Background: The type VI secretion system provides Gram-negative bacteria with a competitive advantage.
Results: The V. cholerae T6SS component VgrG-3 has lysozyme-like activity that is inhibited by the product of the downstream gene tsaB.
Conclusion: VgrG-3 and TsaB are a toxin-antitoxin complex of the V. cholerae T6SS.
Significance: A T6SS effector is characterized along with its cognate antitoxin.
Keywords: Bacterial pathogenesis, Bacterial toxins, Cell wall, Host-pathogen interactions, Protein secretion
Abstract
The type VI secretion system (T6SS) of Gram-negative bacteria has been implicated in microbial competition; however, which components serve purely structural roles, and which serve as toxic effectors remains unresolved. Here, we present evidence that VgrG-3 of the Vibrio cholerae T6SS has both structural and toxin activity. Specifically, we demonstrate that the C-terminal extension of VgrG-3 acts to degrade peptidoglycan and hypothesize that this assists in the delivery of accessory T6SS toxins of V. cholerae. To avoid self-intoxication, V. cholerae expresses an anti-toxin encoded immediately downstream of vgrG-3 that inhibits VgrG-3-mediated lysis through direct interaction.
Introduction
VgrG (valine-glycine repeat G) proteins, along with hemolysin-coregulated protein (Hcp)2 represent two hallmark proteins of the type VI secretion system (T6SS), a virulence mechanism present in most Gram-negative bacteria (1). Hcp forms hexameric rings that stack upon each other to form a membrane spanning nanotube with an internal diameter of 4 nm (2). Based on structural homology to the T4 bacteriophage tail spike, VgrG proteins are thought to form a trimeric tip on the distal end of the Hcp nanotube, forming a poison dagger (3, 4). This structure is forced through the membranes of both host and target cells through the mechanical force generated by a contractile sheath surrounding the Hcp nanotube (5). Puncture of a neighboring cell by the Hcp nanotube could provide a conduit through which effector proteins are delivered to the target cell, much like viral DNA is delivered to bacteria by the bacteriophage T4 tail spike; however, this scenario seems unlikely as x-ray crystallographic studies indicate that the trimeric VgrG complex forms a closed cap on the Hcp nanotube (3).
Alternatively, Hcp tubes injected into neighboring cells might utilize VgrG proteins as effectors. The T6SS of Vibrio cholerae encodes three VgrG genes (vgrG-1, vgrG-2, vgrG-3), with each sharing the phage late control gene D protein (GPD) and phage-related base plate assembly protein domains of T4 bacteriophage gp27 and gp5, respectively. In addition to their structural role in penetration of host membranes, VgrG-1 and VgrG-3 each encode additional functional domains on their C termini. These C-terminal extensions have putative effector functions that could be important for virulence toward adjacent cells. Previous work has shown that the C-terminal extension of VgrG-1 exhibits actin cross-linking activity that, upon phagocytosis, results in toxicity toward murine macrophages and the social amoeba Dictyostelium discocoideum (6). VgrG extensions in other bacterial species have also been shown to contain effector domains; VgrG-1 of Aeromonas hydrophila has ADP-ribosylating activity (7) and a VgrG (COG5529) in SPI-21 of Salmonella enterica contains a pyocin domain (1, 8). It has been noted that the C-terminal extension of V. cholerae VgrG-3 encodes a peptidoglycan binding domain (PG1); however, the physiological role of this domain is yet to be determined (1).
Recent reports have demonstrated the importance of the T6SS in V. cholerae and other Gram-negative bacteria in microbial competition (9–12). Here, we show that the C-terminal extension of VgrG-3 is used to hydrolyze the cell wall of Gram-negative bacteria, thereby conferring a competitive advantage to V. cholerae against other Gram-negative competitors. In addition, the product of the downstream gene, tsaB (type six secretion antitoxin B) is shown to directly inhibit VgrG-3 in a toxin-antitoxin manner.
MATERIALS AND METHODS
Bioinformatics Analysis
The amino acid sequence of V. cholerae N16961 VgrG-3 (Uniprot ID Q9KN42_VIBCH) was analyzed using HMMER to identify conserved domains.
Strains and Culture Conditions
A V. cholerae V52 strain in which hapA, rtxA, and hlyA had been deleted by in-frame mutation was used in this study and is denoted as V52 wt. For periplasmic expression, proteins were provided with a Sec secretion signal and expressed from pBAD24-LS-based constructs in the Escherichia coli cloning strain TOP10 (Invitrogen). The expression strain BL21*DE3 (Invitrogen) was used for large-scale expression of recombinant proteins. For lysis assays and peptidoglycan isolation, a rifampin-resistant isolate of the E. coli K12 strain MG1655 was employed. All cultures were grown in Luria Bertani broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl) at 37 °C with shaking.
Molecular Cloning
For recombinant expression, VgrG-3 (residues 1–1017) and VgrG-3C (VgrG-3 residues 727–1017) were cloned between the NdeI and XhoI sites of pET28a (Invitrogen) yielding an in-frame N-terminal 6× His-tag followed by a thrombin cleavage sequence upstream of the Vibrio gene. TsaB was also cloned between the NdeI and XhoI sites of pET28a but was fused to the C-terminal 6× His-tag, and the predicted N-terminal signal peptide (residues 1–27) was omitted to ensure retention of the recombinant protein in the cytoplasm. The constructs were transformed into E. coli BL21*DE3 for expression.
The periplasmic expression vector pBAD24-LS was constructed by inserting the N-terminal signal sequence of E. coli dsbA downstream of the AraC promoter of pBAD24. The VgrG-3 gene was divided into two functional regions, the VgrG core (VgrG-3N, residues 1–726) and the C-terminal extension (VgrG-3C, residues 727–1017) and cloned in-frame with the secretion signal using PstI and XbaI restriction sites. The resulting constructs were transformed into E. coli TOP10 (Invitrogen) for functional analyses.
Purification of Recombinant Proteins
Recombinant VgrG-3 and TsaB constructs were purified from 4-liter expression cultures by nickel affinity. Briefly, the cell pellets were lysed in resuspension buffer (20 mm HEPES, 100 mm NaCl, pH 8) with 10 units of Dnase I (Fermentas) and complete protease inhibitor mixture (Roche Applied Science) using a French pressure cell (Thermo Scientific, French Press Cell Disruptor). Insoluble cellular debris were pelleted at 25,000 × g, and the lysate was filtered and applied to a HisTrap FF column equilibrated with binding buffer (20 mm HEPES, 500 mm NaCl, 20 mm imidazole, pH 8) on an AKTA basic FPLC system (GE Healthcare). Following extensive washing, the His-tagged proteins were eluted with a 20-ml gradient of 20–500 mm imidazole. Pure fractions were pooled and dialyzed against resuspension buffer to remove imidazole and decrease salinity.
Zymography
Peptidoglycan (PG) was isolated from E. coli MG1655-Rif according to the method of Hoyle and Beveridge (13). Briefly, four liters of volume of overnight culture was pelleted and resuspended in water to a density of 200 g/liter, then added dropwise to an equal volume of boiling 8% SDS. The mixture was boiled for 1–3 h before ultracentrifugation at 100,000 × g at room temperature to sediment PG. The pellet was washed with distilled water four to five times to remove SDS and then lyophilized to dryness to determine the yield. The crude PG preparation was mixed to 0.1% w/v in 12% SDS-PAGE. Samples for zymography were prepared in 1× Laemmli buffer and electrophoresed at 200 V for 1 h. After electrophoresis, the gel was washed with water to remove SDS and then equilibrated in renaturation buffer (10 mm Tris-HCl, pH 7, 0.1% Triton X-100). Fresh renaturation buffer was added, and the gel was incubated at 37 °C overnight with agitation. To visualize degraded PG, the gel was washed quickly three times with water and then stained with methylene blue stain (0.1% methylene blue, 0.01% KOH) for 3 h followed by water washing until bands were clearly visible. To assess the optimal buffer conditions for VgrG-3 degradation of PG, purified recombinant protein was run on a zymogram and incubated in variations of renaturation buffer as indicated in Fig. 4.
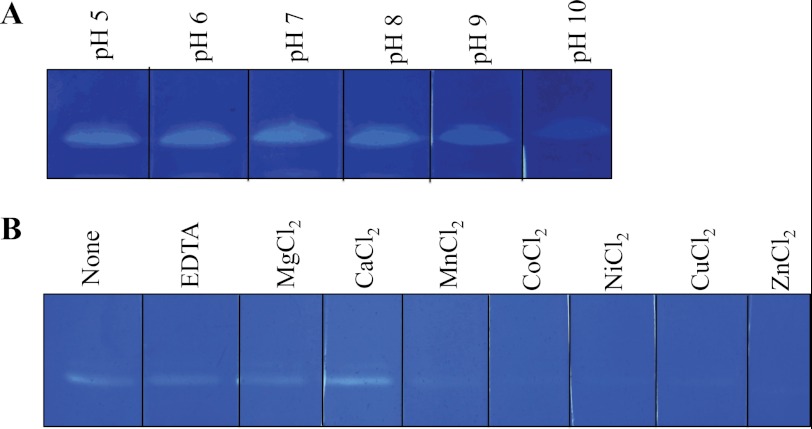
FIGURE 4.
Zymogram analysis demonstrating the effects of pH (A) and divalent cations on lytic activity of VgrG-3C (B). VgrG-3C within a SDS-polyacrylamide gel containing 0.1% (w/v) E. coli PG was renatured in 25 mm Tris, pH 7.0, 0.1% Triton X-100 with the indicated additive at a concentration of 10 mm. The atomic number of the divalent cations increases from left to right.
Periplasmic Targeting
Overnight cultures of E. coli TOP10 (Invitrogen) cells harboring the pBAD24-LS::vgrG-3 plasmid or relevant controls were diluted to A600 ∼ 1.0 in either LB + 0.2% d-glucose (inhibiting) or LB + 0.2% l-arabinose (inducing) and transferred to a 96-well plate. The A600 was measured in an xMark microplate spectrophotometer (Bio-Rad), and then the plate was incubated at 37 °C on a vibrating shaker for 60 min, and the A600 was measured once more. The relative change in A600 was calculated as (A600 60 min − A600 0 min)/A600 0 min.
Microscopy
Overnight cultures of E. coli TOP10 cells harboring the pBAD24-LS::vgrG-3 plasmid or relevant controls were diluted to A600 ∼ 1.0 in LB + 0.2% d-glucose. After washing with PBS, bacteria were stained with 5(6)-FAM NHS ester (5-(and 6)-carboxyfluorescein succinimidyl ester) (Invitrogen) in PBS + 0.2% d-glucose for 30 min at 37 °C. After washing with PBS + 0.2% glucose, bacteria were resuspended in LB broth supplemented with 0.2% d-glucose and 20 μm propidium iodide (Live/Dead®; Invitrogen). Bacteria were directly transferred onto an objective slide prepared with a thin LB agar pad supplemented with 0.2% l-arabinose. Samples were analyzed with a Leica DMI6000B fluorescence microscope (Leica Microsystems; Wetzlar, Germany) for 2 h in an incubation chamber (TokaiHit) at 37 °C. Images were analyzed using NIH ImageJ software.
Killing Assay
Assessment of bacterial competition was conducted as described in Ref. 14. Briefly, predator and prey bacteria strains were mixed at a 10:1 ratio, spotted onto LB plates, and incubated at 37 °C for 4 h. After incubation, spots were harvested, and serial dilutions were plated to determine the surviving prey cfu/ml. Results are the mean of three experiments, with each performed in duplicate. Where applicable, 0.2% l-arabinose was added to the agar medium to induce expression from the pBAD24 plasmid or 0.2% d-glucose was added to repress basal expression.
Chemical Cross-linking Studies
Purified recombinant VgrG-3C and TsaB were mixed at a 1:1 mass ratio in resuspension buffer (each at 0.5 mg/ml) and cross-linked with 2 mm dithiobis(succinimidyl propionate) (Pierce) according to the manufacturer's instructions. The samples were mixed with SDS-PAGE sample buffer with and without β-mercaptoethanol to cleave the cross-linker, separated by 12% SDS-PAGE, and stained with Coomassie Blue. Dominant bands from the lane corresponding to cross-linked VgrG-3C-TsaB were submitted for LC-MS/MS for protein identification (Institute of Biomolecular Design, University of Alberta).
Immunoblotting
To assess the effect of VgrG-3 on secretion of Hcp, overnight cultures of bacterial strains were diluted 1:100 in LB broth containing appropriate antibiotics and incubated at 37 °C with shaking until they reached late mid-logarithmic growth phase (A600 ∼ 0.6). l-Arabinose (0.1%) was added to induce expression of the PBAD promoter in pBAD24. Bacteria were pelleted at high speed in a tabletop microcentrifuge for 5 min. Supernatants were filtered through 0.22-μm low protein-binding PVDF syringe filters (Millipore) and concentrated by TCA/acetone precipitation. Protein pellets were resuspended in 1× Laemmli buffer, subjected to SDS-PAGE (10% acrylamide), and then transferred to a nitrocellulose membrane for immunoblot analysis. Antisera used include a mouse monoclonal antibody against DnaK (Enzo Life Sciences, diluted 1:15,000), polyclonal rabbit anti-Hcp (6), antiserum (diluted 1:500). Secondary antibodies used were goat anti-mouse horseradish peroxidase (HRP) and goat anti-rabbit-HRP (both from Santa Cruz Biotechnology, diluted 1:3000).
For detection of VgrG-3C and TsaB in cross-linking studies, polyclonal antisera were raised against the recombinant proteins in rabbits at the Southern Alberta Cancer Research Institute (Calgary, Canada). Sera were diluted 1:5,000 and detected with goat anti-rabbit-HRP (Santa Cruz Biotechnology, diluted 1:3,000).
Size Exclusion Chromatography
A Superdex 75 pg 30/100 (GE Healthcare) column was equilibrated with 20 mm Tris-HCl, 100 mm NaCl, pH 8. Purified recombinant VgrG-3C, TsaB, or a mixture of the two (100 μg each) was separated at a flow-rate of 0.5 ml/min and the A230 chromatograms were compared. To confirm the identity of the proteins in each peak, fractions corresponding to peak apexes were analyzed by SDS-PAGE and immunoblotting with anti-His6 (Santa Cruz Biotechnology, 1:1,000) and anti-VgrG-3 (Fig. 7B) (6).
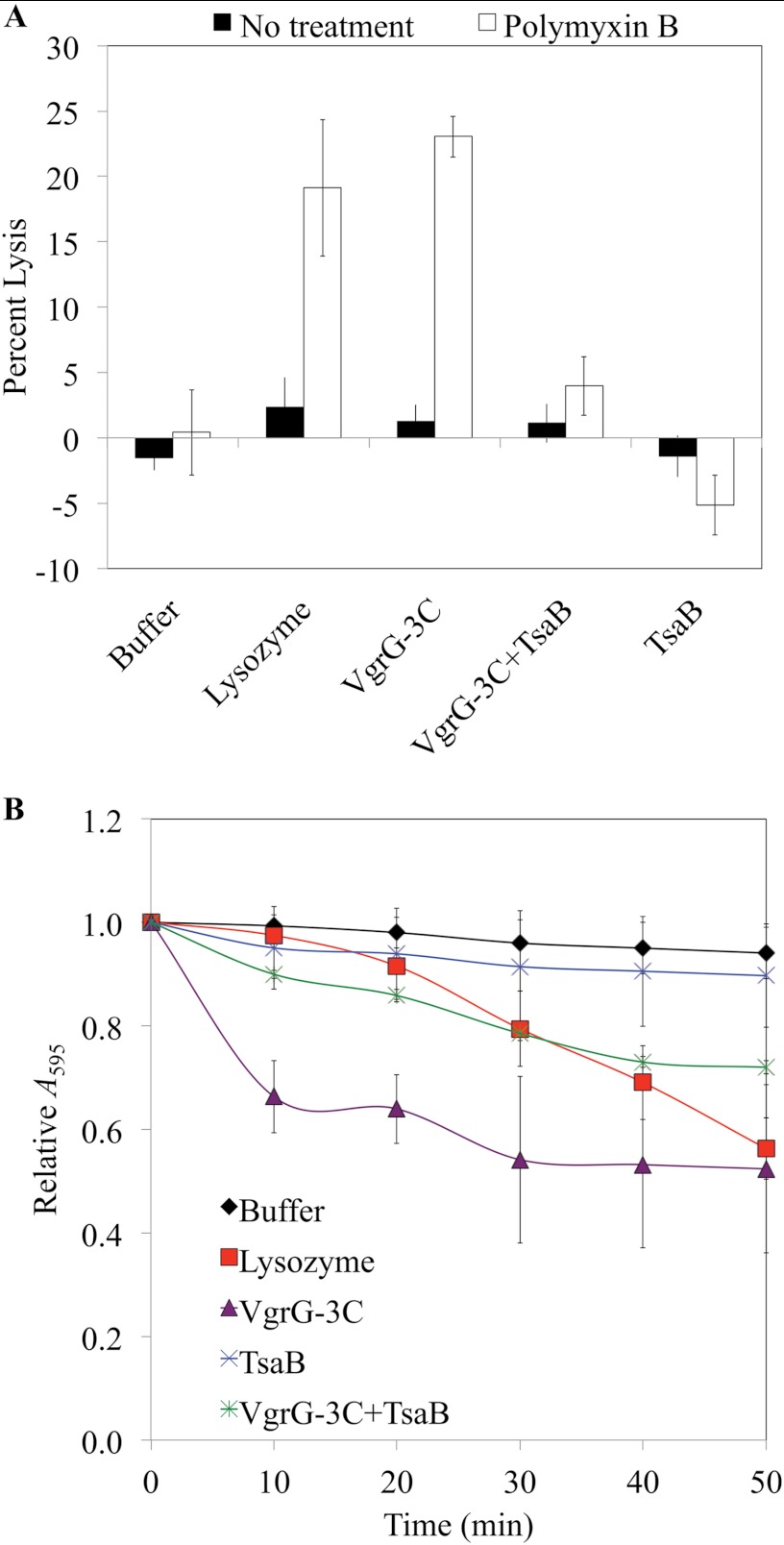
FIGURE 7.
Addition of exogenous VgrG-3 C-terminal domain (VgrG-3C) leads to lysis of E. coli in the presence of polymyxin B and degradation of PG. A, mid-logarithmic phase E. coli were resuspended in 20 mm Tris-HCl, pH 7.0, and incubated with purified proteins, and the A600 was measured at 0 and 5 min. Permeabilization of the outer membrane with 40 μg/ml polymyxin B led to lysis in the presence of lysozyme or VgrG-3C. Preincubation of VgrG-3C with a 3-fold molar excess of the immunity protein TsaB for 10 min on ice protects against this effect. Results represent the average of four independent experiments each performed in duplicate. Bars represent the S.E. of all data points. B, 10 μg of purified recombinant VgrG-3C or TsaB were added to 2 mg E. coli PG and the A595 was monitored over 50 min in a microplate reader at 37 °C. For VgrG-3C +TsaB, 10 μg of each protein were mixed together and incubated on ice for 10 min prior to adding to the substrate. Bars represent the S.E. of six data points from two experiments.
Lysis Assays
Mid-log cultures were harvested from overnight LB-agar plates and resuspended to an A600 ∼ 1.0 in 20 mm Tris-HCl, pH 7. Aliquots of 100 μl were transferred to a 96-well plate and the A600 at 0 min was measured in an xMark microplate spectrophotometer (Bio-Rad). A volume of 10 μl of buffer or 1 mg/ml VgrG-3C, lysozyme, or VgrG-3C + TsaB was added to wells as appropriate, followed immediately by the addition of 1 μl of 4 mg/ml polymyxin B. The plate was incubated for 5 min at 37 °C on a vibrating shaker, and the A600 was measured again. Percent lysis was calculated as ((A600 at 0 min − A600 at 5 min)/A600 at 0 min) × 100%.
Turbidity Assay
Digestion of purified E. coli PG by VgrG-3C was monitored by mixing 10 μg of protein with 100 μg of PG in 100 μl of 20 mm HEPES, pH 6.8 in a 96-well plate then incubating at 37 °C in an xMark microplate spectrophotometer (Bio-Rad) and measuring A595 over a 60 min period. Buffer alone served as negative control, whereas lysozyme was used as a positive control. The experiment was performed twice in quadruplicate.
RESULTS
VgrG-3 Plays a Structural Role in the V. cholerae T6SS
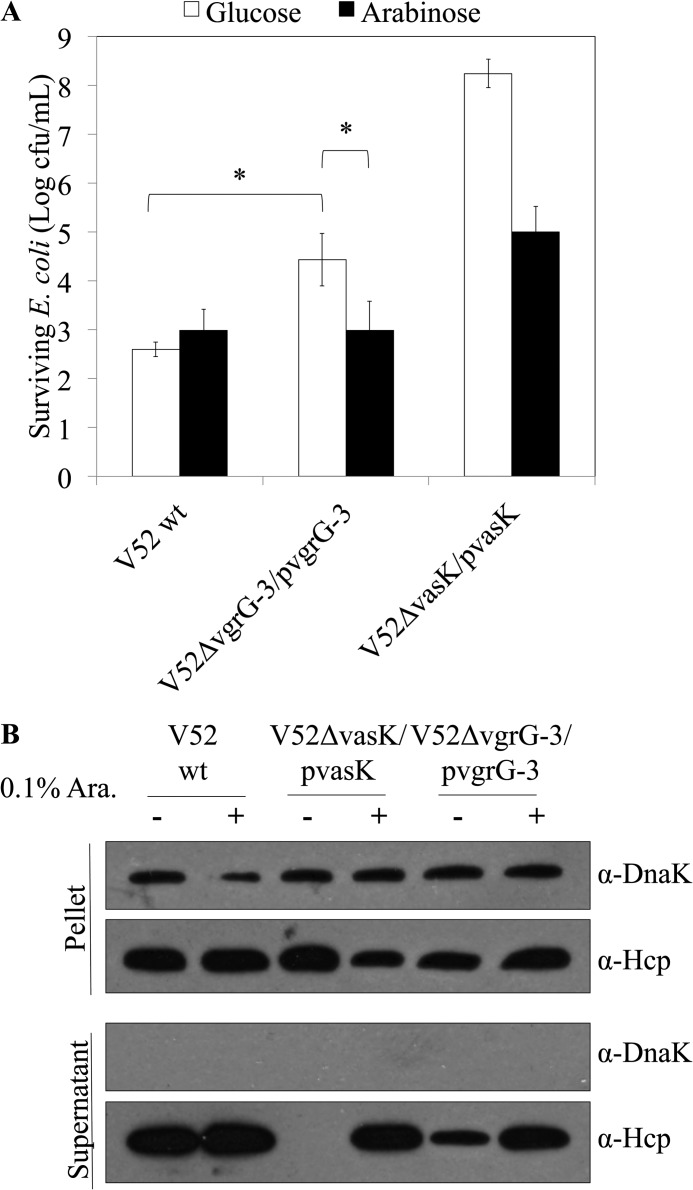
Bioinformatic analysis of the full-length amino acid sequence of VgrG-3 indicated the presence of a peptidoglycan-binding motif within the C-terminal extension; however, unlike many characterized PG hydrolases, no catalytic domain could be identified (Fig. 1). To determine whether VgrG-3 plays a role in microbial competition, our standard killing assay was performed (Fig. 2). Competition between E. coli MG1655 and wild-type V52 resulted in ∼ 103 surviving E. coli cfu/ml of the 107 cfu/ml initially spotted. Interestingly, in-frame deletion of vgrG-3 led to a statistically significant reduction in killing with ∼104 surviving E. coli cfu/ml (p < 0.01). In contrast, competition between E. coli and a V52 strain in which a critical inner membrane T6SS component, vasK, is deleted resulted in ∼ 108 surviving E. coli cfu/ml. Expression of vasK or vgrG-3 in trans from the arabinose-inducible pBAD24 complemented the killing defect partially for vasK and completely for vgrG-3. To explore whether deletion of vgrG-3 results in a loss of T6SS function, immunoblot analysis of Hcp secretion was performed on pellets and supernatants of the strains tested in the killing assay. As is shown by immunoblot analysis, deletion of vgrG-3 resulted in a decrease in secreted Hcp levels, which could be restored by complementation in trans (Fig. 2B). These data indicate that VgrG-3 is required for maximal T6SS function.
FIGURE 1.

HMMER diagram of predicted VgrG-3 domains. Phage late control gene D protein (green) and phage base plate assembly protein (red) domains are within the VgrG core and share homology with the bacteriophage T4 tail spike. The dark blue domain belongs to the PG-binding 1 family. A region of low complexity is indicated in light blue, whereas a C-terminal coiled-coil is indicated in light green. The brackets above define the portions of the gene used to generate VgrG-3N and VgrG-3C constructs.
FIGURE 2.
The bacterial competition assay between V. cholerae V52 and E. coli MG1655 shows an intermediate killing phenotype when vgrG-3 is deleted. A, V52 derivatives were mixed with E. coli in a 10:1 ratio and incubated for 4 h on an LB plate prior to harvesting and serial dilution to enumerate surviving E. coli. Complementation from the plasmid is induced in the presence of arabinose (Ara.) and silenced in the presence of glucose. Asterisks indicate a statistically significant decrease in surviving E. coli relative to the V52ΔvgrG-3-pvgrG-3 glucose condition (two-tailed t test, p < 0.01). B, deletion of vgrG-3 results in reduced Hcp secretion in V. cholerae V52. Mid-logarithmic cultures were pelleted, and the supernatant TCA was precipitated for immunoblot analysis. Samples were adjusted for cell density and blotted for DnaK (loading and lysis control) and Hcp.
The C-terminal Extension of VgrG-3 Contains Lytic Activity
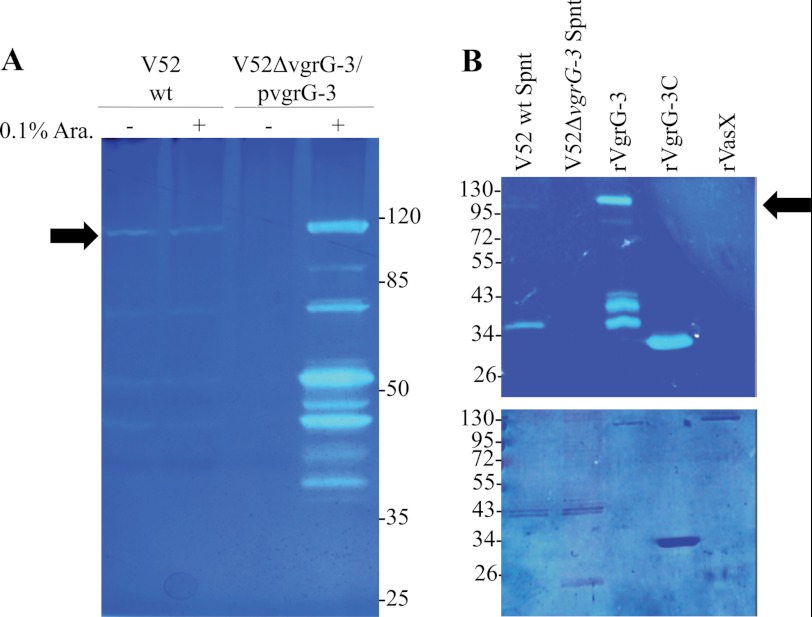
To determine whether VgrG-3 degrades peptidoglycan, cell lysates and supernatants of wild-type V. cholerae and the in-frame vgrG-3 deletion mutant were assayed in an E. coli PG zymogram. A zone of clearing corresponding to the molecular weight of VgrG-3 (112 kDa) was present in the wild-type lysate and supernatant but was absent when vgrG-3 was deleted (Fig. 3). The identity of this lytic enzyme as VgrG-3 is further validated in Fig. 3A as this zone of clearing could be complemented in the deletion strain by arabinose-induced overexpression of episomally encoded vgrG-3. In addition, several bands unique to the wild type and complemented samples were observed and predicted to be degradation products based on their absence in the uninduced V52ΔvgrG-3-pvgrG3 lane. To further validate the PG hydrolytic activity of VgrG-3 and ascribe it to the C-terminal extension, both full-length VgrG-3 and a C-terminal truncation, including residues 727–1017 (VgrG-3C), were purified and analyzed on a PG zymogram. These zymograms confirmed that VgrG-3 produces a zone of clearing on a PG zymogram and that this activity is contained within the C-terminal extension (Fig. 3B). It is important to note that zones of clearing also appear at ∼30 and 35 kDa in the lane corresponding to full-length VgrG-3, although they are not detected in the Coomassie-stained gel below. MALDI-TOF analyses of each of these bands identified 10 peptides from VgrG-3 (12.6% coverage) identifying them as breakdown products of low abundance (Fig. 3B, bottom) and high PG-degrading activity (Fig. 3B, top). Lastly, VasX, a T6SS toxin with no predicted PG-binding activity, was included as a negative control (15).
FIGURE 3.
VgrG-3 degrades peptidoglycan. A, total cell lysates of V52 wild type (wt), and V52ΔvgrG-3/pvgrG-3 under inducing and non-inducing conditions (± arabinose (Ara.)) were analyzed for PG degradation by zymography. Protein samples were run on SDS-PAGE containing 0.1% E. coli peptidoglycan sacculi and stained with methylene blue to visualize zones of clearing (upper panel). The arrow indicates the molecular weight of VgrG-3. B, zymogram analysis of culture supernatants and of purified VgrG-3 constructs and analyzed by zymography as described in A. The same samples were run on a standard SDS-PAGE and stained with Coomassie Blue as a loading control (lower panel). The asterisk indicates a VgrG-3 degradation product with lytic activity as indicated by MALDI-TOF analysis.
Zymography was chosen for the characterization of VgrG-3 enzymatic activity as it allows the qualitative comparison of the effects of a range of conditions (16). In this study, the purified recombinant C-terminal extension of VgrG-3 (VgrG-3C) was run in each lane of a zymogram gel and renatured in buffers at different pHs (5.0–9.0) or at pH 7 in the presence of various cations as the activity of many peptidoglycan hydrolases has been shown to be dependent on cation cofactors (17, 18). The results of these analyses (Fig. 4) indicate that VgrG-3C is most active between pH 5 and 8, and in the presence of Ca2+, whereas it is inhibited by alkaline pH and larger divalent cations such as Cu2+ and Zn2+. Incubation with the cation scavenger EDTA did not diminish activity significantly, indicating that the cation cofactor is not essential but may play an accessory role by assisting in folding or stabilization of the active site of the enzyme (17).
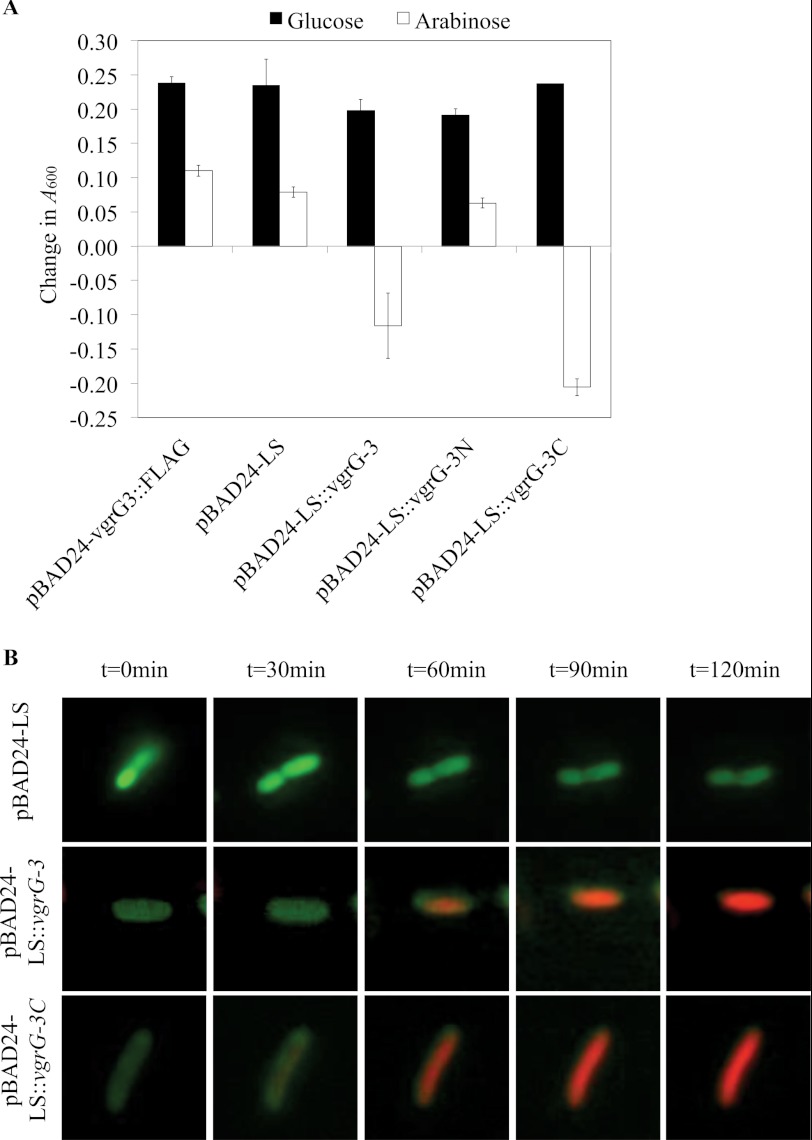
Delivery of VgrG-3 or VgrG-3C to the Periplasm Is Toxic to E. coli
VgrG-3, its N-terminal core domain (VgrG-3N), and its C-terminal extension (VgrG-3C) (Fig. 1) were tagged with an N-terminal SPI periplasmic secretion signal peptide and expressed in E. coli. As a control, a cytosolic FLAG-tagged vgrG-3 construct (pBAD24-vgrG-3::FLAG) was included. Comparison of change in optical density at 600 nm (A600) for >60 min indicated that VgrG-3 is not toxic when localized within the E. coli cytosol, although induction with arabinose does slow growth. However, induction of VgrG-3 or VgrG-3C with the secretion signal resulted in cell lysis, as observed by a decrease in A600, whereas induction and secretion of the VgrG-3N showed a reduction in growth similar to empty vector control (pBAD24-LS) (Fig. 5A). For a better understanding of the effect of VgrG-3-mediated toxicity, live/dead staining was performed on E. coli cells expressing the periplasmic localized VgrG-3 and visualized by live cell fluorescent microscopy over a 2-h time period (Fig. 5B). All E. coli cells were stained with 5(6)-FAM NHS ester (green) and, upon cell death, their membranes became permeable, allowing the influx of propidium iodide (red). Stained bacteria were placed onto an objective slide prepared with a thin LB agar pad containing arabinose to induce expression. Cells carrying the empty pBAD24-LS vector remained healthy during a 2-h incubation on inducing agar (82% of 44 cells remained green); however, cells expressing VgrG-3 or VgrG-3C began to die after 1 h, as indicated by a color change from green to red. After 2 hr induction ∼62% of 68 cells expressing full-length VgrG-3 turned red, whereas ∼64% of 58 cells expressing VgrG-3C turned red. Notably, we did not observe any change in cell shape or rupture of the cell membrane.
FIGURE 5.
Periplasmic expression of VgrG-3 or its C-terminal domain in E. coli results in cell death. A, quantitative assay in which cell lysis is measured by a drop in optical density. A600 was measured at 0 and 60 min post-induction (arabinose) or post-repression (glucose) of the indicated plasmid construct. Bars indicate S.D. of quadruplicate data of a representative experiment. B, time-lapse microscopy of E. coli carrying an arabinose-inducible plasmid (pBAD24-LS) with full-length VgrG-3 or VgrG-3C. The cells are stained with 5(6)-FAM NHS ester (colors healthy cells green), and PI (stains permeabilized cells red). For clarity, a single representative cell is shown for each condition.
The Antitoxin TsaB Directly Binds to and Inhibits VgrG-3C
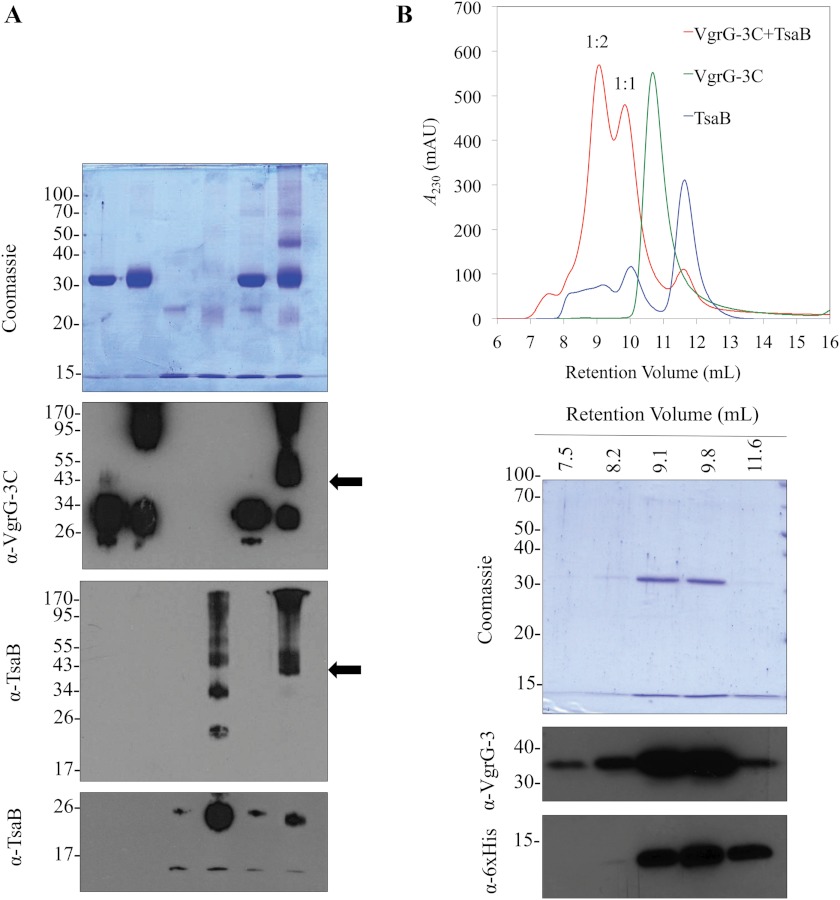
Zheng and colleagues (9) reported that they were unable to delete the ORF VCA0124 (tsaB) located downstream of vgrG-3 and proposed that it was essential for immunity to a toxic effector. To test whether TsaB acts as an antitoxin for VgrG-3C, direct interaction of the two proteins was assessed through cross-linking (Fig. 6A) and size exclusion chromatography (Fig. 6B) studies. Chemical cross-linking of proximal lysine residues demonstrated that TsaB tends to form homo-oligomers. This finding was corroborated by the appearance of multiple peaks on size exclusion chromatography of the purified protein. In addition, a polyclonal rabbit antisera that resulted from inoculation with the purified TsaB shows much greater affinity for multimeric TsaB than monomeric (Fig. 6A), indicating that the multimer was recognized by the rabbit immune system and is strongly antigenic. When mixed with VgrG-3C, a unique band was observed with a molecular weight of approximately that of a VgrG-3C/TsaB heterodimer. Immunoblot analysis using sera specific for VgrG-3C and for TsaB, as well as MALDI-TOF MS analysis confirmed that this band is a heterodimer (eight peptides, 37.5% coverage of VgrG-3C, and two peptides, 22.2% coverage of TsaB). Similarly, size exclusion chromatography of VgrG-3C preincubated with TsaB generated two unique peaks that are composed of both proteins (Fig. 6B). Analysis of the apparent molecular weights of the two unique peaks indicates that both 1:1 and 1:2 VgrG-3C:TsaB stoichiometries exist under the conditions tested. It is important to note that results of chemical cross-linking (Fig. 6A) did not clearly indicate the presence of two oligomeric states. This may be due to poor resolution of the gel in the 40–60 kDa range, as the immunoblots indicate a smear in this region that could indicate the presence of both oligomeric forms. Alternatively, the spacer length of dithiobis(succinimidyl propionate) may create a bias toward the VgrG-3C/TsaB interaction rather than the TsaB/TsaB interaction because the 47-kDa band in the Coomassie stained gel corresponds to the 1:1 heteromeric form. Nonetheless, the co-elution of VgrG-3C with TsaB at a unique retention volume indicates the formation of a stable noncovalent interaction. Taken together, these data suggest that VgrG-3C and TsaB form a stable interaction.
FIGURE 6.
VgrG-3C and TsaB form a stable interaction in solution. (A) VgrG-3C and TsaB (500 μg/ml of each protein) were covalently linked by addition of 2 mm dithiobis(succinimidyl propionate) and the formation of a heterodimer was assessed by SDS-PAGE and immunoblots for VgrG-3C and TsaB. Arrows indicate the molecular weight of the VgrG-3C:TsaB heterodimer. Anti-TsaB blots are presented in two exposures to highlight the heterodimer band as well as the oligomeric state of native TsaB. B, size exclusion chromatogram overlay of VgrG-3C (green), TsaB (blue), and VgrG-3C premixed with TsaB (red). Purified proteins (500 μg/ml) were separated on a Superdex 75 size exclusion column at a flow rate of 0.4 ml/min, and A230 was monitored. VgrG-3C and TsaB were mixed at a 1:1 mass ratio for ∼30 min prior to chromatography. Void volume of 7 ml was used to calculate apparent MW and infer the stoichiometry of the dominant peaks (red) corresponding to the VgrG-3C:TsaB hetero-oligomers. Samples from each peak present in the VgrG-3C+TsaB chromatogram (red) were analyzed by SDS-PAGE and immunoblots to determine the composition of each peak.
To determine the effect of this interaction on peptidoglycan degradation, purified recombinant VgrG-3C was added to E. coli cells whose outer membranes were permeabilized by polymyxin B to allow access of the toxin to its periplasmic substrate, and the A600 was monitored to determine the degree of lysis (Fig. 7A). Buffer alone served as a negative control, whereas lysozyme was added as a positive control. As expected, up to 25% lysis occurred when VgrG-3C or lysozyme was added to permeabilized cells. This lysis was not observed in nonpermeabilized cells, indicating that VgrG-3 must be delivered to the periplasm to exert its toxic effect. Similar results were observed when Pseudomonas aeruginosa PA14, Vibrio alginolyticus, or Vibrio parahemeolyticus were used as target cells (data not shown). In contrast, preincubation of VgrG-3C with TsaB abrogated the observed lytic effect, indicating that interaction with TsaB inhibits the toxic activity of VgrG-3C. Using another approach, purified E. coli PG was also incubated with recombinant VgrG-3C and TsaB (Fig. 7B). Addition of VgrG-3C resulted in a rapid drop in optical density (A595) that could be inhibited in part by pre-incubation with TsaB. Given the two oligomeric forms of VgrG-3C/TsaB as observed through size exclusion chromatography using the same preincubation conditions, we hypothesize that the incomplete inhibition of PG-degrading activity may arise from unsaturation of VgrG-3C. Zhang et al. (19) describe a similar interaction between Tae4 and Tai4, a toxin-antitoxin pair of the Enterobacter cloacae T6SS, in which an antitoxin dimer is required to inactivate the Tae4 toxin. These results indicate that TsaB has a direct inhibitory effect on the lytic activity of VgrG-3C.
DISCUSSION
Bacterial secretion systems must traverse several barriers to transfer molecules from one cell to another. In the case of T6SS, an effector must pass through the host inner membrane, peptidoglycan layer, and outer membrane before coming in contact with a target cell. For the effector to act on an inner membrane or cytoplasmic target in a prey bacterium, it must also pass through the outer membrane and peptidoglycan layer of the target cell. In this study, we demonstrated that the C-terminal extension of V. cholerae VgrG-3 degrades the Gram-negative cell wall and thereby might confer a competitive advantage against neighboring Gram-negative bacteria encountered in the environment or in the human gastrointestinal tract. According to the data presented in Fig. 2, VgrG-3 plays a direct role in Hcp secretion and microbial competition. This conflicts with previous studies (9) that found that deletion of vgrG-3 does not affect T6SS function. The discrepancy may arise from the higher numbers of bacteria used in the microbial competition assays of this study, which allow for greater sensitivity in detection of surviving E. coli. Thus, the precise role for VgrG-3 in the V. cholerae merits discussion.
The current model of T6SS antimicrobial activity is that of a poison dagger. An Hcp nanotube decorated with VgrG is driven into the periplasm of a neighboring cell through mechanical force (5). Cell puncture provides VgrG-3 access to the peptidoglycan layer of the target cell where it can exert its toxic effect. The cell lysis assay (Fig. 7A) demonstrated that addition of the recombinantly purified VgrG-3 C terminus to semipermeabilized E. coli cells resulted in lysis; however, nonpermeabilized cells were insensitive to VgrG-3-mediated lysis. Similarly, heterologous expression and targeting to the E. coli periplasm resulted in a decrease in optical density, whereas, cytosolic expression did not (Fig. 5). Analysis of VgrG C-terminal extension sequences found that VgrG-3 of V cholerae has predicted lysozyme activity (1). Our results support the peptidoglycan-binding activity of the C-terminal extension of VgrG-3 indicated by bioinformatic analyses (Fig. 1). Cell wall-degrading enzymes have also been identified in T6SS of P. aeruginosa (10) and Burkholderia sp. (20); however, these function purely as effectors and are not fused to VgrG. P. aeruginosa is equipped with two T6SS-delivered PG hydrolases, one that cleaves the sugar backbone (Tse3) and a second one that cleaves the peptide chains (Tse1). Interestingly, in silico analyses such as BlastP or HMMER failed to ascribe catalytic function to the VgrG-3 C terminus, indicating that VgrG-3 may contain a previously not recognized PG hydrolase motif.
Because VgrG-3 must transit through the periplasm of the predator cell to be delivered into the prey periplasm, V. cholerae must produce an antitoxin to prevent digestion of its own peptidoglycan or that of neighboring sister cells. TsaB has been shown to display some of the hallmarks of a type II toxin-antitoxin system: (i) the start codon of tsaB overlaps with the stop codon of vgrG-3, (ii) TsaB forms a direct interaction with the C-terminal extension of VgrG-3 (Fig. 6), (iii) the pI of TsaB is acidic (4.66), whereas the pI of VgrG-3C is nearly neutral (6.25), and (iv) TsaB forms dimers and higher order oligomers in solution (21, 22). More importantly, this work has shown that the interaction of TsaB with VgrG-3C prevents PG hydrolysis (Fig. 7). It remains to be determined whether the role of TsaB is primarily to protect the VgrG-3-producing cell against self-intoxication, to protect against VgrG-3-mediated toxicity injected by a neighboring cell, or both. If TsaB interacts with VgrG-3 in the producing cell, a mechanism for dissociation of the antitoxin upon toxin secretion must exist (21). Li et al. (22) describe a similar toxin-antitoxin system in the Pseudomonas T6SS in which the toxin Tse2 interacts directly with Tsi2 in the cytoplasm of the producing cell. Further research will determine whether T6SS toxin-antitoxin systems comprise a new class of these genetic elements.
The gp5 of bacteriophage T4, to which all VgrGs are homologous, is produced as a protoxin in which the PG-degrading domain is released, whereas the prodomain remains attached to the phage base plate. Kanamaru et al. (23) propose that the C-terminal prodomain assists in delivery of the toxin to the prey periplasm. Similarly, attachment of a lytic C-terminal domain to the VgrG core might allow entry of the effector into the periplasm through mechanical puncture of the prey by the T6SS needle. It remains to be determined whether the C-terminal domain of VgrG-3 dissociates from the core at any point during its delivery. Some evidence that this might be the case can be found in the PG zymograms (Fig. 3B), in which the ∼35-kDa recombinant VgrG3 breakdown products (*) appear as active, if not more active, than the full-length VgrG3 observed at ∼120 kDa.
This study has examined a potential role of VgrG-3 in microbial competition through attack; however, VgrG-3 might also facilitate ejection of the T6SS complex through the peptidoglycan layer as is the case for the lytic transglycosylases in T3 and T4SS (22). This may explain the decreased virulence of the vgrG-3 deletion mutant observed in the killing assay (Fig. 2A) and reduced secretion of the T6SS hallmark protein Hcp (Fig. 2B). Alternatively, the secretion defect observed for V52ΔvgrG-3 might imply that VgrG-3 is required for optimal assembly of the T6SS needle, similar to the deletion of vgrG-2 which results in a nonfunctional T6SS (6, 9, 15, 25).
In addition to having a role in microbial competition, it is also possible that VgrG-3 aids V. cholerae in other aspects of its life cycle. VgrG-3 might also target compounds related to peptidoglycan such as mucin or chitin whose β1–4-linked N-acetylglucosamine (26) may serve as substrate for VgrG-3 peptidoglycan hydrolase activity. Potential roles for this activity abound given the life cycle of V. cholerae. For instance, in the environment, V. cholerae populates marine estuaries by associating with small aquatic crustaceans called copepods whose chitinaceous shells are digested as a carbon source. Also, upon infecting a human, V. cholerae must penetrate the mucosal layer of the small intestine to establish infection and release major virulence factors such as cholera toxin into the mucosal epithelium. VgrG-3-mediated cleavage of the glycans linked to mucins could facilitate the passage of Vibrio through the mucosa. Therefore, VgrG-3 has the potential to play diverse roles in the life cycle of V. cholerae.
Acknowledgments
We thank Sarah Miyata and Cory Brooks for helpful discussions, Marcia Craig for critically reviewing the manuscript, and the laboratory of Joel Weiner for the gift of the pBAD24-LS plasmid. The Leica Microsystems microscope was purchased with funds from the Canadian Foundation for Innovation (LOFS1–21139).
This work was supported by Canadian Institute for Health Research Operating Grant MOP-84473 and Alberta Innovates–Health Solutions (funded by the Alberta Heritage Foundation for Medical Research Endowment Fund).
- Hcp
- hemolysin-coregulated protein
- T6SS
- type VI secretion system(s)
- PG
- peptidoglycan
- 5(6)-FAM NHS ester
- 5-(and 6)-carboxyfluorescein succinimidyl ester.
REFERENCES
- 1. Pukatzki S., McAuley S. B., Miyata S. T. (2009) The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12, 11–17 [DOI] [PubMed] [Google Scholar]
- 2. Ballister E. R., Lai A. H., Zuckermann R. N., Cheng Y., Mougous J. D. (2008) In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. U.S.A. 105, 3733–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., Burley S. K., Almo S. C., Mekalanos J. J. (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hachani A., Lossi N. S., Hamilton A., Jones C., Bleves S., Albesa-Jové D., Filloux A. (2011) Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J. Biol. Chem. 286, 12317–12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Basler M., Pilhofer M., Henderson G. P., Jensen G. J., Mekalanos J. J. (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma A. T., McAuley S., Pukatzki S., Mekalanos J. J. (2009) Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suarez G., Sierra J. C., Erova T. E., Sha J., Horneman A. J., Chopra A. K. (2010) A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blondel C. J., Jiménez J. C., Contreras I., Santiviago C. A. (2009) Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10, 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng J., Ho B., Mekalanos J. J. (2011) Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6, e23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Russell A. B., Hood R. D., Bui N. K., LeRoux M., Vollmer W., Mougous J. D. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., Silverman J. M., Ohlson B. B., Hicks K. G., Plemel R. L., Li M., Schwarz S., Wang W. Y., Merz A. J., Goodlett D. R., Mougous J. D. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 7, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Basler M., Mekalanos J. J. (2012) Type 6 secretion dynamics within and between bacterial cells. Science 337, 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoyle B. D., Beveridge T. J. (1984) Metal binding by the peptidoglycan sacculus of Escherichia coli K-12. Can J. Microbiol. 30, 204–211 [DOI] [PubMed] [Google Scholar]
- 14. MacIntyre D. L., Miyata S. T., Kitaoka M., Pukatzki S. (2010) The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U.S.A. 107, 19520–19524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miyata S. T., Kitaoka M., Brooks T. M., McAuley S. B., Pukatzki S. (2011) Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun. 79, 2941–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilkesman J., Kurz L. (2009) Protease Analysis by Zymography: A review on tehcniques and patents. Recent Pat. Biotechnol. 3, 175–184 [DOI] [PubMed] [Google Scholar]
- 17. Schmelcher M., Waldherr F., Loessner M. J. (2012) Listeria bacteriophage peptidoglycan hydrolases feature high thermoresistance and reveal increased activity after divalent metal cation substitution. Appl. Microbiol. Biotechnol. 93, 633–643 [DOI] [PubMed] [Google Scholar]
- 18. Son B., Yun J., Lim J. A., Shin H., Heu S., Ryu S. (2012) Characterization of LysB4, an endolysin from the Bacillus cereus-infecting bacteriophage B4. BMC Microbiol. 12, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang H., Gao Z. Q., Wang W. J., Liu G. F., Xu J. H., Su X. D., Dong Y. H. (2013) Structure of the type VI effector-immunity complex (Tae4-Tai4) provides novel insights into the inhibition mechanism of the effector by its immunity protein. J. Biol. Chem. 288, 5928–5939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Russell A. B., Singh P., Brittnacher M., Bui N. K., Hood R. D., Carl M. A., Agnello D. M., Schwarz S., Goodlett D. R., Vollmer W., Mougous J. D. (2012) A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe. 11, 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamaguchi Y., Park J. H., Inouye M. (2011) Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79 [DOI] [PubMed] [Google Scholar]
- 22. Li M., Le Trong I., Carl M. A., Larson E. T., Chou S., De Leon J. A., Dove S. L., Stenkamp R. E., Mougous J. D. (2012) Structural basis for type VI secretion effector recognition by a cognate immunity protein. PLoS Pathog. 8, e1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanamaru S., Gassner N. C., Ye N., Takeda S., Arisaka F. (1999) The C-terminal fragment of the precursor tail lysozyme of bacteriophage T4 stays as a structural component of the baseplate after cleavage. J Bacteriol. 181, 2739–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zahrl D., Wagner M., Bischof K., Bayer M., Zavecz B., Beranek A., Ruckenstuhl C., Zarfel G. E., Koraimann G. (2005) Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151, 3455–3467 [DOI] [PubMed] [Google Scholar]
- 25. Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. (2007) Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moran A. P., Gupta A., Joshi L. (2011) Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut. 60, 1412–1425 [DOI] [PubMed] [Google Scholar]