Background: Canavanine (Can) is a highly toxic arginine (Arg) analogue found in some plant seeds.
Results: Replacement of all Arg residues with Can in MazF-bs(can), an mRNA interferase, resulted in a higher RNA cleavage specificity.
Conclusion: Enzymatic function of a protein can be modulated by Arg-to-Can replacement.
Significance: For the first time, a new functional protein was created by complete Arg-to-Can replacement.
Keywords: Amino Acid, Aminoacyl tRNA Synthetase, Enzymes, RNA-binding Protein, Toxins
Abstract
Replacement of a specific amino acid residue in a protein with nonnatural analogues is highly challenging because of their cellular toxicity. We demonstrate for the first time the replacement of all arginine (Arg) residues in a protein with canavanine (Can), a toxic Arg analogue. All Arg residues in the 5-base specific (UACAU) mRNA interferase from Bacillus subtilis (MazF-bs(arg)) were replaced with Can by using the single-protein production system in Escherichia coli. The resulting MazF-bs(can) gained a 6-base recognition sequence, UACAUA, for RNA cleavage instead of the 5-base sequence, UACAU, for MazF-bs(arg). Mass spectrometry analysis confirmed that all Arg residues were replaced with Can. The present system offers a novel approach to create new functional proteins by replacing a specific amino acid in a protein with its analogues.
Introduction
There are a large number of nonnatural amino acid analogues (1). It is quite intriguing to replace all residues of a specific amino acid in a protein with its analogues, because it may create novel functional proteins with altered structures. However, this is highly challenging because most amino acid analogues are highly toxic to the cells. To circumvent this problem, chemically modified aminoacylated tRNAs have been used in a cell-free system (2, 3). Alternatively, an orthogonal aminoacyl tRNA synthetase/tRNA pair from other species was incorporated into bacteria (4, 5) or eukaryotes (6). One such highly toxic analogue is l-canavanine (Can),3 an Arg analogue (Fig. 1, A and B) that is found as an insecticide in certain leguminous plants such as jack bean (7). Its incorporation into cellular proteins leads to the production of functionally aberrant proteins, leading to failure of various cellular functions, causing eventual cell death (7). In a previous attempt, 18 of 200 Arg residues in vitellogenin, an egg yolk protein, in locusts were replaced with Can, resulting in the production of an aberrantly structured protein (7), whereas one-third of 3 Arg residues in diptericin A, an antibacterial protein from the fly, Phormia terranovoe, was replaced with Can, resulting in loss of the antibacterial activity (8). In another attempt, 21% of Arg residues in the lysozyme molecule from the tobacco hornworm, Manduca sexta, were replaced with Can, resulting in loss of 49.5% of the catalytic activity (9). None of these attempts, however, could achieve the complete replacement of all Arg residues in a protein with Can.
FIGURE 1.
The structures of l-canavanine and l-arginine and the tertiary structure and multiple alignment of MazF-bs. A and B, structure of l-canavanine (A) and l-arginine (B). C, alignments of MazF-bs with other MazF homologues from Staphylococcus aureus (MazF-sa) and E. coli (MazF-ec), PemK from E. coli (PemK-ec), ChpBK from E. coli (ChpBK-ec), MazF-mt1 to -mt7 from Mycobacterium tuberculosis, MazF-mx from Myxococcus xanthus, and MazF-hw from Haloquadra walsbyi. White and black boxes indicate β-sheets and α-helices on the basis of the secondary structure of MazF-bs, respectively. ClustalW2 was used for multiple sequence alignments. All Arg residues in MazF-bs are indicated by black shading and homologous residues to Arg by gray shading. D, crystal structure of MazF-bs dimer (Protein Data Bank ID code: 1NE8) imaged in PyMOL. Green and blue colors show as an individual monomer, and Arg residues are colored in magenta.
RNA-mediated mRNA interference with the use of antisense RNA, miRNA, and siRNA has been well documented, including their important roles in gene regulation from bacteria to human cells (10, 11) and a possible use for the treatment of human diseases (12). More recently, it has been shown that mRNA interference is also mediated by proteins using sequence-specific endoribonucleases, called mRNA interferases (13). The first such enzyme reported was MazF-ec from Escherichia coli consisting of 111 residues, which cleaves RNA specifically at ACA sequences (14). The x-ray structure of its complex with the cognate antitoxin, MazE, has been determined, consisting of one MazE dimer with two MazF dimers (15). Since then, a number of MazF homologues have been discovered from bacteria and archaea (16) (Fig. 1C). Most recently, a 7-base specific MazF homologue from a superhalophilic archaeon from a hypersaline pool on the Sinai Peninsula (MazF-hw) was found to cleave RNA at UUACUCA, which can be used for regulating specific gene expression in E. coli (17). In the present paper, we attempted to replace all 7 Arg residues in MazF-bs, a 5-base specific RNA interferase from Bacillus subtilis (18) with Can using the Single Protein Production (SPP) system (19, 20).
EXPERIMENTAL PROCEDURES
Strain Construction
E. coli BL21(DE3) (ΔargHΔtrpCΔhisB) was constructed from E. coli BL21(DE3) (ΔtrpCΔhisB) (19) by P1 transduction using the ΔargH strain from the Keio collection (21).
Plasmid Construction
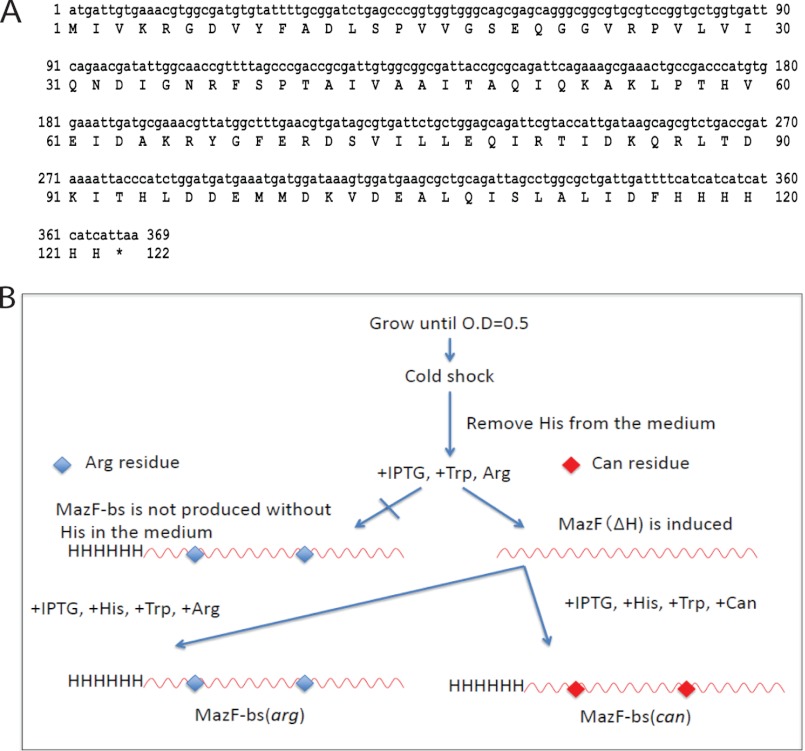
The gene for MazF-bs with a C-terminal His tag (Fig. 2A) was synthesized (Genescript). The gene was designed for the optimal codon usage in E. coli and to have no ACA sequences. The gene was cloned into pColdIII (SP-4) (19, 20).
FIGURE 2.
Schematic procedures for the production of MazF-bs(arg) and MazF-bs(can). A, DNA sequence of the mazF-bs gene. The gene is designed to be ACA-less and codon-optimized for E. coli. The amino acid sequence of MazF-bs is shown under the DNA sequence. B, dual inducible SPP system. The BL21(DE3) (ΔargHΔtrpCΔhisB) cells were transformed with pColdIIImazF-bs together with pACYCmazF(ΔH) and grown in a 1-liter culture of M9-glucose medium in the presence of Arg (20 μg/ml), His (20 μg/ml), and Trp (20 μg/ml) at 37 °C. When the A600 value reached 0.5, the culture was chilled in an ice-water bath for 5 min and incubated at 15 °C for 1 h to acclimate the cells to cold shock conditions. Cells were harvested and washed twice with M9 medium. The cells were resuspended in 50 ml of M9-glucose medium containing Arg (20 μg/ml) and Trp (20 μg/ml) but without His. IPTG (0.5 mm) was added to induce the only expression of MazF(ΔH) followed by an additional 2-h incubation at 15 °C. Cells were harvested and washed twice with M9 medium. The cells were resuspended in 50 ml of M9-glucose medium containing His (20 μg/ml), and Trp (20 μg/ml), Can (100 μg/ml), and IPTG (0.5 mm) to incorporate Can into MazF-bs. The cell culture was incubated at 15 °C for additional 24 h to induce MazF-bs(can). When Arg (20 μg/ml) was added instead of Can, MazF-bs(arg) was produced.
Protein Expression and Purification
The BL21(DE3) (ΔargHΔtrpCΔhisB) cells were transformed with pColdIIImazF-bs together with pACYCmazF(ΔH) and grown in a 1-liter culture of M9-glucose medium in the presence of Arg (20 μg/ml), His (20 μg/ml), and Trp (20 μg/ml) at 37 °C. When the A600 value reached 0.5, the culture was chilled in an ice-water bath for 5 min and incubated at 15 °C for 1 h to acclimate the cells to cold shock conditions. Cells were harvested and washed twice with M9 medium. The cells were resuspended in 50 ml of M9-glucose medium containing Arg (20 μg/ml) and Trp (20 μg/ml) but without His. Isopropyl β-d-1-thiogalactopyranoside (IPTG; 0.5 mm) was added to induce the only expression of MazF(ΔH) followed by an additional 2-h incubation at 15 °C. Cells were harvested and washed twice with M9 medium. The cells were resuspended in 50 ml of M9-glucose medium containing His (20 μg/ml), and Trp (20 μg/ml), Can (100 μg/ml; Sigma) and IPTG (0.5 mm) to incorporate Can into MazF-bs. The cell culture was incubated at 15 °C for additional 24 h to induce MazF-bs(can) (Fig. 2B). Cells were collected by centrifugation and subjected to SDS-PAGE followed by Coomassie Blue staining. MazF-bs(arg) and MazF-bs(can) were purified from BL21(DE3) (ΔargHΔtrpCΔhisB) cells carrying pColdIIImazF-bs with use of nickel-nitrilotriacetic acid resin (Qiagen) following the manufacturer's protocol. The MazF-bs(can) and MazF-bs(arg) were further purified by ion-exchange chromatography using DEAE-Sepharose (GE Healthcare).
Circular Dichroism (CD) Analysis
CD analysis was carried out using an Aviv model 62DS spectropolarimeter (Aviv Associates, Inc., Lakewood, NJ) Spectra were recorded in 2.0-nm steps between 260 and 200 nm at 4 °C with an integration time of 4 s at each wavelength, and the base line was corrected against buffer alone. Protein melting was examined at 208 nm with increasing temperature, from 0 to 90 °C, in 0.3 °C steps. Protein solutions were equilibrated at each temperature point for 1.5 min, and the temperature was increased with an average rate of 0.1 °C/min. The path length of the cell used was 0.1 cm, and all measurements were carried out in 10 mm Tris-HCl (pH 7.8).
Cleavage of MS2 Phage RNA by MazF-bs(can)
MS2 phage RNA (70 nm; Roche Applied Science) was incubated with 0.5 μm MazF-bs(arg) or 0.5 μm MazF-bs(can) in a reaction mixture (10 μl) containing 10 mm Tris-HCl (pH 7.8), 1 mm dithiothreitol, and the Protector RNase inhibitor at 37 °C for 0, 1, 5, 10, and 30 min. After denaturation in urea, the products were analyzed by electrophoresis on a 1.2% agarose gel.
Primer Extension Analysis
Primer extension analysis of the cleavage sites was carried out as described previously (22). Briefly, 0.7 μm MS2 phage RNA was incubated with 0.5 μm MazF-bs(arg) or MazF-bs(can) in the presence of CspA protein, an RNA chaperone (20 μm) at 37 °C for 10 min in a reaction mixture (10 μl) in 10 mm Tris-HCl (pH 7.8), containing 0.2 μl of the Protector RNase inhibitor (Roche Applied Science). Primer extension was carried out at 47 °C for 1 h. The reactions were stopped by 2× stop solution (90% formamide, 50 mm EDTA, 0.05% bromphenol blue, and 0.05% xylene cyanol FF). The samples were incubated at 90 °C for 5 min prior to electrophoresis on a 6% polyacrylamide gel containing 8 m urea.
Cleavage of Synthetic RNA by MazF-bs(arg) and MazF-bs(can)
Four 13-base ribonucleotides (CUCXUACAUAUCA) were synthesized, where the 4th base (X) was A, U, G, or C. Three additional 13-base RNA ribonucleotides (CUCUUACAUYUCA) were synthesized, where the Y position was replaced with U, G, or C. These ribonucleotides were used as substrates. The labeled substrates (0.2 μm) with [γ-32P]ATP using T4 kinase (New England Biolabs) were incubated with 0.1 μm MazF-bs(arg) or MazF-bs(can) for 10 min at 37 °C in a reaction mixture (10 μl) in 10 mm Tris-HCl (pH 7.8) containing 0.2 μl of the Protector RNase inhibitor. The reactions were stopped by the use of 2× stop solution. To analyze the cleavage of the synthetic RNAs, the products were analyzed by electrophoresis on a 20% polyacrylamide gel containing 8 m urea with a molecular mass ladder.
Kinetics Analysis
A 13-base ribonucleotide (CUCAUACAUAUCA) was used as a substrate. The substrate in various concentrations (0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, and 4.0 μm) was incubated with 1 nm MazF-bs(arg) or 5 nm MazF-bs(can) in a reaction mixture (10 μl) in 10 mm Tris-HCl (pH 7.8) containing 0.2 μl of the Protector RNase inhibitor. The reaction with MazF-bs(arg) was incubated for 5, 10, 15, and 20 min. The reaction with MazF-bs(can) was incubated for 30, 60, 90, and 120 min. The reaction was stopped by the use of 2× stop solution, and the sample mixtures were incubated at 90 °C for 5 min prior to electrophoresis on a 20% polyacrylamide gel containing 8 m urea. The cleavage products were analyzed by ImageJ software.
Competitive Analysis
A 13-base ribonucleotide (CUCUUACAUAUCA) was used as substrate, and three other 13-base ribonucleotides (CUCUUACAUUUCA, CUCUUACAUCUCA, and CUCUUACAUGUCA) in which only the 10th bases are different from the substrate (shown in bold) were used to examine whether these ribonucleotides are able to inhibit the cleavage of the substrate. The concentration of the substrate analogues was fixed at 1 μm, whereas the substrate concentrations were used at 1.0 and 4.0 μm. The substrate with and without the substrate analogues in a 10-μl reaction mixture containing 10 mm Tris-HCl (pH7.8) and 0.2 μl of Protector RNase inhibitor was incubated with 5 nm MazF-bs(can) at 37 °C for 30, 60, 90, and 120 min. The reaction was stopped by the use of 2× stop solution, and the reaction mixtures were incubated at 90 °C for 5 min prior to electrophoresis on a 20% polyacrylamide gel containing 8 m urea. The cleavage products were analyzed by ImageJ.
RESULTS
Production of MazF-bs(can) by the SPP System
To replace all 7 Arg residues in MazF-bs with Can, we applied the SPP system (19, 20) with use of an Arg auxotroph. In the SPP system, E. coli cells are converted into a bioreactor producing only a target protein, in which an ACA-specific mRNA interferase, MazF-ec, from E. coli is induced to eliminate all cellular mRNAs but the ACA-less mRNA for the target protein. Therefore, the use of the SPP system enables us to avoid the cytotoxicity of Can to replace all Arg residues in MazF-bs with Can. Thus, all ACA sequences in the MazF-bs mRNA are changed to other sequences without altering the MazF-bs amino acid sequence. For the complete replacement of all Arg residues with Can, it is also important to block the biosynthesis of Arg in the cells using an Arg auxotroph. For this, the argH deletion mutation from the Keio collection (21) was transduced into E. coli BL21(DE3) ΔtrpC, ΔhisB (19). The gene for MazF-bs designed to be ACA-less and codon-optimized for E. coli (Fig. 2A) was synthesized and cloned into pColdIII(SP-4) vector (19, 20), yielding pColdIIImazF-bs. E. coli BL21(DE3)ΔargHΔtrpCΔhisB cells were co-transformed with pColdIIImazF-bs and pACYCmazF(ΔH). In MazF(ΔH), His-28 and Gly-27 in MazF-ec were replaced with Arg and Lys, respectively, which has no effect on the MazF-ec mRNA interferase activity (19). Because MazF(ΔH) thus obtained does not contain His residues, this protein can still be synthesized in E. coli BL21(DE3)ΔargHΔtrpCΔhisB cells in the absence of His in the medium containing Trp, Arg, and IPTG as reported previously (19, 20). Under this condition, cell growth is completely arrested, and MazF(ΔH) eliminates all ACA-containing cellular mRNAs. Note that in contrast to MazF-ec produced from pACYCmazF(ΔH), MazF-bs used in the present study contains a C-terminal extension containing 6 His residues so that in the absence of His in the medium, the production of MazF-bs is completely blocked. Using this condition, Arg (20 μg/ml) was replaced with Can (100 μg/ml) in the medium in the presence of Trp and His (Fig. 2B).
Complete Replacement of Arg Residues in MazF-bs with Can
After a 24-h incubation using the SPP system in the presence of Can, a new band was induced at 14 kDa, and purified protein is shown in Fig. 3A. This protein termed (MazF-bs(can)) was subsequently purified by nickel-nitrilotriacetic acid affinity chromatography and DEAE ion-exchange column chromatography (Fig. 3A). If all 7 Arg residues were replaced with Can, the molecular mass of MazF-bs(can) should be larger by 13.8 Da (1.97 Da ×7) than that of MazF-bs(arg). The mass spectrometry analysis revealed that MazF-bs(can) was larger by 13.4 Da than MazF-bs(arg) (Fig. 3B), indicating that 97% of Arg residues were replaced with Can or that in four of five MazF-bs molecules the complete replacement was achieved. In the remaining one molecule, all but 1 Arg residue of 7 were replaced with Can.
FIGURE 3.
Analysis of the secondary structures of MazF-bs(can) and MazF(can). A, purified MazF-bs(arg) (lane 1) and MazF-bs(can) (lane 2) shown by 17% SDS-PAGE stained with Coomassie Blue. B, total mass measurement by MALDI-TOF (Applied Biosystems) of MazF-bs(arg) and MazF-bs(can), respectively. C, CD spectra of the secondary structures of MazF-bs(arg) and MazF-bs(can). D, thermal stabilities of MazF-bs(arg) and MazF-bs(can). The open and filled circles represent MazF-bs(can) and MazF-bs(arg), respectively.
Structural Analysis of MazF-bs(can) by Circular Dichroism (CD) Spectroscopy
The secondary structures of purified MazF-bs(arg) and MazF-bs(can) were analyzed by CD spectroscopy. MazF-bs(arg) showed minimum peaks around at 208 and 222 nm, which are characteristic for α-helical structures (23). MazF-bs(can) also showed a minimum peak at 208 nm which is higher than that for MazF-bs(arg), whereas the signal at 222 nm for MazF-bs(can) was lower than that for MazF-bs(arg) (Fig. 3C), indicating that α-helix contents of MazF-bs(can) slightly increased from 27.5 to 29.7%, whereas its β-sheet content decreased from 39.5 to 37.2%. Next, the thermal stability was examined for both proteins between 4 and 90 °C by measuring the change in ellipticity at 222 nm in the CD spectra. Notably, the Tm for MazF-bs(can) was lower by ∼4 °C than that for MazF-bs(arg) (Fig. 3D). MazF-bs(can) is likely folded in a manner very similar to MazF-bs(arg); however, the substitution of Arg with Can appears to affect the α-helical structures. The hydrogen bonds between Arg-5 and Ala-112 and a salt bridge between Arg-87 and Glu-20 have been shown to stabilize the dimer formation (24). Although both Arg and Can contain a guanidine group, the replacement of the methylene group in Arg with oxygen in Can results in the reduction of the pKa value from 12.48 to 7.01 (7). Therefore, the salt bridge in MazF-bs(arg) is likely to weaken substantially when all of the Arg residues are replaced with Can, resulting in a less thermo-stable protein. Note that the pI value of MazF-bs changed from 6.34 to 5.86 as a result of the Arg-to-Can replacement.
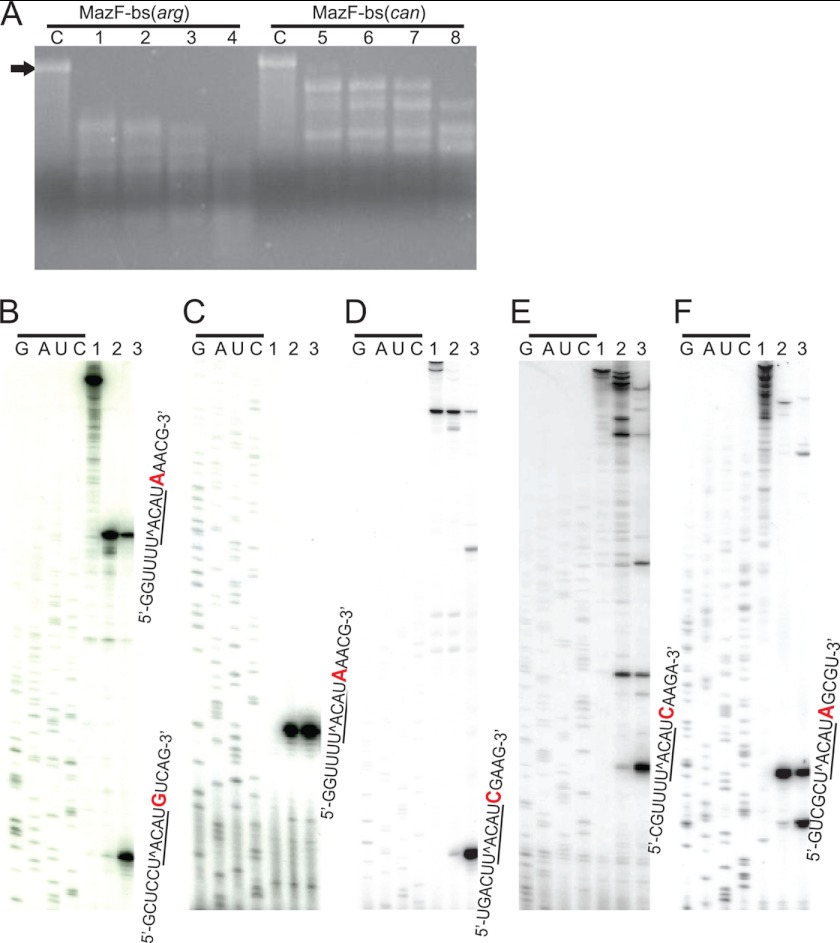
Specificity Alteration of a 5-Base to a 6-Base Recognition for RNA Cleavage
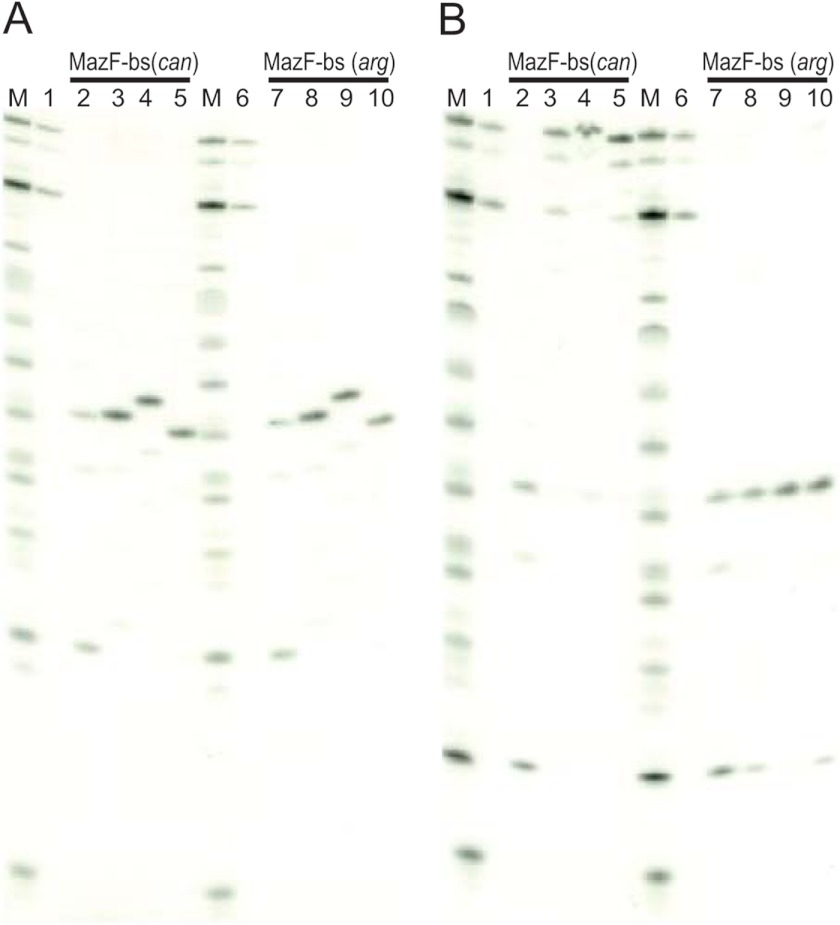
Next, we analyzed the endoribonuclease activity of MazF-bs(can) using 3.5-kb MS2 phage RNA as a substrate (17). Because the RNA cleavage patterns were found to be quite different between the two enzymes (Fig. 4A), in vitro primer extension experiments were carried out to determine the exact cleavage site sequences. As shown in Fig. 4A, after incubation of the RNA with MazF-bs(can) and MazF-bs(arg) at 37 °C for 10 min, MazF-bs(arg) cleaved MS2 phage RNA at all U↑ACAU sites as expected (↑ indicates the cleavage site) (18), whereas MazF-bs(can) appears to cleave the MS2 RNA at U↑ACAU sites, only when these sites contain one extra A residue at the 3′ end (Fig. 4, B–F), indicating that MazF-bs(can) acquired a higher RNA cleavage specificity from a 5-base to a 6-base cutter. To further confirm this notion, we synthesized 13-base RNA substrates covering all of the possible 7-base sequences having an extra base at both sides of UACAU and confirmed that MazF-bs(can) is specific for U↑ACAUA (Fig. 5). Lanes 2 (MazF-bs(can)) and 7 (MazF-bs(arg)) in Fig. 5A show an extra band corresponding to the product cleaved after the 1st C residue in addition to the cleavage product after the 5th U residue C↑UCUU↑ACAUAUCA (↑ indicates the cleavage sites), whereas no cleavage products are observed with three other ribonucleotides (CUCAUACAUAUCA, CUCGUACAUAUCA, and CUCCUACAUAUCA) for both MazF-bs(can) and MazF-bs(arg) (bases which are replaced are shown in bold). Furthermore, lane 2 in Fig. 5B using MazF-bs(can) with CUCUUACAUAUCA shows an extra cleavage product (cleaved after the 1st C residue) in addition to the product after the 5th U residue. Lanes 7, 8, and 10 in Fig. 5B using MazF-bs(arg) also show an extra cleavage product corresponding to C↑UCUUACAUAUCA, C↑UCUUACAUGUCA, and C↑UCUUACAUCUCA. These cleavages were not observed with MazF-bs(can). It is unknown at present why these substrates were cleaved by MazF-bs(arg) after the 1st C residue.
FIGURE 4.
Acquirement of a higher RNA cleavage specificity in MazF-bs(can). A, analysis of cleavage sites in MS2 phage RNA by MazF-bs(arg) or MazF-bs(can). Lane C represents a control reaction in which no protein was added; MS2 RNA was incubated with MazF-bs(arg) or MazF-bs(can) for 1, 5, 10, and 30 min (lanes 1–4, MazF-bs(arg); lanes 5–8, MazF-bs(can)). A black arrow indicates a full-length (3.5 kb) of MS2 phage RNA. B–F, analysis of MazF-bs(can) cleavage sites in MS2 phage RNA by in vitro primer extension. Each panel represents different UACAU sites in MS2 RNA. Lane 1, MS2 RNA incubated with purified CspA. Lanes 2 and 3, MS2 RNA incubated with MazF-bs(can) and MazF-bs(arg) in the presence of CspA, an RNA chaperone, respectively. G, A, U, and C with an upper black bar indicate the sequence ladder for each reaction primer. Ribonucleotide sequences in each panel (B–F) indicate the cleavage sequences for MazF-bs(can) and MazF-bs(arg). The U↑ACAU sequences with an under bar indicate the cleavage sites by MazF-bs(arg); one extra ribonucleotide, A, at the 3′ end is colored in red, which is required for the cleavage by MazF-bs(can).
FIGURE 5.
Identification of a change of RNA cleavage specificity in MazF-bs(can). A, 13-base ribonucleotides (CUCXUACAUAUCA) synthesized, where the 4th base (X) was U, A, G, or C, were incubated with MazF-bs(can) or MazF-bs(arg) (lanes 2–5 and 7–10, respectively). Lanes 1 and 6 represent control reactions in which no protein was added. Lanes 2 and 7 show an extra band corresponding to the product cleaved after the first C residue in addition to the cleavage product after the 5th U residue C↑UCUU↑ACAUAUCA (↑ indicates the cleavage sites), whereas no cleavage products were observed with three other ribonucleotides (CUCAUACAUAUCA, CUCGUACAUAUCA, and CUCCUACAUAUCA) for both MazF-bs(can) and MazF-bs(arg) (bases that are replaced are shown in bold). B, 13-base ribonucleotides (CUCUUACAUYUCA) synthesized, where the Y position was A, U, G, or C, were incubated with MazF-bs(can) or MazF-bs(arg) (lanes 2–5, and 7–10, respectively). Lanes 1 and 6 represent control reactions in which no protein was added. Lane 2 shows an extra cleavage product (cleaved after the 1st C residue) in addition to the product cleaved after the 5h U residue. Lanes 7, 8, and 10 also show an extra cleavage product corresponding to C↑UCUUACAUAUCA, C↑UCUUACAUGUCA, and C↑UCUUACAUCUCA. These cleavages were not observed with MazF-bs(can).
Kinetic Study
Using UACAUA as a substrate, the Km value and the Kcat/Km value of MazF-bs(arg) were determined to be 2.0 ± 0.2 μm and 1.0 ± 0.2 × 10−2, respectively. Although the Kcat/Km value of MazF-bs(can) is ∼5% of that of MazF-bs(arg), the Km value for MazF-bs(can) was almost identical to that of MazF-bs(arg) (see Table 1). The difference in the Kcat/Km values is likely due to the charging status of the guanidino groups of Can residues in MazF-bs(can). Because MazF-bs(can) became 6-base-specific, cleaving at U↑ACAUA, but not UACAUG, UACAUC, and UACAUU (Fig. 5), we next examined whether the cleavage of U↑ACAUA is inhibited by substrate analogues having different bases at the 6th position (UACAUU, UACAUC, and UACAUG), which are not cleavable by MazF-bs(can), and we found that there was no inhibition of the cleavage reaction by UACAUG, UACAUC, and UACAUU, indicating that the A residue at the 6th position plays a critical role for the substrate binding to the enzyme (Table 2). We found that there was no inhibition of the cleavage reaction by UACAUG, UACAUC, and UACAUU, even if the inhibitor-to-substrate ratio increased (Table 2).
TABLE 1.
Kinetic constants for MazF-bs(arg) and MazF-bs(can)
| MazF | Vmax | Km | Kcat | Kcat/Km |
|---|---|---|---|---|
| μm/min | μm | min−1 | μm−1 ·min−1 | |
| MazF-bs(arg) | 4.2 ± 1.1 × 10−2 | 2.0 ± 0.2 | 2.2 ± 0.4 × 10−2 | 1.0 ± 0.2 × 10−2 |
| MazF-bs(can) | 8.4 ± 1.2 × 10−3 | 1.8 ± 0.3 | 8.4 ± 1.2 × 10−4 | 5.0 ± 1.0 × 10−4 |
TABLE 2.
Relative cleavage activity of MazF-bs(can) using CUCUUACAUAUCA as a substrate in the presence and the absence of substrate analogues having different bases at the 6th position (CUCUUACAUCUCA, CUCUUACAUGUCA, and CUCUUACAUUUCA)
| Substrate | CUCUUACAUAUCA only | +CUCUUACAUCUCA | +CUCUUACAUGUCA | +CUCUUACAUUUCA |
|---|---|---|---|---|
| CUCUUACAUAUCA:substrate analogue = 1:1 | 1.0 | 1.1 | 1.0 | 1.1 |
| CUCUUACAUAUCA:substrate analogue = 4:1 | 1.0 | 1.1 | 1.1 | 1.2 |
DISCUSSION
Amino acid analogues are toxic for cells because they are incorporated into cellular proteins producing structurally and functionally abnormal proteins, which results in cell growth arrest and eventual cell death. Therefore, the simple addition of an amino acid analogue into a culture medium does not yield a protein in which all of the residues of a specific amino acid in the protein are replaced with its analogue. Furthermore, to achieve the complete replacement of all of the residues of a particular amino acid in a protein, it is important to suppress the biosynthesis of that particular amino acid completely. Therefore, to achieve the complete replacement of a specific amino acid in a protein, there are two essential requirements: first, the incorporation of an amino acid analogue into any other cellular proteins but the target protein has to be completely prevented. Second, the de novo biosynthesis of that particular amino acid should be completely inhibited.
To achieve the replacement of all 7 Arg residues in MazF-bs with Can, we used the SPP system for the first requirement so that Can incorporation into cellular proteins but MazF-bs was prevented while maintaining the biosynthetic function of the cells. The second requirement was achieved by using an Arg auxotroph. The use of SPP system for the replacement of all Arg residues in a protein with Can seems to be crucial because Can incorporation into other cellular proteins likely affects their functions, leading to severe inhibitory effects on various biosynthetic reactions including protein synthesis.
The present system, therefore, can be used for other toxic amino acid analogues as far as they can be recognized by E. coli aminoacyl-tRNA synthetases. The second requirement for the present system is the use of an amino acid auxotroph to avoid the incorporation of a natural amino acid into a target protein. The use of the SPP system in combination with amino acid auxotroph strains thus opens a new avenue to create proteins of unprecedented novel structures and functions without genetic manipulation of tRNAs and aminoacyl tRNA synthetases.
The most intriguing outcome from the present experiments is that MazF-bs(can) was no more able to cleave RNA at the original MazF-bs(arg) 5-base sequence, UACAU, requiring one extra A residue at the 3′ end. The Km value of MazF-bs(can) using UACAUA as a substrate is almost identical to that of MazF-bs(arg) using UACAU as a substrate, suggesting that the substrate binding affinity in MazF-bs(can) was compensated by an extra A residue at the 3′ end of the substrate. Notably, however, the cleavage activity of MazF-bs(can) was reduced to ∼5% of MazF-bs(arg) (Table 1). The MazF-bs functions as a dimer, and its interphase is predicted to be involved with RNA binding and catalysis (24). Because of a total of 7 Arg residues in MazF-bs, Arg-25, Arg-81, and Arg-87 are located in the interphase between the two monomers in a dimer (Fig. 1D), some or all these 3 residues may have critical roles in the specific RNA sequence recognition and the enzymatic activity. At present it is not known whether the other 4 Arg residues also play roles in MazF-bs function. It remains to be determined whether some of these Arg residues can be replaced with Lys without losing MazF-bs function.
Acknowledgments
We thank Drs. Sangita Phadtare and Vikas Nanda for critical reading of the manuscript and Drs. Zhuoxin Yu and Sung-gun Kim for the CD data analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1GM081567.
- Can
- l-canavanine (l-2-amino-4-(guanidinooxy) butyric acid)
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- SPS
- Single Protein Production.
REFERENCES
- 1. Hendrickson T. L., de Crécy-Lagard V., Schimmel P. (2004) Incorporation of nonnatural amino acids into proteins. Annu. Rev. Biochem. 73, 147–176 [DOI] [PubMed] [Google Scholar]
- 2. Noren C. J., Anthony-Cahill S. J., Griffith M. C., Schultz P. G. (1989) A general method for site-specific incorporation of unnatural amino acids into proteins. Science 244, 182–188 [DOI] [PubMed] [Google Scholar]
- 3. Stigers D. J., Watts Z. I., Hennessy J. E., Kim H. K., Martini R., Taylor M. C., Ozawa K., Keillor J. W., Dixon N. E., Easton C. J. (2011) Incorporation of chlorinated analogues of alphatic amino acids during cell-free protein synthesis. Chem. Commun. 47, 1839–1841 [DOI] [PubMed] [Google Scholar]
- 4. Bae J. H., Rubini M., Jung G., Wiegand G., Seifert M. H., Azim M. K., Kim J. S., Zumbusch A., Holak T. A., Moroder L., Huber R., Budisa N. (2003) Expansion of the genetic code enables design of a novel “gold” class of green fluorescent proteins. J. Mol. Biol. 328, 1071–1081 [DOI] [PubMed] [Google Scholar]
- 5. Hancock S. M., Uprety R., Deiters A., Chin J. W. (2010) Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J. Am. Chem. Soc. 132, 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gautier A., Nguyen D. P., Lusic H., An W., Deiters A., Chin J. W. (2010) Genetically encoded photocontrol of protein localization in mammalian cells. J. Am. Chem. Soc. 132, 4086–4088 [DOI] [PubMed] [Google Scholar]
- 7. Rosenthal G. A., Reichhart J. M., Hoffmann J. A. (1989) l-Canavanine incorporation into vitellogenin and macromolecular conformation. J. Biol. Chem. 264, 13693–13696 [PubMed] [Google Scholar]
- 8. Rosenthal G. A., Lambert J., Hoffmann D. (1989) Canavanine incorporation into the antibacterial proteins of the fly, Phormia terranovae (Diptera), and its effect on biological activity. J. Biol. Chem. 264, 9768–9771 [PubMed] [Google Scholar]
- 9. Rosenthal G. A., Dahlman D. L. (1991) Studies of l-canavanine incorporation into insectan lysozyme. J. Biol. Chem. 266, 15684–15687 [PubMed] [Google Scholar]
- 10. Davis B. M., Waldor M. K. (2007) RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol. Microbiol. 65, 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu H. Y., Guo S., Xi J., Yan Z., Fu N., Zhang X., Menzel C., Liang H., Yang H., Zhao M., Zeng R., Chen W., Pääbo S., Khaitovich P. (2011) MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 7, e1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Angaji S. A., Hedayati S. S., Poor R. H., Madani S., Poor S. S., Panahi S. (2010) Application of RNA interference in treating human diseases. J. Genet. 89, 527–537 [DOI] [PubMed] [Google Scholar]
- 13. Yamaguchi Y., Inouye M. (2011) Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9, 779–790 [DOI] [PubMed] [Google Scholar]
- 14. Zhang J., Zhang Y., Inouye M. (2003) Characterization of the interactions within the mazEF addiction module of Escherichia coli. J. Biol. Chem. 278, 32300–32306 [DOI] [PubMed] [Google Scholar]
- 15. Kamada K., Hanaoka F., Burley S. K. (2003) Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol. Cell 11, 875–884 [DOI] [PubMed] [Google Scholar]
- 16. Yamaguchi Y., Park J. H., Inouye M. (2011) Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79 [DOI] [PubMed] [Google Scholar]
- 17. Yamaguchi Y., Nariya H., Park J. H., Inouye M. (2012) Inhibition of specific gene expressions by protein-mediated mRNA interference. Nat. Commun. 3, 607. [DOI] [PubMed] [Google Scholar]
- 18. Park J. H., Yamaguchi Y., Inouye M. (2011) Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 585, 2526–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vaiphei S. T., Mao L., Shimazu T., Park J. H., Inouye M. (2010) Use of amino acids as inducers for high-level protein expression in the single-protein production system. Appl. Environ. Microbiol. 76, 6063–6068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suzuki M., Roy R., Zheng H., Woychik N., Inouye M. (2006) Bacterial bioreactors for high yield production of recombinant protein. J. Biol. Chem. 281, 37559–37565 [DOI] [PubMed] [Google Scholar]
- 21. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Construction of Escherichia coli K-12, in-frame single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y., Zhang J., Hara H., Kato I., Inouye M. (2005) Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 280, 3143–3150 [DOI] [PubMed] [Google Scholar]
- 23. Greenfield N. J. (2006) Determination of the folding of proteins as a function of denaturants, osmolytes or ligands using circular dichroism. Nat. Protoc. 1, 2733–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gogos A., Mu H., Bahna F., Gomez C. A., Shapiro L. (2003) Crystal structure of YdcE protein from Bacillus subtilis. Proteins 53, 320–322 [DOI] [PubMed] [Google Scholar]