Background: The type VI secretion system (T6SS) is a bacterial device similar to bacteriophages.
Results: Pseudomonas aeruginosa HsiB and HsiC proteins form a T6SS subcomplex, which polymerizes into tubules via the C terminus of HsiC.
Conclusion: HsiB and HsiC form a structure with similarity to the bacteriophage tail sheath.
Significance: TssB and TssC members of the T6SS form a complex resembling the bacteriophage tail sheath and mimic the gp18 protein.
Keywords: Bacterial Pathogenesis, Bacteriophage, Microbiology, Protein Secretion, Protein-Protein Interactions, Pseudomonas aeruginosa
Abstract
Protein secretion systems in Gram-negative bacteria evolved into a variety of molecular nanomachines. They are related to cell envelope complexes, which are involved in assembly of surface appendages or transport of solutes. They are classified as types, the most recent addition being the type VI secretion system (T6SS). The T6SS displays similarities to bacteriophage tail, which drives DNA injection into bacteria. The Hcp protein is related to the T4 bacteriophage tail tube protein gp19, whereas VgrG proteins structurally resemble the gp27/gp5 puncturing device of the phage. The tube and spike of the phage are pushed through the bacterial envelope upon contraction of a tail sheath composed of gp18. In Vibrio cholerae it was proposed that VipA and VipB assemble into a tail sheathlike structure. Here we confirm these previous data by showing that HsiB1 and HsiC1 of the Pseudomonas aeruginosa H1-T6SS assemble into tubules resulting from stacking of cogwheel-like structures showing predominantly 12-fold symmetry. The internal diameter of the cogwheels is ∼100 Å, which is large enough to accommodate an Hcp tube whose external diameter has been reported to be 85 Å. The N-terminal 212 residues of HsiC1 are sufficient to form a stable complex with HsiB1, but the C terminus of HsiC1 is essential for the formation of the tubelike structure. Bioinformatics analysis suggests that HsiC1 displays similarities to gp18-like proteins in its C-terminal region. In conclusion, we provide further structural and mechanistic insights into the T6SS and show that a phage sheathlike structure is likely to be a conserved element across all T6SSs.
Introduction
The type VI secretion system (T6SS)5 (1) is one of many protein secretion nanomachines used by Pseudomonas aeruginosa to successfully infects its host (2). P. aeruginosa is an opportunistic Gram-negative bacterial pathogen that infects a wide range of tissues in humans, including ear, eyes, skin, urinary tract, blood, and lungs, and is fatal for cystic fibrosis patients (3). In cystic fibrosis, the infection is chronic, and the T6SS seems to play an active role in the persistence of this microorganism and its successful colonization of the lung at the expense of other bacteria and fungi (4).
In P. aeruginosa, three gene clusters encoding T6SS components have been identified in most sequenced strains to date. These are called H1-, H2-, and H3-T6SS and are located at distinct places on the chromosome (5). The best characterized system to date is H1-T6SS because its expression is active in mutant strains deleted for the retS regulatory gene (4). A more general observation is that bacterial T6SS genes are mostly expressed during host contact/colonization and thus are not suitable for in vitro studies (5, 6). RetS is a histidine kinase-like sensor and has been shown to repress the GacS/GacA two-component system regulatory cascade that induces expression of the small RNAs, RsmY and RsmZ, thus relieving the translation repression exerted by RsmA on H1-T6SS transcripts (7–9).
The T6SS is composed of about 12 core components, and here we will use the H1-T6SS names as general nomenclature for clarity (4, 5). The IcmF, DotU, and Lip proteins are likely to form a cell envelope platform. In the T6SS of enteroaggregative Escherichia coli and Agrobacterium tumefaciens, it was shown that IcmF (TssM) and DotU (TssL) are inner membrane proteins interacting with each other, whereas IcmF also interacts with the outer membrane lipoprotein Lip (TssJ) (10–12). In enteroaggregative E. coli, an additional component called SciZ or TagL anchors the complex to the peptidoglycan (13). This component is not present in all T6SSs described so far, but some DotU-like proteins, as in the case of the H1-T6SS DotU1, carry a peptidoglycan-binding domain at their C terminus, thus performing this anchoring function. It is noteworthy that the DotU and IcmF protein families have been shown to be involved in the type IV secretion system (14), but these are the sole components conserved between T6SS and the type IV secretion system, and all other T6SS components described are system-specific.
Although not finding further similarities with the type IV secretion system, it was shown earlier that the T6SS shares extensive similarities with the tail of contractile bacteriophages, such as the T4 phage. Most remarkably, the VgrG protein family structurally resembles the gp27-gp5 complex, which forms the puncturing device of the bacteriophage (15). This complex is made of two trimers, gp273-gp53 with the gp5 trimer assembling into a rigid helix of β-strands, which is the spike of the device and is required for puncturing the bacterial outer membrane. VgrG proteins form a trimer with structural resemblance to the gp273-gp53 complex, although one domain, which corresponds to a lysozyme domain within gp5 (16, 17), is lacking. In the T4 phage, the puncturing device sits at the tip of the tail tube, which is made of the gp19 protein (18). It was reported that despite weak amino acid sequence similarities the structure of the Hcp1 protein could be superimposed with the structure of the tail tube protein of the phage λ, gpV (17). Furthermore, the Hcp1 structure is seen as a hexamer with an external diameter of 85 Å, and this structure could be nicely fitted onto the top of a gp27 trimer, thus mimicking the connection between gp27 and the gp19 tail tube.
Thus, it is clear that T6SS and bacteriophage tail components evolved from a common ancestor, but the poor amino acid sequence similarities suggest that they diverged a very long time ago. Thus, it is expected that other T6SS components may be related to phage components, but such similarity might only be observable when comparing resolved or predicted protein structures. This has been the case for example with the baseplate phage protein gp25 and the H1-T6SS component HsiF1 (TssE) (19) and should be expected for HsiB1 (TssB) (20), HsiC1 (TssC), HsiG1 (TssF), and HsiH1 (TssG). In Vibrio cholerae, the VipA and VipB proteins, members of the TssB and TssC family, respectively, have been shown to form tubule-like structures that could be compared with the tail sheathlike structure of contractile bacteriophages (21–23). In the present study, we showed that the HsiB1 and HsiC1 proteins could be co-purified and as in V. cholerae form exquisite tubule and cogwheel structures. We demonstrated which domains of these proteins are required for interaction and which are essential for formation of the tubules. This interaction is highly specific, and heterocomplex formation between components of the H1-T6SS and the H2-T6SS was not observed. The similarity was also established using bioinformatics approaches and suggests that the C terminus of HsiC1 shows resemblance with phage tail sheath proteins, known as gp18 in the case of the bacteriophage T4 (24).
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Bacterial strains used in this study are described in Table 1. P. aeruginosa strains were grown in tryptone soy broth supplemented with antibiotics where appropriate (15 μg/ml tetracycline). E. coli strains were grown in Luria-Bertani (LB) broth supplemented with antibiotics where appropriate (50 μg/ml streptomycin, 50 or 100 μg/ml ampicillin, 50 μg/ml kanamycin, 50 μg/ml gentamicin, 30 μg/ml chloramphenicol, and 15 μg/ml tetracycline).
TABLE 1.
Strains used in this study
| Strain | Relevant characteristics | Source/Ref. |
|---|---|---|
| E. coli | ||
| One-shot® TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu) | Invitrogen |
| XL1 blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| CC118 λpir | Host strain for pKNG101 replication; D (ara-leu) araD DlacX74 galE galK-phoA20 thi-1 rpsE rpoB argE (Am) recA1 Rfr (λpir) | 28 |
| DHM1 | cya-854 recA1 gyrA96 (NaI) thi1 hsdR17 spoT1 rfbD1 glnV44(AS) | 26 |
| B834(DE3) | F − ompT hsdSB (rB − mB −) gal dcm met (DE3) | 58 |
| P. aeruginosa | ||
| PAK | Wild-type P. aeruginosa | Laboratory collection |
| PAKΔretS | In-frame deletion of retS (PA4856) in P. aeruginosa PAK wild-type strain | 34 |
| PAK ΔretSΔhsiC1 | In-frame deletion of hsiC1 (PA0084) in PAKΔretS | This study |
| PAK ΔretSΔhsiC1+C1 | PAK ΔretSΔhsiC1with hsiC1 integrated at the att site using mini-CTC-C1 | This study |
| PAK ΔretSΔhsiC1+C2 | PAK ΔretSΔhsiC1with hsiC2 integrated at the att site using mini-CTC-C2 | This study |
Plasmids
All plasmids and oligonucleotides used in this study are listed in Tables 2 and 3, respectively. All constructs were confirmed by sequencing (GATC Biotech, Germany) prior to use. The plasmids pET28-B1C1 and pET28-B1C1N were constructed as follows. The gene pair hsiB1/hsiC1 or hsiB1/hsiC1N was amplified in tandem by PCR from genomic DNA of P. aeruginosa PAO1 using primer pairs 381/660 and 381/838, respectively. PCR products were cloned into pET28a, resulting in the recombinant plasmid pET-B1C1 encoding HsiB1 with an N-terminal histidine tag together with untagged HsiC1 and pETB1C1N encoding HsiB1 with an N-terminal histidine tag together with untagged HsiC1(1–212), respectively.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source/Ref. |
|---|---|---|
| pRK2013 | Tra+Mob+ helper strain for mobilization of non-self-transmissible plasmids, KmR | 29 |
| Mini-CTX-1 | Plasmid for the integration of genes into the att site of the P. aeruginosa chromosome, TcR | 59 |
| Mini-CTX-C1 | Mini-CTX-1 encoding hsiC1 under a plac promoter for integration into the att site of the P. aeruginosa chromosome for the purpose of complementation, TcR | This study |
| Mini-CTX-C2 | Mini-CTX-1 encoding hsiC2 under a plac promoter for integration into the att site of the P. aeruginosa chromosome, TcR | This study |
| pET28a | Protein expression vector used for expression of N-terminal 6-histidine fusion proteins | Novagen |
| pET28-B1C1 | pET28a expressing HsiB1 with an N-terminal histidine tag in tandem with untagged HsiC1 | This study |
| pET28-B1C1N | pET28a expressing HsiB1 with an N-terminal histidine tag in tandem with untagged HsiC1 (1–212) | This study |
| pKT25 | BTH vector for fusion of target proteins to Bordetella pertussis cya gene T25 fragment; Plac::cya1–675 p15 ori, KmR | 26 |
| pUT18C | BTH vector for fusion of target proteins to B. pertussis cya gene T18 fragment; Plac::cya675–1197-MCS pUC ori, ApR | 26 |
| pKT25-zip | Fusion of zip encoding leucine zipper from GCN4 to cya gene T25 fragment in pKT25, KmR | 26 |
| pUT18C-zip | Fusion of zip encoding leucine zipper from GCN4 to cya gene T18 fragment in pUT18C, ApR | 26 |
| pKT25-hsiB1 | Fusion of hsiB1 to cya gene T25 fragment in pKT25, KmR | This study |
| pUT18C-hsiB1 | Fusion of hsiB1 to cya gene T18 fragment in pUT18C, ApR | This study |
| pKT25-hsiC1 | Fusion of hsiC1 to cya gene T25 fragment in pKT25, KmR | This study |
| pUT18C-hsiC1 | Fusion of hsiC1 to cya gene T18 fragment in pUT18C, ApR | This study |
| pKT25-hsiB2 | Fusion of hsiB2 to cya gene T25 fragment in pKT25 KmR | This study |
| pUT18C-hsiB2 | Fusion of hsiB2 to cya gene T18 fragment, ApR | This study |
| pKT25-hsiC2 | Fusion of hsiC2 to cya gene T25 fragment in pKT25, KmR | This study |
| pUT18C-hsiC2 | Fusion of hsiC2 to cya gene T18 fragment in pUT18C, ApR | This study |
| pUT18C-hsiC1N | Fusion of hsiC1 encoding the first 212 aa of HsiC1 to cya gene T18 fragment in pUT18C, ApR | This study |
TABLE 3.
Oligonucleotides used in this study
FW, forward; RV, reverse.
| Oligo | Oligonucleotide sequence (5′ → 3′) | Application |
|---|---|---|
| 306 | GGAAGCACTACCAGCAGTC | HsiC1 (PA0084) mutagenesis |
| 307 | TCAGGCCTCTTCGGCCATGCTGGAATC | HsiC1 (PA0084) mutagenesis |
| 308 | ATGGCCGAAGAGGCCTGAGGCATTCAA | HsiC1 (PA0084) mutagenesis |
| 309 | GGATGTTCCAGCCGTACTTG | HsiC1 (PA0084) mutagenesis |
| 310 | CCTGCTCGAGTATTACGTTCG | HsiC1 (PA0084) mutagenesis |
| 311 | AACAGTTCTTCGGCGATCAT | HsiC1 (PA0084) mutagenesis |
| 655 | GCTCTAGAGATGGCCAAAGAAGGCTCG | BTH HsiB2 |
| 656 | CGGAATTCTCAGGCGTCCTGGGAGGG | BTH HsiB2 |
| 657 | GCTCTAGAGATGAGCACCAGTGCCGCA | BTH HsiC2 |
| 658 | CGGAATTCTTACTCTTTGTCCAGCTT | BTH HsiC2 |
| 659 | GCTCTAGAGATGGCCGAATTGAGCACC | BTH HsiC1 |
| 660 | CGGAATTCTCAGGCCTCTTTGGCCGA | BTH HsiC1, pET-B1C1 (RV) |
| 838 | CGGAATTCTCAGTTGGACAGTTCCTGCCA | BTH HsiC1 (1–212) |
| 729a | GCTCTAGAGATGGGAAGCACTACCAGCAGTCAG | BTH HsiB1 |
| 729 | GGGGTACCTTACGCCTGCGGCTCGTC | BTH HsiB1 |
| 381 | CGGGATCCCATATGGGAAGCACTACCAGCAGTC | pET-B1C1/pET-B1C1(1–212) (FW) |
| 838 | CGGAATTCTCAGTTGGACAGTTCCTGCCA | pET-B1C1(1–212) (RV) |
| 1271 | GAGGGCCCTTCACACAGGAAACAGCTATGGCCGAATTGAGCACC | Mini-CTX-C1 (FW) |
| 1272 | CGAATTCTCAGGCCTCTTTGGCCGA | Mini-CTX-C1 (RV) |
| 1419 | GGGGTACCTCACTCGGCCTCGGCGAAGTCGTAGTGGAACTGGTT | Mini-CTX-C2 (FW) |
| 1421 | CGGACTAGTTCATCCTCCTCCCTCTTTGTCCAGCTT | Mini-CTX-C2 (RV) |
Deletion mutants in P. aeruginosa were constructed using the suicide vector pKNG101 (25). DNA regions upstream and downstream of the hsiC1 gene were amplified from P. aeruginosa PAK genomic DNA using oligonucleotide pairs listed in Table 3. The resulting overlapping PCR products, which were then merged by PCR, were cloned into pCR2.1 and subcloned into pKNG101 using restriction sites ApaI and BamHI, yielding the suicide vector pKNG-ΔhsiC1.
All plasmids for bacterial two-hybrid (BTH) analysis were constructed as follows. The gene of interest was amplified from P. aeruginosa PAO1 genomic DNA, adding restriction sites XbaI and KpnI using the oligonucleotides listed in Table 3. The resulting PCR product was ligated into BTH plasmids pKT25 and pUT18C, leading to plasmids expressing in-frame fusions of the protein of interest to the T25 or T18 subunit of adenylate cyclase, respectively (26).
The plasmids mini-CTX-C1 and mini-CTX-C2 were constructed as follows. The region encoding the plac promoter was amplified from pCR2.1 (Invitrogen), adding restriction sites for subcloning into mini-CTX-1, and ligated into mini-CTX-1 (20). The hsiC1 and hsiC2 genes were amplified from P. aeruginosa genomic DNA using primers 1271/1272 and 1419/1421, respectively, and ligated into mini-CTX-1 downstream of the plac promoter, resulting in the plasmids mini-CTX-C1 and mini-CTX-C2 for conjugation and integration into the chromosome of PAKΔretSΔhsiC1.
Antibodies and Reagents
Polyclonal anti-VgrG1a and anti-Tse3 and peptide antibody against Hcp1 have been described previously (16). Anti-VgrG1a and anti-Hcp1 were used at a dilution of 1:1000, whereas anti-Tse3 was used at a dilution of 1:250. Monoclonal RNA polymerase antibody (W0023, Neoclone) directed against the β subunit of RNA polymerase and polyclonal anti-HsiB1 antibodies (20) were used at a dilution of 1:1000.
Monoclonal anti-polyhistidine antibody (H1029, Sigma) was used at a dilution of 1:5000. Primary antibodies were incubated with nitrocellulose membranes (Whatman) for 1–2 h followed by a 45-min incubation at room temperature with secondary antibody as appropriate (goat anti-rabbit-HRP (A6154, Sigma) or rabbit anti-mouse-HRP (A 9044, Sigma)) at a dilution of 1:5000. Western blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce) and a Las3000 Fuji imager.
Construction of Deletion Mutants in P. aeruginosa
P. aeruginosa deletion mutants were constructed as described previously (27) using the suicide plasmid pKNG101 (25, 28). To create PAKΔretSΔhsiC1, the plasmid pKNG-ΔhsiC1 was constructed as described above, maintained in the E. coli strain CC118 λpir, and mobilized in P. aeruginosa PAKΔretS using E. coli 1047 carrying the conjugative plasmid pRK2013 (29). Clones in which double recombination events occurred, resulting in the deletion of hsiC1, were selected on sucrose plates as described previously (25, 27). Deletion of hsiC1 was verified by PCR.
Co-purification of HsiB1 and HsiC1
Expression vectors pET-B1C1 and pET-B1C1N were transformed into E. coli B834(DE3) cells for production and purification of recombinant proteins. Cells were grown at 37 °C to an A600 of 0.6, and expression was subsequently induced using 1 mm isopropyl β-d-thiogalactoside (Sigma) for 16 h at 18 °C. Cells were pelleted at 4000 × g for 15 min at 4 °C and lysed by passage through a French press in lysis buffer (50 mm Tris, 500 mm NaCl, 5 mm DTT, 1 mm EDTA supplemented with Complete® protease inhibitor mixture). The cell extract was clarified at 40,000 × g for 45 min at 4 °C, and supernatants were applied to Ni+-nitrilotriacetic acid columns (Hi-Trap, GE Healthcare) for purification of His-tagged HsiB1 in complex with HsiC1 (no tag) by Ni+-nitrilotriacetic acid affinity chromatography using an imidazole gradient. Eluted fractions were analyzed by SDS-PAGE, Coomassie staining, and immunoblotting for the presence of HsiB1. The identity of HsiB1, HsiC1, and HsiC1(1–212) in the eluted fraction was also confirmed by peptide mass fingerprinting using trypsin digestion and a MALDI mass spectrometer (Protein and Nucleic Acid Chemistry Facility, Cambridge, UK). The obtained protein complexes were further purified prior to analysis by electron microscopy by gel filtration on a HiLoad 16/60 Superdex 75 column (GE Healthcare) using 50 mm Tris, 250 mm NaCl, 1 mm EDTA, 5 mm DTT, pH 8.
Preparation of Supernatant from P. aeruginosa Culture
P. aeruginosa strains were grown in tryptone soy broth overnight, subcultured to an A600 of 0.1, and grown to early stationary phase at 37 °C under agitation. Cells were separated from culture supernatants by centrifugation at 4000 × g at 4 °C. Cells were directly resuspended in 1× Laemmli buffer (30). Concentrated P. aeruginosa culture supernatant (10×) was prepared as follows. Proteins of the culture supernatant were precipitated using 6 m trichloroacetic acid (TCA; Sigma) at a final TCA concentration of 10%. Protein pellets were washed in 90% acetone, dried, suspended in 1× Laemmli buffer, and boiled at 95 °C for 10 min before SDS-PAGE.
Bacterial Competition Assay
The bacterial competition assay was carried out as described previously (20). Overnight cultures of indicated P. aeruginosa strains were incubated with overnight cultures of equivalent bacterial numbers of E. coli containing the plasmid pCR2.1 (carrying part of the lacZ gene) on LB agar for 5 h at 37 °C. Subsequently, patches of bacteria were recovered and resuspended in LB broth, and dilution series ranging from 100 to 10−7 were plated in triplicate on LB supplemented with 100 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal; Invitrogen), which allowed for colorimetric detection of lacZ-positive E. coli. For semiquantitative analysis of the amount of surviving E. coli, the β-galactosidase activity of each patch of the 10−2 dilution was measured using ortho-nitrophenyl β-galactoside (Sigma) as a substrate as described previously (31). Arbitrary units representing survival of E. coli were calculated using a modified formula used to calculate Miller units of β-galactosidase activity (31) and assuming equal β-galactosidase activity in any E. coli cell carrying the pCR2.1 vector.
Bacterial Two-hybrid Assay
Protein-protein interactions were analyzed using the BTH system as described previously (26). In brief, DNA fragments encoding the protein of interest were amplified by PCR, adding appropriate restriction sites, using P. aeruginosa PAK genomic DNA. DNA fragments encoding the proteins or protein domains of interest were cloned into plasmids pKT25 and pUT18C, which each encode for complementary fragments of the adenylate cyclase enzyme, as described previously (26), resulting in recombinant plasmids encoding an N-terminal fusion of the protein of interest with the T25 or T18 subunit of adenylate cyclase. Recombinant pKT25 and pUT18C plasmids were transformed simultaneously into the E. coli DHM1 strain, which is devoid of adenylate cyclase, and transformants were spotted onto MacConkey agar plates (Difco) supplemented with 1% maltose in the presence of 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 1 mm isopropyl β-d-thiogalactoside. Positive interactions were identified as red colonies on MacConkey agar plates after a 48-h incubation at 30 °C. The positive controls used in the study were pUT18C and pKT25 derivatives encoding the leucine zipper from GCN4, which readily dimerizes.
β-Galactosidase Assay
For quantitative analysis of BTH interactions, β-galactosidase activity of co-transformants scraped from MacConkey plates was measured as described previously, and activity was calculated in Miller units (31).
Negative Stain Electron Microscopy
Purified samples of HsiB1C1 (∼0.1 mg/ml) were negatively stained with 2% uranyl acetate on glow-discharged continuous carbon-coated 300-mesh copper grids (TAAB Laboratories). Electron microscopy was performed on a Philips CM200 FEG electron microscope operating at 200 kV. Images were recorded on a TVIPS F415 4000 × 4000 charge-coupled device camera at 38,000 magnification with a pixel size of 2.32 Å at the specimen level. Single particle analysis was performed using Boxer (EMAN2) (32) for particle selection and IMAGIC-5 software for reference-free and multireference alignments, multivariate statistical analysis, and classification (33).
Bioinformatics Analysis
P. aeruginosa gene and protein sequences were retrieved from the Pseudomonas Genome Database. Secondary structure prediction was carried out using web-based PSIPRED. Homology detection and structure prediction were carried out using the HHpred server from the Max Planck Institute for Developmental Biology.
RESULTS
HsiB1/HsiC1 Encoded by the H1-T6SS Form a Stable Complex
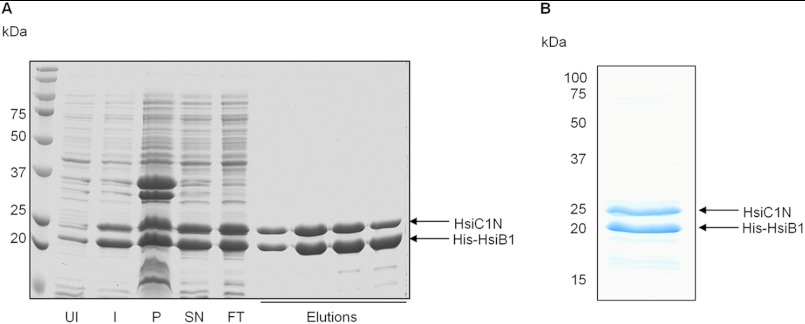
In V. cholerae the T6SS core components VipA and VipB were shown to form cogwheel-like structures in vitro that resemble the structure of the bacteriophage tail sheath composed of gp18 subunits (21, 22). Each of the three P. aeruginosa T6SSs encodes a VipA and a VipB homologue, namely HsiB1 and HsiC1 (H1-T6SS), HsiB2 and HsiC2 (H2-T6SS), and HsiB3 and HsiC3 (H3-T6SS) (5). Note that HsiC1, HsiC2, and HsiC3 have pairwise sequence identities between 38 and 46%, whereas for HsiB1, HsiB2, and HsiC3, it ranges between 25 and 35%. To investigate whether HsiB1 and HsiC1 interact and form comparable tubular structures, they were co-expressed from pET-B1C1 in E. coli B834(DE3) cells. HsiB1 was expressed with an N-terminal 6-histidine tag, whereas HsiC1 was untagged. Protein production was induced upon addition of isopropyl β-d-thiogalactoside, and His-tagged protein was purified by Ni+ affinity chromatography. Eluted fractions were analyzed for the presence of HsiB1 and HsiC1 by SDS-PAGE followed by Coomassie staining (Fig. 1, A and B, left panel) and immunoblotting (Fig. 1B, middle and right panels). The two bands in the eluted fractions were identified as HsiB1 and HsiC1 using antibodies directed against HsiB1 (Fig. 1B) and peptide mass fingerprinting using a trypsin digest (data not shown). Co-purification of HsiB1 and HsiC1 strongly suggests that HsiB1 and HsiC1 form a stable complex.
FIGURE 1.
HsiB1 forms a stable complex with HsiC1. A, co-purification of HsiB1 and HsiC1. HsiB1 was expressed with an N-terminal His tag from pET-B1C1, which also expressed untagged HsiC1. His-HsiB1 was purified by Ni+ affinity chromatography in complex with untagged HsiC1. Samples were analyzed for the presence of both HsiB1 and HsiC1 by SDS-PAGE followed by Coomassie staining. UI, uninduced cells; I, 16 h after protein induction; SN, supernatant or soluble fraction; P, pellet or insoluble fraction; FT, flow-through; Elutions, fractions collected after elution of proteins by imidazole gradient. B, identification of components of the purified HsiB1C1 complex. The purified protein complex was analyzed by SDS-PAGE followed by Coomassie staining (left panel). HsiB1 was further identified by immunoblotting using anti-HsiB1 (middle panel) and monoclonal anti-His antibody (right panel), respectively. The molecular mass is indicated on the left in kDa. Note that the identity of the two apparent bands was also confirmed by mass spectrometry; the lower band is HsiB1 and the upper band is HsiC1 (not shown).
The HsiB1/C1 Interaction Is Specific
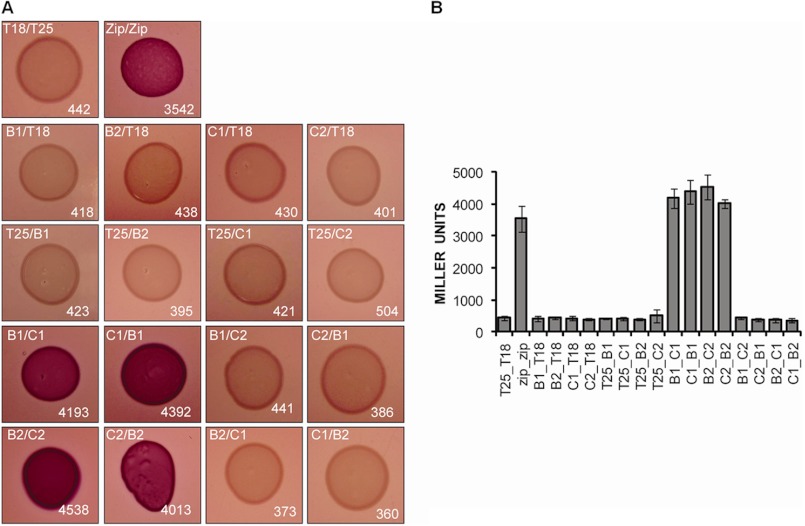
Because P. aeruginosa has three T6SSs, each of which encodes a VipA and VipB homologue, we wondered whether the interaction between HsiB and HsiC proteins is specific or whether cross-interactions are possible between proteins encoded by different T6SS clusters. We used the bacterial two-hybrid system in which proteins of interest are fused to either the T18 or the T25 subunit of adenylate cyclase. Upon interaction of the two target proteins, adenylate cyclase activity is restored, and positive interactions are characterized by dark red colonies on MacConkey agar plates. These interactions can be quantified by measurement of β-galactosidase activity. The leucine zipper protein (zip), which readily forms a dimer, is used as a positive control (26). Although HsiB1 interacted strongly with HsiC1 (β-galactosidase activity, 4193 ± 302 Miller units; positive control (zip/zip), 3542 ± 412 Miller units; negative control (T25/T18), 442 ± 73 Miller units) and HsiB2 interacted strongly with HsiC2 (β-galactosidase activity, 4538 ± 381 Miller units), no interaction was detectable between HsiB1 and HsiC2 (β-galactosidase activity, 441 ± 39 Miller units) or HsiB2 and HsiC1 (β-galactosidase activity, 373 ± 64 Miller units) (Fig. 2). Comparable results were obtained independent of which protein was fused to the T25 or T18 subunit of adenylate cyclase (data not shown). These results indicate that the interaction between the VipA and VipB homologues of each P. aeruginosa T6SS is specific within the system, and no cross-interaction occurs.
FIGURE 2.
The interaction between HsiB1 and HsiC1 is specific. Bacterial two-hybrid analysis of possible interactions among HsiB1, HsiC1, HsiB2, and HsiC2 is shown. Various combinations of recombinant pKT25 and pUT18C plasmids harboring proteins of interest were co-transformed into E. coli DHM1, and β-galactosidase activity of co-transformants was measured after plating on MacConkey agar plates. Plasmid combinations are annotated in the order pKT25/pUT18C fusion. Experiments were carried out in triplicates, and error bars represent the S.D. Zip, leucine zipper domain of the yeast transcription factor GCN4 (positive control); T18, empty vector pUT18C; T25, empty vector pKT25. For T25/T18 fusion proteins, B1 represents HsiB1, C1 represents HsiC1, B2 represents HsiB2, and C2 represents HsiC2. A, images of colonies formed by co-transformants on MacConkey Agar plates (dark red colonies indicate a positive interaction). Plasmid combinations are indicated in the top left corner of every panel in the order T25/T18 fusion protein. The strength of interaction was investigated by measuring the β-galactosidase activity of cells in the respective colonies, and the average activity in Miller units is indicated in the bottom right corner of each image and is represented in B. B, graphic representation of the β-galactosidase activity of co-transformants after incubation on MacConkey agar. Experiments were carried out in triplicates, and error bars represent the S.D. Plasmid combinations are indicated in the order pKT25/pUT18C and abbreviations apply as described above.
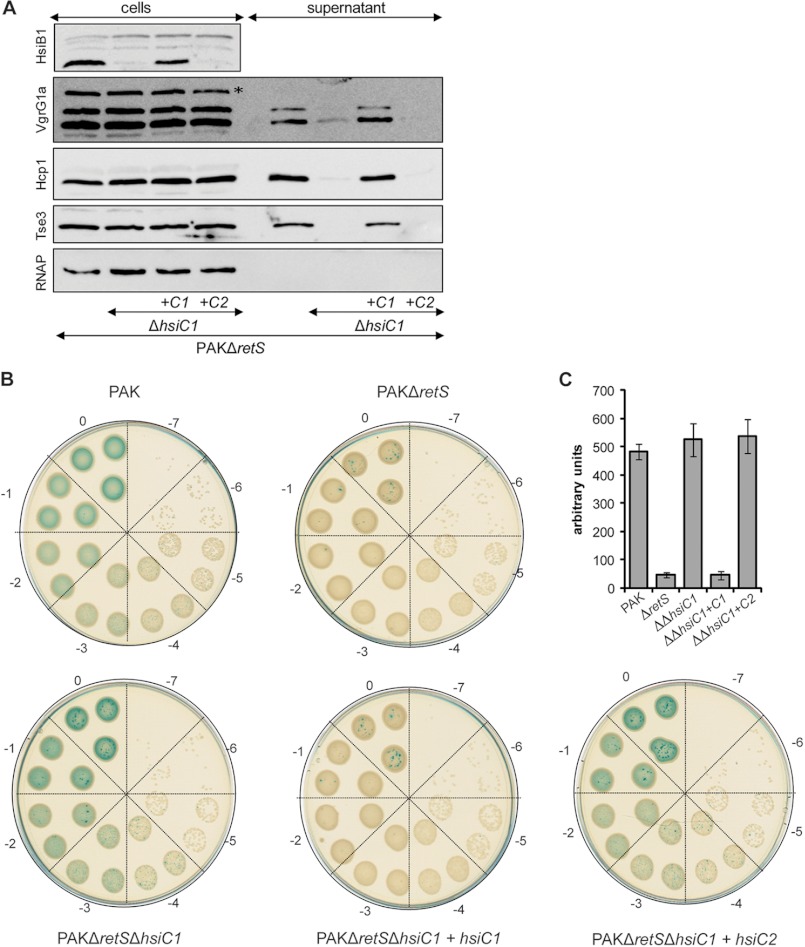
The observation that HsiB2 does not strongly interact with HsiC1 and HsiB1 does not strongly interact with HsiC2 suggests that there is no redundancy between multiple T6SSs within one organism. To further confirm this result, we constructed a deletion mutant of hsiC1 in PAKΔretS, a strain in which the H1-T6SS is constitutively active (34). We then monitored the ability of P. aeruginosa to secrete Hcp1, VgrG1a/c, and the toxin Tse3 as a marker for a functional H1-T6SS (4) (Fig. 3A). Monitoring of cytosolic RNA polymerase in both whole cell lysates and the supernatant fraction assured that no significant cell lysis had occurred (Fig. 3A, bottom panel). PAKΔretSΔhsiC1 secreted significantly less Hcp1, VgrG1a, and Tse3 than the parental strain in line with the hypothesis that HsiC1 is an essential H1-T6SS core component (Fig. 3A, second panel). This reduction in Hcp1, VgrG1a, and Tse3 secretion was fully recovered after reintroducing hsiC1 into the chromosome of PAKΔretSΔhsiC1 using the complementing plasmid mini-CTX-C1. However, expression of HsiC2 from mini-CTX-C2 did not restore secretion of Hcp1, VgrG1a, or Tse3 in PAKΔretSΔhsiC1, indicating that HsiC2 cannot fulfill the function of HsiC1.
FIGURE 3.
HsiC2 cannot fulfill the function of HsiC1. The H1-T6SS component hsiC1 was deleted in PAKΔretS, resulting in PAKΔretSΔhsiC1. The genes hsiC1 and hsiC2 were integrated into the chromosome of PAKΔretSΔhsiC1 using mini-CTX-C1 (PAKΔretSΔhsiC1+C1) and mini-CTX-C2 (PAKΔretSΔhsiC1+C2), respectively. A, expression of HsiC2 does not restore a functional H1-T6SS in PAKΔretSΔhsiC1. The secretion profile of PAKΔretSΔhsiC1, PAKΔretSΔhsiC1+C1, and PAKΔretSΔhsiC1+C2 was compared with that of PAKΔretS (H1-T6SS induced). Strains were grown to late exponential phase before culture supernatants were separated from bacterial cells, and secretion of VgrG1a, Hcp1, and Tse3 was monitored by Western blotting using polyclonal antibodies directed against VgrG1a/c (second panel), Hcp1 (third panel), and Tse3 (fourth panel). RNA polymerase was monitored in both whole cell lysates and culture supernatants using monoclonal antibody directed against the β-subunit of RNA polymerase (RNAP) (150 kDa). The absence of RNA polymerase in the culture supernatant ensured the absence of cell lysis. Anti-VgrG1a antibody also detects VgrG1b in whole cell extract that is secreted independently of a functional H1-T6SS and is indicated by *. Stability of the sheath component HsiB1 was monitored in whole cell lysates using polyclonal anti-HsiB1 antibody (first panel). B, impact of hsiC1 or hsiC2 expression in PAKΔretSΔhsiC1 on complementing H1-T6SS-mediated bacterial killing. Overnight cultures of the P. aeruginosa strains PAK (H1-T6SS-negative), PAKΔretS (constitutive H1-T6SS expression), PAKΔretSΔhsiC1, PAKΔretSΔhsiC1+C1, and PAKΔretSΔhsiC1+C2 were mixed with equivalent numbers of E. coli carrying a plasmid expressing β-galactosidase. Mixed cultures were spotted onto LB agar and incubated for 5 h. Dilution series of recovered bacteria ranging from 0 (undiluted) to −7 (107-fold diluted) were spotted onto LB containing X-gal. The level of visible blue color indicates survival of E. coli. C, semiquantitative analysis of bacterial killing. Graphic representation of the β-galactosidase activity recovered from bacterial patches is shown in B. Bacteria were recovered from the patches of the 10−2 dilution, and β-galactosidase activity was determined using ortho-nitrophenyl β-galactoside as a substrate. Error bars represent S.D. of triplicates of three independent patches. ΔretS, PAKΔretS; ΔΔhsiC1, PAKΔretSΔhsiC1; ΔΔhsiC1+C1, PAKΔretSΔhsiC1; ΔΔhsiC1+C2, PAKΔretSΔhsiC1+C2.
Additionally, it is known from studies in V. cholerae that VipA and VipB require each other for their stability (22). We hence checked the stability of HsiB1 in PAKΔretSΔhsiC1 whole cell lysates by immunoblotting using the anti-HsiB1 antibody. We observed a reduction in HsiB1 level in PAKΔretSΔhsiC1 compared with the parental strain PAKΔretS (Fig. 3A, top panel). This instability of HsiB1 was reversed upon expression of hsiC1, confirming that HsiC1 is required for the stability of HsiB1. Furthermore, production of HsiC2 did not stabilize HsiB1 (Fig. 3A, top panel).
A very sensitive assay to monitor T6SS activity is based on the ability of P. aeruginosa to kill other Gram-negative bacteria, such as E. coli, in an H1-T6SS-dependent manner due to the activity of the Tse1–3 toxins (35, 36). Using this method, we further assessed the potential ability of HsiC2 to partially compensate for the absence of HsiC1. The E. coli strain harbors a plasmid that allows α-complementation of the β-galactosidase enzyme (pCR2.1), and resulting bacterial colonies appear blue on LB plates supplemented with X-gal. After incubation of PAKΔretS on solid medium together with the E. coli strain, hardly any blue colonies were observed when a dilution series of the mixed bacterial culture was spotted onto LB agar supplemented with X-gal. In contrast, after incubation of E. coli with a P. aeruginosa strain that lacked an active H1-T6SS (PAK wild-type), little to no E. coli killing was observed as seen by the significant number of blue colonies detectable (Fig. 3B). Incubation of E. coli with the hsiC1 mutant led to the recovery of many blue E. coli, showing that deletion of hsiC1 renders the H1-T6SS non-functional as already seen by its lack of ability to secrete Hcp1, VgrG1a/c, and Tse3 (Fig. 3A). This lack of ability of an hsiC1 mutant to efficiently kill E. coli was complemented by reintroduction of hsiC1 into the chromosome of the hsiC1 mutant using mini-CTX-C1 but not by expression of hsiC2 using mini-CTX-C2 (Fig. 3B). Semiquantitative analysis (see “Experimental Procedures”) of the recovered E. coli following incubation with P. aeruginosa reflects the qualitative result (Fig. 3C). Taken together, these data indicate that the evolutionarily distinct T6SSs in P. aeruginosa function independently, and no cross-talk between the individual components occurs as shown here with HsiB/HsiC.
The HsiB1C1 Complex Forms Tubules and Cogwheels
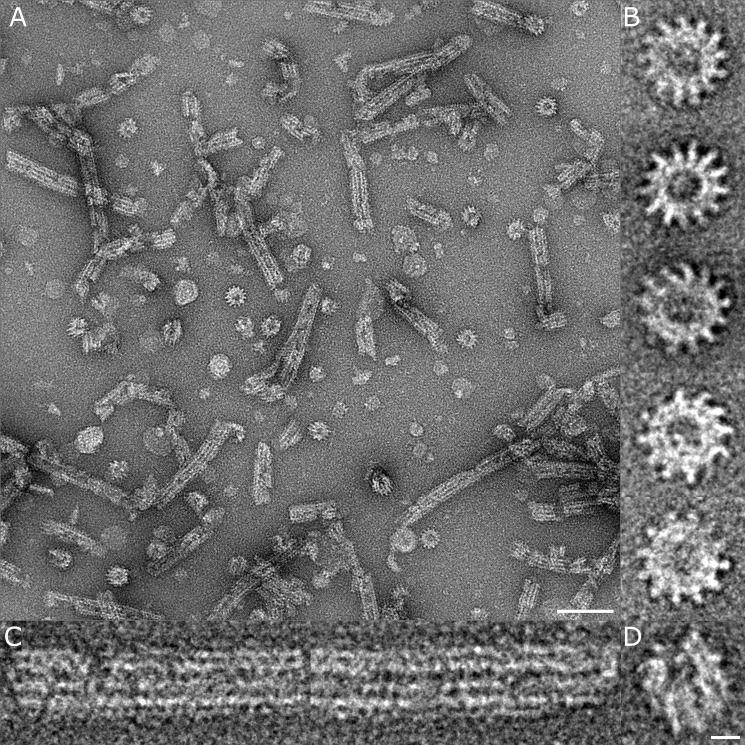
As discussed above, recombinant HsiB1 and HsiC1 forms a complex when co-expressed in E. coli (Fig. 1). The protein complex eluted from the Ni+ affinity chromatography column was further purified by size exclusion chromatography (see “Experimental Procedures”) before analysis by electron microscopy. Negatively stained HsiB1C1 complexes form a variety of tubular and cogwheel structures (Fig. 4). Individual raw images of cogwheels show predominantly 12-fold symmetry, although we also observed cogwheels with 13-fold symmetry (Fig. 4B). The tubular structures show discontinuous staining, although the predominant feature is four horizontal striations (Fig. 4C). Several oblique views show that the tubule structures are made up of layers of cogwheels (Fig. 4D), suggesting that the cogwheel particles we observed are actually fragmented tubules. Measurements of individual raw images of both cogwheels and tubules give an outer diameter of the tubule of ∼250–330 Å with an internal diameter of ∼100 Å (Fig. 5, A and B). We performed an initial single particle image analysis separately on individual cogwheels and short fragments of tubular side views. From the symmetry analysis (37) of only translational centered cogwheels, we found possible oligomeric arrangements of 12- and 13-fold symmetry (Fig. 5, A, B, D, and E). Interestingly, both projections show similar dimensions, suggesting that an additional protein unit within the tubule ring can be accommodated. A summation of 300 individual raw images of tubular assemblies shows a striking striated pattern comprising four layers (Fig. 5, C and F) representing the spokes of cogwheel structures in projection. Based on these observations, we also drew a sketch of a cogwheel and tubule with a 12-fold symmetry (Fig. 5G). The structures we observed are very similar to those reported for V. cholerae T6SS core components VipA and VipB both in overall shape and dimensions (21, 22), confirming that HsiB1/HsiC1 are equivalent structural orthologues in P. aeruginosa.
FIGURE 4.
Negative stain electron microscopy images of the HsiB1C1 complex shows tubular and cogwheel structures. A, micrograph of negatively stained HsiB1C1 complexes showing both tubular and cogwheel-like structures. The scale bar corresponds to 100 nm. B, examples of raw images of cogwheels exhibiting both 13- and 12-mer arrangements with a central hole of ∼100 Å. C, examples of tubules that show a four-layered striation with discontinuous staining. D, oblique view of a short tubule structure that clearly shows the central channel. The scale bar corresponds to 10 nm (applies also to B and C).
FIGURE 5.
Dimensions and image processing of the HsiB1C1 complex. A and B, top view of negatively stained HsiB1C1 cogwheel structures showing 12- and 13-fold symmetry, respectively. C, side view of single negatively stained HsiB1C1 tubule structure. The bars in both panels correspond to 250, 100, and 300 Å, respectively, and the scale bars correspond to 10 nm. D and E, class sums of negatively stained HsiB1C1 cogwheel structures showing 12- and 13-fold symmetry, respectively. F, total sum of 300 individual raw images of mixed tubular assemblies and shortened tubular structures treated as single particles. A four-layered striated structure is consistent with cogwheel structures in projection. The scale bar corresponds to 35 nm. G, sketch of a cogwheel and tubule with a 12-fold symmetry.
We also carried out immuno-EM labeling with an anti-His antibody against His-tagged HsiB1 (supplemental Fig. 1B) and supplemental Experimental Procedures. We observed a number of tubular clusters bound by the antibody, indicating that the N-terminal histidine tag for HsiB1 is surface-accessible on the tubules. To confirm this, we also used a 5-nm gold particle to specifically label available histidine tags. We found gold particles decorating the tubular structures, confirming that the N terminus of HsiB1 is surface-accessible and that HsiB1 is part of the observed tubular structures (supplemental Fig. 1C).
The HsiB1C1 Combination Shows Sequence and Structural Similarity to gp18
As discussed above, the macromolecular tubules formed by HsiB1 in complex with HsiC1 show remarkable structural resemblance to the tail sheath of the T4 bacteriophage (38). However, no significant similarity can be identified at the amino acid sequence level using standard alignment programs, such as ClustalW and TCoffee. In fact, sequence similarity among sheath proteins of various phages is also rather limited. In contrast, HsiC1, HsiC2, and HsiC3 have pairwise sequence identities between 38 and 46%, and their secondary structure predictions are almost identical (supplemental Fig. 2), suggesting a common mechanism of sheath protein assembly among the three evolutionarily distinct T6SSs in P. aeruginosa. Homology detection and structure prediction by HMM-HMM comparison (HHpred) (39, 40) reveals that HsiC1, HsiC2, and HsiC3 show similarity at their C terminus to members of the protein family PFO4984 (including Protein Data Bank codes 3HXL and 3LML), which represents tail sheath proteins from various bacteriophages (Fig. 6). The similarity encompasses amino acids 332–489 at the C terminus of HsiC1, amino acids 325–485 at the C terminus of HsiC2, and amino acids 323–487 at the C terminus of HsiC3. This observation is the first direct bioinformatics evidence that proteins of this family (i.e. TssC) are related to bacteriophage sheath components and furthermore strengthens the model of the T6SS as an inverted phage.
FIGURE 6.
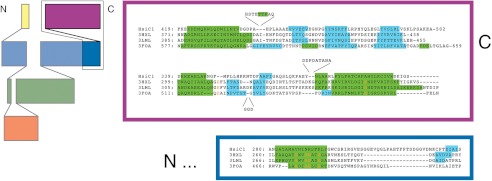
Structural conservation between HsiC and phage tail sheath proteins. On the left is a topology diagram of the domain organization of gp18 proteins (adapted from Leiman and Shneider (57)). The C-terminal half of HsiC shows homology to the C-terminal part of gp18, which is framed in black. On the right is a structure-based sequence alignment of HsiC1 (top sequence) to the bacteriophage gp18 proteins DSY3957 (second sequence), Lin1278 (third sequence), and T4 gp18 (bottom sequence). Predicted (HsiC1 and Protein Data Bank code 3FOA beyond residue 510) and observed (gp18 proteins) secondary structure elements are highlighted in green (helices) and cyan (strands). The colored boxes correspond to the domains framed in black on the left. Residues conserved in at least three sequences are shown in orange. Despite the marginal sequence similarity, the secondary structure elements are largely conserved.
The N Terminus of HsiC1 Is Sufficient for Interaction with HsiB1
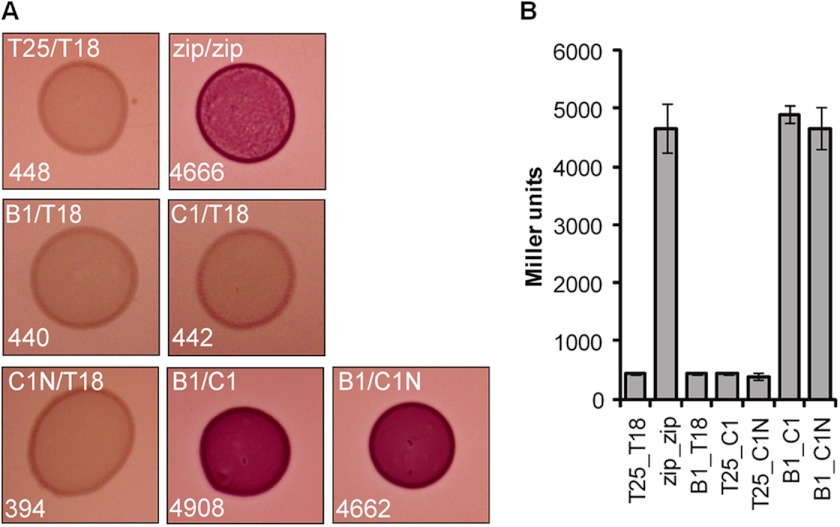
VipA/VipB and HsiB1/HsiC1 belong to the TssB/TssC family of proteins. The interaction between TssB-like and TssC-like proteins of the T6SS has been demonstrated in other organisms, such as Francisella tularensis (IglA/IglB) (41) and Burkholderia cenocepacia (BcsLB/BcsKC) (42). A conserved α-helix has been identified in IglA (TssB-like protein) that is crucial for the interaction with IglB (TssC-like protein) (43). This α-helix is also found in HsiB1, HsiB2, and HsiB3 (20). Furthermore, analysis of the TssC-like protein BcsKC encoded by B. cenocepacia has shown that its first 212 aa are sufficient for its interaction with BcsLB (TssB-like protein) (42). Comparison of the secondary structure prediction of BcsKC with that of HsiC1 revealed an almost identical secondary structure (data not shown). Based on these results, we investigated whether the first 212 residues of HsiC1 are sufficient for its interaction with HsiB1 and whether this complex can still form a macromolecular structure. To analyze the impact of the deletion of the C terminus of HsiC1 on the interaction with HsiB1, we used the bacterial two-hybrid system as described above. The DNA fragment encoding HsiC1(1–212) was cloned into pUT18C, resulting in pUT18C-C1N expressing an N-terminal fusion of the T18 adenylate cyclase subunit to the first 212 aa of HsiC1. Bacterial two-hybrid analysis showed that HsiC1 and HsiC1(1–212) interact to the same extent with HsiB1 as shown by red colony formation on MacConkey agar and a β-galactosidase activity of 4908 ± 148 and 4662 ± 352 Miller units, respectively (Fig. 7). In conclusion, the N-terminal region of HsiC1 is sufficient for its interaction with HsiB1, suggesting a conserved mechanism of interaction between TssB-like and TssC-like components of different T6SSs.
FIGURE 7.
The N-terminal 212 residues of HsiC1 are sufficient for interaction with HsiB1. HsiC1(1–212) is sufficient for interaction with HsiB1 as determined by BTH. Various combinations of recombinant pKT25 and pUT18C plasmids harboring proteins of interest were co-transformed into E. coli DHM1, and β-galactosidase activity of co-transformants was measured after plating on MacConkey agar plates. Plasmid combinations are annotated in the order pKT25/pUT18C fusion. Experiments were carried out in triplicates, and error bars represent the S.D. zip, leucine zipper domain of the yeast transcription factor GCN4 (positive control); T18, empty vector pUT18C; T25, empty vector pKT25. For the T25/T18 fusion proteins, B1 represents HsiB1, C1 represents HsiC1, and C1N represents HsiC1(1–212). A, images of colonies formed by co-transformants on MacConkey Agar plates (dark red colonies indicate a positive interaction). Plasmid combinations are indicated in the top left corner of every image in the order T25/T18 fusion protein. The strength of interaction was investigated by measuring the β-galactosidase activity of cells in the respective colonies, and the average activity in Miller Units is indicated in the bottom left corner of each image and is represented in B. B, graphic representation of the β-galactosidase activity of co-transformants after incubation on MacConkey agar. Experiments were carried out in triplicates, and error bars represent the S.D. Plasmid combinations are indicated in the order pKT25/pUT18C, and abbreviations apply as described above.
The C Terminus of HsiC1 Is Required for Cogwheel and Tubule Formation
Because the N-terminal 212 amino acids of HsiC1 are sufficient for the interaction between HsiB1 and HsiC1 as detected using a bacterial two-hybrid assay, we investigated whether HsiB1 and HsiC1(1–212) can also be co-purified as a complex when co-expressed in E. coli. HsiB1 and HsiC1(1–212) were produced in E. coli B834(DE3) cells from pET-B1C1N, leading to expression of 6-histidine-tagged HsiB1 and untagged HsiC1(1–212) upon addition of isopropyl β-d-thiogalactoside. Cell lysates were applied to a Ni+ affinity chromatography column, and eluted fractions were analyzed for the presence of HsiB1 and HsiC1(1–212) using SDS-PAGE followed by Coomassie staining (Fig. 8A). The two apparent bands were identified as HsiB1 and HsiC1(1–212) by peptide mass fingerprinting. Histidine-tagged HsiB1 was further confirmed by immunoblotting using monoclonal anti-histidine antibody and polyclonal anti-HsiB1 antibody (data not shown). The eluted fractions containing both HsiB1 and HsiC1(1–212) were collected and further purified using gel filtration chromatography (Fig. 8B) before the potential protein complex was analyzed by electron microscopy. No macromolecular structures were visible under the electron microscope (supplemental Fig. 1A). This indicates that although HsiB1 and HsiC1(1–212) can still form a complex the C terminus of HsiC1 is required for the formation of tubular structures as observed with HsiB1C1.
FIGURE 8.
The N-terminal 212 residues of HsiC1 are sufficient for complex formation with HsiB1. Co-purification of HsiB1 and HsiC1(1–212) (indicated as HsiC1N) is shown. HsiB1 was expressed with an N-terminal His tag from pET-B1C1N, which also expressed the untagged N-terminal region of HsiC1 (HsiC1(1–212)). His-HsiB1 was purified by Ni+ affinity chromatography in complex with untagged HsiC1(1–212). A, co-purification of HsiB1 and HsiC1(1–212). Fractions collected after elution of proteins by imidazole gradient were analyzed by SDS-PAGE followed by Coomassie staining. Uninduced (UI) and induced (I) whole cell lysates were analyzed. SN, supernatant or soluble fraction; P, pellet or insoluble fraction; FT, flow-through; Elutions, fractions collected after elution of proteins by imidazole gradient. B, pooled fractions analyzed in A containing both HsiB1 and HsiC1(1–212) were collected and further purified by size exclusion chromatography. The resulting protein complex was analyzed by SDS-PAGE and Coomassie staining. Analysis of the two apparent bands by mass spectrometry identified the lower band as His-HsiB1 and the upper band as HsiC1(1–212). The molecular mass is indicated on the left of each image in kDa.
DISCUSSION
It has become clear over the last few years that if one wants to understand the T6SS mechanism one should follow the bacteriophage trail. The bacteriophage structure is essentially made of a baseplate that includes as many as 12 distinct proteins (38). In the T6SS, at least one protein, HsiF/TssE, has been shown to be a baseplate homologue (19). A hub is assembled at the center of the baseplate, or puncturing device, that consists of the gp5 and gp27 proteins that are homologous to the T6SS component VgrG (17, 44). The puncturing device is pushed through the baseplate by a tube that results from the polymerization of gp19, the homologue of Hcp in the T6SS. The unidirectional movement of the tail tube is a direct consequence of the contraction of the gp18-containing tail sheath surrounding the tube. In the T6SS, it has been proposed from previous studies in V. cholerae that two proteins, VipA and VipB, form tubular complexes (21, 22), which could resemble the bacteriophage tail sheath. In this study, we showed that this is a general feature of the T6SS because HsiB1 and HsiC1 from P. aeruginosa form similar structures, which we observed by negative stain electron microscopy. It is also striking that these structures are very similar to the P. aeruginosa R-type pyocins (45). These pyocins are part of the family of contractile tail systems and are close relatives of the phage P2 tail (46). Although we observed tubule structures primarily with 12-fold symmetry, we observed a small number of 13-fold symmetric structures. It is possible that this heterogeneity is due to the co-expression and assembly of the recombinant complex and/or the biochemical conditions used for purification. It is notable that we observed fragmented structures by EM, suggesting that the HsiBC tubules are only partially assembled under these conditions and can form aggregates. Nevertheless, our images provide compelling evidence to show that the HsiBC complex can form distinct tubule structures with an inner diameter of ∼100 Å that comprise repeated layers of symmetrically arranged complexes.
gp18 encoded by the bacteriophage T4 is a 659-amino acid-long protein with a molecular mass of 71 kDa, and its structure has been partly resolved (24). This protein polymerizes into a tail sheath that includes 138 of these subunits. The sheath is 925 Å long in its extended version (420 Å in the contracted form), forms a six-start helix, and surrounds the rigid gp19 tube, which also contains 138 subunits (47). Such structure results from the assembly of rings of six gp18 subunits stacked on top of each other and that are slightly rotated by 17.2° from one ring to the other in the extended version. A top view of the sheath reconstructed by cryoelectron microscopy shows a cogwheel-like structure with an apparent 12-fold symmetry (38). The sheath is contractile and could be in an extended or contracted form. In the extended version, the baseplate is seen as a hexagonal dome-shaped structure (47), whereas in its contracted version, the baseplate has a star-shaped structure (48). By comparing the shape of the density corresponding to gp18 in the extended and contracted versions, three domains were identified whose position is slightly modified in each respective conformation. Previous studies showed that the C terminus of gp18 is located toward the center of the ring and is involved in interaction within the sheath, whereas the N terminus is associated with the cogwheel spikes and is not involved in interaction (49). The contraction of the sheath is likely induced upon phage landing and contact with the lipopolysaccharides (LPS) on the target bacterial cell. This induces a change in the baseplate, from dome-shaped to star-shaped, that is thus at the origin of the force exerted onto the sheath and that results in its contraction (50). A similar suggestion could be made for the T6SS because the assembled HsiBC tubule structure we observed could change conformation once in contact with the inner face of the cytoplasmic membrane. In contrast to the phage, the contracted T6SS structure will then be disassembled and recycled by the ATPase activity of the T6SS component ClpV as has been shown for the VipA and VipB homologues in V. cholerae (21, 22, 51, 52).
Thus, it is a possibility that gp18 is a combination of TssB and TssC members. The length of HsiB1 and HsiC1 is 172 and 498 aa, respectively, and the molecular mass is 18.8 and 55.77 kDa, respectively. The molecular mass of the combined HsiBC (670-aa) complex would be 74.5 kDa compared with the 71 kDa of the 659-aa-long gp18. Although the gp18-containing tubules are quite similar to the TssB/TssC-like tubules, only weak amino acid homologies could be observed between the T6SS components and gp18. The three-dimensional structure of gp18 has been obtained from truncated versions of the protein that lacked the ability to polymerize (53–55). In particular, it was shown that the Ile507–Gly530 sequence is key to the assembly into tubules. The three-dimensional structure of a soluble version (aa 1–510) of gp18 shows three distinct domains. Some of these domains are made by a distal region in the primary sequence, such as domain II, which includes residues 88–97 and 189–345, whereas domain III includes residues 20–87 and 346–510 (24). From this observation, it is clear that the homologies between gp18 and the HsiBC complex cannot be found by simply looking at a linear connection between HsiB and HsiC amino acid sequences and that structure determination of the HsiBC complex will be needed to further elucidate these comparisons. For example, using the amino acid sequence of gp18 to perform a BLAST analysis only retrieved the tail sheath proteins that are found in close relatives of phage T4 and not proteins constituting the sheath of other phages. However, using HHpred analysis, we have been able to match the C-terminal region of HsiC1 (aa 282–488) with a domain whose structure is available, Protein Data Bank code 3HXL (aa 262–437) (Fig. 6) and that corresponds to the product of a prophage infecting Desulfitobacterium hafniense. This is now classified as a domain from the gp18 family and is the first clear evidence that HsiC and gp18 share similarities. Furthermore, we showed that the deletion of this region in HsiC1 eliminates the ability of the HsiB1C1 complex to form the tubules and cogwheels. This is again in agreement with the observations that the C terminus of gp18 is important for the formation of T4 polysheaths and that the gp18 C terminus appears in the cryo-EM reconstruction at the center of the cogwheels, making interactions within the sheath. In the case of HsiB1, the only relevant region that could be identified is a stretch of 11 residues between positions 106 and 116 that displays 55% identity with a coat protein from filamentous phage ph75 (56). This stretch of amino acids is also very well conserved in HsiB2 and HsiB3, suggesting that it might be key to the function.
It is of course highly relevant that the genes encoding the TssB and TssC homologues are found successively in tandem in almost all the T6SS clusters, which suggests that both genes could have fused into one single gene, such as the one encoding gp18. Conserved Pfam domains in the TssB and TssC families of proteins are also identified as DU770 and DUF877. It is interesting to note that these domains are found combined in a number of proteins, such as the 622-aa-long γ proteobacterium HdN1, thus supporting the hypothesis that they could have been fused or split during evolution.
In conclusion, our data further support that the TssB/TssC family of proteins forms a macromolecular complex that assembles into tubules resembling the phage tail sheath. Further demonstration that this complex is structurally similar to gp18 is currently underway by attempting to resolve the three-dimensional structure of the HsiBC complex in which HsiC is truncated from the C terminus. This has proven to work in the case of gp18. Obtaining the three-dimensional structure of a soluble HsiBC complex will be a major step forward and will help refine the model that is presented in this work.
Acknowledgments
We thank Alida Haworth for help with image processing and Abderrahman Hachani and Cerith Jones for helpful discussions.

This article contains supplemental Experimental Procedures and Figs. 1 and 2.
- T6SS
- type VI secretion system
- BTH
- bacterial two-hybrid
- zip
- leucine zipper protein
- HMM
- hidden Markov model
- aa
- amino acids.
REFERENCES
- 1. Silverman J. M., Brunet Y. R., Cascales E., Mougous J. D. (2012) Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66, 453–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bleves S., Viarre V., Salacha R., Michel G. P., Filloux A., Voulhoux R. (2010) Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int. J. Med. Microbiol. 300, 534–543 [DOI] [PubMed] [Google Scholar]
- 3. Jones A. M., Dodd M. E., Morris J., Doherty C., Govan J. R., Webb A. K. (2010) Clinical outcome for cystic fibrosis patients infected with transmissible Pseudomonas aeruginosa: an 8-year prospective study. Chest 137, 1405–1409 [DOI] [PubMed] [Google Scholar]
- 4. Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., Goodman A. L., Joachimiak G., Ordoñez C. L., Lory S., Walz T., Joachimiak A., Mekalanos J. J. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Filloux A., Hachani A., Bleves S. (2008) The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 6. Bingle L. E., Bailey C. M., Pallen M. J. (2008) Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11, 3–8 [DOI] [PubMed] [Google Scholar]
- 7. Brencic A., Lory S. (2009) Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72, 612–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goodman A. L., Merighi M., Hyodo M., Ventre I., Filloux A., Lory S. (2009) Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 23, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moscoso J. A., Mikkelsen H., Heeb S., Williams P., Filloux A. (2011) The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13, 3128–3138 [DOI] [PubMed] [Google Scholar]
- 10. Aschtgen M. S., Thomas M. S., Cascales E. (2010) Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP … what else? Virulence 1, 535–540 [DOI] [PubMed] [Google Scholar]
- 11. Felisberto-Rodrigues C., Durand E., Aschtgen M. S., Blangy S., Ortiz-Lombardia M., Douzi B., Cambillau C., Cascales E. (2011) Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 7, e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma L. S., Narberhaus F., Lai E. M. (2012) IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J. Biol. Chem. 287, 15610–15621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aschtgen M. S., Gavioli M., Dessen A., Lloubès R., Cascales E. (2010) The SciZ protein anchors the enteroaggregative Escherichia coli type VI secretion system to the cell wall. Mol. Microbiol. 75, 886–899 [DOI] [PubMed] [Google Scholar]
- 14. Sexton J. A., Miller J. L., Yoneda A., Kehl-Fie T. E., Vogel J. P. (2004) Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect. Immun. 72, 5983–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. (2007) Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hachani A., Lossi N. S., Hamilton A., Jones C., Bleves S., Albesa-Jové D., Filloux A. (2011) Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J. Biol. Chem. 286, 12317–12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., Burley S. K., Almo S. C., Mekalanos J. J. (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pukatzki S., McAuley S. B., Miyata S. T. (2009) The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12, 11–17 [DOI] [PubMed] [Google Scholar]
- 19. Lossi N. S., Dajani R., Freemont P., Filloux A. (2011) Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system, in Pseudomonas aeruginosa. Microbiology 157, 3292–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lossi N. S., Manoli E., Simpson P., Jones C., Hui K., Dajani R., Coulthurst S. J., Freemont P., Filloux A. (2012) The archetype Pseudomonas aeruginosa proteins TssB and TagJ form a novel sub-complex in the bacterial Type VI secretion system. Mol. Microbiol. 86, 437–456 [DOI] [PubMed] [Google Scholar]
- 21. Basler M., Pilhofer M., Henderson G. P., Jensen G. J., Mekalanos J. J. (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A. (2009) Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Filloux A. (2009) The type VI secretion system: a tubular story. EMBO J. 28, 309–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aksyuk A. A., Leiman P. G., Kurochkina L. P., Shneider M. M., Kostyuchenko V. A., Mesyanzhinov V. V., Rossmann M. G. (2009) The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 28, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaniga K., Delor I., Cornelis G. R. (1991) A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109, 137–141 [DOI] [PubMed] [Google Scholar]
- 26. Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vasseur P., Vallet-Gely I., Soscia C., Genin S., Filloux A. (2005) The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151, 985–997 [DOI] [PubMed] [Google Scholar]
- 28. Herrero M., de Lorenzo V., Timmis K. N. (1990) Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172, 6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Figurski D. H., Helinski D. R. (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 31. Miller J. H. (1992) A Short Course in Bacterial Genetics: a Laboratory Manual and Handbook for Escherichia coli and Related Bacteria, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 33. van Heel M., Gowen B., Matadeen R., Orlova E. V., Finn R., Pape T., Cohen D., Stark H., Schmidt R., Schatz M., Patwardhan A. (2000) Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 33, 307–369 [DOI] [PubMed] [Google Scholar]
- 34. Goodman A. L., Kulasekara B., Rietsch A., Boyd D., Smith R. S., Lory S. (2004) A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7, 745–754 [DOI] [PubMed] [Google Scholar]
- 35. Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., Silverman J. M., Ohlson B. B., Hicks K. G., Plemel R. L., Li M., Schwarz S., Wang W. Y., Merz A. J., Goodlett D. R., Mougous J. D. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Russell A. B., Hood R. D., Bui N. K., LeRoux M., Vollmer W., Mougous J. D. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dube P., Tavares P., Lurz R., van Heel M. (1993) The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 12, 1303–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leiman P. G., Arisaka F., van Raaij M. J., Kostyuchenko V. A., Aksyuk A. A., Kanamaru S., Rossmann M. G. (2010) Morphogenesis of the T4 tail and tail fibers. Virol. J. 7, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Söding J. (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 [DOI] [PubMed] [Google Scholar]
- 40. Söding J., Biegert A., Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Bruin O. M., Ludu J. S., Nano F. E. (2007) The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aubert D., MacDonald D. K., Valvano M. A. (2010) BcsKC is an essential protein for the type VI secretion system activity in Burkholderia cenocepacia that forms an outer membrane complex with BcsLB. J. Biol. Chem. 285, 35988–35998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bröms J. E., Lavander M., Sjöstedt A. (2009) A conserved α-helix essential for a type VI secretion-like system of Francisella tularensis. J. Bacteriol. 191, 2431–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kanamaru S., Leiman P. G., Kostyuchenko V. A., Chipman P. R., Mesyanzhinov V. V., Arisaka F., Rossmann M. G. (2002) Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557 [DOI] [PubMed] [Google Scholar]
- 45. Williams S. R., Gebhart D., Martin D. W., Scholl D. (2008) Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl. Environ. Microbiol. 74, 3868–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakayama K., Takashima K., Ishihara H., Shinomiya T., Kageyama M., Kanaya S., Ohnishi M., Murata T., Mori H., Hayashi T. (2000) The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to λ phage. Mol. Microbiol. 38, 213–231 [DOI] [PubMed] [Google Scholar]
- 47. Leiman P. G., Chipman P. R., Kostyuchenko V. A., Mesyanzhinov V. V., Rossmann M. G. (2004) Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell 118, 419–429 [DOI] [PubMed] [Google Scholar]
- 48. Kostyuchenko V. A., Chipman P. R., Leiman P. G., Arisaka F., Mesyanzhinov V. V., Rossmann M. G. (2005) The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 12, 810–813 [DOI] [PubMed] [Google Scholar]
- 49. Arisaka F., Takeda S., Funane K., Nishijima N., Ishii S. (1990) Structural studies of the contractile tail sheath protein of bacteriophage T4. 2. Structural analyses of the tail sheath protein, Gp18, by limited proteolysis, immunoblotting, and immunoelectron microscopy. Biochemistry 29, 5057–5062 [DOI] [PubMed] [Google Scholar]
- 50. Moody M. F. (1973) Sheath of bacteriophage T4. 3. Contraction mechanism deduced from partially contracted sheaths. J. Mol. Biol. 80, 613–635 [DOI] [PubMed] [Google Scholar]
- 51. Basler M., Mekalanos J. J. (2012) Type 6 secretion dynamics within and between bacterial cells. Science 337, 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bönemann G., Pietrosiuk A., Mogk A. (2010) Tubules and donuts: a type VI secretion story. Mol. Microbiol. 76, 815–821 [DOI] [PubMed] [Google Scholar]
- 53. Efimov A. V., Kurochkina L. P., Mesyanzhinov V. V. (2002) Engineering of bacteriophage T4 tail sheath protein. Biochemistry 67, 1366–1370 [DOI] [PubMed] [Google Scholar]
- 54. Kuznetsova T. A., Efimov A. V., Aijrich L. G., Kireeva I. Y., Marusich E. I., Cappuccinelli P., Fiori P., Rappelli P., Kurochkina L. P., Poglazov B. F., Mesyanzhinov V. V. (1998) Properties of recombinant bacteriophage T4 tail sheath protein and its deletion fragments. Biochemistry 63, 702–709 [PubMed] [Google Scholar]
- 55. Poglazov B. F., Efimov A. V., Marco S., Carrascosa J., Kuznetsova T. A., Aijrich L. G., Kurochkina L. P., Mesyanzhinov V. V. (1999) Polymerization of bacteriophage T4 tail sheath protein mutants truncated at the C-termini. J. Struct. Biol. 127, 224–230 [DOI] [PubMed] [Google Scholar]
- 56. Pederson D. M., Welsh L. C., Marvin D. A., Sampson M., Perham R. N., Yu M., Slater M. R. (2001) The protein capsid of filamentous bacteriophage PH75 from Thermus thermophilus. J. Mol. Biol. 309, 401–421 [DOI] [PubMed] [Google Scholar]
- 57. Leiman P. G., Shneider M. M. (2012) in Viral Molecular Machines (Rossmann M. G., Rao V. B., eds) Vol. 726, pp. 93–114, Springer, New York [Google Scholar]
- 58. Wood W. B. (1966) Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J. Mol. Biol. 16, 118–133 [DOI] [PubMed] [Google Scholar]
- 59. Hoang T. T., Kutchma A. J., Becher A., Schweizer H. P. (2000) Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72 [DOI] [PubMed] [Google Scholar]