagged with the N-terminal or C-terminal part of YFP (35). The sequenced constructs were used for transient transfection of A. thaliana protoplasts (34), and bimolecular fluorescence was investigated using an Olympus Bx61 fluorescence microscope.
GST Pulldown and in Vitro Kinase Assays
Two μg of GST-BIN2 and MKK4-His6-Myc proteins were incubated in 500 μl of 1× PBS buffer for 1 h on ice. Then 20 μl of glutathione-Sepharose beads (GE Healthcare) were added and incubated further for 2 h at 4 °C on a rotating wheel. After centrifugation at 1000 rpm for 1 min, the beads were washed with PBS containing 0.05% Tween 20. 4× SDS loading buffer (200 mm Tris/HCl, pH 6.8, 400 mm DTT, 8% SDS, 40% glycerol, and 0.01% bromphenol blue) was added to a final concentration of 1×, and Western blotting was performed using anti-Myc tag antibody (Santa Cruz Biotechnology). The in vitro kinase assays were performed as described previously (29). Briefly, 0.5 μg (unless otherwise stated) of purified recombinant proteins was mixed together and the reaction was started by the addition of reaction mixture containing the kinase buffer (20 mm HEPES, pH = 7.4; 15 mm MgCl2; 5 mm EGTA; 1 mm DTT), 1 μm ATP, and 10 μCi of [γ-32P]ATP (unless otherwise stated). The reaction was performed in a total volume of 20 μl at room temperature for 30 min. The reaction was stopped by adding 6 μl of 4× SDS loading buffer (67 mm Tris, pH 6.8, 133 mm DTT, 2.7% SDS, 13% glycerol, 0.01% bromphenol blue). Kinase activities were analyzed by SDS-PAGE followed by autoradiography.
Mass Spectrometry
After Coomassie Brilliant Blue staining of the SDS-PAGE gel, protein bands corresponding to GST-BIN2 and GST-MKK4 were cut in 1–2-mm3 pieces and subsequently washed in consecutive baths of acetonitrile, ultrapure water, and again in acetonitrile to remove SDS-PAGE buffer and Coomassie Brilliant Blue dye, and the steps proceeded further according to the protocol given in detail in the supplemental data.
RESULTS
BR Signaling Deficiency Induces Stomata Patterning Defects in A. thaliana
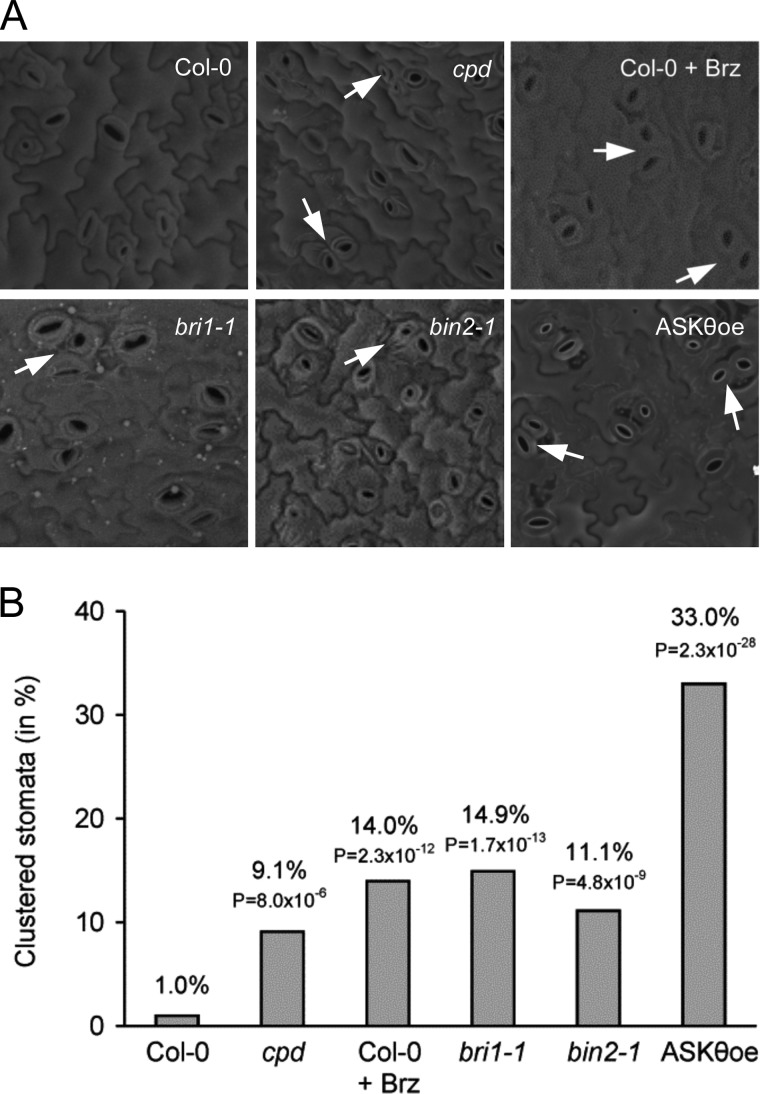
To investigate whether BRs impact on cell patterning during stomata differentiation, we analyzed the abaxial epidermis of the first leaf pair of BR-deficient and BR signaling-deficient mutants by scanning electron microscopy. First we examined cpd, a BR biosynthetic mutant, which has previously been reported to display duplicated guard cells (18). Fig. 1 shows that although wild-type A. thaliana seedlings exhibited the characteristic one-cell spacing patterning in stomatal distribution (1, 36), stomata in cpd clustered at a strongly increased rate (of 9.1%). To verify these results, wild-type seedlings were grown for 10 days on medium containing Brz, an inhibitor of BR biosynthesis (37). As shown in Fig. 1, Brz treatment had the same effects on stomata distribution. It induced stomatal patterning defects; stomata clustering occurred at a rate of 14.0%. To investigate whether this was a phenotype induced by BR deficiency or whether it was due to defective BR signaling, we analyzed the BR signaling-deficient mutants bri1-1 (38) and bin2-1 (39), as well as plants overexpressing ASKθ, an A. thaliana GSK3/Shaggy-like kinase that acts redundantly with BIN2 in BR signaling (29). Interestingly, in plants, in which BR downstream responses are constitutively impaired, a high frequency of clustered stomata (bri1-1, 14.9%; bin2-1, 11.1%; ASKθoe, 33%) was observed. These increases were statistically highly significant as compared with wild type, as indicated by the low p values derived from a two-tailed χ2-test (Fig. 1B). Thus, in summary, BR deficiency and more specifically defects in BR signaling disrupt the coordinated cell fate determination of stomata versus pavement cells.
FIGURE 1.
BR deficiency and BR signaling deficiency induces stomata patterning defects in A. thaliana. A, seedlings of wild-type Col-0, cpd, bri1-1, bin2-1, and ASKθoe were grown vertically on A. thaliana salts plates for 10 days, and stomatal distribution was analyzed by scanning electron microscopy (SEM). For Brz treatment, 1μm Brz was directly added to A. thaliana salts medium. B, the frequency of clustering events was calculated by counting at least 100 stomata per line or treatment. All lines showed statistically highly significantly increased stomata clustering as compared with untreated wild type as indicated by the low p values of a χ2-test.
BIN2 and Its Homologues Can Phosphorylate MKK4 in Vitro
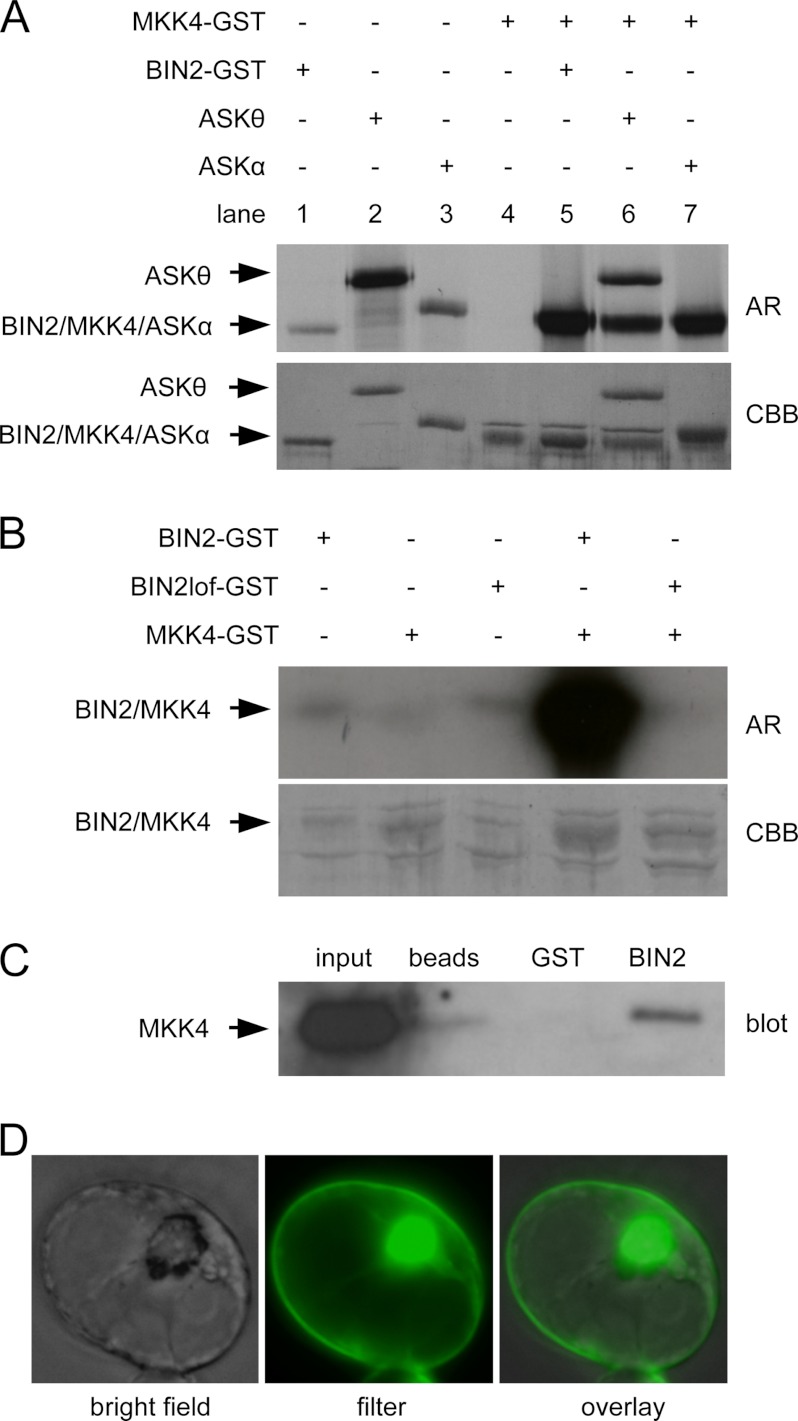
The stomata patterning defects of bin2-1 and ASKθoe plants suggested that a cross-talk of BR signaling with factors controlling stomata cell fate specification occurred either at the level or downstream of BIN2 and redundantly acting GSK3/Shaggy-like kinases. In a candidate gene approach, we thus tested whether GSK3/Shaggy-like kinases may directly phosphorylate players of the YDA-MKK4/5-MPK3/6 module. In vitro kinase assays were performed using recombinant MPK3, MPK6, and MKK4 and the GSK3/Shaggy-like kinases BIN2, ASKθ, and ASKα. Although MPK3 and MPK6 could not be phosphorylated (data not shown and see Fig. 4, lane 1), GST-BIN2 and its two homologues were able to efficiently phosphorylate GST-MKK4 (Fig. 2A, lanes 5–7). To further verify this result, a BIN2 loss-of-function (GST-BIN2lof) protein, generated by a point mutation of lysine 69 to arginine, was tested for its ability to phosphorylate MKK4. Fig. 2B illustrates the results of these kinase assays and shows that although wild-type BIN2 exhibited strong phosphorylation activity against MKK4, the BIN2lof protein was unable to catalyze MKK4 phosphorylation. Moreover in vitro pulldown assays were performed that showed that recombinant BIN2, expressed and purified as a GST fusion protein from E. coli, was able to interact with recombinant c-Myc-tagged MKK4 as detected by pulldowns using GST beads and Western blot analysis (Fig. 2C).
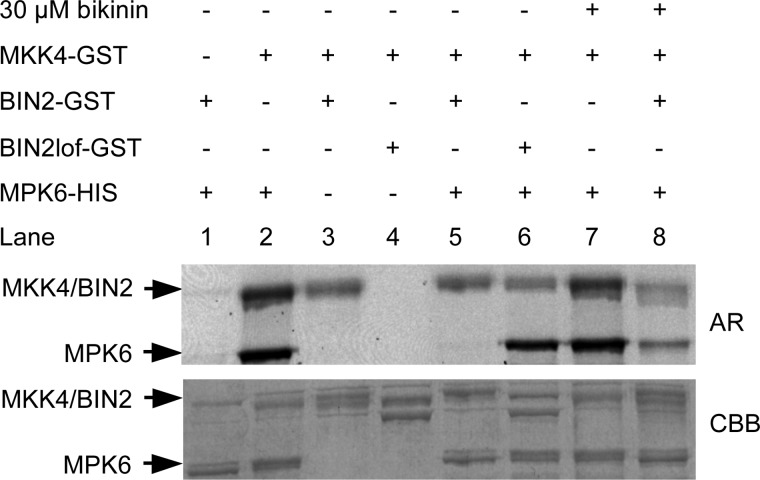
FIGURE 4.
BIN2 phosphorylation of MKK4 inhibits its activity against MPK6 in vitro. In vitro kinase assays were performed using recombinant GST-BIN2, GST-BIN2lof, GST-MKK4, and His6-MPK6 proteins. The proteins were incubated together in kinase reaction mixture containing 500 μm ATP and 20 μCi of radioactively labeled [γ-32P]ATP. Samples were then separated by SDS-PAGE and subjected to autoradiography (AR). Lanes 1 and 2 show phosphorylation of MPK6 by BIN2 and MKK4, respectively, whereas lanes 3 and 4 show phosphorylation of MKK4 by BIN2 and BIN2lof, respectively. In lanes 5 and 6, BIN2 or BIN2lof was preincubated with MKK4 in the kinase buffer containing 500 μm ATP and 20 μCi of [γ-33P]ATP at room temperature for 30 min prior to the addition of MPK6. In lane 8, BIN2 and 30 μm bikinin were incubated together for 10 min prior to the addition of MKK4, MPK6, and the reaction mixture. Lane 7 is a control for bikinin specificity in which MKK4 was incubated with 30 μm bikinin for 10 min prior to adding MPK6 and reaction mixture. CBB, Coomassie Brilliant Blue.
FIGURE 2.
BIN2 interacts with and phosphorylates MKK4. A, in vitro kinase assays using GST-BIN2, GST-ASKθ, or GST-ASKα as well as MKK4 were performed. Proteins were incubated in kinase buffer in the presence of [γ-33P]ATP as a co-substrate. Subsequently, samples were separated by SDS-PAGE and subjected to autoradiography (AR). Lanes 1–4 show the autophosphorylation of the kinases. In lanes 5–7, phosphorylation of MKK4 by the ASKs is visible. CBB, Coomassie Brilliant Blue. B, in vitro kinase assays using MKK4 as substrate with wild type and a loss-of-function version of GST-BIN2 (GST-BIN2 and GST-BIN2lof, respectively). Stains with Coomassie Brilliant Blue are shown as loading controls. For this assay, [γ-32P]ATP was used as a co-substrate. C, in vitro pulldown assays using GST-BIN2 and MKK4-His6-MYC proteins were performed. Recombinant MKK4-His6-MYC protein was incubated alone, with GST, or with GST-BIN2 in reaction buffer on ice, and the proteins were then pulled down with GST beads. MKK4-His6-MYC (input) and pulled down proteins were detected with anti-Myc antibody. D, bimolecular fluorescence complementation assay showing a representative protoplast transformed with ASKθ-cYFP and MKK4-nYFP constructs.
To investigate whether GSK3/Shaggy-like kinases can interact with MKK4 in planta, bimolecular fluorescence complementation assays (35) were carried out in A. thaliana protoplasts. For this purpose, ASKθ-YFP was chosen because it is expressed to comparably high levels as MKK4-YFP in protoplasts (whereas BIN2 is expressed at very low levels in this system (supplemental Fig. 1)). ASKθ fused to the C-terminal portion of YFP and MKK4 fused to the N-terminal portion of YFP were co-expressed in protoplasts. As shown in Fig. 2D, yellow fluorescence was observed diffusely in the nucleus when protoplasts were co-transformed with ASKθ and MKK4. In contrast the controls (protoplast transformed with one of the constructs only) did not emit detectable fluorescence (data not shown).
BIN2 Specifically Phosphorylates MKK4 and MKK5 in Vitro
MKK4 acts redundantly with MKK5 in controlling MPK3 and MPK6 activity to negatively regulate stomata development (20). Thus, the question arose whether MKK5 may also be a substrate of BIN2 and whether MKK4- or MKK5-unrelated MKKs would also be phosphorylated. For this analysis, two additional MKKs were chosen: MKK7, which has recently been shown to play a role in stomata development, albeit by presently unknown means (21), and MKK2, which is one of the most distant relatives of MKK4 in sequence similarity (40). In vitro kinase assays were performed in which the activity of BIN2 was investigated against recombinant GST-tagged versions of MKK2, MKK4, MKK5, and MKK7. Interestingly, BIN2 exhibited strong activity against MKK4 and MKK5, whereas its activity against MKK2 or MKK7 was negligible (Fig. 3). Therefore BIN2 interacts with and phosphorylates MKK4 and its closest homologue MKK5, but does not accept MKK2 and MKK7 as substrates in vitro.
FIGURE 3.
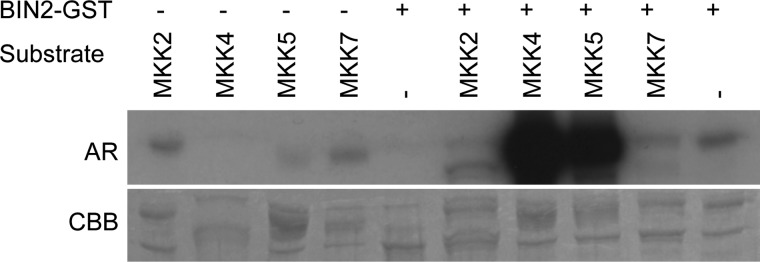
BIN2 specifically phosphorylates MKK4 and MKK5. In vitro kinase assays using GST-BIN2 were performed to analyze its activity against GST-MKK2, GST-MKK4, GST-MKK5, and GST-MKK7. Stains with Coomassie Brilliant Blue (CBB) are shown as loading controls in the lower panel. AR, autoradiography.
BIN2 Phosphorylation of MKK4 Negatively Regulates Its Activity against MPK6 in Vitro
The fact that in the dominant bin2-1 mutant and in ASKθoe plants stomata clustering phenotypes occurred, which are reminiscent of plants in which MKK4 and/or MKK5 expression is silenced (20), suggested a model in which GSK3-mediated phosphorylation inhibits MKK4. To test this hypothesis, in vitro kinase assays were performed in which the activity of GST-MKK4 was investigated using recombinant His6-MPK6 as a substrate in the presence or absence of GST-BIN2. In the absence of BIN2, MKK4 phosphorylated MPK6 as expected, whereas this phosphorylation was abolished when BIN2 was added to the reaction (Fig. 4, lanes 2 and 5). When an inactive BIN2 variant (BINlof) was used (lane 6) or when BIN2 was inhibited by adding the GSK3/Shaggy-like kinase inhibitor bikinin (33) to the reaction (lane 8), phosphorylation of MPK6 by MKK4 was restored, proving that BIN2 phosphorylation negatively regulates MKK4 activity in vitro.
Identification of BIN2 Phosphorylation Sites in MKK4 by Mass Spectrometry Analysis
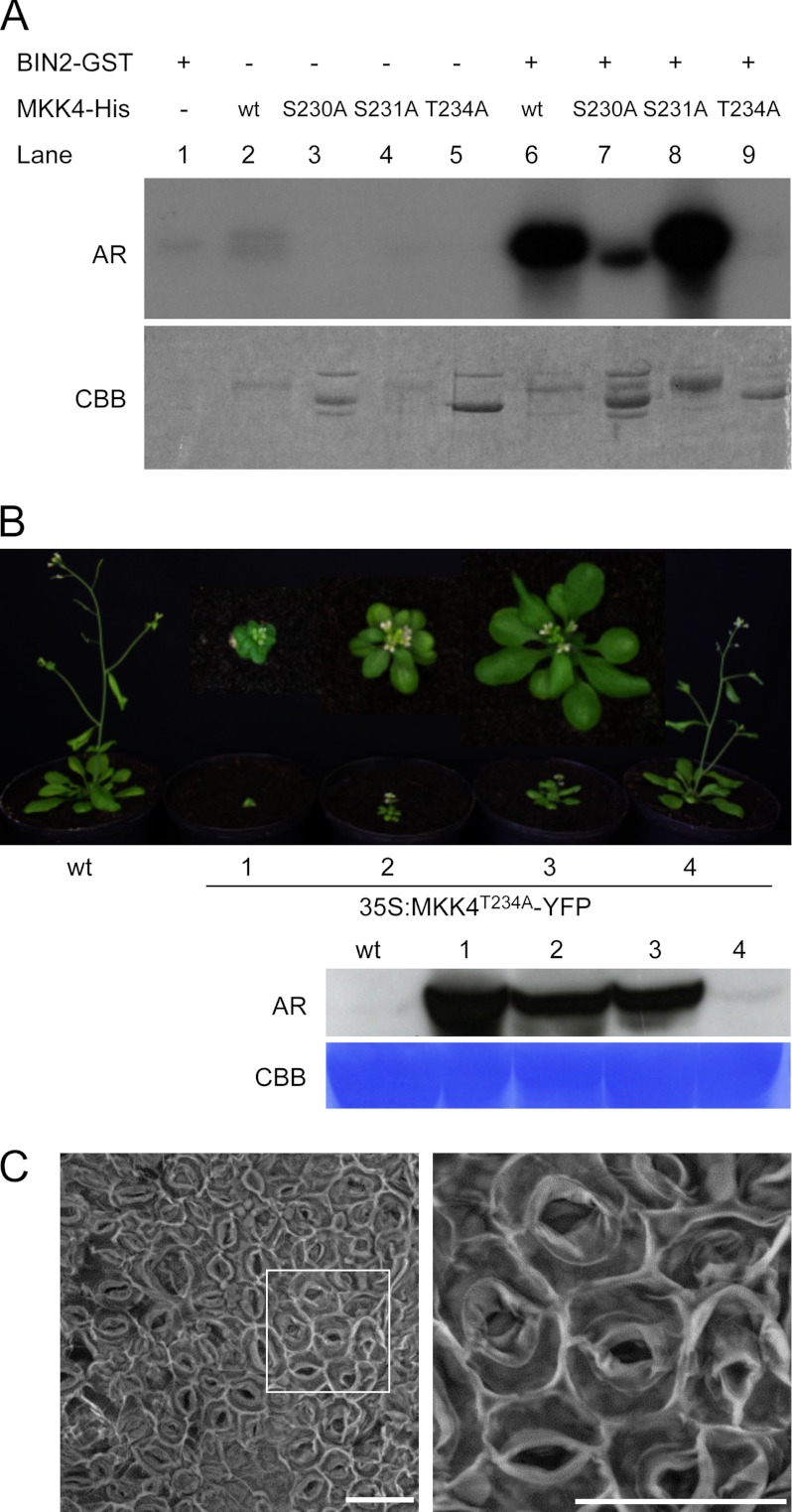
To identify putative BIN2 phosphorylation sites in MKK4, a mass spectrometric analysis was performed. Purified GST-MKK4 fusion protein was incubated with GST-BIN2 in the presence of ATP. The reaction products were separated by SDS-PAGE, and a band corresponding to GST-MKK4 was isolated from the gel and digested with trypsin. The resulting peptides were subsequently subjected to mass spectrometry (MS) analysis. Phosphopeptides that increased in abundance following phosphorylation by BIN2 contained phosphoserine at position 6, phosphothreonine at position 234, and phosphoserine at position 230 or 231, which are located in the activation loop (MS analysis did not allow us to distinguish which of both serines was phosphorylated; for mass spectra, see supplemental Fig. 2). These sites were mutated to alanine to create single mutants, and kinase assays were performed with the mutants. Although BIN2-mediated phosphorylation of the S6A mutant (supplemental Fig. 3) and a S231A variant was similar to wild type (Fig. 5A, lane 8), phosphorylation of the S230A and T234A mutants was strongly reduced (Fig. 5A, lanes 7 and 9). Thus, Ser-230 and Thr-234 are essential for phosphorylation of MKK4 by BIN2 in vitro. Interestingly, mutation of Thr-234 also to other amino acids such as Glu or Asp abolished activity of MKK4 against MPK6 in vitro (supplemental Fig. 4), showing that a Thr at this site is of high importance for MKK4 activity in vitro.
FIGURE 5.
Effects of BIN2 target site mutations on MKK4 activity in vitro and in planta. A, in vitro kinase assays using recombinant GST-BIN2 and GST-tagged wild-type or mutant variants of MKK4. AR, autoradiography; CBB, Coomassie Brilliant Blue; B, top, wild-type plants and T1 plants expressing 35S:MKK4T234A-YFP. Bottom, Western blot analysis of the studied lines with anti-YFP antibody. C, SEM image of the abaxial cotyledon epidermis of a representative plant, which strongly expresses 35S:MKK4T234A-YFP, 12 days after germination. The scale bar represents 30 μm.
Overexpression of T234A Induces Phenotypes Reminiscent of Plants in Which Activities of MKK4 and MKK5 Are Lost
In an aim to investigate the effect of modification of Thr-234 on MKK4 activity in planta, we generated a MKK4T234A-YFP fusion construct and overexpressed it under control of the constitutive 35S promoter in A. thaliana plants. T1 plants were selected and were analyzed for MKK4T234A-YFP expression by Western blotting. Very interestingly, representative plants, which expressed MKK4T234A-YFP to high levels, were characterized by striking developmental defects, such as severe dwarfism, round leaves, sterility, and premature death (Fig. 5B). Because these phenotypes clearly resembled phenotypes described for plants in which MKK4 and MKK5 is silenced (20), we assessed whether 35S:MKK4T234A-YFP lines may also be affected in stomata distribution by scanning electron microscopy. The result is shown in Fig. 5C and illustrates that MKK4T234A-YFP overexpression resulted in drastic stomata patterning defects, with the epidermis of cotyledons almost solely composed of stomata in highly expressing lines.
MKK4 Expression at the Subcellular Level
To gain further insight into the role of MKK4, its subcellular localization was investigated. A YFP fusion to the C terminus of full-length MKK4 was expressed under control of the 35S promoter (35S:MKK4-YFP), and plant lines stably expressing this construct were generated (Fig. 6A). A subsequent analysis of MKK4-YFP subcellular expression revealed that MKK4 is localized in the cytoplasm as well as in the nucleus, both when driven by the 35S promoter (analyzed in planta: Fig. 6B) as well as when driven by the endogenous MKK4 promoter (analyzed in protoplasts: supplemental Fig. 5). BR treatment or treatment with Brz did not alter MKK4 subcellular localization when analyzed in protoplasts (supplemental Fig. 5).
FIGURE 6.
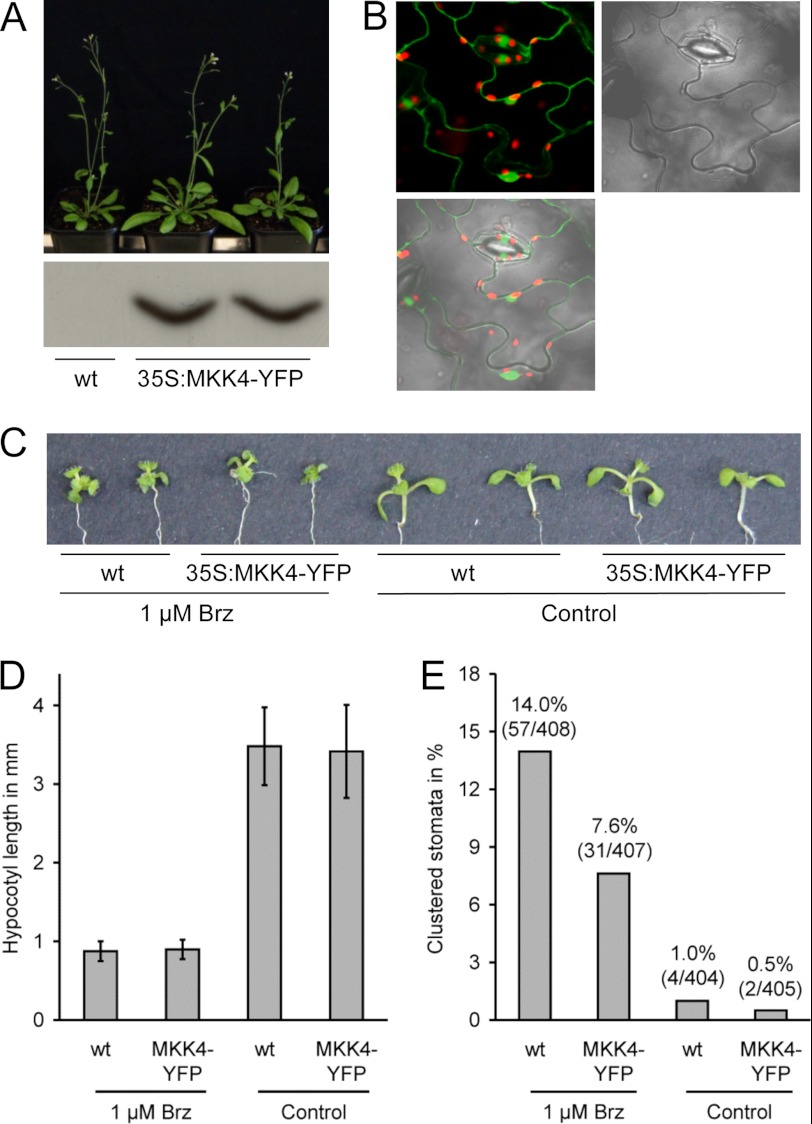
BIN2 acts upstream of MKK4 in stomata patterning. A, top, wild-type and homozygous lines expressing 35S:MKK4-YFP. Bottom, Western blot analysis of the studied lines with anti-YFP antibody. B, analysis of MKK4-YFP subcellular localization in A. thaliana plants stably expressing 35S:MKK4-YFP. C, phenotypes of seedlings with or without 1 μm Brz. D, hypocotyl length of seedlings used for quantification of stomata clustering. Error bars indicate S.D. E, the frequency of stomata clustering in wild-type and 35S:MKK4-YFP plants in the presence or absence of 1 μm Brz as determined by light microscopy. The results are shown in percentages. p values derived of a χ2-test are given.
MKK4 or MKK5 Overexpression Suppresses Stomata Clustering in BR-deficient Plants
The data presented above suggested a model in which, in a BR-deficient situation, BIN2 and redundantly acting GSK3/Shaggy-like kinases would phosphorylate MKK4/MKK5 to reduce their activity and thereby promote stomata clustering. Therefore an increase in the level of MKK4 should (partially) rescue the stomata clustering phenotype of BR-deficient plants. To investigate this hypothesis, we treated wild-type Col-0 and a representative MKK4-YFP-overexpressing plant line with Brz. As shown in Fig. 6, C and D, wild-type and 35S:MKK4-YFP plants reacted to Brz treatment as expected; a BR-deficient phenotype was induced, characterized among other traits by shortened hypocotyls and dark green, downwardly curled leaves. However, importantly, when the frequency of stomata clustering was determined, a statistically highly significant difference was found between the two lines. Although 14.0% of the stomata of Brz-treated wild-type seedlings was present in clusters, only 7.6% of the stomata of 35S:MKK4-YFP plants grown under the same conditions was clustered (p value of a χ2-test: 0.0035). In the absence of Brz, both lines showed a very low clustering frequency of less than 1.0% (Fig. 6E). 35S:MKK5-YFP-expressing plants similarly showed an increased resistance to Brz application in regard to stomata clustering (6.4%; p value of a χ2-test: 0.0061), whereas 35S:MKK2-MYC-expressing plants behaved like wild-type in this assay (supplemental Fig. 6). This result shows that MKK4-YFP and MKK5-YFP overexpression suppresses stomata clustering in BR-deficient plants.
DISCUSSION
Stomata are cellular epidermal valves, which plants have evolved to control water and gas exchange (41, 42). In A. thaliana, stomata development is preceded by asymmetric cell divisions, and stomata distribution follows the one-cell spacing rule, reflecting the coordination of cell fate specification. Stomatal patterning is considered to involve the transmission of spatial cues from the stomata to the adjacent cell, which are required to correctly orient the plane of the spacing division (41). Thus, spacing is thought to result from cell-cell signaling, and it is known that the MAPK signaling module YDA-MKK4/5-MPK3/6 is necessary for transducing signals to control cell division and cell fate decisions during stomata development and in embryogenesis (20, 21, 43, 44).
BRs are essential plant hormones that regulate diverse aspects of growth and development, including cell elongation, cell division, and cell differentiation (12, 45, 46). Previous findings suggested that BRs also participate in cellular patterning (15), and here further evidence is provided that supports this role. In addition our results also provide a possible mechanistic explanation how BRs may influence stomata patterning. We show that BIN2 directly interacts with and phosphorylates MKK4 to negatively regulate its activity in phosphorylating MPK6 in vitro. Interestingly, a regulatory effect of BIN2 on the YDA-MKK4/5-MPK3/6 module was described very recently in a different study. The authors showed that BIN2 phosphorylates YDA to reduce its activity (47). Together these results indicate that BIN2 targets both MAPKKK and MAPKK in the MPK3/6 module for suppression, which would allow for a dynamic cellular repatterning in response to developmental or environmental cues perceived and transduced by BRs.
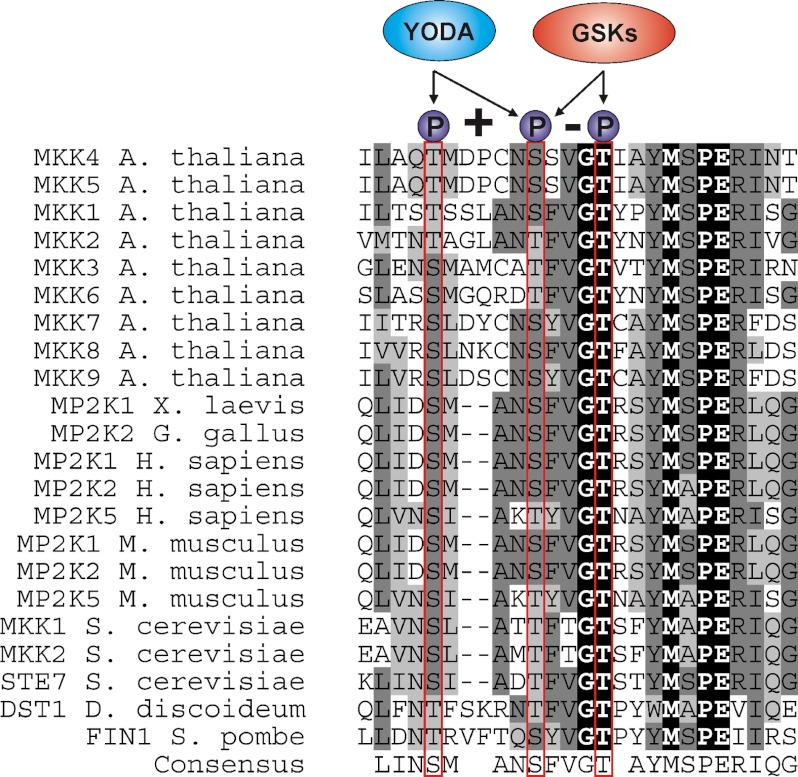
The BR response pathway signals through the inhibition of GSK3/Shaggy-like kinase activities including BIN2, and here we provide evidence that BIN2 and homologues control the activity of MAPKKs. GSK3/Shaggy-like kinases are a group of highly conserved, constitutively active S/T kinases implicated in numerous signaling pathways and controlling metabolism, cell fate determination, and tissue patterning in various organisms (48–50). In mammalian systems substrates of GSK3s include glycogen synthase, β-catenin, cyclin D1, c-Jun, and Smad1 (48–50). This work is the first across all biological kingdoms to report that a GSK3/Shaggy-like kinase can phosphorylate a MAPKK. BIN2 phosphorylates MKK4 at Ser-230 and Thr-234 in vitro, which form a classical GSK3 phosphorylation motif ((S/T)XXX(S/T) (51)). Because mutation of Thr-234 completely abrogates BIN2 phosphorylation of MKK4, it is possible that Thr-234 is the main phosphorylation site, and Ser-230 may be less important. Our results show that phosphorylation of Thr-234 by BIN2 rendered MKK4 inactive. Furthermore, a point mutation of this site to other amino acids resulted in a kinase-dead protein (supplemental Fig. 4), highlighting the importance of a Thr at position 234. Indeed Thr-234 is highly conserved in eukaryotic MAPKKs, suggesting a functional relevance in controlling MAPKK activities (Fig. 7). Ser-230 is part of the activation loop of MKK4, which is suggested to be phosphorylated by the upstream kinases MEKK1 and YDA to activate it in stress signaling and stomatal patterning pathways, respectively (20, 52). It seems possible that GSK3-mediated phosphorylation of Thr-234 may sterically hinder phosphorylation of the active loop by upstream MAPKKK. In a different scenario a competition for phosphorylation at Ser-230 between upstream MAPKKK and BR-regulated GSK3/Shaggy-like kinases could take place. It will be interesting to investigate whether MKK4 and/or MKK5 phosphorylation by BIN2 at Thr-234 overrides upstream regulatory events in this MAPK module.
FIGURE 7.
Amino acid alignment of MAPKKs from different eukaryotes. Dashes indicate gaps introduced to maximize alignment of conserved residues. Residues corresponding to Thr-224, Ser-230, and Thr-234 are marked in red. X. laevis, Xenopus laevis; G. gallus, Gallus gallus; H. sapiens, Homo sapiens; M. musculus, Mus musculus; S. cerevisiae, Saccharomyces cerevisiae; D. discoideum, Dictyostelium discoideum; S. pombe, Schizosaccharomyces pombe.
An overexpression of MKK4T234A, which is inactive against MPK6 in vitro, resulted in phenotypes reminiscent of plants in which the activities of either MKK4/5 or MPK3/6 are lost (20). This suggests that also in plants MKK4T234A is inactive because in a situation of constitutive overexpression, it may outcompete endogenous, active MKK4/5 in binding to MPK3/6, which would interfere with their activation and result in the observed developmental defects. It will be interesting to determine the physiological significance of BIN2-mediated Ser-230 and/or Thr-234 phosphorylation events and investigate whether modifications at these sites may allow for a dynamic regulation of MKK4 and MKK5 activity by the BR and YDA-MKK4/5-MPK3/6 signaling pathways in planta.
Cross-talk of BR signaling with the YDA-MKK4/5-MPK3/6 cascade at the receptor level has previously been reported with BAK1 playing a dual role as a co-receptor of both BRI1 and FLS2 (53–55). Moreover very recently BIN2 was shown to also directly control SPCH activity (31). Our study now provides further indications for a cross-communication of the two pathways, opening up the possibility of a web of interactions, which enables BRs to dictate cellular patterning and other responses signaled by the YDA-MKK4/5-MPK3/6 MAPK module.
Acknowledgments
We thank Armin Djamei for providing constructs containing the MKK2, MKK4, MKK5, and MKK7 cDNAs. We also thank Miklos Szekeres and Tobias Sieberer for very helpful suggestions and critical comments on the manuscript and Renata Milčevičova and Irene Ziegler for technical assistance. The staff of the Wissenschaftszentrum Weihenstephan (WZW) and Max F. Perutz Laboratories (MFPL) plant facilities provided excellent plant care.
This study was supported by the Austrian Science Fund FWF (Projects P19948 and P22734) and by a Ph.D. fellowship of the Higher Education Commission of Pakistan (to M. K.) and a Hertha-Firnberg fellowship of the FWF (to B. P.).

This article contains supplemental Materials and Methods and Figs. 1–6.
- BR
- brassinosteroid
- Brz
- brassinazole
- GSK3
- glycogen synthase kinase 3
- BIN2
- brassinosteroid-insensitive 2
- MKK
- MAP kinase kinase
- MPK3/6
- mitogen-activated protein kinase 3/6
- MAPKK
- MAPK kinase
- MAPKKK
- MAPK kinase kinase
- BRI1
- brassinosteroid-insensitive 1
- ASKθ/α
- A. thaliana Shaggy-like kinase θ/α
- ASKθoe
- A. thaliana Shaggy-like kinase θ overexpressor mutant
- YDA
- YODA
- Col-0
- Columbia-0
- Brz
- brassinozole
- lof
- loss-of-function.
REFERENCES
- 1. Bergmann D. (2006) Stomatal development: from neighborly to global communication. Curr. Opin. Plant Biol. 9, 478–483 [DOI] [PubMed] [Google Scholar]
- 2. Costa S., Shaw P. (2006) Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature 439, 493–496 [DOI] [PubMed] [Google Scholar]
- 3. Jaillais Y., Chory J. (2010) Unraveling the paradoxes of plant hormone signaling integration. Nat. Struct. Mol. Biol. 17, 642–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grove M. D., Spencer G. F., Rohwedder W. K., Mandava N. B., Worley J. F., Warthen J. D., Steffens G. L., Flippen-Anderson J. L., Cook J. C. (1979) A unique growth promoting steroid from Brassica napus pollen. Nature 281, 216–217 [Google Scholar]
- 5. Poppenberger B., Rozhon W., Khan M., Husar S., Adam G., Luschnig C., Fujioka S., Sieberer T. (2011) CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 30, 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim T. W., Wang Z. Y. (2010) Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61, 681–704 [DOI] [PubMed] [Google Scholar]
- 7. Vert G., Nemhauser J. L., Geldner N., Hong F., Chory J. (2005) Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21, 177–201 [DOI] [PubMed] [Google Scholar]
- 8. Azpiroz R., Wu Y., LoCascio J. C., Feldmann K. A. (1998) An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10, 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu Y., Bao F., Li J. (2000) Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24, 693–701 [DOI] [PubMed] [Google Scholar]
- 10. Fukuda H. (1997) Programmed cell death during vascular system formation. Cell Death Differ. 4, 684–688 [DOI] [PubMed] [Google Scholar]
- 11. Iwasaki T., Shibaoka H. (1991) Brassinosteroids act as regulators of tracheary-element differentiation in isolated zinnia mesophyll cells. Plant Cell Physiol. 32, 1007–1014 [Google Scholar]
- 12. Clouse S. D., Sasse J. M. (1998) Brassinosteroids: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451 [DOI] [PubMed] [Google Scholar]
- 13. González-García M. P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A. I. (2011) Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849–859 [DOI] [PubMed] [Google Scholar]
- 14. Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M. J., Chory J., Savaldi-Goldstein S. (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138, 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuppusamy K. T., Chen A. Y., Nemhauser J. L. (2009) Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 106, 8073–8076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geisler M., Nadeau J., Sack F. D. (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12, 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nadeau J. A., Sack F. D. (2003) Stomatal development: cross talk puts mouths in place. Trends Plant Sci. 8, 294–299 [DOI] [PubMed] [Google Scholar]
- 18. Szekeres M., Németh K., Koncz-Kálmán Z., Mathur J., Kauschmann A., Altmann T., Rédei G. P., Nagy F., Schell J., Koncz C. (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182 [DOI] [PubMed] [Google Scholar]
- 19. Bergmann D. C., Lukowitz W., Somerville C. R. (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497 [DOI] [PubMed] [Google Scholar]
- 20. Wang H., Ngwenyama N., Liu Y., Walker J. C., Zhang S. (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lampard G. R., Lukowitz W., Ellis B. E., Bergmann D. C. (2009) Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21, 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lampard G. R., Macalister C. A., Bergmann D. C. (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113–1116 [DOI] [PubMed] [Google Scholar]
- 23. Chang L., Karin M. (2001) Mammalian MAP kinase signalling cascades. Nature 410, 37–40 [DOI] [PubMed] [Google Scholar]
- 24. Colcombet J., Hirt H. (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 413, 217–226 [DOI] [PubMed] [Google Scholar]
- 25. Rodriguez M. C., Petersen M., Mundy J. (2010) Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649 [DOI] [PubMed] [Google Scholar]
- 26. Clough S. J., Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 27. Lincoln C., Britton J. H., Estelle M. (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hellens R. P., Edwards E. A., Leyland N. R., Bean S., Mullineaux P. M. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832 [DOI] [PubMed] [Google Scholar]
- 29. Rozhon W., Mayerhofer J., Petutschnig E., Fujioka S., Jonak C. (2010) ASKθ, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 62, 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dal Santo S., Stampfl H., Krasensky J., Kempa S., Gibon Y., Petutschnig E., Rozhon W., Heuck A., Clausen T., Jonak C. (2012) Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell 24, 3380–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gudesblat G. E., Schneider-Pizoń J., Betti C., Mayerhofer J., Vanhoutte I., van Dongen W., Boeren S., Zhiponova M., de Vries S., Jonak C., Russinova E. (2012) SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14, 548–554 [DOI] [PubMed] [Google Scholar]
- 32. Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J. L., Hirt H. (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15, 141–152 [DOI] [PubMed] [Google Scholar]
- 33. De Rybel B., Audenaert D., Vert G., Rozhon W., Mayerhofer J., Peelman F., Coutuer S., Denayer T., Jansen L., Nguyen L., Vanhoutte I., Beemster G. T., Vleminckx K., Jonak C., Chory J., Inzé D., Russinova E., Beeckman T. (2009) Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16, 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cardinale F., Meskiene I., Ouaked F., Hirt H. (2002) Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14, 703–711 [PMC free article] [PubMed] [Google Scholar]
- 35. Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 [DOI] [PubMed] [Google Scholar]
- 36. Nadeau J. A., Sack F. D. (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700 [DOI] [PubMed] [Google Scholar]
- 37. Asami T., Min Y. K., Nagata N., Yamagishi K., Takatsuto S., Fujioka S., Murofushi N., Yamaguchi I., Yoshida S. (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123, 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li J., Chory J. (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938 [DOI] [PubMed] [Google Scholar]
- 39. Li J., Nam K. H., Vafeados D., Chory J. (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127, 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant. Sci. 7, 301–308 [DOI] [PubMed] [Google Scholar]
- 41. Bergmann D. C., Sack F. D. (2007) Stomatal development. Annu. Rev. Plant Biol. 58, 163–181 [DOI] [PubMed] [Google Scholar]
- 42. Dong J., Bergmann D. C. (2010) Stomatal patterning and development. Curr. Top. Dev. Biol. 91, 267–297 [DOI] [PubMed] [Google Scholar]
- 43. Bergmann D. C. (2004) Integrating signals in stomatal development. Curr. Opin. Plant Biol. 7, 26–32 [DOI] [PubMed] [Google Scholar]
- 44. Lukowitz W., Roeder A., Parmenter D., Somerville C. (2004) A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116, 109–119 [DOI] [PubMed] [Google Scholar]
- 45. Altmann T. (1998) Recent advances in brassinosteroid molecular genetics. Curr. Opin. Plant Biol. 1, 378–383 [DOI] [PubMed] [Google Scholar]
- 46. Nakaya M., Tsukaya H., Murakami N., Kato M. (2002) Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol. 43, 239–244 [DOI] [PubMed] [Google Scholar]
- 47. Kim T. W., Michniewicz M., Bergmann D. C., Wang Z. Y. (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cohen P., Frame S. (2001) The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2, 769–776 [DOI] [PubMed] [Google Scholar]
- 49. He X., Saint-Jeannet J. P., Woodgett J. R., Varmus H. E., Dawid I. B. (1995) Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature 374, 617–622 [DOI] [PubMed] [Google Scholar]
- 50. Xu C., Kim N. G., Gumbiner B. M. (2009) Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle 8, 4032–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Frame S., Cohen P. (2001) GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., Boller T., Ausubel F. M., Sheen J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983 [DOI] [PubMed] [Google Scholar]
- 53. Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Chinchilla D., Rathjen J. P., de Vries S. C., Zipfel C. (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. U.S.A. 109, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Belkhadir Y., Jaillais Y., Epple P., Balsemão-Pires E., Dangl J. L., Chory J. (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. U.S.A. 109, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Z. (2012) Brassinosteroids modulate plant immunity at multiple levels. Proc. Natl. Acad. Sci. U.S.A. 109, 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]