Background: The 26 S proteasome requires ATP hydrolysis to degrade ubiquitinated proteins.
Results: Ubiquitin conjugates activate ATP hydrolysis, provided they contain loosely folded domains and their ubiquitin chains bind to the 26 S-associated DUBs.
Conclusion: By stimulating ATP hydrolysis, ubiquitinated proteins induce their own degradation.
Significance: Regulation of ATPase activity by Usp14 and Uch37 links proteolysis and deubiquitination and enhances the specificity of the proteasome for ubiquitin conjugates.
Keywords: ATP-dependent Protease, Deubiquitination, Proteasome, Protein Degradation, Ubiquitin
Abstract
Degradation of ubiquitinated proteins by 26 S proteasomes requires ATP hydrolysis, but it is unclear how the proteasomal ATPases are regulated and how proteolysis, substrate deubiquitination, degradation, and ATP hydrolysis are coordinated. Polyubiquitinated proteins were shown to stimulate ATP hydrolysis by purified proteasomes, but only if the proteins contain a loosely folded domain. If they were not ubiquitinated, such proteins did not increase ATPase activity. However, they did so upon addition of ubiquitin aldehyde, which mimics the ubiquitin chain and binds to 26 S-associated deubiquitinating enzymes (DUBs): in yeast to Ubp6, which is essential for the ATPase activation, and in mammalian 26 S to the Ubp6 homolog, Usp14, and Uch37. Occupancy of either DUB by a ubiquitin conjugate leads to ATPase stimulation, thereby coupling deubiquitination and ATP hydrolysis. Thus, ubiquitinated loosely folded proteins, after becoming bound to the 26 S, interact with Ubp6/Usp14 or Uch37 to activate ATP hydrolysis and enhance their own destruction.
Introduction
In eukaryotic cells, the majority of proteins are targeted for degradation by the 26 S proteasome through the attachment of a polyubiquitin (Ub)2 chain. The 26 S proteasome is an ATP-dependent proteolytic complex composed of a cylindrical 20 S proteolytic core particle and one or two 19 S regulatory particles. The 20 S contains the proteolytic sites in its central chamber, and substrates can only access this chamber through a central gated channel at either end (1). However, due to the narrow gate, substrates require unfolding and linearization before they can enter the 20 S. The 19 S particle catalyzes these processes and triggers gate opening, which is essential for substrate entry. The 19 S contains the subunits Rpn10 and Rpn13 to which polyubiquitinated proteins bind initially (2, 3), as well as six AAA-ATPase subunits, Rpt1–6. ATP binding or hydrolysis is essential for multiple steps in proteasome function and ATP binding to the 19 S promotes association with the 20 S particle (4), increases the affinity for ubiquitinated substrates (5), and triggers opening of the gated channel for substrate entry into the 20 S particle. ATP hydrolysis supports the unfolding of globular proteins and substrate translocation through the ATPase ring into the 20 S core particles.
The efficient degradation of ubiquitinated substrates requires these different 19 S activities as well as removal and disassembly of the Ub chain. To further understand proteasome function, we have systematically studied how ATP hydrolysis is regulated and whether ATP hydrolysis by the purified 26 S is coupled to substrate binding or degradation. In bacteria and archaea, protein degradation occurs independently of Ub and involves multiple ATP-hydrolyzing protease complexes (e.g. Lon, HslUV, ClpAP, PAN-20 S), which, similar to the 26 S, contain hexameric AAA-ATPase complexes (6–8) that have been useful models to clarify 26 S mechanisms. These ATPases are all activated upon binding of protein substrates, such as the largely unstructured protein casein. For example, PAN, the proteasome-regulatory ATPase complex in archaea and evolutionary precursor of the six ATPases in eukaryotic proteasomes, hydrolyzes ATP 2–4-fold faster upon binding casein or oligopeptides containing the targeting sequence ssrA (6). This ability of unfolded substrates to allosterically activate ATP hydrolysis and to promote their own degradation enhances the specificity of the proteolytic process and reduces the wasteful utilization of ATP. In the present studies, we examined whether the 26 S ATPases show a similar activation by unfolded polypeptides or ubiquitinated substrates.
Once Ub conjugates bind to the 26 S proteasome, they enhance its degradative capacity by opening further the gated channel into the 20 S (9–11). Ubiquitinated proteins initially bind to the 19 S subunits Rpn10 and Rpn13, but the facilitation of substrate entry occurs subsequently when the Ub chain interacts with the active site of the 26 S-associated deubiquitinating enzymes (DUB) (11). Accordingly, gate opening can also be stimulated by Ub aldehyde, a transition state inhibitor of the 26 S-associated DUB, Ubp6 in yeast, and its mammalian homolog Usp14 (11). However, the mammalian 26 S also contains another Ub aldehyde-sensitive deubiquitinating enzyme, Uch37/UchL5, and we have investigated here whether it also plays a role in regulating proteasomal function. These 26 S-associated DUBs remove and disassemble the Ub chain (12), although the precise roles of Usp14 and Uch37 are currently unclear. It is now well established that Ubp6/Usp14 is a major regulator of proteasome function (11, 13) that serves as a timing device for proteolysis and links its two main functions, substrate deubiquitination and degradation. For example, the occupancy of Ubp6/Usp14 by a Ub chain promotes 20 S gate-opening by somehow altering the functioning of the ATPase subunits (11). These findings emphasize several important, unresolved questions about 26 S function. 1) Although Uch37 can function in the absence of protein degradation by the proteasome (14), it is unclear how deubiquitination and proteolysis are coupled during conjugate degradation. 2) It also is unclear how the rates of ATP hydrolysis by the six 19 S ATPase subunits, Rpt1–6, are regulated and 3) how ATP hydrolysis is coupled to substrate deubiquitination and translocation.
After Ub conjugates bind to the 19 S receptors, those destined for degradation undergo an ATP-dependent transition to a “tightly bound” state, which is no longer dependent on the Ub chain and seems to involve direct interaction of an unfolded domain of the substrate with the proteasomal ATPases (5). Thus, efficient degradation of a protein by the 26 S requires both a Ub chain and also a loosely folded domain of the polypeptide (5, 15, 16). We show here that substrate occupancy of a proteasome-associated DUB (either Ubp6/Usp14 or Uch37) by the Ub chain of the substrate and simultaneous binding of a loosely folded domain of the substrate to the 26 S then stimulates proteasomal ATP hydrolysis. In this way, ubiquitinated substrates can activate the degradative capacity of the proteasome and promote their own degradation, and the key 26 S functions, proteolysis, deubiquitination, and ATP hydrolysis, seem to be coordinated.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Antibodies
The yeast strains sub61 (wt), snu61 (Δubp6), and sJH155 (ubp6C118A), for 26 S proteasome purification as well as the expression plasmids for Ubp6 (pDL74) and Ubp6C118A (pJH1), were kindly provided by Dan Finley (Harvard Medical School). Anti-Uch37 antibodies were obtained from Epitomics, anti-HA was from Sigma, and anti-Usp14 was from Santa Cruz Biotechnology.
Purification of 26 S Proteasomes and Synthesis of Ubiquitin Conjugates
Mammalian rabbit muscle 26 S proteasomes were affinity purified as described before in the presence of 150 mm NaCl (17), whereas yeast 26 S particles were purified with the same method but without the addition of NaCl after harvesting the cells at an A600 of 4 (11, 17).
The E3 ligases E6AP and Nedd4 were used for the generation of poly-Ub conjugates as described before (18). In short, autoubiquitination of the immobilized E3 occurred in the presence of Ub, ATP, E1, and the E2 UbcH5b. After the completion of the autoubiquitination reaction, E1, E2, Ub, and the remaining nucleotides were removed by washing five times. Polyubiquitinated Sic1 was generated as described previously (19, 20). For the binding assay (5), the Ub conjugates remained immobilized on the resin and for the proteasome activity measurements (11), Ub conjugates were eluted from the resin.
Measurement of Proteasome Binding to Ubiquitin Conjugates and Proteasome Activity
The initial and tighter binding of 26 S proteasomes to Ub conjugates were measured as described previously (5). ATP hydrolysis by proteasome particles was measured using the malachite green assay that detects the release of free phosphate (21) or the coupled enzymatic assay using pyruvate kinase and lactate dehydrogenase (22), which assays the production of ADP. ATPase activity and hydrolysis of the small peptide Suc-GGL-amc (Bachem) by proteasomes were measured in the presence of 25 mm Hepes/KOH, pH 8, 2.5 mm MgCl2, 125 mm potassium acetate, 0.025% Triton X-100, 1 mm ATP, 1 mm DTT, 0.1 mg/ml BSA (Sigma) as reported previously (11). Stimulation of ATPase as well as gate opening were assayed under identical conditions using 5–10 nm of 26 S proteasomes for the malachite green-based assay or 15 nm 26 S particles for the enzymatic coupled assay together with a 100-fold excess of poly-Ub conjugates or unmodified E3 ligases over 26 S particles unless stated otherwise. Casein, DHFR, and lysozyme (Sigma) were used at final concentrations of 1 μm and Ub aldehyde (Boston Biochem) at 500 nm. Lysozyme was partially denatured using 800 mm guanidine hydrochloride to generate its molten globular form (23). The Ub5-DHFR was a kind gift from Millennium Pharmaceuticals (Cambridge, MA). Each experiment includes the at least three independent experiments with three replicates for each condition. Every experiment included control reactions containing all reagents with and without the addition of the 26 S particles to detect phosphate and ATPase contaminations in our assays. All values are the means of these experiments ± S.E. after background subtraction.
RESULTS
Ubiquitin Conjugates Stimulate 26 S Proteasome ATPase Activity
Because the bacterial ATP-dependent proteases (8) and the archaeal proteasome regulatory ATPase, PAN, hydrolyze ATP faster upon binding of protein substrates or small peptides (ssrA) that target proteins for degradation (6), we examined whether the primary substrates of the 26 S proteasomes, ubiquitinated proteins, also increase its ATPase activity, as suggested previously (10). We used the Ub-like domain affinity method to purify 26 S proteasomes from rabbit muscle or yeast (11, 17). This purification method releases most proteasome-bound Ub conjugates, and the resulting 26 S particles contain only a very small amount of associated ubiquitinated proteins (supplemental Fig. S1A) (17). To obtain Ub conjugates in chemical amounts, Sepharose-bound GST-E6AP or GST-Nedd4 (supplemental Fig. S1B) was allowed to autoubiquitinate by incubating it with E1, ATP, Ub, and the E2 UbcH5b, which were removed after the reaction by washing. We then incubated the proteasomes with ATP and ubiquitinated E6AP or unmodified E6AP and ATP hydrolysis was measured using either the Malachite Green assay (Fig. 1A) (21) or a coupled pyruvate kinase/lactate dehydrogenase enzymatic assay (Fig. 1B) (22), which gave similar results as reported before (22, 24).
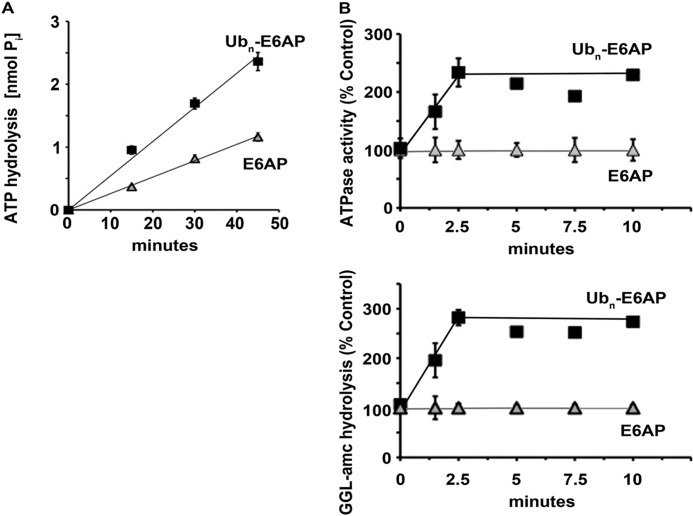
FIGURE 1.
Ub conjugates stimulate ATPase activity of 26 S proteasomes. A, ATP hydrolysis by rabbit muscle 26 S proteasomes was measured in the presence of E6AP or ubiquitinated E6AP. Production of free phosphate (Pi) was detected using the malachite green assay. B, the rabbit muscle 26 S ATPase activity and peptide hydrolysis were measured simultaneously in the presence of E6AP or ubiquitinated E6AP, which were added at the outset (time 0), and the increase in activity was observed with time. Proteasomal ATPase activity at the outset was taken as 100% and was measured using the coupled pyruvate kinase/lactate dehydrogenase enzymatic assay (upper panel), and peptidase activity using suc-GGL-amc (lower panel). All values are the means of at least three experiments ± S.E.
The addition of polyubiquitinated E6AP but not unmodified E6AP increased the rate of ATP hydrolysis 2–3-fold (Fig. 1) in accordance with the prior report (10). A similar stimulation was seen with other ubiquitinated proteins, including the ubiquitinated substrate Ub5-DHFR (see Fig. 5A), which contains a short homogenous Ub chain. By contrast, addition of Ub (Fig. 2B) or even monoubiquitinated proteins (supplemental Fig. S1C) failed to activate ATP hydrolysis. Because ATP hydrolysis by the homologous ATP-dependent proteases and PAN is stimulated by casein (6), we tested whether this loosely folded protein also stimulates the 26 S. However, unlike those AAA-ATPases, the 26 S ATPases were not stimulated by casein (Fig. 2, A and B), even though this protein without ubiquitination can be degraded by 26 S proteasomes (25). Thus, only polyubiquitinated proteins appear capable of stimulating ATP hydrolysis. These conjugates caused a similar activation of ATP hydrolysis, when the degradative capacity of the proteasome was blocked with MG132 (supplemental Fig. S1D). Thus, this effect appears to be signaled by Ub conjugate binding and not proteolysis (see below).
FIGURE 5.
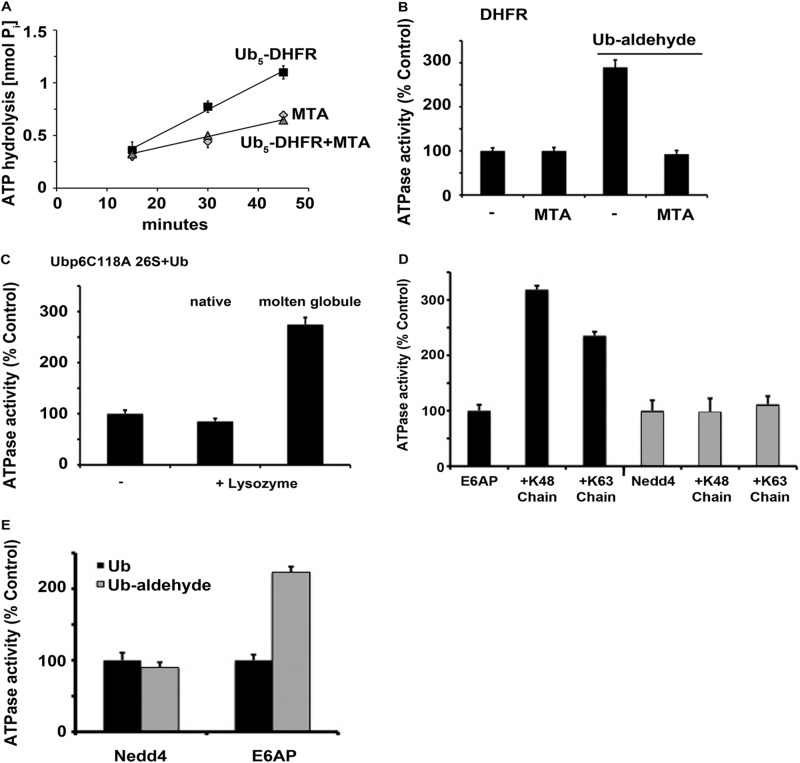
Loosely folded (but not tightly folded) substrates stimulate ATP hydrolysis. A, ATP hydrolysis by rabbit muscle 26 S proteasomes was measured as described in Fig. 1 in the presence of Ub5-DHFR with and without methotrexate (MTA). 26 S ATPase activity in the presence of MTA was used as a control. B, ATP hydrolysis was measured in the presence of DHFR with and without Ub aldehyde or methotrexate (MTA). ATPase activity in the presence of DHFR was taken as 100%. C, ATP hydrolysis by 26 S particles purified from yeast ubp6C118A mutant strain was measured in the presence of Ub and native or molten globular lysozyme, which was pretreated with 800 mm guanidine hydrochloride. ATPase activity in the presence of Ub was taken as 100%. D, ATP hydrolysis by rabbit muscle 26 S proteasomes was measured in the presence of non-ubiquitinated E6AP (black bars) or Nedd4 (gray bars) or after both Ub ligases were allowed to autoubiquitinate with Lys-48 or Lys-63 linked tetra-Ub. ATPase activity in the presence of E6AP was taken as 100%. E, ATP hydrolysis was measured in the presence of non-ubiquitinated E6AP or Nedd4 together with Ub (black bars) or Ub aldehyde (gray bars). ATPase activity in the presence of the unmodified E3s was taken as 100%. All values are the means of at least three experiments ± S.E.
FIGURE 2.
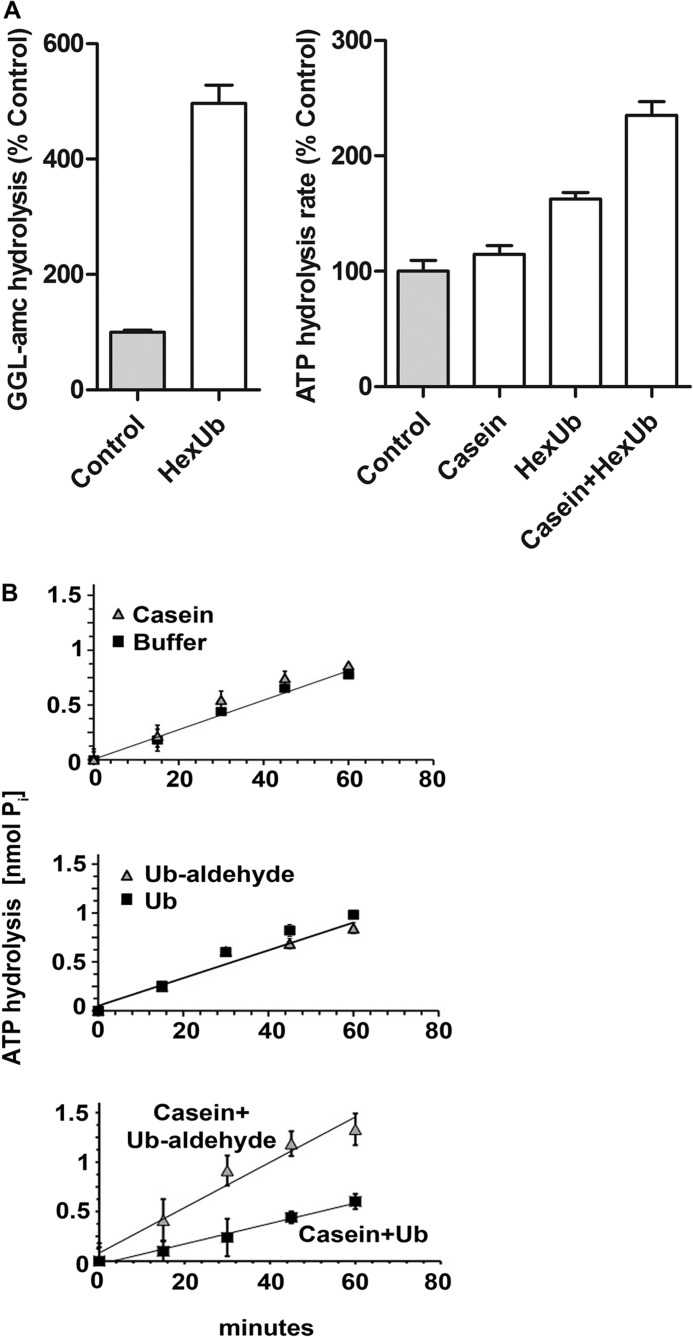
The stimulation of ATPase activity requires binding of the substrate to two sites on the 26 S proteasome. A, peptide hydrolysis by MEF 26 S proteasomes was measured in the presence or absence of hexaubiquitin (HexUb; left panel). Peptidase activity in the absence of hexaubiquitin was taken at 100%. Rates of ATP hydrolysis by 26 S proteasomes were measured in the presence of casein (1 μm), linear hexaubiquitin (1 μm), or a combination of the two, by measuring the accumulation of free phosphate over time (right panel). Rate of ATP hydrolysis in the absence of casein or hexaubiquitin was taken as 100%. B, ATPase activity of rabbit 26 S proteasomes was measured by the release of free phosphate over time in the presence of casein (top panel). ATPase activity was measured in the presence of Ub or Ub aldehyde (middle panel). ATPase activity was measured in the presence of casein plus Ub or casein plus Ub aldehyde (bottom panel). The variations in the basal rate of ATP hydrolysis between the top/middle panel and the bottom panel are caused by using different 26 S preparations. All values are the means of at least three experiments ± S.E.
After addition of the Ub conjugates to the 26 S proteasome (Fig. 1B), ATPase activity increased rapidly and was maximal within 2.5 min. The time course of the activation of ATP hydrolysis was indistinguishable from that for the stimulation of 20 S gate opening by Ub conjugates, which was measured by following the enhanced hydrolysis of fluorogenic peptide substrates (Fig. 1B). Because of these similar time courses, the mechanisms for the increase in ATP hydrolysis and gate opening by Ub conjugates seemed similar or linked.
Ubiquitin Aldehyde with a Protein Substrate also Activates 26 S ATPases
It had been reported previously that free, unanchored Ub chains, similar to Ub aldehyde, can stimulate gate opening (9, 10). Therefore, we tested whether linear Ub6 chains, which stimulate gate opening, similar to Lys-48 chains (Fig. 2A), might also promote ATP hydrolysis. We observed a small but reproducible increase in ATPase activity, which was further enhanced in the presence of casein (Fig. 2A). Thus, ATP hydrolysis can be activated even if the ubiquitin chain and protein are not covalently linked. These Ub chains presumably bind to the Ub receptors, Rpn10 and Rpn13 as well as the 26 S-associated DUBs (Usp14, Uch37, and Rpn 11) and perhaps other sites. However, the increase in gate opening is triggered by the interaction of the Ub chain with the DUB, Ubp6/Usp14, and also requires nucleotide binding to the ATPases (11). Therefore, we tested whether Ub chains stimulate ATP hydrolysis by occupancy of the active site of the proteasome-associated DUBs using the inhibitor Ub aldehyde, which only binds to Uch37 and Usp14. Although this transition state inhibitor-stimulated peptide hydrolysis by the 26 S complex (11), by itself it failed to increase the rates of ATP hydrolysis (Fig. 2B). Thus, the mechanisms for the stimulation of the ATPases and gate opening by Ub conjugates must differ in important respect(s).
Because the nonubiquitinated substrate, casein, and Ub aldehyde are unable by themselves to enhance ATP hydrolysis, we tested whether the addition of this largely structureless protein substrate together with Ub aldehyde could promote ATP hydrolysis. As shown in Fig. 2B, casein together with Ub aldehyde increased ATP hydrolysis in a similar fashion as ubiquitinated proteins and as the combination of free chains and casein (Fig. 2A). Thus, Ub conjugates probably stimulate ATP hydrolysis by interacting simultaneously with one of the 26 S-associated DUBs, either Uch37 or Usp14, as well as with the ATPase subunits, which (similar to PAN) bind unfolded proteins. In other words, there are two requirements for substrate activation of ATP hydrolysis, and Ub aldehyde can replace the Ub chain attached to the substrate.
Stimulation of Yeast 26 S ATPases Requires Ubp6, but Not Its Enzymatic Activity
A similar ATPase stimulation by Ub conjugates or by Ub aldehyde and casein was observed with mammalian 26 S proteasomes and with yeast particles, which contain only one DUB sensitive to Ub aldehyde, Ubp6. Therefore, the increase in ATP hydrolysis requires occupancy of the active site of Ubp6/Usp14 by Ub aldehyde or a Ub chain in addition to a loosely folded domain in the protein. We then determined whether the catalytic activity of Ubp6 is essential for the regulation of ATP hydrolysis or if it functions allosterically, as Ubp6 does in regulating gate opening and overall proteolysis (11, 13). These experiments used the active site mutant, Ubp6C118A, which, unlike the wt enzyme, lacks enzymatic activity but surprisingly binds Ub with high affinity (11). As shown in Fig. 5C, a similar activation was observed with yeast 26 S particles bearing the mutant Ubp6C118A upon addition of molten globular lysozyme in the presence of Ub. Consequently, this effect of the Ub chain is an allosteric action, and Ub aldehydes (and with the C118A mutant Ub) are acting by binding to the active site of Ubp6 and not by inhibiting its DUB activity.
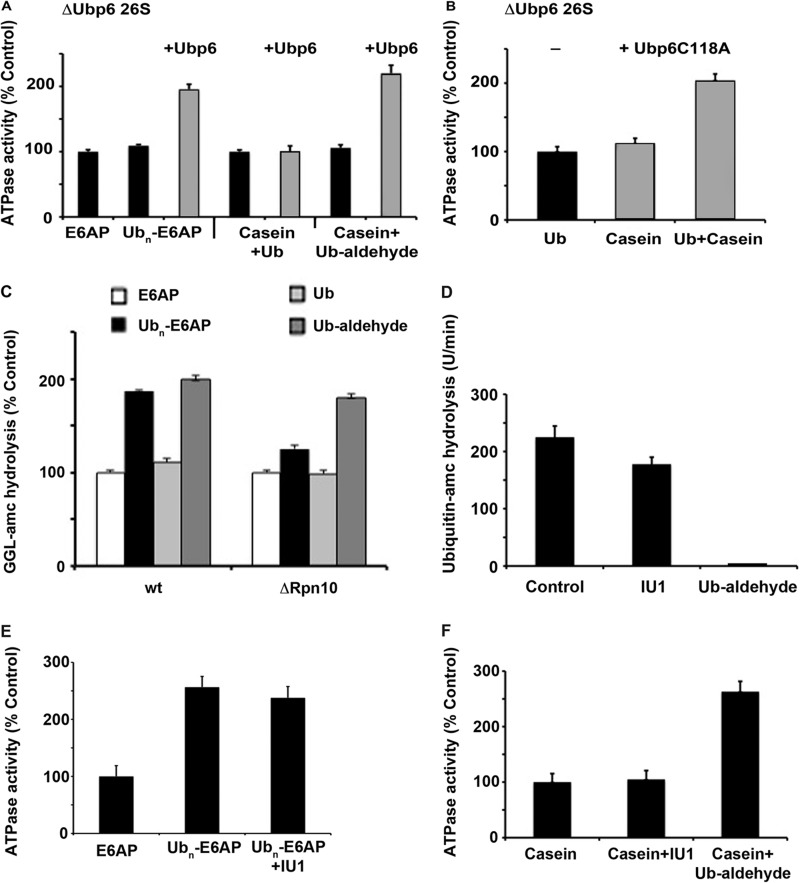
To further test these conclusions, we purified 26 S particles from a yeast strain lacking ubp6 and examined directly whether the stimulation of ATPase activity requires this DUB. In contrast to the ATPase activity of proteasomes from wt yeast, the ATPases of these Ubp6-deficient particles could not be stimulated by ubiquitinated E6AP or by Ub aldehyde plus casein (Fig. 3A). To confirm that this defect was due to the lack of Ubp6, we tried to reconstitute this stimulation by the addition of purified recombinant Ubp6p to these particles. Although Ubp6p by itself did not affect ATPase activity (supplemental Fig. S2), it restored the stimulation by both ubiquitinated proteins and Ub aldehyde plus casein (Fig. 3A).
FIGURE 3.
The stimulation of ATPase activity by Ub conjugates in yeast proteasomes requires Ubp6. A, ATP hydrolysis by ΔUbp6 yeast 26 S proteasomes was measured in the presence of E6AP or ubiquitinated E6AP or casein plus Ub or casein plus Ub aldehyde. The stimulation of ATP hydrolysis was restored by addition of purified Ubp6p (gray bars) in the presence of ubiquitinated E6AP or casein plus Ub aldehyde. ATP hydrolysis in the presence of E6AP was taken as 100%. B, ATP hydrolysis by ΔUbp6 yeast 26 S proteasomes was measured in the presence of Ub, casein, or casein plus Ub. The stimulation of ATP hydrolysis was restored by addition of purified Ubp6C118Ap (gray bars), which lacks DUB activity in the presence of casein plus Ub. ATP hydrolysis in the presence of Ub was taken as 100%. C, peptide hydrolysis of GGL-amc in wt and Δrpn10 yeast 26 S proteasomes was measured in the presence of E6AP, ubiquitinated E6AP, Ub, and Ub aldehyde. D, total ubiquitin-amc hydrolysis by wt 26 S particles (1 nm) was measured. IU1 (50 μm) shows only a slight inhibition, whereas ubiquitin aldehyde (500 nm) completely blocks DUB activity. E, 26 S ATPase activity (10 nm) was measured in the presence of E6AP (500 nm), ubiquitinated E6AP, or ubiquitinated E6AP after the proteasomes where pretreated with 50 μm IU1. F, 26 S ATPase activity (10 nm) was measured in the presence of casein (1 μm), casein plus IU1 (50 μm), or casein plus ubiquitin aldehyde (500 nm). All values are the means of at least three experiments ± S.E.
These findings confirm that the acceleration of ATP hydrolysis by Ub conjugates, such as the enhancement of gate opening (11), does not require the enzymatic activity of Ubp6 but only occupancy of its active site. As noted above, this active site mutant, unlike the wt Ubp6, also binds tightly free Ub, and addition of Ub also stimulated gate opening in the mutant (11). Furthermore, the ATPase activity of the 26 S containing Ubp6C118A, unlike the wt, could be activated by Ub plus casein (Fig. 3B). Although occupancy of the active site of Ubp6 is thus essential for the stimulation of both gate opening and ATP hydrolysis, this effect, in addition, requires a loosely folded polypeptide (see below), either attached or “in trans” with Ub aldehyde.
These observations indicate that although the Ub conjugates initially bind to the receptor subunits Rpn10 and Rpn13 (2, 3, 5), they subsequently activate proteolysis by binding to the DUB Ubp6, which also the target of Ub aldehyde. Accordingly, proteasomes isolated from a yeast deletion strain lacking Rpn10, showed reduced levels of gate opening by Ub conjugates (9). However, the stimulation of peptide hydrolysis by Ub aldehyde was not decreased (Fig. 3C). Thus, Ub aldehyde, unlike ubiquitinated proteins, bypasses the need for the Ub receptors Rpn10 and Rpn13 on the 26 S to activate the proteasome.
In Mammalian 26 S, Occupancy of Uch37 also Increases Gate Opening and ATPase Activity
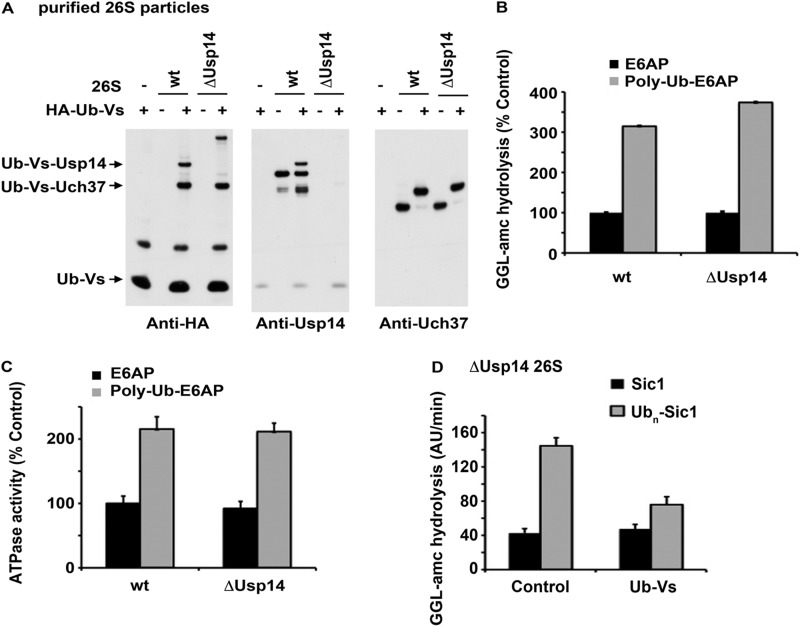
In addition to Usp14, the mammalian 26 S contains another DUB, Uch37, which is sensitive to Ub aldehyde. It is currently unclear whether Uch37 and Usp14 have distinct or overlapping functions. Therefore, we tested whether Uch37 can also mediate the stimulation of gate opening and ATPase activity by Ub conjugates (and Ub aldehyde plus casein). We purified 26 S particles from wt and Usp14−/− MEF cells and labeled the 26 S-associated DUBs by incubating them with HA-Ub-vinylsulfone (26) to assay for the presence of Usp14 and Uch37 in our preparations. As expected, Usp14 was not detectable in those from the Usp14−/− cells, and Uch37 was present in similar amounts in both wt and mutant particles (Fig. 4A). The DUB activity of these particles, assayed with Ub-amc, was consistently lower by ∼30% after loss of Usp14 (supplemental Fig. S3A). Importantly, the stimulation of gate opening (Fig. 4B) and also ATPase activity (Fig. 4C) by ubiquitinated proteins or gate opening by Ub aldehyde (s u p p l em e n t a l F i g . S 3B) was similar in the particles lacking Usp14 as in the wild type. Consequently, Uch37 must also have the capacity to support the substrate-induced activation of the ATPases.
FIGURE 4.
Uch37 also can promote the stimulation of ATP hydrolysis 26 S proteasomes by Ub conjugates. A, 26 S associated DUBs in proteasomes from wt and Usp14−/− MEF cells were labeled with HA-Ub vinylsulfone and detected by Western blotting with anti-HA, anti-Usp14, and anti-Uch37 antibodies. B, stimulation of gate opening (i.e. enhanced peptide hydrolysis) by ubiquitinated E6AP was measured in wt and ΔUsp14 proteasomes. C, stimulation of 26 S ATP hydrolysis was measured by the addition of ubiquitinated E6AP in wt and ΔUsp14 proteasomes. D, stimulation of gate opening (i.e. enhanced peptide hydrolysis) by ubiquitinated Sic1 was measured in ΔUsp14 proteasomes with and without blocking of Uch37 active site with Ub vinylsulfone (Ub-Vs). All values are the means of at least three experiments ± S.E. AU, arbitrary units.
To confirm that Uch37 can support the stimulation of 26 S activity by Ub conjugates, we blocked the Uch37 active site of proteasomes isolated from Usp14−/− MEF cells with HA-Ub-vinylsufone, which eliminated proteasome-associated DUB activity almost completely (supplemental Fig. S3C). Although basal gate opening (measured by peptidase activity) by the 26 S was not affected by HA-Ub-vinylsulfone treatment, the stimulation of gate opening by Ub conjugates (Fig. 4D) or HA-Ub-aldehyde (supplemental Fig. S3D) was largely suppressed. Thus, in the absence of Usp14, the binding of Ub conjugates or Ub-aldehyde to Uch37 is necessary to trigger gate opening.
In addition, we used the Usp14-specific inhibitor IU1 (19), which caused a small (<25%) reduction in total Ub-amc hydrolysis (Fig. 3D) by wt proteasomes. However, this agent did not affect the stimulation of 26 S ATPase activity by Ub conjugates (Fig. 3E). Unlike Ub aldehyde, IU1 by itself did not stimulate 26 S ATPase activity in combination with casein (Fig. 3F). Thus, the catalytic activity of Usp14 is not important for the activation of the 26 S (as was also shown by the yeast Ubp6 Cys→Ala mutants), but only its occupancy by Ub conjugates or Ub aldehyde.
Incidentally, during these experiments, we detected with HA-Ub-vinylsulfone, another DUB in the 26 S particles purified from ΔUsp14 MEF cells, which from its molecular weight most likely is Usp5. This DUB seems to be induced in ΔUsp14 MEF cells (supplemental Fig. S4) and is not 26 S-associated but binds directly to the Ub-like domain column used to isolate the 26 S proteasomes (17). Also, its presence in our 26 S preparations did not affect our conclusion about ubiquitin conjugate-activating proteasome function because Usp5 only cleaves free Ub chains and is inactive against ubiquitinated proteins. (However, because of its presence and the high level of DUB activity, tetra-Ub chains are very rapidly destroyed in these preparations and their effects cannot be studied meaningfully.)
Stimulation of ATP Hydrolysis Requires a Loose Domain in the Ubiquitinated Substrate
To become committed to proteolysis (5) and to be efficiently degraded, a ubiquitinated protein must also contain a loosely folded or unfolded domain (15, 16). We therefore tested whether a loosely folded domain is also essential for the stimulation of ATPase activity. Ubiquitinated DHFR was used as the substrate because DHFR assumes a tightly folded conformation upon binding of the inhibitor, methotrexate, which prevents tight association with the 26 S (5) and degradation (27). Although penta-ubiquitinated DHFR stimulated ATP hydrolysis 2-fold, methotrexate binding prevented this effect (Fig. 5A). Furthermore, although the nonubiquitinated DHFR alone had no effect on ATPase activity, when present together with Ub aldehyde, it promoted ATP hydrolysis (Fig. 5B). This stimulation by DHFR plus Ub aldehyde could also be blocked completely by the addition of methotrexate (Fig. 5B). Thus, only a loosely folded form of this protein could support the activation.
To further define this influence of substrate conformation on ATPase activity, we used lysozyme because it can be partially denatured to a molten globular state by treatment with 800 mm guanidine hydrochloride (23). When the native and molten globular lysozymes were added to yeast Ubp6C118A 26 S proteasomes in the presence of Ub, only the loosely folded molten globular form increased ATP hydrolysis (Fig. 5C). These experiments confirm that the activation of the ATPase requires both a Ub chain (or Ub aldehyde) occupying a proteasomal DUB and a loosely folded (i.e. a molten globule) conformation on the polypeptide.
The Increase in ATPase Activity Follows the Tight Binding of Ub Conjugates
Ub conjugates initially bind to the proteasome through their Ub chains, which associate with the 19 S receptor proteins, Rpn10 and Rpn13 (2, 3). For a ubiquitinated substrate to be committed to proteolysis, it must then undergo an ATP-dependent transition to a more tightly bound state, which no longer requires the Ub chain and is resistant to high salt (5). This transition also requires a loose conformation in the protein, presumably to interact with the ATPases. Because of their similar requirements, we attempted to clarify the relationship between transition from Ub-dependent to the no longer Ub-dependent tight, salt-resistant binding step and the activation of ATP hydrolysis. We therefore compared the capacity of different conjugates to undergo this transition and activate ATP hydrolysis. Because Ub conjugates containing a Lys-48 or Lys-63 chain are degraded at similar rates by purified 26 S (18, 28), we tested whether E6AP, which forms a Lys-48 chain, can also stimulate 26 S ATPase when linked to a Lys-63 chain. This E3 was allowed to autoubiquitinate with ATP, E1, UbcH5b and Lys-63-linked tetra-Ub in place of Ub. The resulting Lys-63 poly-ubiquitinated E6AP, similar to the Lys-48-E6AP, became tightly bound to the 26 S (supplemental Fig. S5A) and stimulated ATP hydrolysis (Fig. 5D). Surprisingly, ubiquitinated E6AP was not degraded to any significant extent (supplemental Fig. S6B) by the 26 S for unclear reasons. These findings confirm that the capacity to stimulate ATP hydrolysis can be uncoupled from conjugate degradation but appears linked to the capacity of the unfolded domains of the substrate to directly bind to the 26 S ATPases.
Surprisingly, when we used another homologous to the E6-AP carboxyl terminus ligase, Nedd4, which forms a Lys-63-linked chain on itself (18), the ubiquitinated Nedd4 did not stimulate ATPase activity (Fig. 5D), and it did not become tightly bound to the 26 S (supplemental Fig. S5B). These differences from E6AP cannot be due to the Lys-63 chain because when Nedd4 was allowed to auto-ubiquitinate with Lys-48-linked tetra-Ub (Fig. 5D), it initially bound through its Ub chain to Rpn10 and 13, but Nedd4 still failed to become tightly bound or to activate the ATPases (supplemental Fig. S5B). Therefore, the conformation of the Nedd4 (rather than the nature of the Ub linkages) is the reason it fails to stimulate the ATPase (Tables 1 and 2). Accordingly, non-ubiquitinated E6AP stimulated the ATPase activity in combination with Ub aldehyde, whereas Nedd4 did not (Fig. 5E). The different effects of ubiquitinated E6AP and Nedd4 provide further evidence that the stimulation of ATP hydrolysis requires that the substrate must have transitioned from the Ub-dependent to the tighter, Ub-independent (salt-resistant,) mode because only ubiquitinated proteins that could become bound in this manner (e.g. Ubn-E6AP) promoted ATPase activity (see legend of Table 1).
TABLE 1.
Conditions that allow tight binding of ubiquitin conjugates to the 26 S stimulate its ATPase activity
nd, not determined; MTA, methotrexate; −, not observed; +, observed.
| Type of ubiquitin conjugate | Tight binding | Stimulation of ATPase | Stimulation of gate opening |
|---|---|---|---|
| Lys-48 polyubiquitinated E6AP | + | + | + |
| Lys-63 polyubiquitinated E6AP | + | + | nd |
| Lys-63 polyubiquitinated Nedd4 | − | − | + |
| Lys-48 polyubiquitinated Nedd4 | − | − | nd |
| Ub5-DHFR | + | + | + |
| Ub5-DHFR+MTA | − | − | + |
TABLE 2.
ΔUbp6 and ATPase mutants can bind substrates tightly but do not allow stimulation of 26 S ATPase or gate opening
| Type of proteasome | Tight binding | Stimulation of ATPase | Stimulation of gate opening |
|---|---|---|---|
| wt | + | + | + |
| ΔUbp6 | + | − | − |
These observations enabled us to determine whether tight binding of conjugates to the proteasome (5) is essential for the enhanced ATPase activity or whether the stimulation of ATP hydrolysis is required for the substrate to become bound in this manner. We compared these processes in 26 S particles from wt and ΔUbp6 yeast strains. As noted above, proteasomes lacking Ubp6 did not show an increase in ATPase activity when bound to polyubiquitinated E6AP (Fig. 3A). The Ub receptors on these particles bind Ub conjugates at 4 °C with high affinity, but they can be washed off easily by the addition of the Ub binding Ub interacting motif from S5a or salt (supplemental Fig. S5C) (5, 17). However, after an additional incubation at 30 °C in the presence of ATP, both the wt and ΔUbp6 proteasomes became tightly bound and could no longer be displaced by the Ub interacting motif or salt. Therefore, the transition to Ub-independent binding does not require Ubp6 and thus can occur without activation of the ATPases. Together, these observations show that conjugates can become tightly bound and activate ATP hydrolysis without themselves being degraded and imply that the transition to the no longer Ub-dependent binding step is an essential prerequisite for the stimulation of ATP hydrolysis and proteolysis.
DISCUSSION
Activation of ATP Hydrolysis by Loosely Folded Ubiquitinated Substrates
Ubiquitination is generally viewed only as a mechanism to target proteins to specific binding sites, primarily to the 26 S proteasome for degradation. The present observations and related findings (9–11) demonstrate that Ub chains on proteins also have important regulatory functions. Thus, when an appropriate Ub conjugate is bound, the degradative capacity of the proteasome is stimulated by enhancing both 20 S gate opening (9–11) and ATP hydrolysis (10). The coordinated activation of these processes must increase the likelihood of substrate degradation, whereas greater gate opening must enhance the chances of an unfolded polypeptide entering into the 20 S, and the increased ATPase activity can promote substrate translocation and unfolding.
Activation of ATP hydrolysis by protein substrates is a general feature the AAA-ATPases that regulate proteolysis in bacteria, archaea, and mitochondria (6–8, 10). These complexes exhibit basal ATPase activity until they associate with an appropriate substrate, which leads to a transitory increase in their activity that presumably lasts until the substrate is digested. Thus, ATP consumption is maximal only when needed to promote the digestion of the bound substrate, and termination of this activated state upon substrate digestion must reduce nonspecific degradation of cell proteins.
Unlike the 26 S ATPases, the bacterial AAA-ATPases and the archaeal PAN complex are activated by unfolded polypeptides, which by themselves have no effect on proteasomal ATP consumption (Fig. 2) (24). Stimulation of the 26 S ATPases requires two interactions: not only an unfolded domain in the substrate (presumably with the ATPase) but also a poly-Ub chain or Ub aldehyde for occupancy of the active site of one of the 26 S-associated DUBs. Although a Ub chain is initially necessary for high affinity binding to the 19 S particle, the loosely folded domain in the polypeptide appears necessary for the subsequent ATP-dependent transition of the conjugate to the more tightly bound state, which then is no longer dependent on Ub (Fig. 6) (5). This two-step mechanism for tight substrate association and commitment to proteolysis differs in an important respect from the two-site requirement for the stimulation of the ATPases. This latter activation step requires chain interaction with the 19 S DUBs. Because the deletion of Ubp6 in yeast blocks the stimulation of ATP hydrolysis and gate opening (11) but still allows tight conjugate binding, the transition to the tight binding state must occur first. Then, the acceleration of ATP hydrolysis follows when the Ub chain becomes associated with Ubp6/Usp14 or Uch37.
FIGURE 6.
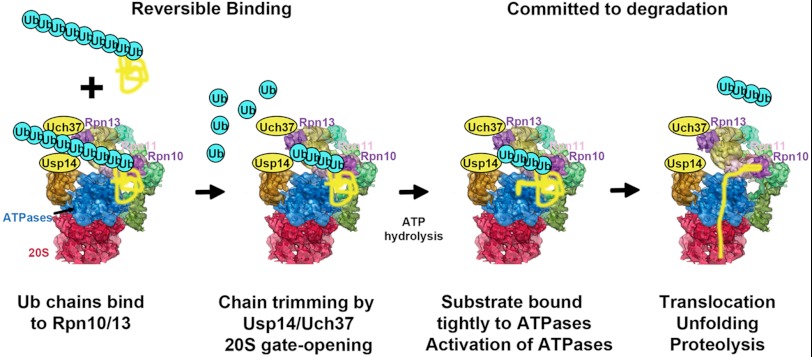
Ubiquitin conjugates activate their own degradation by the 26 S proteasome. Ub conjugates initially bind reversibly to Rpn10 and Rpn13. The Ub chain interacts then with the 19 S-associated DUBs Usp14 and Uch37 trimming the chain from its distal end leading to gate opening substrates with a loosely folded domain suitable for the initiation of degradation also stimulate ATP hydrolysis when binding directly to the ATPase subunits. This transient increase in ATP hydrolysis and gate opening allows for maximum rates of degradation while the polypeptide is hydrolyzed.
Activation of 26 S ATPase and Substrate Selection
This regulation of ATP hydrolysis by Ub conjugates must also comprise a substrate selection mechanism that increases the specificity of the 26 S proteasome for certain types of ubiquitinated proteins. For example, the 19 S binds many, perhaps all polyubiquitinated proteins with high affinity through its receptor subunits, Rpn10 and 13, but only those conjugates that can become bound in the tighter, Ub-independent (salt-resistant) manner and then activate the ATPases are substrates that are likely to be degraded (5). The two interactions leading to this tight binding and ATPase activation allows for the selection of Ub conjugates that can be efficiently degraded and for tightly folded proteins to be deubiquitinated and released from the proteasome. Such ubiquitinated tightly folded proteins may undergo enzymatic unfolding by the p97-VCP-Cdc48 ATPase complex, which seems to partially unfold many ubiquitinated substrates before delivery to the 26 S (29). Presumably, this AAA-ATPase serves to expose unfolded domains in the substrate that are required for tight binding and stimulation of the proteasomal ATPases.
This strict regulation of proteasomal ATPase activity should also help prevent wasteful energy expenditure on conjugates that can't be efficiently degraded. It is noteworthy that Ub conjugates that cannot be degraded and cannot stimulate ATP hydrolysis (e.g. methotrexate-bound DHFR) can still stimulate gate opening (supplemental Fig. S6A) (9). Because only unfolded, linearized polypeptides can enter the 20 S through its narrow central channel, increased gate opening per se, without the activation of the ATPases probably has relatively little effect on the degradation of globular proteins in cells. Because the increase in ATPase activity occurs only with ubiquitinated substrates that have become tightly bound through their polypeptide, this transition seems to function as a selectivity step, during which the folding state can be monitored to determine whether the substrate is degradable.
Although linked to substrate deubiquitination and degradation, the activation of ATP hydrolysis can be easily dissociated from these fates, e.g. with MG132, which blocks proteolysis but does not influence ATP hydrolysis (supplemental Fig. S1D). A similar activation of the PAN ATPase occurs in the absence of proteasomes. Thus, this response is linked to substrate binding and triggers the subsequent unfolding translocation.
Substrate Occupancy of Either Ubp6/Usp14 or Uch37 Enhances ATPase Activity
As shown by mutations and in vitro reconstitution, the stimulation of gate opening and ATPase activity by the yeast 26 S requires the presence of Ubp6. Thus, in addition to removing the Ub chain, Ubp6 links the two main 19 S activities, substrate deubiquitination and the ATP-driven unfolding and translocation. Because an identical stimulation was induced by Ub chains and Ub aldehyde, gate-opening and the increase in ATPase activity must occur when the 26 S-associated DUBs are actively disassembling the Ub chain. However, as shown with the Ub aldehyde (or the inactive Cys→Ala Ubp6), the enzymatic activity of Ubp6 is not required to stimulate gate opening or the ATPases. Thus, occupancy of the active site of Ubp6 by the Ub chain or Ub aldehyde induces these effects allosterically.
There is growing evidence that chain removal by Usp14/Ubp6 functions as a timing device (32) that provides an interval during which the substrate can be degraded. Our findings indicate that Usp14/Ubp6 also determines the duration of proteasome activation by substrates. Consequently, prolonging substrate association with the DUB may enhance the likelihood that a substrate is degraded by extending the duration of maximal gate opening and ATPase activation. Recently, a small molecule inhibitor of Usp14 activity has been shown to stimulate the degradation of certain proteins (19). Interestingly, this inhibitor, unlike Ub aldehyde, does not activate ATP hydrolysis, but, by slowing substrate deubiquitination, it may allow a more prolonged occupancy of Usp14 by Ub chains and thus should extend the activation of the ATPases and gate opening, thereby enhancing the likelihood that a Ub conjugate is digested.
An unexpected and important finding in these studies was that although Rpn11 lacks these regulatory functions, the third DUB present in the 26 S of higher eukaryotes, Uch37 (33–35), has similar regulatory roles as Usp14 because loss of Usp14 does not prevent the stimulation by Ub aldehyde of ATPase activity or gate opening, in contrast to the loss of Ubp6 in yeast. However, modification of the active site of Uch37 with Ub-vinylsulfone prevented this activation. Although both Usp14 and Uch37 can activate gate opening and ATP hydrolysis, it remains unclear whether the two DUBs have identical roles in conjugate deubiquitination and the advantage in higher eukaryotes of having two DUBs with overlapping regulatory functions on the proteasome.
These findings also raise the important question of how occupancy of either Usp14 or Uch37 can increase gate-opening or ATPase function, because these DUBs are located in quite different sites within the proteasome. Ubp6/Usp14 is bound to Rpn1 and Uch37 to Rpn2 and Rpn13 (36–39).
A New Step in the Sequence of Events Leading to Conjugate Degradation
These findings and related studies emphasize that the degradation of Ub conjugates is a tightly regulated, selective process that involves multiple ATP-driven steps. The regulation of ATP hydrolysis by two 19 S-associated DUBs indicates one mechanism by which these various enzymatic processes are linked to ensure efficient degradation. The two-step mechanism for tight substrate binding and the two-site requirement for the stimulation of 26 S activity also imply a specific order of events in processing of Ub conjugates, in which activation of the ATPases is a key new step (Fig. 6). Based on these findings, the likely sequence leading to proteolysis consists of the following. 1) Initially, the Ub conjugate binds through its Ub chain to Rpn10 and/or Rpn13 (2, 3). 2) Deubiquitination may begin leading to release of tightly folded proteins. 3) However, if a loosely folded domain is present in the polypeptide and ATP is hydrolyzed, the polypeptide becomes tightly bound to the 26 S ATPase. 4) When the Ub chain also interacts with and is trimmed by Ubp6/Usp14 and/or Uch37, the ATPases and gate opening are allosterically activated. 4) The resulting increased ATP consumption drives substrate unfolding, translocation into the 20 S, and proteolysis. 5) The 19 S then reverts to the basal, low ATPase state until another ubiquitinated substrate is bound. In this cycle, the kinetic competition between substrate deubiquitination and release versus its becoming tightly bound and committed to degradation seems to comprise a critical “life-death” decision step that leads to the activation of ATP hydrolysis, translocation, and proteolysis (Fig. 6).
Acknowledgments
We are grateful to Lisa Bacis and Mary Dethavong for valuable assistance and to Dan Finley and Suzanne Elsasser for providing the yeast strains and the Ubp6 expression plasmids.
This work was supported by grants from the National Institutes of Health/NIGMS (R01 GM051923-17), the Amyotropic Lateral Sclerosis Association (K2Y740), and the Fidelity Biosciences Research Initiative.

This article contains supplemental Figs. S1–S6.
- Ub
- ubiquitin
- DUB
- deubiquitinating enzyme
- PAN
- Proteasome-activating nucleosidase
- MEF
- mouse embryonic fibroblast
- DHFR
- Dihydrofolate reductase.
REFERENCES
- 1. Groll M., Bajorek M., Köhler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. (2000) A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 2. Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. (2004) Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279, 26817–26822 [DOI] [PubMed] [Google Scholar]
- 3. Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., Dikic I. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu C. W., Li X., Thompson D., Wooding K., Chang T. L., Tang Z., Yu H., Thomas P. J., DeMartino G. N. (2006) ATP binding and ATP hydrolysis play distinct roles in the function of 26 S proteasome. Mol. Cell 24, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peth A., Uchiki T., Goldberg A. L. (2010) ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol. Cell 40, 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benaroudj N., Zwickl P., Seemüller E., Baumeister W., Goldberg A. L. (2003) ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell 11, 69–78 [DOI] [PubMed] [Google Scholar]
- 7. Hwang B. J., Woo K. M., Goldberg A. L., Chung C. H. (1988) Protease Ti, a new ATP-dependent protease in Escherichia coli, contains protein-activated ATPase and proteolytic functions in distinct subunits. J. Biol. Chem. 263, 8727–8734 [PubMed] [Google Scholar]
- 8. Waxman L., Goldberg A. L. (1986) Selectivity of intracellular proteolysis: protein substrates activate the ATP-dependent protease (La). Science 232, 500–503 [DOI] [PubMed] [Google Scholar]
- 9. Bech-Otschir D., Helfrich A., Enenkel C., Consiglieri G., Seeger M., Holzhütter H. G., Dahlmann B., Kloetzel P. M. (2009) Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nat. Struct. Mol. Biol. 16, 219–225 [DOI] [PubMed] [Google Scholar]
- 10. Li X., Demartino G. N. (2009) Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem. J. 421, 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peth A., Besche H. C., Goldberg A. L. (2009) Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 36, 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koulich E., Li X., DeMartino G. N. (2008) Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol. Biol. Cell 19, 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 [DOI] [PubMed] [Google Scholar]
- 14. Yao T., Song L., Jin J., Cai Y., Takahashi H., Swanson S. K., Washburn M. P., Florens L., Conaway R. C., Cohen R. E., Conaway J. W. (2008) Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol. Cell 31, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prakash S., Tian L., Ratliff K. S., Lehotzky R. E., Matouschek A. (2004) An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 11, 830–837 [DOI] [PubMed] [Google Scholar]
- 16. Takeuchi J., Chen H., Coffino P. (2007) Proteasome substrate degradation requires association plus extended peptide. EMBO J. 26, 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Besche H. C., Haas W., Gygi S. P., Goldberg A. L. (2009) Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 48, 2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 19. Lee B. H., Lee M. J., Park S., Oh D. C., Elsasser S., Chen P. C., Gartner C., Dimova N., Hanna J., Gygi S. P., Wilson S. M., King R. W., Finley D. (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saeki Y., Isono E., Toh-E A. (2005) Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 399, 215–227 [DOI] [PubMed] [Google Scholar]
- 21. Lanzetta P. A., Alvarez L. J., Reinach P. S., Candia O. A. (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100, 95–97 [DOI] [PubMed] [Google Scholar]
- 22. Koodathingal P., Jaffe N. E., Kraut D. A., Prakash S., Fishbain S., Herman C., Matouschek A. (2009) ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J. Biol. Chem. 284, 18674–18684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mizuguchi M., Arai M., Ke Y., Nitta K., Kuwajima K. (1998) Equilibrium and kinetics of the folding of equine lysozyme studied by circular dichroism spectroscopy. J. Mol. Biol. 283, 265–277 [DOI] [PubMed] [Google Scholar]
- 24. Henderson A., Erales J., Hoyt M. A., Coffino P. (2011) Dependence of proteasome processing rate on substrate unfolding. J. Biol. Chem. 286, 17495–17502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tanaka K., Waxman L., Goldberg A. L. (1983) ATP serves two distinct roles in protein degradation in reticulocytes, one requiring and one independent of ubiquitin. J. Cell Biol. 96, 1580–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Borodovsky A., Kessler B. M., Casagrande R., Overkleeft H. S., Wilkinson K. D., Ploegh H. L. (2001) A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20, 5187–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnston J. A., Johnson E. S., Waller P. R., Varshavsky A. (1995) Methotrexate inhibits proteolysis of dihydrofolate reductase by the N-end rule pathway. J. Biol. Chem. 270, 8172–8178 [DOI] [PubMed] [Google Scholar]
- 28. Hofmann R. M., Pickart C. M. (2001) In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 276, 27936–27943 [DOI] [PubMed] [Google Scholar]
- 29. Beskow A., Grimberg K. B., Bott L. C., Salomons F. A., Dantuma N. P., Young P. (2009) A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. J. Mol. Biol. 394, 732–746 [DOI] [PubMed] [Google Scholar]
- 30. Goldberg A. L., Dice J. F. (1974) Intracellular protein degradation in mammalian and bacterial cells. Annu. Rev. Biochem. 43, 835–869 [DOI] [PubMed] [Google Scholar]
- 31. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hamazaki J., Iemura S., Natsume T., Yashiroda H., Tanaka K., Murata S. (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 25, 4524–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qiu X. B., Ouyang S. Y., Li C. J., Miao S., Wang L., Goldberg A. L. (2006) hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 25, 5742–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yao T., Song L., Xu W., DeMartino G. N., Florens L., Swanson S. K., Washburn M. P., Conaway R. C., Conaway J. W., Cohen R. E. (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 8, 994–1002 [DOI] [PubMed] [Google Scholar]
- 36. Lasker K., Förster F., Bohn S., Walzthoeni T., Villa E., Unverdorben P., Beck F., Aebersold R., Sali A., Baumeister W. (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. U.S.A. 109, 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lander G. C., Estrin E., Matyskiela M. E., Bashore C., Nogales E., Martin A. (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 39. Chen X., Lee B. H., Finley D., Walters K. J. (2010) Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol. Cell 38, 404–415 [DOI] [PMC free article] [PubMed] [Google Scholar]