Background: ER stress impairs progression through G1 and G2 phases of the cell cycle.
Results: G2 arrest is enhanced in p53 mutant cells but is not enhanced by expression of the p53/47 isoform.
Conclusion: Early G2 arrest in ER stress is a response to translation attenuation.
Significance: Understanding cell cycle regulation in ER stress has implications for rational cancer therapy.
Keywords: Cell Cycle, Checkpoint Control, ER Stress, p53, Translation, CHK1, GADD34
Abstract
Cell cycle checkpoints ensure that proliferation occurs only under permissive conditions, but their role in linking nutrient availability to cell division is incompletely understood. Protein folding within the endoplasmic reticulum (ER) is exquisitely sensitive to energy supply and amino acid sources because deficiencies impair luminal protein folding and consequently trigger ER stress signaling. Following ER stress, many cell types arrest within the G1 phase, although recent studies have identified a novel ER stress G2 checkpoint. Here, we report that ER stress affects cell cycle progression via two classes of signal: an early inhibition of protein synthesis leading to G2 delay involving CHK1 and a later induction of G1 arrest associated both with the induction of p53 target genes and loss of cyclin D1. We show that substitution of p53/47 for p53 impairs the ER stress G1 checkpoint, attenuates the recovery of protein translation, and impairs induction of NOXA, a mediator of cell death. We propose that cell cycle regulation in response to ER stress comprises redundant pathways invoked sequentially first to impair G2 progression prior to ultimate G1 arrest.
Introduction
Failure to maintain proteostasis, the state of healthy protein maturation, underlies the pathology of many diseases including diabetes and cancer (1). The folding of secretory proteins is especially vulnerable to such failure because cells must balance the activity of ribosomes in the cytoplasm with the efficiency of protein folding within the lumen of the ER3 (2). In pathological states where protein synthesis exceeds protein maturation, the cell faces failure of its secretory pathway caused by the aggregation of aberrant protein conformers within this organelle and is said to experience “ER stress.” It has been appreciated for more than a decade that this impairs cell cycle progression (3–9), but if ER stress is especially severe or prolonged, cell death is triggered through multiple pathways (10–12). In the context of diabetes, ER stress has been shown to contribute to the exhaustion of β-cell mass in the face of persisting insulin resistance through either elevated levels of apoptosis or failure to permit the expansion of β-cell number (13–17). In contrast, in models of cancer, intact ER stress signaling has been shown to enable solid tumor growth (18, 19). This reflects the benefits afforded by reducing metabolic activity to match limiting nutrient supplies. In solid tumors and other relatively ischemic tissues, a relative deficiency of energy is the major cause of ER stress, and so restricting cell growth can protect the tissue from damage.
Early observations revealed that following prolonged ER stress cells arrest in the G1 phase of the cell cycle and that this correlates with loss of cyclin D1 (3, 4). Part of the loss of cyclin D1 is mediated by reduced rates of protein translation due to phosphorylation of eIF2α by the ER stress-responsive kinase PERK (3, 4). In addition, ER stress has been shown to favor cyclin D1 degradation by the proteasome (8). However, loss of cyclin D1 alone cannot adequately account for the G1 arrest seen during ER stress because isolated activation of PERK with depletion of cyclin D1 fails to induce G1 arrest in the absence of additional signals (9). Multiple lines of evidence have implicated p53 in regulating G1 during ER stress, and induction of the cyclin-dependent kinase inhibitor p21 appears central to this (5–7, 20–23). However, the effect of ER stress on p53 remains unclear with reports of ER stress-induced activation of p53 (22, 24) contrasting with reports of net p53 export from the nucleus leading to its inactivation (5–7).
More recently, we and others described ER stress-dependent inhibition of G2 cell cycle progression independent of effects on G1 (9, 23). When PERK is overexpressed in Drosophila melanogaster, tissue growth is impaired through impaired G2 cell cycle progression (9). We reported that partial loss of Drosophila CHK1 (grapes) could rescue the impairment of tissue growth caused by PERK overexpression and that in mammalian cells CHK1 activation occurs during ER stress as a result of impaired protein translation (9). In parallel, others described an ER stress-induced G2 checkpoint correlated with translation of a short isoform of p53 (23). The mRNA encoding p53 contains at least two internal ribosomal entry sites that can generate either full-length p53 protein or an N-terminally truncated p53/47 isoform lacking the first transactivation domain (25, 26). Although it has been proposed that p53/47 functions as a dominant negative inhibitor of p53, recent work suggests that it can promote G2 arrest by inducing 14-3-3σ (23).
We set out to study the roles of p53 and CHK1 in the regulation of cell cycle progression during ER stress. Herein, we show that ER stress affects cell cycle progression via two classes of signal: an early inhibition of protein synthesis leading to G2 delay mediated by CHK1 and a later induction of G1 arrest associated with both the induction of p53 target genes and loss of cyclin D1. We show that substitution of p53/47 for p53 impairs the ER stress G1 checkpoint, attenuates the recovery of protein translation, and impairs induction of NOXA, a mediator of cell death. We propose that cell cycle regulation in response to ER stress comprises redundant pathways invoked sequentially first to impair G2 progression prior to ultimate G1 arrest.
EXPERIMENTAL PROCEDURES
Expression Plasmids
The coding sequence of p53/p47 was subcloned from pcDNA3.1.p53/p47ΔIRES (a gift from Prof. Robin Fahraeus, INSERM, France) into pEGFP-C3 (Clontech) between HindIII and SalI restriction sites. The coding sequence of full-length p53 was subcloned from pcDNA3.1.p53wt (Prof. Robin Fahraeus, INSERM, France) into pEGFP-C1 between BglII and SalI sites.
Cell Culture
HCT116 p53+/+ and p53−/− cells stably transfected with inducible CHK1 Tet-On small interfering hairpin RNA (shRNAi) or scrambled control shRNAi were a kind gift from Dr. Giovanna Damia (Milan, Italy) (27). HCT116, COS-7, HeLa, and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS. ATR+/+ (GM02188) and ATR−/− (DK0064) B lymphocytes (28) and ATM+/+ (GM03491) and ATM−/− (GM02052) fibroblasts (29) were a gift from Prof. Ashok Venkitaraman (University of Cambridge, UK). ATR+/+ and ATR−/− lymphoblastic cells were grown in RPMI 1640 medium supplemented with 20% FBS and 0.2% glutamine. Ataxia-telangiectasia mutated (ATM) cells were maintained in DMEM with 15% FBS, 1% non-essential amino acids, and 0.2% Hanks' balanced salt solution. H1299 cells were a kind gift from Prof. Robin Fahreaus (23) and were cultured in RPMI 1640 medium with 10% FBS. Adherent cell lines were passaged using trypsin-EDTA, whereas suspension cells were directly split into fresh medium.
Immunoblot and Immunohistochemistry
CHOP, BiP, and eIF2α were detected as described previously (12). Commercial antibodies used were CHK1 (catalog number 2345, Cell Signaling Technology), P317-CHK1 (catalog number 2344, Cell Signaling Technology), cyclin D1 (mouse monoclonal DCS6, catalog number 2926, Cell Signaling Technology), H2AX (catalog number 2995, Cell Signaling Technology), γH2AX (catalog number 2577, Cell Signaling Technology), GADD34 (catalog number 10449-1-AP, Proteintech, UK), ATM (catalog number 2873, Cell Signaling Technology), ATM- and Rad3-related (ATR) (catalog number 2790, Cell Signaling Technology), P345-CHK1 (catalog number 2341, Cell Signaling Technology), P-CHK2 (catalog number 2661, Cell Signaling Technology), CHK2 (catalog number 2662, Cell Signaling Technology), Mcl-1 (catalog number 4572, Cell Signaling Technology), p21 (Ab7960, Abcam, UK), NOXA (Ab36833, Abcam, UK), PUMA (catalog number 4976, Cell Signaling Technology), BAX (catalog number 2772, Cell Signaling Technology), total p53 (catalog number 9282, Cell Signaling Technology), p53 Ab6 DO1 (OP34T, Calbiochem), p53 pAb1801 (Ab28, Abcam, UK), and actin (Ab8227, Abcam, UK).
Whole cell lysates were sheared directly in the SDS loading buffer using a 26-gauge needle. For nuclear and cytoplasmic fractions, the cell pellet was resuspended in 5 volumes of 10 mm HEPES, pH 7.9, 50 mm NaCl, 500 mm sucrose, 0.1 mm EDTA, 0.5% Triton X-100, 1 mm DTT, 1 mm PMSF, 1× CompleteTM protease inhibitor mixture (Roche Applied Science), 10 mm tetrasodium pyrophosphate, 15.5 mm β-glycerophosphate, 100 mm NaF) and incubated on ice for 5 min. Nuclei were pelleted by centrifugation at 800 × g for 10 min, and the supernatant was taken as the cytosolic fraction. Nuclei were washed in 10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 EGTA, 1 mm DTT with protease inhibitors, and then soluble nuclear proteins were extracted in 4 pellet volumes of 10 mm HEPES, pH 7.9, 500 mm NaCl, 0.1 mm EDTA, 0.1 mm EGTA, 0.1% Nonidet P-40, 1 mm DTT with protease inhibitors by vortexing for 15 min at 4 °C. The non-extractable proteins and DNA were pelleted by centrifugation at 16,000 × g for 10 min, and the supernatant was taken as extractable nuclear proteins.
Fluorescence-activated Cell Sorting (FACS)
Cells were cultured in DMEM with 10% (v/v) FBS and 1 μg/ml doxycycline for 48 h, then replated, and cultured for a further for 0, 16, and 24 h with 500 nm thapsigargin. Cells were recovered by trypsinization; fixed with 70% (v/v) ethanol; and incubated with PBS, RNase (5 mg/ml) (MP Biomedicals, Illkirch Cedex, France), and propidium iodide (20 μg/ml) (Invitrogen) at 37 °C for 30 min. The cells were analyzed using a CyAn fluorescence-activated cell sorting instrument (Dako, Stockport, UK) using FlowJo software (TreeStar, Ashland, OR).
Drosophila Stocks
The UAS-dPERK-WT flies have been described previously (9). For gene silencing in Drosophila, all RNAi lines used were obtained from the Vienna Drosophila RNAi Center, Austria: grp RNAi line v12680, atm/tefu RNAi line v22502, atr/mei-41 line v11251, and chk2/lok lines v44981 and v44980. For expression in the eye imaginal disc posterior to the morphogenetic furrow, the GMR-Gal4 line BL1104 was purchased from the Bloomington Stock Center. All stocks were in a w1118 background and maintained at 18 °C using standard techniques unless otherwise stated. All crosses were set up at 18 °C.
ATF6-Luciferase Reporter
HCT116 cells were grown in 6-well plates for 16 h at 37 °C 5% CO2. Twenty-four hours prior to lysis, the cells were co-transfected with 2 μg of a plasmid encoding firefly luciferase under the control of a UPR response element (p5×ATF6-Luc) and 50 ng of pRL-TK (Renilla luciferase) transfection efficiency control reporter plasmids (both kind gifts from Dr. Timothy Weaver, Cincinnati Children's Hospital Medical Center, Cincinnati, OH). The cells were transfected for 6 h and then induced with 500 nm thapsigargin for the designated time periods. Transfected cells were lysed and assayed for firefly and Renilla luciferase activity using the Dual-Luciferase kit (Promega). Both firefly and Renilla luciferase activities were measured using a GloMax luminometer (Promega), and firefly luciferase activity was calculated relative to Renilla luciferase activity. All measurements were performed in triplicate and repeated on at least three independent occasions.
Metabolic Labeling
To determine the rate of new protein synthesis, cells were grown in DMEM in the presence of 500 nm thapsigargin for the designated time points. The cells were then washed in PBS and metabolically labeled with 1.3 MBq of [35S]Met/Cys for 15 min in DMEM. Cells were then rinsed and lysed in 50 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1× CompleteTM protease inhibitor mixture (Roche Applied Science). Twenty micrograms of cell lysate were separated by 10% SDS-PAGE and quantified by autoradiography using a Cyclone phosphorimaging system (PerkinElmer Life Sciences).
qPCR
The following primers were purchased from Invitrogen: cyclin D1: forward, 5′-TGT TCG TGG CCT CTA AGA TGA AG-3′; reverse, 5′-AGG TTC CAC TTG AGC TTG TTC AC-3′; p21: forward, 5′-ATG TGT CCT GGT TCC CGT TTC-3′; reverse, 5′-CAT TGT GGG AGG AGC TGT GA-3′; GADD34: forward, 5′-ATG TAT GGT GAG CGA GAG GC-3′; reverse, 5′-GCA GTG TCC TTA TCA GAA GGC-3′; CHOP: forward, 5′-ACC AAG GGA GAA CCA GGA AAC G-3′; reverse, 5′-TCA CCA TTC GGT CAA TCA GAG C-3′; actin: forward, 5′-CTC TTC CAG CCT TCC TTC CT-3′; reverse, 5′-AGC ACT GTG TTG GCG TAC AG-3′.
For each reaction, 2× Qiagen SYBR Green PCR reagent, 1.2 mm forward primer, and 1.2 mm reverse primer were used in a total volume of 24 μl with RNase-free water. To each reaction, 1 μl of cDNA (diluted 1:100) was added to make a final volume of 25 μl. The qPCR was performed on an ABI 2000 analyzer with the following program: 95 °C for 15 min; then 95 °C for 1 min, 58 °C for 30 s, and 72 °C for 30 s repeated for 40 cycles; and 4 °C for 10 min. Absolute changes in transcription (Ct) were measured, and further analyses were performed based on primer efficiency in accordance with Pfaffl calculations (30).
XBP1s qPCR
Splicing of XBP1 mRNA was measured by qPCR as described previously (31). Briefly, primers were designed to span the 26-base pair intron of XBP1 mRNA: forward, 5′-TGC TGA GTC CGC AGC AGG TG-3′; reverse, 5′-GCT GGC AGG CTC TGG GGA AG-3′). qPCR was carried out at 95 °C for 3 min followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 62 °C for 15 s, and extension at 72 °C for 30 s using IQ SYBR Green Supermix (Bio-Rad). Each assay was run in triplicate, and arbitrary mRNA concentrations were calculated manually using the relative standard curve method. Relative mRNA concentrations of actin and RPL13A housekeeping genes were used to calculate the normalized expression of the XBP1s mRNA.
Statistical Analysis
Statistical analysis by analysis of variance with a Bonferroni post hoc test or Student's t test where appropriate was performed using the GraphPad Prism program (GraphPad Software Inc.). Statistically significant changes (p < 0.05) are indicated.
RESULTS
Translation Attenuation Activates CHK1
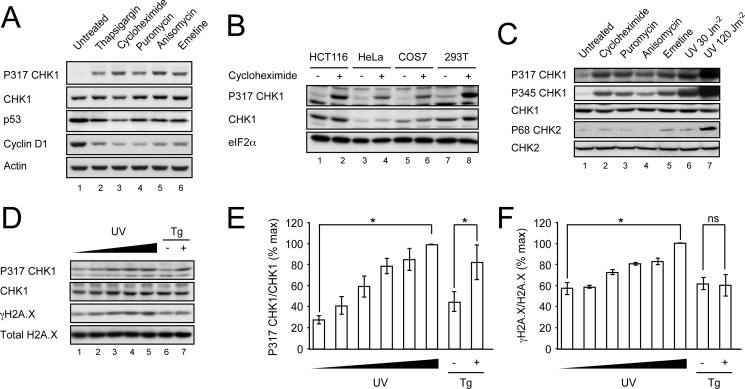
We showed previously that acute ER stress leads to the activation of CHK1 through phosphorylation of serine 317 (9). This was abolished in cells lacking the ER stress-sensing kinase PERK or expressing a mutant eIF2α that could not be phosphorylated by PERK. Moreover, activation of PERK in isolation in the absence of other ER stress signals was sufficient to induce CHK1 phosphorylation. An important consequence of eIF2α phosphorylation is the inhibition of protein translation (32), and we showed that pharmacological inhibition of translation with cycloheximide was sufficient to activate CHK1 (9). Because cycloheximide inhibits translation by binding to the ribosome E-site and thus blocking the translocation step of elongation (33) whereas eIF2α phosphorylation limits the abundance of eIF2α·GTP·Met-tRNA ternary complex (34), we reasoned that the precise nature of the ribotoxic stress is unlikely to be important in this response. To test this, we treated HCT116 colon carcinoma cells with a variety of ribotoxins including puromycin, which causes premature chain termination; anisomycin, a blocker of peptide bond formation; and emetine, an inhibitor of translocation that stabilizes the ribosome in a conformation distinct from that of cycloheximide (35, 36). Each agent induced phosphorylation of CHK1 on serine 317 in a manner indistinguishable from the ER stress inducer thapsigargin (Fig. 1A). Treatment with each compound for 1 h caused a similar depletion of cyclin D1 and p53. Comparable phosphorylation of CHK1 was observed in a number of cell lines treated with cycloheximide, suggesting that these results are generalizable to other cells and tissues (Fig. 1B). Activation of CHK1 involves phosphorylation on serines 317 and 345, and so we probed for both phosphorylation events and obtained similar results (Fig. 1C). However, we were unable to detect significant phosphorylation of CHK2 except following exposure to lethal doses of UV radiation (Fig. 1C). This suggests that ribotoxic stress causes selective activation of CHK1 or that our current reagents are insufficiently sensitive to detect weak activation of CHK2.
FIGURE 1.
Translation attenuation activates CHK1. A, HCT116 cells were treated for 1 h with thapsigargin (500 nm), cycloheximide (50 μg/ml), anisomycin (5 μg/ml), puromycin (75 μg/ml), or emetine (100 μg/ml). Postnuclear lysates were separated by 10% SDS-PAGE, transferred to nitrocellulose, and immunoblotted for P317-CHK1, total CHK1, p53, cyclin D1, and actin. B, HCT116, HeLa, COS-7, and 293T cells were treated with cycloheximide (50 μg/ml) for 1 h and analyzed by immunoblot for total P317-CHK1, total CHK1, and eIF2α. C, HCT116 cells were treated with cycloheximide (50 μg/ml), puromycin (75 μg/ml), anisomycin (5 μg/ml), or emetine (100 μg/ml) for 1 h or irradiated with 30 or 120 J/m2 UV light. Total cell lysates were analyzed by immunoblot for total P317-CHK1, P345-CHK1, total CHK1, P68-CHK2, and total CHK2. D, HCT116 cells were irradiated with UV light at 0, 8, 10, 12, or 15 J/m2 or incubated in the presence or absence of 500 nm thapsigargin (Tg) for 1 h. Total cell lysates were analyzed by immunoblot for P317-CHK1, total CHK1, P139-H2AX (γH2AX), and H2AX. E, graphical representation of data from D showing P317-CHK1 band intensity corrected for total CHK1 band intensity and represented as percent maximum signal. F, graphical representation data from D showing γH2AX band intensity corrected for total H2AX. Data are mean ± S.E. (error bars). n = 3; *, p < 0.05; ns, not significant.
The mechanism of CHK1 activation by ribotoxic stress is unknown. Following DNA damage, CHK1 is phosphorylated primarily by the kinase ATR (37). There is good evidence that ER stress can cause DNA damage through macromolecular oxidation, although PERK has been shown to limit this (18). We sought to detect evidence of DNA damage in response to ER stress following a brief treatment with thapsigargin. As a positive control, we titrated the level of UV irradiation such that it would induce levels of CHK1 phosphorylation similar to that seen with thapsigargin (Fig. 1D). At comparable levels of CHK1 activation, UV irradiation caused dose-dependent C-terminal phosphorylation of histone H2AX (γH2AX) indicative of DNA damage, but this was not seen with thapsigargin (Fig. 1D, compare lanes 4 and 5 with lane 7). When three independent repeats were analyzed densitometrically, UV irradiation was shown to induce phosphorylation of both CHK1 and H2AX, whereas thapsigargin only increased the phosphorylation of CHK1 (Fig. 1, E and F).
ATR and ATM Are Not Required for CHK1 Activation during Translation Attenuation
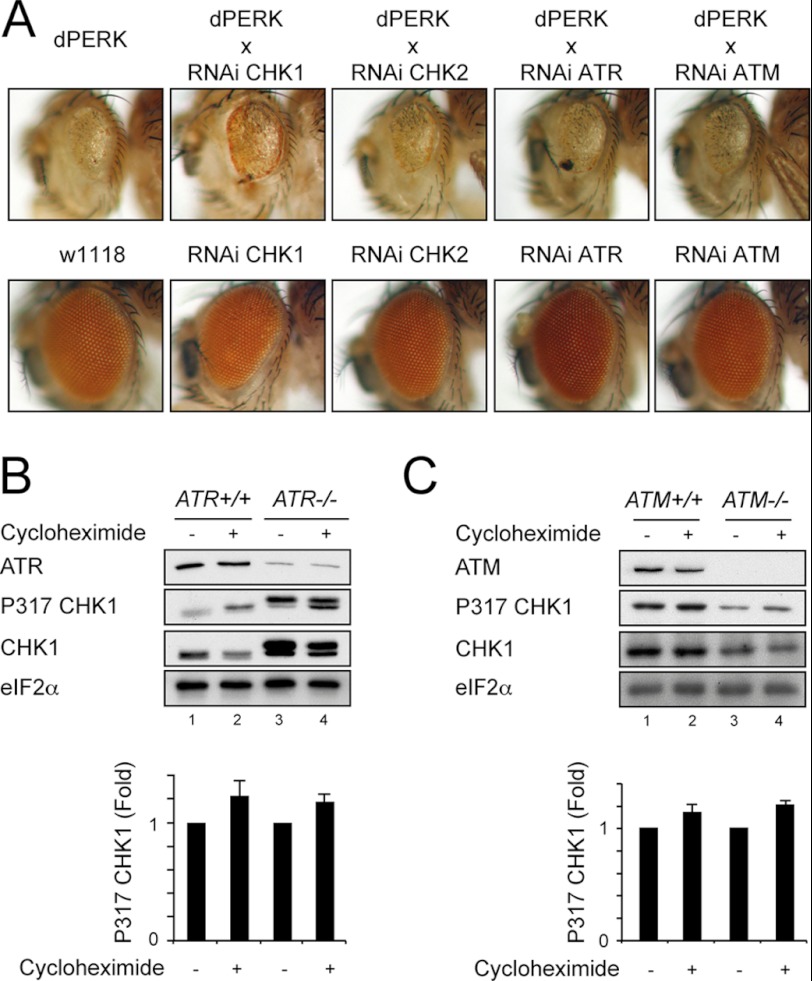
To determine whether ATR or ATM was required for activation of CHK1 in response to inhibition of protein translation, we next used a Drosophila model of PERK signaling. We previously described the consequences of the overexpression of PERK within the eye imaginal discs of developing Drosophila and its inhibitory effect on eye morphogenesis (9). The wild type Drosophila eye is composed of regularly arrayed light-sensitive pigmented ommatidia (Fig. 2A, lower panels), but when eye development is impaired by persistent activation of dPERK, the eye is rendered small and depigmented (Fig. 2A upper left panel). Depletion of grapes/CHK1 by shRNAi caused a modest rescue of eye development with increased pigmentation similar to that reported previously (Fig. 2A and Ref. 9). In contrast, depletion of CHK2, ATR, or ATM had no effect on eye development. These findings suggest that neither ATR nor ATM is absolutely required to mediate the effects of PERK overexpression on eye morphogenesis.
FIGURE 2.
ATR and ATM are not required for CHK1 activation during translation attenuation. A, representative light micrographs of eyes from GMR-GAL4>dPERK Drosophila or GMR-GAL4>dPERK animals expressing UAS-shRNAi against grp/CHK1, CHK2, dATR, and dATM (upper row) or w1118 strain control expressing GMR-GAL4 driver alone or GMR-GAL4 with each of the specified shRNAi (lower row). All flies were raised at 18 °C. B, ATR+/+ and ATR−/− lymphoblasts were treated for 1 h with cycloheximide (50 μg/ml), and whole cell lysates were separated by 10% SDS-PAGE, transferred to nitrocellulose, and immunoblotted for ATR, P317-CHK1, total CHK1, and eFI2α. A graphical summary of P317-CHK1 from three independent repeats normalized to untreated for each genotype is shown. C, ATM+/+ and ATM−/− fibroblasts were treated for 1 h with cycloheximide (50 μg/ml), and postnuclear lysates were separated by 10% SDS-PAGE, transferred to nitrocellulose, and immunoblotted for ATM, P317-CHK1, total CHK1, and eFI2α. A graphical summary of P317-CHK1 from three independent repeats normalized to untreated for each genotype is shown. Data in B and C are mean ± S.E. (error bars).
Next, we examined the effects of translation attenuation induced by cycloheximide on CHK1 phosphorylation in cells with either mutant ATR or ATM (Fig. 2, B and C). ATR-deficient cells and wild type controls were treated with cycloheximide for 1 h. In mutant cells that expressed only low levels of ATR (28), hyperphosphorylation of CHK1 on serine 317 was detectable in unstressed cells (Fig. 2B, compare lanes 1 and 3). Nevertheless, the relative phosphorylation of CHK1 following treatment with cycloheximide was similar in control and ATR-deficient cells (Fig. 2B, lane 4). Although classically thought to phosphorylate CHK2 rather than CHK1, ATM has been reported to phosphorylate CHK1 in some circumstances (38). We therefore tested for CHK1 activation in ATM-deficient cells (Fig. 2C). The deficient cells showed lower basal levels of both total and phosphorylated CHK1. However, the relative phosphorylation of CHK1 following treatment with cycloheximide was similar in control and ATM-null cells. It has been suggested that ATM antagonizes the responses to ER stress induced by tunicamycin (39). The failure of the ATM genotype to alter CHK1 phosphorylation in response to cycloheximide suggests that no such relationship exists between ATM and direct inhibition of protein translation. Taken together, these results indicate that CHK1 is activated by the inhibition of protein translation during ER stress in the absence of detectable DNA damage and that neither ATR nor ATM is absolutely required for this response.
G2 Delay during Endoplasmic Reticulum Stress Is Enhanced in p53 Mutant Cells
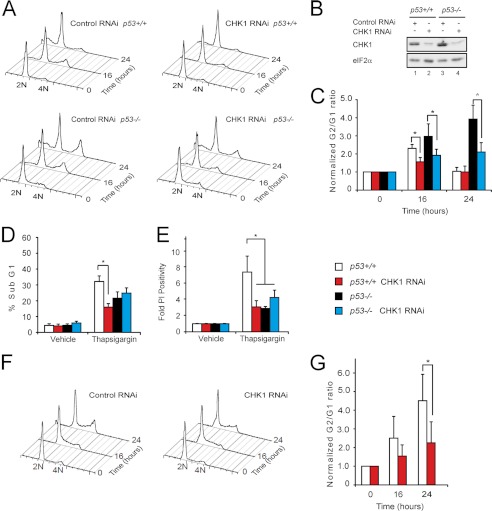
There is good evidence that p53 plays a role in the G1 arrest seen during ER stress (5–7, 20–23). Because in Drosophila p53 does not regulate the cell cycle but instead links genotoxic stress with cell death, it is unsurprising that we previously were unable to observe induction of G1 arrest in Drosophila by PERK (9). This contrasted with mammalian cells in which we observed an early G2 delay followed ultimately by G1 arrest. We reasoned that in mammalian cells the activation of distinct parallel pathways might explain the transient nature of the G2 delay. To test this, we used HCT116 cells that express either control scrambled shRNAi or CHK1-directed shRNAi (27). Although ∼50% of cell lines have impaired p53 function, HCT116 cells are suitable to study p53 function because this pathway remains intact. Targeted insertion of antibiotic resistance cassettes into exon 2 of the p53 gene has previously generated a derivative cell line lacking full-length p53 (HCT116 p53−/−) (40). We previously reported that the G2 delay observed in parental HCT116 cells treated with thapsigargin, which induces ER stress and causes marked translational attenuation, could be abrogated by expression of shRNAi against CHK1 (9). We repeated these experiments with p53−/− HCT116 derivative cells (Fig. 3, A–C, and Table 1). Immunoblotting confirmed the degree of CHK1 depletion (Fig. 3B). In the absence of functional p53, the early G2 delay persisted and became more prominent (Fig. 3A, lower left panel). Once again, induction of CHK1 shRNAi reduced this response (Fig. 3A, lower right panel). This is consistent with previous evidence that p53 plays an important role in the induction of G1 cell cycle arrest (5–7, 20–23), although it appears at odds with a recent report that ER stress-induced G2 delay might involve a short isoform of p53 (23).
FIGURE 3.
G2 delay during endoplasmic reticulum stress is enhanced in p53 mutant cells. A, representative FACS histograms of HCT116 cells of the indicated genotypes expressing either scrambled or CHK1 shRNAi and treated with thapsigargin (500 nm) for 0, 16, or 24 h; permeabilized; and then stained with propidium iodide. B, cell lysates were prepared from HCT116 cells of the indicated genotypes expressing either scrambled or CHK1 shRNAi. These were separated by 10% SDS-PAGE, transferred to nitrocellulose, and immunoblotted for total CHK1. eFI2α served as a loading control. C, graphs of G2/G1 normalized to untreated cells from A. Data are mean ± S.E. (error bars). n = 5; *, p < 0.05. D, graphs of sub-G1 cells from A. Data are mean ± S.E. (error bars). n = 5; *, p < 0.05. E, experiments were repeated identically, but cells were stained with propidium iodide without prior fixation to identify late cell death. Data are mean ± S.E. (error bars). n = 5; *, p < 0.05. F, representative FACS histograms of p53 wild type HCT116 cells expressing either scrambled or CHK1 shRNAi and treated with thapsigargin (500 nm) for 0, 16, or 24 h; permeabilized; and then stained with propidium iodide. G, graphs of G2/G1 normalized to untreated cells from F. Data are mean ± S.E. (error bars). n = 3; *, p < 0.05.
TABLE 1.
Cell cycle indices for experiments presented in Fig. 3
| Time | Cell cycle phases |
||||
|---|---|---|---|---|---|
| <G1 | G1 | G1S | G2 | ||
| h | % | ||||
| Thapsigargin | |||||
| p53+/+ | |||||
| Control RNAi | 0 | 4.4 ± 1.1 | 66.4 ± 5.1 | 22.0 ± 4.4 | 9.1 ± 1.1 |
| 16 | 12.3 ± 2.1 | 46.5 ± 2.9 | 28.1 ± 3.0 | 13.8 ± 1.4 | |
| 24 | 32.2 ± 3.5 | 46.3 ± 3.2 | 16.7 ± 1.6 | 6.3 ± 1.5 | |
| CHK1 RNAi | 0 | 4.4 ± 1.3 | 62.2 ± 4.1 | 24.5 ± 3.9 | 9.2 ± 0.5 |
| 16 | 7.9 ± 1.4 | 62.4 ± 3.8 | 17.4 ± 3.1 | 12.6 ± 0.7 | |
| 24 | 16.0 ± 2.2 | 63.3 ± 4.2 | 11.2 ± 1.9 | 8.8 ± 1.9 | |
| p53−/− | |||||
| Control RNAi | 0 | 4.9 ± 1.2 | 49.9 ± 2.1 | 36.1 ± 1.9 | 7.5 ± 2.0 |
| 16 | 13.4 ± 2.7 | 42.6 ± 4.0 | 26.3 ± 1.6 | 17.8 ± 3.9 | |
| 24 | 22.5 ± 4.6 | 32.9 ± 3.9 | 21.2 ± 2.2 | 19.3 ± 2.9 | |
| CHK1 RNAi | 0 | 6.3 ± 1.2 | 54.0 ± 2.3 | 32.2 ± 1.9 | 6.7 ± 0.8 |
| 16 | 19.4 ± 1.9 | 49.3 ± 3.8 | 20.2 ± 2.4 | 11.9 ± 2.4 | |
| 24 | 25.1 ± 3.5 | 45.2 ± 3.2 | 16.4 ± 1.7 | 12.2 ± 4.0 | |
| Cycloheximide | |||||
| p53+/+ | |||||
| Control RNAi | 0 | 3.1 ± 0.9 | 64.8 ± 6.1 | 28.7 ± 4.4 | 7.0 ± 1.6 |
| 16 | 4.3 ± 0.5 | 49.2 ± 1.3 | 40.5 ± 1.0 | 8.0 ± 0.7 | |
| 24 | 4.4 ± 1.3 | 43.7 ± 3.2 | 41.8 ± 2.8 | 11.6 ± 0.9 | |
| CHK1 RNAi | 0 | 2.3 ± 0.8 | 68.2 ± 4.3 | 24.7 ± 2.7 | 7.8 ± 1.1 |
| 16 | 5.2 ± 1.7 | 55.5 ± 2.7 | 35.8 ± 1.9 | 5.6 ± 1.2 | |
| 24 | 5.6 ± 2.0 | 53.3 ± 3.4 | 36.9 ± 1.4 | 5.6 ± 3.6 | |
As we have shown previously (9), depletion of CHK1 led to reduced cell death in response to thapsigargin treatment as reported by reduced sub-G1 material (Fig. 3, A and D, and Table 1). This effect was also observed when death was determined by permeability to propidium iodide in independent experiments (Fig. 3E). Of note, in the latter experiments, lower levels of cell death were also observed in p53-null cells.
When wild type HCT116 cells were treated with cycloheximide to inhibit protein translation, progressive G2 arrest was observed, and once again depletion of CHK1 attenuated this response (Fig. 3, F and G, and Table 1). This suggests that the inhibition of translation is sufficient to induce G2 arrest, but additional signals, perhaps mediated by p53, are required for effects on G1.
p53 Genotype Affects CHOP and GADD34 Protein Accumulation
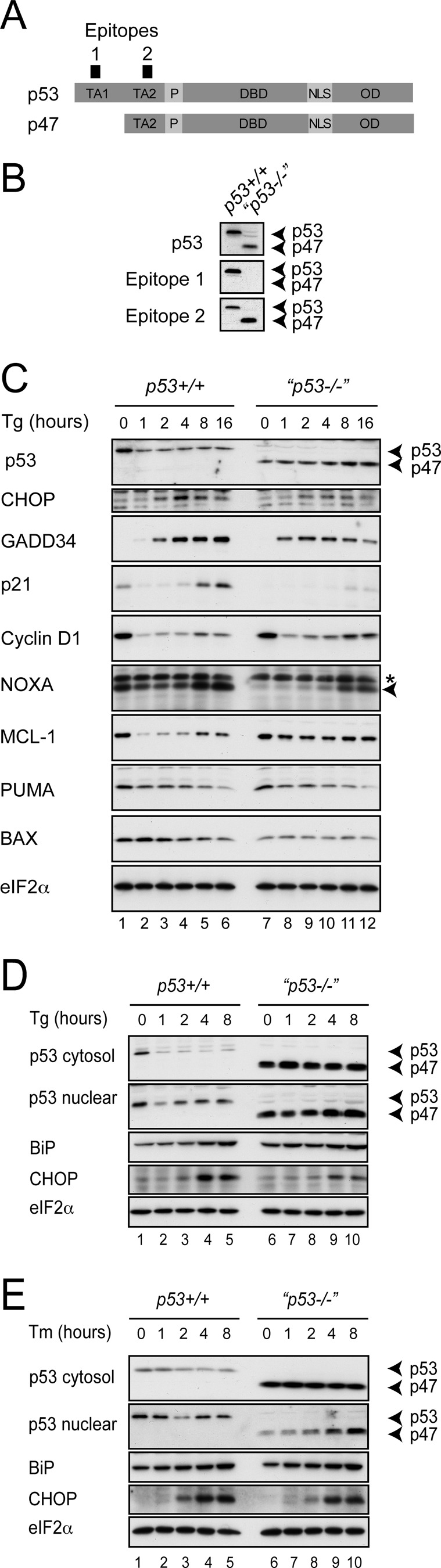
Multiple isoforms can be generated from the p53 gene either by differential splicing or by the initiation of protein translation at alternate AUGs (41). Indeed, the full-length p53 mRNA has been shown to contain at least two separate internal ribosome entry sites that generate distinct isoforms (41). One of these, denoted “full-length p53” or simply p53, contains two N-terminal transactivation domains (TA1 and TA2), whereas initiation at an alternate internal AUG can generate a p53/47 isoform that lacks TA1 (42) (Fig. 4A). When we examined HCT116 p53+/+ and p53−/− lines, we noted high levels of an N-terminally truncated isoform to be expressed by the insertional mutant cells (Fig. 4, B–E). Although the insertional mutant cells have been widely reported as p53-deficient, a similar 47-kDa p53-immunoreactive species has been reported previously in these cells (43). Using monoclonal antibodies selective for epitopes within either TA1 or TA2, we established that the shorter isoform lacks TA1, indicating that it most likely represents the p53/47 isoform (Fig. 4B).
FIGURE 4.
p53 genotype affects CHOP and GADD34 protein accumulation. A, schematic representation of domain structure of p53 and p53/47 isoforms. TA, transactivation domain; P, polyproline-rich domain (PXXP); DBD, DNA binding domain; NLS, nuclear localization sequence; OD, oligomerization domain; Epitope 1 within TA1, p53 Ab6 DO1 antibody; Epitope 2 within TA2, p53 pAb1801 antibody. B, postnuclear lysates from HCT116 cells of the indicated genotypes were analyzed by immunoblot using a polyclonal antibody against full-length p53 (polyclonal rabbit; catalog number 9282, Cell Signaling Technology), Epitope 1 (p53 Ab6 DO1), and Epitope 2 (p53 pAb1801). C, postnuclear lysates from HCT116 cells of the indicated genotypes that had been treated with thapsigargin (Tg) at 500 nm for the indicated times were analyzed by immunoblot for total p53, CHOP, GADD34, p21, cyclin D1, NOXA (note that the upper band is a nonspecific signal marked by an asterisk, and the specific band marked by an arrowhead), Mcl-1, PUMA, BAX, and eIF2α. D and E, postnuclear supernatant (cytosol) and high salt nuclear protein extract (nuclear) from HCT116 cells of the indicated genotypes treated with 400 nm thapsigargin (Tg) or 2.5 μg/ml tunicamycin (Tm) were analyzed by immunoblot; cytosol was blotted for p53, BiP, and eIF2α; and nuclear proteins were blotted for p53 and CHOP.
Next, we examined the levels of p53 in whole cell lysates from the full-length p53 cells and from the mutant cells referred to hereafter as p53−/−. Treatment with the ER stress-inducing agent thapsigargin caused a rapid drop in total p53 levels in wild type cells, whereas the p53/47 isoforms progressively increased in the mutant cells (Fig. 4C). Although p53/47 is able to homo-oligomerize and hetero-oligomerize with full-length p53, it lacks interaction with mdm2, perhaps explaining the increased stability we observed during ER stress (44). Of note, we were unable to detect the generation of p53/47 in the p53+/+ cells despite a report that ER stress leads to the preferential translation of this isoform (23). This may indicate inadequate sensitivity of detection or that p53/47 generation is not a universal feature of ER stress. To confirm successful activation of PERK, we probed for CHOP and GADD34, two downstream targets of the PERK arm of the unfolded protein response (12). Both proteins increased over the expected time course, although consistently both CHOP and GADD34 levels were lower in the p53/47-expressing cells (Fig. 4, C and D).
We then examined expression of modulators of the cell cycle. p21 is a target of p53 that arrests cells in G1. In the p53+/+ cells, we observed an initial fall of this protein followed by a marked induction (Fig. 4C). In contrast, no p21 expression was detected in the p53−/− cells. Conversely, cyclin D1, a regulatory subunit of CDK4 and CDK6 necessary for G1/S progression, fell rapidly in both cell lines followed by a recovery that was more marked in the p53−/− cells.
We next examined the expression of p53 targets involved in the regulation of cell death. Activation of apoptosis during genotoxic stress involves the transcriptional up-regulation of the proapoptotic BH3-only proteins PUMA and NOXA, which inhibit the function of antiapoptotic Bcl-2 family members including Mcl-1. The loss of antiapoptotic factors releases BAX and BAK to oligomerize within the mitochondrial membrane, thus triggering cytochrome c release and apoptosis via the intrinsic pathway (45). PUMA levels fell monotonically during ER stress in both cell types (Fig. 4C). However, NOXA levels were markedly higher under basal conditions in the p53+/+ cells, initially falling on induction of ER stress followed by a marked up-regulation after 8 h. In the p53−/− cells, NOXA levels were almost undetectable under basal conditions and rose only modestly after 8 h of ER stress. NOXA can target prosurvival Mcl-1 for degradation (46); accordingly, the levels of the antiapoptotic protein Mcl-1 fell in p53+/+ cells during ER stress but remained constant in the p53−/− cells (Fig. 4C).
There are contradictory reports regarding the effect of ER stress on the levels of p53 with reports both of increases (22, 24) and decreases with enhanced nuclear export (5–7). We therefore compared the level of p53 in cytosolic and nuclear protein extracts during ER stress (Fig. 4D). In accordance with the reduction of p53 levels seen during ER stress in whole cell lysates, we observed marked falls in p53 levels in the cytosol (Fig. 4D). However, when we examined nuclear extracts, we detected a rise in p53 level following the original fall. In contrast, p53/47 levels remained unchanged in the cytosol but progressively increased in the nucleus. Similar effects on p53 were seen when using tunicamycin to induce ER stress (Fig. 4E). The effect of the p53 genotype on CHOP was less marked, whereas no effect on GADD34 expression was seen with tunicamycin (not shown). This might reflect the known differences in the effect on translation seen with these agents: thapsigargin induces intense translational attenuation via phosphorylation of eIF2α, whereas tunicamycin has far weaker translational effects (12, 32).
Induction of mRNA Encoding CHOP and GADD34 Does Not Depend upon p53 Expression
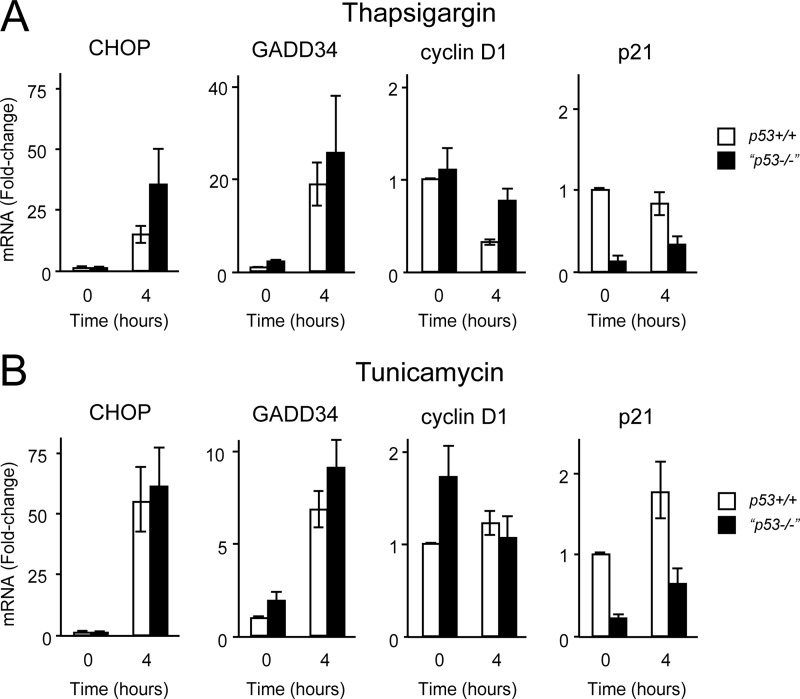
We reasoned that lower expression of CHOP and GADD34 proteins in p53-deficient cells might suggest that they were transcriptional targets of p53. We therefore treated p53+/+ and p53−/− cells with thapsigargin for 4 h and measured expression of CHOP and GADD34 by quantitative real time PCR (Fig. 5A). We simultaneously measured expression levels of p21 as a classical target of p53. As expected, p21 levels were significantly lower in p53−/− cells compared with control cells. Surprisingly, neither CHOP nor GADD34 showed lower expression in response to thapsigargin in p53−/− cells. To confirm this, we repeated the experiments with the glycosylation inhibitor tunicamycin to induce ER stress and once more saw no less induction of CHOP and GADD34 in p53−/− cells compared with controls (Fig. 5B). This suggested that in HCT116 cells neither CHOP nor GADD34 is likely to be transactivated by p53, although formal proof of this requires further study.
FIGURE 5.
CHOP and GADD34 are not direct targets of p53. A, HCT116 cells of the specified genotypes were treated with thapsigargin (500 nm) for 4 h, and mRNA was extracted, reverse transcribed using oligo(dT) primers, analyzed by quantitative PCR, normalized to actin mRNA, and expressed as -fold change relative to HCT116 p53+/+ untreated cells. Data are mean -fold change ±S.E. (error bars). n = 3 independent experiments performed in triplicate. B, experiment performed as in A but with tunicamycin (2.5 μg/ml) in place of thapsigargin.
Recovery of Translation Is Reduced in p53 Mutant Cells
Following the initial response to ER stress wherein protein translation is inhibited through the phosphorylation of eIF2α by PERK, GADD34 is induced transcriptionally under the control of the transcription factors CHOP and ATF4 (12). In complex with PP1, GADD34 selectively dephosphorylates eIF2α, enabling the recovery of protein synthesis (47–49). This process is necessary for the synthesis of target proteins of the unfolded protein response and ultimately the adaptation to ER stress. However, when ER stress is prolonged, the recovery of protein translation mediated by CHOP and GADD34 is itself toxic by increasing the load of client proteins entering the ER, resulting in worse protein aggregation (12).
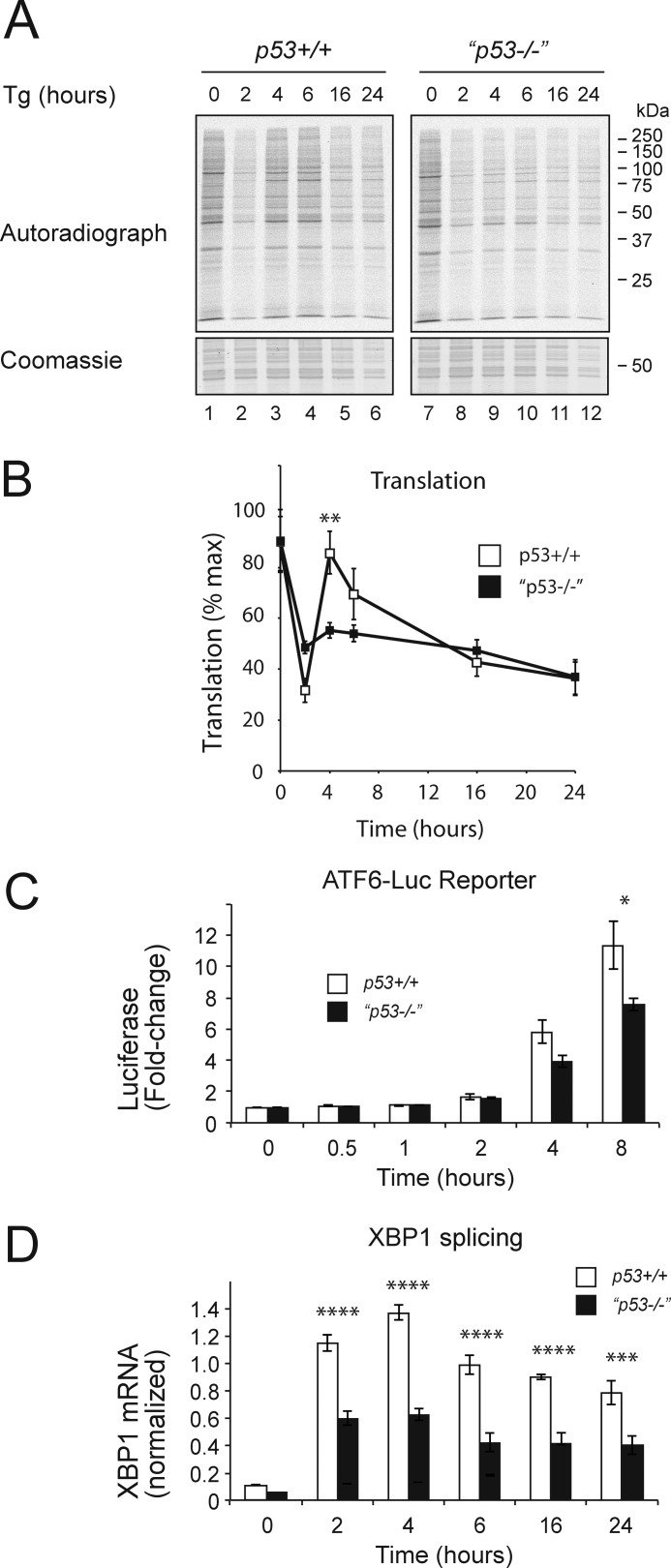
The observed differences in CHOP and GADD34 protein levels between wild type and p53−/− cells despite similar levels of mRNA suggested differences in either protein synthesis or stability. To test this, we used metabolic labeling to determine the level of new protein translation in each genotype during treatment with thapsigargin to induce ER stress (Fig. 6, A and B). As expected, in wild type cells, protein translation recovered between 4 and 6 h and correlated with the induction of GADD34 in wild type cells (Figs. 4C and 6A). At later time points, protein synthesis fell once more, suggesting either additional regulation of translation or a nonspecific agonal response. In contrast, the p53 mutant cells with their lower levels of CHOP and GADD34 failed to recover protein translation even transiently. In this light, the more efficient recovery of cyclin D1 and the sustained expression of Mcl-1 were noteworthy because both are short lived proteins. The level of cyclin D1 mRNA did not increase with thapsigargin treatment, suggesting that its mRNA may be translated efficiently even when global protein synthesis is attenuated (Fig. 4C).
FIGURE 6.
Recovery of translation is reduced in p53 mutant cells. A, HCT116 cells of the specified genotypes were treated with thapsigargin (Tg) (500 nm) for the indicated times and labeled with 35S-labeled methionine and cysteine for 15 min, and then equal quantities of postnuclear lysate were separated by 10% SDS-PAGE, fixed, stained, dried, and exposed to a phosphorimaging screen. B, pooled data from three independent repeats of A expressed as percent maximum signal. Data are mean ± S.E. (error bars). n = 3. C, HCT116 cells of the specified genotypes were co-transfected with p5×ATF6-Luc encoding firefly luciferase under the control of a UPR response element and with pRL-TK Renilla luciferase as a normalization control. Data presented as firefly luciferase activity normalized to Renilla luciferase are mean ± S.E. (error bars). n = 3 independent repetitions. D, XBP1s mRNA level normalized to the mRNA levels of actin and RPL13A mRNA (normalized expression). HCT116 cells of the specified genotypes were treated with thapsigargin (500 nm) for the indicated times. mRNA levels were determined by qPCR. Data are mean ± S.E. (error bars). n = 3 independent repetitions; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 (analysis of variance with Bonferroni post hoc test).
We have shown previously that the recovery of protein translation mediated by GADD34 can exacerbate the degree of ER stress (12). Moreover, genetic manipulations that decreased the recovery of protein translation including deletion of the CHOP gene and mutating GADD34 led to lower levels of accumulating misfolded protein aggregates within the ER lumen and consequently to reduced ER stress signaling. Our observation that CHOP and GADD34 mRNA levels were not decreased in p53−/− cells appeared to argue against a difference in the degree of ER stress between the genotypes (Fig. 5). However, both CHOP and GADD34 are targets of the transcription factor ATF4, which could potentially confound this interpretation. ATF4 is translated most efficiently during periods of eIF2α phosphorylation when most other proteins are translated less efficiently (50). It was therefore possible that efficient ATF4 translation caused by lower rates of translation in stressed p53−/− cells might mask a difference in the degree of ER stress. To address this, we examined ATF6 activation in both cells types. ATF6 is an ER membrane protein that is activated during ER stress by site-specific cleavage to liberate a soluble transcription factor (51). We measured ATF6 activity using a firefly luciferase reporter, p5×ATF6-Luc (52), and controlled for alterations of global translation rate by simultaneously measuring constitutively synthesized Renilla luciferase, a protein with a half-life similar to that of firefly luciferase (Fig. 6C). Using this system, we observed a reduction in ATF6-luciferase reporter activity in the p53−/− cells consistent with lower levels of ER stress in these mutant cells compared with the wild type controls. Furthermore, activation of the ER stress sensor IRE1 was measured following treatment with thapsigargin by quantifying the splicing of the mRNA encoding XBP1 (Fig. 6D). Because splicing of XBP1 mRNA is independent of new protein translation (53), the observed difference between p53+/+ and p53−/− cannot be explained as defective synthesis of the reporter and strongly suggests that p53−/− cells experience lower levels of ER stress than do wild type cells.
GADD34 Stability Is Affected by p53 Genotype
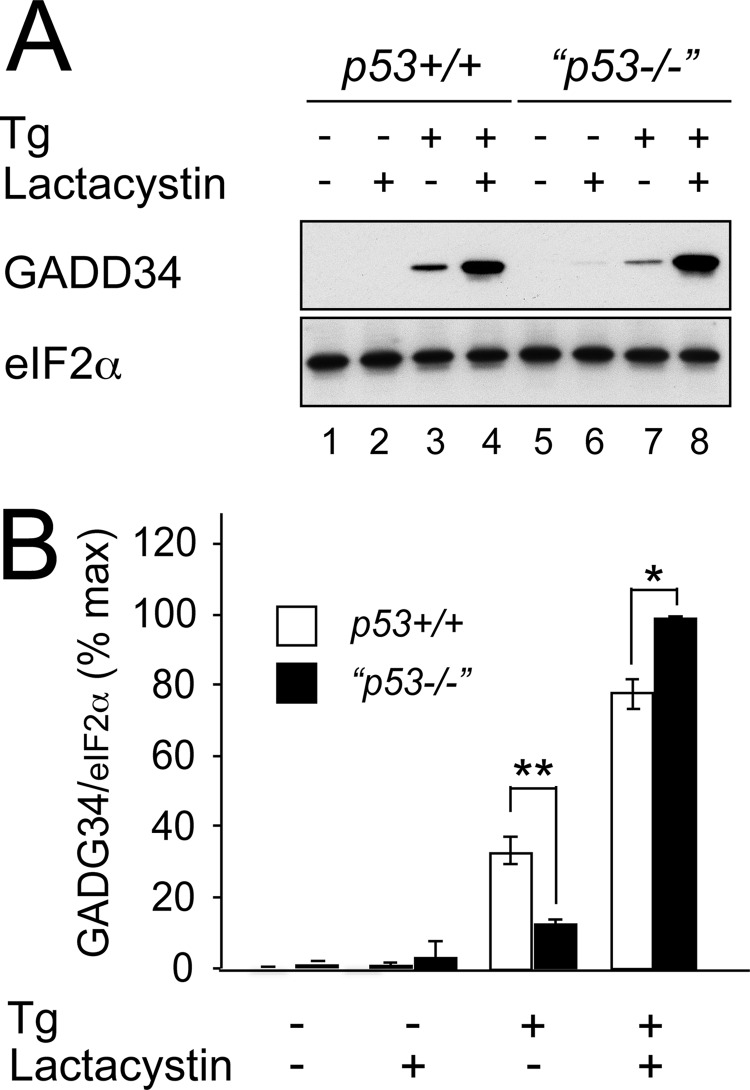
Although p53 is known to perform a homeostatic role in ribosome biogenesis (54), we detected no significant difference in basal translation rates between unstressed wild type and p53−/− cells (Fig. 6A, compare lanes 1 and 7). We therefore considered whether differences in GADD34 stability might contribute to the differences in GADD34 levels. To assess further the contribution of protein stability to the GADD34 level, we inhibited the proteasome with lactacystin (Fig. 7, A and B). Although treatment with thapsigargin induced higher levels of GADD34 in wild type cells (Fig. 7A, compare lanes 3 and 7), the addition of lactacystin increased GADD34 levels to a significantly greater level in p53−/− cells (Fig. 7, A and B). This suggested that differences in GADD34 translation were not the primary cause of its higher expression in wild type cells and that GADD34 was subject to increased proteasomal degradation in p53 mutant cells.
FIGURE 7.
GADD34 stability is affected by p53 genotype. A, HCT116 cells of the specified genotypes were treated with or without thapsigargin (Tg) (500 nm) for 24 h and/or with lactacystin (5 μm). Cell lysates were analyzed by 10% SDS-PAGE, transferred to nitrocellulose, and immunoblotted for GADD34 and eIF2α. B, graphical representation of A. Immunoblots were analyzed using ImageJ software. Data presented as percent GADD34 band intensity relative to eIF2α loading control are mean ± S.E. (error bars). n = 3 independent repeats; *, p < 0.05; **, p < 0.01.
Taken together, this series of experiments revealed that p53-expressing cells experience elevated levels of ER stress when treated with thapsigargin because of early recovery of protein translation mediated by GADD34. The difference in GADD34 protein level between wild type and p53−/− cells reflects increased GADD34 instability in p53-expressing cells. Because GADD34 expression increases protein synthesis during ER stress, minor differences in GADD34 stability are likely to have a profound effect on translation rates through a positive feedback mechanism whereby GADD34-mediated recovery of translation leads to increased GADD34 synthesis and further recovery of translation.
DISCUSSION
In this study, we examined the relationship among ER stress, protein translation, and cell cycle progression. Our observations combined with previous published work led us to propose a two-signal model for the regulation of cell proliferation during ER stress. The first signal represents inhibition of protein translation mediated via the phosphorylation of eIF2α by PERK, leading to impaired G2/M cell cycle progression. This appears to require activation of CHK1 by a mechanism that is yet to be fully determined, but neither ATR nor ATM is essential for this activation during ER stress. The second signal, which induces G1 at later stages of ER stress, requires full-length p53. The loss of cyclin D1 that occurs early during ER stress and is mediated by activation of PERK also appears to contribute to this checkpoint but is not sufficient (3, 4, 8, 9).
This model offers a conceptual framework that explains several features of the cell cycle response to ER stress. First, the G2 delay that we and others recently described is observed during intense ER stress that results in the marked attenuation of protein translation, for example following treatment with thapsigargin (this study and Refs. 9 and 23). The transient nature of the G2 delay in wild type cells can be explained as the combined effect of translational recovery mediated by GADD34 and the delayed induction of p53-dependent factors including p21 that induce G1 arrest. In Drosophila models, activation of PERK has been shown to induce a G2 delay combined with cell death, but there is no evidence of G1 arrest (9).4 This may reflect species differences in p53 function because Drosophila p53 does not regulate cell cycle progression but does regulate cell death in response to genotoxicity (55).
Evidence that during ER stress a p53/47 N-terminally truncated isoform of p53 can be generated through altered choice of translation initiation site and induce G2 delay is consistent with this model because it too represents a response to translation attenuation mediated by PERK (23). Although the p53/47 isoform is unable to up-regulate expression of p21 or to induce apoptosis in response to DNA damage (56), recent evidence suggests that it can contribute to G2 arrest through induction of 14-3-3σ (23). This may contribute to the exaggerated G2 arrest that we observed in p53 mutant cells, which overexpress p53/47. However, because these cells do not display basal G2 delay and lose the ER stress G2 checkpoint when CHK1 is depleted by shRNAi, it seems likely that p53/47 is only one part of the response and may be subordinate to other components including CHK1. Full-length p53 is regulated through direct phosphorylation by CHK1 (57), but it remains to be determined whether this is the case for p53/47.
The literature currently contains a number of apparent contradictions pertaining to the relationship between p53 and ER stress. For example, it has been shown that ER stress can lead to the accumulation of active p53 (22, 24), whereas other studies have shown nuclear export and degradation of p53 in response to ER stress (5–7). In our current study, we observed a biphasic response with an initial loss of p53 in response to ER stress in both the nucleus and cytosol followed by a recovery of p53 within nuclear fractions at later time points. The nature and intensity of the ER stress may also play a role because we observed a loss of p53 most markedly in cells experiencing intense eIF2α phosphorylation in response to thapsigargin or during ribotoxic stress, whereas tunicamycin, which induces ER stress more gradually and therefore has less dramatic effects on the rate of protein translation, caused a less dramatic change in p53. It is worth noting that induction of mRNA of the p53 target gene p21 was more marked at 4 h of treatment with tunicamycin than with thapsigargin. This suggests that the relative importance of signal 1 (translation attenuation) and signal 2 (p53 activation) is likely to be highly context-specific with differing effects from intense rapid and mild chronic ER stress.
Our study has not specifically addressed the signals linking ER stress with cell death, but because altered protein translation mediated by PERK and target genes of p53 have both been implicated in triggering apoptosis during ER stress, it is noteworthy that marked differences were seen in the expression of death-related genes between wild type and p53 mutant cells. As reported previously, we observed induction of the proapoptotic BH3-only protein NOXA in a p53-dependent fashion at later stages of ER stress (22) and the early loss of the antiapoptotic Bcl-2 family member Mcl-1. It has been suggested that phosphorylation of eIF2α during ER stress contributes to cell death by blocking the expression of Mcl-1 in insulinoma cells (58). However, we saw no loss of Mcl-1 in p53−/− cells despite a more pronounced attenuation of protein translation in these cells, suggesting that active degradation induced by NOXA may be the dominant regulatory mechanism.
Acknowledgment
We thank Professor Ashok Venkitaraman (University of Cambridge) for insightful comments and critical reading of the manuscript.
This work was supported in part by Diabetes UK, the Medical Research Council (MRC) (UK), and the British Lung Foundation.
Elke Malzer and Stefan J. Marciniak, unpublished data.
- ER
- endoplasmic reticulum
- PERK
- protein kinase RNA-like endoplasmic reticulum kinase
- shRNAi
- small interfering hairpin RNA
- ATM
- ataxia-telangiectasia mutated
- ATR
- ATM- and Rad3-related
- CHOP
- CCAAT/enhancer-binding protein homologous protein
- BiP
- binding immunoglobulin protein
- PUMA
- p53-upregulated modulator of apoptosis
- BAX
- Bcl-2-associated X protein
- BAK
- Bcl-2 antagonist/killer
- UPR
- unfolded protein response
- qPCR
- quantitative PCR
- XBP1s
- spliced form of XBP1
- dPERK
- Drosophila PERK
- Ab
- antibody
- pAb
- polyclonal antibody.
REFERENCES
- 1. Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Adapting proteostasis for disease intervention. Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 2. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 3. Brewer J. W., Hendershot L. M., Sherr C. J., Diehl J. A. (1999) Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. U.S.A. 96, 8505–8510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brewer J. W., Diehl J. A. (2000) PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 97, 12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qu L., Huang S., Baltzis D., Rivas-Estilla A. M., Pluquet O., Hatzoglou M., Koumenis C., Taya Y., Yoshimura A., Koromilas A. E. (2004) Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3β. Genes Dev. 18, 261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pluquet O., Qu L. K., Baltzis D., Koromilas A. E. (2005) Endoplasmic reticulum stress accelerates p53 degradation by the cooperative actions of Hdm2 and glycogen synthase kinase 3β. Mol. Cell. Biol. 25, 9392–9405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baltzis D., Pluquet O., Papadakis A. I., Kazemi S., Qu L. K., Koromilas A. E. (2007) The eIF2α kinases PERK and PKR activate glycogen synthase kinase 3 to promote the proteasomal degradation of p53. J. Biol. Chem. 282, 31675–31687 [DOI] [PubMed] [Google Scholar]
- 8. Raven J. F., Baltzis D., Wang S., Mounir Z., Papadakis A. I., Gao H. Q., Koromilas A. E. (2008) PKR and PKR-like endoplasmic reticulum kinase induce the proteasome-dependent degradation of cyclin D1 via a mechanism requiring eukaryotic initiation factor 2α phosphorylation. J. Biol. Chem. 283, 3097–3108 [DOI] [PubMed] [Google Scholar]
- 9. Malzer E., Daly M. L., Moloney A., Sendall T. J., Thomas S. E., Ryder E., Ryoo H. D., Crowther D. C., Lomas D. A., Marciniak S. J. (2010) Impaired tissue growth is mediated by checkpoint kinase 1 (CHK1) in the integrated stress response. J. Cell Sci. 123, 2892–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12, 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakagawa T., Zhu H., Morishima N., Li E., Xu J., Yankner B. A., Yuan J. (2000) Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 403, 98–103 [DOI] [PubMed] [Google Scholar]
- 12. Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., Ron D. (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song B., Scheuner D., Ron D., Pennathur S., Kaufman R. J. (2008) Chop deletion reduces oxidative stress, improves β cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Investig. 118, 3378–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ozcan U., Cao Q., Yilmaz E., Lee A. H., Iwakoshi N. N., Ozdelen E., Tuncman G., Görgün C., Glimcher L. H., Hotamisligil G. S. (2004) Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306, 457–461 [DOI] [PubMed] [Google Scholar]
- 15. Nakatani Y., Kaneto H., Kawamori D., Yoshiuchi K., Hatazaki M., Matsuoka T. A., Ozawa K., Ogawa S., Hori M., Yamasaki Y., Matsuhisa M. (2005) Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J. Biol. Chem. 280, 847–851 [DOI] [PubMed] [Google Scholar]
- 16. Laybutt D. R., Preston A. M., Akerfeldt M. C., Kench J. G., Busch A. K., Biankin A. V., Biden T. J. (2007) Endoplasmic reticulum stress contributes to β cell apoptosis in type 2 diabetes. Diabetologia 50, 752–763 [DOI] [PubMed] [Google Scholar]
- 17. Thomas S. E., Dalton L. E., Daly M. L., Malzer E., Marciniak S. J. (2010) Diabetes as a disease of endoplasmic reticulum stress. Diabetes Metab. Res. Rev. 26, 611–621 [DOI] [PubMed] [Google Scholar]
- 18. Bobrovnikova-Marjon E., Grigoriadou C., Pytel D., Zhang F., Ye J., Koumenis C., Cavener D., Diehl J. A. (2010) PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene 29, 3881–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koumenis C., Naczki C., Koritzinsky M., Rastani S., Diehl A., Sonenberg N., Koromilas A., Wouters B. G. (2002) Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol. Cell. Biol. 22, 7405–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J., Marechal V., Levine A. J. (1993) Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13, 4107–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jin A., Itahana K., O'Keefe K., Zhang Y. (2004) Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24, 7669–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li J., Lee B., Lee A. S. (2006) Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J. Biol. Chem. 281, 7260–7270 [DOI] [PubMed] [Google Scholar]
- 23. Bourougaa K., Naski N., Boularan C., Mlynarczyk C., Candeias M. M., Marullo S., Fåhraeus R. (2010) Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Mol. Cell 38, 78–88 [DOI] [PubMed] [Google Scholar]
- 24. Zhang F., Hamanaka R. B., Bobrovnikova-Marjon E., Gordan J. D., Dai M. S., Lu H., Simon M. C., Diehl J. A. (2006) Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. J. Biol. Chem. 281, 30036–30045 [DOI] [PubMed] [Google Scholar]
- 25. Ray P. S., Grover R., Das S. (2006) Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 7, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Candeias M. M., Powell D. J., Roubalova E., Apcher S., Bourougaa K., Vojtesek B., Bruzzoni-Giovanelli H., Fåhraeus R. (2006) Expression of p53 and p53/47 are controlled by alternative mechanisms of messenger RNA translation initiation. Oncogene 25, 6936–6947 [DOI] [PubMed] [Google Scholar]
- 27. Ganzinelli M., Carrassa L., Crippa F., Tavecchio M., Broggini M., Damia G. (2008) Checkpoint kinase 1 down-regulation by an inducible small interfering RNA expression system sensitized in vivo tumors to treatment with 5-fluorouracil. Clin. Cancer Res. 14, 5131–5141 [DOI] [PubMed] [Google Scholar]
- 28. Alderton G. K., Joenje H., Varon R., Børglum A. D., Jeggo P. A., O'Driscoll M. (2004) Seckel syndrome exhibits cellular features demonstrating defects in the ATR-signalling pathway. Hum. Mol. Genet. 13, 3127–3138 [DOI] [PubMed] [Google Scholar]
- 29. Gilad S., Khosravi R., Shkedy D., Uziel T., Ziv Y., Savitsky K., Rotman G., Smith S., Chessa L., Jorgensen T. J., Harnik R., Frydman M., Sanal O., Portnoi S., Goldwicz Z., Jaspers N. G., Gatti R. A., Lenoir G., Lavin M. F., Tatsumi K., Wegner R. D., Shiloh Y., Bar-Shira A. (1996) Predominance of null mutations in ataxia-telangiectasia. Hum. Mol. Genet. 5, 433–439 [DOI] [PubMed] [Google Scholar]
- 30. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Schadewijk A., van't Wout E. F., Stolk J., Hiemstra P. S. (2012) A quantitative method for detection of spliced X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic reticulum (ER) stress. Cell Stress Chaperones 17, 275–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harding H. P., Zhang Y., Ron D. (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274 [DOI] [PubMed] [Google Scholar]
- 33. Schneider-Poetsch T., Ju J., Eyler D. E., Dang Y., Bhat S., Merrick W. C., Green R., Shen B., Liu J. O. (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marciniak S. J., Ron D. (2006) Endoplasmic reticulum stress signaling in disease. Physiol. Rev. 86, 1133–1149 [DOI] [PubMed] [Google Scholar]
- 35. Rabinovitz M., Fisher J. M. (1962) A dissociative effect of puromycin on the pathway of protein synthesis by Ehrlich ascites tumor cells. J. Biol. Chem. 237, 477–481 [PubMed] [Google Scholar]
- 36. Ingolia N. T., Lareau L. F., Weissman J. S. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao H., Piwnica-Worms H. (2001) ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21, 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gatei M., Sloper K., Sorensen C., Syljuäsen R., Falck J., Hobson K., Savage K., Lukas J., Zhou B. B., Bartek J., Khanna K. K. (2003) Ataxia-telangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J. Biol. Chem. 278, 14806–14811 [DOI] [PubMed] [Google Scholar]
- 39. He L., Kim S. O., Kwon O., Jeong S. J., Kim M. S., Lee H. G., Osada H., Jung M., Ahn J. S., Kim B. Y. (2009) ATM blocks tunicamycin-induced endoplasmic reticulum stress. FEBS Lett. 583, 903–908 [DOI] [PubMed] [Google Scholar]
- 40. Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 41. Grover R., Ray P. S., Das S. (2008) Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle 7, 2189–2198 [DOI] [PubMed] [Google Scholar]
- 42. Grover R., Sharathchandra A., Ponnuswamy A., Khan D., Das S. (2011) Effect of mutations on the p53 IRES RNA structure: implications for de-regulation of the synthesis of p53 isoforms. RNA Biol. 8, 132–142 [DOI] [PubMed] [Google Scholar]
- 43. Murray-Zmijewski F., Lane D. P., Bourdon J. C. (2006) p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 13, 962–972 [DOI] [PubMed] [Google Scholar]
- 44. Olivares-Illana V., Fåhraeus R. (2010) p53 isoforms gain functions. Oncogene 29, 5113–5119 [DOI] [PubMed] [Google Scholar]
- 45. Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Willis S. N., Chen L., Dewson G., Wei A., Naik E., Fletcher J. I., Adams J. M., Huang D. C. (2005) Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 19, 1294–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Novoa I., Zeng H., Harding H. P., Ron D. (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brush M. H., Weiser D. C., Shenolikar S. (2003) Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1α to the endoplasmic reticulum and promotes dephosphorylation of the α subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 23, 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ma Y., Hendershot L. M. (2003) Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J. Biol. Chem. 278, 34864–34873 [DOI] [PubMed] [Google Scholar]
- 50. Lu P. D., Harding H. P., Ron D. (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haze K., Yoshida H., Yanagi H., Yura T., Mori K. (1999) Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Irving J. A., Ekeowa U. I., Belorgey D., Haq I., Gooptu B., Miranda E., Pérez J., Roussel B. D., Ordóñez A., Dalton L. E., Thomas S. E., Marciniak S. J., Parfrey H., Chilvers E. R., Teckman J. H., Alam S., Mahadeva R., Rashid S. T., Vallier L., Lomas D. A. (2011) The serpinopathies studying serpin polymerization in vivo. Methods Enzymol. 501, 421–466 [DOI] [PubMed] [Google Scholar]
- 53. Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 [DOI] [PubMed] [Google Scholar]
- 54. Krastev D. B., Slabicki M., Paszkowski-Rogacz M., Hubner N. C., Junqueira M., Shevchenko A., Mann M., Neugebauer K. M., Buchholz F. (2011) A systematic RNAi synthetic interaction screen reveals a link between p53 and snoRNP assembly. Nat. Cell Biol. 13, 809–818 [DOI] [PubMed] [Google Scholar]
- 55. Steller H. (2000) Drosophila p53: meeting the Grim Reaper. Nat. Cell Biol. 2, E100–E102 [DOI] [PubMed] [Google Scholar]
- 56. Ma W., Lin Y., Xuan W., Iversen P. L., Smith L. J., Benchimol S. (2012) Inhibition of p53 expression by peptide-conjugated phosphorodiamidate morpholino oligomers sensitizes human cancer cells to chemotherapeutic drugs. Oncogene 31, 1024–1033 [DOI] [PubMed] [Google Scholar]
- 57. Polager S., Ginsberg D. (2009) p53 and E2f: partners in life and death. Nat. Rev. Cancer 9, 738–748 [DOI] [PubMed] [Google Scholar]
- 58. Allagnat F., Cunha D., Moore F., Vanderwinden J. M., Eizirik D. L., Cardozo A. K. (2011) Mcl-1 downregulation by pro-inflammatory cytokines and palmitate is an early event contributing to β-cell apoptosis. Cell Death Differ. 18, 328–337 [DOI] [PMC free article] [PubMed] [Google Scholar]